Abstract
In this study, we report the mitochondrial genome of the black-tailed dasyure (Murexia melanurus) of New Guinea. The circular genome is 17,736 bp in length and has an AT content of 60.5%. Its gene content – 13 protein-coding genes (PCGs), 2 ribosomal (rRNA) genes, 21 transfer RNA (tRNA) genes, a tRNA pseudogene (tRNALys), and a non-coding control region (CR) – and gene arrangement are consistent with previous marsupial mitogenome assemblies.
Keywords: Mitochondrial genome, marsupial, dasyure, Dasyuridae, New Guinea, Murexia
The marsupial family Dasyuridae (or carnivorous marsupials) includes ∼75 species native to mainland Australia and Tasmania, New Guinea, and other adjacent islands (Baker and Dickman 2018). The dasyurid subfamily Phascogalinae comprises three genera: Antechinus and Phascogale of Australia, and Murexia of New Guinea. Although morphological similarities exist, the current evidence suggests these genera derived from a common ancestor some 12.5 million years ago (Mutton et al. 2019). Murexia spp. have not received as much attention in evolutionary studies as their Australian counterparts within Phascogalinae, and genetic information on the various constituent species within Murexia is poorly represented. Here, we describe the complete mitochondrial genome of the black-tailed dasyure (Murexia melanurus).
Murexia melanurus genomic DNA was extracted from ear tissue (voucher specimen ABTC46020) collected from Tibi, Papua New Guinea. Paired-end short-insert (200 bp) DNA libraries were sequenced by BGI (Hong Kong, China), to generate ∼30× genome coverage. Raw data were filtered using Flexbar v3.4.0 (Roehr et al. 2017). To remove microbial contaminants, we used bowtie2 v2.3.4.1 (Langmead and Salzberg 2012) to map reads to all bacterial and fungal sequences in NCBI Genomes, retaining 99.68% of reads. Two to 95 million reads were assembled using NOVOPlasty v2.7.2 (Dierckxsens et al. 2017), with the ND1 coding sequence from a partial M. melanurus mitogenome (GenBank: KJ868127) (Mitchell et al. 2014) as a seed sequence and the parameters ‘Type = mito, K-mer = 39, Genome range = 16,000–22,000’. The subset with the lowest number of reads which yielded a circular genome was retained (here: 12 M reads; 0.26% were assembled into a 17,736 bp contig at 179× coverage). Geneious Prime v2019.1.3 (Biomatters Ltd., Auckland, New Zealand) was used to align 95 M reads against the assembled contig and generate a consensus genome sequence, with a 75% masking threshold. The genome was annotated using GenBank features of the northern quoll (Dasyurus hallucatus; accession no. NC_007630). Various genome features were compared to the Virginia opossum (Didelphis virginiana), a seminal species in the marsupial mitochondrial genetics literature [e.g. see Janke et al. (1994) and Nilsson (2009)].
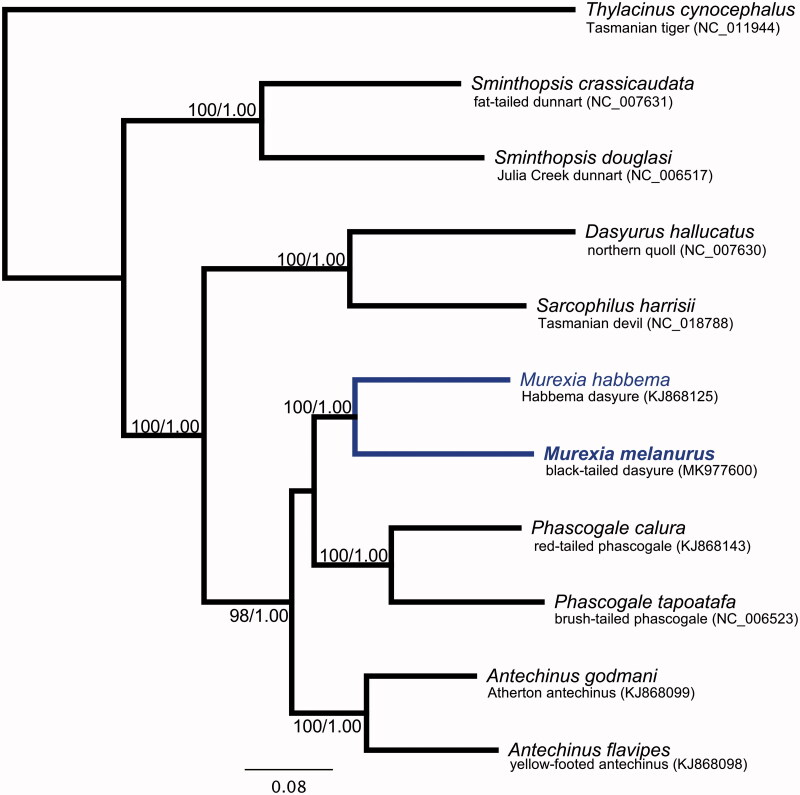
The M. melanurus mitochondrial genome (GenBank: MK977600) is 17,736 bp and has a base composition of 32% A, 28.5% T, 14.1% G, and 25.4% C. As in other marsupials, the genome has 13 protein-coding genes (PSGs), 2 ribosomal (rRNA) genes, and 21 transfer RNA (tRNA) genes. The genome shares unique features with all marsupial mitogenomes reported to date. These include: an ‘ACWNY’ tRNA gene re-arrangement (Paabo et al. 1991); a tRNALys pseudogene (Janke et al. 1994; Dorner et al. 2001); and lack of an anticodon for aspartic acid, which is likely rescued by RNA-editing (Janke and Paabo 1993). In agreement with a recent molecular appraisal (Mitchell et al. 2014; Westerman et al. 2016), phylogenetic analysis revealed that M. melanurus and Murexia habbema are sisters, to the exclusion of the closely-related genera Phascogale and Antechinus (Figure 1). Maximum-likelihood (ML; estimated using IQ-TREE (Nguyen et al. 2015)) and Bayesian Interference (BI; implemented in MrBayes v3.2.7 (Ronquist and Huelsenbeck 2003)) gave the same tree topology.
Figure 1.
Phylogenetic tree of black-tailed dasyure (Murexia melanurus; indicated in bold blue font), nine other species in the marsupial family Dasyuridae, and the outgroup species Thylacinus cynocephalus. Because there are no complete mitogenomes from the genera Antechinus and Murexia in GenBank, phylogenetic reconstruction was performed with coding sequences of 12 protein-coding genes (excluding ND6). The number at each node is ML/BI bootstrap support value.
Acknowledgements
We thank Leanne Wheaton (South Australian Museum) for providing the M. melanurus ear tissue sample.
Geolocation information
Geospatial coordinates for the black-tailed dasyure (Murexia melanurus) ear tissue collection: 6°11′S, 143°9′E
Disclosure statement
The authors declare that they have no competing interests.
References
- Baker A, Dickman C. 2018. Secret lives of carnivorous marsupials. Clayton (Australia): CSIRO Publishing. [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucl Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner M, Altmann M, Paabo S, Morl M. 2001. Evidence for import of a lysyl-tRNA into marsupial mitochondria. MBoC. 12(9):2688–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke A, Feldmaier-Fuchs G, Thomas WK, von Haeseler A, Paabo S. 1994. The marsupial mitochondrial genome and the evolution of placental mammals. Genetics. 137(1):243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke A, Paabo S. 1993. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucl Acids Res. 21(7):1523–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KJ, Pratt RC, Watson LN, Gibb GC, Llamas B, Kasper M, Edson J, Hopwood B, Male D, Armstrong KN, et al. 2014. Molecular phylogeny, biogeography, and habitat preference evolution of marsupials. Mol Biol Evol. 31(9):2322–2330. [DOI] [PubMed] [Google Scholar]
- Mutton TY, Phillips MJ, Fuller SJ, Bryant LM, Baker AM. 2019. Systematics, biogeography and ancestral state of the Australian marsupial genus Antechinus (Dasyuromorphia: Dasyuridae). Zool J Linn Soc. 186(2):553–568. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson MA. 2009. The structure of the Australian and South American marsupial mitochondrial control region. Mitochondrial DNA. 20(5–6):126–138. [DOI] [PubMed] [Google Scholar]
- Paabo S, Thomas WK, Whitfield KM, Kumazawa Y, Wilson AC. 1991. Rearrangements of mitochondrial transfer RNA genes in marsupials. J Mol Evol. 33(5):426–430. [DOI] [PubMed] [Google Scholar]
- Roehr JT, Dieterich C, Reinert K. 2017. Flexbar 3.0 - SIMD and multicore parallelization. Bioinformatics. 33(18):2941–2942. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574. [DOI] [PubMed] [Google Scholar]
- Westerman M, Krajewski C, Kear BP, Meehan L, Meredith RW, Emerling CA, Springer MS. 2016. Phylogenetic relationships of Dasyuromorphian marsupials revisited. Zool J Linn Soc. 176(3):686–701. [Google Scholar]