Abstract
Background
Untreated nephropathy can progress to renal failure. The traditional Mongolian remedy Narenmandula regulates the kidney “yang.” This study aimed to identify key microRNAs (miRNAs) targeted by Narenmandula in a rat model of nephropathy.
Methods
Fifteen rats exhibiting normal renal function were randomized to three study arms. Nephropathy was induced in n = 10 rats using doxorubicin hydrochloride, followed by either Narenmandula treatment (treatment group) or no treatment (control group). In n = 5 rats, no doxorubicin was given and renal function remained unchanged (healthy group). Microarray analysis identified miRNAs which were differentially expressed (DE-miRNAs) between groups. Target genes of DE-miRNAs were predicted using miRWalk version 2.0, followed by enrichment analysis using DAVID, and construction of the miRNA coregulatory network using Cytoscape.
Results
Nephropathy was successfully induced, with doxorubicin resulting in differential expression of 3645 miRNAs (1324 upregulated and 2321 downregulated). Narenmandula treatment induced differential expression of a total of 159 miRNAs (102 upregulated and 57 downregulated). Upregulated DE-miRNAs (e.g., miR-497-5p, miR-195-5p, miR-181a-5p, miR-181c-5p, and miR-30e-5p) and downregulated DE-miRNAs (e.g., miR-330-3p and miR-214-3p) regulated a high number of target genes. Moreover, the miRNA pairs (e.g., miR-195-5p—miR-497-5p, miR-181a-5p—miR-181c-5p, and miR-30e-5p—miR-30a-5p) coregulated a high number of genes. Enrichment analysis indicated functional synergy between miR-30e-5p—miR-30a-3p, miR-34a-5p—miR-30e-5p, miR-30e-5p—miR-195-3p, and miR-30a-3p—miR-195-3p pairs.
Conclusion
Narenmandula may modulate doxorubicin-induced nephropathy via targeting miR-497-5p, miR-195-5p, miR-181a-5p, miR-181c-5p, miR-30e-5p, miR-330-3p, miR-214-3p, miR-34a-5p, miR-30a-3p, and miR-30a-5p.
1. Introduction
Chronic nephropathy is a progressive disorder, which when untreated can result in renal failure, including consequent uremia and death [1, 2]. The main risk factors for nephropathy are glomerular immunoglobulin A (IgA) deposition, analgesic consumption, xanthine oxidase deficiency, and cytotoxic chemotherapeutic drugs [3]. Globally, nephropathy impacts quality of life in millions of patients, and thousands of these require life-saving renal transplantation [4]. A more comprehensive understanding of the pathogenesis of nephropathy will contribute to development of more effective therapies and improved outcomes for nephropathy patients.
In recent years, nephropathy mechanisms have been partially elucidated by several studies. For example, in reflux nephropathy (RN), angiotensin-converting enzyme (ACE) overexpression induces tubulointerstitial damage via increased extracellular matrix (ECM) component production; ACE inhibition may thus suppress renal fibrosis in this context [5]. By suppressing activation of the v-akt murine thymoma viral oncogene homolog (AKT)/phosphatase and tensin homolog (PTEN) pathway, miR-21 inhibition restrains tubular cell and podocyte fibrogenesis in IgA nephropathy (IgAN) [6, 7]. Furthermore, miR-146a plays an anti-inflammatory role in the context of diabetic nephropathy (DN) [8], and miR-18a-5p modulates autophagy by regulating the ataxia telangiectasia mutated (ATM) gene, which may be prophylactic or therapeutic in the context of DN [9]. Moreover, miRNAs have exhibited utility as biomarkers and/or therapeutic targets in the context of renal disorders such as IgA nephropathy and DN [10–12]. Nevertheless, pathogenesis of nephropathies remains incompletely understood.
The traditional Mongolian remedy Narenmandula (also known as Sheng Yang Shi Yi Wei Wan) is a combination of pomegranate, cinnamon, cardamom, humble pie, yellow essence, safflower, wintergreen fruit, bamboo, asparagus, Bletilla, and Tribulus terrestris [13]. The major traditional uses of Narenmandula include regulating the kidney “yang”, promoting capillary refill and digestion, and relieving diarrhea and edema [14]. Although polysaccharides present in Narenmandula exhibit antioxidant activity [15], potential mechanisms of Narenmandula in opposing nephropathy have not been investigated.
In the present study, doxorubicin hydrochloride was used to induce nephropathy in rats, followed by treatment with Narenmandula. Microarray-based miRNA expression profiling of healthy, treatment, and control groups was followed by differential expression analysis, prediction of genes targeted by differentially expressed miRNAs (DE-miRNAs), enrichment analysis, and miRNA coregulatory network construction. Results regarding key miRNAs modulated by Narenmandula may contribute to identifying novel therapeutic targets in the context of nephropathy.
2. Materials and Methods
2.1. Model Establishment and Sample Acquisition
Study protocols were approved by the Ethical Committee of Affiliated Hospital of Inner Mongolia University for the Nationalities, Tongliao, Inner Mongolia Autonomous of China. Rats were obtained from Yisi Laboratory Animal Technology Co., Ltd. (Jilin, Changchun, China). After a 5-day acclimation period, 15 rats exhibiting normal total 24 h urine protein content were selected for randomization to a healthy group (receiving 6.5 mL/kg normal saline), treatment group (receiving 6.5 mg/kg doxorubicin hydrochloride, Shenzhen Arcandor's Pharmaceutical Co., Ltd., Guangdong, Shenzhen, China) followed after a 4-day period by 3.0 g Narenmandula (Tong Kang Pharmaceutical Co., Ltd., Hebei, Anguo, China) once per day for 21 consecutive days), and a control group (receiving doxorubicin followed by distilled water rather than Narenmandula). Each group contained n = 5 rats. Saline and doxorubicin were administered intravenously by injection into the tail caudal vein, while Narenmandula was administered intragastrically. After the final dose of Narenmandula or water, all rats were fasted for 12 h prior to anesthetization with pentobarbital. Animals were then euthanized via abdominal aortic phlebotomy.
2.2. Microarray Analysis and Data Preprocessing
An miRNA 4.0 Array (Affymetrix, Santa Clara, CA) was used to determine whole blood miRNA expression profiles of rats in each group. Expression data have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE123776). The Robust Multichip Average (RMA) algorithm in R package “affy” [16] was used in conjunction with CEL result files to conduct background correction (integrating probe and probeset signals) and normalization (minimizing the effect of between-sample biological variability).
2.3. Model Validation: Comparing Healthy and Treatment Groups
R package “limma” [17] was used to determine whether any miRNAs were differentially expressed between the healthy and treatment groups, using as screening criteria |log2 fold-change (FC)| > 0.58 and p value ≤0.05. Rodent nephropathy model success was defined as the presence of significant DE-miRNAs.
2.4. Comparing Treatment and Control Groups and Prediction of Target Genes of Identified DE-miRNAs
Differential expression analysis was again performed using R package “limma” and identical criteria. Genes targeted by DE-miRNAs were predicted using miRWalk version 2.0 [18]. Using combined predictive results from 12 databases, including miRWalk [19], miRanda [20], MicroT4 [21], miRDB [22], miRBridge [23], PICTAR2 [24], PITA [25], miRMap [26], miRNAMap [27], RNA22 [28], TargetScan [29], and RNAhybrid [30], DE-miRNA-target gene pairs were screened. Pairs predicted by ≥six databases were retained for downstream analysis.
2.5. Target Gene Enrichment Analysis
Using DAVID version 6.8 (with classification stringency set to medium) [31], Gene Ontology (GO) [32] and Kyoto Encyclopedia of Genes and Genomes (KEGG) [33] terms were predicted for target genes of miRNAs differentially expressed between treatment and control groups. The threshold for significance was set at p value <0.05.
2.6. Construction of the miRNA Coregulatory Network
Based on shared targets of miRNA pairs, an miRNA coregulatory network was constructed using Cytoscape version 3.4 [34]. For miRNA pairs coregulating the highest number of target genes, GO enrichment analysis was conducted. The threshold for significance was set at p value <0.05. Functional synergy was defined as significant enrichment of biological process (BP) terms between common targets of an miRNA pair [35].
3. Results
3.1. Model Validation: Comparing Healthy and Treatment Groups
When comparing healthy and treatment groups, 3645 miRNAs were significantly differentially expressed (1324 upregulated and 2321 downregulated in the treatment group; Figure 1), suggesting successful induction of nephropathy.
Figure 1.
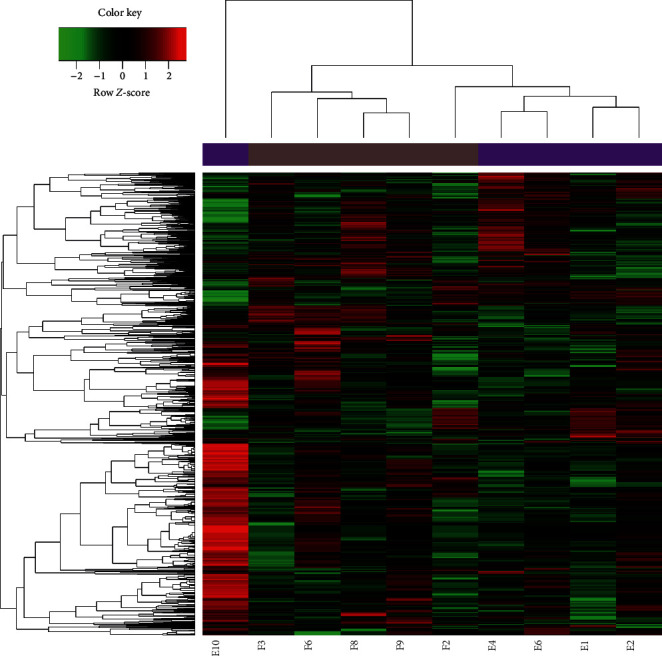
Clustered heat map of miRNAs differentially expressed between healthy (E) and treatment (F) groups.
3.2. Comparing Treatment and Control Groups and Prediction of Target Genes of Identified DE-miRNAs
When comparing treatment and control groups, 159 miRNAs were significantly differentially expressed (102 upregulated and 57 downregulated in the treatment group; Figure 2). Based on FC, the top ten most upregulated and downregulated DE-miRNAs are shown in Table 1.
Figure 2.
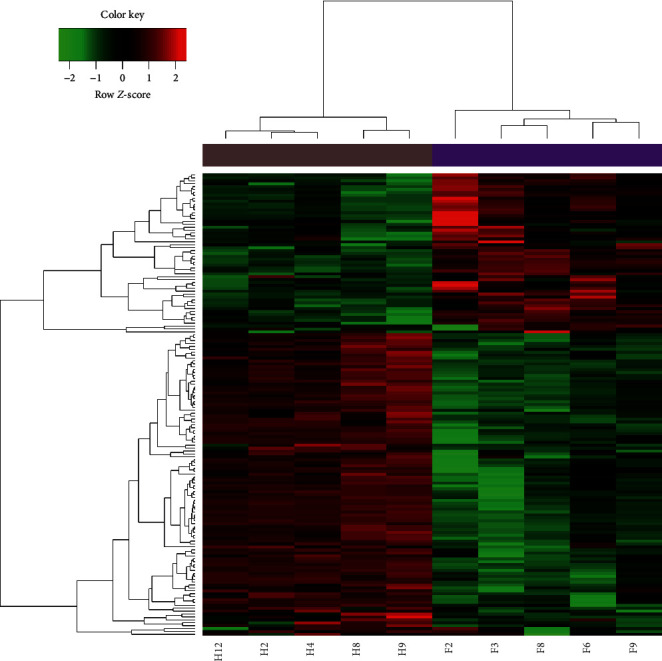
Clustered heat map of miRNAs differentially expressed between treatment (F) and control (H) groups.
Table 1.
Top ten miRNAs differentially expressed between treatment and control groups.
| miRNA | Log fold-change | p value | Type |
|---|---|---|---|
| rno-miR-34a-5p | 3.507493097 | 7.83E − 07 | Up |
| rno-miR-375-3p | 2.8912011 | 1.88E − 04 | Up |
| rno-miR-203b-3p | 2.820823524 | 2.69E − 06 | Up |
| rno-miR-132-3p | 2.752535606 | 2.39E − 06 | Up |
| rno-miR-196a-5p | 2.702999341 | 3.79E − 04 | Up |
| rno-miR-146b-5p | 2.669461069 | 8.91E − 07 | Up |
| rno-miR-203a-3p | 2.582396551 | 1.19E − 06 | Up |
| rno-miR-183-5p | 2.487984128 | 6.41E − 04 | Up |
| rno-miR-30e-3p | 2.303984656 | 5.21E − 07 | Up |
| rno-miR-155-5p | 2.244312291 | 3.55E − 06 | Up |
| rno-miR-32-3p | −2.110700334 | 3.19E − 05 | Down |
| rno-miR-423-5p | −1.948686272 | 8.55E − 05 | Down |
| rno-miR-466b-5p | −1.935924295 | 2.38E − 04 | Down |
| rno-miR-214-3p | −1.894326482 | 5.88E − 07 | Down |
| rno-miR-760-3p | −1.734602185 | 4.45E − 06 | Down |
| rno-miR-138-5p | −1.722493294 | 1.31E − 03 | Down |
| rno-miR-652-5p | −1.569177177 | 8.48E − 05 | Down |
| rno-miR-92b-5p | −1.515952809 | 5.95E − 07 | Down |
| rno-miR-500-3p | −1.453944308 | 3.84E − 05 | Down |
| rno-miR-328a-5p | −1.439955519 | 1.31E − 07 | Down |
Based on output from 12 databases, 17002 miRNA-gene pairs (involving 97 upregulated miRNAs) and 5582 miRNA-gene pairs (involving 53 downregulated miRNAs) were identified. Based on the number of target genes, the top ten upregulated DE-miRNAs (e.g., miR-497-5p, miR-195-5p, miR-181a-5p, miR-181c-5p, and miR-30e-5p) and downregulated DE-miRNAs (e.g., miR-330-3p and miR-214-3p) with the highest number of target genes are shown in Table 2.
Table 2.
Top ten differentially expressed miRNAs with the highest number of target genes.
| Upregulated | Number | Downregulated | Number |
|---|---|---|---|
| rno-miR-497-5p | 446 | rno-miR-330-3p | 408 |
| rno-miR-15b-5p | 384 | rno-miR-214-3p | 390 |
| rno-miR-195-5p | 357 | rno-miR-93-5p | 320 |
| rno-miR-16-5p | 354 | rno-miR-320-3p | 284 |
| rno-miR-181a-5p | 341 | rno-miR-140-3p | 252 |
| rno-miR-181c-5p | 335 | rno-miR-32-3p | 248 |
| rno-miR-30e-5p | 333 | rno-miR-465-5p | 216 |
| rno-miR-128-3p | 332 | rno-miR-351-5p | 204 |
| rno-miR-30c-5p | 331 | rno-miR-324-3p | 200 |
| rno-miR-30b-5p | 330 | rno-miR-339-5p | 185 |
3.3. Target Gene Enrichment Analysis
Biological process and KEGG biological pathway enrichment analysis of genes targeted by the top ten most significantly up- and downregulated DE-miRNAs (Figure 3) revealed that genes targeted by upregulated miRNAs are mainly involved in response to hormone stimulus (GO BP) and ubiquitin-mediated proteolysis (KEGG pathway) (Figure 3(a)) and that genes targeted by downregulated miRNAs are mainly involved in the phosphorus metabolic process (GO BP) and Wnt signaling pathway (KEGG pathway) (Figure 3(b)).
Figure 3.
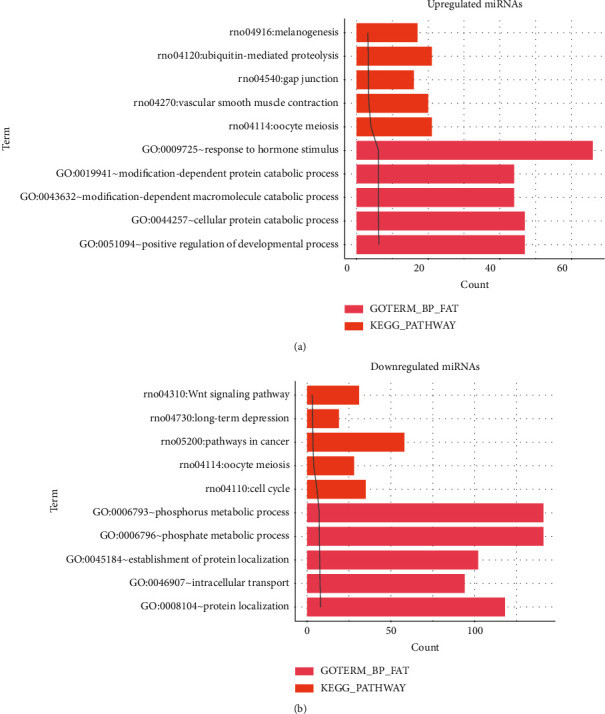
Top five enriched biological processes and KEGG pathways represented by target genes of upregulated miRNAs (a) and downregulated miRNAs (b). GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological process.
3.4. Construction of the miRNA Coregulatory Network
Shared genes targeted by miRNA pairs were identified (the top ten miRNA pairs coregulating the highest number of genes, such as miR-195-5p—miR-497-5p, miR-181a-5p—miR-181c-5p, and miR-30e-5p—miR-30a-5p, are shown in Table 3). Subsequently, an miRNA coregulatory network was constructed (Figure 4). To limit the size of the network, only miRNA coregulatory pairs with target gene count larger than ten were incorporated. Finally, GO BP enrichment of genes targeted by the top ten miRNA pairs coregulating the highest number of genes (including miR-30e-5p—miR-30a-3p, miR-34a-5p—miR-30e-5p, miR-30e-5p—miR-195-3p, and miR-30a-3p—miR-195-3p) (Table 4) demonstrated the presence of functional synergy between these miRNA pairs.
Table 3.
Top ten miRNA pairs coregulating the highest number of target genes.
| miR1 | miR2 | Number |
|---|---|---|
| rno-miR-15b-5p | rno-miR-497-5p | 355 |
| rno-miR-195-5p | rno-miR-497-5p | 334 |
| rno-miR-497-5p | rno-miR-16-5p | 332 |
| rno-miR-195-5p | rno-miR-16-5p | 326 |
| rno-miR-15b-5p | rno-miR-195-5p | 323 |
| rno-miR-30b-5p | rno-miR-30c-5p | 319 |
| rno-miR-15b-5p | rno-miR-16-5p | 317 |
| rno-miR-181c-5p | rno-miR-181a-5p | 313 |
| rno-miR-30e-5p | rno-miR-30a-5p | 309 |
| rno-miR-30b-5p | rno-miR-30a-5p | 300 |
Figure 4.
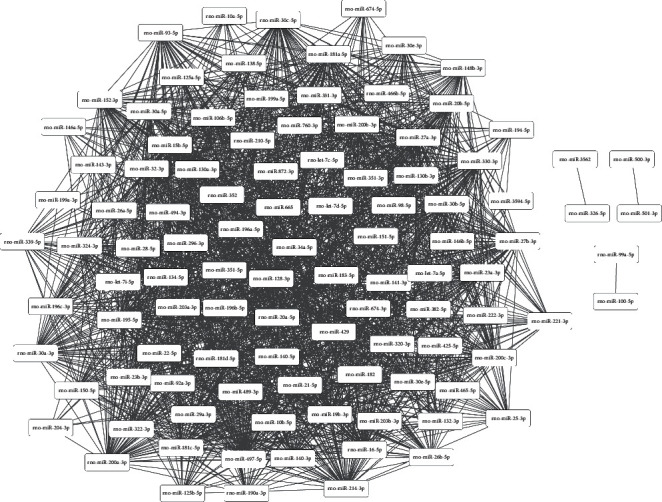
Constructed miRNA coregulatory network. Edge thickness indicates the number of common targets.
Table 4.
Enriched biological processes represented by gene targets of the top ten miRNA coregulatory pairs.
| miR1 | miR2 | Cogene | Co-GO-BP |
|---|---|---|---|
| rno-miR-30a-3p | rno-miR-30e-5p | 355 | 28 |
| rno-miR-195-3p | rno-miR-32-3p | 334 | 115 |
| rno-miR-30e-5p | rno-miR-32-3p | 332 | 120 |
| rno-miR-30a-3p | rno-miR-465-5p | 326 | 121 |
| rno-miR-30e-5p | rno-miR-465-5p | 323 | 94 |
| rno-miR-32-3p | rno-miR-465-5p | 319 | 67 |
| rno-miR-34a-5p | rno-miR-30e-5p | 317 | 99 |
| rno-miR-30e-5p | rno-miR-195-3p | 313 | 131 |
| rno-miR-195-3p | rno-miR-465-5p | 309 | 54 |
| rno-miR-30a-3p | rno-miR-195-3p | 300 | 55 |
GO, Gene Ontology; BP, biological process.
4. Discussion
The present study demonstrated that 159 miRNAs (102 upregulated and 57 downregulated) were significantly differentially expressed between treatment and control groups. Upregulated miRNAs (e.g. miR-497-5p, miR-195-5p, miR-181a-5p, miR-181c-5p, and miR-30e-5p) and downregulated miRNAs (e.g. miR-330-3p and miR-214-3p) targeted the largest number of genes. Furthermore, miRNA pairs (e.g., miR-195-5p—miR-497-5p, miR-181a-5p—miR-181c-5p, and miR-30e-5p—miR-30a-5p) coregulated the largest number of target genes. Finally, enrichment analysis suggested the existence of functional synergy between certain miRNA coregulatory pairs (e.g., miR-30e-5p—miR-30a-3p, miR-34a-5p—miR-30e-5p, miR-30e-5p—miR-195-3p, and miR-30a-3p—miR-195-3p).
Melatonin is known to alleviate endothelial-to-mesenchymal transition (EMT) of glomerular endothelial cells during DN by altering expression of miR-497 [36] (upregulated by Narenmandula in the present study). Overexpression of miR-497 can significantly repress proliferation, migration, and invasiveness of renal cancer cells and may serve as a prognostic factor and therapeutic target for this tumor type [37]. The present study demonstrates upregulation of miR-195 by Narenmandula. Decreased miR-195 expression prevents mesangial cell apoptosis, and miR-195 may also exert antiapoptotic effects during early DN [38]. The immunosuppressive drug cyclosporine A (CsA) is nephrotoxic, producing renal injury and fibrosis, but miR-181c (upregulated by Narenmandula) may help protect renal tissues against this [39]. However, miR-181a inhibition alleviates 5-fluorouracil- (5-FU-) associated nephrotoxicity and may represent a promising target for treatment of chemotherapy-induced nephrotoxicity [40]. Furthermore, miR-181a overexpression contributes to clear cell renal cell carcinoma (ccRCC) progression via modulating Krüppel-like factor 6 (KLF6) expression; targeting miR-181a may have therapeutic potential in this context [41, 42]. Also upregulated by Narenmandula in the present study, miR-30e exhibits low-level expression in DN, and overexpression can prevent progression of DN to renal fibrosis [43]. We speculate that miR-497-5p, miR-195-5p, miR-181a-5p, miR-181c-5p, and miR-30e-5p may be implicated in protective effects of Narenmandula in the context of nephropathy.
Regarding miRNAs demonstrated by the present study to be downregulated by Narenmandula, morin-induced miR-330 expression prevents abnormal fructose-induced renal insulin signal transduction and is a novel candidate for treating renal injury [44]. The twist/miR-214/E-cadherin axis in renal tubular epithelial cells (RTECs) plays roles in EMT, suggesting that anti-miR-214 therapy may slow development of renal fibrosis [45]. Cross-talk between PTEN and miR-214 attenuates glomerular hypertrophy, implying that miR-214 may be a promising target for treatment of DN [46]. Thus, miR-330-3p and miR-214-3p may also contribute to nephropathy-alleviating effects of Narenmandula.
Regarding miRNA pairs coregulating a large number of target genes, suppression of miR-30a-3p and miR-30c-2-3p promotes expression of hypoxia-inducible factor-2α (HIF-2α), which weakens HIF-1α-mediated ccRCC inhibition [47]. Furthermore, urinary miR-30a-5p (as well as miR-490 and miR-196a) levels are associated with focal segmental glomerulosclerosis (FSGS) activity, and miR-30a-5p predicts the response of active FSGS to steroid treatment [48]. Via the miR-34a-5p/sirtuin 1 (SIRT1)/HIF-1α pathway, long intergenic noncoding RNA (lincRNA) 1700020I14Rik suppresses expression of renal fibrosis biomarkers, inhibits mesangial cell proliferation under diabetic conditions, and is involved in progression of DN [49]. We thus speculate that Narenmandula impacts nephropathy via targeting of miR-34a-5p, miR-30a-3p, and miR-30a-5p.
In conclusion, key miRNAs, including miR-497-5p, miR-195-5p, miR-181a-5p, miR-181c-5p, miR-30e-5p, miR-330-3p, miR-214-3p, miR-34a-5p, miR-30a-3p, and miR-30a-5p, may be associated with protective effects of Narenmandula in nephropathy. However, Narenmandula-modulated expression of key DE-miRNAs in rat model renal tissues has not yet been orthogonally verified (e.g., using real-time PCR). Moreover, we did not measure urinary albumin or the albumin/creatinine ratio (ACR) in order to elucidate mechanisms by which key Narenmandula-modulated DE-miRNAs may oppose nephropathy. Additional research is therefore required to confirm our findings.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81641149).
Data Availability
Datasets generated by this study are available from the Gene Expression Omnibus (GEO) database (accession number GSE123776).
Additional Points
(1) There were 159 dysregulated miRNAs between treatment and control groups. (2) Several miRNA coregulatory pairs exhibit functional synergy. (3) Narenmandula may modulate nephropathy via targeting key miRNAs.
Ethical Approval
The present study was approved by the Ethical Committee of Affiliated Hospital of Inner Mongolia University for the Nationalities, Tongliao, Inner Mongolia Autonomous of China.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- 1.Lemley K. V. Glomerular pathology and the progression of chronic kidney disease. American Journal of Physiology-Renal Physiology. 2016;310:F1385–F1388. doi: 10.1152/ajprenal.00099.2016. [DOI] [PubMed] [Google Scholar]
- 2.Ni W.-J., Tang L.-Q., Wei W. Research progress in signalling pathway in diabetic nephropathy. Diabetes/Metabolism Research and Reviews. 2015;31(3):221–233. doi: 10.1002/dmrr.2568. [DOI] [PubMed] [Google Scholar]
- 3.Hajivandi A., Amiri M. World kidney day 2014: kidney disease and elderly. Journal of Parathyroid Disease. 2014;2:3–4. [Google Scholar]
- 4.Jha V., Garcia-Garcia G., Iseki K., et al. Chronic kidney disease: global dimension and perspectives. The Lancet. 2013;382(9888):260–272. doi: 10.1016/s0140-6736(13)60687-x. [DOI] [PubMed] [Google Scholar]
- 5.Chertin B., Solari V., Reen D., Farkas A., Puri P. Up-regulation of angiotensin-converting enzyme (ACE) gene expression induces tubulointerstitial injury in reflux nephropathy. Pediatric Surgery International. 2002;18(7):635–639. doi: 10.1007/s00383-002-0857-5. [DOI] [PubMed] [Google Scholar]
- 6.Bao H., Hu S., Zhang C., et al. Inhibition of miRNA-21 prevents fibrogenic activation in podocytes and tubular cells in IgA nephropathy. Biochemical and Biophysical Research Communications. 2014;444(4):455–460. doi: 10.1016/j.bbrc.2014.01.065. [DOI] [PubMed] [Google Scholar]
- 7.Wang J., Gao Y., Ma M., et al. Effect of miR-21 on renal fibrosis by regulating MMP-9 and TIMP1 in kk-ay diabetic nephropathy mice. Cell Biochemistry and Biophysics. 2013;67(2):537–546. doi: 10.1007/s12013-013-9539-2. [DOI] [PubMed] [Google Scholar]
- 8.Bhatt K., Lanting L. L., Jia Y., et al. Anti-inflammatory role of microRNA-146a in the pathogenesis of diabetic nephropathy. Journal of the American Society of Nephrology. 2016;27(8):2277–2288. doi: 10.1681/asn.2015010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X. H., Ding D. F., Yong H. J., et al. Resveratrol transcriptionally regulates miRNA-18a-5p expression ameliorating diabetic nephropathy via increasing autophagy. European Review for Medical and Pharmacological Sciences. 2017;21(21):4952–4965. [PubMed] [Google Scholar]
- 10.Simpson K., Wonnacott A., Fraser D. J., Bowen T. MicroRNAs in diabetic nephropathy: from biomarkers to therapy. Current Diabetes Reports. 2016;16:p. 35. doi: 10.1007/s11892-016-0724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szeto C.-C., Wang G., Ng J. K.-C., et al. Urinary miRNA profile for the diagnosis of IgA nephropathy. BMC Nephrology. 2019;20:p. 77. doi: 10.1186/s12882-019-1267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun I. O., Lerman L. O. Urinary microRNA in kidney disease: utility and roles. American Journal of Physiology-Renal Physiology. 2019;316(5):F785–F793. doi: 10.1152/ajprenal.00368.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S.Q., HaiYi Z. Research of the quality standards of Sheng yang Shi yi Wei wan. Chinese Journal of Ethnic Medicine. 2009;15:47–48. [Google Scholar]
- 14.Chen Y. Research progression of Sheng yang Shi Yi Wei Wan. Northern Pharmacy. 2012;9:p. 41. [Google Scholar]
- 15.Wu-lan-ge-ri-le W., Liang X. U., Qin C., Zhen-Guo M. A., Zhang J. M., Hu-Shan B. A. Isolation and purification of polysaccharides from Mongolian medicine Narenmandula and its antioxidant activity. Chinese Journal of Pharmaceutical Analysis. 2013;33:1285–1288. [Google Scholar]
- 16.Gautier L., Cope L., Bolstad B. M., Irizarry R. A. affy--analysis of affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 17.Ritchie M. E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43(7):p. e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parveen A., Gretz N., Dweep H. Obtaining miRNA-target interaction information from miRWalk2.0. Current Protocols in Bioinformatics. 2016;55 doi: 10.1002/cpbi.14. [DOI] [PubMed] [Google Scholar]
- 19.Dweep H., Gretz N., Sticht C. miRWalk database for miRNA-target interactions. RNA Mapping. 2014;1182:p. 289. doi: 10.1007/978-1-4939-1062-5_25. [DOI] [PubMed] [Google Scholar]
- 20.Enright A. J., Bino J., Ulrike G., Thomas T., Sander C., Marks D. S. MicroRNA targets in Drosophila. Genome Biology. 2004;5:p. R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maragkakis M., Vergoulis T., Alexiou P., et al. DIANA-microT web server upgrade supports fly and worm miRNA target prediction and bibliographic miRNA to disease association. Nucleic Acids Research. 2011;39(suppl):W145–W148. doi: 10.1093/nar/gkr294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong N., Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Research. 2015;43(D1):p. D146. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsang J. S., Ebert M. S., Van Oudenaarden A. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Molecular Cell. 2010;38(1):140–153. doi: 10.1016/j.molcel.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blin K., Dieterich C., Wurmus R., Rajewsky N., Landthaler M., Akalin A. DoRiNA 2.0--upgrading the doRiNA database of RNA interactions in post-transcriptional regulation. Nucleic Acids Research. 2015;43:160–167. doi: 10.1093/nar/gku1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nature Genetics. 2007;39(10):1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 26.Vejnar C. E., Zdobnov E. M. miRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Research. 2012;40(22):p. 11673. doi: 10.1093/nar/gks901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu S.-D., Chu C.-H., Tsou A.-P., et al. miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes. Nucleic Acids Research. 2008;36:165–169. doi: 10.1093/nar/gkm1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miranda K. C., Huynh T., Tay Y., et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 29.Shin C., Nam J.-W., Farh K. K.-H., Chiang H. R., Shkumatava A., Bartel D. P. Expanding the MicroRNA targeting code: functional sites with centered pairing. Molecular Cell. 2010;38(6):789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krüger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Research. 2006;34:451–454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dennis G., Jr, Sherman B. T., Douglas A. H., Yang J., Gao W. DAVID: database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4:p. P3. doi: 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- 32.Balakrishnan R., Harris M. A., Huntley R., Van Auken K., Michael Cherry J. A guide to best practices for gene ontology (GO) manual annotation. Database. 2013;2013 doi: 10.1093/database/bat054.bat054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J., Chen L., Kong X., Huang T., Cai Y. D. Analysis of tumor suppressor genes based on gene ontology and the KEGG pathway. PLoS One. 2014;9 doi: 10.1371/journal.pone.0107202.e107202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohl M., Wiese S., Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods in Molecular Biology, Data Mining in Proteomics. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 35.Xu J., Li C.-X., Li Y.-S., et al. MiRNA-miRNA synergistic network: construction via co-regulating functional modules and disease miRNA topological features. Nucleic Acids Research. 2011;39(3):825–836. doi: 10.1093/nar/gkq832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu F., Zhang S., Xu R., Gao S., Yin J. Melatonin attenuates endothelial-to-mesenchymal transition of glomerular endothelial cells via regulating miR-497/ROCK in diabetic nephropathy. Kidney and Blood Pressure Research. 2018;43(5):1425–1436. doi: 10.1159/000493380. [DOI] [PubMed] [Google Scholar]
- 37.Zhao X., Zhao Z., Xu W., Hou J., Du X. Down-regulation of miR-497 is associated with poor prognosis in renal cancer. International Journal of Clinical and Experimental Pathology. 2015;8(1):758–764. [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y. Q., Wang X. X., Yao X. M., et al. Abated microRNA-195 expression protected mesangial cells from apoptosis in early diabetic renal injury in mice. Journal of Nephrology. 2012;25(4):p. 566. doi: 10.5301/jn.5000034. [DOI] [PubMed] [Google Scholar]
- 39.Sun W., Min B., Du D., et al. miR-181c protects CsA-induced renal damage and fibrosis through inhibiting EMT. Febs Letters. 2017;591(21):p. 3588. doi: 10.1002/1873-3468.12872. [DOI] [PubMed] [Google Scholar]
- 40.Liu X.-Y., Zhang F.-R., Shang J.-Y., et al. Renal inhibition of miR-181a ameliorates 5-fluorouracil-induced mesangial cell apoptosis and nephrotoxicity. Cell Death & Disease. 2018;9:p. 610. doi: 10.1038/s41419-018-0677-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei Z., Ma X., Li H., et al. Up-regulation of miR-181a in clear cell renal cell carcinoma is associated with lower KLF6 expression, enhanced cell proliferation, accelerated cell cycle transition, and diminished apoptosis. Urologic Oncologyy: Seminars and Original Investigations. 2017;36:93.e23–93.e37. doi: 10.1016/j.urolonc.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 42.Lai Y., Zhao L., Hu J., et al. microRNA-181a-5p functions as an oncogene in renal cell carcinoma. Molecular Medicine Reports. 2018;17 doi: 10.3892/mmr.2018.8899. [DOI] [PubMed] [Google Scholar]
- 43.Zhao D., Jia J., Shao H. miR-30e targets GLIPR-2 to modulate diabetic nephropathy: in vitro and in vivo experiments. Journal of Molecular Endocrinology. 2017;59(2):181–190. doi: 10.1530/jme-17-0083. [DOI] [PubMed] [Google Scholar]
- 44.Gu T. T., Song L., Chen T. Y., et al. Fructose down-regulates miR-330 to induce renal inflammatory response and insulin signaling impairment: attenuation by morin. Molecular Nutrition & Food Research. 2017;61:p. 1600760. doi: 10.1002/mnfr.201600760. [DOI] [PubMed] [Google Scholar]
- 45.Liu M., Liu L., Bai M., et al. Hypoxia-induced activation of Twist/miR-214/E-cadherin axis promotes renal tubular epithelial cell mesenchymal transition and renal fibrosis. Biochemical and Biophysical Research Communications. 2018;495(3):p. 2324. doi: 10.1016/j.bbrc.2017.12.130. [DOI] [PubMed] [Google Scholar]
- 46.Wang X., Shen E., Wang Y., et al. Cross talk between miR-214 and PTEN attenuates glomerular hypertrophy under diabetic conditions. Scientific Reports. 2016;6 doi: 10.1038/srep31506.31506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathew L. K., Lee S. S., Skuli N., et al. Restricted expression of miR-30c-2-3p and miR-30a-3p in clear cell renal cell carcinomas enhances HIF2 activity. Cancer Discovery. 2014;4:53–60. doi: 10.1158/2159-8290.cd-13-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang W., Zhang C., Chen H., et al. Evaluation of microRNAs miR-196a, miR-30a-5P, and miR-490 as biomarkers of disease activity among patients with FSGS. Clinical Journal of the American Society of Nephrology. 2014;9(9):p. 1545. doi: 10.2215/cjn.11561113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manish K. T., Doxtater K., Keramatnia F., et al. Role of lncRNAs in ovarian cancer: defining new biomarkers for therapeutic purposes. Drug Discovery Today. 2018;23:1635–1643. doi: 10.1016/j.drudis.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets generated by this study are available from the Gene Expression Omnibus (GEO) database (accession number GSE123776).


