Abstract
Eria corneri is a perennial epiphytic orchid distributed in southeastern China with high value of ornamental and medicinal. In this study, the complete chloroplast genome sequence of E. corneri was determined from Illumina pair-end sequencing data. The complete chloroplast genome sequence of E. corneri is 150,538 base pairs (bp) in length, including one large single-copy region (LSC, 85,941 bp), one small single-copy region (SSC, 13,099 bp), and a pair of inverted repeat regions (IRs) of 25,749 bp. Besides, the complete chloroplast genome contains 132 genes, including 77 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that E. corneri was most closely related to Calanthe triplicata and Calanthe davidii. Our study provides a foundation for the identification and genotyping of Eria species.
Keywords: Chloroplast genome, phylogenetic analysis, Orchidaceae, Eria corneri
The genus Eria is regarded as one of the largest genera in Orchidaceae family, with approximately 370 species worldwide, distributed in tropical Asia to Oceania, and there are 43 species in China, produced in southeastern provinces (Chen et al. 1999; Pridgeon et al. 2009). E. corneri is a perennial epiphytic orchid that grows on trees or rocks at an altitude of 500 to 1500 meters (Chen et al. 1999). Pharmacological researches showed that some chemical constituents of E. corneri, such as erianin and confusarin, have good antitumor and anti-oxidation activities (Bhuiya et al. 2017). In this study, we report the complete chloroplast genome of E. corneri. On the one hand, it will contribute to the species identification, germplasm diversity, and genetic engineering of the Eria genus (Lin et al. 2015). On the other hand, this is of great significance to the research and development of E. corneri at the genetic level.
The samples were collected from Qinglong Waterfall Scenic Area, Yongtai County, Fuzhou City, Fujian province, China (25°42′46.14′´N, 118°50′12.68″E), and deposited at Herbarium of Fujian Agriculture and Forestry University (specimen code: YT-BZML). The genomic DNA was extracted from fresh leaves using a modified CTAB method (Doyle and Doyle 1987) and sequenced by the BGISEQ-500 platform. The clean reads were used to assemble the complete chloroplast genome by the GetOrganelle pipe-line (Jin et al. 2018), with the chloroplast genome of Liparis nervosa as the reference sequences. The assembled chloroplast genome was annotated using the Geneious R11.15 (Kearse et al. 2012). Finally, we obtained a complete chloroplast genome of E. corneri and submitted to GenBank with accession number MN477202.
The complete chloroplast genome of E. corneri is 150,538 bp in length, containing a large single-copy (LSC) region of 85,941 bp, a small single-copy (SSC) region of 13,099 bp, and two inverted repeat (IR) regions of 25,749 bp. The new sequence has a total of 132 genes, including 77 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The overall GC-content of the whole plastome is 43.3%, whereas the corresponding values of the LSC, SSC, and IR regions are 57.09%, 8.70%, and 17.1%, respectively.
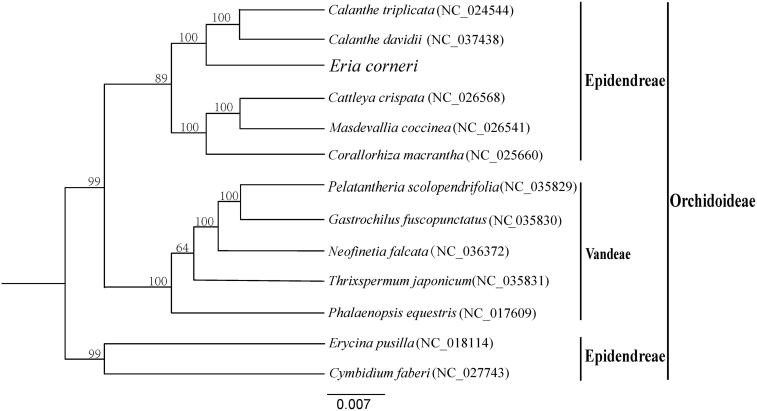
To confirm the phylogenetic position of E. corneri, a molecular phylogenetic tree was constructed based on 13 complete chloroplast genome sequences of Orchidaceae (Calanthe triplicata, Calanthe davidii, E. corneri, Cattleya crispata, Masdevallia coccinea, Corallorhiza macrantha, Pelatantheria scolopendrifolia, Gastrochilus fuscopunctatus, Neofinetia falcata, Thrixspermum japonicum, Phalaenopsis equestris, Erycina pusilla, Cymbidium faberi). All sequences were aligned with the HomBlock pipeline (Bi et al. 2018) and subsequently checked manually in Bioedit v5.0.9 (Hall 1999). Then, the phylogenetic tree constructed by RAxML (Stamatakis 2014) with 1000 ultrafast bootstrap (UFBoot) replicates (Minh et al. 2013; Chernomor et al. 2016). The results showed that E. corneri was mostly related to Calanthe triplicata and Calanthe davidii (Figure 1).
Figure 1.
A phylogenetic tree was constructed based on 13 complete chloroplast genome sequences of Orchidaceae. All the sequences were downloaded from NCBI GenBank.
Disclosure statement
There are no conflicts of interest for all the authors including the implementation of research experiments and writing this article.
References
- Bhuiya NMMA, Hasan M, Al Mahmud Z, Qais N, Kabir MSH, Ahmed F, Uddin MMN. 2017. In vivo analgesic, antipyretic and anti-inflammatory potential of leaf extracts and fractions of Eria javanica. Journal of Complementary and Integrative Medicine. 14(1). [DOI] [PubMed] [Google Scholar]
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: A multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22. [DOI] [PubMed] [Google Scholar]
- Chen XQ, Jin ZH, Luo YB. 1999. Orchidaceae flora of China. Beijing: Science Press. 19:15–16. [Google Scholar]
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65(6):997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acid S. 41:95–98. [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for denovo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479. [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CS, Chen JJ, Huang YT, Chan MT, Daniell H, Chang WJ, Hsu CT, Liao DC, Wu FH, Lin SY, et al. 2015. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci Rep. 5(1):9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN. 2009. Genera Orchidacearum. Oxford (UK): Oxford University Press. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]