Abstract
Microcycas calocoma is the monotypic species from the critical endangered and endemic cycad genus Microcycas in Cuba, an important taxon to study the evolution of extant gymnosperms. Here we report the complete chloroplast sequences of M. calocoma and characterize the genome structure of this species. The genome size of M. calocoma is 165,667 bp in length which contains 135 genes, including 88 protein-coding genes, eight ribosomal RNA genes and 39 transfer RNA genes. The GC content of this genome is 39.6%.
Phylogenomic study shows M. calocoma is mostly closely related to the cycad genus Zamia, which corresponds to previous studies based on multiple nuclear genes and combined plastid and nuclear evidence. The plastome information of M. calocoma offered by this study can contribute to further comparative chloroplast genome in cycads/gymnosperms as well as conservation genetic studies of M. calocoma.
Keywords: Cycad, Cuba, Critical Endangered, Microcycas calocoma, chloroplast genome
Microcycas calocoma (Miq.) A.DC was first described by F.A.W. Miquel as the name Zamia calocoma (Miquel 1852) and formally named as M. calocoma by De Candolle under the monotypic genus Microcycas (De Candolle 1868). Based on the assessment of IUCN red list, this species is considered to be Critically Endangered, with a geographical range restricted to a total of five known localities in Western Cuba in the province of Pinar del Rio (Bösenberg 2010).
On account of the particular phylogenetic position of cycads, the cycad plastomes can be informative to trace the lineage history of extant gymnosperms and to study the plastid structure evolution in seed plants. However, Microcycas is the only genus among extant cycads whose chloroplast genome information is unavailable. Here, we assembled and characterized the complete chloroplast (cp) genome of M. calocoma in order to provide information for further conservation genetics within M. calocoma as well as comparative plastid genomic studies in seed plants.
Fresh leaf tissues were collected in Nong Nooch Tropical Botanical Garden, Thailand from cultivated plants (Acc. 456, BK) introduced from wild-collected plants in Consolacion del Sur of Cuba (22°33′11″N, 83°32′16″ W). Total genomic DNA was extracted by modified CTAB method (Doyle 1991). A total of 2 G raw data from Illumina Hiseq Platform (Novogene, Beijing, China) were screened and assembled into contigs by Get_Organelle pipeline (Jin et al. 2018) with published Zamia furfuracea (JX416857) as reference. The resulted contigs were reordered and further trimmed according to the Z. furfuracea reference to obtain the complete chloroplast genome. We applied PGA (Qu et al. 2019) to annotate the Microcycas plastid genome by using Z. furfuracea as reference. The annotations were double-checked in Geneious v11.0.3 (Kearse et al. 2012) and deposited in NCBI under the accession MN428318. To infer the intergenic relationship of all extant cycads, we used MAFFT v7 (Katoh and Standley 2017) to align an 11-plastome sequence dataset including all cycad genera and Ginkgo. ML tree inference was implemented in IQTREE 1.6.12 (Nguyen et al. 2015) with 1000 bootstrap replicates.
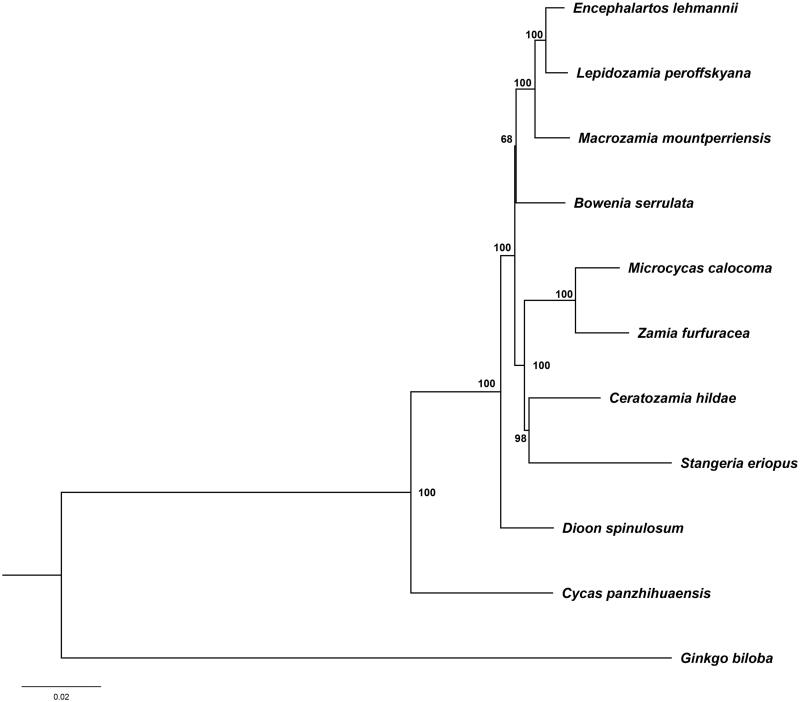
Based on the assembling results, the complete chloroplast genome of M. calocoma has typical quadripartite structure with a length of 165,667 bp, which contains 135 genes (88 protein-coding genes, eight rRNA genes and 39 tRNA genes). Among them, there are 10 genes (rps16, atpF, rpoC1, petB, petD, rpl16, rps12, rpl2, ndhB, ndhA) which have a single intron, while two genes (clpP and ycf3) were found to have two introns. This genome has two inverted repeat (IR) sequences, consistent with all other reported cycad plastomes (Wu and Chaw 2015). The overall GC content of M. calocoma genome is 39.6%, with the number 42% in IR, 38.8% and 37.1% in LSC (large single copy) and SSC (small single copy) regions, respectively. Phylogenomic analysis revealed a well-resolved tree within cycads (Figure 1). Microcycas was strongly supported as a sister with Zamia which agrees with the previous phylogenetic inferences of cycads based on both multiple single-copy nuclear genes (Salas-Leiva et al. 2013) and concatenated chloroplast and nuclear datasets (Liu et al. 2018).
Figure 1.
Intergeneric phylogenetic relationships across cycads based on the maximum-likelihood (ML) analysis of whole sequence evidence. Numbers on nodes represent for the bootstrap values based on 1000 replicates from IQTREE. Accession numbers: Bowenia serrulata NC_026036; Ceratozamia hildae NC_026037; Cycas panzhihuaensis KX_138991; Dioon spinulosum NC_027512; Encephalartos lehmannii LC_049336; Lepidozamia peroffskyana NC_027513; Macrozamia mountperriensis NC_027511; Stangeria eriopus LC_049067; Zamia furfuracea JX_416857; Ginkgo biloba JN_867585.
The complete chloroplast genome of M. calocoma in this study can contribute to further conservation of genetic studies of this species and also provide essential data for plastid comparative genomic studies across cycads/gymnosperms.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
References
- Bösenberg JD. 2010. Microcycas calocoma. The IUCN Red List of Threatened Species; https://www.iucnredlist.org/species/42107/10647674 [accessed 2019 Aug 02]. 2010e.T42107A10647674.
- De Candolle A. 1868. Microcycas calocoma, Prodromus Systemalis Naturalis Regnis Vegetabilis. 16:538. [Google Scholar]
- Doyle J. 1991. DNA protocols for plants Vol. 57 In: Hewitt, G.M., Johnston, A.W.B. and Young, J.P.W., Eds., Molecular techniques in taxonomy. Switzerland: Springer; p. 283–293. [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv. :256479. doi: 10.1101/256479 [DOI] [Google Scholar]
- Katoh K, Standley DM. 2017. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol Biol Evol 30:772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England). 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang S, Nagalingum NS, Chiang Y-C, Lindstrom AJ, Gong X. 2018. Phylogeny of the gymnosperm genus Cycas L. (Cycadaceae) as inferred from plastid and nuclear loci based on a large-scale sampling: evolutionary relationships and taxonomical implications. Mol Phylogenet Evol. 127:87–97. [DOI] [PubMed] [Google Scholar]
- Miquel FAW. 1852. Sur une espèce nouvelle de Zamia des Indes occidentales, introduite dans l'ètablissement Van Houtte, à Gand. Flore Des Serres et Des Jardins de L'Europe. 7:141–142. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Meth. 15:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Leiva DE, Meerow AW, Calonje MA, Griffith MP, Francisco-Ortega J, Nakamura K, Stevenson DW, Lewis CA, Namoff SM. 2013. Phylogeny of the cycads based on multiple single-copy nuclear genes: congruence of concatenated parsimony, likelihood and species tree inference methods. Ann Bot. 112:1263–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Chaw SM. 2015. Evolutionary stasis in Cycad plastomes and the first case of plastome GC-biased gene conversion. Genome Biol Evol. 7:2000–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]