Abstract
Primary mitochondrial complex I deficiency is the most common defect of the mitochondrial respiratory chain. It is caused by defects in structural components and assembly factors of this large protein complex. Mutations in the assembly factor NDUFAF5 are rare, with only five families reported to date. This study provides clinical, biochemical, molecular and functional data for four unrelated additional families, and three novel pathogenic variants. Three cases presented in infancy with lactic acidosis and classic Leigh syndrome. One patient, however, has a milder phenotype, with symptoms starting at 27 months and a protracted clinical course with improvement and relapsing episodes. She is homozygous for a previously reported mutation, p.Met279Arg and alive at 19 years with mild neurological involvement, normal lactate but abnormal urine organic acids. We found the same mutation in one of our severely affected patients in compound heterozygosity with a novel p.Lys52Thr mutation. Both patients with p.Met279Arg are of Taiwanese descent and had severe hyponatremia. Our third and fourth patients, both Caucasian, shared a common, newly described, missense mutation p.Lys109Asn which we show induces skipping of exon 3. Both Caucasian patients were compound heterozygotes, one with a previously reported Ashkenazi founder mutation while the other was negative for additional exonic variants. Whole genome sequencing followed by RNA studies revealed a novel deep intronic variant at position c.223-907A>C inducing an exonic splice enhancer. Our report adds significant new information to the mutational spectrum of NDUFAF5, further delineating the phenotypic heterogeneity of this mitochondrial defect.
Keywords: Complex I, Leigh Syndrome, Mitochondrial Disease, Splicing, Hyponatremia, NDUFAF5
Introduction
Leigh syndrome (LS; MIM# 256000) or subacute necrotizing encephalopathy is a heterogeneous disorder secondary to mutations in mitochondrial (mtDNA) as well as nuclear (nDNA) encoded genes [1]. The central diagnostic criteria are bilateral foci in basal ganglia as well as in the brain stem with subthalamic nuclei involvement [2, 3]. The onset usually begins in infancy or early childhood, with mainly unspecific symptoms manifesting as hypotonia, nystagmus or recurrent vomiting. Associated neurological features include seizures, tremor, dystonia, ataxia and muscle weakness [1]. Disease progression is usually rapid with death in the first few years of life. However individuals with atypical Leigh syndrome displaying delayed symptom onset and survival into adulthood have been reported [2].
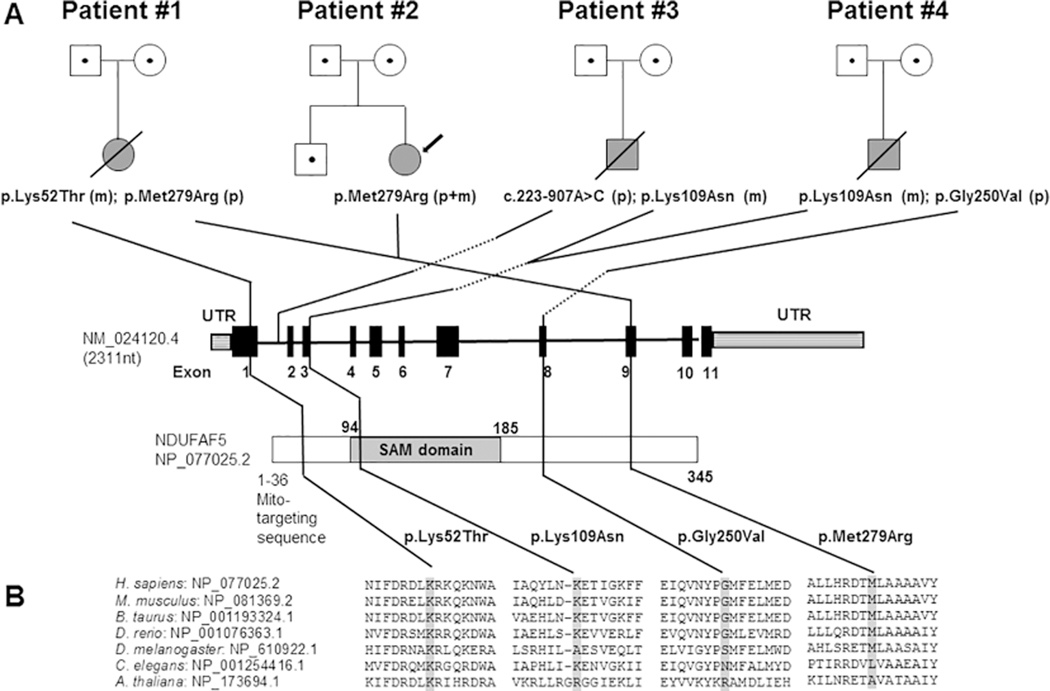
Biochemical abnormalities in complex I represent the most frequent laboratory finding in Leigh syndrome patients [4]. The complex is comprised of 44 unique subunits, seven of which are encoded by mtDNA and 37 by nDNA [5, 6]. The main function of complex I is to establish the mitochondrial electrochemical gradient, powering ATP production. For this, electrons enter the L-shaped protein complex at the NADH binding site located on the matrix or peripheral arm (N-module), while proton pumping itself is restricted to the hydrophobic inner mitochondrial membrane (P-module) arm. The connector of the two arms (Q module) comprises the Co-Q binding site [7]. Assembly of this dynamic complex is aided by proteins, which temporarily associate with either single complex I subunits or developing complex I modules but are not part of the holoenzyme [8]. Of the 15 putative assembly factors for complex I, nine have been implicated in disease and at least seven of them [NDUFAF2 [9], NDUFAF3 [10], NDUFAF4 [11], NDUFAF5 [12], NDUFAF6 [13], NDUFAF8 [14], FOXRED1 [15]] have been implicated in Leigh syndrome [16]. While their roles in complex I assembly have been well established, recent studies on protein-protein interactions suggest many unrecognized additional functional properties, which may in part explain the phenotypic heterogeneity [17, 14, 18]. NDUFAF5, for example, directly associates with the complex I accessory subunit NDUFAB1 (mitochondrial acyl carrier protein-ACP), which also plays a role in the mitochondrial fatty acid synthesis type II (FASII) machinery. Since NDUFAF5 has an S-adenosylmethionine-dependent methyltransferase (SAM) domain, the assembly factor has been suggested to be involved in posttranslational modification of some of its associated proteins. Surprisingly, NDUFAF5 has not been shown to be a methylase, but rather an arginine hydroxylase of the complex I subunit NDUFS7 [18, 6]. Phenotypic heterogeneity has also been attributed in the past to the fact that NDUFAF5 has two isoforms, which are expressed in most tissues. While isoform 1 is comprised of 11 exons, isoform 2 is an alternative splice transcript that does not include exon 5; therefore, mutations in exon 5 are expected to be less deleterious, as they affect only isoform 1 rather than isoform 1 and 2.
Only five families, all with homozygous mutations in NDUFAF5 (C20orf7), have been described to date (Supplemental Figure 1, Supplemental Table 1).
The connection was initially established in 2008 via homozygosity mapping in a child from a consanguineous Egyptian family with lethal neonatal mitochondrial disease and the missense mutation p.Leu229Pro in exon 7 [19]. In another report, molecular studies in a consanguineous Moroccan family with atypical Leigh syndrome revealed a missense mutation p.Leu159Phe in exon 5, affecting only isoform 1, which was thought to explain the milder course of disease (20). In a third report, a homozygous Ashkenazi founder mutation p.Gly250Val in exon 8 was identified in two families with classical Leigh syndrome and a combined biochemical complex I and IV defect [12]. The report indicated that the high frequency of the mutation, with a carrier rate of 1/290, should prompt testing in all Leigh syndrome patients in that ethnic group prior to muscle biopsy. Most recently a report on exome sequencing in patients with neurodevelopmental delay describes a Chinese infant with brain stem involvement on MRI who died at eight months due to respiratory failure and was found to be compound heterozygous for p.Met279Arg and p.Arg49Gly mutations [20].
Here we present clinical, biochemical, molecular and functional studies in four new patients with NDUFAF5 defects and report three novel mutations. The p.Lys52Thr amino acid change is predicted to be severely damaging and found together with a previously reported mutation. The p.Lys109Asn change, found in two of our patients, has a carrier frequency of 1/500 in Caucasians and an unclear in silico pathogenicity prediction for the amino acid substitution. However our studies clearly show p.Lys109Asn to be associated with severely decreased NDUFAF5 mRNA and protein level. Finally, our studies for c.223–907A>C, a novel deep intronic variant, show aberrant splicing and decreased levels of NDUFAF5. This manuscript provides additional information about this rare disorder and may help to diagnose complex I deficiency for families with previously negative molecular results.
1. Materials and Methods
Whole Exome Sequencing
Whole exome sequencing was performed on a clinical basis by Ambry Genetics (patient 1 and 3 both as a trio), Baylor Genetics (patient 2, patient only and mutations confirmed in parents via Sanger Sequencing) and GeneDx (patient 4).
Whole Genome Sequencing
Whole Genome Sequencing was performed for patient 3 on a clinical basis by the Illumina iHope Program. Sequencing was performed on Illumina HiSeq with an average read depth of 30-fold coverage.
Next Generation Sequencing Data Analysis
Data was analyzed using CLC Bio Biomedical Workbench.
Cell Culture
Primary human skin fibroblasts were established from 3mm punch biopsies. Tissue was grown on plates coated with fibronectin (10 μg/ml of PBS) (LifeTechnologies) in alpha MEM with 1x nonessential amino acids (NEAA) (LifeTechnologies), Primocin (Invivogen) and 15% FBS (Hyclone-Fetal Clone 3). Established cultures were maintained either in DMEM-F12 or DMEM-5mM Glucose, 1x NEAA, 1x Antibiotic/Antimycotic (all LifeTechnologies) and 10% FBS (Hyclone-Fetal Clone 3) at 37°C in 5% CO2.
Immunoblotting
Cells were lysed on ice in RIPA buffer (Pierce) supplemented with protease inhibitor cocktail (50ul/ml) (Sigma) for 30min at 4°C and cleared at 16,000 rpm for 20min. Protein concentration was determined in triplicate via DC protein assay (Biorad). Lysates were stored in Nupage LSD sample buffer (LifeTechnologies) at −20°C. SDS-PAGE was performed using 1.5mm NuPage 4–12% Bis-Tris Gels and NuPage MES running buffer (LifeTechnologies) according to manufacturer’s recommendations employing the use of sample reducing agent and antioxidant (LifeTechnologies). Proteins were blotted onto PVDF membrane (LifeTechnologies) with 1x Transfer buffer (LifeTechnologies) containing 15% Methanol and antioxidant at 30V for 2hours. Blocking was done for 30min at room temperature (RT) with Fast Blocking buffer (Pierce). The same blot was used for all 4 antibodies employed in Figure 3D and a second blot for the antibodies shown in figure 3E. Membranes were incubated consecutively with primary antibodies as follows: 1:2000 rb_anti-C20orf7_38kD (Abcam, ab192235), 1:40,000 ms_anti-beta Actin_42kD (Abcam, ab184220), 1:1000 rb_anti-NDUFS3_27kD (Abcam, ab177471), 1:10,000 rb_anti-TOMM20_16kD (Abcam, ab186735) for Figure 3D and 1:500 ms_OXPHOS human_WB antibody cocktail (Abcam, ab110411) plus 1:2000 ms_antiNDUFB8 (Abcam, ab110242), 1:40,000 anti-beta Actin (Abcam, ab8227) each at 4°C overnight, washed for 5 min with 1xTBS-T and incubated with 1:20,000 HRP-conjugated goat anti-rabbit (Abcam, ab6721) or 1:20,000HRP-conjugated goat anti-mouse IgG (Abcam, ab97023) at RT for 1hr. Membranes were then washed 3x with 1x TBS-T at RT for 5 min each and proteins were detected with SuperSignal Chemi-luminescent substrate (LifeTechnologies) and imaged on CL-Xposure film (LifeTechnologies). Post imaging membranes were washed for 5 hours in 1x TBS-T and briefly rinsed in fast blocking buffer before incubation with the next primary antibody.
Measurements of oxygen consumption rate by Seahorse
Basal and maximal oxygen consumption rate (OCR) measurements were performed on a Seahorse Bioscience XFe-24 bioanalyzer using the Seahorse XF Cell Mito Stress Test Kit (Agilent). The kit uses sequential addition of substrates and inhibitors of the mitochondrial respiratory chain. After measuring basal respiration, oligomycin is added to block the mitochondrial ATP synthase followed by addition of FCCP, uncoupling mitochondrial respiration from ATP production. In the last step rotenone and antimycin A are added to inhibit complex I and III to stop all mitochondrial respiration. Cells were seeded in XF24-well microplates in growth medium at 3 × 104 cells/well. The following day growth medium was replaced with unbuffered assay medium supplemented with glucose, pyruvate and L-glutamine. Cells were equilibrated prior to Mitostress testing with unbuffered medium for 45 minutes at 37°C in a CO2-free incubator. Data was analyzed using XF Cell Mito Stress Test Report Generator. Upon assay completion cells were rinsed in ice cold PBS and lysed with RIPA buffer supplemented with protease inhibitor cocktail. Protein levels were determined for each well in triplicate using DC protein assay (Biorad). Values were used for normalization of OCR as per ug/ul of protein.
RT-PCR and Sanger Sequencing
RNA was extracted from patient 3 and age matched control fibroblasts using the PureLink RNA Mini Kit (Ambion) according to the manufacturer protocol including DNAse treatment. Parental RNA was extracted from whole blood drawn into PAX tubes using the PAXgene blood RNA kit (Qiagen) and DNAse treated. RNA concentration was assessed using Nanodrop. RT-PCR was performed using the SuperScript III one step RT-PCR kit with Platinum Taq (Invitrogen) with forward primers in the 5’UTR (5’GCACAAAAAGCGCCGGCAAT3’) or exon 1 (5’AGGGAAGTCACCTCTGGTGT3’) respectively and with reverse primers in exon 3 (5’TGTGCAATGTAACCTCTTCCA3’) or exon 4 (5’TTCTGCAATGTCAGCTTGGA3’) respectively. Primers were chosen using Primer-Blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast). Amplicons were run on a 2% Agarose gel and gel purified using the Zymoclean™ Gel DNA Recovery Kit (Zymo Research). Purified amplicons were Sanger Sequenced using the same primers (Retrogen) and analyzed using Sequencher 5.4 Software.
Quantification of NDUFAF5 Expression
RNA was extracted from patient and control fibroblasts using the Qiagen RNeasy kit according to the manufacturer protocol, and the samples were treated with DNase to produce pure RNA. Quantitative PCR for NDUFAF5 was performed on CFX Connect Real-Time PCR Detection System using iTaq Universal SYBR Green One-Step Kit (BioRad). qPCR was performed using IDT PrimeTime qPCR primers with forward primers in exon 1 (5’CCGGGATTTGAAAAGGAAACAG3’), exon 2 (5’CGCAGACCGTGTATATGACAT3’), exon 7 (5’CACATTTCTCCTTTCACTGCTG3’), and exon 11 (5’CCAGATCTATTACATGATAGGATGGA3’) respectively and reverse primers in exon 2 (5’GGTATGTCATATACACGGTCTGC3’) in exon 4 (5’TTCTGCAATGTCAGCTTGGA3’), exon 8 (5’TCCAGGATAGTTAACTTGAATTTCATC3’) and exon 12 (5’AGCTCTCCAAATGACACAGTT3’). Primers were designed against NM_024120.
Variant Submission:
Variants have been submitted to the Leiden Open Variation Database. http://www.lovd.nl/3.0/home
2. Results
Patient #1
The patient was born via NSVD at term with a birth weight of 2.5kg. The parents were of Taiwanese descent and family history was otherwise noncontributory. Early developmental milestones and growth were normal. At six months of age, the patient began having swallowing difficulties and around the same time a head tilt, horizontal nystagmus and blepharospasm were noted. By eight months, the patient started having intermittent episodes of vomiting. An initial neurology evaluation confirmed the presence of nystagmus and revealed mild global hypotonia. Brain magnetic resonance imaging (MRI) was suggestive of Leigh syndrome showing symmetric lesions in the thalamus and midbrain as well as dysgenesis of the corpus callosum (Figure 1.1).
Figure 1.
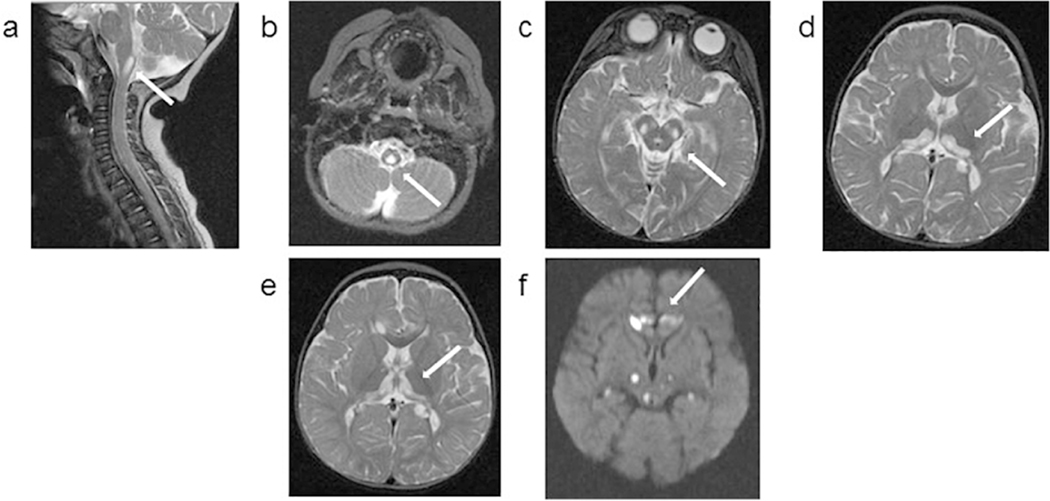
1.1. Patient 1: Axial MRI images obtained at 10 months (upper panel) and 13 months (lower panel) of age. Images a, b, c, d and e are T2 weighted, and image f is a diffusion weighted sequence (DWI). At 10 months of age, nonenhancing cystic appearing focus at the cervical medullary junction was noted, which had similar signal intensity to CSF in all imaging modalities (a, b). Bilateral symmetrical hyperintensities are noted in the midbrain (c) and thalami with a semicircular shape (d). In the MRI obtained 3 months later, the extension of thalamic lesions are noted (e). New bilateral and symmetrical areas of restricted diffusion are also noted in frontal lobes, and thalami (f).
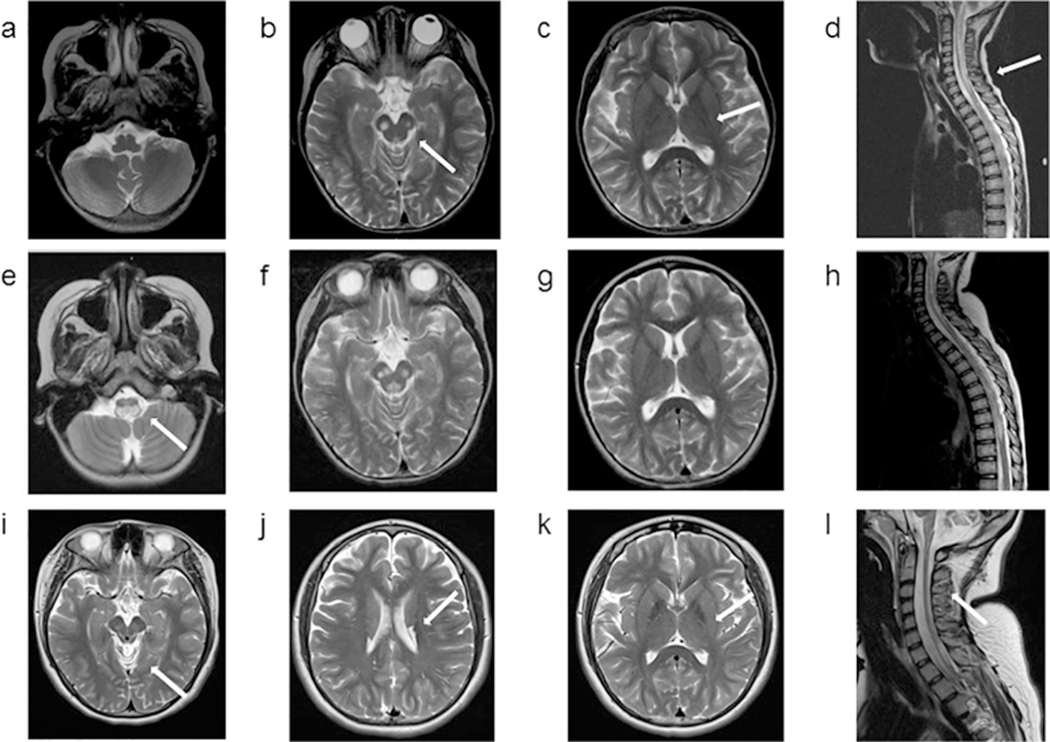
1.2. Patient 2: Axial MRI images obtained at 7 years (upper panel), 4 months later (middle panel) and at 14 years (lower panel) of age. All images are T2 weighted images. At 7 years of age, bilateral symmetrical hyperintensities are noted in the cerebral peduncles as well as in the lower medulla and extensively throughout the spinal cord. Bilateral thalami are unaffected. In the MRI obtained 4 months later, extensive abnormal foci of T2 prolongation in the cerebral peduncles, and increased signal is seen within the medulla extending into the cervicomedullary junction. The MRI at 14 years of age shows new cystic appearing lesions in the bilateral caudate, which had similar signal intensity to CSF in all imaging modalities. Interval atrophy of the cervical spinal cord with persistent intramedullary signal abnormality is seen but similar to the past, bilateral thalami remain unaffected. Diffuse posterior fossa atrophy is noted (not shown).
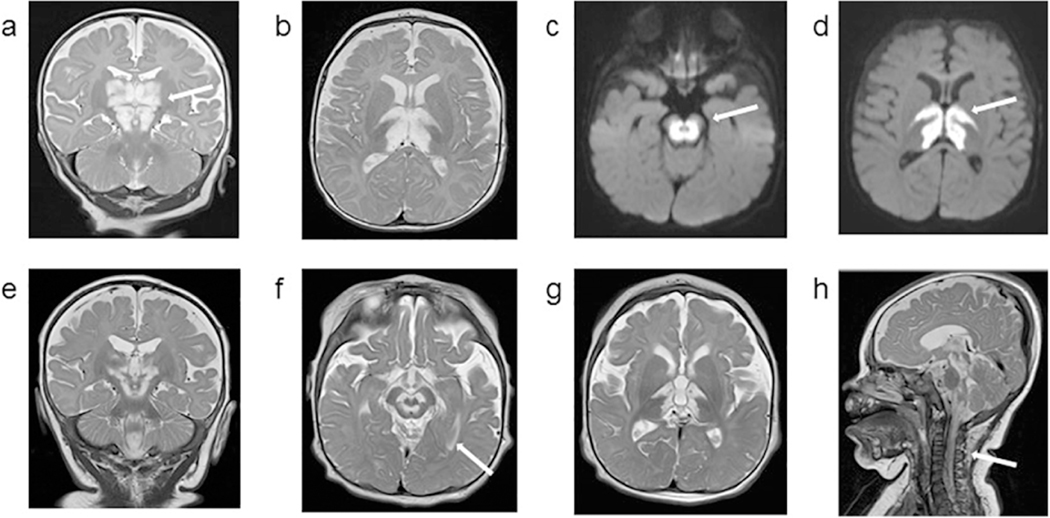
1.3. Patient 3: Axial MRI images obtained at 4 months (upper panel) and 8 months (lower panel) of age. Images a, b, e, f, g and h are T2 weighted images, and images c and d are diffusion weighted sequence (DWI). MRI obtained at 4 months shows extensive abnormal symmetrical T2 hyperintense signals involving large portions of the thalamus and midbrain, with restricted diffusion. There is also abnormal symmetrical T2 hyperintense signal with restricted diffusion in the bilateral posterior limbs of the internal capsules. The cerebellum and cerebellar white matter appeared spared. At 8 months of age, diffuse abnormal high signal involving the thalami, brainstem and proximal cervical spinal cord are noted. Since the prior study, T2 hyperintense signal is less extensive in the thalami, midbrain and internal capsule, but progression of disease in the pons and medulla with associated new areas of restricted diffusion is detected. Extensive abnormal T2 signal throughout the cervical and thoracic spinal cord to the level of T12, with most involvement in the cervical cord and cervicomedullary junction.
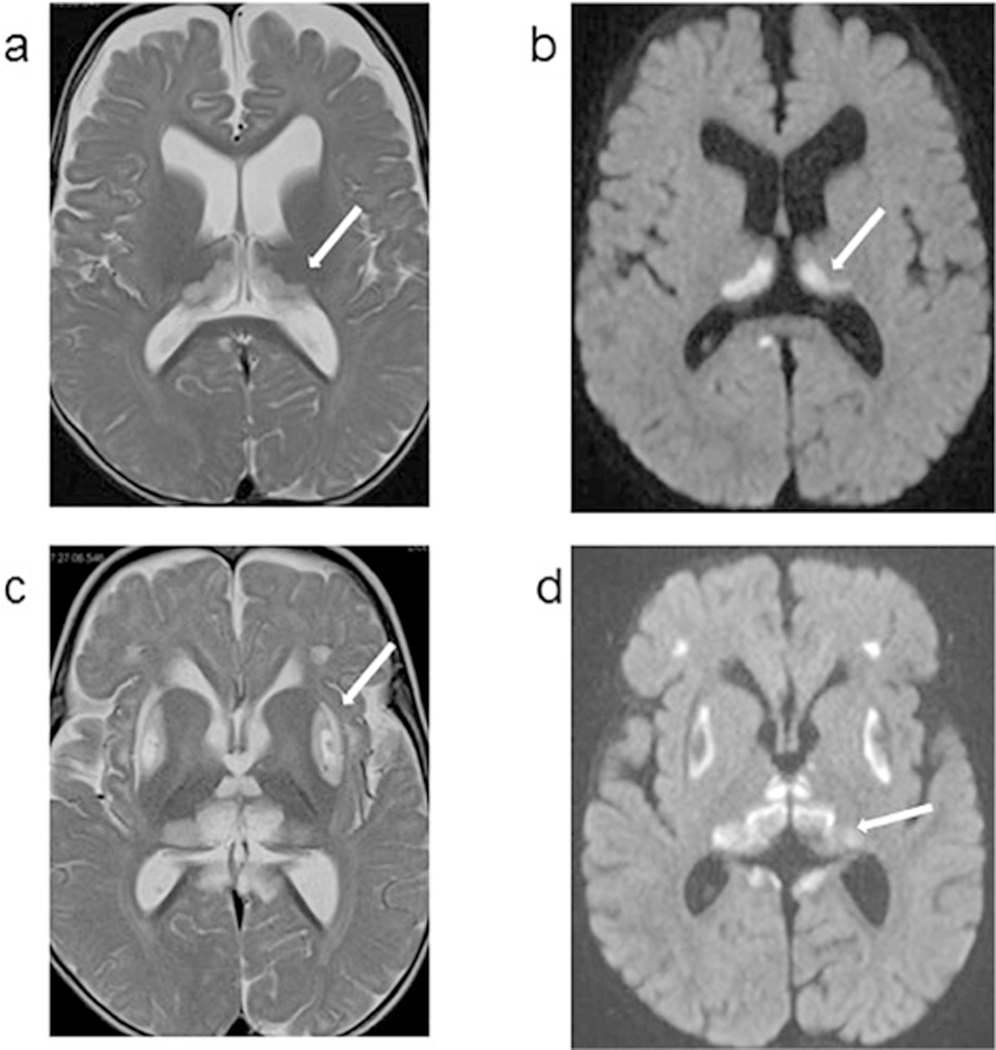
1.4. Patient 4: Axial MRI images obtained at 9 months (left panel) and 12 months (right panel) of age. Images a and c are T2 weighted images, and images b and d are diffusion weighted sequence (DWI). At 9 months, bilateral symmetrical hyperintensities are noted in thalami, with a semicircular shape. these thalamic bilateral signal abnormalities also show restricted diffusion on the DWI sequence. In the MRI obtained 3 months later, the putamen is also involved with cystic lesions. Small bilateral and symmetrical hyperintense areas are also noted in the frontal lobes and areas surrounding quadrigeminal cistern. The thalamic lesions have evolved since prior exam likely due to evolution from acute infarction with restricted diffusion to a chronic insult.
Initial metabolic evaluation, at ten months, revealed lactic acidosis at 4.5 mmol/l (normal <2.2), elevated 3-hydroxybutyrate at 14.1 mg/dl (normal, <2.81) and abnormal urine organic acids (increased 3-OH-butyrate, fumarate, malate and 2-ketoglutarate). Treatment with thiamine was started with no clinical improvement. The patient’s appetite progressively decreased and feeding difficulties became more severe. At twelve months, her height and weight were below the third percentile while her head circumference was in the 21st percentile. Developmentally, she could pull to stand, walked with support and was babbling.
Full sequence of mtDNA as well as POLG1 and SURF1 sequences were normal. This was followed by whole exome sequencing, which revealed compound heterozygous mutations in NDUFAF5 at position c.155A>C; p.Lys52Thr and c.836T>G; p.Met279Arg (Figure 2A). The p.Met279Arg mutation has been previously reported in compound heterozygosity with p.Arg49Gly in a Chinese infant with hypotonia, seizures, hyponatremia and abnormal MRI revealing brainstem lesions and hypoplasia of the corpus callosum who died of respiratory failure at eight months of age [20]. Six out of six interrogated pathogenicity prediction tools deem this mutation as pathogenic (Supplemental Table 2) which is in part due to the fact that the canonical methionine residue is highly conserved throughout evolution (Figure 2B). The p.Lys52Thr is novel and five out of six in silico tools predict the mutation as damaging (Supplemental Table 2) with high conservation throughout evolution (Figure 2B). p.Lys52Thr has been reported eleven times and p.Met279Arg seventeen times in the gnomAD database with approximately 250,000 alleles interrogated. Both mutations segregate exclusively with East Asian ethnic background, with an allele frequency of approximately 1/1600 and 1/1100 respectively but are never observed in a homozygous manner (Supplemental Table 2). After molecular diagnosis of NDUFAF5-associated Leigh’s disease was made, treatment with coenzyme Q10 and levocarnitine were added.
Figure 2:
A. Pedigrees of four unrelated families with mutations in NDUFAF5 and Leigh syndrome. Open symbols with dots indicate mutation carriers, grey shading indicates Leigh syndrome; (p) indicates a paternally (m) a maternally inherited mutation. The S-adenosylmethionine-dependent methyltransferase domain is abbreviated as “SAM domain” B. The conservation between 7 species is demonstrated by protein sequence alignment with the mutation shaded in grey.
At sixteen months, due to continued feeding difficulties and failure to thrive, the patient was admitted for nutritional support and evaluation for gastric tube placement. Soon thereafter she developed hyponatremia (sodium 121 mEq/L [normal 132–143]), associated with decreased urine output (<0.6 ml/kg/h), increased urine sodium (173 mEq/L) and urine osmolarity (506 MOS/kg). She was diagnosed with SIADH and was treated with water restriction, and later-on with salt supplementation. Cortisol, ACTH, TSH, T4 and β-natriuretic peptide (BNP) values were normal. She also developed hypertension, of unclear etiology, that required treatment with propranolol and hydrochlorothiazide. During this thirty day hospitalization she experienced further deterioration, worsening of her hypotonia and regression of motor milestones. A G-tube was placed, and the patient was discharged on G-Tube feedings.
Over the following months, the patient was stable and had some improvement in her neurological status, until the age of twenty-five months when she contracted a respiratory infection which prompted rapid developmental regression. Her condition continued to worsen and at twenty-seven months she presented to the emergency room with fever and respiratory distress followed by cardiopulmonary arrest. She was resuscitated but had persistent metabolic acidosis and died five days later. Parents did not consent to biopsies, therefore functional studies were not possible.
Patient #2:
The patient is the second child of a non-consanguineous Taiwanese couple. A brother, 15 months her senior, is healthy and there was no additional contributory family history. The pregnancy was complicated by gestational diabetes and delivery was preterm at thirty weeks with a birthweight of 2.3kg, after induction secondary to oligohydramnios. She was considered normal at birth and met all her early milestones. Initial symptoms started suddenly at twenty-seven months of age with strabismus, followed by ptosis and vomiting during an intercurrent illness. This prompted an admission where brain MRI studies showed hyperintense lesions in the medulla and upper cervical cord (Figure 1.2). Urine organic acids, biotinidase activity, as well as lactic acid and amino acids in blood and CSF were normal. A suspicion of mitochondrial disease prompted muscle biopsy, where histology revealed increase in number and size of lipid droplets but there were no ragged red fibers or other abnormal stains. Electron microscopy showed normal mitochondrial distribution, number and shape. Electron transport chain studies on mitochondria, isolated from fresh muscle, were also normal (Table 1). Molecular studies on DNA isolated from muscle were negative for mtDNA common point mutations and deletions/duplications. The patient was started on a mitochondrial cocktail including coenzyme Q10 200 mg/day, riboflavin 30 mg/day, thiamine 10 mg/day and carnitine 360 mg/day. Her overall condition slowly improved, but at 35 months, during an intercurrent illness, organic acids were again abnormal with large increase in 3-hydroxybutyrate and acetoacetate, and mild elevations of 3-methylglutaconic, fumarate and malate, suggestive of a mitochondrial disorder. The patient continued improving and over time her parents discontinued the mitochondrial cocktail and were lost to follow-up. At age seven years she was hospitalized due to frequent emesis, inability to walk and a skin rash on her head, neck, trunk and upper extremities, with severe itching. The rash had been resistant to treatment with steroids and antihistamines; symptoms worsened over several months with pain and infected lesions secondary to scratching. Simultaneously, the patient had developed significant fatigue, muscle weakness, headaches, pain in legs and back, as well as tremors and contractures in her hands and lost the ability to ambulate or feed herself. Physical exam revealed extensive erythematous skin rash with superimposed infected lesions in face neck and upper body. She had strabismus but no other focal defects. Muscle masses and strength were decreased while patellar reflexes were brisk, with positive ankle clonus. Ophthalmological testing revealed nystagmus as well as optic neuritis. She also had poor food intake and a feeding tube was placed. An MRI of the brain and spine showed worsening of the signal abnormalities in the brainstem with foci of symmetrical abnormalities in the cerebral peduncles and extensive abnormal T2 signal in cervical and thoracic spine (Figure 1.2). At this time, she was also found to have hyponatremia with sodium levels as low as 116 mEq/L secondary to cerebral salt wasting, which responded to treatment with salt supplementation. The patient was started on treatment with gabapentin for her severe pain and pruritus, with mild improvement. CSF studies for myelin basic protein and oligoclonal bands were negative. Three months later she was readmitted due to vertigo and weakness. On admission she was found to have hypertension, with systolic BP > 170 mmHg leading to seizures requiring treatment with IV labetalol. She was transitioned to propranolol and captopril with good blood pressure control but a few days later the patient had a respiratory arrest. She had to be intubated and underwent tracheostomy eight days later. Due to gastrostomy feeding intolerance, a gastrojejunostomy tube was placed. During this admission, hyponatremia was again identified despite treatment with salt supplementation and the patient developed neurogenic bladder requiring catheterizations. After a forty day admission she was transferred to a chronic care facility with tracheostomy and continuous ventilation support, on treatment with captopril (7.5 mg TID), thiamine (200 mg/day), riboflavin (50 mg/day), CoQ10 (300 mg/day) and carnitine (660 mg TID or 90 mg/kg/day). Her condition slowly improved, her ventilation support was weaned off over one year and the tracheostomy was decannulated the year after. Over this time the patient regained her muscle strength but was still unable to walk. Intellectually she was back to baseline, attending school at an age appropriate grade while requiring special assistance due to vision and ambulation problems. Her hypertension and hyponatremia resolved, and captopril and sodium supplementation were discontinued. Pruritus and skin lesions as well as lower extremity pain improved on treatment with gabapentin.
Table 1.
Clinical results for mitochondrial respiratory chain enzyme analyses in patient 2 and 3. Patient 2 was investigated in muscle as well as fibroblasts. Values are normalized by citrate synthase and expressed as percentage of the control mean. N/D indicates that this particular assay had not been performed.
| Complex Interrogated | Enzyme Analyzed | Patient 2 (age: 3 years) Muscle mitochondria | Patient 2 (age: 7 years) Fibroblast | Patient 3 Muscle Homogenate |
|---|---|---|---|---|
| Complex I | NADH-Ferricyanide-Dehydrogenase | N/D | N/D | 25% |
| NADH-CoQ Oxidoreductase | 115% | 131% | N/D | |
| Complex I→III | NADH-Cyt. C Reductase (Rotenone Sensitive | N/D | N/D | 40% total (26% rot.sensitive) |
| Complex II | Succinate Deyhydrogenase | N/D | N/D | 43% |
| Succinate-CoQ Oxidoreductase | N/D | 73% | N/D | |
| Complex II→III | Succinate-Cyt.c oxidoreductase (Antimycin Sensitive) | 125% | 66% | 38% |
| Complex III | Decylubiquinol Cytochrome C Reductase | N/D | 67% | N/D |
| Complex IV | Cytochrome c Oxidase | 81% | 108% | 34% |
| Citrate Synthase | 85% of control mean | 120% of control mean | 200% of control mean |
At ten years of age the patient was hospitalized with an episode of seizures and severe hyponatremia of 117 mEq/L as well as fever and increased work of breathing that resolved with careful adjustment of the electrolyte imbalance. She was discharged after three days in stable condition and sodium supplementation was continued. Additional testing done at the time included a microarray, pyruvate dehydrogenase assay, as well as Sanger sequencing for mtDNA and multiple nuclear encoded mitochondrial genes, which were all negative.
From age ten years until her current age of nineteen years, she had an overall stable clinical course, with intermittent worsening at times during intercurrent illness or exertion resulting in acute altered level of consciousness with fatigue and difficulty to arouse. Her blood pressure has been managed with enalapril and later with lisinopril, GI motility improved and was able to tolerate GT as well as some oral feeds. Vision difficulties worsened as she developed bilateral optic neuropathy. Urinary incontinence is currently intermittent, but she does not require frequent catheterizations. Her rash and pain have been under control with gabapentin. She remains wheel chair bound and was diagnosed with hip dislocation and scoliosis. Her intellect continues to be normal and she has graduated from high school planning to attend a community college.
Exome sequencing performed at age thirteen revealed the same NDUFAF5 mutation, p.Met279Arg, found in patient 1. In this case the mutation is homozygous, with both parents being carriers (Figure 2). p.Met279Arg has been reported seventeen times in the gnomAD database with all seventeen alleles segregating with East Asian ethnicity and an allele frequency of 0.0009 in that ethnic group (Supplemental Table 2). Since patient 1 and 2 are of Taiwanese descent we investigated the allele frequency of p.Met279Arg in 220 alleles from Taiwanese controls with negative results, ruling out the possibility that this mutation represents a common Taiwanese polymorphism.
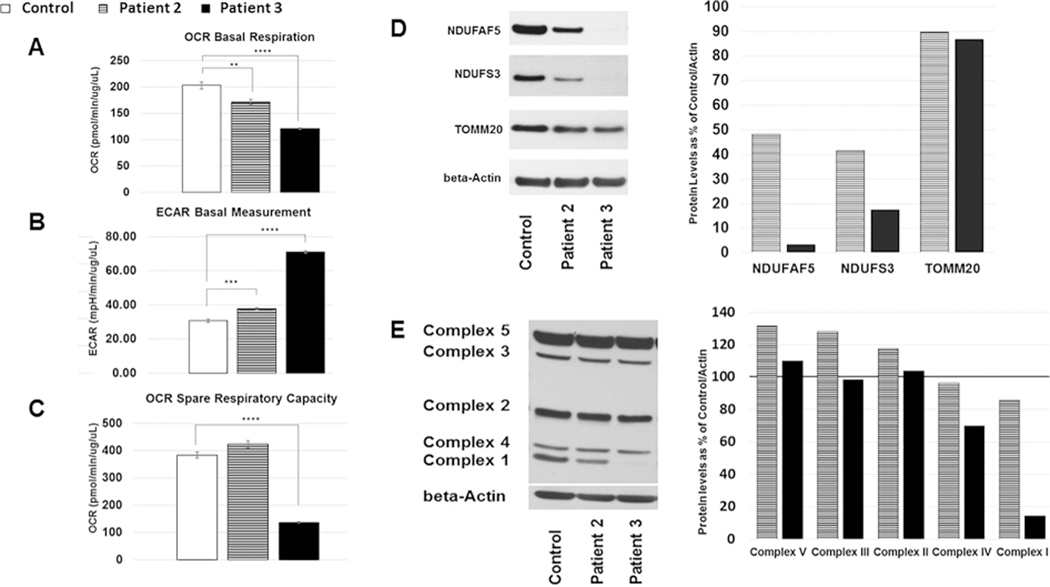
At this time, respiratory chain enzyme analysis via spectro-photometric methods were performed in fibroblasts in a commercial laboratory (CIDEM), with normal results for complex II through IV and unexpectedly high activity for complex I. (Table 1). Follow-up testing on whole cells using the Oroboros system was also reported as normal (Supplemental Table 2). Cellular respiration capacity interrogated using the Seahorse Bioanalyzer showed low basal respiration and high extracellular acidification rate indicating a respiratory chain defect (Figure 3A and B) while spare respiratory capacity was normal (Figure 3C). We then proceeded to assess whether the p.Met279Arg variant may have affected NDUFAF5 protein levels by performing SDS polyacrylamide gel electrophoresis (SDS-PAGE) on fibroblast cell lysates. Results showed levels of approximately 50% of NDUFAF5 compared to control (Figure 3D). We also interrogated protein levels for the complex I subunit NDUFS3, representing the peripheral arm of complex I, and found decreased levels at 40% of control (Figure 3D). To investigate the possibility of mitochondrial proliferation we assessed the levels of the outer mitochondrial membrane marker TOMM20, which were essentially normal, indicating normal levels of mitochondrial content (Figure 3D).
We also interrogated protein levels of all five mitochondrial complexes via SDS-PAGE and an OXPHOS antibody cocktail that uses anti-NDUFB8 to interrogate the membrane arm of complex I (Figure 3E) with essentially normal results indicating that p.Met279Arg is a mild mutation.
Figure 3:
A-C Results of experiments on fibroblasts assessed on a Seahorse Bioscience XF24 Flux Analyzer. A. Oxygen Consumption Rate (OCR) at baseline B. Extracellular Acidification Rate (ECAR) at baseline. C. Spare Respiratory Capacity (defined as maximal respiration minus basal respiration). Data was analyzed using XF Cell Mito Stress Test Report Generator. Values were normalized per ug/ul of protein. Unpaired t-test with unequal variance was performed for statistical analysis (mean +/− SEM). D-E Results of experiments on fibroblast protein lysates assessed using SDS-Page and applicable antibodies D. anti-NDUFAF5, anti-NDUFS3 and anti-TOMM20 with anti-beta-Actin as the loading control. Quantification of horizontal slices was performed using Image J and displayed as % of control, Images show cropped bands. E. Oxphos antibody cocktail interrogating stability of complex I with anti-NDUFB8, complex II with anti-SDHB, complex III with anti-UQCRC2, complex IV with anti-COX II, and complex V with anti-ATP5A, anti-beta-Actin was used as the loading control. Image was cropped for beta Actin only. Quantification of data displayed as % of control. Striped shading indicates patient 2, black shading patient 3.
Patient #3
This patient was the only child of a non-consanguineous couple with no significant family history. He was born via C-section due to breech presentation at 39 weeks with a birth weight of 3.5 kg. At 1 month of age he was diagnosed with gastroesophageal reflux, treated with ranitidine. The patient met all milestones until three months when parents were concerned about irritability, decreased sleep and arching, followed by loss of head control and social smile. At four months, he was admitted due to concern for seizures. Physical examination showed a normocephalic, non-dysmorphic irritable infant with severe truncal hypotonia but increased tone and deep tendon reflexes in extremities with positive ankle clonus. MRI showed symmetrical T2 hyperintense signals involving portions of the thalamus and midbrain. A video telemetry study was diagnostic for infantile spasms and he was started on adrenocorticotrophic hormone (ACTH). The metabolic service was consulted, mitochondrial diseases or a biotin-thiamine transporter defect were considered, and treatment with CoQ-10, biotin and thiamine were started. Laboratory results included increased lactic acid and alanine in serum at 2.9 (0.7–2.1 mmol/L) and 455 (143–439 umol/L) respectively, as well as in CSF at 4.5 (0.7–1.9 mmol/L) and 57 (6–41 umol/L). Urine organic acids showed mild increase in lactate, pyruvate, 3-OH-butyrate, fumarate and malate with large increase in 2-ketoglutarate and citrate. Carnitine, acylcarnitines, ammonia and biotinidase were normal as well as a chromosomal microarray.
While on ACTH the patient developed hypertension, which over time required treatment with hydralazine, amlodipine and enalapril. He was discharged after eight days but was readmitted at five months due to increased irritability, reflux, poor oral intake and suspected new seizure activity. Examination was also concerning for mild hyperventilation, with respiratory alkalosis and lack of pupillary response to light. A follow-up video telemetry electroencephalogram displayed generalized background slowing, multifocal epileptiform activity, asymmetry of the sleep architecture and epileptic infantile spasms. Ophthalmological evaluation was consistent with cortical blindness and a swallowing study was positive for aspiration, prompting initiation of NG tube feedings until a gastrostomy was placed at six months. A muscle biopsy was obtained at the time of gastrostomy placement. Modified trichrome staining of muscle revealed some myofibers with prominent granular staining, consistent with ragged red fibers, although electron microscopy examination did not show abnormal mitochondrial number or structure. Electron transport chain studies on muscle were diagnostic for a mitochondrial complex I defect (Table 1). Seahorse respiration studies on patient fibroblasts revealed decreased basal respiration and spare respiratory capacity as well as increased extracellular acidification rates (Figure 3A–C).
His seizures persisted and vigabatrin was added while ACTH treatment was weaned and discontinued. However, he continued with intermittent hypertension requiring medication after discontinuation of ACTH. At eight months, he was taken to the hospital due to difficulty breathing and a cyanotic episode. He was found to have hypoxia and respiratory acidosis requiring endotracheal intubation; repeat brain MRI confirmed progression of disease (Figure 1.3) and family decided not to continue aggressive care. The patient was extubated and died shortly thereafter. At six months of age, clinical whole exome sequencing for the parental-patient trio was negative. Clinical whole genome sequencing on the trio performed by the Illumina iHope program which was also negative. Data was obtained with parental consent (CHOC approved IRB 130990); re-analysis of clinical sequencing data on a research basis revealed a potential disease candidate variant in exon 3 of NDUFAF5 at position c.327G>C, p.Lys109Asn (maternal) (all nomenclature refers to NM_024120, CCDS13118.1, NP_077025 ISOFORM 1) (Figure 2A), which had bypassed the clinical filtering criteria. This variant is reported 117 times in the gnomAD database but never in homozygous fashion. Its allele frequency is 0.002 in the Ashkenazi Jewish population and 0.0007 in Non-Finnish Europeans. The variant is classified as possibly pathogenic by three out of six in silico pathogenicity prediction tools (Supplemental Table 2).
ClinVar has two entries concerning this variant. An entry from 8/2016 (Praxis fuer Humangenetik, Tuebingen, Germany) classifies the variant as “uncertain significance” while the second entry from 5/2017 (GeneDx) classifies the variant as likely pathogenic. SDS-PAGE for NDUFAF5 on patient fibroblasts revealed significantly decreased levels (5% of control), further substantiating our suspicion that the patient’s phenotype was due to mutations in this gene (Figure 3D). We then interrogated several other complex I subunits (NDUFS3 (Figure 3D) and NDUFB8 as the complex I interrogating component of the human mitochondrial oxphos cocktail (Figure 3E) which were also low, a finding consistent with a complex I assembly defect [21].
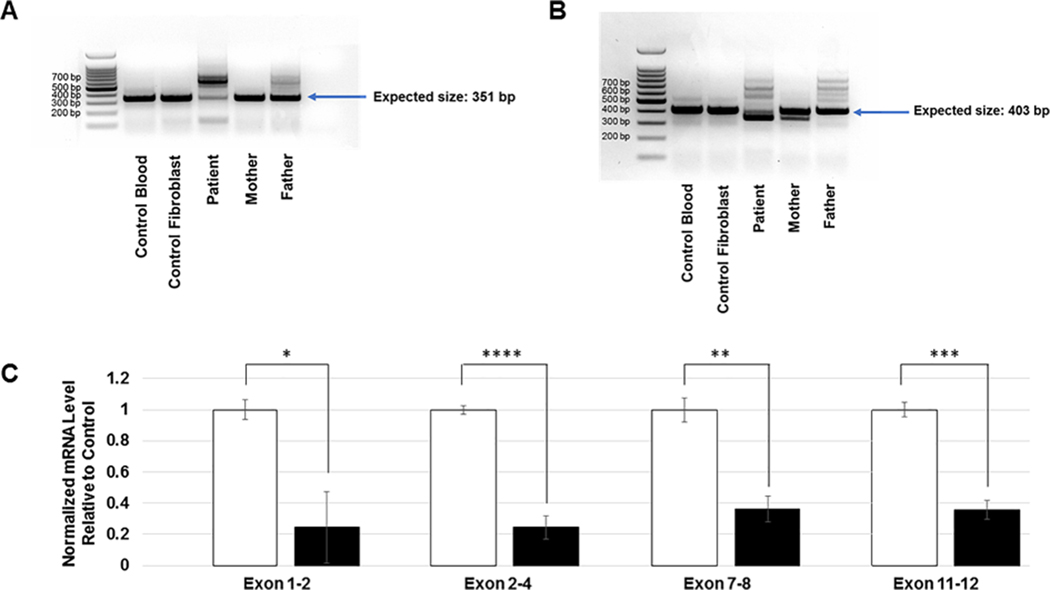
To investigate the location of a potential second intronic NDUFAF5 mutation we filtered the patient’s 7,181,082 WGS variants for NDUFAF5, which resulted in 45 hits. One variant fit inheritance of rare recessive disease (solely inherited from, and not homozygous in, the father) at position c.223–907A>C (20:13,767,051 A>C) which to our knowledge has not been reported. This intronic variant results in the creation of a new, exonic splice enhancer (ESE) site. Human Splicing Finder version 3.1 (http://www.umd.be/HSF3) predicts the formation of four splice enhancer motifs: Cgccctt, tcatCg,catCgc and atCgcc due to the c.223–907A>C variant. To test whether the new ESE site induced aberrant splicing, we generated RNA from controls, patient fibroblasts, as well as from blood for both parents. We performed one step RT-PCR with a forward primer in the 5’UTR and a reverse primer in exon 3 with an expected amplicon size of 351bp. Amplification of RNA extracted from control blood, control fibroblasts (patient age matched) and mother’s blood resulted in a single band of the expected size while additional, higher molecular weight bands were observed for the patient and the father (Figure 4A).
Figure 4:
A. RT-PCR of RNA from control, patient 3 and patient 3 parents using forward primers in 5’UTR and reverse primers in Exon 3 B. RT-PCR of RNA from control, patient 3 and patient 3 parents using forward primers in 5’UTR and reverse primers in Exon 4. C. qPCR demonstrating normalized mRNA levels for the respective NDUFAF5 exons interrogated. RNA was extracted from patient 3 fibroblasts and age matched control. Beta-Actin was used as an internal standard. Unpaired t-test with unequal variance was performed for statistical analysis, error bars indicate standard deviation.
We then re-amplified the father’s and patient’s RNA using a forward primer in exon 1 and the same reverse primer as mentioned above in exon 3 and sequenced the most prominent band upon agarose gel purification (Supplemental Figure 2A). Results showed inclusion of a 258bp cryptic exon spliced between exon 1 and 2, with the new splice acceptor site at position c.223–964(A)_963(G) (Supplemental Figure 2B). The new ESE site formed by the paternal intronic mutation is the 56th base pair into the cryptic exon and is represented in the Sanger Sequencing of the gel purified band as a homozygous C since it represents only the paternal allele (Supplemental Figure 2C). Extending 31 amino acids into the cryptic exon the open reading frame ends in a STOP codon (Supplemental Figure 2B and C). These results indicate that the paternal mutation causes abnormal splicing of intron 1.
We then interrogated whether the maternal mutation at position c.327G>C, p.Lys109Asn also results in abnormal splicing. The strength of the canonical intronic splice donor site GT is modulated by the last two base pairs of an exon with the most highly conserved sequence being AG. Since the maternal c.327G>C mutation changes the last basepair of exon 3 to a C we expected weakening of the splice donor. We therefore performed one step RT-PCR with the forward primer in the 5’UTR and a reverse primer in exon 4 with an expected amplicon size of 403bp. Amplification of RNA extracted from control blood, control fibroblasts (patient age matched) and father’s blood resulted in a single band of the expected size while an additional, lower molecular weight band were observed for the patient and the mother (Figure 4B). We sequenced the lower molecular weight band from the patient upon agarose gel purification. Results were consistent with skipping of exon 3 which results in a STOP codon at the end of exon 4 for isoform #1. We then assessed NDUFAF5 mRNA levels on patient fibroblast RNA via qPCR using primers interrogating multiple exons and found NDUFAF5 mRNA levels to be decreased to 25 – 40% of normal. This decrease is consistent with abnormal splicing studies found associated with both variants (Figure 4C).
Patient #4
The patient was born to Ashkenazi Jewish parents at 36 weeks gestation by C-section secondary to maternal hypertension and non-identical twin gestation. The pregnancy was a product of IVF without complications. Birth weight was 2.32 kg and there were no neonatal complications. His development lagged behind his twin brother, and at five months of age a large head size prompted an ultrasound which did not show hydrocephalus. He developed torticollis, horizontal nystagmus, and head bobbing and was given the diagnosis of spasmus nutans by ophthalmology. By six months he was starting to roll over but was unable to sit up on his own or crawl like his brother. Nystagmus continued, and an MRI at nine months revealed areas symmetrical T2/flair hyperintensity in the thalami (Figure 1.4). He was admitted to the hospital for further evaluation. Physical exam showed a mildly hypotonic, delayed nine-month-old responsive male with active smile who was mildly dysmorphic. Height and weight were at 50th percentile, but his head circumference was 49 cm (>99th percentile). His head was asymmetric with a left-sided bulge and asymmetric ears, both attributable to fetal deformation. His eyes had horizontal nystagmus, his nose was anteverted, and his chin and mouth were small. A tentative diagnosis of Co-factor Responsive Basal Ganglia Disease/ SLC19A3 defect was made, and he was started on high dose biotin, pantothenic acid and thiamine supplementation. Organic acids, amino acids, lactic, pyruvic and very long chain fatty acids were normal. At eleven months of age he developed swallowing and feeding difficulties. At twelve months, he was admitted to the pediatric intensive care unit with metabolic acidosis triggered by vomiting. New multifocal serpiginous areas of restricted diffusion were found on MRI (Figure 1.4). Serum lactic acid was elevated at 3.08 (nl; 0.36–1.29 nmol/L), while CSF lactic and pyruvic were also elevated at 3.2 (nl; 1.1–2.8 mmol/L) and 2.60 (nl 0.50–1.70 mg/dL) respectively. Leigh syndrome was suggested and whole exome testing was ordered. He was started on levocarnitine supplementation and underwent a laparoscopic fundoplication with gastrostomy tube placement. Patient improved weight gain which was followed by slow improvement in his development. He was able to wave good bye, was babbling, walking in a walker, playing with toys and sitting up, but he was unable to crawl.
Exome sequencing revealed compound heterozygous mutations in NDUFAF5 at position c.327G>C, p.Lys109Asn (maternal) and c.749G>T, p.Gly250Val (paternal). At this time, a mitochondrial “cocktail”, including: CoQ-10 and a vitamin B complex was added. At sixteen months, patient began showing periods of apnea lasting up to twenty seconds. He was later admitted to the PICU for presumed respiratory infection, stridor and fever. Parents declined intubation or extreme measures and opted for palliative care. He died in his sleep at home at seventeen months of age.
p.Lys109Asn is the same mutation as observed for patient 3 of this report, where we establish pathogenicity via functional studies, however samples were not available for this patient. The pathogenicity of p.Gly250Val was reported previously and represents the common Ashkenazi Jewish founder mutation [12].
3. Discussion
NDUFAF5 mutations were first associated with mitochondrial respiratory complex I disease in 2008 [19], with only three additional reports published to date. The phenotypes have been heterogeneous, including severe disease with prenatal onset and neonatal demise, intermediate presentation with death in childhood, and milder disease with survival into adulthood [19, 22, 12, 20]. This variability may have been further confounded by the fact that three out of the four reports pertain to consanguineous families, with potential additional recessive mutations in the regions of homozygosity.
Our studies in four additional non-consanguineous families contribute to the delineation of the clinical, biochemical and molecular spectrum associated with NDUFAF5. In patients 1, 3 and 4 we observe a classical Leigh syndrome phenotype, with onset before six months of age and death between eight and twenty-seven months. Lactic acid was elevated in patients 1,3 and 4, urine organic acids were abnormal in patient 1 and 3 and complex I deficiency on muscle was identified for patient 3. All three patients had strikingly similar thalamic and brain stem lesions on MRI. These 3 patients were compound heterozygous, with patients 3 and 4 sharing the same novel missense mutation at position p.Lys109Asn. Our functional data in patient 3 strongly supports the pathogenicity of p.Lys109Asn despite the fact that complementation assays were not performed in this study. mRNA studies via RT-PCR showed that the mutation induces skipping of exon 3 resulting in low NDUFAF5 mRNA levels, likely secondary to nonsense-mediated decay. Western blot revealed severely decreased NDUFAF5 protein and moderately low levels in other complex I subunits suggestive of a complex I assembly defect. Additionally, muscle biopsy showed mild mitochondrial proliferation with normal electron microscopy, while complex I activity was low, consistent with the genetic defect. In concordance, fibroblast studies displayed decreased basal respiration and spare respiratory capacity with increased acidification rates. Moreover, in patient 4, p.Lys109Asn was found in conjunction with a previously described deleterious NDUFAF5 variant which further substantiates its pathogenicity (12).
The p.Lys109Asn mutation found in patients 3 and 4, has been reported a total of 117 times in the gnomAD database but never in a homozygous state. The mutation is predominantly found in European (non-Finnish) and Ashkenazi Jewish, where the carrier rate is 1/230 [23]. In agreement, patient 3 was Caucasian and patient 4 was of Ashkenazi Jewish descent. The relatively high allele frequency in conjunction with an indeterminate pathogenicity prediction score (3 out of 6 damaging) caused the mutation to be filtered-out as a VUS in our patient 3. Our data therefore has the potential to prompt re-analysis of WES and WGS SNPs and aid in diagnosing other complex I deficiency patients with previously negative results.
The delineation of the molecular pathology for patient 3 also demonstrates that muscle biopsy remains an important component of the mitochondrial disease diagnostic toolbox. Without enzymatic studies we would not have interrogated the pathogenicity of the p.Lys109Asn variant in this patient, particularly since we started our functional studies prior to the observation of p.Lys109Asn in patient 4. Our findings also add to the literature emphasizing the importance of whole genome sequencing as to increase diagnostic yield for rare disease [24]. Initial attempts to demonstrate a second mutation for patient 3 via RT-PCR were negative and sequencing of the entire NDUFAF5 cDNA was normal due to the low levels of the unstable, mutant RNA. Segregation analysis of WGS intronic variants coupled with the use of Human Splicing Finder afforded us the design of accurate primers. This step led to amplification of a low level transcript revealing the insertion of a cryptic exon [25].
Patient 2 of this publication displays an attenuated phenotype, similar to the brothers with a homozygous p.Leu159Phe mutation reported by Gerards et al [22]. These patients presented with spasticity at the age of three years, followed by dystonia and developmental delays and were reported to be alive in their late twenties. The authors suggest that the remaining complex I activity on muscle at 36% and mature assembled complex I at 40%, were responsible for the attenuated presentation, which may be attributable to the fact that the p.Leu159Phe mutation only affects isoform 1 of NDUFAF5 while isoform 2 is presumably normal. The p.Met279Arg mutation reported for our patient 2, does affect both isoforms of NDUFAF5 and therefore isoform specificity cannot explain the mild phenotype. While in silico tools predict this mutation to be deleterious, we found that the Seahorse studies showed only mild abnormalities at baseline with normal spare respiratory capacity. Additionally, ETC activity in muscle and fibroblasts were normal, consistent with a mild phenotype. While about 50% of remaining NDUFAF5 protein was present in the patient’s fibroblasts, additional protein studies suggest that the p.Met279Arg mutation selectively affects complex I assembly intermediates. In particular the level of the matrix arm subunit NDUFS3 of complex I was decreased, with only 40% remaining while the membrane arm associated accessory protein NDUFB8 was found at 80–90% of control on SDS-PAGE. We therefore suspect that the p.Met279Arg variant in homozygous state affects NDUFAF5 function involving complex I assembly, but not sufficiently to translate into respiratory deficiency. Gene complementation studies may further elucidate the effects of the p.Met279Arg mutation. In addition to the genotype, clinical outcome in patients with mitochondrial defects may be influenced by several factors, including the susceptibility and exposure to infections, rapid treatment during intercurrent illnesses and or response to medications. It is difficult to assess how these factors contributed to the mild disease in this patient.
In addition to the delineation of a novel mutation prevalent in Caucasians, our report points to a potential NDUFAF5 founder mutation in the East Asian population. The p.Met279Arg variant is found 17 times in gnomAD and segregates exclusively with East Asian heritage. Patient 1 and 2, both of Taiwanese descent, share the p.Met279Arg amino acid change and patient 2 is homozygous despite a lack of consanguinity. Additionally, the same change has been previously reported in a Chinese infant in conjunction with p.Arg49Gly and a phenotype similar to our patient 1 [20].
Interestingly patients 1 and 2, with p.Met279Arg, developed significant hyponatremia. In patient 1, hyponatremia became apparent at sixteen months of age and was secondary to SIADH, while in patient 2 it was first documented at ten years of age and was found to be secondary to cerebral salt wasting syndrome (CSWS). Interestingly, the patient described by Tong et al. , who carried the same mutation, presented with low sodium at 8 months of age, but the mechanism was not discussed in the paper [20]. Hyponatremia has been previously reported in association with mitochondrial disease, but the etiology is difficult to ascertain, with SIADH and CSWS being the most commonly suspected causes. A retrospective review in seven patients with MELAS, confirmed episodic hyponatremia with increased urine sodium in four of them. However, the etiology could only be clarified in two patients, being secondary to relative adrenal insufficiency and acute renal failure combined with paralytic ileus [26]. A review of hyponatremia associated with neuromyelitis optica, a disease with significant overlap with the mitochondrial optic neuropathies, offers several explanations why differential diagnosis is difficult and that SIADH and CSWS could occur in the same patient [27–29]. The authors speculate that hypothalamic lesions are mostly associated with SIADH since cell damage in the hypothalamus can lead to leakage of ADH into circulation. In spinal cord lesions however, impaired sympathetic vascular innervation may initially lead to hypovolemia and secretion of ADH but in later stages lead to damage of the renal sympathetic pathway which may impair sodium retention [27].
All three patients with p.Met279Arg had MRI lesions involving the brainstem and two of them also in the spinal cord, consistent with the above mentioned hypothesis. Unfortunately, specific images for the hypothalamus were not obtained, so lesions in that region could not be accurately evaluated. Three of our patients (patient 1–3) presented also with hypertension, a finding previously associated with Leigh Syndrome and spinal cord lesions [30, 31]. Involvement of the medulla oblongata, which integrates cardiorespiratory reflexes, has been suggested as a cause of the hypertension through dysregulation of central autonomic activity [30].
5. Conclusion and Clinical Significance
Our report adds clinical, biochemical and molecular descriptions for four new cases with NDUFAF5 defects including three novel mutations. In two of them, our functional data establishes the pathogenicity via RNA studies. Our findings suggest that complex I deficiency secondary to mutations in NDUFAF5 is likely underreported. Exome filtering criteria relying on pathogenicity prediction of missense variants via in silico tools are often too stringent and will miss important mutations. Additionally, in silico prediction for splice variants is often restricted to the splice junction which in combination with a lack of intronic data fails to interrogate pathogenic variants in deep intronic regions. Our report also adds to the literature of mitochondrial disease associated with hypertension and hyponatremia which was found in most of our patients.
Supplementary Material
Acknowledgements
The authors thank the families for their commitment to participation in these studies and all health care providers who cared for the patients and families.
Funding
This work was supported by the National Institutes of Health [5T32GM008243-32] to MTS; the Fry Family Foundation [40031028] to MTS, SSE and AES; and the Brian and Caris Chan Family Foundation and Campbell Family Foundation of Caring to RYW. WGS studies in patient 3 were funded by the Illumina iHope program.
Abbreviations
- ACP
Acyl carrier protein
- ACTH
Adrenocorticotropic hormone
- ADH
Anti-diuretic hormone
- BNP
β-natriuretic peptide
- BN-PAGE
Blue native polyacrylamide gel electrophoresis
- BP
Blood pressure
- bp
Base pair
- cDNA
Complementary DNA
- CSF
Cerebrospinal fluid
- CSWS
Cerebral salt wasting syndrome
- DNA
Deoxyribonucleic acid
- DNase
Deoxyribonuclease
- ECAR
Extracellular acidification rate
- ED
Emergency department
- ESE
Exonic splice enhancer
- ETC
Electron transport chain
- FASII
Fatty acid synthesis type II
- FBS
Fetal bovine serum
- gnomAD
Genome aggregation database
- GT
Gastrostomy tube
- G-tube
Gastrostomy tube
- HRP
Horse radish peroxidase
- IRB
Institutional review board
- IVF
In vitro fertilization
- MELAS
Mitochondrial encephalopathy, lactic acidosis, and stroke-like symptoms
- MEM
Minimal essential medium
- MRI
Magnetic resonance imaging
- mRNA
Messenger ribonucleic acid
- mtDNA
Mtochondrial DNA
- NADH
Nicotinamide adenine dinucleotide + hydrogen
- nDNA
Nuclear DNA
- N/D
Not done
- NEAA
Non-essential amino acids
- NG
Nasogastric
- NR
Not reported
- NSVD
Normal spontaneous vaginal delivery
- OCR
Oxygen consumption rate
- OXPHOS
Oxidative phosphorylation
- PICU
Pediatric intensive care unit
- PVDF
Polyvinylidene difluoride
- PBS
Phosphate buffered saline
- qPCR
Quantitative polymerase chain reaction
- RNA
Ribonucleic acid
- RT
Room temperature
- RT-PCR
Reverse transcriptase polymerase chain reaction
- SAM
S-adenosylmethionine-dependent methyltransferase
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SIADH
Syndrome of inappropriate anti-diuretic hormone
- SNP
Signal nucleotide polymorphism
- TSH
Thyroid stimulating hormone
- UTR
Untranslated region
- VUS
Variant of undetermined significance
- WES
Whole exome sequencing
- WGS
Whole genome sequencing
Footnotes
Conflict of Interest
The authors do not report any conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finsterer J. Leigh and Leigh-Like Syndrome in Children and Adults. Pediatric Neurology 2008, 39, 223–235, doi: 10.1016/j.pediatrneurol.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 2.Rahman S; Blok RB; Dahl HH; Danks DM; Kirby DM; Chow CW; Christodoulou J; Thorburn DR Leigh syndrome: clinical features and biochemical and DNA abnormalities. Ann Neurol 1996, 39, 343–351, doi: 10.1002/ana.410390311. [DOI] [PubMed] [Google Scholar]
- 3.Bekiesinska-Figatowska M; Mierzewska H; Jurkiewicz E. Basal ganglia lesions in children and adults. European Journal of Radiology 2013, 82, 837–849, doi: 10.1016/j.ejrad.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Fassone E; Rahman S. Complex I deficiency: clinical features, biochemistry and molecular genetics. Journal of Medical Genetics 2012, 49, 578–590, doi: 10.1136/jmedgenet-2012-101159. [DOI] [PubMed] [Google Scholar]
- 5.Wallace DC Mitochondrial diseases in man and mouse. Science 1999, 283, 1482–1488. [DOI] [PubMed] [Google Scholar]
- 6.Rhein VF; Carroll J; Ding S; Fearnley IM; Walker JE NDUFAF7 Methylates Arginine-85 in the NDUFS2 Subunit of Human Complex I. J Biol Chem 2013, doi: 10.1074/jbc.M113.518803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LAZAROU M; THORBURN D; RYAN M; MCKENZIE M. Assembly of mitochondrial complex I and defects in disease. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2009, 1793, 78–88, doi: 10.1016/j.bbamcr.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Guerrero-Castillo S; Baertling F; Kownatzki D; Wessels HJ; Arnold S; Brandt U; Nijtmans L. The Assembly Pathway of Mitochondrial Respiratory Chain Complex I. Cell Metabolism 2017, 25, 128–139, doi: 10.1016/j.cmet.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Herzer M; Koch J; Prokisch H; Rodenburg R; Rauscher C; Radauer W; Forstner R; Pilz P; Rolinski B; Freisinger P; et al. Leigh disease with brainstem involvement in complex I deficiency due to assembly factor NDUFAF2 defect. Neuropediatrics 2010, 41, 30–34, doi: 10.1055/s-0030-1255062. [DOI] [PubMed] [Google Scholar]
- 10.Baertling F; Sánchez-Caballero L; Timal S; van den Brand M; Ngu LH; Distelmaier F; Rodenburg RJ; Nijtmans LG Mutations in mitochondrial complex I assembly factor NDUFAF3 cause Leigh syndrome. Molecular Genetics and Metabolism 2017, 120, 243–246, doi: 10.1016/j.ymgme.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Baertling F; Sánchez-Caballero L; van den Brand MAM; Wintjes LT; Brink M; van den Brandt FA; Wilson C; Rodenburg RJT; Nijtmans LGJ NDUFAF4 variants are associated with Leigh syndrome and cause a specific mitochondrial complex I assembly defect. European Journal of Human Genetics : EJHG 2017, 25, 1273–1277, doi: 10.1038/ejhg.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saada A; Edvardson S; Shaag A; Chung WK; Segel R; Miller C; Jalas C; Elpeleg O. Combined OXPHOS complex I and IV defect, due to mutated complex I assembly factor C20ORF7. J Inherit Metab Dis 2012, 35, 125–131, doi: 10.1007/s10545-011-9348-y. [DOI] [PubMed] [Google Scholar]
- 13.McKenzie M; Tucker EJ; Compton AG; Lazarou M; George C; Thorburn DR; Ryan MT Mutations in the gene encoding C8orf38 block complex I assembly by inhibiting production of the mitochondria-encoded subunit ND1. Journal of molecular biology 2011, 414, 413–426, doi: 10.1016/j.jmb.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Floyd BJ; Wilkerson EM; Veling MT; Minogue CE; Xia C; Beebe ET; Wrobel RL; Cho H; Kremer LS; Alston CL; et al. Mitochondrial Protein Interaction Mapping Identifies Regulators of Respiratory Chain Function. Molecular cell 2016, 63, 621–632, doi: 10.1016/j.molcel.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fassone E; Duncan AJ; Taanman J-W; Pagnamenta AT; Sadowski MI; Holand T; Qasim W; Rutland P; Calvo SE; Mootha VK; et al. FOXRED1, encoding an FAD-dependent oxidoreductase complex-I-specific molecular chaperone, is mutated in infantile-onset mitochondrial encephalopathy. Hum Mol Genet 2010, 19, 4837–4847, doi: 10.1093/hmg/ddq414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Formosa LE; Dibley MG; Stroud DA; Ryan MT Building a complex complex: Assembly of mitochondrial respiratory chain complex I. Seminars in cell & developmental biology 2017, doi: 10.1016/j.semcdb.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Stroud DA; Surgenor EE; Formosa LE; Reljic B; Frazier AE; Dibley MG; Osellame LD; Stait T; Beilharz TH; Thorburn DR; et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature 2016, 538, 123–126, doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- 18.Rhein VF; Carroll J; Ding S; Fearnley IM; Walker JE NDUFAF5 Hydroxylates NDUFS7 at an Early Stage in the Assembly of Human Complex I. J Biol Chem 2016, 291, 14851–14860, doi: 10.1074/jbc.M116.734970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugiana C; Pagliarini DJ; McKenzie M; Kirby DM; Salemi R; Abu-Amero KK; Dahl H-HM; Hutchison WM; Vascotto KA; Smith SM; et al. Mutation of C20orf7 Disrupts Complex I Assembly and Causes Lethal Neonatal Mitochondrial Disease. The American Journal of Human Genetics 2008, 83, 468–478, doi: 10.1016/j.ajhg.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong W; Wang Y; Lu Y; Ye T; Song C; Xu Y; Li M; Ding J; Duan Y; Zhang Le; et al. Whole-exome Sequencing Helps the Diagnosis and Treatment in Children with Neurodevelopmental Delay Accompanied Unexplained Dyspnea. Scientific reports 2018, 8, 5214, doi: 10.1038/s41598-018-23503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piekutowska-Abramczuk D; Assouline Z; Mataković L; Feichtinger RG; Koňařiková E; Jurkiewicz E; Stawiński P; Gusic M; Koller A; Pollak A; et al. NDUFB8 Mutations Cause Mitochondrial Complex I Deficiency in Individuals with Leigh-like Encephalomyopathy. American Journal of Human Genetics 2018, 102, 460–467, doi: 10.1016/j.ajhg.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerards M; Sluiter W; van den Bosch BJC; Wit L.E.A.de; C. MH; Frentzen M; Akbari H; Schoonderwoerd K; Scholte HR; Jongbloed RJ; et al. Defective complex I assembly due to C20orf7 mutations as a new cause of Leigh syndrome. Journal of Medical Genetics 2010, 47, 507–512, doi: 10.1136/jmg.2009.067553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lek M; Karczewski KJ; Minikel EV; Samocha KE; Banks E; Fennell T; O’Donnell-Luria AH; Ware JS; Hill AJ; Cummings BB; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291, doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lionel AC; Costain G; Monfared N; Walker S; Reuter MS; Hosseini SM; Thiruvahindrapuram B; Merico D; Jobling R; Nalpathamkalam T; et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genetics in medicine : official journal of the American College of Medical Genetics 2018, 20, 435–443, doi: 10.1038/gim.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desmet F-O; Hamroun D; Lalande M; Collod-Beroud G; Claustres M; Beroud C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Research 2009, 37, e67–e67, doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubota H; Tanabe Y; Takanashi J.-i.; Kohno Y. Episodic hyponatremia in mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes (MELAS). J Child Neurol 2005, 20, 116–120. [DOI] [PubMed] [Google Scholar]
- 27.Jin S; Long Z; Wang W; Jiang B. Hyponatremia in neuromyelitis optica spectrum disorders: Literature review. Acta Neurologica Scandinavica 2018, 138, 4–11, doi: 10.1111/ane.12938. [DOI] [PubMed] [Google Scholar]
- 28.McClelland CM; van Stavern GP; Tselis AC Leber hereditary optic neuropathy mimicking neuromyelitis optica. Journal of neuro-ophthalmology : the official journal of the North American Neuro-Ophthalmology Society 2011, 31, 265–268, doi: 10.1097/WNO.0b013e318225247b. [DOI] [PubMed] [Google Scholar]
- 29.Yu-Wai-Man P; Votruba M; Burté F; La Morgia C; Barboni P; Carelli V. A neurodegenerative perspective on mitochondrial optic neuropathies. Acta Neuropathologica 2016, 132, 789–806, doi: 10.1007/s00401-016-1625-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohmeier K; Distelmaier F; van den Heuvel LP; Rodenburg RJT; Smeitink J; Mayatepek E; Hoehn T. Fatal hypertensive crisis as presentation of mitochondrial complex I deficiency. Neuropediatrics 2007, 38, 148–150, doi: 10.1055/s-2007-985903. [DOI] [PubMed] [Google Scholar]
- 31.Narita T; Yamano T; Ohno M; Takano T; Ito R; Shimada M. Hypertension in Leigh syndrome--a case report. Neuropediatrics 1998, 29, 265–267, doi: 10.1055/s-2007-973572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.