Abstract
The dry root (Radix Fici Hirtae) of Ficus hirta has been used as a traditional herbal medicine in Ling nan regions of China for a long time. As its large market demand, the wild resources of F. hirta have sharply reduced. It is necessary to conduct the study of conservation genetics. However, there is still lack of complete genome information for the research on evolutionary biology, population genetics and phylogeography of this species. Here, we sequenced the complete chloroplast (CP) genome of F. hirta using Next Generation Sequencing technology (NGS). The CP genome of F. hirta is 160,374 bp in length, which contains a large single-copy (LSC) region of 88,446 bp, a small sing-copy (SSC) region of 18,134 bp, and two inverted repeat (IRa and IRb) regions of 26,897 bp. A total of 130 genes were successfully annotated containing 85 protein-coding genes, 37 tRNA genes and 8 rRNA genes. Phylogenetic analysis support genus Ficus is monophyletic and F. hirta is closely related to F. carica within this genus.
Keywords: Ficus hirta, complete chloroplast genome, phylogenetic analysis
The dry root (Radix Fici Hirtae) of Ficus hirta Vahl is a traditional herbal medicine in Ling nan regions of China with a long history for application. Radix Fici Hirtae has the effects of strengthening the spleen, nourishing the lung, removing dampness and relaxing muscles (Ma and Zhang 2010). In addition to medical values, Radix Fici Hirtae is drug-food homologous, whose food ingredient is favored by the Ling nan residents of China (Shi et al. 2013; Wu et al. 2013). With the development of all kinds of products sourced from Radix Fici Hirta, the wild resources of F. hirta have reduced increasingly (Dong et al. 2014). It is necessary to conduct management and protection of wild resources of F. hirta to prevent the resources depletion. Although, Radix Fici Hirta has been widely researched and applied, they mostly focused on quality standard, chemical composition, pharmacological action etc. (Luo and Jiang 2014), there is still lack of study on resources conservation of F. hirta, such as the aspects of evolutionary biology, population genetics, phylogeography etc. Therefore, we sequenced the complete chloroplast genome of F. hirta and aimed to obtain much more genetic information of this species.
A sample of F. hirta was collected in XinYi city, China (N:22° 18' 09''; E:111°10' 32''), and deposited at the herbarium of School of Traditional Chinese Medicine, Southern Medical University (specimen code: CYR-1). CTAB method was used to extract total genomic DNA (Yang et al. 2014). Illumina paired-end (PE) library was pre-pared and sequenced on an illumina Hiseq4000 platform (Novogene biotechnology Co.Ltd, Bejing, China). The chloroplast genome was assembled using SPAdes version 3.11.1 (Bankevich et al. 2012) with the chloroplast genome of Ficus carica as a reference (GenBank accession: KY635880). The chloroplast genome of F. hirta was annotated using Geneious version 11.0.4 (Kearse et al. 2012) and Plastid Genome Annotator (PGA) (Qu et al. 2019) and deposited in GenBank (Accession NO.: MN364706).
The complete chloroplast genome of F. hirta is 160,374 bp in length, including a large single-copy (LSC) region of 88,446 bp, a small single-copy(SSC) region of 18,134 bp, and two inverted repeat (IRa and IRb) regions of 26,897 bp. The GC content of cp genome of F. hirta is 36.0%. The CP genome comprises a total of 130 genes, including 85 protein-coding, 8 ribosomal RNA (rRNA) genes, and 37 transfer RNA (tRNA) genes. Fifteen genes (trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC and ndhA) contain one intron, two genes (clpP, ycf3) have two introns. A trans-splicing gene was found (rps12 gene).
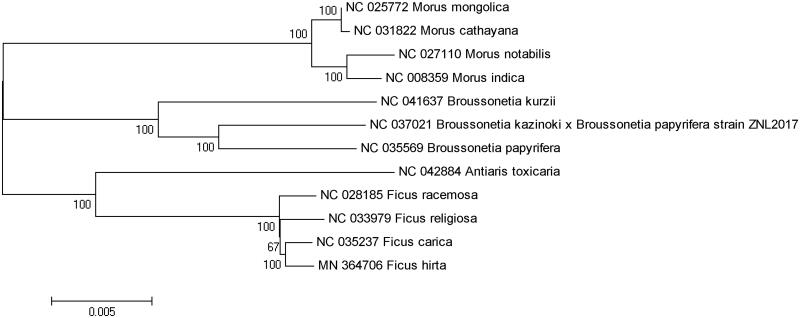
The phylogenetic reconstruction including the represents of genus Ficus, and closely related genus Antiaris, Broussonetia, Morus (Moraceae) was conducted. 11 chloroplast genome sequences downloaded from GenBank and one (F. hirta) were used for phylogenetic analysis (Figure 1). All sequences were aligned with the MAFFT v7.037 (Katoh and Standley 2013). Phylogenetic reconstruction was performed with MEGA v7.0 based on the neighbour-joining (NJ) analysis (Tamura et al. 2011). The results support genus Ficus is monophyletic and F. hirta is closely related to F. carica within this genus (Figure 1).
Figure 1.
Neighbour-joining (NJ) phylogenetic tree constructed from 12 complete chloroplast genome sequences of Moraceae family based on K2-P distance with 1000 bootstrap replicates. The bootstrap support values are indicated at the nodes.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong QS, Yan ZG, Wei SG, Ke F, Wu QH. 2014. Study on the biological characters of the seed of Ficus hirta. Jiangsu Agr Sci. 42:278–280. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment soft-ware version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo YD, Jiang LR. 2014. Research progress on quality control, production process and pharmacological action of Yao medicine Radix Ficus Hirta. China Modern Med. 31:193–196. [Google Scholar]
- Ma J, Zhang HW. 2010. Ling Nan herbs. Bejing: Science Press. [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and fexible batch annotation of plastomes. Plant Methods. 15:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi TX, Lin YY, Luo M, Wei P, Liu DH, Lin HY, Xu RX, Wei G. 2013. HPLC fingerprint of Radix Fici in medicated diet. Modern Food Sci Tech. 29:401–404. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum like-lihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WK, Chen FH, Yan QR, Song W. 2013. Study on folk medicine used by Hakka in Meizhou. Zhongguo Zhong Yao Za Zhi. 38:3984–3987. [PubMed] [Google Scholar]
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Res. 14:1024–1031. [DOI] [PubMed] [Google Scholar]