Abstract
Chromatin remodeling impacts the structural neighborhoods and regulates gene expression. However, the role of enhancer-guided chromatin remodeling in the gene regulation remains unclear. Here, using RNA-seq and ChIP-seq, we identified for the first time that neurotensin (NTS) serves as a key oncogene in uveal melanoma and that CTCF interacts with the upstream enhancer of NTS and orchestrates an 800 kb chromosomal loop between the promoter and enhancer. Intriguingly, this novel CTCF-guided chromatin loop was ubiquitous in a cohort of tumor patients. In addition, a disruption in this chromosomal interaction prevented the histone acetyltransferase EP300 from embedding in the promoter of NTS and resulted in NTS silencing. Most importantly, in vitro and in vivo experiments showed that the ability of tumor formation was significantly suppressed via deletion of the enhancer by CRISPR-Cas9. These studies delineate a novel onco-enhancer guided epigenetic mechanism and provide a promising therapeutic concept for disease therapy.
INTRODUCTION
Chromosomal remodeling plays a key role in the maintenance of homeostasis, and aberrant chromosomal remodeling dysregulates downstream gene expression and leads to the occurrence of a variety of diseases (1). Spatial organization is an inherent property for accommodating the 2 meters of DNA in the nucleus. During this organization, topologically associating domains (TADs) function as a fundamental structural unit which guides regulatory elements to their cognate promoters (2). A disruption in the TAD boundary near the EHPA4 locus results in abnormal development of the limb bud (3). In addition, chromosomal loops between the Tfap2c and Bmp7 genes are split into two structural and functional domains. The transformation of these two phases is essential in heart development (4). In a genomic analysis of over 9000 tumor cases, it was found that CTCF/cohesin-binding sites are frequently mutated in cancers (5). Thus, the role of chromosomal remodeling in physiology and pathogenesis is potentially interesting.
As a key component of chromosomal remodeling, enhancers are noncoding regulatory elements that stimulate transcription through looping-mediated interactions with promoters and are activated in specific cellular contexts by different combinations of sequence-specific transcription factors (TFs) (6). Active enhancers are generally considered to function via 3D chromosomal interactions (7), which adopt a signature chromatin structure and can be identified by mapping an open chromatin status or histone acetylation markers (8). Aberrant oncogene expression in these events is thought to be dependent on looping interactions between the oncogene promoter and cis-regulatory elements from the translocation partner locus (9,10). For example, in medulloblastoma, somatic structural variants juxtapose GFI1- or GFI1B-coding sequences proximal to active enhancer elements, including super enhancers, instigating oncogenic activity (11). In addition, recurrent tandem duplications intersecting with a TAD boundary mediate the de novo formation of a 3D contact domain comprising IGF2 and a lineage-specific super enhancer, resulting in IGF2 activation in colorectal tumors (12). Therefore, the exploration of enhancer-associated chromatin remodeling has attracted increasing attention in diversified biological processes, especially tumorigenesis.
Under most situations, chromatin remodeling factors show their functions in gene expression through DNA methylations, histone modifications and chromatin insulations (13,14). In this process, CCCTC-binding factor (CTCF) can directly bind to the promoters, silencers, and insulators of genes involved in mRNA splicing, differentiation, apoptosis, imprinting and X chromosome inactivation (15). For example, our previous study demonstrated that the N-terminus of CTCF could form a chromosomal loop with polycomb repressive complex-2 (PRC2) and maintain the imprinting of human insulin-like growth factor II (IGF-2) (16). In addition, CTCF-cohesion loops disrupt polycomb-dependent chromosome interactions in embryonic stem cells (17). Moreover, CTCF-mediated chromatin loops have been revealed to regulate alternative splicing (18) and the tumorigenesis of leukemia and medulloblastoma (19–21). Intriguingly, CTCF can hijack distal enhancers by CTCF dimers to form chromosomal loops and regulate gene expression (22,23); however, the role of the enhancer-guided chromatin remodeling in tumorigenesis remains to be fully elucidated.
In this study, we identified for the first time that neurotensin (NTS) is an important initiator of tumorigenesis in malignant uveal melanoma (UM). We also revealed that CTCF-EP300-guided chromatin remodeling triggered oncogene transcription and tumorigenesis. Our data represent a novel mechanism of chromatin remodeling in tumorigenesis and provide a promising therapeutic method in which the targeted correction of abnormal chromosomal interactions is efficient in tumor therapy.
MATERIALS AND METHODS
Cell lines and cell culture
The human UM cell lines OCM1, OCM1α and OM431 were kindly provided by Professor John F. Marshall (Tumor Biology Laboratory, Cancer Research UK Clinical Center, John Vane Science Centre, London, UK). The human normal melanocyte cell line PIG1 was obtained from the Department of Ophthalmology, Peking University Third Hospital. The ARPE19 and HEK293T cell lines were purchased from the ATCC. OCM1 and OCM1α cell lines are derived from distant metastases, and other UM cell lines are derived from primary ocular tumors. The usage and methods of the cell lines were approved by the Ethical and Institutional Review Board of Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine. All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, USA) supplemented with 10% certified heat-inactivated fetal bovine serum (FBS; Gibco, USA), penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37°C in a humidified 5% CO2 atmosphere. These cells were characterized by short tandem repeat (STR) markers and were confirmed to be mycoplasma-free (last tested in June 2019).
ChIP
ChIP assays were performed as previously described (24). One hundred million cells were fixed with 1% formaldehyde and sonicated for 8 min (10 s on and 15 s off) on ice with a 2-mm microtip at 40% output control and 90% duty cycle settings. To perform ChIP, sonicated chromatin (150 μl) was diluted 10-fold, and Protein G Agarose (60 μl) (Millipore, USA) was added with shaking at 4°C for 2 h. Then, the mixture was briefly centrifuged at 1000 rpm for 5 min at 4°C, and the supernatant was collected into a new tube. Antibodies against CTCF and EP300 obtained from Abcam, Inc. (Abcam, USA) were added to the supernatant overnight at 4°C. Protein A and Protein G Magnetic Beads (60 μl) (Millipore, USA) were used to pull down the protein at 4°C for 6 h. The DNA was released from the bound chromatin after crosslinking reversal and proteinase K treatment and precipitated and diluted in 100 μl of 0.2 M glycine. The PCR conditions were 95°C for 5 min, followed by 34 cycles at 95°C for 30 s, 30 s at the optimal annealing temperature, and 72°C for 30 s of extension. The PCR products were separated on an 8% polyacrylamide–urea gel. The PCR primers used in this study are listed in Supplementary Table S2.
Tissue specimens
UM tissues were collected from affected eyes during enucleation at the Department of Ophthalmology, Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine. A normal ocular control sample was obtained from enucleation resulting from a traumatic accident. Fresh tissue samples were snap-frozen in liquid nitrogen and stored at −80°C. All samples were pathologically confirmed. Patient consent and approval from the Institutional Research Ethics Committee were obtained prior to surgery. Detailed patient information is listed in Supplementary Tables S2 and S3.
RNA extraction, library construction and Illumina sequencing (RNA-seq)
Total RNA was extracted from the UM (OCM1, OCM1a and OM431) and normal (PIG1 and AREP19) cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). We confirmed the RNA integrity on a 2100 Bioanalyzer (Agilent Technologies, USA). We measured the RNA concentration on a Qubit 2.0 fluorometer with the Qubit RNA Assay Kit (Life Technologies, Carlsbad, CA, USA). We prepared the libraries from 100 ng of total RNA using an Illumina TruSeq RNA Sample Prep Kit (San Diego, CA, USA). In total, libraries were sequenced using the Illumina HiSeq 2500 platform (San Diego, CA, USA). The mRNA abundance levels of the unigenes identified using TopHat v2.0.9 and Cufflinks were normalized by the fragments per kilobase of exon model per million mapped reads (FPKM), and the log2 fold changes between two samples were tested statistically to determine whether an individual gene's expression was altered significantly. We used the criteria of a false discovery rate (FDR) < 0.01 and a fold change <0.25 or >4.0 (< −2 or >2 in the log2 ratio value, P value < 0.05) to identify the differentially expressed genes.
FAIRE
FAIRE was performed as previously described (25). Briefly, crosslinking was performed with a final concentration of 1% formaldehyde followed by the addition of 2.5 M glycine to quench the formaldehyde. The samples were then centrifuged and washed. The fixed cells were then resuspended in 1 ml of lysis buffer and sonicated to achieve an average DNA fragment of ∼100–500 bp. The supernatant was incubated with 1 μl of DNase-free RNaseA. Subsequently, phenol/chloroform/isoamyl alcohol (Sangon, Shanghai, China) was added, and the sample was vortexed for 10 s followed by centrifugation at 12 000 g for 5 min. The aqueous (top) layer was then transferred, and the extraction process was repeated three times, pooling all the aqueous solutions. Subsequently, 1/10 volume of 3 M sodium acetate, 2 volumes of 95% ethanol, and 1 μl of 20 mg/ml glycogen were added, and the solution was incubated at −80°C for at least 30 min. Notably, the clinical samples were collected in DMEM and homogenized by using precooled mechanical trituration. A single-cell suspension was then prepared and examined under a microscope. The DNA was then purified using a DNA Clean-up Kit (AxyPrep). The PCR primers used in this study are listed in Supplementary Table S2.
3C assay
The 3C assay was performed as described previously (16). Briefly, 1.0 × 107 cells were crosslinked with 2% formaldehyde and quenched with 0.125 M glycine. Cells were lysed with cell lysis buffer (10 mM Tris [pH 8.0], 10 mM NaCl, 0.2% NP-40, and protease inhibitors), and nuclei were collected. The nuclei were resuspended in 1× restriction enzyme buffer in the presence of 0.3% sodium dodecyl sulfate (SDS) and incubated at 37°C for 1 h. Triton X-100 was then added to a final concentration of 1.8% to sequester the SDS. An aliquot of nuclei (2 × 106) was digested with 800 U of the restriction enzyme Pst I at 37°C overnight. Next, 1.6% SDS was added, and the mixture was incubated at 65°C for 20 min to stop the reaction. Chromatin DNA was diluted with T4 ligation reaction buffer, and 2 μg DNA was ligated with 4000 U of T4 DNA ligase (Takara, Japan) at 16°C for 4 h (final DNA concentration, 2.5 μg/ml). After treatment with 10 mg/ml proteinase K at 65°C overnight to reverse crosslinks and with 0.4 μg/ml RNase A for 30 min at 37°C, DNA was extracted with phenol–chloroform, ethanol precipitated and used for PCR amplification of the ligated DNA products. Notably, the clinical samples were collected in DMEM and homogenized by using precooled mechanical trituration. A single-cell suspension was then prepared and examined under a microscope. Patient consent was obtained, and patient information is listed in Supplementary Table S3. All 3C lysates were purified by nested PCR to achieve a final product of 100–350 bp in length for further analysis. For result analysis, qPCR was done for quantification and RT-PCR was performed for validating specificity of primers.
Western blot analysis
Cells were harvested and rinsed with PBS. For clinical samples, three ocular melanoma tissues and three normal uveas were collected. The cohort information is listed in Supplementary Table S4. Cell and tissue extracts were prepared with lysis buffer and centrifuged. Protein samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride membranes. After blocking with 5% BSA for 2 h at room temperature, the membrane was incubated overnight at 4°C. The membrane was then incubated with a secondary antibody conjugated to a fluorescent tag (Invitrogen). The band signals were visualized and quantified using the Odyssey Infrared Imagining System (LI-COR, USA). The following antibodies were used in this study: anti-NTS (Abcam, ab233107, USA), anti-EP300 (Abcam, ab10485, USA), anti-CTCF (Abcam, ab70303, USA), anti-SMC1 (Abcam, ab9262, USA) and GAPDH (Sigma-Aldrich).
Co-IP
Co-IP was performed according to manufacturer's protocol provided in the Nuclear Complex Co-IP Kit (Active Motif, USA). Briefly, nuclear extracts were prepared by suspending cells in 1× hypotonic buffer for 15 min on ice. The cells were homogenized and centrifuged for 30 s at 14 000 g. IP was performed with 300 μg nuclear protein and 5 μg anti-CTCF and anti-EP300 antibodies (Abcam, CA) in 500 μl IP incubation buffer at 4°C overnight. The reaction mixtures were incubated with PureProteome™ Protein A and Protein G Magnetic Beads (50 μl) (Millipore, USA) at 4°C for 1 h on a rotator. The immunoprecipitated complexes were washed twice with IP wash buffer supplemented with 1 mg/ml BSA and without BSA. The washed beads were incubated with 2× reduction loading buffer and boiled at 100°C for 5 min. The protein released from the components of the complexes was examined by SDS-PAGE and Western blot analysis.
Tumor xenograft and metastasis models in nude mice
Animal protocols were approved by the Animal Care and Use Committee at Shanghai Jiao Tong Medical College. Male 5-week-old nude mice were deeply anesthetized. Next, 3 × 106 cells (OCM1 siNC, OCM1 siNTS, OCM1 sgRNA NC, OCM1-sgRNA1 and sgRNA2) in a 0.2-ml volume of sterile saline solution were subcutaneously injected into the right flank. Tumor growth was monitored using a caliper every 7 days. Tumor volume was calculated using the following formula: length (mm) × width (mm)2/2. Four mice from each group were sacrificed, and the tumors were weighed. For the metastasis model, male 5-week-old nude mice were deeply anesthetized. Cells were pretransfected with a lentivirus encoded by the plvx-luciferase-mCherry-Blasticidin vector (OCM1 sgRNA NC, OCM1-sgRNA1 and sgRNA2). Next, 1 × 106 cells in a 0.1-ml volume of sterile saline solution were injected through the caudal vein. Bioluminescence was detected after 40 days by in vivo small animal imaging systems, the numbers of nude mice presented with stronger luminescence signal in sgRNA groups have been calculated as compared with NC group.
Wound healing and colony formation assays
A wound healing assay was performed after seeding 1 million cells into a six-well plate. The cells were incubated in FBS-free medium. A wound was made by manually scraping the cell monolayer with a 200 μl pipet tip. Images were taken at the indicated times. For colony formation assays, 1000 cells were resuspended in 2.0 ml of DMEM and seeded into a six-well plate. The cells were then incubated for 7–14 days. The colonies were stained with 0.005% crystal violet and then photographed.
Subarachnoid injection
The animal experiments were approved by the Shanghai Jiao Tong University Animal Care and Use Committee and conducted following the animal policies of Shanghai Jiao Tong University in accordance with the guidelines established by the National Health and Family Planning Commission of China. The cells were harvested by trypsinization and washed twice with PBS (GIBCO). BALB/c nude mice (male, 6 weeks old) were used for the study. Each animal was first anesthetized with the topical anesthetic Benoxil. Methocel eye drops were used to avoid drying of the eyes. Injections were performed using a surgical microscope. Two microliters of sterile phosphate-buffered saline containing 2 × 104 OCM1 cells were injected into the vitreous of each eye through the sclera using a Hamilton syringe. After the injection, the eyes were treated with antibiotic eye drops. All mice were cervically dislocated 40 days after implantation for the tumor formation analysis and 72 days after intravitreal injection for the survival analysis.
CRISPR/Cas9-mediated deletions
Four single guide RNAs (sgRNAs) were cloned separately into lenti-Guide-Puro plasmids. To delete the 400 bp CTCF binding peak-containing region from the genome, COM1 cells were transfected with plasmids containing guide RNAs (and Cas9) targeting the left or right side of the region to be deleted. Colonies were derived from single cells and tested for the loss of the risk region. The control group was transfected with the gRNA-empty vector using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions.
TCGA dataset
To validate the potential role of NTS in UM, we queried the TCGA (http://www.cbioportal.org) and GEPIA (gepia.cancer-pku.cn); R2 (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi) provided the transcriptional landscape and follow-up information on 78 UM samples.
Statistical analysis
We performed the statistical analyses with GraphPad Prism 8 software. Biological triplicates were performed where indicated. Data are presented as the mean ± SD and the differences between two groups were calculated by unpaired two-tailed Student's t-test. Survival plots were generated by Kaplan-Meier curve, and P values were calculated by the log-rank test. P-value <0.05 was considered statistically significant. Asterisks denote statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001).
RESULTS
An open chromatin status was found at the NTS locus in tumor cells and clinical tissues
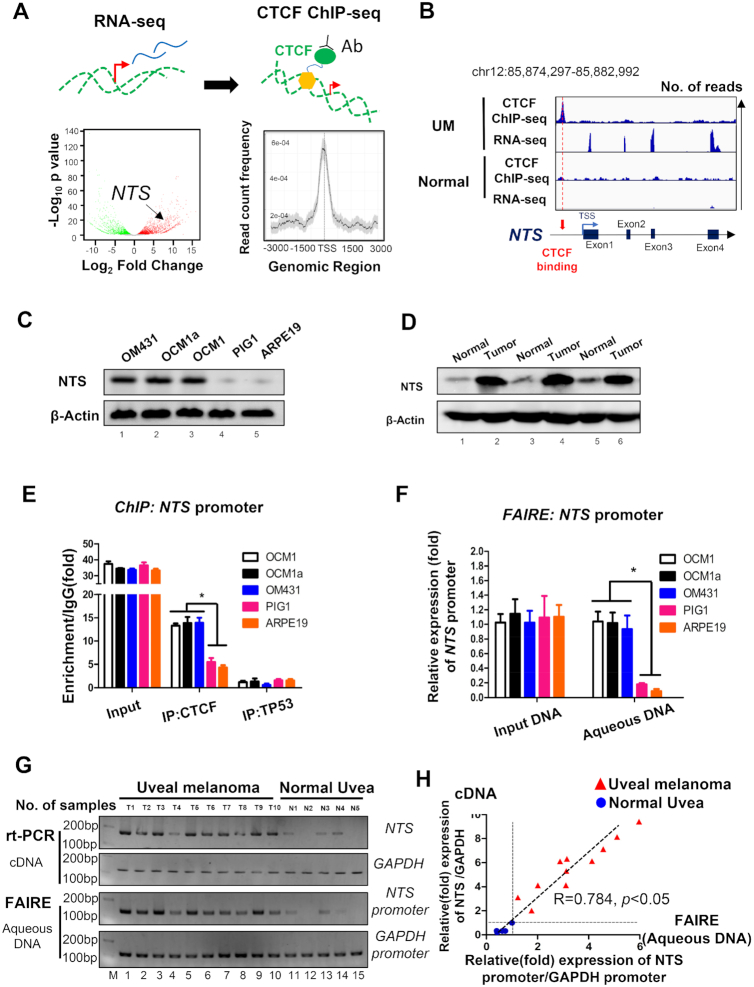
To explore the role of enhancer-guided chromatin remodeling in gene regulation, a potential target gene needs to be identified. As CTCF serves a key regulator in high order chromosomal organization and transcriptional regulation, we were interested in locating specific CTCF binding site through genome-wide sequencing and transcriptome screening. Firstly, thorough RNA-seq (UM and normal controls, GEO accession number: GSE137675), we identified 5714 genes presented with differential expression, including 3506 upregulated and 2208 downregulated genes (|Fold Change| > 1.5, P <0.05 , Figure 1A, left). In addition, using CTCF chromatin immunoprecipitation (ChIP)-seq assay (UM and normal controls, GEO accession number: GSE154736, GSE137311 and GSE60024), we observed 19 811 CTCF binding sites in UM cells, which were mainly distributed nearby TSS region (Figure 1A, right, Supplementary Table S1). By combining RNA-seq and CTCF ChIP-seq assays, we observed a significant enrichment of the CTCF signal at the NTS promoter and a strongly expressed peak at four NTS exons in UM, while the expressed peak and CTCF signal at the NTS locus disappeared in normal retinal pigment epithelium (RPE) cells (Figure 1B; Supplementary Figure S1A–D). To verify the bioinformatics analysis described above, we examined NTS expression in UM cells and tissues. Western blot assays showed that NTS was highly expressed in UM cells (OCM1, OCM1a, and OM431) (Figure 1C) and tissues (Figure 1D), while normal control cells (ARPE19 and PIG1) and tissues showed weak NTS expression. ChIP assays also indicated that CTCF was significantly bound to the promoter of NTS in tumor cells compared to normal control cells (Figure 1E). Next, we tested the chromatin status of the NTS promoter through formaldehyde-assisted isolation of regulatory elements (FAIRE) assays and found that the CTCF-bound NTS promoter presented an open chromatin status in tumor cells, while the non-CTCF-bound NTS promoter showed a closed chromatin status in normal cells (Figure 1F). Importantly, we observed a similar open chromatin status in most clinical tumor tissues (Figure 1G). In addition, we found that NTS expression was strongly positively correlated with the open chromatin status of the NTS promoter region (R = 0.784, P < 0.05) (Figure 1H). These data indicate that CTCF is likely to be involved in the regulation of the NTS gene. We thus chose this locus to explore the potential role of CTCF-guided enhancer-associated chromatin remodeling in gene regulation.
Figure 1.
Open chromatin status at the NTS locus in tumor cells and clinical tissues. (A) Searching for a potential CTCF-guided enhancer locus. Left: RNA-seq screening of upregulated genes in UM (GSE137675); Right: Locating the wild-type CTCF-binding site in tumor cells. Genes selected should meet: 1) upregulated in UM cells; 2) with CTCF bind in the promoter. (B) Upper panel:NTS was highly expressed, with CTCF binding at the promoter region in OCM1 cells. Lower panel: NTS expression was untestable and without CTCF binding at the promoter region in normal RPE cells. (C, D): Western blot analysis demonstrated that NTS is upregulated in UM cell lines (D) and clinical tumor samples (E). (E) ChIP assay of the CTCF status in the NTS promoter in UM (OCM1, OCM1a and OM431) and normal control (ARPE19 and PIG1) cells. Triplicates were performed. Data are presented as the mean ± SD and the differences between two groups were calculated by unpaired two-tailed Student's t-test. * P < 0.05. (F) Formaldehyde-assisted isolation of regulatory elements (FAIRE) assay of the CTCF status in the NTS promoter in UM (OCM1, OCM1a and OM431) and normal control (ARPE19 and PIG1) cells. Triplicates were performed. Data are presented as the mean ± SD and the differences between two groups were calculated by unpaired two-tailed Student's t-test. * P <0.05. (G) RT-PCR and FAIRE assays were performed in 10 UM and 5 normal uveal samples to test NTS expression and the chromatin status of the NTS promoter. GAPDH and its promoter were selected as controls. (H) The correlation (R = 0.784, P< 0.05) between NTS expression and the open status in the NTS promoter.
NTS is a novel regulator that promotes the tumorigenesis of UM
Although NTS is a well-known oncogene in other tumors, no data have shown its role in the tumorigenesis of UM. Thus, we next explored the role of NTS in UM. Real-time PCR and Western blot assays both demonstrated that NTS expression was significantly decreased after small interfering RNA (siRNA) interference (Supplementary Figure S2). The NTS-interfered group presented fewer and smaller colonies and could be partially rescued after the exogenous supplementation of recombinant NTS at a concentration of 100 ng/μl (Figure 2A, B, Supplementary Figure S3). The wound healing assay demonstrated a similar inhibition of cell migration after NTS silencing, which could also be rescued by the exogenous addition of recombinant NTS (100 ng/μl, Supplementary Figure S4A–C). In the subarachnoid transplantation model, the bioluminescence signal was weaker after NTS interference (Figure 2C). Similarly, in the subcutaneous xenograft model, the tumor volume in the NTS-silenced group was significantly reduced (Figure 2D, Supplementary Figure S4D). Next, by exploring The Cancer Genome Atlas (TCGA) database, we investigated the overall survival probability in the NTS high expression group and the low expression group in patients with UM. As expected, Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn) showed that high NTS levels positively correlated with a poor prognosis (P = 0.000047) (Figure 2E). These data show that NTS plays an oncogenic role in the tumorigenesis of UM.
Figure 2.
NTS is a novel oncogene in UM. (A, B) Colony formation analysis demonstrated that NTS-silenced cells exhibited fewer colonies and could be rescued by recombinant NTS supplementation in OCM1 and OCM1a cells. Data are presented as the mean ± SD and the differences between two groups were calculated by unpaired two-tailed Student's t-test. (C) Animal imaging system demonstrated the suppressive effects on tumor bioluminescent signals in NTS-silenced OCM1 cells in orthotopic xenografts. * P <0.05, ** P < 0.01. (D) Suppressive effects on tumor volume in NTS-silenced OCM1 cells in a subcutaneous xenograft model. * P < 0.05, ** P < 0.01. (E) The TCGA database of UM demonstrated prolonged survival time in patients with low NTS expression.
CTCF orchestrates a novel intrachromosomal loop in the NTS locus
Next, we examined whether CTCF could hijack enhancers to form promoter-enhancer interactions. To avoid local chromosomal aberrations, such as chromosomal translocation, duplication or deletion, a karyotype assay demonstrated that the NTS locus was located at chromosome 12p21.31 in a non-aberrant chromosome (Supplementary Figure S5). By focusing on the seven CTCF-binding sites (Supplementary Figure S6) and two negative sites (site 2, –99 kb from TSS; and site 6, –670 kb from TSS) of the NTS locus, we performed a chromosome conformation capture (3C) assay and showed that the NTS promoter interacted frequently with a chromosomal region at an intron in the LRRIQ1 gene that is located nearly 800 kb upstream in tumor cells (Figure 3A, 1/7 interaction), however, we did not detect chromatin interaction in testing CTCF-binding sites of normal control cells. We next set this new region as a bait and tested its interaction with variant CTCF binding sites. Similarly, we also showed that the NTS promoter interacted with above LRRIQ1 bait in tumor cells, and we did not detect additional chromatin interactions in all testing CTCF-binding sites in normal control cells. (Supplementary Figure S7A).
Figure 3.
A novel intrachromosomal loop in the NTS locus. (A) A chromosomal conformation capture (3C) assay was performed to detect intrachromosomal interactions between the NTS promoter and enhancer regions in OCM1, OCM1a and OM431 cells. Right panel: the intrachromosomal interactions between 1–2, 1–3, 1–4 and 1–5 by PCR. M, DNA marker. Numbers under 1–5: distance from the translation start site (TSS). The interaction frequency was determined by normalizing the 3C PCR signal to that of the positive control (input DNA). *P < 0.05 compared to negative control human Schwann cells (HSCs), arising retinal pigment epithelia 19 (ARPE19) cells and pigmented (PIG1) cells. (B) Schematic diagram of variant primer sets in 3C assay. L1–L4: primers in LRRIQ1 intronic region, N1-N5: primers in NTS promoter region. (C) The interaction frequency was determined in OCM1, OCM1a and OM431 cells by normalizing the 3C PCR signal to that of the positive control (input DNA). *P < 0.05 compared to negative control retinal pigment epithelial (RPE) cells and pigmented (PIG1) cells. L2 was set as 3C bait. (D). Identification of the NTS upstream interacting region as an NTS enhancer. Enhancer activity was measured as the relative luciferase units in 293T cells. The NTS promoter and its enhancer inserted upstream of pGL2-promoter-Luc; Mock, empty pGL2-promoter-Luc vector; 293T, wild-type 293T cells. For comparison, the luciferase expression of the mock insert at 48 h was arbitrarily set as 1 in the calculation. *P < 0.05 compared to mock luciferase expression. (E) ChIP assay of the CTCF status in the enhancer (LRRIQ1 intron) in UM (OCM1, OCM1a and OM431) and normal control (ARPE19 and PIG1) cells. * P < 0.05. (F) A 3C assay was performed to detect the existence of chromosomal looping in clinical samples.
To confirm this new NTS-LRRIQ1 interaction, we further examined the chromatin loop between NTS promoter and LRRIQ1 intronic region using different primer sets (L1–L4 and N1–N5) along these two loci (Figure 3B). As expected, we observed NTS-LRRIQ1 interactions using L2a-N4 and L2b-N5 primer sets in tumors (Figure 3C). However, all NTS-LRRIQ1 chromatin interactions were not detected in other tested primer sets (Supplementary Figure S8A-C) and normal control cells (Figure 3A and C; Supplementary Figure S8D, panels 1–3). These NTS-LRRIQ1 interactions were then confirmed by DNA sequencing (Figure 3C, Supplementary Figure S9-S10). In addition, we also showed that this new enhancer had no significant interactions with other distal region by using Hi-C data of normal RPE cells (26) (Supplementary Figure S7B). More specifically, virtual 4C plot suggested that this enhancer did not present with significant interactions in normal control cells (Supplementary Figure S7C). These data provide an alternative explanation that the NTS enhancer hijacking might not occur in normal cells.
To address the role of the upstream region, we cloned a 446 bp fragment of LRRIQ1 intron and tested whether its activity regulates gene expression. As expected, luciferase activity showed that this fragment of LRRIQ1 intron significantly augmented promoter activity as compared to negative control or empty vector (mock). The NTS promoter fragment was used as positive control (Figure 3D). In addition, we found CTCF enrichment in the LRRIQ1 intron of tumor cells, while weak CTCF signals were detected in normal cells (Figure 3E). Importantly, 70% of clinical tumor tissues presented an NTS-LRRIQ1 interaction in our tested samples (Figure 3F). These data show that the LRRIQ1 intron acts as a distal enhancer that regulates NTS and that the CTCF hijacks an enhancer to form a novel intrachromosomal loop in the NTS locus.
Interrupted chromosomal looping abolishes NTS expression by deleting the enhancer
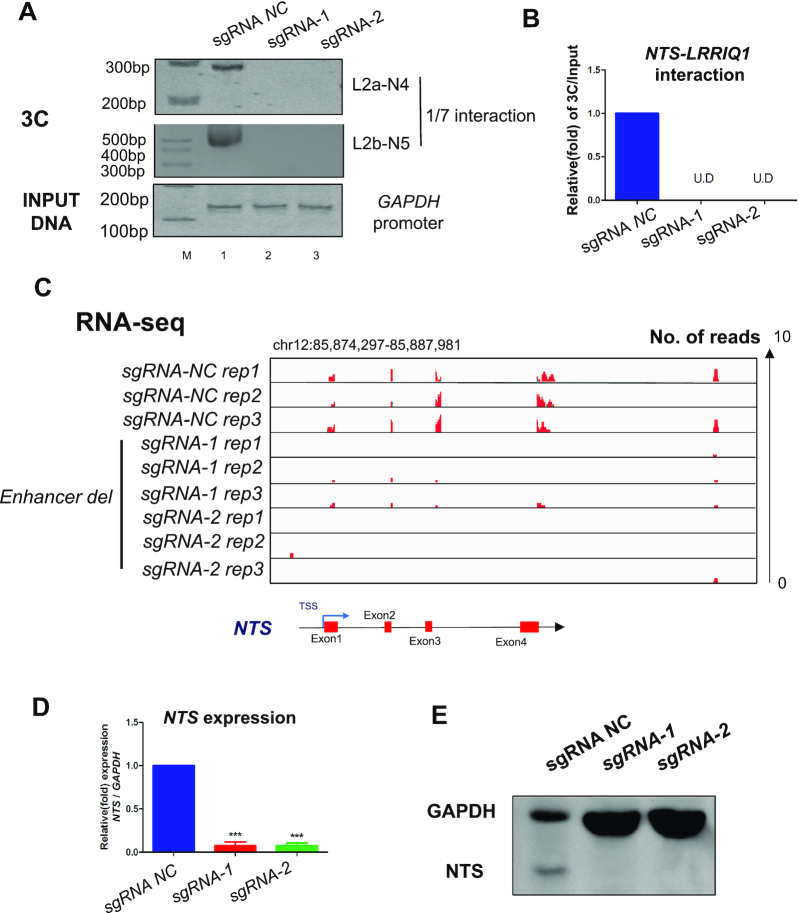
To further investigate the role of intrachromosomal looping in the regulation of NTS, we next deleted the 1.186 kb enhancer region using the CRISPR/Cas9 method. As shown in Supplementary Figure S11A, the enhancer was deleted from the genome containing the full-length CTCF-binding site. Next, we found that intrachromosomal looping between the NTS promoter and the upstream enhancer was abolished (Figure 4A, lanes 2, 3; Figure 3B). To determine whether the deletion could control the transcription of NTS, we tested the expression of NTS through RNA-seq. As expected, the downregulated expression of NTS was detected in enhancer-deleted tumor cells (Figure 4C). To confirm the above bioinformatics data, we then use qPCR and western blots to examine the expression of NTS in mRNA (Figure 4D) and protein (Figure 4E) levels. In consistent with RNA-seq data, we observed a significant decrease in expression of NTS. In addition, we found that the expression of LRRIQ1 remained unchanged (Supplementary Figure S12). Taken together, these data indicate that intrachromosomal looping between the promoter and enhancer controls the expression of endogenous NTS.
Figure 4.
Interrupted chromosomal looping abolishes NTS expression by deleting the enhancer. (A, B) A chromosomal conformation capture assay was performed to capture intrachromosomal interactions between the NTS promoter and enhancer regions in OCM1 and OCM1a cells after deleting its enhancer. (C) RNA-seq was performed to observe the expression of NTS after enhancer deletion in OCM1 and OCM1a cells. (D) Real-time PCR was performed to detect NTS mRNA expression after enhancer deletion in OCM1 and OCM1a cells. Data are presented as the mean ± SD and the differences between two groups were calculated by unpaired two-tailed Student's t-test. *P< 0.05, ***P< 0.001. (E) Western blot was performed to detect NTS protein expression after enhancer deletion in OCM1 and OCM1a cells.
Deletion of the enhancer inhibits tumorigenesis in vitro and in vivo
We next tested whether the interrupted chromosomal looping induced by enhancer deletion could inhibit tumorigenesis. KEGG analysis showed that UM-related pathways had significantly downregulated in enhancer-deleted tumor cells, including the MAPK, Rap1 and PI3K–Akt pathways (Figure 5A). We also observed that the number of colonies was significantly decreased in enhancer-deleted tumor cells (Figure 5B). To examine the role of intrachromosomal looping in vivo, we established subcutaneous xenograft, orthotopic xenograft and metastatic models in nude mice using enhancer-deleted OCM1 tumor cells. We then evaluated the size of the resultant tumors every 7 days for 28 days in subcutaneous xenografts. As expected, the tumor volume in the enhancer-deleted group was significantly reduced compared with the empty vector group (Figure 5C, Supplementary Figure S13A). In orthotopic xenografts, we injected luciferase-labeled enhancer-deleted OCM1 cells into the subarachnoid space. Small animal bioluminescence imaging demonstrated weaker intensity of the tumor signal captured in the enhancer-deleted group compared with the control group (Figure 5D, E). After enucleation, the weight of the tumor was also significantly reduced in the enhancer-deleted group (Figure 5F). Most importantly, after deleting the enhancer, the survival rate of the mice was significantly extended (Figure 5G). In the metastatic tumor model, there were fewer metastatic loci in the lungs of nude mice at 15 weeks in the enhancer-deleted group compared with the control group (Figure 5H, Supplementary Figure S13B). These data suggest that the NTS-LRRIQ1 intrachromosomal loop is a key oncogenetic regulator in NTS-mediated tumorigenesis both in vitro and in vivo.
Figure 5.
Deletion of the enhancer inhibits tumorigenesis in vitro and in vivo. (A) KEGG analysis showing tumor cell growth after enhancer deletion. X axe: Gene ratio (number of counts/total genes); Y axe: Altered signaling pathways, tumorigenesis related pathways were shown in the data. The size of dot represents gene counts and the color represent P value. (B) Plate clone formation assay demonstrated that tumor formation ability was significantly reduced in enhancer-deleted OCM1 cells. (C) Suppressive effects on tumor volume in enhancer-deleted OCM1 cells in a subcutaneous xenograft model. Data are presented as the mean ± SD and the differences between two groups were calculated by unpaired two-tailed Student's t-test. * P < 0.05, ** P < 0.01. (D, E) Animal imaging system demonstrated the suppressive effects on tumor bioluminescent signals in enhancer-deleted OCM1 cells in orthotopic xenografts. (F) Suppressive effects on weight in enhancer-deleted OCM1 cells in orthotopic xenografts. Data are presented as the mean ± SD and the differences between two groups were calculated by unpaired two-tailed Student's t-test. * P < 0.05, ** P < 0.01. (G) Prolonged survival time in enhancer-deleted OCM1 cells in orthotopic xenografts. (H) Fewer metastatic spots in enhancer-deleted OCM1 cells in the metastatic model.
CTCF recruits the histone acetyltransferase EP300 to the NTS promoter and enhancer
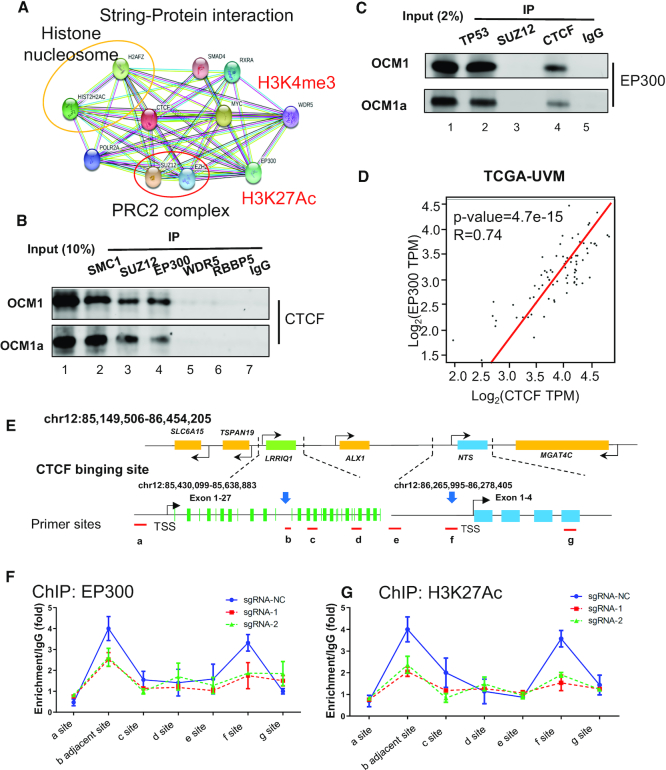
Next, we explored which modification coordinated with this novel NTS-LRRIQ1 chromosomal loop and controlled NTS activation. Through a STRING protein interaction (www.string-db.org) network, we determined that CTCF could bind to histone nucleosomes (e.g. H2AFZ, HIST2H2AC), polycomb-group proteins (e.g. PRC2, SUZ12, and EZH2), and H3K4me3 (WDR5)- and H3K27Ac (EP300)-related modifiers (Figure 6A). Given that the histone modifications H3K4me3 and H3K27Ac represent transcriptional activation, we then tested whether CTCF could bind to these modified sites. SMC1 and SUZ12 antibodies were used as CTCF-binding positive controls (Figure 6B, lanes 2 and 3). As expected, coimmunoprecipitation (co-IP) assays showed that the CTCF protein could be pulled out by baiting the EP300 protein (Figure 6B, lane 4), while the WDR5, RBBP5 and IgG groups exhibited weak binding (Figure 6B, lanes 5–7). In turn, another co-IP experiment showed that EP300 could also be pulled out by baiting the CTCF protein (Figure 6C, lane 4). The EP300-interacting protein TP53 was used as a positive control (Figure 6C, lane 2), and the SUZ12 and IgG groups were used as negative controls (Figure 6C, lane 3). Intriguingly, the TCGA database showed that the expression of CTCF might correlated (R = 0.74, P < 0.001) with EP300 expression in clinical tumor tissues (Figure 6D). We next examined the EP300 binding status and H3K27Ac level after deleting the enhancer (Figure 6E). The ChIP assay showed that the EP300 binding (Figure 6F) and H3K27Ac modification levels (Figure 6G) at the NTS promoter (f site) and enhancer (b site, adjacent to the deleted region) were significantly decreased in enhancer-deleted tumor cells. These data suggested that CTCF could recruit the acetyltransferase EP300 to the NTS promoter and enhancer.
Figure 6.
CTCF recruits the histone acetyltransferase EP300 to the NTS promoter and enhancer. (A) A STRING protein interaction (www.string-db.org) network identified the potential CTCF-binding proteins, including histone nucleosomes (e.g., H2AFZ, HIST2H2AC), polycomb-group proteins (e.g. PRC2 SUZ12, and EZH2), and H3K4me3 (WDR5)- and H3K27Ac (EP300)-related modifiers. (B, C) Interaction between CTCF and EP300 detected by coimmunoprecipitation. (E) The CTCF protein was pulled out by baiting the EP300 protein, while the binding between WDR5 and RBBP5 could not be detected. IP of the SMC1 and SUZ12 groups was performed as a positive control. IgG was used as a negative control. (F) EP300 could also be pulled out by baiting the CTCF protein, and IP of the TP53 group was performed as a positive control. (D) Gene expression correlation analysis from the TCGA database indicated that EP300 expression strongly correlated with CTCF (R = 0.74, P = 4.7e–15). (E) Schematic diagram of the ChIP site. Sites a-e are distributed in the LRRIQ1 region, and site b is the enhancer and CTCF-binding site. Sites E–G are distributed in the NTS genomic region. Site f is the NTS promoter and another CTCF-binding site. (F–G): ChIP assay of the EP300 and H3K27Ac status in the NTS promoter and its enhancer region after enhancer deletion in OCM1 and OCM1a cells.
Silencing EP300 abolishes H3K27 acetylation at the NTS locus and inhibits NTS expression
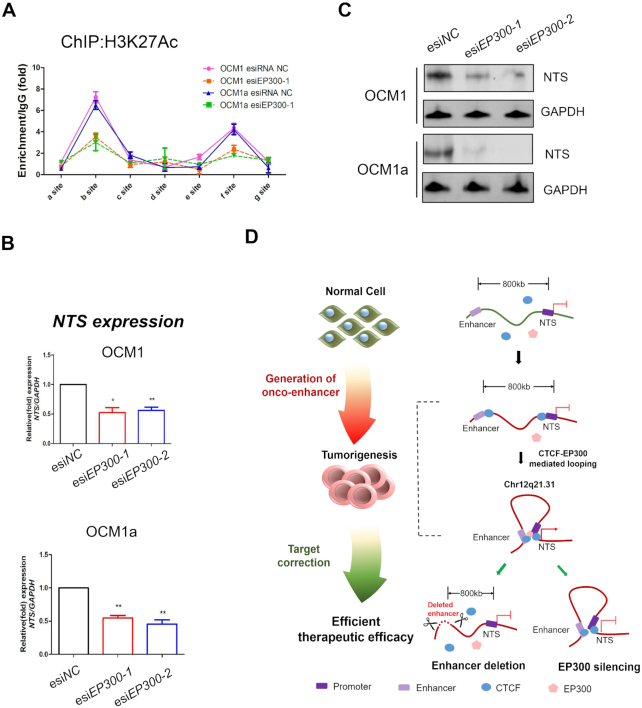
To examine the role of EP300 in intrachromosomal looping and NTS expression, we designed two endoribonuclease-prepared siRNAs (esiRNAs) to silence EP300. As expected, the mRNA (Supplementary Figure S14A) and protein (Supplementary Figure S14B) expression levels of EP300 were diminished after interference with two esiRNAs. Next, we found that EP300 interacted with the NTS promoter (Supplementary Figure S14C, f site, esiNC groups) and its upstream enhancer fragment (∼ -800 kb) (Supplementary Figure S14C, b site, esiNC groups). We also observed a high level of H3K27 acetylation at the NTS promoter and enhancer in wild-type tumor cells by quantitative ChIP assays (Figure 7A, esiNC groups). However, after knocking down EP300, both EP300 binding and the H3K27Ac signal were significantly decreased at the NTS promoter and enhancer (Figure 7A, esiEP300–1 groups). In addition, we found that the mRNA and protein levels of NTS were significantly reduced in EP300-silenced tumor cells (Figure 7B, C). We also found that the NTS-LRRIQ1 intrachromosomal loop remained after EP300 silencing (Supplementary Figure S15). These data indicate that EP300 is not involved in loop formation and might play a key histone acetyltransferase to control NTS expression.
Figure 7.
Silencing EP300 abolishes H3K27 acetylation at the NTS locus and inhibits NTS expression. (A) ChIP assay of the H3K27Ac status in the NTS promoter and its enhancer region after siRNA EP300 transfection in UM cells. (B) qRT-PCR showed that NTS was silenced after silencing EP300 in UM cells. *P< 0.05, **P< 0.01. (C) Western blot was performed to detect NTS protein expression after silencing EP300 in UM cells. (D) Schematic diagram of the research model. CTCF interacts with distal enhancers to form chromosomal loops and recruits the histone acetyltransferase EP300 to activate the oncogene NTS and contributes to tumor progression. The targeted correction of abnormal NTS-LRRIQ1 chromosomal interactions significantly inhibited tumorigenesis by deleting oncogenic enhancers or inactivating histone acetyltransferases.
DISCUSSION
High-order chromosomal organization is an inherent property for the cellular genomic arrangement to accommodate ∼2 m of DNA into the ∼5 μm nucleus (27). During tumor formation, numerous oncogenes are activated by hijacking a distal enhancer, which has been identified as an essential oncogenic driver required for the maintenance of cancer cell identity. For example, the enhancer reorganization of MYC promoted tumorigenesis in high-risk pediatric neuroblastomas (8). In addition, enhancer hijacking activates GFI1 family oncogenes in medulloblastoma by somatic structural variants (11). To explore the mechanism underlying the role of enhancer-guided 3D chromosomal loops in tumorigenesis, we for the first time identified novel CTCF-EP300-guided enhancer-triggered chromatin remodeling and oncogene transcription at the NTS locus. The targeted correction of abnormal NTS-LRRIQ1 chromosomal interactions significantly inhibited tumorigenesis by deleting an oncogenic enhancer or inactivating a histone acetyltransferase, thereby leading to a novel mechanism for responding to the critical role of novel onco-enhancer in chromatin remodeling and tumorigenesis (Figure 7D).
It should be noted that CTCF is a key architectural component of the genome that anchors long-range interactions that play a vital role in imprinting maintenance (28), B cell differentiation (29) and cancer formation (27). For example, the CTCF-mediated KCNQ1 and CDKN1C intrachromosomal conformation could significantly inhibit tumor formation, while aberrant chromosomal looping may lead to imprinting abnormalities and activation of the oncogenic long noncoding RNA (lncRNA) KCNQ1OT1, thereby accelerating tumorigenesis (16). In this study, we investigated whether CTCF-mediated chromosomal interactions may also play a leading role in the tumorigenesis of UM. To our knowledge, this is the first example whereby a CTCF-guided intrachromosomal interaction contributes to UM tumorigenesis. Since the epigenetic deregulation of variant oncogenes remains unclear, further studies should focus on the identification of other chromatin loops to determine the mechanism of deregulated gene expression in tumorigenesis.
Although global depletion of CTCF loops result in minor changes to gene expression (30), in this study, we found that nearly 2,000 genes were significant changed after enhancer deletion. An alternative explanation is that enhancer deletion-caused NTS silencing plays a critical role in regulation of other gene expression. NTS has been proven to involve in numerous signal pathways, including Wnt/β-Catenin pathway, IL-8 pathways, and epithelial-mesenchymal transition pathways (31–33), NTS silencing can lead to alteration of a series of cascade effect and subsequently induce dramatic gene changes. Thus, it would be of great interest to explore the novel downstream pathway of NTS in uveal melanoma.
It should also be noted that CTCF is a typical genomic insulator involved in numerous biological processes. Typically, CTCF can induce DNA methylation and recruit the PRC2 complex, inducing histone K27 trimethylation in the promoter, to occlude target gene expression (34,35). In addition, CTCF has also been proven to be associated with gene activation via the removal of H3K27me3 and retains an open chromatin status (36). Here, we first found that CTCF could interact with the histone acetyltransferase EP300, recruiting it to the promoter of the target gene NTS and leading to gene activation. Although we cannot theoretically eliminate other genetic or epigenetic causes in the regulation of NTS, this is the first study to imply that the expression of NTS is highly dependent on histone H3K27 acetylation at the promoter region of NTS. Thus, it would be of great interest to focus on the identification of other factors to better understand NTS transcriptional regulation. Further studies should focus on the identification of other factors that can modify histone acetylation to regulate NTS expression.
It should be emphasized that CTCF establishes a chromosomal loop between enhancer and promoter through DNA binding and self-dimerization. The transcriptional modifiers subsequently be recruited into specific locus and interact with CTCF dimer for further gene activation or silencing. In IGF2/H19 imprinted locus, CTCF dimer initially forms an intrachromosomal loop and recruits PRC2 complex for maintaining IGF2 imprinting (16,37). In this study, we noticed that the knockdown of EP300 did not appear to cause as severe a reduction in NTS expression as loss of the enhancer. Thus, it provides a possibility that CTCF dimer firstly orchestrates an enhancer-promoter looping and secondly recruits the EP300, subsequently, looping-based enhancer activity and EP300-based epigenetic modification co-regulate the NTS expression. Abolishment of EP300 diminishes factor-based regulated layer but remains the looping-based enhancer activity. However, after the enhancer deletion, two regulated layers are completely interrupted. These two separated phenomena is likely to provide an alternative explanation for the role of enhancer activity in mediating enhancer-promoter looping, at least in this case, further studies should focus on the identification of the co-regulated mechanism between enhancer activity and epigenetic factor modification in NTS expression of tumorigenesis.
It should be noted that the NTS gene (neurotensin) is a secreted tridecapeptide (38) that is widely distributed throughout the central nervous system and may function as a neurotransmitter (39). It was proven to be involved in dopamine-associated pathophysiological events, in the maintenance of gut structure and function, and in the regulation of fat metabolism (40). Moreover, activated NTS can also contribute to tumor formation in hepatocarcinogenesis (41), ovarian cancer (42) and colon cancer (43). However, thus far, evidence does not indicate the regulatory role of the NTS gene in the formation of UM. In this report, however, we clearly demonstrate that NTS contributes to tumor growth and metastasis and is considered a novel oncogene in UM.
In summary, our results reveal a completely novel model of tumorigenesis in which CTCF interacts with distal enhancers to form chromosomal loops and recruits the histone acetyltransferase EP300 to activate the oncogene and accelerate tumorigenesis, thereby providing a novel therapeutic method in which the targeted correction of onco-enhancer triggered chromatin remodeling is efficient for tumor therapy. Most importantly, many chromosomal interactions play a key role in the pathogenesis of various diseases, thereby suggesting new inspirations for exploring the novel therapeutic concept that the targeted correction of abnormal chromosomal interactions is promising for disease therapy.
Supplementary Material
ACKNOWLEDGEMENTS
Authors’ contributions: P.W.C. and J.Y. designed and performed the experiments and drafted the manuscript; R.B.J., X.Y.W., T.Y.D. and X.Y.Z. were responsible for sample collection and data analysis; R.B.J. and S.F.G. discussed the manuscript. H.Z. and X.Q.F. discussed, revised and approved the manuscript. All the authors approved this manuscript.
Contributor Information
Peiwei Chai, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Ninth People's Hospital, Shanghai JiaoTong University School of Medicine, Shanghai, P. R. China.
Jie Yu, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Ninth People's Hospital, Shanghai JiaoTong University School of Medicine, Shanghai, P. R. China.
Ruobing Jia, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Ninth People's Hospital, Shanghai JiaoTong University School of Medicine, Shanghai, P. R. China.
Xuyang Wen, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Ninth People's Hospital, Shanghai JiaoTong University School of Medicine, Shanghai, P. R. China.
Tianyi Ding, Institute for Regenerative Medicine, Shanghai East Hospital, School of Life Science and Technology, Tongji University, Shanghai, P. R. China; Frontier Science Research Center for Stem Cells, Tongji University, Shanghai, 200092, P. R. China.
Xiaoyu Zhang, Institute for Regenerative Medicine, Shanghai East Hospital, School of Life Science and Technology, Tongji University, Shanghai, P. R. China; Frontier Science Research Center for Stem Cells, Tongji University, Shanghai, 200092, P. R. China.
Hongyan Ni, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Ninth People's Hospital, Shanghai JiaoTong University School of Medicine, Shanghai, P. R. China.
Renbing Jia, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Ninth People's Hospital, Shanghai JiaoTong University School of Medicine, Shanghai, P. R. China.
Shengfang Ge, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Ninth People's Hospital, Shanghai JiaoTong University School of Medicine, Shanghai, P. R. China.
He Zhang, Institute for Regenerative Medicine, Shanghai East Hospital, School of Life Science and Technology, Tongji University, Shanghai, P. R. China; Frontier Science Research Center for Stem Cells, Tongji University, Shanghai, 200092, P. R. China.
Xianqun Fan, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Ninth People's Hospital, Shanghai JiaoTong University School of Medicine, Shanghai, P. R. China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Plan [2017YFE0196300]; National Natural Science Foundation of China [31870748]; ShuGuang Project of Shanghai Municipal Education Commission and Shanghai Education Development Foundation [17SG19]; Outstanding Yong Medical Scholar of Shanghai Municipal Commission of Health and Family Planning [2017YQ067]; Outstanding Yong Scholar Grant of Tongji University [PA2019000239]; Startup Funding of Frontier Science Research Center for Stem Cells & Shanghai East Hospital of Tongji University. Funding for open access charge: National Key Research and Development Plan of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Monahan K., Rudnick N.D., Kehayova P.D., Pauli F., Newberry K.M., Myers R.M., Maniatis T.. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-alpha gene expression. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:9125–9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lupianez D.G., Spielmann M., Mundlos S.. Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet. 2016; 32:225–237. [DOI] [PubMed] [Google Scholar]
- 3. Spielmann M., Lupianez D.G., Mundlos S.. Structural variation in the 3D genome. Nat. Rev. Genet. 2018; 19:453–467. [DOI] [PubMed] [Google Scholar]
- 4. Tsujimura T., Takase O., Yoshikawa M., Sano E., Hayashi M., Takato T., Toyoda A., Okano H., Hishikawa K.. Control of directionality of chromatin folding for the inter- and intra-domain contacts at the Tfap2c-Bmp7 locus. Epigenet. Chromatin. 2018; 11:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Katainen R., Dave K., Pitkanen E., Palin K., Kivioja T., Valimaki N., Gylfe A.E., Ristolainen H., Hanninen U.A., Cajuso T. et al.. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet. 2015; 47:818–821. [DOI] [PubMed] [Google Scholar]
- 6. Haller F., Bieg M., Will R., Korner C., Weichenhan D., Bott A., Ishaque N., Lutsik P., Moskalev E.A., Mueller S.K. et al.. Enhancer hijacking activates oncogenic transcription factor NR4A3 in acinic cell carcinomas of the salivary glands. Nat. Commun. 2019; 10:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jia Y., Chng W.J., Zhou J.. Super-enhancers: critical roles and therapeutic targets in hematologic malignancies. J. Hematol. Oncol. 2019; 12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zimmerman M.W., Liu Y., He S., Durbin A.D., Abraham B.J., Easton J., Shao Y., Xu B., Zhu S., Zhang X. et al.. MYC drives a subset of high-risk pediatric neuroblastomas and is activated through mechanisms including enhancer hijacking and focal enhancer amplification. Cancer Discov. 2018; 8:320–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cuddapah S., Jothi R., Schones D.E., Roh T.Y., Cui K., Zhao K.. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009; 19:24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cui H., Niemitz E.L., Ravenel J.D., Onyango P., Brandenburg S.A., Lobanenkov V.V., Feinberg A.P.. Loss of imprinting of insulin-like growth factor-II in Wilms' tumor commonly involves altered methylation but not mutations of CTCF or its binding site. Cancer Res. 2001; 61:4947–4950. [PubMed] [Google Scholar]
- 11. Northcott P.A., Lee C., Zichner T., Stutz A.M., Erkek S., Kawauchi D., Shih D.J., Hovestadt V., Zapatka M., Sturm D. et al.. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014; 511:428–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weischenfeldt J., Dubash T., Drainas A.P., Mardin B.R., Chen Y., Stutz A.M., Waszak S.M., Bosco G., Halvorsen A.R., Raeder B. et al.. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 2017; 49:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bell A.C., West A.G., Felsenfeld G.. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999; 98:387–396. [DOI] [PubMed] [Google Scholar]
- 14. Chao W., Huynh K.D., Spencer R.J., Davidow L.S., Lee J.T.. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002; 295:345–347. [DOI] [PubMed] [Google Scholar]
- 15. Zhang H., Jiao W., Sun L., Fan J., Chen M., Wang H., Xu X., Shen A., Li T., Niu B. et al.. Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell Stem Cell. 2013; 13:30–35. [DOI] [PubMed] [Google Scholar]
- 16. Zhang H., Niu B., Hu J.F., Ge S., Wang H., Li T., Ling J., Steelman B.N., Qian G., Hoffman A.R.. Interruption of intrachromosomal looping by CCCTC binding factor decoy proteins abrogates genomic imprinting of human insulin-like growth factor II. J. Cell Biol. 2011; 193:475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rhodes J.D.P., Feldmann A., Hernandez-Rodriguez B., Diaz N., Brown J.M., Fursova N.A., Blackledge N.P., Prathapan P., Dobrinic P., Huseyin M.K. et al.. Cohesin disrupts polycomb-dependent chromosome interactions in embryonic stem cells. Cell Rep. 2020; 30:820–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S.. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011; 479:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo Y.A., Chang M.M., Huang W., Ooi W.F., Xing M., Tan P., Skanderup A.J.. Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers. Nat. Commun. 2018; 9:1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marshall A.D., Bailey C.G., Champ K., Vellozzi M., O’Young P., Metierre C., Feng Y., Thoeng A., Richards A.M., Schmitz U. et al.. CTCF genetic alterations in endometrial carcinoma are pro-tumorigenic. Oncogene. 2017; 36:4100–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walker C.J., Miranda M.A., O’Hern M.J., McElroy J.P., Coombes K.R., Bundschuh R., Cohn D.E., Mutch D.G., Goodfellow P.J.. Patterns of CTCF and ZFHX3 mutation and associated outcomes in endometrial cancer. J. Natl. Cancer Inst. 2015; 107:djv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee H.K., Willi M., Wang C., Yang C.M., Smith H.E., Liu C., Hennighausen L.. Functional assessment of CTCF sites at cytokine-sensing mammary enhancers using CRISPR/Cas9 gene editing in mice. Nucleic Acids Res. 2017; 45:4606–4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Willi M., Yoo K.H., Reinisch F., Kuhns T.M., Lee H.K., Wang C., Hennighausen L.. Facultative CTCF sites moderate mammary super-enhancer activity and regulate juxtaposed gene in non-mammary cells. Nat. Commun. 2017; 8:16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fan J., Xing Y., Wen X., Jia R., Ni H., He J., Ding X., Pan H., Qian G., Ge S. et al.. Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol. 2015; 16:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gaulton K.J., Nammo T., Pasquali L., Simon J.M., Giresi P.G., Fogarty M.P., Panhuis T.M., Mieczkowski P., Secchi A., Bosco D. et al.. A map of open chromatin in human pancreatic islets. Nat. Genet. 2010; 42:255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Darrow E.M., Huntley M.H., Dudchenko O., Stamenova E.K., Durand N.C., Sun Z., Huang S.C., Sanborn A.L., Machol I., Shamim M. et al.. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. PNAS. 2016; 113:E4504–E4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nichols M.H., Corces V.G.. A CTCF Code for 3D Genome Architecture. Cell. 2015; 162:703–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ling J.Q., Li T., Hu J.F., Vu T.H., Chen H.L., Qiu X.W., Cherry A.M., Hoffman A.R.. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science (New York, N.Y.). 2006; 312:269–272. [DOI] [PubMed] [Google Scholar]
- 29. Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y., Xu X., Lv X., Hugnot J.P., Tanay A. et al.. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017; 171:557–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rao S.S.P., Huang S.C., Glenn St Hilaire B., Engreitz J.M., Perez E.M., Kieffer-Kwon K.R., Sanborn A.L., Johnstone S.E., Bascom G.D., Bochkov I.D. et al.. Cohesin loss eliminates all loop domains. Cell. 2017; 171:305–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brunetti J., Falciani C., Lelli B., Minervini A., Ravenni N., Depau L., Siena G., Tenori E., Menichetti S., Pini A. et al.. Neurotensin branched peptide as a tumor-targeting agent for human bladder cancer. Biomed. Res. Int. 2015; 2015:173507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qiu S., Pellino G., Fiorentino F., Rasheed S., Darzi A., Tekkis P., Kontovounisios C.. A review of the role of neurotensin and its receptors in colorectal cancer. Gastroenterol. Res. Pract. 2017; 2017:6456257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu S., Tian H., Niu X., Wang J., Li X., Jiang N., Wen S., Chen X., Ren S., Xu C. et al.. Neurotensin and its receptors mediate neuroendocrine transdifferentiation in prostate cancer. Oncogene. 2019; 38:4875–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bergmaier P., Weth O., Dienstbach S., Boettger T., Galjart N., Mernberger M., Bartkuhn M., Renkawitz R.. Choice of binding sites for CTCFL compared to CTCF is driven by chromatin and by sequence preference. Nucleic Acids Res. 2018; 46:7097–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Junier I., Dale R.K., Hou C., Kepes F., Dean A.. CTCF-mediated transcriptional regulation through cell type-specific chromosome organization in the beta-globin locus. Nucleic Acids Res. 2012; 40:7718–7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weth O., Paprotka C., Gunther K., Schulte A., Baierl M., Leers J., Galjart N., Renkawitz R.. CTCF induces histone variant incorporation, erases the H3K27me3 histone mark and opens chromatin. Nucleic Acids Res. 2014; 42:11941–11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lleres D., Moindrot B., Pathak R., Piras V., Matelot M., Pignard B., Marchand A., Poncelet M., Perrin A., Tellier V. et al.. CTCF modulates allele-specific sub-TAD organization and imprinted gene activity at the mouse Dlk1-Dio3 and Igf2-H19 domains. Genome Biol. 2019; 20:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brown J., Sagante A., Mayer T., Wright A., Bugescu R., Fuller P.M., Leinninger G.. Lateral hypothalamic area neurotensin neurons are required for control of orexin neurons and energy balance. Endocrinology. 2018; 159:3158–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Woodworth H.L., Beekly B.G., Batchelor H.M., Bugescu R., Perez-Bonilla P., Schroeder L.E., Leinninger G.M.. Lateral hypothalamic neurotensin neurons orchestrate dual weight loss behaviors via distinct mechanisms. Cell Rep. 2017; 21:3116–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barchetta I., Cimini F.A., Capoccia D., Bertoccini L., Ceccarelli V., Chiappetta C., Leonetti F., Di Cristofano C., Silecchia G., Orho-Melander M. et al.. Neurotensin is a lipid-induced gastrointestinal peptide associated with visceral adipose tissue inflammation in obesity. Nutrients. 2018; 10:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu J., Ren X., Chen Y., Liu P., Wei X., Li H., Ying G., Chen K., Winkler H., Hao X.. Dysfunctional activation of neurotensin/IL-8 pathway in hepatocellular carcinoma is associated with increased inflammatory response in microenvironment, more epithelial mesenchymal transition in cancer and worse prognosis in patients. PLoS One. 2013; 8:e56069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu J., Agopiantz M., Poupon J., Wu Z., Just P.A., Borghese B., Segal-Bendirdjian E., Gauchotte G., Gompel A., Forgez P.. Neurotensin receptor 1 antagonist SR48692 improves response to carboplatin by enhancing apoptosis and inhibiting drug efflux in ovarian cancer. Clin. Cancer Res. 2017; 23:6516–6528. [DOI] [PubMed] [Google Scholar]
- 43. Wang X., Wang Q., Ives K.L., Evers B.M.. Curcumin inhibits neurotensin-mediated interleukin-8 production and migration of HCT116 human colon cancer cells. Clin. Cancer Res. 2006; 12:5346–5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.