Abstract
Bougainvillea spectabilis is an important ornamental plant in tropical and subtropical regions, whose complex and diverse germplasm has brought great difficulties to variety selection and identification. Here, to provide data support for the identification of the relationship between cultivars of B. spectabilis, we sequenced the chloroplast genome of B. spectabilis. The chloroplast genome is 154,520 bp in length, including a large single-copy (LSC) region of 88,101 bp and small single-copy (SSC) region of 17,729 bp, and a pair of invert repeats (IR) regions of 24,171 bp. The chloroplast genome contains 129 genes, including 83 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The phylogenetic analysis based on 14 chloroplast genome demonstrated a close relationship between B. spectabilis and other plant species in Caryophyllales. The chloroplast genome will help for further of taxonomy research and provide data support for the identification of genetic relationship among different cultivars of B. spectabilis.
Keywords: Bougainvillea spectabilis, chloroplast genome, phylogeny, Nyctaginacea
Bougainvillea belong to Nyctaginaceae, which native to South America, widely cultivated in tropical and subtropical regions (Xu et al. 2008; Zhao et al. 2019). Bougainvillea has 18 species, among which B. spectabilis, B. glabra and B. peruviana are the most ornamental value (Zhou et al. 2011). Their bracts have gorgeous color, long florescence and strong resistance, which are of high value in urban landscape greening and ornamental horticulture. In addition, there are more than 400 varieties of Bougainvillea have been registered on the international authority, and the genetic background of these varieties are complex (Li et al. 2011; Zhao et al. 2019). However, few studies on the genetic relationship among varieties of Bougainvillea (Yin et al. 2001). Therefore, we reported the complete chloroplast genome of B. spectabilis, which will be helpful to support for the identification of the relationship between varieties of B. spectabilis.
The plant material of B. spectabilis was collected from Xiamen botanical garden, Xiamen City, Fujian province, China (118°6′59.09″E, 24°27′14.50″N). The voucher specimen is kept at the Herbarium of College of Forestry, Fujian Agriculture and Forestry University (specimen code FAFU17028) (Chen et al. 2019).
DNA extraction from fresh leaf tissue, with 500 bp randomly interrupted by the Covaris ultrasonic breaker for library construction (Xiang et al. 2019). The constructed library was sequenced PE150 by Illumina Hiseq Xten platform, approximately 2GB data was generated. Illumina data were filtered by script in the cluster (default parameter: -L 5, -p 0.5, -N 0.1). Complete chloroplast genome of B. spectabilis (GeneBank accession: KJ001129) is used as reference, chloroplast genome of B. spectabilis was assembled by GetOrganelle pipe-line (https://github.com/Kinggerm/GetOrganelle), it can get the chloroplast-like reads, and the reads were viewed and edited by Bandage (Wick et al. 2015). Assembled choroplast genome annotation was based on comparison with Portulaca oleracea by Geneious v 11.1.5 (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. 2012). The annotation result was draw with the online tool OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. 2013).
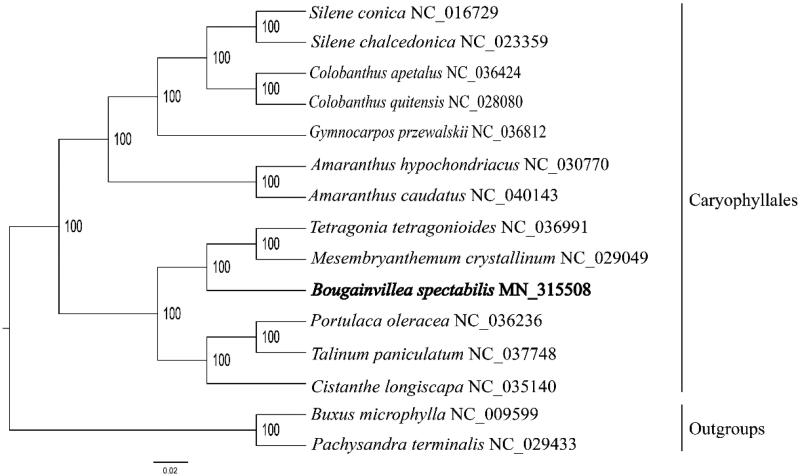
The complete chloroplast genome sequence of B. spectabilis (GenBank accession: MK840978) was154,520 bp in length, with a large single-copy (LSC) region of 88,101 bp, a small single-copy (SSC) region of 17,729bp, and a pair of inverted repeats (IR) regions of 24,171 bp. Complete chloroplast genome contain 129 genes, there were 83 protein-coding genes, 37 tRNA genes, 8 rRNA genes.The complete genome GC content was 36.5%. In order to reveal the phylogenetic position of B. spectabilis with other members of Nyctaginaceae, a phylogenetic analysis was performed based on 14 complete chloroplast genomes of Nyctaginaceae, and two taxa (Alnus cordata, Betula cordifolia) as outgroups. Them all downloaded from NCBI GenBank. The sequences were aligned by MAFFT v7.307 (Katoh and Standley 2013), and phylogenetic tree constructed by RAxML (Stamatakis 2014) The phylogenetic tree showed that B. spectabilis is sister to Tetragonia tetragonioides and Mesembryanthemum crystallinum, which forms a base clade of Caryophyllales with strong support (Figure 1).
Figure 1.
Phylogenetic analysis of 12 species of Caryophyllales and two taxa (Buxus microphylla, Pachysandra terminalis) as outgroup based on chloroplast genome sequences by RAxML, bootstrap support value near the branch.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Chen DQ, Xiang S, Liu ZJ, Zou SQ. 2019. The complete chloroplast genome sequence of Kandelia obovata (Rhizophoraceae). Mitochondrial DNA Part B. 4(2):3494–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment soft-ware version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FY, Huang YJ, Wu SH. 2011. ISSR analysis of germplasm resources of Bougainvillea spectabilis Willd. Chin J Trop Crop. 32(9):1692–1696. [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar genome DRAW—a suite of tools for generating physical maps of chloroplast and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Liu X-D, Sun W-H, Lan S-R, Liu Z-J, Zou S-Q. 2019. The complete chloroplast genome sequence of Euscaphis japonica (Staphyleaceae). Mitochondrial DNA Part B. 4(2):3484–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SX, Wang LS, Shu QY, Su MH, Huang Q, Zhang W, Liu G. 2008. Progress of study of the biology of the resource plant Bougainvillea. Chin Bull Bot. 25(04):483–490. [Google Scholar]
- Yin JM, Ou WJ, Zhang X. 2001. Protein analysis of interspecific sibship of Bougainvillea. Chin J Trop Crop. 22(04):61–66. [Google Scholar]
- Zhao T, Chang SX, Leng QY, Xu SS, Yin J, Niu J. 2019. Development of SSR molecular markers based on transcriptome sequencing of Bougainvillea. Mol Plant Breed. 17(13):4331–4341. [Google Scholar]
- Zhou Q, Huang KF, Ding YL, Guo HZ. 2011. Investigation and taxonomic identification on introduced ornamental varieties in Bougainvillea in China. Acta Agriculturae Jiangxi. 23(5):53–56. [Google Scholar]