Abstract
Spondylus violaceus is a species of the bivalve family Spondylidae, commonly known as thorny oyster. Here, we report the complete mitochondrial genome of S. violaceus. The mitochondrial genome of S. violaceus is 30,160 bp in length, which contains 12 protein-coding genes, 2 ribosomal RNA genes and 19 transfer RNA genes. The overall AT content (58.31%) was higher than GC content (41.69%). This is the first record of complete mitochondrial genome in the family Spondylidae, providing the basis for future taxonomic and phylogenetic studies of this bivalve family.
Keywords: Spondylus violaceus, mitochondrial genome, phylogenetic analysis
Spondylus violaceus belongs to the genus Spondylus within the bivalve family Spondylidae, which is commonly known as thorny oyster due to its similar appearance to oysters and long spines on the shells (Koyama et al. 2006). Mitochondrial DNAs have been extensively used for studying population structure, phylogeography and phylogenetic relationships at various taxonomic levels (Curole and Kocher 1999; Saccone et al. 1999). To date, the mitochondrial genomes of nearly 200 bivalve species have been sequenced and annotated, none of which came from the family Spondylidae. In this study, we successfully assembled and annotated the mitochondrial genome of S. violaceus, representing the first complete mitochondrial genome in the family Spondylidae.
The sample of S. violaceus was collected from Tung Ping Chau Bay near Tung Chung village (Guangdong province, China). The DNA sample of S. violaceus was extracted by using phenol/chloroform/isoamyl alcohol method (Sambrook et al. 1989). The DNA sample was deposited at the Key Laboratory of Marine Genetics and Breeding (Ministry of Education), Ocean University of China (Specimen code: OUC-MGB-2018-SF-07). Whole genomic sequencing (WGS) of this sample was performed using the Illumina NovaSeq 6000 sequencing platform. We assembled the mitochondrial genome based on the WGS data using the software NOVOPlasty (Dierckxsens et al. 2017). The genome sequence was further analyzed using MITOS software (Bernt et al. 2013) to retrieve gene information.
The complete mitogenome of S. violaceus was 30,160 bp in length and encoded 12 protein-coding genes (PCGs), 2 rRNAs and 19 tRNAs, lacking the atp8 gene as in most bivalves (Wu et al. 2009). The mitogenome sequence was submitted to the GenBank under the accession number of MN019127. The 12 PCGs were cytochrome oxidase subunit (I, II, III), NADH dehydrogenase subunit (1, 2, 3, 4, 5, 6 and 4 L), ATP synthase subunit (6), and cytochrome b apoenzyme. The small subunit ribosomal RNA (12S rRNA) and large subunit ribosomal RNA (16S rRNA) were annotated with sizes of 879 and 1726 bp, respectively. The length of 19 tRNAs ranged from 65 bp to 75 bp. The overall base composition of the mitochondrial genome was 22.46% for A, 35.85% for T, 28.65% for G, and 13.04% for C, demonstrating a bias of higher AT content (58.31%) than GC content (41.69%). All genes of S. violaceus were encoded on the same strand, consistent with previous findings for most bivalves (Milbury and Gaffney 2005; Xu et al. 2011).
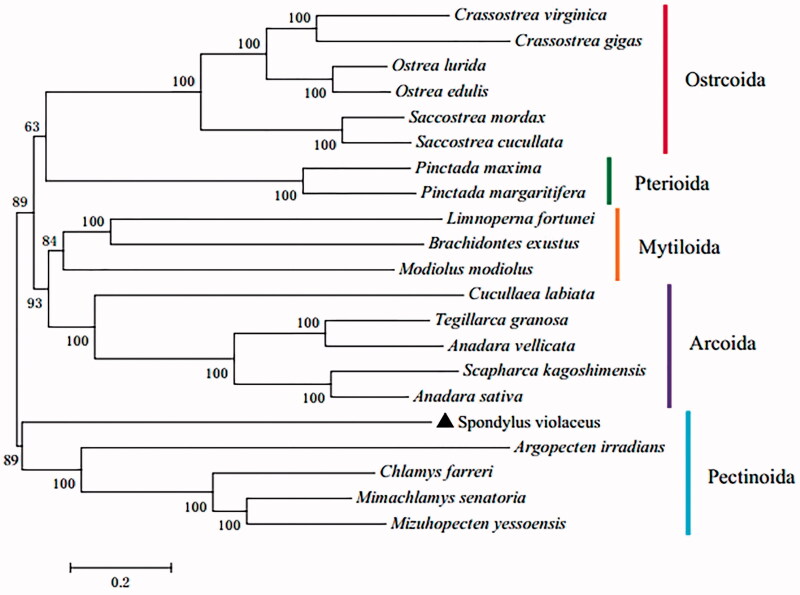
To assess the phylogenetic relationship of S. violaceus, we downloaded the mitochondrial genome sequences of 20 species (representing five orders under the subclass Pteriomorphia) from NCBI database to construct genome-wide sequence alignment. A phylogenic tree was constructed by the neighbor-joining (NJ) algorithm with 1000 bootstrap trials. Despite the morphological resemblance with oysters, the phylogenetic tree provided strong molecular evidence that S. violaceus belongs to the order Pectinoida (Figure 1), consisting with previously inferred phylogenetic relationships of the family Spondylidae based on several nuclear/mitochondria-derived genes (Bieler et al. 2014).
Figure 1.
The phylogenetic tree of S. violaceus and 20 other bivalves based on complete mitochondrial genomes. The accession numbers of downloaded sequences are as follows: Crassostrea virginica (AY905542.2), Crassostrea gigas (NC_001276.1), Ostrea lurida (NC_022688.1), Ostrea edulis (JF274008.1), Saccostrea mordax (FJ841968.1), Saccostrea cucullata (NC_027724.1), Pinctada maxima (NC_018752.1), Pinctada margaritifera (NC_021638.1), Limnoperna fortunei (NC_028706.1), Brachidontes exustus (NC_024882.1), Modiolus modiolus (NC_033537.1), Cucullaea labiata (NC_029848.1), Tegillarca granosa (NC_026081.1), Anadara vellicata (KP954700.1), Scapharca kagoshimensis (NC_025509.1), Anadara sativa (NC_024927.1), Argopecten irradians (EU023915.1), Chlamys farreri (EF473269.1), Mimachlamys senatoria (NC_022416.1), and Mizuhopecten yessoensis (FJ595959.1).
Disclosure statement
The authors report no competing interests declared.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319. [DOI] [PubMed] [Google Scholar]
- Bieler R, Mikkelsen PM, Collins TM, Glover EA, González VL, Graf DL, Harper EM, Healy J, Kawauchi GY, Sharma PP, et al. 2014. Investigating the bivalve tree of life an exemplar-based approach combining molecular and novel morphological characters. Invert Syst. 28:32–115. [Google Scholar]
- Curole JP, Kocher TD. 1999. Mitogenomics: digging deeper with complete mitochondrial genomes. Trends Ecol Evol (Amst). 14:394–398. [DOI] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Chounan R, Uemura D, et al. 2006. Hepatoprotective effect of a hot-water extract from the edible thorny oyster Spondylus varius on carbon tetrachloride-induced liver injury in mice. J Agric Chem Soc Jpn. 70:729–731. [DOI] [PubMed] [Google Scholar]
- Milbury CA, Gaffney PM. 2005. Complete mitochondrial DNA sequence of the Eastern oyster Crassostrea virginica. Mar Biotechnol. 7:697–712. [DOI] [PubMed] [Google Scholar]
- Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A. 1999. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene. 238:195–209. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Lab Press. [Google Scholar]
- Wu X, Xu X, Yu Z, Kong X. 2009. Comparative mitogenomic analyses of three scallops (Bivalvia: Pectinidae) reveal high level variation of genomic organization and a diversity of transfer RNA gene sets. BMC Res Notes. 2:60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Kanno M, Yu H, Li Q, Kijima A. 2011. Complete mitochondrial DNA sequence and phylogenetic analysis of Zhikong scallop Chlamys farreri (Bivalvia: Pectinidae). Mol Biol Rep. 38:3067–3074. [DOI] [PubMed] [Google Scholar]