Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein acquired a D614G mutation early in the pandemic that confers greater infectivity and is now the globally dominant form. To determine whether D614G might also mediate neutralization escape that could compromise vaccine efficacy, sera from spike-immunized mice, nonhuman primates, and humans were evaluated for neutralization of pseudoviruses bearing either D614 or G614 spike. In all cases, the G614 pseudovirus was moderately more susceptible to neutralization. The G614 pseudovirus also was more susceptible to neutralization by receptor-binding domain (RBD) monoclonal antibodies and convalescent sera from people infected with either form of the virus. Negative stain electron microscopy revealed a higher percentage of the 1-RBD “up” conformation in the G614 spike, suggesting increased epitope exposure as a mechanism of enhanced vulnerability to neutralization. Based on these findings, the D614G mutation is not expected to be an obstacle for current vaccine development.
Keywords: vaccine, mRNA, LNP, COVID-19, SARS-CoV-2, nucleoside-modified, neutralization, Spike, electron micrograph, D614G
Graphical Abstract
Serum from SARS-CoV-2 spike-vaccinated mice, NHPs and humans, and convalescent patients, along with receptor-binding domain (RBD)-specific monoclonal antibodies neutralize the widespread G614-containing virus at greater levels than the original D614 version. Structural data demonstrate that the G614 spike is in a more open conformation with extended RBDs.
Introduction
There is an urgent worldwide need to develop a safe and effective vaccine for coronavirus disease 2019 (COVID-19). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiologic agent of COVID-19, is a novel coronavirus that was first reported in Wuhan, China in December 2019 and four months later was declared a pandemic by the World Health Organization. Vaccine efforts began shortly after the first sequence of the virus was made available in January 2020 (Lurie et al., 2020). Presently, at least 30 SARS-CoV-2 vaccines are in clinical trials (https://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-vaccine-tracker). Because of the urgency and time required to develop a vaccine, most COVID-19 vaccines entering phase 3 trials are based on an early index strain of the virus. These vaccines focus on the viral spike protein with the goal of eliciting protective neutralizing antibodies (Jackson et al., 2020; Mulligan et al., 2020).
The trimeric spike protein mediates virus attachment and entry into host cells (Yuan et al., 2020). Each monomer is comprised of an S1 subunit, which contains the receptor-binding domain (RBD), and an S2 subunit that mediates membrane fusion (Wrapp et al., 2020). The RBD is the primary target of neutralizing antibodies, although the N-terminal domain and other regions of spike are also known to possess neutralization epitopes (Brouwer et al., 2020; Corbett et al., 2020; Liu et al., 2020; Pinto et al., 2020; Shi et al., 2020; Wec et al., 2020; Wu et al., 2020; Yuan et al., 2020).
Early in the pandemic, the virus acquired a D614G mutation in the spike protein that rapidly increased in frequency and is now the dominant form of the virus globally (Biswas and Majumder, 2020; Gong et al., 2020; Isabel et al., 2020; Islam et al., 2020; Korber et al., 2020a; Koyama et al., 2020a; Koyama et al., 2020b; Mercatelli and Giorgi, 2020). The pattern of spread, combined with increased infectivity in vitro, suggests the mutation gave the virus a fitness advantage for transmission (Korber et al., 2020a; Zhang et al., 2020), despite the fact that this mutation is outside of the RBD in the S1 subunit of spike. The mutation is also associated with higher virus loads in respiratory secretions, but it does not appear to increase disease severity (Korber et al., 2020a). A critical question is whether this mutation also mediates neutralization escape that could reduce the effectiveness of vaccines in general and especially ones based on the D614 version of the spike immunogen.
Here, we addressed this question by using sera from spike-immunized mice, nonhuman primates (NHPs), and humans. Neutralization was measured in 293T/ACE2 cells by using lentivirus particles pseudotyped with full-length SARS-CoV-2 spike containing either D614 or G614. Because this was the only difference between the pseudoviruses, any change in phenotype can be directly attributed to D614G.
Results
Mice, rhesus macaques, and humans were immunized with the nucleoside-modified mRNA-LNP vaccine platform (Alameh et al., 2020; Pardi et al., 2018a). Four different variants of the spike protein were used as mRNA-encoded immunogens (Table S1, related to Figure 1 ): (1) monomeric secreted RBD protein (Laczkó et al., 2020) in macaques; (2) trimeric RBD protein made by adding a T4 fibritin-derived “foldon” trimerization domain to increase immunogenicity (Güthe et al., 2004) in humans; (3) prefusion diProline stabilized, first developed with the MERS spike immunogen (Pallesen et al., 2017), full-length cell-associated spike protein (Kirchdoerfer et al., 2016; Kirchdoerfer et al., 2018; Wrapp et al., 2020) in mice; and (4) Wuhan index strain (Lurie et al., 2020) with a mutated furin cleavage-site cell-associated trimeric spike (Laczkó et al., 2020) in macaques. The furin mutant potentially stabilizes the full-length spike and maintains the association of the S1 and S2 subunits (Kirchdoerfer et al., 2016). Pseudoviral neutralization titers were calculated as 50% and 80% inhibitory doses (ID50 and ID80, respectively). Assays were performed with nearly equivalent amounts of input virus doses for both forms of the virus, i.e., relative light units (RLU) in virus control wells were 501–840× background in all assays.
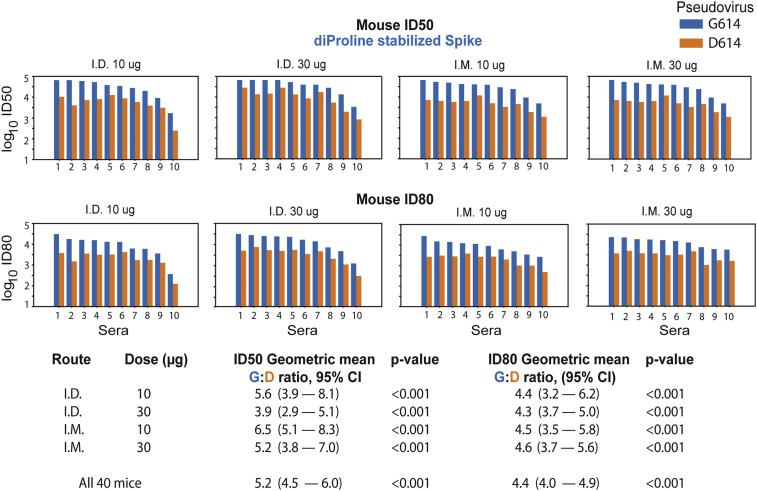
Figure 1.
The G614 Spike Is Neutralized More Potently than the D614 Spike by Mouse Sera
The G614 spike is neutralized more potently than the D614 spike by sera from mice immunized twice at a four-week interval with nucleoside-modified mRNA-LNPs encoding the Wuhan sequence of spike (D614) with diProline stabilization mutations. Sera (10 animals/group) obtained four weeks after the second immunization were tested for neutralization against pseudoviruses with the D614 and G614 variants of spike. Log10 of the inhibitory dose to reduce infection by 50% (ID50) and by 80% (ID80) are shown; higher values indicate increased neutralization activity. Each pair of bars represents serum from one animal; for each serum, the blue bar shows the neutralization titer against the G614 form, and the orange bar is the titer against the original D614 form. Top panels are ID50; bottom panels are ID80. Summary statistics for each group are shown at the bottom. The geometric means for the ratio of G614:D614 neutralizing antibodies are shown. Log10 of values of the ID50 and ID80 titers were used in a paired t test to calculate the p value and the 95% CI of geometric mean for the ratio of G614:D614.
A total of 40 mice were immunized twice at a four-week interval with nucleoside-modified mRNA encoding the diProline stabilized cell surface D614 spike (Kirchdoerfer et al., 2016; Kirchdoerfer et al., 2018; Pallesen et al., 2017; Wrapp et al., 2020) in four equal-sized groups, with varying dose and route of vaccine administration. This is the immunogen being employed in the Moderna vaccine in phase 3 clinical trials (mRNA-1273, NCT04470427) (Jackson et al., 2020). Immunizations were performed by the intradermal (i.d.) and intramuscular (i.m.) routes with 10- and 30-μg doses. Preimmune and serum four weeks after the second immunization were analyzed for neutralization of pseudoviruses with the D614 and G614 sequences. Preimmune sera from all mice scored negative. At four weeks after the second immunization, a relative increase in neutralizing antibody (nAb) titer for G614 over D614 was observed for each animal (Figures 1 and S1A related to Figure 1; Tables S2A and S2B, related to Figure 1); analyses were done both as a single group and separately by dose and route. Across all routes and doses (N = 40), the geometric mean for the G614:D614 ratio was 5.2-fold (95% confidence interval [CI) 4.5–6.0) for ID50 and 4.4 (95% CI 4.0–4.9) for ID80. Similar patterns were seen across the 10-μg and 30-μg doses and for both i.m. and i.d. routes. Figure 1 shows the geometric mean for the ratio of G614:D614 nAb titers across route and doses, which ranged from 3.9 to 6.6 ID50 and 4.3 to 4.6 ID80. We found no exception to the increased sensitivity of G614 spike to vaccine sera, even though the D614 form was used for immunization, with p < 0.001 for all comparisons. The maximum percent inhibition was 100% for all animals against both viral strains.
Eleven rhesus macaques were immunized with the nucleoside-modified mRNA-LNP vaccine platform using two different immunogens by the i.m. route (50 μg). The first encoded the Wuhan index strain sequence (Lurie et al., 2020) cell surface spike protein with a mutated furin cleavage site (furin mut). The second encoded the RBD domain as a secreted monomer. Sera obtained at baseline and four weeks after the second immunization were assessed for neutralization of the D614 and G614 variants (Figures 2 S1B, and S1C, related to Figure 2; Table S2C, related to Figure 2). Baseline (preimmune) sera were all negative. nAb titers in all postimmunization sera were higher against the G614 variant virus than the D614 (Wuhan index strain) virus for both immunogens. For the furin mut spike immunogen (N = 5), the geometric mean for the G614:D614 ID50 nAb titer ratio was 5.4-fold (95% CI, 3.0–9.8) and for ID80 was 2.9-fold (95% CI 2.5–3.3) (Figure 2). For the soluble RBD (N = 6), the G614:D614 ID50 nAb titer ratio had a geometric mean of 6.45-fold (p < 0.001) and for ID80 was 3.22-fold (p < 0.001) (Figure 2). Similar to the data in immunized mice, all postimmune NHP sera neutralized both the D614 and G614 viruses to 100%. The observation that the G variant was more sensitive to antibodies induced by both a D614-containing cell surface spike trimer and an RBD-secreted monomer suggests that the G614 mutation increases RBD-mediated neutralization despite the fact that D614G lies in S1 outside the RBD.
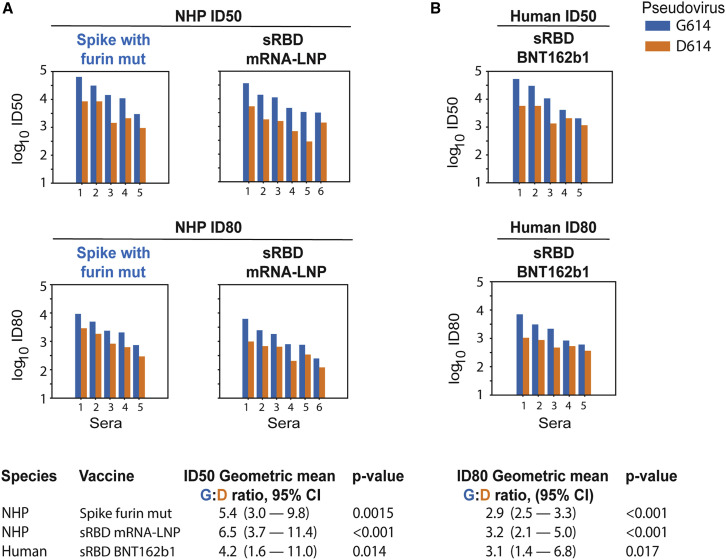
Figure 2.
The G614 Spike Is Neutralized More Potently than the D614 Spike by NHP Sera
The G614 spike is neutralized more potently than the D614 spike by sera from NHPs (rhesus macaques) immunized with nucleoside-modified mRNA-LNPs encoding RBD and full-length spike immunogens and humans immunized with RBD trimers. Sera from macaques immunized twice at a four-week interval with the Wuhan sequence of spike (D614) with a mutated furin cleavage site (n = 5) or secreted RBD monomers (n = 6) obtained four weeks after the second immunization were tested for neutralization against pseudoviruses with the D614 and G614 variants of spike (A). Also shown are sera from five humans immunized twice at a three-week interval with nucleoside-modified mRNA-LNPs encoding a secreted RBD trimer (B). Each pair of bars represents one macaque or human. Top panels are ID50; bottom panels are ID80. For each serum, the blue bar shows the neutralization sensitivity of the G614 form, and the orange bar shows the original D614 form. The geometric means for the ratio of G614:D614 neutralizing antibody titers measured in sera are provided in the summary at the bottom. Log10 of values of the ID50 and ID80 titers were used in a paired t test to calculate the p value and the 95% CI of geometric mean for the ratio of G614:D614. Overall response levels were comparable between the two different immunogens in the NHP and between NHPs and humans.
Preliminary results from a phase 1/2 clinical trial using the nucleoside-modified mRNA-LNP vaccine platform that delivered a secreted RBD trimer were recently published and demonstrated potent ELISA binding and neutralization in all subjects at all tested doses: 10, 30, and 50 μg (Mulligan et al., 2020). Virus neutralizing titers after the second immunization of 10 and 30 μg were 1.8-fold and 2.8-fold greater, respectively, than were convalescent sera from SARS-CoV-2-infected patients (Mulligan et al., 2020), although the nAb titers in convalescent patients exhibited a wide range, and the patients were older than vaccinated subjects.
Sera obtained preimmunization and seven days after the second immunization with 50 μg (N = 3), 30 μg (N = 1), and 10 μg (N = 1) of human subjects from a phase 1 trial performed in Germany using the same vaccine were analyzed for neutralization of the D614 and G614 spike variant pseudoviruses. Similar to the data in murine and NHP immunizations (Figures 1, 2A, and S1A–S1C, related to Figures 1 and 2A; Tables S2A–S2C, related to Figures 1 and 2A), the G614 pseudovirus was more potently neutralized than the D614 pseudovirus by the same serum samples (Figures 2B and S1D related to Figure 2B; Table S2D, related to Figure 2B). The geometric mean of the G614:D614 ID50 nAb titer ratio was 4.22-fold (p = 0.014) and for ID80 was 3.1-fold (p = 0.017). No neutralizing activity was detected in the corresponding preimmune sera. As with the vaccinated mice and NHP, all human vaccine sera achieved complete neutralization of the virus (100% maximum percent inhibition [MPI]).
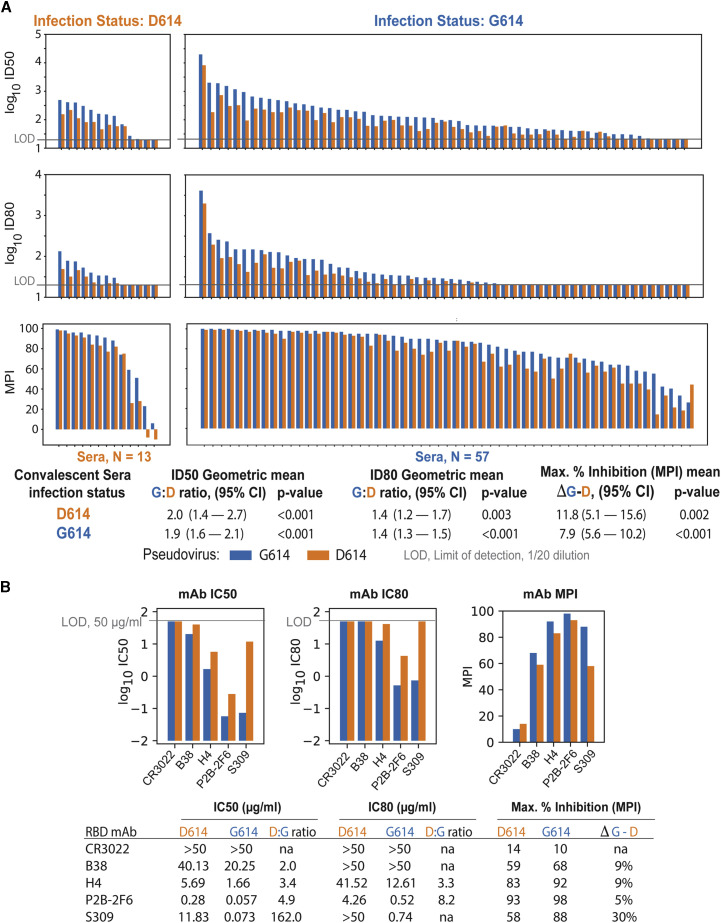
We also compared the neutralization susceptibility of D614 and G614 pseudoviruses by using convalescent serum samples from people known to be infected with either the D614 or G614 variant of the virus. As shown in Figure 3 A and Table S2E, related to Figure 3A, the G614 pseudovirus was moderately more sensitive to neutralization by sera from people infected with either variant; this difference was remarkably consistent across the 70 sera tested and was highly statistically significant for ID50, ID80, and MPI. Additional clinical data were available for these sera, enabling us to explore the possibility that the infecting strain (D614 verses G614), viral load, or declining antibody levels over time in convalescent sera (Ibarrondo et al., 2020) might impact the level of the antibody responses. We used a mixed-effect model to predict log ID50. We modeled the different levels of responses in sera from different individuals (N = 70) as a random effect and used fixed effects to model the contribution the number of days between diagnosis and sera sampling, the real-time PCR cycle threshold (Ct) values, the pseudovirus tested, and the D- and G-infecting strain. Although the relationship between the pseudovirus tested and the ID50 was highly significant (Figure S2, related to Figure 3A), consistent with the paired statistic in Figure 3A, we found that none of the other factors, including interactions between the D- or G-infecting virus and pseudovirus assayed, were statistically significant predictors.
Figure 3.
G614 Spike-Pseudotyped Virus Is Neutralized More Potently than D614 Spike-Pseudotyped Virus by Human Sera and mAbs
G614 spike-pseudotyped virus is neutralized more potently than D614 spike-pseudotyped virus by sera from people infected with either the D614 or G614 variant of spike or RBD-specific mAbs.
(A) Each blue/orange pair of bars represents convalescent serum sampled from one person. Left, people infected with D614. Right, people infected with G614.
(B) MAbs were assayed at 3-fold dilutions starting at 50 μg/mL for a total of eight dilutions. IC50 and IC80 values are in μg/mL, where a lower bar height corresponds to greater neutralization potency. MPI was calculated as the percentage of neutralization at the highest mAb concentration tested.
Additional comparisons of the neutralization susceptibility of D614 and G614 pseudoviruses were made with RBD-specific monoclonal antibodies (mAbs) derived from SARS-CoV-1- and SARS-CoV-2-infected individuals. Antibody CR3022, a weak RBD binder that is reported to be non-neutralizing (ter Meulen et al., 2006; Yuan et al., 2020), was negative against both pseudoviruses (Figure 3B). Antibody B38 was weakly neutralizing against both viruses but 2-fold more potent against G614. Antibodies H4, P2B-2F6, and S309 were 3–142 times more potent against G614. These results are further evidence that G614 is more sensitive to neutralization by RBD-directed antibodies.
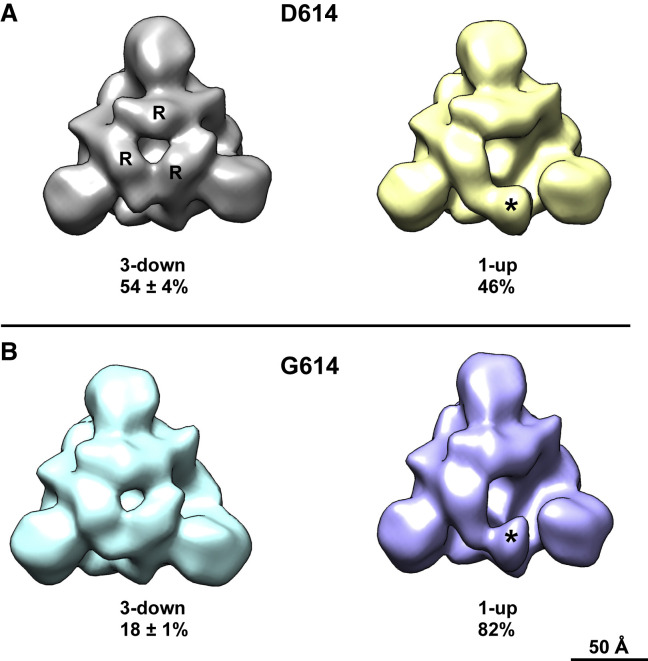
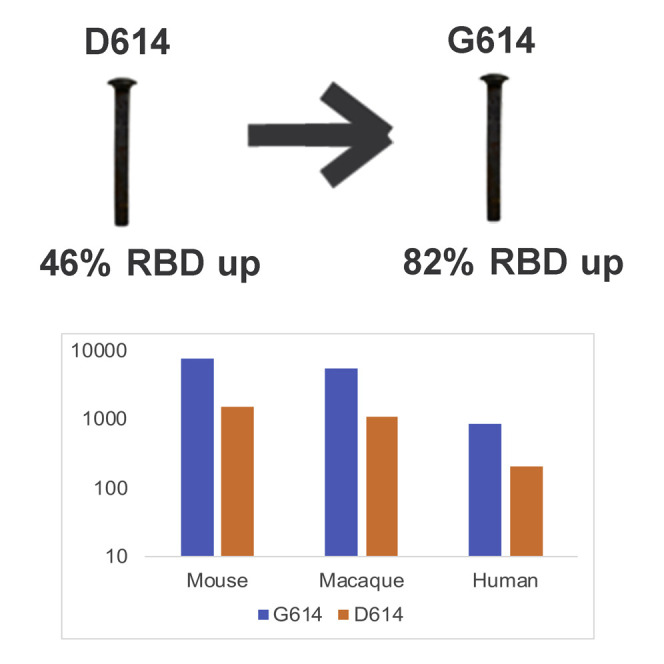
Sera from macaques and humans immunized with RBD-only immunogens more potently neutralized G614 pseudovirus even though the D614G mutation was not in the RBD, and this observation suggested that the mutation induced a structural change in the expressed spike that increased the exposure of neutralizing epitopes on the RBD. To evaluate this possibility, we used negative stain electron microscopy (NSEM) and single-particle reconstruction coupled with 3D classification to determine the structure and variability of the two variants in furin-deficient spike ectodomain constructs. The D614 variant showed two classes of structures called “3-down” or “1-up” in reference to their RBD positions (Figure 4 A). The D614 variant demonstrated a roughly equal proportion of the 3-down to 1-up states (54% versus 46%), consistent with cryo-EM data reported by others (Walls et al., 2020). The G614 variant also demonstrated 3-down and 1-up structures (Figure 4B); however, the 1-up state was more heavily populated, 82%. Similar shifts in spike population, with the G614 variant showing a higher propensity for “up” RBD states, have also been reported in other studies in different spike constructs ((Gobeil et al., 2020); Yurkovetskiy et al., 2020; Zhang et al., 2020). This shift toward the 1-up state demonstrates an allosteric effect of the D614G mutation on RBD dynamics and suggests a mechanism for the enhanced neutralization susceptibility of the G614 variant.
Figure 4.
Negative Stain Electron Microscopy Reconstructions and 3D Classification of Expressed Spike Constructs
Negative stain electron microscopy reconstructions of expressed spike constructs after 3D classification. View is looking down the 3-fold trimer axis onto the S1 domain.
(A) D614 variant showing the 3-RBD-down structure on the left with individual RBDs labeled (R), and the 1-RBD-up structure on the right with the up RBD labeled (asterisk). The fraction of particle images that sorted into each class is indicated below, expressed as average ± standard deviation, n = 3 each.
(B) G614 variant also showing 3-down and 1-up structures.
Discussion
Soon after the G614 mutation in spike appeared early in the pandemic, it rapidly replaced the D614 variant in many countries (Korber et al., 2020a). This mutation is associated with increased infectivity (Korber et al., 2020a; Zhang et al., 2020) when tested in in vitro model systems. Over 100 vaccines using various platforms and immunogens are being developed to combat COVID-19 and end the devastating financial, societal, and health burdens. Currently, over 30 vaccines are in clinical testing, some of which have entered phase 3 trials. Most SARS-CoV-2 vaccines were originally designed by using the D614 variant of the spike protein, which was present in the first sequence of SARS-CoV-2 from Wuhan (Lurie et al., 2020). The most critical finding that will ease the concern for most current vaccines in clinical trials is our data showing that the SARS-CoV-2 spike protein with the G614 mutation does not escape neutralization but rather is neutralized at a higher level by serum from vaccinated mice, NHPs, and humans that used immunogens derived from the D614 variant of the virus. Consistent with this finding, our data also show that the G614 variant of the virus was more sensitive to neutralization by RBD-specific mAbs and serum samples from people known to be infected with either variant.
D614 is on the surface of the spike protomer and has the potential to influence the conformation and flexibility of the spike protein. The recently published cryo-EM structure of the SARS-CoV-2 spike demonstrates that the D614 sidechain can form a hydrogen bond with the neighboring protomer T859 amino acid (Wrapp et al., 2020). This interaction could be critical, because it could bridge residues from the S1 region of one protomer to the S2 region of an adjacent protomer. This interaction would bracket the furin and S2 cleavage sites (Gobeil et al., 2020). Potentially, it could reduce shedding of S1 from viral-membrane-bound S2, and the introduction of G614 could increase S1 release. Our structural data demonstrate that, although the D614G mutation is located in the SD2 subdomain and distal from the RBD region, in the context of a soluble ectodomain construct, this mutation leads to an increased proportion of the 1-RBD-up conformation (Figure 4). A recent publication demonstrated a similar effect of the G614 mutation to increase the number of RBDs in the up position (Yurkovetskiy et al., 2020). By using an alternative structural analysis method, extensive microsecond timescale atomistic molecular dynamics simulations, reveal that in the G form the interprotomer interactions in the spike trimer become more symmetric than do the D form. This equalization of interprotomer energetics results in a higher population of 1-up spike conformations, leading to increased encounter between RBD and ACE2 receptor and greater exposure of RBD domain for neutralization (Mansbach et al., 2020).
Our results in immunized mice, NHPs, and human subjects immunized with nucleoside-modified mRNA-LNP vaccines with various spike immunogens; humans known to be infected with either the D614 or G614 variant of the virus; and with RBD-specific monoclonal antibodies conclusively demonstrate a modest but highly consistent increase in neutralization-susceptibility of the G614 variant. A clinical trial (Sahin et al., 2020) and a preclinical analysis (Corbett et al., 2020) also using nucleoside-modified mRNA-LNP COVID-19 vaccines have been published after submission of this manuscript. They both analyzed neutralization of D614 and G614 viruses with assays that utilized pseudotyped vesicular stomatitis virus. Both observed no statistical reduction in neutralization of the G614 variant in comparison with D614 but did not directly compare neutralization of each virus by serum from individual subjects or animals. Therefore, we reanalyzed the published data from sera from the 24 mice vaccinated with an mRNA vaccine that encodes a SARS-CoV-2 spike protein stabilized in the prefusion conformation (Corbett et al., 2020) with a paired t test statistic; the greater sensitivity of the G614 form to sera from the vaccinated mice was supported by this test (p = 0.025). Assays performed in Erica Ollmann-Saphire’s laboratory demonstrate equivalent or better neutralization of G614-bearing pseudovirus in comparison with D614-bearing pseudovirus using convalescent sera from six COVID-19 subjects, but it was not known whether the individuals were infected with the D614 or G614 variant (Korber et al., 2020b). More recently, the D614G mutation was shown to render live SARS-CoV-2 virus more susceptible to neutralization by sera from D614 spike-immunized mice (Hou et al., 2020).
A conclusion from our data is that the virus became modestly more susceptible to neutralization by host antibody responses as a consequence of acquiring a mutation that provides a fitness advantage for transmission. Previous work has shown that the G614 virus variant is more infectious in vitro (Korber et al., 2020a; Zhang et al., 2020), and recent evidence from studies in animal models supports a transmission fitness advantage (Hou et al., 2020). The fact that the mutation is outside of the RBD and fusion machinery and affects both infectivity and neutralization is common for viral fusion proteins. Thus, coronavirus spike proteins are prefusion stabilized by similar types of mutations (Kirchdoerfer et al., 2016). Also, a variety of mutations distinct from neutralization epitopes in HIV envelope is known to alter antigenicity (Medina-Ramírez et al., 2017).
In summary, we demonstrate here that vaccinated mice, NHPs, and humans using the nucleoside-modified mRNA-LNP vaccine platform encoding four different SARS-CoV-2 spike immunogens generate antibody responses that not only recognize the G614 mutation that has taken over the pandemic (Korber et al., 2020a; Zhang et al., 2020) but also have stronger titers of neutralization to this virus variant. The mechanism appears to be that the mutation increases the up formation of the RBD in the spike trimer, increasing the exposure of neutralization epitopes. Over 30 vaccines are currently in clinical trial testing. Most of the immunogens in these trials were either derived from the initial D614 virus or contain D614G in the spike. Although the G614 variant has replaced the original D614 sequence in the SARS-CoV-2 spike throughout much of the world, our finding that this is not an escape mutation and, in fact, is better neutralized by sera from mice, NHPs, and humans immunized with vaccines derived from the D614 viral spike alleviates a major concern regarding the current efforts to develop an effective SARS CoV-2 vaccine.
Limitations of Study
Potential drawbacks to our studies include the following: (1) although we studied four different variations of the spike immunogen, we only used a single type of vaccine platform, nucleoside-modified mRNA-LNP platform (reviewed in Alameh et al., [2020] and Pardi et al. [2018b]). This platform is recognized as an outstanding new approach that has entered phase 3 clinical trials by two separate pharmaceutical/biotech companies, Moderna and Pfizer/BioNTech. The results of phase 1 clinical trials demonstrated that all immunized subjects safely developed neutralizing responses (Jackson et al., 2020; Mulligan et al., 2020). (2) We performed pseudovirus neutralization assays. Although these assays are considered excellent methods to measure neutralization and are used for the development of many viral vaccines, live virus neutralization or animal challenges would offer additional methods to measure effective vaccine response to the G614 variant. A new study directly compared two different pseudovirus neutralization assays, including lentiviral used in this study, with live virus neutralization (Schmidt et al., 2020). Highly significant correlations were observed between the assays demonstrating no significant differences in the use of live virus versus pseudovirus neutralization assays for both convalescent plasma and human mAbs, which is perhaps not surprising given that neutralization in both assays is based on entry inhibition. As with the pseudovirus assay, assays with live SARS-CoV-2 are typically performed with viruses that are produced and assayed in cell lines; neutralization assays that use natural target cells of the respiratory system are technically challenging, low throughput, and difficult to standardize. (3) Our structural studies were performed in the context of a furin cleavage-deficient spike ectodomain. Although this soluble ectodomain has been shown to be a good mimic of the native spike, and the shift in the proportion of RBD “up” conformation between the D614 and G614 variants suggest an allosteric effect of the D614G mutation on the RBD conformations, the structures of the native spike could have some differences from what we observe in the context of the ectodomain. (4) Our experiments do not rule out possible differences in spike incorporation as another mechanism for the increased neutralization-susceptibility of the D614G spike pseudovirus.
STAR★Methods
Key Resources Table
| REAGENTS or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CR3022 | Peter Kwong, VRC/NIH | ter Meulen et al., 2006; Yuan et al., 2020 |
| B38 | Peter Kwong, VRC/NIH | Wu et al., 2020 |
| H4 | Peter Kwong, VRC/NIH | Wu et al., 2020 |
| P2B-2F6 | Peter Kwong, VRC/NIH | (Ju et al., 2020a) |
| S309 | Peter Kwong, VRC/NIH | Pinto et al., 2020 |
| Chemicals and Recombinant Proteins | ||
| M1-Pseudouridine 5'-triphosphate | TriLink | N-1081 |
| CleanCap | TriLink | N-7413 |
| Cellulose | Sigma-Aldrich | 11363-250G |
| MEGAscript™ T7 Transcription Kit | ThermoFisher Scientific | AMB13345 |
| QuikChange Lightning Site-Directed Mutagenesis Kit | Agilent | 210518 |
| FreeStyle 293 Expression Medium | GIBCO | 12338018 |
| Hyclone SFM4HEK293 | Cytiva | SH30521.02 |
| Opti-MEM I | GIBCO | 31985-070 |
| Turbo293 | Speed BioSystems | PXX1002 |
| 8% Glutaraldehyde | Electron Microscopy Sciences | 16019 |
| 300mesh Cu carbon coated | Electron Microscopy Sciences | CF300-Cu |
| Uranyl formate | Electron Microscopy Sciences | 22450 S-888 |
| Experimental Models: Organisms/strains | ||
| BALB/c mouse | Charles Rivers Laboratories | N/A |
| Rhesus macaques | BioQUAL | N/A |
| Experimental Models: Cell Lines | ||
| 293T/ACE2 cells | Drs. Mike Farzan and Huihui Mu at Scripps | 293T/ACE2 cells |
| HEK293T/17 | ATCC | N/A |
| Freestyle 293-F cells | GIBCO | R79007 |
| Recombinant DNA | ||
| pTEV-di-proline SARS-CoV-2 Spike-A101 | Weissman Lab | N/A |
| pTEV full length S-furin mutant-A101 | Weissman Lab | N/A |
| pTEV-RBD-A101 | Weissman Lab | N/A |
| VRC7480.D614G | VRC and Montefiori lab | N/A |
| pCMV ΔR8.2 | VRC | Naldini et al., 1996 |
| pHR’ CMV Luc | VRC | Naldini et al., 1996 |
| ραH-S-GSAS/PP | Wrapp et al., 2020 | N/A |
| ραH-S-GSAS | Acharya Lab | N/A |
| ραH-S-GSAS/D614G | Acharya Lab | N/A |
| Software and Algorithms | ||
| Image Lab | Bio-Rad | Version 6.0 |
| Relion | (Scheres, 2012; Scheres, 2016b) | Version 3.1 |
| UCSF Chimera | (Pettersen et al., 2004) | http://www.cgl.ucsf.edu/chimera/ |
| Data and Software Availablity | ||
| Additional Supplemental Items | Mendeley Data | https://doi.org/10.17632/cn5b3hc5zd.1 |
Resource Availability
Lead Contact
Further information and requests for supporting data, resources, and reagents should be directed to and will be fulfilled upon request by the Lead Contacts: Drew Weissman (drew@pennmedicine.upenn.edu).
Materials Availability
Reagents from this study are available upon request.
Data and Code Availability
Data from this study are available upon request.
Experimental Models and Subject Details
Mice
BALB/c mice aged 8 weeks old were purchased from Charles Rivers Laboratories and immunized after 1 week of acclimation at the University of Pennsylvania. Both sexes were used in each experiment. All protocols were approved by the University of Pennsylvania IACUC.
Nonhuman Primate
NHP studies were performed at Bioqual, Inc, Rockville, MD following their and University of Pennsylvania’s IACUC approval. Animals were 3-7 years of age and approximately equal numbers of male and female were used.
Human
Serum from subjects enrolled in a phase 1/2 clinical trial of a nucleoside-modified mRNA-LNP vaccine encoding trimeric SARS-CoV-2 RBDs (BNT162b1)(Mulligan et al., 2020) were obtained (NCT04368728). Subjects were 18-55 years of age and both male and female.
Serum samples from people known to be infected with either the D614 or G614 form of SARS-CoV-2 were collected following informed consent from healthcare workers at Sheffield Teaching Hospitals NHS Foundation Trust, Sheffield, UK as part of the COVID-HERO SARS-CoV-2 seroprevalence study (Research Ethics Committee reference 20/HRA/2180).
Cell Lines
293T/ACE2 cells were kindly provided by Drs. Mike Farzan and Huihui Mu at Scripps. Cells were grown in DMEM + glutamine + 10% FCS at 37°C with 5% CO2.
Method Details
Ethics Statement
The clinical trial described in this manuscript was randomized double-blinded and the sera collections were approved by the appropriate IRBs. All protocols, experimentation and animal manipulation adhered to the Guide for the Care and Use of Laboratory Animals by the Committee on Care of Laboratory Animal Resources Commission on Life Sciences, National Research Council under IACUC approved protocols.
mRNA Production
N1-methylpseudouridine modified mRNA was produced as previously described (Freyn et al., 2020) using T7 RNA polymerase (MegaScript, ThermoFisher Scientific, Waltham, MA, USA) on Not-I/AlfII double digested and linearized plasmids encoding codon-optimized di-proline modified pre-fusion SARS-CoV-2 Spike (Wuhan Hu-1 complete genome, GenBank: MN908947.3), full length S-furin mutant protein (RRAR furin cleavage site abolished), and RBD. In vitro transcribed (IVT) mRNAs were co-transcriptionally capped using the CleanCap system (TriLink Biotechnologies, San Diego, CA, USA), and purified using a cellulose-based chromatography method (Baiersdörfer et al., 2019). All IVT mRNAs were analyzed on agarose (1.4% w/v, 1X TAE buffer) for integrity, and subjected to additional quality control to rule out double stranded RNA (dsRNA) contamination and endotoxin contamination prior to formulations into lipid nanoparticles (LNPs), as described (Pardi et al., 2015).
Production of mRNA-LNP
Lipid nanoparticles (LNP) used in this study contain an ionizable lipid proprietary to Acuitas /DSPC/Cholesterol/PEG-Lipid (Lin et al., 2013). Encoding mRNA was encapsulated in LNP using a self-assembly process in which an aqueous solution of mRNA at 4.0 pH was rapidly mixed with a solution containing the aforementioned lipids premixed and dissolved in ethanol. The proprietary lipid and LNP composition are described in US patent US10,221,127. All LNP were characterized post-production at Acuitas pharmaceutical (Vancouver, BC, Canada) for their size, polydispersity using a Malvern Zetasizer (Zetasizer Nano DS, Malvern, UK), encapsulation efficiency, and shipped on dry ice and stored at −80°C until use.
Administration of Test Articles (Immunization) and Blood Collection
8-12 week old BALB/c mice were immunized with either 10 or 30 μg of mRNA-LNP via the intramuscular (I.M.) or intradermal (I.D.) routes of administration using a 29G X1/2’’ Insulin syringe. Mice received a booster injection on day 28 (4 weeks). Blood was collected at day 0, 28 and 56 through the retro-orbital route using non heparinized micro hematocrit capillary tubes (ThermoFisher Scientific, Waltham, MA, USA). Serum was separated by centrifugation (10 000 g, 5 min) using a non-refrigerated Eppendorf 5424 centrifuge (Eppendorf, Enfield, CT, USA), heat-inactivated (56°C) for 30 min, and stored at −20°C until analysis.
Administration of mRNA/LNPs to Rhesus Macaques
Fifty micrograms of either mRNA-LNP encoding an unstabilized transmembrane (TM) Spike protein with a mutant furin cleavage site or a monomeric soluble RBD were administered I.M. in two sites in the left and right quadriceps on weeks 0, and 4. Animals were anesthetized with ketamine prior to blood draws from the femoral vein. Serum samples were analyzed in 5 macaques immunized with mRNA-LNP encoding unstabilized TM Spike with a mutant furin cleavage site and in 6 animals immunized with mRNA-LNP encoding soluble RBD. All studies were performed at Bioqual, Inc, Rockville, MD following IACUC approval.
Clinical Trial Samples
Serum from subjects enrolled in a phase 1/2 clinical trial of a nucleoside-modified mRNA-LNP vaccine encoding trimeric SARS-CoV-2 RBDs (BNT162b1)(Mulligan et al., 2020) were obtained (NCT04368728). Five subjects that received 2 immunizations at a 3-week interval with 10, 30, or 50 μg of mRNA were used. All subjects were considered a single group, as similar serologic data was obtained for each dose and the comparison and calculation of statistical significance was performed within each sample. Serum was obtained prior to the first immunization and 7 days after the second immunization.
Serum samples from people known to be infected with either the D614 or G614 form of SARS-CoV-2 were collected following informed consent from healthcare workers at Sheffield Teaching Hospitals NHS Foundation Trust, Sheffield, UK as part of the COVID-HERO SARS-CoV-2 seroprevalence study (Research Ethics Committee reference 20/HRA/2180). Details of previous RT-PCR confirmed diagnosis of SARS-CoV-2 were recorded. SARS-CoV-2 sequences were generated using samples collected for routine clinical diagnostic use in individuals presenting with COVID-19 at Sheffield Teaching Hospitals NHS Foundation Trust, Sheffield, UK. This work was performed under approval by the Public Health England Research Ethics and Governance Group for the COVID-19 Genomic UK consortium (R&D NR0195). Extracted nucleic acid from nasal or throat swabs was used for whole genome sequencing of SARS-CoV-2 (Oxford Nanopore Technologies (ONT), Oxford, UK) using the ARTIC network protocol (https://artic.network/ncov-2019). Following base calling, data were demultiplexed using ONT Guppy using a high accuracy model. Reads were filtered based on quality and length (400 to 700bp) and mapped to the Wuhan reference genome (MN908947). Variants, including the A-to-G change at nucleotide 23,403 leading to D614G, were called using nanopolish (https://github.com/jts/nanopolish) and consensus sequences compared to the reference.
Monoclonal Antibodies
SARS-CoV-2 RBD-binding mAbs CR3022, B38, H4, P2B-2F6, and S309 were obtained from Dr. Peter Kwong at the Vaccine Research Center, National Institutes of Health, USA. CR3022 was isolated from a convalescent SARS-CoV patient, exhibits cross-reactive binding to SARS-CoV-2 RBD distal from the receptor binding motif, and does not neutralize SARS-CoV-2 even at concentrations as high as 400 μg/mL (ter Meulen et al., 2006; Yuan et al., 2020). B38 and H4 were isolated from a COVID-19 subject, bind different epitopes on RBD that partially overlap, and exhibit potent SARS-COV-2 neutralizing and ACE2-blocking activity (Wu et al., 2020). P2B-2F6 was isolated from a COVID-19 subject in Shenzhen, China, exhibits potent SARS-CoV-2 neutralizing activity and blocks ACE2 binding (Ju et al., 2020a). S309 was isolated from a recovered SARS-CoV-infected subject, potently cross-neutralizes SARS-CoV and SARS-CoV-2, binds outside the receptor binding motif of RBD and, based on cryo-EM structure, is not predicted to interfere with ACE2 binding (Pinto et al., 2020).
SARS-CoV-2 Pseudovirus Neutralization Assay
SARS-CoV-2 neutralization was assessed with Spike-pseudotyped viruses in 293T/ACE2 cells as a function of reductions in luciferase (Luc) reporter activity. 293T/ACE2 cells were kindly provided by Drs. Mike Farzan and Huihui Mu at Scripps. Cells were maintained in DMEM containing 10% FBS and 3 μg/mL puromycin. An expression plasmid encoding codon-optimized full-length Spike of the Wuhan-1 strain (VRC7480), was provided by Drs. Barney Graham and Kizzmekia Corbett at the Vaccine Research Center, National Institutes of Health (USA). The D614G amino acid change was introduced into VRC7480 by site-directed mutagenesis using the QuikChange Lightning Site-Directed Mutagenesis Kit from Agilent Technologies (Catalog # 210518). The mutation was confirmed by full-length Spike gene sequencing. Pseudovirions were produced in HEK293T/17 cells (ATCC cat. no. CRL-11268) by transfection using Fugene 6 (Promega Cat#E2692) and a combination of Spike plasmid, lentiviral backbone plasmid (pCMV ΔR8.2) and firefly Luc reporter gene plasmid (pHR’ CMV Luc)(Naldini et al., 1996) in a 1:17:17 ratio. Transfections were allowed to proceed for 16-20 h at 37°C. Medium was removed, monolayers rinsed with growth medium, and 15 mL of fresh growth medium added. Pseudovirus-containing culture medium was collected after an additional 2 days of incubation and was clarified of cells by low-speed centrifugation and 0.45 μm micron filtration and stored in aliquots at −80°C. TCID50 assays were performed on thawed aliquots to determine the infectious dose for neutralization assays (RLU 500-1000x background, background usually averages 50-100 RLU). All neutralization data is available at: https://doi.org/10.17632/cn5b3hc5zd.1
For neutralization, a pre-titrated dose of virus was incubated with 8 serial 3-fold or 5-fold dilutions of serum samples or mAbs in duplicate in a total volume of 150 μl for 1 h at 37°C in 96-well flat-bottom poly-L-lysine-coated culture plates (Corning Biocoat). Cells were suspended using TrypLE express enzyme solution (Thermo Fisher Scientific) and immediately added to all wells (10,000 cells in 100 μL of growth medium per well). One set of 8 control wells received cells + virus (virus control) and another set of 8 wells received cells only (background control). After 66-72 h of incubation, medium was removed by gentle aspiration and 30 μL of Promega 1X lysis buffer was added to all wells. After a 10 min incubation at room temperature, 100 μl of Bright-Glo luciferase reagent was added to all wells. After 1-2 min, 110 μl of the cell lysate was transferred to a black/white plate (Perkin-Elmer). Luminescence was measured using a PerkinElmer Life Sciences, Model Victor2 luminometer. Neutralization titers are the serum dilution (ID50/ID80) or mAb concentration (IC50/IC80) at which relative luminescence units (RLU) were reduced by 50% and 80% compared to virus control wells after subtraction of background RLUs. Maximum percent inhibition (MPI) is the % neutralization at the lowest serum dilution or highest mAb concentration tested. Serum samples were heat-inactivated for 30 min at 56°C prior to assay.
Protein Expression and Purification
SARS-CoV-2 ectodomain constructs (Wrapp et al., 2020) were produced and purified as follows. Genes encoding residues 1-1208 of the SARS-CoV-2 S (GenBank: MN908947) with a “GSAS” substitution at the furin cleavage site (residues 682–685), with and without proline substitutions of residue K986 and V987 (S-GSAS/PP or S-GSAS), a C-terminal T4 fibritin trimerization motif, an HRV3C protease cleavage site, a TwinStrepTag and an 8XHisTag were synthesized and cloned into the mammalian expression vector ραH. The S-GSAS template was used to include the D614G mutation (S-GSAS(D614G)). Plasmids were transiently transfected into FreeStyle-293F cells using Turbo293 (SpeedBiosystems). Protein was purified on the sixth day post-transfection from filtered supernatant using StrepTactin resin (IBA), followed by size-exclusion chromatography (SEC) purification using a Superose 6 10/300 GL column (GE healthcare) equilibrated in 2mM Tris, pH 8.0, 200 mM NaCl, 0.02% sodium azide buffer (Figure S3 related to Figure 4).
Negative-Stain Electron Microscopy
Samples of S-GSAS and S-GSAS (D614G) ectodomain constructs were diluted to 100 μg/mL with room-temperature buffer containing 20 mM HEPES pH 7.4, 150 mM NaCl, 5% glycerol and 7.5 mM glutaraldehyde, and incubated 5 min; then glutaraldehyde was quenched for 5 min by addition of 1M Tris stock to a final concentration of 75 mM. A 5-μl drop of sample was applied to a glow-discharged, carbon-coated grid for 10-15 s, blotted, stained with 2% uranyl formate, blotted and air-dried. Images were obtained with a Philips EM420 electron microscope at 120 kV, 82,000 × magnification, and a 4.02 Å pixel size. The RELION program (Scheres, 2016a) was used for particle picking, 3D classification, and 3D refinements. The number of particle images that sorted into each class during the classification provides an estimate of the fraction for each state. We have previously shown this method has adequate resolution to resolve and compare up and down conformations (Henderson et al., 2020).
Statistical Methods
For a given experiment, neutralizing antibody titers for G614 and D614 were measured from serum on each animal and compared using a paired t test of the logarithm of the antibody titers. This formally tests whether the ratio of the titer is different from 1. All statistical tests were performed at the 0.05 level in the R software (v3.6.1) (Team, 2019).
To explore whether the infecting form of the virus impacted the serum potency, we modeled the logarithm of ID50 as a mixed effect model, using a random effect to account for the different levels of responses in sera from different individuals and (Table S3, related to Figure 3) effects to model the contribution the number of days between diagnosis and sera sampling, the RT PCR cycle threshold (Ct) values, the pseudovirus tested and the D and G infecting strain. Observations below the Limit Of Detection (LOD) were replaced by LOD/. The resulting model only includes the effect of the pseudo-strain at a much higher level of confidence (see fixed-effect ANOVA that tests the statistical significance of the pseudo-strain effect). In this analysis, neither viral load (as measured by CT), days between infection and testing, nor interactions between the infecting virus strain and the pseudovirus were statistically significant predictors for log ID50.
Acknowledgments
We would like to acknowledge members of the Sheffield COVID-19 Genomics Group for help during SARS-CoV-2 sequencing and the COVID-HERO research team for help with participant recruitment and sample processing. We thank Peter Kwong (Vaccine Research Center, NIH) for providing the monoclonal antibodies. We also thank James Theiler for statistical advice. BioNTech is the sponsor of the clinical trial study that the human samples were obtained from; N.P. was supported by NIAID (1R01AI146101); U.S. and C.B. work for BioNTech; M.L. was supported by NIAID (R01AI123738); D.W. was supported by grants from NIAID (AI142596 and AI124429), IPCAVD (U19AI142596), and BioNTech; B.H. was funded by a contract from the state of North Carolina funded by the Coronavirus Aid, Relief, and Economic Security Act (CARES Act) and by NIH grant IPCAVD-U19AI142596; P.A. was funded by NIAID (R01 - AI145687) and by a contract from the state of North Carolina funded by the Coronavirus Aid, Relief, and Economic Security Act (CARES Act); D.M. was funded by NIH (HHSN272201800004C), the Gates Foundation (OPP1146996), and by a contract from the state of North Carolina funded by the Coronavirus Aid, Relief, and Economic Security Act (CARES Act); B.K. was funded by the Los Alamos National Laboratory-directed research (LDRD) project 20200706ER; The COG-UK CONSORTIUM was supported by the Medical Research Council (MRC) part of UK Research & Innovation (UKRI), the National Institute of Health Research (NIHR), and Genome Research Limited, operating as the Wellcome Sanger Institute. T.I.d.S. was supported by a Wellcome Trust Intermediate Clinical Fellowship (110058/Z/15/Z). We also acknowledge support from NIAID-NIH contract HHSN272201800004C Option 28. This research has been funded in whole or part with federal funds under a contract from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Contract Number 75N93019C00050.
Author Contributions
M.-G.A., C.C.L.B., R.J.E., L.S., S.S., K.M., S.G., C.M., and N.P. conducted the experiments. D.W., T.d.S., P.C., H.H., R.B., N.H., P.J.C.L., Y.T., P.A.S., M.-G.A., M.G.L., P.A., B.F.H., B.K., and D.C.M. designed the experiments, C.B. and U.S. performed the phase 1 clinical trial. T.d.S., P.C., H.H., and R.B. obtained and supplied samples from infected patients. D.W., B.K., P.A.S., P.A., and D.C.M. wrote the paper. All authors reviewed and edited the manuscript.
Declaration of Interests
In accordance with the University of Pennsylvania policies and procedures and our ethical obligations as researchers, we report that N.P. and D.W. are named on patents that describe the use of nucleoside-modified mRNA as a platform to deliver therapeutic proteins and vaccines. We have disclosed those interests fully to the University of Pennsylvania, and we have in place an approved plan for managing any potential conflicts arising from licensing of our patents.
Published: December 1, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2020.11.012.
Supplemental Information
References
- Alameh M.G., Weissman D., Pardi N. Messenger RNA-Based Vaccines Against Infectious Diseases. Curr. Top. Microbiol. Immunol. 2020 doi: 10.1007/82_2020_202. [DOI] [PubMed] [Google Scholar]
- Baiersdörfer M., Boros G., Muramatsu H., Mahiny A., Vlatkovic I., Sahin U., Karikó K. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol. Ther. Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas N.K., Majumder P.P. Analysis of RNA sequences of 3636 SARS-CoV-2 collected from 55 countries reveals selective sweep of one virus type. Indian J. Med. Res. 2020;151:450–458. doi: 10.4103/ijmr.IJMR_1125_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyn A.W., Ramos da Silva J., Rosado V.C., Bliss C.M., Pine M., Mui B.L., Tam Y.K., Madden T.D., de Souza Ferreira L.C., Weissman D. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020;28:1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobeil S., Janowska K., McDowell S., Mansouri K., Parks R., Manne K., Stalls V., Kopp M., Henderson R., Edwards R.J. D614G mutation alters SARS-CoV-2 spike conformational dynamics and protease cleavage susceptibility at the S1/S2 junction. bioRxiv. 2020 doi: 10.1101/2020.10.11.335299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y.N., Tsao K.C., Hsiao M.J., Huang C.G., Huang P.N., Huang P.W., Lee K.M., Liu Y.C., Yang S.L., Kuo R.L. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg. Microbes Infect. 2020;9:1457–1466. doi: 10.1080/22221751.2020.1782271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güthe S., Kapinos L., Möglich A., Meier S., Grzesiek S., Kiefhaber T. Very fast folding and association of a trimerization domain from bacteriophage T4 fibritin. J. Mol. Biol. 2004;337:905–915. doi: 10.1016/j.jmb.2004.02.020. [DOI] [PubMed] [Google Scholar]
- Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Gobeil S.M.C., Kopp M., Li D., Parks R., Hsu A.L. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020;27:925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., Leist S.R., Schäfer A., Nakajima N., Takahashi K. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020 doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. Rapid Decay of Anti-SARS-CoV-2 Antibodies in Persons with Mild Covid-19. N. Engl. J. Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel S., Graña-Miraglia L., Gutierrez J.M., Bundalovic-Torma C., Groves H.E., Isabel M.R., Eshaghi A., Patel S.N., Gubbay J.B., Poutanen T. Evolutionary and structural analyses of SARS-CoV-2 D614G spike protein mutation now documented worldwide. Sci. Rep. 2020;10:14031. doi: 10.1038/s41598-020-70827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam O.K., Al-Emran H.M., Hasan M.S., Anwar A., Jahid M.I.K., Hossain M.A. Emergence of European and North American mutant variants of SARS-CoV-2 in South-East Asia. Transbound. Emerg. Dis. 2020 doi: 10.1111/tbed.13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., mRNA-1273 Study Group An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8:15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Foley B., Giorgi E.E., Bhattacharya T., Parker M.D. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.29.069054v1. [DOI] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98:495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Weeraratne D., Snowdon J.L., Parida L. Emergence of Drift Variants That May Affect COVID-19 Vaccine Development and Antibody Treatment. Pathogens. 2020;9:E324. doi: 10.3390/pathogens9050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaño D., Amanat F., Muramatsu H., Oguin T.H., 3rd A Single Immunization with Nucleoside-Modified mRNA Vaccines Elicits Strong Cellular and Humoral Immune Responses against SARS-CoV-2 in Mice. Immunity. 2020;53:724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P.J., Tam Y.Y., Hafez I., Sandhu A., Chen S., Ciufolini M.A., Nabi I.R., Cullis P.R. Influence of cationic lipid composition on uptake and intracellular processing of lipid nanoparticle formulations of siRNA. Nanomedicine (Lond.) 2013;9:233–246. doi: 10.1016/j.nano.2012.05.019. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Huang Y., Rapp M.A., Wang Q., Luo Y., Sahi V., Figueroa A. Potent Neutralizing Monoclonal Antibodies against Multiple Epitopes on the SARS-CoV-2 Spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Lurie N., Saville M., Hatchett R., Halton J. Developing Covid-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- Mansbach R.A., Chakraborty S., Nguyen K., Montefiori D., Korber B., Gnanakaran S. The SARS-CoV-2 Spike Variant D614G Favors an Open Conformational State. bioRxiv. 2020 doi: 10.1101/2020.07.26.219741. 2020.07.26.219741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Ramírez M., Sanders R.W., Sattentau Q.J. Stabilized HIV-1 envelope glycoprotein trimers for vaccine use. Curr. Opin. HIV AIDS. 2017;12:241–249. doi: 10.1097/COH.0000000000000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli D., Giorgi F.M. Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A. Phase 1/2 Study to Describe the Safety and Immunogenicity of a COVID-19 RNA Vaccine Candidate (BNT162b1) in Adults 18 to 55 Years of Age: Interim Report. medRxiv. 2020 doi: 10.1101/2020.06.30.20142570v1. [DOI] [Google Scholar]
- Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D. Concurrent human antibody and TH1 type T-cell responses elicited by a COVID-19 RNA vaccine. medRxiv. 2020 doi: 10.1101/2020.07.17.20140533. [DOI] [Google Scholar]
- Scheres S.H.W. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol. 2016;579:125–157. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- Scheres S.H.W. The Resolution Revolution: Recent Advances In cryoEM. 2016. Processing of Structurally Heterogeneous Cryo-EM Data in RELION; pp. 125–157. [DOI] [PubMed] [Google Scholar]
- Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.H., Michailidis E., Lorenzi J.C.C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217:217. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Team R.C. R: A language and environment for statistical computing. Computing. 2019;1 doi: 10.1890/0012-9658(2002)083[3097:CFHIWS]2.0.CO;2. [DOI] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6, e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T., Farzan M., Choe H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020 doi: 10.1101/2020.06.12.148726. 2020.06.12.148726. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from this study are available upon request.