Abstract
To characterize the molecular structure of IncR plasmid-related sequences, comparative genomic analysis was conducted using 261 IncR plasmid backbone-related sequences. Among the sequences, 257 were IncR plasmids including the multidrug-resistance IncR plasmid pR50-74 from Klebsiella pneumoniae strain R50 of this work, and the other four were from bacterial chromosomes. The IncR plasmids were derived from different bacterial genera or species, mainly Klebsiella pneumoniae (70.82%, 182/257), Escherichia coli (11.28%, 29/257), Enterobacter cloacae (7.00%, 18/257), and Citrobacter freundii (3.50%, 9/257). The bacterial chromosomes carrying IncR plasmid backbone sequences were derived from Proteus mirabilis AOUC-001 and Klebsiella pneumoniae KPN1344, among others. The IncR backbone sequence of P. mirabilis AOUC-001 chromosome shows the highest identity with that of pR50-74. Complex class 1 integrons carrying various copies of ISCR1-sdr-qnrB6-△qacE/sul1 (ISCR1-linked qnrB6 unit) were identified in IncR plasmids. In addition to two consecutive copies of qnrB6-qacE-sul1, the other resistance genes encoded on pR50-74 are all related to mobile genetic elements, such as IS1006, IS26, and the class 1 integron. This study provides a clear understanding of the mobility and plasticity of the IncR plasmid backbone sequence and emphasizes the important role of ISCR in the recruitment of multicopy resistance genes.
1. Introduction
IncR was first characterized as a new plasmid incompatibility group in 2009 [1]. The first sequenced IncR single-replicon plasmid was pEFER, though IncR often coexists with other replicons such as IncC, IncN, IncHI, and IncFII [2]. There are an increasing number of reports about IncR plasmids carrying various resistance genes, such as blaKPC-2, blaDHA-1, blaNDM-1, blaVIM-1, qnrS1, or armA, in clinical Enterobacterales, especially in Klebsiella pneumoniae strains from different geographical regions [3–5]. Importantly, the pool of resistance genes carried by IncR replicon plasmids may spread to transferable plasmids through transposition or plasmid recombination events, contributing to the high plasticity of multiple-replicon plasmids [2].
Integrons and insertion sequence common region (ISCR) elements were originally named common regions (CRs), which represent a family of mobile genetic elements (MGEs) that are similar to the insertion sequence (IS) element [6]. ISCR can transpose adjacent DNA sequences by rolling circle replication, and this process has been demonstrated experimentally using MGEs IS91 and IS1294 [7]. ISCR elements are notable for their close association with a wide variety of antibiotic resistance genes. Complex class 1 integrons are large, flexible genetic elements that consist of different class 1 integrons followed by ISCR [8]. As complex class 1 integrons are powerful gene-capturing tools that can mobilize extremely large sections of DNA encoding a variety of resistance genes related to chloramphenicol, trimethoprim, quinolone, and β-lactam antibiotics [7], these elements play important roles in the recruitment and spread of resistance genes.
Based on sequencing of a multidrug-resistance IncR plasmid from a K. pneumoniae strain carrying a complex class 1 integron with two consecutive ISCR1-linked qnrB6 units, comparative genome analysis of the IncR plasmid with various structures of the ISCR1-related complex class 1 integron was performed in this study.
2. Materials and Methods
2.1. Bacterium Genome Sequencing and Annotation
K. pneumoniae R50 was isolated from an anal swab sample of a rabbit from a farm in Wenzhou, South China. Genomic DNA was extracted from K. pneumoniae R50 using an AxyPrep Bacterial Genomic DNA Miniprep Kit (Axygen Scientific, Union City, CA, USA) and sequenced by Illumina HiSeq-2500 and Pacific Bioscience sequencers at Annoroad Gene Technology Co., Ltd. (Beijing, China). Sequencing reads of approximately 10-20 kb in length were assembled with Canu v1.7 [9]. Error correction of tentative complete circular sequences of the plasmids and the chromosome was performed using Pilon version 1.18 [10] with two FASTQ read sets derived from the HiSeq-2500 sequencing platform.
Open reading frames (ORFs) and pseudogenes were predicted using Prokka with default parameters, combined with BLASTP searches against the UniProtKB/Swiss-Prot and RefSeq databases [11–14]. The rRNA gene sequences were annotated by RNAmmer [15], and the tRNA sequences were annotated by tRNAscan-SE 2.0 [16]. Annotation of resistance genes, mobile genetic elements, and other features was carried out using online databases including CARD [17], ISfinder [18], INTEGRALL [19], ResFinder [20], and the Tn Number Registry [21]. The number of iteron tandem repeats was predicted by Tandem Repeats Finder [22]. Gene organization diagrams were drawn in Inkscape 0.48.1 (https://inkscape.org/en/).
2.2. Antimicrobial Susceptibility Testing
Minimum inhibitory concentration (MIC) assessment of 16 antimicrobial agents was performed using the agar dilution method in accordance with the guidelines of Clinical and Laboratory Standards Institute (CLSI document M100-S27, 2017).
2.3. Comparative Genomics and Phylogenetic Analysis
Plasmid pHN84KPC (KY296104) was selected as reference because it contains the most complete IncR backbone, and gene sequences with coverage > 50% and identity > 80% to those of pHN84KPC were considered effective homologous backbone genes [5]. Sequences containing the backbone genes resD, repB, parAB, and umuCD were used for the phylogenetic analysis. Multiple sequence alignments were performed using MAFFT v7 [23], and an unrooted maximum likelihood phylogenetic tree was reconstructed using the generalized time-reversible model of evolution with uniform rates of substitution in MEGA7 [24]. The phylogenetic tree was visualized using the interactive web platform iTOL [25]. The sequence retrieval, statistical analysis, and other bioinformatics tools used in this study were applied with python scripts.
2.4. Nucleotide Accession Numbers
The complete sequences of the K. pneumoniae R50 chromosome (CP040362) and plasmid pR50-74 (CP040363) have been submitted to NCBI GenBank.
3. Results and Discussion
3.1. Main Features of K. pneumoniae R50
The K. pneumoniae R50 genome consists of one 5,210,287 bp circular chromosome encoding 4236 ORFs and one plasmid (named pR50-74) of 74,011 bp in length encoding 89 ORFs (Table S1). Based on analyses of seven housekeeping genes, including gapA, infB, mdh, pgi, phoE, rpoB, and tonB [26], K. pneumoniae R50 was classified into the multilocus sequence type ST948. To the best of our knowledge, this is the first sequenced K. pneumoniae strain of this multilocus sequence type. R50 was further classified into the K5 capsular serotype based on the capsular locus of the wzi-5 allele. A total of 13 antibiotic resistance genes are encoded in the K. pneumoniae R50 genome, with 4 and 9 in the chromosome and the plasmid pR50-74, respectively. The resistance genes are mainly related to the antibiotics of florfenicol, tetracycline, quinolone, aminoglycoside, rifampicin, trimethoprim, streptomycin, and sulfonamides (Table 1), in accordance with the results of antimicrobial susceptibility testing of the bacterium (Table 2).
Table 1.
Antibiotic resistance gene of R50.
| Resistance to | Gene | Genetic context | Identity% |
|---|---|---|---|
| Beta-lactams | blaSHV-1 | Chromosome | 100 |
| Aminoglycosides | aadA16 | Plasmid | 98 |
| aac(6)-Ib | Plasmid | 95 | |
| Quinolones | oqxA | Chromosome | 99 |
| oqxB | Chromosome | 99 | |
| qnrB6 | Plasmid | 100 | |
| Sulfonamides | sul1 | Plasmid | 100 |
| Rifapentine | arr3 | Plasmid | 100 |
| Trimethoprim | dfrA27 | Plasmid | 100 |
| Tetracyclines | tetD | Plasmid | 99 |
| tetR | Plasmid | 99 | |
| Fosfomycin | fosA | Chromosome | 96 |
| Florfenicol | floR | Plasmid | 99 |
Table 2.
Antibiotic resistance profile of R50.
| Drug | MIC (μg/ml) | Phenotype |
|---|---|---|
| Ampicillin | 32 | Resistant |
| Spectinomycin | 32 | |
| Florfenicol | 1024 | Resistant |
| Chloramphenicol | 512 | Resistant |
| Nalidixic acid | 32 | Resistant |
| Gentamicin | ≤0.25 | Sensitive |
| Tobramycin | 4 | Sensitive |
| Tetracyclines | 512 | Resistant |
| Kanamycin | 4 | Sensitive |
| Streptomycin | 4 | Sensitive |
| Amikacin | ≤1 | Sensitive |
| Cephalothin | ≤0.03 | |
| Azithromycin | ≤1 | Sensitive |
| Sisomicin | 2 | |
| Rifampin | >1024 | Resistant |
| Erythromycin | 64 |
3.2. Structural Characterization of IncR Plasmid Backbone Sequences
The backbone sequences of the most sequenced IncR plasmids usually contain vagCD (toxin-antitoxin system), resD (multimer resolution), repB (replication initiation), parAB (partition), umuCD (stability), and retA (maintenance) genes [2]. Plasmid pR50-74 with two replicons, R and FIA(HI1), carries seven of these genes but lacks vagCD, which are generally located upstream of resD on IncR plasmids.
To characterize the presence of backbone genes among IncR plasmids, we used the single-replicon plasmid pHN84KPC as a reference because it contains the most complete IncR backbone regions [27]. A total of 261 genome sequences containing the IncR replicon gene repB, showing >90% identity and >80% coverage with that of pHN84KPC, were retrieved from GenBank. The range of identity and coverage of repB gene in 261 sequences to that of pHN84KPC was 93.6%-100% and 98%-100%, respectively. Among these 266 sequences, 257 are from plasmids and 4 from chromosomes. The plasmid sequences are from 20 different species: K. pneumoniae (70.82%; 182/257), Escherichia coli (11.28%; 29/257), Enterobacter cloacae (7.00%; 18/257), Citrobacter freundii (3.50%; 9/257), and 16 other species (8.17%; 21/257) (Table S2). In addition to repB, other backbone genes were also screened. Approximately 64.20% (165/257) of the plasmids contain all 9 backbone genes; 14.00% (36/257) of the plasmids only lack vagCD genes, and 7.00% (18/257) of the plasmids lack both vagCD and retA genes (Figure 1). The results demonstrate that vagCD and retA are less conservative than other backbone genes. Interestingly, among the 4 chromosome sequences, one from K. pneumoniae KPN1344 (CP033901) and one from Proteus mirabilis AOUC-001 (CP015347) harbor all 9 backbone genes; the two from E. cloacae strains 339389L (CP026536) and FDAARGOS_77 (CP026975) only contain 3 or 5 backbone genes, respectively.
Figure 1.
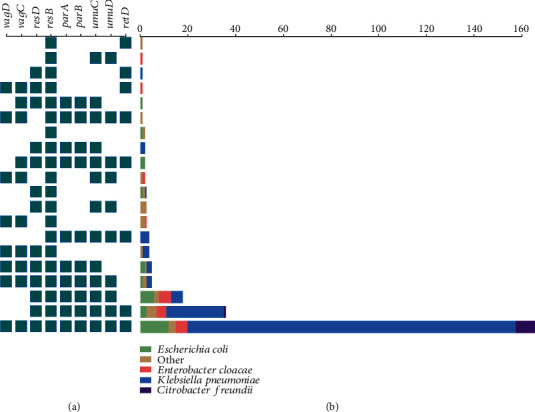
Distribution of plasmids with different numbers of backbone genes. The small squares on the left represent the presence of corresponding backbone genes. (b) The bars represent the number of plasmids and the corresponding species.
The IncR backbone region of AOUC-001 and pR50-74 were almost identical (95% BLAST coverage and 99% nucleotide identity) to each other. However, the IncR backbone sequence in the KPN1344 chromosome can be separated into three parts, vagCD, resD, and repB-parAB-umuCD-retA, among which the second and third parts are approximately 40 kb apart (Figure 2). Comparison result of the sequence of these three parts combined with the 257 IncR plasmids mentioned above revealed that the highest homologous sequence to it was the backbone sequence of plasmid AR_0126 (CP021741) from K. pneumoniae (Figure 2). Further comparative genomics analysis was applied to AOUC-001, pR50-74, pHN84KPC, KPN1344, and AR_0126. The backbone region of pHN84KPC is flanked by two IS elements, ISKpn19 and IS1X3, which may promote the mobility of the sequence. AOUC-001 and pR50-74 share a nearly identical backbone sequence, except that the latter lacks vagCD genes. Furthermore, an identical traC-clsB-mercury operon sequence downstream of the retA gene of both AOUC-001 and pR50-74 is also present on other IncR plasmids, such as pKP6402 (AP018752) from K. pneumoniae (Figure 2). The backbone sequence of AOUC-001 is flanked by ISKpn14 and IS6100; however, no adjacent direct repeat sequence was identified. Nonetheless, vagCD genes are missing from the plasmid AR_0126, and the region upstream of resD is an approximately 3 kb fragment harboring ccdAB, which is different from pHN84KPC and other IncR plasmids. This result suggests that the IncR plasmid backbone sequence in KPN1344 and AR_0126 may have derived from the same ancestor and underwent deletion of vagCD as well as insertion of a 40 kb fragment between resD and repB during evolution.
Figure 2.
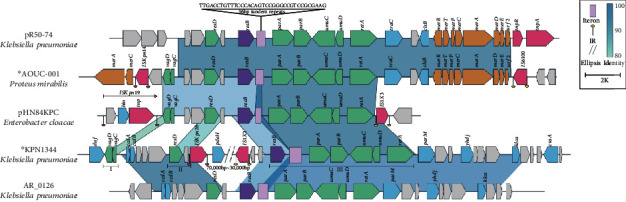
Linear comparison of the backbone regions of pR50-74, AOUC-001, PHN84KPC, KPN1344, and AR_0126. Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on functional classification. The colors of the shadow areas represent different identities (80%-100%). Numbers under double diagonal indicate nucleotide positions for corresponding plasmids. Chromosome sequences are marked with asterisks.
The sequence differences in IncR backbones among pHN84KPC, pR50-74, KPN1344, AOUC-001, and AR_0126 are mainly in the regions encoding vagCD, resD, repB, and iterons which are direct repeat DNA sequences that play an important role in regulating plasmid copy number in bacterial cells [28]. The iteron regions of these five sequences are all composed of the same 36 bp tandem repeats, only with repeat number difference (Figure 2). The numbers of tandem repeats for pHN84KPC, pR50-74, KPN1344, AOUC-001, and AR_0126 were 10.5, 15.5, 22.5, 15.5, and 16.5, respectively. The identities of the vagCD genes on KPN1344 and AOUC-001 compared to those on pHN84KPC are 86% and 89%, respectively. In addition, the identities of the resD genes of pR50-74, KPN1344, AOUC-001, and AR_0126 compared to those of pHN84KPC are 100%, 86%, 100%, and 88%, respectively. Moreover, phylogenetic analysis of concatenated backbone genes resD, repB, parAB, and umuCD among 215 sequences (Table S3), including the two chromosomes, indicated that the IncR backbone sequence of AOUC-001 is evolutionarily closer to that of pla74k (Figure 3). It could be deduced that the IncR backbone of pR50-74 and AOUC-001 likely has the same origin. It was previously reported that MGEs, like Tn1331, mediate the evolution of type B plasmids carrying IncN3-like segment from type A plasmids carrying IncR backbone [29]. Papagiannitsis et al. reported that an MDR plasmid pKP1780 carries IncR backbone segment flanked by two-copy IS elements [30]. The MGEs on both sides of the backbone enable extensive reorganization among plasmids and chromosomes.
Figure 3.
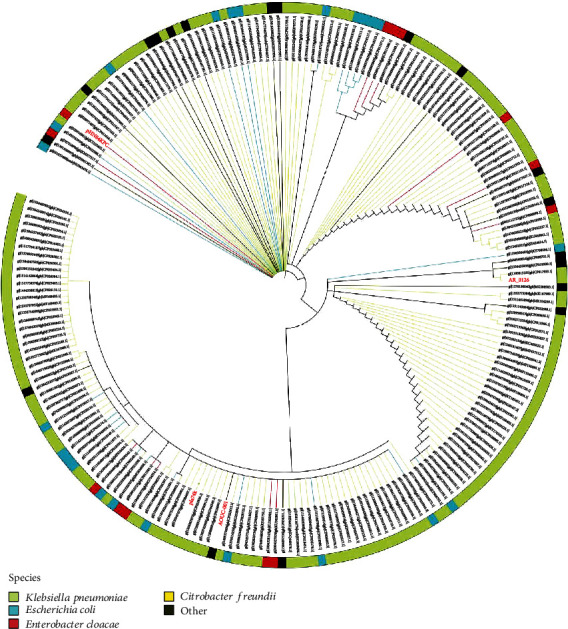
Phylogeny of backbone sequences of IncR plasmids.
3.3. Comparative Analysis of the MDR Region of pR50-74-Related IncR Plasmids
The multidrug resistance (MDR) region of pR50-74 carries floR, tetD, qnrB, aac(6′)-Ib-cr5, arr3, dfrA27, aadA16, and sul1 (Figure 4), conferring resistance to chloramphenicol/florfenicol, tetracyclines, quinolones, aminoglycosides, rifampicin, trimethoprim, streptomycin, and sulfonamides, respectively. The MDR region can be divided into three fragments, and all of the resistance genes are associated with MGEs (Figure 4). The first fragment is a transposon structure carrying a lysR-floR-virD2 unit that is conserved among the majority of floR-related sequences and often shares the same upstream direct repeats (DRs) and a complete or truncated downstream tnp unit [31]. The second fragment is a complex transposon sequence bracketed by two IS26 in opposite orientation containing tetR, tetD, adhC, and orf2, which are commonly found on other plasmids such as those of two Citrobacter sp. strains, TA3 (AB257967) and TA6 (AB089600), and the Alteromonas sp. strain TA55 (AB089601) isolated from fish [32]. The last fragment is a complex class 1 integron carrying 4 resistance gene cassettes (aac(6′)-Ib-cr5, arr3, dfrA27, and aadA16) and two copies of resistance gene-related ISCR1 units (ISCR1-sdr-qnrB6-△qacE-sul1).
Figure 4.
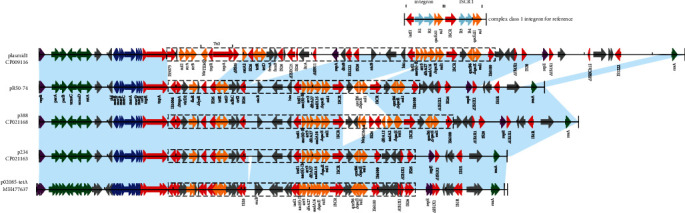
Comparative analysis of the genome structures of pR50-74, plasmid1, p388, p234, and p02085-tetA. Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on functional classification. Shading denotes homologous regions (>95% nucleotide identity).
Four plasmids sharing the highest nucleotide sequence similarities (coverage > 80%, identity ≥ 99%) with pR50-74 were retrieved from the NCBI nucleotide database. Two of the plasmids, p234 (CP021163, 68,573 bp) and p388 (CP021168, 79,064 bp) from E. cloacae, share the highest similarity (100% coverage and 99% identity) with pR50-74. Additionally, plasmid p02085-tetA (MH477637.1, 67,510 bp) from C. freundii shares 98% coverage and 99% identity with pR50-74; the last plasmid (CP009116, 94,760 bp) from the same species (Klebsiella pneumoniae) as pR50-74 shares low similarity (84% coverage and 99% identity) with pR50-74. The entire plasmid sequence of p02085-tetA is almost identical to that of p234, except for the absence of an approximately 1000 bp sequence between IS26 and nicB (Figure 4).
The main difference between pR50-74 and p234 or p388 is a distinct structure of the complex class 1 integron (Figure 4). pR50-74 exhibits a tandem repeat sequence of two copies of the ISCR1-linked qnrB6 unit (ISCR1-sdr-qnrB6-△qacE-sul1) encoded carried by the integron, but p234 has only one copy of the unit. Interestingly, p388 has two different ISCR1-linked resistance gene arrays: one is the ISCR1-linked qnrB6 unit, which is identical to the two units in pR50-74 units, and the other is blaNDM-6-IS26-dfrA12-aadA2-△qacE-sul1. As mentioned above, except for pR50-74, which carries two copies of the ISCR1-linked qnrB6 unit, the other four plasmids have only one. Unlike pR50-74, the upstream region of the tetR-tetD-adhC-orf2 fragment unit in plasmid1 (CP009116) shows insertion of the sul2-strA-strB unit fragment and a Tn2 transposon carrying blaTEM (Figure 4).
3.4. Construction of the Multicopy ISCR1-Mediated Complex Class 1 Integron
The complex class 1 integron of pR50-74 can be divided into two MGE-associated units. One is a typical class 1 integron containing 5′CS (intI1-attI), 3′-CS (qacE△1-sul1-orf5), and a variable region of the gene cassettes (aac6-arr3-dfrA27-aadA16). The other is composed of ISCR1-linked qnrB6 units. Using these two units as a query, 14 sequences containing either or both units of the complex class 1 integron were retrieved from GenBank. These sequences were further clustered into 4 groups. Group 1 includes six plasmids (CP030917, CP028582, CP028553, CP028717, CP019006, and CP018945) that only carry unit one. Group 2 includes six plasmids (MG874044, CP023950, CP009116, CP021163, MH477637, and CP031259) harboring both units. pR50-74, which carries two copies of ISCR1-linked qnrB6 unit, consists of group 3 alone. Group 4 is composed of two plasmids (MG870194 and CP030940), which carry four copies of ISCR1-linked qnrB6 unit, of which MG870194 (from Salmonella enterica) has a class 1 integron identical to that of pR50-74, though aadA16 is absent from CP030940 compared to pR50-74 (Figure S1). No sequence containing three copies of the ISCR1-linked qnrB6 unit was identified. These results suggest that the structure of the complex class 1 integron varies across bacterial species and that these complex class 1 integron sequences, including those with duplicated ISCR1-linked qnrB6 units, share the same ancestor.
As a member of an extended family of IS91-like elements, ISCR elements lack terminal inverted repeats but have distinct terminal sequences designated as oriIS and terIS that indicate unique sites for the initiation and termination, respectively, of the rolling-circle (RC) replication stage of RC transposition [33]. However, there is a degree of inaccuracy with regard to recognition of terIS, causing replication to proceed beyond terIS and into the adjacent sequence. This feature confers ISCR with a critical role in bacterial genetics, and it may be responsible for the mobilization of class 1 integrons [34]. According to the model proposed by Toleman et al. [34], the construction of multicopy ISCR1-linked integrons can be explained in two steps. First, a circular intermediate that carries sdr-qnrB6-△qacE-sul1 is produced by twice aberrant RC replication initiated from the replication origin oriIS of the ISCR1 element. The circular intermediate is then inserted into the integron-like group 1 sequence by homologous recombination at the locus between 41 bp upstream of △qacE and 25 bp downstream of sul1 (Figure 5). This process might occur once or more and generate a single- or multicopy ISCR1-linked complex class 1 integron. A similar process has been reported by Chen et al. [35], in which an ISCR1-linked qnrB2 unit was inserted into an integron-ISCR1 variant to produce an ISCR1-qnrB2-ISCR1 structure.
Figure 5.
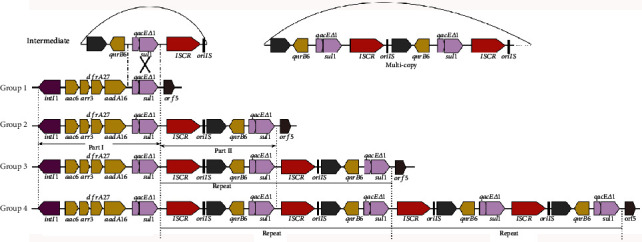
Four groups of sequence diagrams related to the formation of multiple copies of ISCR1-mediated complex class 1 integrons. The proposed circular intermediate of an aberrant RC replication carrying sdr-qnrB6-△qacE-sul1 is shown above. The cross symbol represents a possible homologous recombination event. The sequence in the rectangle represents the starting point of the homologous sequences.
4. Conclusion
The IncR plasmid backbone sequences were identified encoded on both the plasmids and chromosomes in bacteria of a variety of genera. It indicates the wide distribution of IncR plasmids and its backbone sequences. The resistance genes carried on IncR plasmids, such as pR50-74 of this work, are generally related with MGEs. Analyzing the molecular characterization of the complex class 1 integrons, which contain various copies of ISCR1-linked qnrB6 units, allows for a clear understanding of the recruitment and dissemination of resistance genes mediated by MGEs, especially those by ISCRs.
Acknowledgments
This work was funded by grants from the Natural Science Foundation of Zhejiang Province (LY19C060002 and LQ17H190001); Science & Technology Project of Wenzhou City, China (Y20170205); Science & Technology Project of Inner Mongolia Autonomous Region, China (201802125); and the National Natural Science Foundation of China (31500109, 81960381 and 80215049).
Contributor Information
Teng Xu, Email: xuteng@wmu.edu.cn.
Shunfei Lu, Email: lslsf@163.com.
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that there are no conflicts of interest in this work.
Authors' Contributions
Changrui Qian and Xinyi Zhu contributed equally to this work.
Supplementary Materials
Supplementary Figure S1: complex class 1 integrons of group 3 (pR50-74) and group 4 (CP030940, MG870194). Table S1: general features of K. pneumoniae R50 genome. Table S2: species distribution of IncR plasmids. Table S3: bacterial information for evolutionary analysis.
References
- 1.Garcia-Fernández A., Fortini D., Veldman K., Mevius D., Carattoli A. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. The Journal of Antimicrobial Chemotherapy. 2009;63(2):274–281. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- 2.Compain F., Frangeul L., Drieux L., et al. Complete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniae. Antimicrobial Agents and Chemotherapy. 2014;58(7):4207–4210. doi: 10.1128/AAC.02773-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo Q., Spychala C. N., McElheny C. L., Doi Y. Comparative analysis of an IncR plasmid carrying armA, blaDHA-1 and qnrB4 from Klebsiella pneumoniae ST37 isolates. The Journal of Antimicrobial Chemotherapy. 2016;71(4):882–886. doi: 10.1093/jac/dkv444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kocsis E., Gužvinec M., Butić I., et al. blaNDM-1Carriage on IncR plasmid inEnterobacteriaceaeStrains. Microbial Drug Resistance. 2016;22(2):123–128. doi: 10.1089/mdr.2015.0083. [DOI] [PubMed] [Google Scholar]
- 5.Qu D., Shen Y., Hu L., et al. Comparative analysis of KPC-2-encoding chimera plasmids with multi-replicon IncR:IncpA1763-KPC:IncN1 or IncFIIpHN7A8:IncpA1763-KPC: IncN1. Infection and Drug Resistance. 2019;12:285–296. doi: 10.2147/IDR.S189168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stokes H. W., Tomaras C., Parsons Y., Hall R. M. The partial 3'-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid. 1993;30(1):39–50. doi: 10.1006/plas.1993.1032. [DOI] [PubMed] [Google Scholar]
- 7.Toleman M. A., Walsh T. R. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiology Reviews. 2011;35(5):912–935. doi: 10.1111/j.1574-6976.2011.00294.x. [DOI] [PubMed] [Google Scholar]
- 8.Mazel D. Integrons: agents of bacterial evolution. Nature Reviews. Microbiology. 2006;4(8):608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 9.Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H., Phillippy A. M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Research. 2017;27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker B. J., Abeel T., Shea T., et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11, article e112963) doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 12.Boratyn G. M., Camacho C., Cooper P. S., et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Research. 2013;41(W1):W29–W33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boutet E., Lieberherr D., Tognolli M., et al. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods in Molecular Biology. 2016;1374:23–54. doi: 10.1007/978-1-4939-3167-5_2. [DOI] [PubMed] [Google Scholar]
- 14.O'Leary N. A., Wright M. W., Brister J. R., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Research. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagesen K., Hallin P., Rødland E. A., Stærfeldt H. H., Rognes T., Ussery D. W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe T. M., Chan P. P. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Research. 2016;44(W1):W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia B., Raphenya A. R., Alcock B., et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Research. 2017;45(D1):D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Research. 2006;34(90001):D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moura A., Soares M., Pereira C., Leitao N., Henriques I., Correia A. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics. 2009;25(8):1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 20.Zankari E., Hasman H., Cosentino S., et al. Identification of acquired antimicrobial resistance genes. The Journal of Antimicrobial Chemotherapy. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts A. P., Chandler M., Courvalin P., et al. Revised nomenclature for transposable genetic elements. Plasmid. 2008;60(3):167–173. doi: 10.1016/j.plasmid.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh K., Standley D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letunic I., Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research. 2016;44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diancourt L., Passet V., Verhoef J., Grimont P. A. D., Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. Journal of Clinical Microbiology. 2005;43(8):4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jing Y., Jiang X., Yin Z., et al. Genomic diversification of IncR plasmids from China. Journal of Global Antimicrobial Resistance. 2019;19:358–364. doi: 10.1016/j.jgar.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 28.Paulsson J., Chattoraj D. K. Origin inactivation in bacterial DNA replication control. Molecular Microbiology. 2006;61(1):9–15. doi: 10.1111/j.1365-2958.2006.05229.x. [DOI] [PubMed] [Google Scholar]
- 29.Kukla R., Chudejova K., Papagiannitsis C. C., et al. Characterization of KPC-encoding plasmids from Enterobacteriaceae isolated in a Czech hospital. Antimicrobial Agents and Chemotherapy. 2018;62(3) doi: 10.1128/aac.02152-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papagiannitsis C. C., Miriagou V., Giakkoupi P., Tzouvelekis L. S., Vatopoulos A. C. Characterization of pKP1780, a novel IncR plasmid from the emerging Klebsiella pneumoniae ST147, encoding the VIM-1 metallo-β-lactamase. The Journal of Antimicrobial Chemotherapy. 2013;68(10):2259–2262. doi: 10.1093/jac/dkt196. [DOI] [PubMed] [Google Scholar]
- 31.Lu J., Zhang J., Xu L., et al. Spread of the florfenicol resistance floR gene among clinical Klebsiella pneumoniae isolates in China. Antimicrobial Resistance and Infection Control. 2018;7(1):p. 127. doi: 10.1186/s13756-018-0415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furushita M., Akagi H., Kaneoka A., et al. Similarity in the structure of tetD-carrying mobile genetic elements in bacterial strains of different genera isolated from cultured yellowtail. Biocontrol Science. 2016;21(3):183–186. doi: 10.4265/bio.21.183. [DOI] [PubMed] [Google Scholar]
- 33.Tavakoli N., Comanducci A., Dodd H. M., Lett M. C., Albiger B., Bennett P. IS1294, a DNA element that transposes by RC transposition. Plasmid. 2000;44(1):66–84. doi: 10.1006/plas.1999.1460. [DOI] [PubMed] [Google Scholar]
- 34.Toleman M. A., Bennett P. M., Walsh T. R. ISCR elements: novel gene-capturing systems of the 21st century? Microbiology and Molecular Biology Reviews. 2006;70(2):296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y. T., Liao T. L., Liu Y. M., Lauderdale T. L., Yan J. J., Tsai S. F. Mobilization of qnrB2 and ISCR1 in plasmids. Antimicrobial Agents and Chemotherapy. 2009;53(3):1235–1237. doi: 10.1128/AAC.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: complex class 1 integrons of group 3 (pR50-74) and group 4 (CP030940, MG870194). Table S1: general features of K. pneumoniae R50 genome. Table S2: species distribution of IncR plasmids. Table S3: bacterial information for evolutionary analysis.
Data Availability Statement
The data used to support the findings of this study are included within the article.


