Abstract
Transcriptional regulation of DNA repair is of outmost importance for the restoration of DNA integrity upon genotoxic stress. Here we report that the potent environmental carcinogen benzo[a]pyrene (B[a]P) activates a cellular DNA damage response resulting in transcriptional repression of mismatch repair (MMR) genes (MSH2, MSH6, EXO1) and of RAD51, the central homologous recombination repair (HR) component, ultimately leading to downregulation of MMR and HR. B[a]P-induced gene repression is caused by abrogated E2F1 signalling. This occurs through proteasomal degradation of E2F1 in G2-arrested cells and downregulation of E2F1 mRNA expression in G1-arrested cells. Repression of E2F1-mediated transcription and silencing of repair genes is further mediated by the p21-dependent E2F4/DREAM complex. Notably, repression of DNA repair is also observed following exposure to the active B[a]P metabolite BPDE and upon ionizing radiation and occurs in response to a p53/p21-triggered, irreversible cell cycle arrest marking the onset of cellular senescence. Overall, our results suggest that repression of MMR and HR is an early event during genotoxic-stress induced senescence. We propose that persistent downregulation of DNA repair might play a role in the maintenance of the senescence phenotype, which is associated with an accumulation of unrepairable DNA lesions.
INTRODUCTION
Unrepaired or false repaired DNA lesions interfere with replication and transcription, leading to either mutations and cancer, or cell death and senescence. Therefore, a coordinated and faithful DNA damage response and repair is of central importance for maintaining genomic integrity and survival. A significant influence on the outcome of damage repair and processing is exerted by the abundance and regulation of DNA repair factors. Transcriptional induction and activation of DNA repair factors are important regulatory mechanisms contributing to the adaptation of cells to genotoxic stress (1). However, it appears that the inverse strategy, i.e. downregulation of gene expression in response to DNA damage, also plays a role in the fine-tuned regulation of complex DNA repair pathways. In our previous work, we analysed the regulation of DNA repair in response to benzo[a]pyrene (B[a]P) and its active metabolite benzo(a)pyrene 7,8-diol-9,10-epoxide (BPDE). We showed that low (nontoxic) concentrations of BPDE cause AP-1 and p53 dependent upregulation of several nucleotide excision repair (NER) genes, leading to enhanced NER activity, consequently reducing the effectiveness of a challenge dose of this carcinogen (2).
B[a]P is formed by incomplete combustion and released into the atmosphere from industrial production processes and vehicle exhaust emissions. It is also produced during food preparation and tobacco smoking (3). Thus, exposure to B[a]P is ubiquitous in modern life; as an example, the U.S. population has a daily intake of ∼2.2 μg of B[a]P (3). Various studies have shown that B[a]P is mutagenic and associated with developmental damage, immunological impairments, decreased fertility, low birth weight and smaller head circumference in children. A bulk of data in animal models is available showing that B[a]P gives rise to a variety of cancers including alimentary and respiratory tract, liver, kidney, pharynx, and skin. In humans exposed to B[a]P a significantly increased risk of lung cancer has been found. Beside this, B[a]P was reported to be associated with the development of cancers of oesophagus, larynx, mouth, throat, kidney, bladder, pancreas, stomach, cervix and blood. Therefore, already in 1987, B[a]P was classified as a ‘probable human carcinogen’. In 2014, the Environmental Protection Agency (EPA) published a revised health assessment of B[a]P, upgrading its cancer classification to ‘carcinogenic to humans'. Accordingly, B[a]P is listed as a Group 1 carcinogen by the IARC (4).
B[a]P is an incomplete carcinogen, which is metabolized into an ultimate highly electrophilic carcinogenic species (5). Multiple studies showed that BPDE represents the ultimate carcinogenic metabolite of B[a]P (6–9); ∼10% of B[a]P is converted to BPDE (10). B[a]P is metabolized by cytochrome P450 (Cyp1A1) into benzo(a)pyrene-7,8-epoxide and by an epoxide hydrolase to benzo(a)pyrene-7,8-dihydrodiol (11). Additional metabolism by cytochrome P450 results in the ultimate carcinogen benzo(a)pyrene 7,8-diol-9,10-epoxide (BPDE) (11), which binds via the epoxide group at the C-10 position within the bay region of the molecule (12) to the exocyclic N2 position of guanine (13,14), forming a bulky adduct in the DNA. These adducts are removed from DNA by NER (15–18), and human primary endothelial cells, which are impaired in nucleotide excision repair, are sensitive to BPDE (19). Besides NER, translesion synthesis is involved in the bypass of BPDE adducts. Thus, polymerase kappa (POLK) is able to bypass B[a]P-guanine adducts (dG-N2-BPDE) in an error-free manner by inserting dC opposite the lesion (20–22), whereas polymerase eta (POLH) bypasses the adducts in an error-prone manner by inserting dA opposite dG-N2-BPDE (20,23,24). Of note, incorporation of A opposite dG-N2-BPDE matches with the mutation spectrum of BPDE, suggesting POLH plays an important role in BPDE-induced mutagenesis (23).
Microarray-based gene expression studies upon exposure to B[a]P were performed in HepG2, MCF7 and HCT116 cells at early time points (2–48 h), showing induction of DDB2 (25,26). Moreover, XPC expression was induced after BPDE exposure in human mammary epithelial (27) and breast cancer MCF-7 cells (28). In our previous work, we observed transcriptional activation of the p53-regulated NER genes DDB2 and XPC upon exposure of metabolically competent MCF7 cells to B[a]P and in BPDE-exposed human telomerase-immortalised fibroblasts (VH10tert) and primary epithelial lung cells. Additional experiments showed that pre-treatment with low-dose BPDE not only enhanced the expression of the NER factors but also maintained the expression during the subsequent high-dose exposure, ensuring NER capacity and leading to an adaptive response (2). Similar to the above mentioned NER genes, POLH was also induced. Interestingly, transient overexpression of POLH not only reduced the frequency of apoptosis, but also enhanced the mutation frequency. In addition to the activation of NER and POLH, we observed transcriptional repression of the DNA repair genes MSH2, MSH6, EXO1 and RAD51. EXO1, MSH2 and MSH6 are proteins involved in mismatch repair (MMR), which recognize and repair mispaired bases, while RAD51 is a crucial player in homologous recombination (HR), responsible for the repair of DNA double-strand breaks (29).
In this study, our aim was to analyse the molecular mechanisms underlying the repression of these DNA repair genes and the corresponding pathways in B[a]P-treated cells. We show that the repression is caused by dual abrogation of the E2F1 pathway, which occurs by proteasomal degradation of E2F1 and by transcriptional silencing of the E2F1 gene mediated by the DREAM complex. Downregulation of the E2F1 pathway went along with the induction of B[a]P-induced senescence, which indicates that senescence induction and repression of DNA repair are causally related phenomena.
MATERIALS AND METHODS
Cell culture, drug treatment, siRNA-mediated knockdown and pharmacological inhibition
The human diploid VH10tert foreskin fibroblast cell line was immortalised by stable transfection with the telomerase gene (TERT) and kindly provided by Prof. L. Mullenders (Department of Toxicogenetics, Leiden University Medical Centre, the Netherlands). MCF7 breast cancer cells were obtained from CLS Cell Lines Service GmbH, Eppelheim, Germany. VH10tert cells were cultivated in Dulbecco's minimal essential medium (DMEM) containing 10% FCS under nitrogen atmosphere (5% CO2, 5% O2) and MCF7 cells were cultivated in DMEM-F12 containing 5% FCS under normal atmosphere (5% CO2) at 37°C and were regularly checked for mycoplasma contamination. Human primary bronchial epithelial cells (PBECs) were purchased from Provitro (Berlin) and cultivated in Airway epithelial cell growth medium containing 10% fetal bovine serum. DLD1, LoVo and SW480 cells were purchased from ATCC and cultured in RPMI medium supplemented with 10% FCS at 37°C, 6% CO2. Generation and cultivation of SW480-MSH6ko cells have been described (30).
B(a)P was purchased from SIGMA (B1760), activated r-7,t-8-dihydroxy-t-9,10-epoxy-7,8,9,10-tetrahydrobenzo(a)pyrene (BPDE; CAS no. 58917-67-2) was synthesized from trans-7,8-dihydroxy-7,8-dihydrobenzo(a)pyrene (31) by Dr A. Seidel (BIU Biochemical Institute for Environmental Carcinogens, Prof. Dr G. Grimmer-Stiftung, Grosshansdorf, Germany) as described (32). Ionizing radiation was performed within a Gammacell irradiator 2000 (Cs-137 source, Molsgaard Medical, Denmark).
For silencing, predesigned siRNAs specific for p21 (sc-29427, Santa Cruz Biotechnology), E2F1 (L-003259-00-0005, Dharmacon) and E2F4 (sc-29300) were used; control human non-silencing siRNA (Silencer Select Predesigned siRNA Negative Control #1 siRNA; Ambion) was used as negative control. The transfections of siRNAs were performed using Lipofectamine RNAiMAX Reagent (Invitrogen). The p21 inhibitor UC2288 (CAS 532813, Calbiochem) was used at 5 μM, chloroquin (CAS 50635, Sigma Aldrich) at 5 μM, SKP2 E3 Ligase inhibitor III (506305, Calbiochem) at 10–50 μM, TAME (CAS 901473, Selleckchem) at 0.4 mM, MG132 (CAS 133407826, Selleckchem) at 50–500 nM, rapamycine (CAS 53123889, Selleckchem) at 0.5 μM and MRT-68921 (CAS 1190379704, Selleckchem) at 2 μM.
Preparation of RNA and real-time qPCR
Total RNA was isolated using the NucleoSpin® RNA Kit (Macherey-Nagel, Düren, Germany) and 1 μg of total RNA was transcribed into cDNA (Verso cDNA Kit, Thermo Scientific, Dreieich, Germany). qPCR was performed in technical triplicates using the GoTaq® qPCR Master Mix Protocol (Promega, Madison, USA) and the CFX96 Real-Time PCR Detection System (Biorad, München, Germany). The specific primers are listed in Supplementary Table S1. Non-template controls were included in each run, expression was normalized to gapdh and β-actin; the untreated control was set to one. Analysis was performed using CFX Manager™ Software; SD shows intra-experimental variation.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation was performed as described (33) using E2F1 (sc-193, Santa Cruz Biotechnology) and E2F4 (sc-866, Santa Cruz Biotechnology) antibodies. Real-time PCR was performed using specific primers flanking the E2F1 binding site of EXO1, MSH2, MSH6, and RAD51, which are listed in Supplementary Table S1. Furthermore, binding of E2F4 to a DREAM-specific binding site in the promoters of E2F1, MSH2 and MSH6, was perfomed using previously described primers (34) listed in Supplementary Table S1.
Detection of MSH2 promoter activity
The MSH2-CAT promoter plasmid was described before (35). It was co-transfected with RSV-βgal, and CAT and β-gal activity were determined as described (35). The results are expressed as CAT/β-gal activity in relation to the control.
Determination of apoptosis, cell cycle progression, autophagy and senescence
For quantifying drug-induced apoptosis, annexin V-FITC/propidium iodide (PI) double stained cells were analyzed by flow cytometry; to analyse cell cycle distribution, cells were stained with PI and cell cycle distribution was determined by flow cytometry. Detection of autophagy was performed using the Cyto-ID® Autophagy Detection Kit (Enzo Life Science). Flow cytometry was performed using a BD FACSCanto™ II. Senescence was measured either microscopically by ß-Gal staining, or by cytometry-based detection of C12FDG (ImaGene Green™ C12FDG lacZ Gene Expression Kit) as described before (33). Experiments were repeated at least three times, mean values ± SD are shown.
Separation of G1- and G2-cells and isolation of senescent cells
For isolation of senescent cells, the cells were trypsinized, washed with PBS, fixated with 70% EtOH at −20°C and stored at −80°C. Upon de-freezing, 7 × 106 cells/ml were stained with 20 μM DRAQ 5 (ab108410, Abcam) for 10 min at RT. For isolation of senescent cells, the cells were stained with C12FDG (ImaGene Green™ C12FDG lacZ Gene Expression Kit), resuspended in dissociation buffer (Gibco) containing 1% FCS. G1 and G2 cells as well as C12FDG positive and C12FDG negative cells were sorted using a BD FACSAriaIII SORP cell Sorter using a 100 μm nozzle. RNA isolation was performed using the NucleoSpin RNA Plus XS, Micro kit (Machery and Nagel).
Preparation of protein extracts and western blot analysis
Whole-cell and nuclear extracts were prepared as described (36). For detection of phospho-specific antibodies, cells were directly lysed in 1x SDS-PAGE sample buffer and subsequently sonified. Mouse mAb and rabbit pAb were diluted 1:500–1:2000 in 5% BSA, 0.2% Tween–TBS and incubated overnight at 4°C; the specific antibodies are listed in Supplementary Table S2. Protein-antibody complexes were detected by Pierce® ECL Western Blotting Substrate (Thermo Fisher). Ubiqitinated proteins were isolated using the UBIQAPTURE-Q® kit (BML-UW8995 – Enzo Life Sciences).
DNA repair assays and detection of proteasomal activity
MMR activity was analysed using electromobility shift assays (EMSA), as described (37). Efficiency of HR was determined using the qPCR-based HR Assay kit (Norgen Biotek Corporation, ON, Canada) as described (38). Proteasomal activity was measured using the Proteasome 20s activity assay kit (MAK172, Sigma Aldrich).
Quantification and statistical analysis
The data were evaluated using Student's t-test and were expressed as a mean ± SD. *P ≤ 0.05 was considered statistically significant, **P ≤ 0.01 very significant, ***P ≤ 0.001 highly significant. Statistical analyses were performed using GraphPad Prism version 6.01 for Windows, GraphPad Software, La Jolla, CA, USA (www.graphpad.com).
RESULTS
BPDE/B[a]P-induced DNA damage represses MMR and HR repair
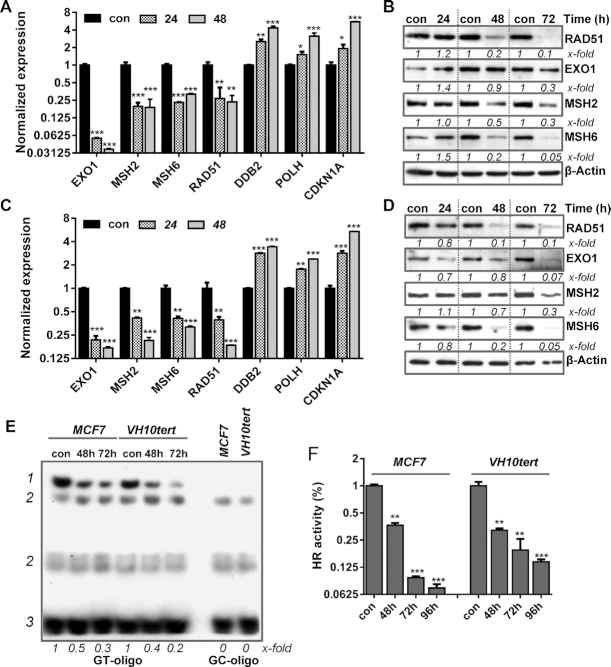
In this study, we utilized MCF7 cells, which are metabolically competent and able to metabolize B[a]P into BPDE. In contrast, VH10tert are not metabolically competent; they were used to verify that the mechanisms identified upon B[a]P exposure of MCF7 cells are caused by BPDE-adducts and not by other metabolites of B[a]P. In our previous work, we reported that B[a]P and the activated metabolite, BPDE, triggers upregulation of the NER system (2). To determine whether B[a]P/BPDE has an impact on other DNA repair pathways, we used qPCR-arrays. We identified several DNA repair genes, which were, however, not upregulated but transcriptionally repressed in MCF7 and VH10tert cells treated with low-dose B[a]P and BPDE, respectively. These downregulated genes encode the MMR proteins EXO1, MSH2, MSH6. RAD51, the main component of the HR, was also downregulated (Figure 1A, C). For verification, we analysed the corresponding proteins. In line with the decrease in mRNA expression, the protein levels of MSH2, MSH6, EXO1 and RAD51 were strongly reduced in response to BPDE/B[a]P-induced DNA damage (Figure 1B, D). To analyse whether the repression of EXO1, MSH2, MSH6 and RAD51 upon B[a]P/BPDE treatment has an effect on MMR, we determined the MMR activity using an electromobility shift assay (EMSA) based on the binding of the MSH2/MSH6 complex to a GT mispair incorporated into a radioactive-labeled oligonucleotide. Specificity of the reaction was shown by using MSH2 and MSH6 deficient cells, as well as CRISPR–Cas9 mediated knockout of MSH6 (Supplementary Figure S1A). A reduction in MSH2/MSH6 binding to the GT mispair was detected using cell lysates of B[a]P treated MCF7 and BPDE treated VH10tert cells (Figure 1E). In addition, analysis of the HR activity of these cells using a qPCR-based HR assay revealed a strong reduction in HR repair activity (Figure 1F). Taken together, our results indicate that B[a]P and BPDE treatment leads to transcriptional repression of MMR and RAD51 gene activity, which results in a functionally compromised MMR and HR repair.
Figure 1.
Time-dependent repression of DNA repair genes after B[a]P treatment in metabolically competent MCF7 breast cancer cells (A) and upon BPDE exposure in VH10tert cells (C) was measured by qPCR. Time-dependent repression of DNA repair proteins upon B[a]P treatment in MCF7 cells (B) and after BPDE exposure in VH10tert cells (D) was measured by immunoblotting. β-Actin was used as internal loading control, x-fold induction was measured densitometrically and is annotated under the respective blot. (E) Binding of MSH2/MSH6 to GT-mismatches was measured via EMSA. GC-oligonucleotides were included as specificity control; 1 = specific band, 2 = unspecific bands, 3 = unbound oligonucleotides, x-fold induction was measured densitometrically and is annotated under blot. (F) HR activity was analysed using a PCR-based HR activity assay. (A, C, F) Experiments were performed in triplicates and differences between treatment and control were statistically analyzed using Student's t test (non labeled = non significant, *P< 0.1 **P< 0.01, ***P< 0.001).
BPDE/B[a]P-induced repression of MMR and HR is caused by abrogated E2F1 signalling
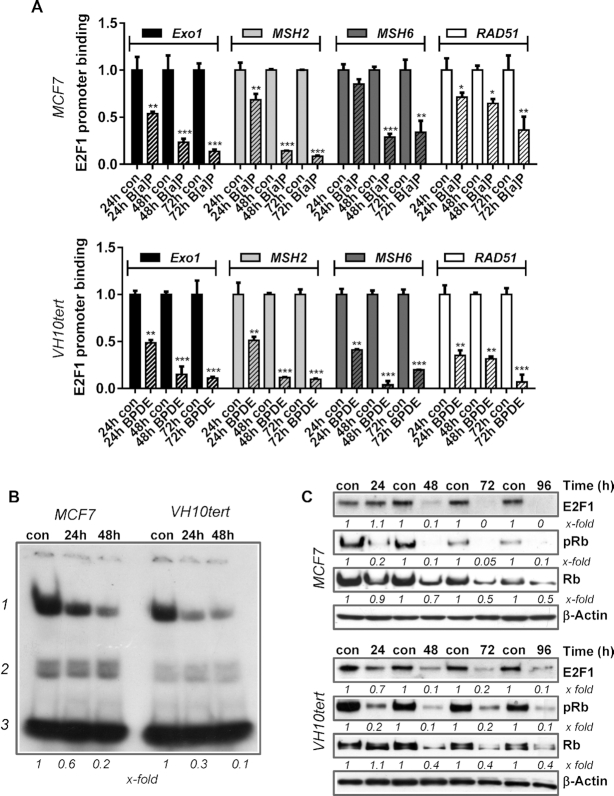
The E2F pathway regulates the replicative status of a cell and is regulated via pocket family proteins, which interact with E2F factors. The pocket protein family consists of p105/Rb, p107 and p130/Rbl2, while the E2F family is composed of several transcription factors, which can either mediate transcriptional activation or repression. The model claims that p130/Rbl2 binds the transcriptional repressor E2F4, whereas p105/Rb binds and thereby inactivates the transcriptional activators E2F1, E2F2 and E2F3 (39). Interestingly, ChIP-Seq and ChIP-on-Chip studies have suggested that EXO1, MSH2, MSH6 and RAD51 might be E2F1 and E2F4 target genes (40,41). Moreover, we showed that E2F1 regulates the expression of MSH2 and MSH6 (and resistance to methylating genotoxins) in mouse embryonic stem cells (42). Therefore, we analysed the binding of E2F1 to the promoters of MSH2, MSH6, EXO1 and RAD51 by ChIP and subsequently performed qPCR 24–72 h after exposure of MCF7 cells to B[a]P and VH10tert cells to BPDE (Figure 2A). Our results revealed strongly reduced binding of E2F1 to these promoters upon treatment. To further support the hypothesis that repression of MSH2, MSH6, EXO1 and RAD51 after B[a]P/BPDE exposure is caused by genotoxin-induced downregulation of E2F1, we silenced E2F1 by RNAi and performed MSH2 promoter assays. Furthermore, we analysed repression of these DNA repair factors upon siRNA mediated silencing of E2F1 in the absence and presence of B[a]P by qPCR (Supplementary Figure S2). The data clearly indicate that silencing of E2F1 causes a decrease in expression of MSH2, MSH6, EXO1 and RAD51 and also reduces MSH2 promoter activity in non-exposed cells. Upon B[a]P exposure, silencing of E2F1 did not further enhance downregulation of MSH2 promoter activity and DNA repair gene repression. Reduced promoter binding of E2F1 was associated with decreased binding of E2F1 to a consensus sequence containing oligonucleotide, as detected by EMSA (Figure 2B). The specificity of the reaction was shown by Supershift-EMSA as well as by siRNA-specific E2F1 silencing (Supplementary Figure S1B). To determine the mechanism leading to the decreased E2F1 binding, we monitored E2F1 and Rb expression levels upon B[a]P/BPDE exposure using immunoblotting. A strong reduction of E2F1 protein was observed following B[a]P/BPDE treatment (Figure 2C), which was paralleled by reduced Rb as well as pRb levels (Figure 2C). Overall, these data indicate that repression of EXO1, MSH2, MSH6 and RAD51 expression after B[a]P/BPDE exposure is caused by genotoxin-induced downregulation of E2F1.
Figure 2.
(A) Binding of E2F1 to the promoters of EXO1, MSH2, MSH6 and RAD51 was analysed by ChIP and subsequent qPCR 24, 48 and 72 h after B[a]P (1 μM) or BPDE (0.25 μM) treatment. The amount of the immunoprecipitated mRNA in the control was set to 1. Experiments were performed in triplicates and differences between treatment and control were statistically analyzed using Student's t test (non labelled = non significant, *P< 0.1 **P< 0.01, ***P< 0.001). (B) E2F1 activity was measured by EMSA 24 and 48 h upon B[a]P in MCF7 cells and upon BPDE in VH10tert cells; 1 = specific band, 2 = unspecific bands, 3 = unbound oligonucleotides, x-fold induction was measured densitometrically and is annotated under blot. (C) Time-dependent expression of E2F1, pRb and Rb was measured by immunoblotting in MCF7 and VH10tert cells upon B[a]P and BPDE treatment respectively. β-Actin was used as internal loading control.
Abrogation of the E2F1 pathway is independent of cell death
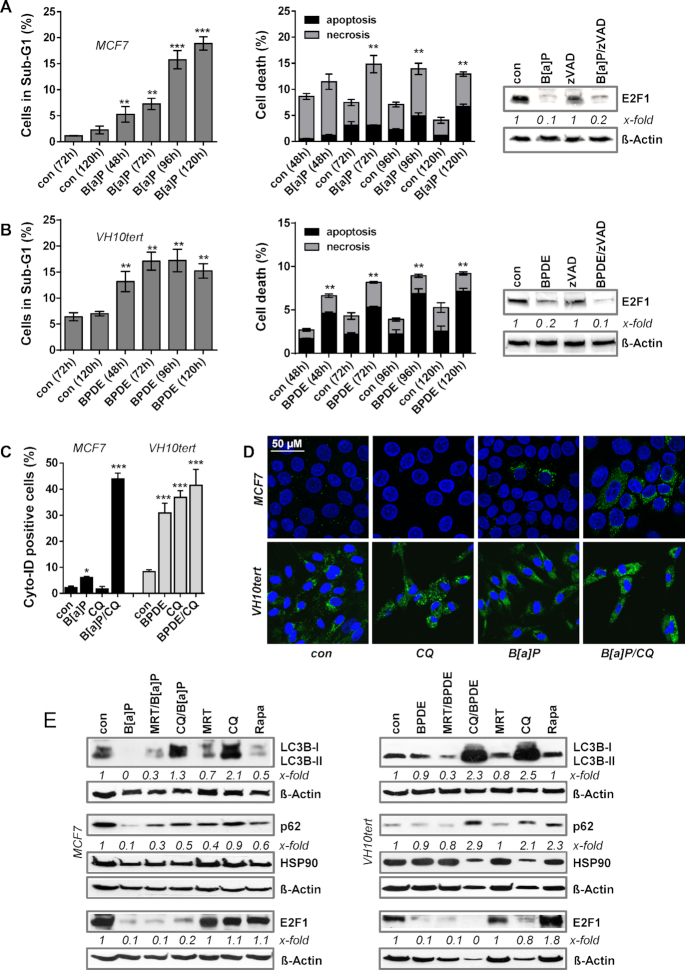
Next, we addressed the question of how B[a]P/BPDE exposure triggers abrogation of the E2F1 pathway. To rule out that the observed effects are unspecific, resulting from genotoxin-induced cytotoxicity, we analysed the sub-G1 fraction and annexin/PI positivity by flow cytometry. Our data revealed that exposure of MCF7 cells to B[a]P and VH10tert cells to BPDE in the applied doses induces cell death only in a minor fraction of the treated cells (Figure 3A, B, left panel). MCF7 cells showed <10% of sub-G1 at early times and up to 72 h after treatment, peaking after 120 h with 20% cell death and VH10tert cells showed ∼15% cell death 96 h after treatment. Using annexinV/PI staining, we observed the majority of MCF7 cells died by necrosis, whereas the majority of VH10tert cells died by apoptosis (Figure 3A/B, middle panel). This difference can be explained by the fact that MCF7 cells are deficient for caspase-3 (43). Moreover, a pan-caspase inhibitor (zVAD) did not affect the B(a)P/BPDE-triggered reduction in E2F1 expression, excluding caspase-dependent E2F1 degradation (Figure 3A, B, right panel). Overall, because of the low proportion of dead cells in the population (<10%) we conclude that the abrogation of the E2F1 pathway is independent of the cell death response.
Figure 3.
(A) MCF7 cells were treated with 1 μM B[a]P and (B) VH10tert cells were treated with 0.25 μM BPDE for different time points. Cell death was measured by flow cytometry using PI staining (left graph) and Annexin V/PI staining (middle graph). Expression of E2F1 upon B[a]P or BPDE exposure was measured by immunoblotting in the presence or absence of 50 μM zVAD (right graph). β-Actin was used as internal loading control, x-fold induction was measured densitometrically and is annotated under the respective blot. (C) Autophagy was measured in MCF7 and VH10tert cells upon exposure to B[a]P and BPDE, respectively, in the presence of chloroquine (CQ) using Cyto-ID staining. (D) MCF7 and VH10tert cells were treated with 1 μM B[a]P and 0.25 μM BPDE for 72 h, respectively, in the absence or presence of CQ. Autophagy was measured by microscopical detection of LC3B-I/II (green fluorescence). TO-PRO III (blue fluorescene) was used for nuclear staining; scale bars equate 50 μm. (E) MCF7 and VH10tert cells were treated for 72 h with 1 μM B[a]P or 0.25 μM BPDE, respectively, in the presence or absence of the autophagy inhibitors MRT and CQ, as well as of the autophagy inducer rapamycine (Rapa). LC3B-I/II, p62 and E2F1 were detected by immunoblotting. HSP90 and β-Actin were used as internal loading control, x-fold induction was measured densitometrically and is annotated under the respective blot. (A–C) Experiments were performed in triplicates and differences between treatment and control were statistically analyzed using Student's t test (non labelled = non significant, *P< 0.1 **P< 0.01, ***P< 0.001).
Downregulation of the E2F1 pathway is independent of autophagy
An important process for protein homeostasis is autophagy, which represents a digestion and recycling of proteins and cell organelles that are unneeded, dysfunctional and required as nutrient source under cellular stress conditions. During autophagy, target proteins are engulfed by autophagosomes, which fuse with lysosomes and thereby form autolysosomes, by which they get digested. In order to test whether B[a]P and BPDE induce autophagy, we studied the formation of autophagic vesicles. Initial flow cytometry-based measurements showed the formation of autophagosomes upon exposure of MCF7 cells to B[a]P and of VH10tert cells to BPDE (Supplementary Figure S3A). Thus, up to 10% of MCF7 and up to 50% of VH10tert cells were Cyto-ID positive upon exposure. Since Cyto-ID positivity is restricted to autophagosomes and is lost in autolysosomes, we inhibited the fusion between autophagosomes and lysosomes using the autophagy-inhibitor chloroquine (CQ). Adding CQ dramatically enhanced Cyto-ID-positivity in B[a]P-treated MCF7 cells to the same level observed in BPDE-treated VH10tert cells (Figure 3C), indicating that the autophagy pathway is activated in response to B[a]P and BPDE treatment. To support these findings, the expression of LC3B, which is part of the autophagosomes, was analysed by confocal microscopy, showing increased LC3B expression upon combined CQ/B[a]P exposure in MCF7 cells and upon CQ, BPDE and combined CQ/BPDE exposure in VH10tert cells (Figure 3D). We should note that already in non-exposed VH10tert cells, strong signs of autophagy were observed, hinting at the cell dependence on autophagy as energy source. This is also reflected by the toxicity of CQ, since treatment of VH10tert cells with CQ alone induced >20% of sub-G1 positive cells. Opposite, CQ was not toxic to MCF7 cells (Supplementary Figure S3B).
To investigate whether the autophagic response is involved in reducing E2F1 protein expression, we analysed the impact of the autophagy inhibitors CQ and MRT68921 (ULK1 inhibitor) as well as of the autophagy inducer rapamycin on E2F1 expression (Figure 3E). The efficiency of the inhibitor treatment was verified by analysing its effect on the expression of lipidated-LC3B-II, which is part of the autophagosomes, and of p62 expression. In both cell models, neither ULK1i- nor CQ-mediated inhibition of autophagy rescued the loss of E2F1. Beside CQ also MRT68921, which blocks rapamycin-dependent autophagy, did not impact the expression of E2F1. Finally, direct induction of autophagy by rapamycin had no impact on E2F1 expression (Figure 3E), although inducing Cyto-ID positivity (Supplementary Figure S3C). In summary, the data indicate that autophagy is not involved in B[a]P/BPDE-dependent abrogation of E2F1 signalling.
Downregulation of the E2F1 pathway depends on ubiquitin-mediated proteasomal degradation
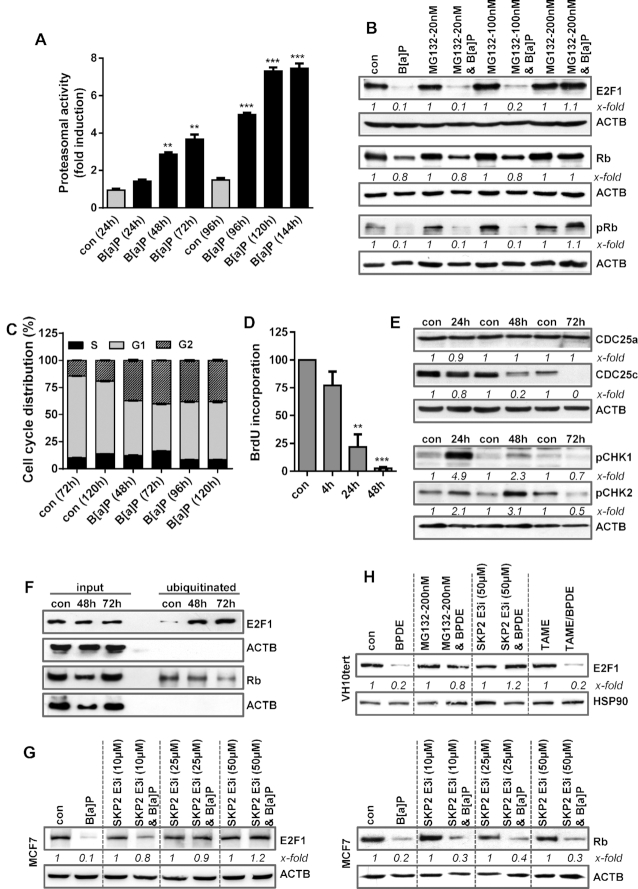
An important mechanism of controlling the E2F1 level is proteasome-dependent degradation (44). To analyse whether B[a]P affects the activity of the 20S proteasome, we measured its activity using a proteasomal activity kit. Treatment with B[a]P enhanced proteasomal activity time-dependently (Figure 4A). To directly address whether B[a]P induces proteasomal degradation of E2F1, MCF7 cells were treated with B[a]P in the presence or absence of the proteasome inhibitor MG132 (Figure 4B). Indeed, E2F1 protein expression was rescued upon efficient proteasome inhibition using 200 nM MG132 (Figure 4B and Supplementary Figure S4A). Similar to E2F1, also Rb protein expression was rescued upon proteasome inhibition (Figure 4B). The results indicate that B[a]P induces proteasome-dependent degradation of E2F1 and Rb. Besides rescuing the degradation of E2F1, proteasome inhibition also partially rescued the expression of MSH2, MSH6, EXO1 and RAD51 mRNA expression (Supplementary Figure S4B).
Figure 4.
(A) MCF7 cells were exposed to 1 μM B[a]P and proteasomal activity was measured. (B) MCF7 cells were treated with 1 μM B[a]P for 72 h in the presence or absence of the proteasomal inhibitor MG132. E2F1, Rb and pRb were detected by immunoblotting (C) Cell cycle distribution was measured in MCF7 cells using PI staining and flow cytometry 48–120 h after B[a]P exposure. (D) DNA synthesis was measured using BrdU incorporation 4–48 h after B[a]P exposure. (E) MCF7 cells were treated with 1 μM B[a]P. 24–72h later, the expression of CDC25a, CDC25c, pCHK1 and pCHK2 was detected by immunoblotting. (F) MCF7 cells were treated with 1 μM B[a]P in the presence of MG132. 48–72 h later, ubiqitinated proteins were isolated using the UBIQAPTURE-Q® kit and subjected to immuoblotting and the presence of E2F1 and Rb was detected. (G) MCF7 cells were treated with 1 μM B[a]P for 72 h in the presence or absence of a SKP2 inhibitor (SKP2 E3i). Expression of E2F1 and Rb was detected by immunoblotting. (H) VH10tert cells were treated with 0.25 μM BPDE for 72 h in the presence or absence of MG132, SKP2 E3i or TAME. E2F1 was detected by immunoblotting. (B, E–H) β-Actin was used as internal loading control. (A, C, D) Experiments were performed in triplicates and differences between treatment and control were statistically analyzed using Student's t test (non labelled = non-significant, *P< 0.1 **P< 0.01, ***P< 0.001). Concerning cell cycle distribution, at all time points a significant (*) decrease of cells in G1 and increase in the G2-phase was observed. (B, E, G, H) x-fold induction was measured densitometrically and is annotated under the respective blot.
E2F1 regulates cell cycle progression, particularly progression from the G1 to the S-phase (44). During the late S/G2-phase and early M-phase, the multi-E3 ubiquitin ligase complexes SCFskp2 and APC/c, respectively, mediate E2F1 poly-ubiquitination to target it for proteasome-dependent degradation, thereby allowing G2/M progression (44). To analyse the effects of B[a]P on cell cycle progression, cell cycle distribution was determined by flow cytometry. B[a]P arrested the cells almost equally in the G1 and the G2-phase (Figure 4C), which was associated with a complete block of replication 48 h post-treatment, as indicated by BrdU incorporation (Figure 4D). Cell cycle arrest upon B[a]P treatment was associated with phosphorylation of CHK1 and CHK2, as well as with degradation of CDC25c (Figure 4E).
To analyse ubiquitination of E2F1, we made use of the UBI-QAPTURE-Q® kit to purify poly-ubiquitinated proteins from cell lysates. To this end, MCF7 cells were treated with B[a]P in the presence of MG132 to inhibit proteasomal degradation of ubiquitinated proteins. Immunoblotting demonstrated an enrichment of E2F1 and Rb upon B[a]P exposure, substantiating for increased ubiquitination of both proteins (Figure 4F). Next, we used inhibitors against the SCF-Skp2 and APC/c complex and analysed their impact on E2F1 degradation. Upon SCF-Skp2 inhibition, the degradation of E2F1, but not of Rb, was rescued (Figure 4G), indicating that the SCF-Skp2 complex is involved in the ubiquitination of E2F1 upon DNA damage, targeting E2F1 for proteasomal degradation. In contrast, APC/c inhibition using TAME, even at high, toxic concentrations had no impact on E2F1 degradation (Supplementary Figure 4C). Similar to MCF7, in VH10tert cells the inhibitors MG132 and SKP2i, but not TAME, protected cells against E2F1 degradation (Figure 4H).
Downregulation of the E2F1 pathway is dependent on E2F4-mediated transcriptional repression
Since only a fraction of cells is arrested in the G2-phase upon B[a]P treatment, additional mechanisms are likely to be involved in the downregulation of the E2F1 pathway in the G1-phase. E2F1 is transcriptionally activated in G1 and reaches the highest expression in the S-phase. An important factor arresting cells in G1 is the cyclin dependent kinase inhibitor 1a (CDKN1A, p21), which inhibits cyclin D-CDK1 and cyclin D-CDK2 complexes thus inhibiting phosphorylation of Rb proteins. This leads to activation of p130-E2F4 and inactivation of p105-E2F1, which causes cell cycle arrest (45,46). Related to its role as CDK inhibitor, p21 is responsible for activation of DREAM, a multi-protein complex mediating transcriptional repression.
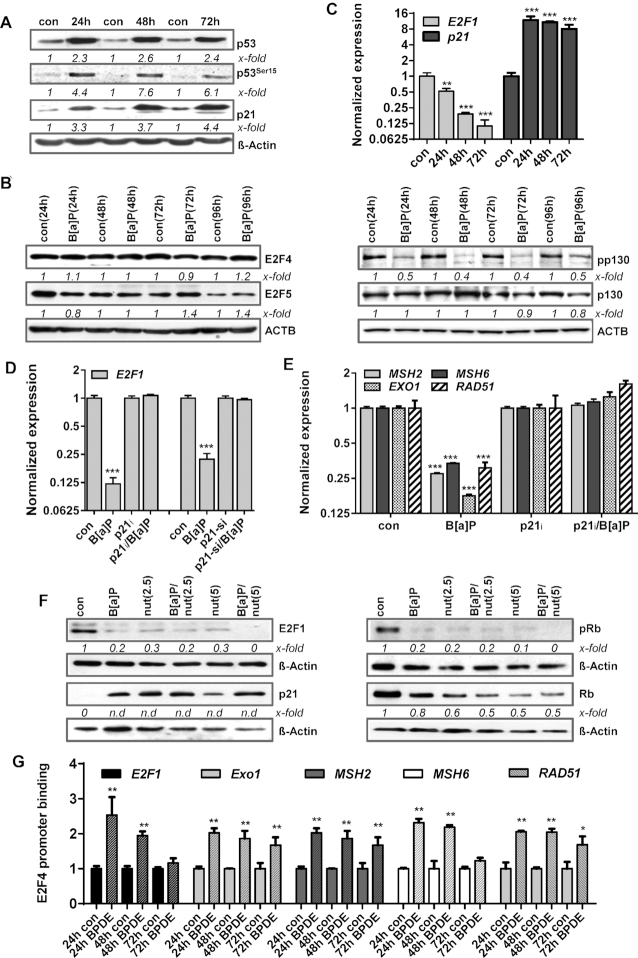
In line with the transcriptional activation of p21 upon B[a]P exposure (Figure 1A), we also observed increased p21 protein expression as part of the B[a]P-induced p53 response (Figure 5A). Analysing a potential DNA damage-induced activation of the DREAM complex, we next determined the expression of the DREAM complex components E2F4, E2F5, p130 and pp130. In contrast to E2F1, the expression of E2F4 and E2F5 was not altered upon B[a]P exposure (Figure 5B, left panel). However, the inactive, phosphorylated form of p130 was lost (Figure 5B, right panel), indicating activation of the DREAM complex following B[a]P treatment.
Figure 5.
(A/B) MCF7 cells were exposed to 1 μM B[a]P for 24–72h. (A) Expression of p53, p53Ser15 and p21 was detected by immunoblotting. (B) MCF7 cells were exposed to 1 μM B[a]P for 24–96 h. Expression of E2F4, E2F5, pp130 and p130 was detected by immunoblotting. (C) Expression of E2F1 and p21 was detected by qPCR. (D) MCF7 cells were exposed to 1 μM B[a]P for 48 h in the presence of the p21 inhibitor UC2288 (p21i) or p21 specific siRNA. Expression of E2F1 and p21 was detected by qPCR. (E) MCF7 cells were exposed to 1 μM B[a]P for 48 h in the presence of the p21 inhibitor UC2288 (p21i). Expression of MSH2, MSH6, EXO1 and RAD51 was detected by qPCR. (F) MCF7 cells were exposed to 1 μM B[a]P for 48 h in the presence or absence of the MDM2 inhibitor nutlin-3A (nut). Expression of E2F1, p21, pRb and Rb was detected by immunodetection. (G) Binding of E2F4 to the promoters of EXO1, MSH2, MSH6 and RAD51 was analysed by ChIP and subsequent qPCR 24–72 h after B[a]P treatment. The amount of the immunoprecipitated mRNA in the control was set to 1. (A, B, F) β-Actin was used as internal loading control, x-fold induction was measured densitometrically and is annotated under the respective blot. n.d: induction factor could not be detected due to missing signal in the control. (C, D, E, G) Experiments were performed in triplicates and differences between treatment and control were statistically analyzed using Student's t test (non labelled = non significant, *P< 0.1 **P< 0.01, ***P< 0.001).
One of the main targets of the DREAM complex is the E2F1 gene. Indeed, upon B[a]P exposure a strong transcriptional repression of E2F1 mRNA expression was observed (Figure 5C). To analyse whether this repression is caused by the p21-DREAM pathway, B[a]P-induced transcriptional repression of E2F1 was analysed upon pharmacological inhibition of p21 and by siRNA-mediated knockdown. Both strategies efficiently rescued the expression of E2F1 mRNA (Figure 5D). p21 inhibition also abolished E2F1 protein repression (Supplementary Figure S4D), further highlighting a critical role of p21. In addition, repression of the E2F1 target genes MSH2, MSH6, EXO1 and RAD51 was also abrogated upon inhibition of p21 (Figure 5E). Similar results were obtained in VH10tert cells upon exposure to BPDE (Supplementary Figure S4E).
Next, we analysed whether p21 induction is sufficient to abrogate the E2F1 pathway using the pharmacological MDM2 inhibitor nutlin-3a. Nutlin-3a binds to the N-terminus of MDM2 and thereby prevents from its interaction with the N-terminal transactivation domain of p53, activating the p53 response in the absence of DNA damage (53). As shown in Figure 5F, nutlin-3a strongly induced p21 expression and abrogated E2F1 and Rb/pRb expression to a similar extent as B[a]P, indicating the activation of p21 is sufficient to completely block E2F1 signalling. To further prove that E2F1, MSH2, MSH6, EXO1 and RAD51 are repressed by the DREAM complex upon B[a]P, we performed ChIP experiments (Figure 5G). The results clearly show an increased binding of E2F4 to the promoters of the analysed genes, indicating that B[a]P triggered repression of E2F1 and of the repair genes MSH2, MSH6, EXO1 and RAD51 is caused by activation of the DREAM complex. To further support this hypothesis, we silenced E2F4 using RNAi and performed MSH2 promoter assays (Supplementary Figure S2). Furthermore, we analyzed the downregulation of E2F1 and of the DNA repair factors upon siRNA mediated silencing of E2F4 in the absence and presence of B[a]P (Supplementary Figure S2). The data clearly indicate that silencing of E2F4 neither altered the expression of E2F1 and the DNA repair factors, nor affected MSH2 promoter activity in non-exposed cells. However, upon B[a]P treatment, silencing of E2F4 significantly prevented the reduction of MSH2 promoter activity and diminished repression of E2F1 and of the DNA repair genes.
Downregulation of the E2F1 pathway represents a senescence-associated mechanism
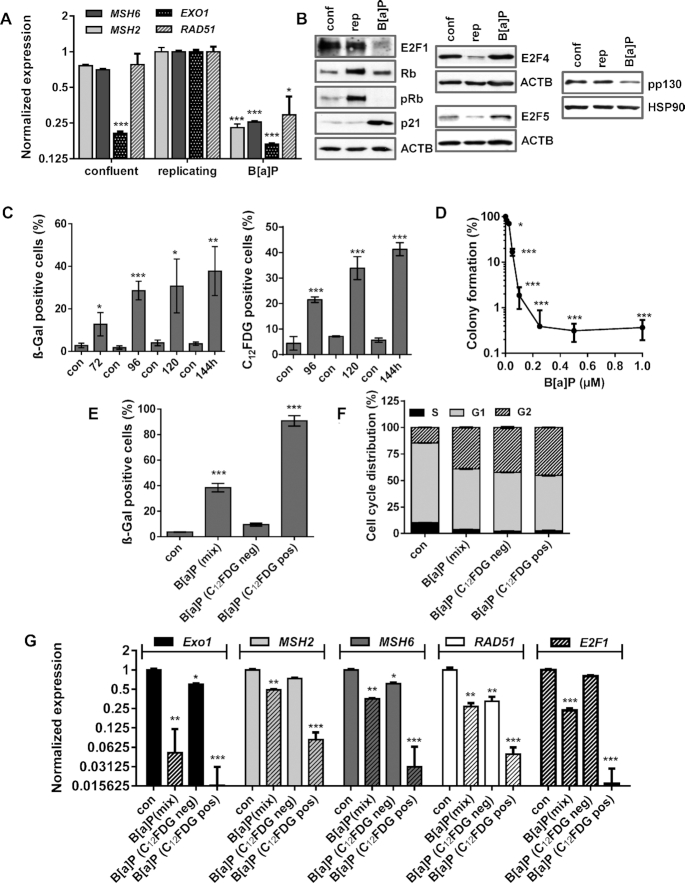
Our data indicate that MSH2, MSH6, EXO1 and RAD51 are repressed upon B[a]P exposure via alterations in the E2F signalling and suggest that two mechanisms are involved: transcriptional repression of E2F1 by the DREAM complex in G1 and proteasomal degradation of E2F1 in G2. Since repression of the DNA repair factors is mediated by the E2F system, which is also inactive in non-proliferating cells, we tested whether the observed gene repression is also caused by DNA damage independent cell cycle arrest. To this end, proliferation of MCF7 cells was blocked by confluency. Although these cells were arrested in the G1-phase (Supplementary Figure S4F), only mild repression of EXO1 and no repression of MSH2, MSH6 and RAD51 was observed (Figure 6A). In contrast, these factors were strongly repressed 72 h after B[a]P treatment (Figure 6A). In addition, reduced expression of E2F1 and activation of the DREAM complex (indicated by reduction in p130 phosphorylation) was exclusively observed in B[a]P-exposed cells. This is in line with our finding that p21 expression was induced upon DNA damage, but not in proliferation-arrested (quiescent) cells (Figure 6B).
Figure 6.
(A/B) MCF7 cells were kept in confluency for one week and replicating cells were either non-exposed or exposed to 1 μM B(a)P for 72 h. (A) Expression of MSH2, MSH6, EXO1 and RAD51 was detected by qPCR. (B) Expression of E2F1, Rb, p21, E2F4, E2F5 and pp130 was detected by immunoblotting. β-Actin and HSP90 were used as internal loading control. (C) Time-dependent induction of senescence upon B[a]P was measured by microscopical detection of ß-Gal positive cells (left panel) and FACS-based detection of C12FDG positive cells (right panel). (D) Clonogenic survival upon B[a]P was measured by the colony formation assay. (E–G) MCF7 cells were exposed to 1 μM B(a)P for 120 h. C12FDG positive and negative cells were separated by FACS. As comparison, non-exposed and B[a]P-exposed cells were included. (E) Frequency of senescence was determined by β-Gal staining in non-exposed cells (con), non-sorted B[a]P-exposed cells (mix), sorted C12FDG positive and negative cells. (F) Cell cycle distribution was measured using PI staining and flow cytometry. (G) Expression of EXO1, MSH2, MSH6 and RAD51 was measured by qPCR in non-exposed cells (con), non-sorted B[a]P-exposed cells (mix), sorted C12FDG positive and negative cells. (A, C–G) Experiments were performed in triplicates and differences between treatment and control were statistically analyzed using Student's t test (non labeled = non-significant, *P< 0.1 **P< 0.01, ***P< 0.001). (F) Concerning cell cycle distribution, in non-sorted B[a]P-exposed cells (mix), sorted C12FDG positive and negative cells a significant (**) decrease of cells in G1 and increase in the G2-phase was observed.
In contrast to quiescence, which is defined as a temporary cell cycle arrest, senescence is considered to be irreversible. To analyse whether B[a]P-induced proliferation arrest is associated with senescence, we measured the induction of senescence. Both microscopical detection of ß-Gal stained cells and flow cytometry-based detection of C12FDG-positive cells showed up to 40% senescent cells (Figure 6C). The irreversibility of the proliferation arrest was checked in colony formation assays, which indicated that only a very low proportion of these cells (<1%) were able to form colonies (Figure 6D).
To further analyse whether repression of E2F1 and the DNA repair factors is associated with B[a]P-induced senescence, senescent cells were separated 120 h after B[a]P treatment via C12FDG staining. As control, non-exposed and non-sorted B[a]P-exposed cells were included. The efficiency of the separation was verified microscopically using ß-Gal staining, showing >90% of ß-Gal positive cells in the senescent population and <10% in the non-senescent population (Figure 6E). Besides the strong difference in ß-Gal positivity, all B[a]P treated cells (mix, sen, non-sen) showed a similar cell cycle distribution, e.g. a redistribution from G1 into G2 (Figure 6F). The expression of EXO1, MSH2, MSH6 and RAD51 was measured by qPCR, showing that predominantly cells displaying the senescent phenotype repressed these repair genes (Figure 6G).
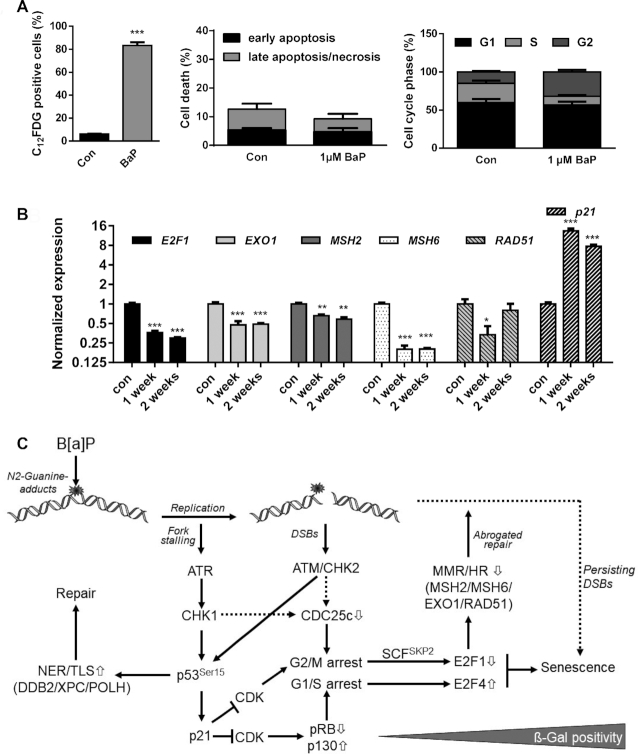
Notably, 120 h (5 days) after B[a]P exposure seems to be an early stage in the development of the senescence phenotype, reaching 30–40% ß-Gal positivity (see Figure 6C). Already 7 days after B[a]P treatment, the amount of senescent MCF7 cells further increased up to ∼80% (Figure 7A, left panel), whereas no signs of toxicity or alterations in the cell cycle distribution were observed (Figure 7A, middle and right panel). This indicates that C12FDG negative cells, which showed a reduced expression of the repair factors, might represent cells at an early state of senescence, preceding alteration in galactosidase activity. Repression of MSH2, MSH6, EXO1 and RAD51 as well as induction of p21 were still observed two weeks after B[a]P exposure (Figure 7B), indicating that this feature is maintained in senescent cells. Induction of senescence was also observed in MCF7 and VH10tert cells upon exposure to the activated B[a]P metabolite, BPDE (Supplementary Figure S5A, B).
Figure 7.
(A) MCF7 cells were exposed to 1 μM B[a]P for one week. Senescence was measured by C12FDG staining (left graph), cell death by Annexin V/PI staining (middle graph) and cell cycle distribution by PI staining (right graph). (B) MCF7 cells were exposed to 1 μM B[a]P for one or two weeks. Expression of E2F1, EXO1, MSH2, MSH6, RAD51 and p21 was detected by qPCR. (A, B) Experiments were performed in triplicates and differences between treatment and control were statistically analyzed using Student's t test (non-labelled = non significant, *P< 0.1 **P< 0.01, ***P< 0.001). (C) Model explaining the interaction between E2F1 signalling abrogation, transcriptional repression of DNA repair and induction of senescence. B[a]P induces genotoxic stress, which activates the DDR resulting either from replicative stress or the formation of DSBs. Activation of ATR, ATM, CHK1 and CHK2 leads to phosphorylation and activation of p53 and, subsequently, to enhanced expression of DNA repair genes DDB2, XPC and POLH and the CDK inhibitor p21. p21 blocks E2F1 activation and activates the DREAM complex, which arrests cells in G1/S and G2/M. In parallel, activation of the DDR triggers degradation of CDC25c, which contributes to the G2/M arrest. In G2-arrested cells, E2F1 is ubiquitinated and degraded by the proteasome, whereas in G1-arrested cells the DREAM complex blocks E2F1 transcription. The dual inhibition of the E2F1 signalling pathway represents the initial step in senescence induction and, in addition, supresses MSH2, MSH6, EXO1 and RAD51, which therefore represents an early mark for B[a]P-induced senescence. It is to suppose that the senescence-associated inhibition of DNA repair further blocks the repair of critical DNA lesions, and therefore might be involved in the maintenance of senescence.
Overall, our results demonstrate that B[a]P-induced DNA damage activates the p53/p21 pathway, which leads to downregulation of E2F1 and E2F1-mediated expression of MMR and HR components and concomitantly to the induction of senescence (Figure 7C).
DISCUSSION
Previously we showed that B[a]P activates DDB2, XPC and POLH, via the sequence-specific transcription factor p53 (2,47). Here, we report that upon B[a]P exposure of metabolically competent cells the MMR genes EXO1, MSH2, MSH6 and the HR gene RAD51 are transcriptionally repressed, which is accompanied by downregulation of the corresponding proteins and DNA repair activity. Of note, inhibition of MMR activity through downregulation of MSH6 upon B[a]P was previously reported for ZR75-1 breast cancer cells (48). We further show that downregulation of these DNA repair factors represents a specific consequence of senescence-accociated abrogation of E2F1 signalling and occurs both in the G1 and the G2-phase through different mechanisms.
DNA repair gene repression is caused by abrogated E2F1 signalling
ChIP experiments revealed that repression of these DNA repair genes was associated with reduced E2F1 binding to the promoters of the respective genes. For MSH2 and MSH6, the data are in line with the previous observations that E2F1 regulates MSH2 and MSH6 in rat cells (49) and that murine stem cells show a higher expression of MSH2 and MSH6 compared to differentiated cells due to increased E2F1 activity (42). Recently, we also observed a repression of these factors upon temozolomide (TMZ) treatment of glioblastoma cells (33). Whereas upon TMZ treatment downregulation of E2F1 signalling was caused by disruption of the E2F1/DP1 complex (33), a dramatic decrease in the E2F1 protein was observed upon B[a]P exposure pointing to drug-specific mechanisms of E2F1 regulation. This is in contrast to other reports showing E2F1 stabilisation in response to genotoxic stress (50–52). We should note, however, that in these studies toxic B[a]P concentrations were used, whereas the decrease in the E2F1 protein level reported here was observed upon sub-toxic (senescence-inducing) B[a]P concentrations. The reduced E2F1 expression was independent of cell death and autophagy.
B[a]P abrogates E2F1 signalling in the G2-phase by proteasomal degradation
During cell cycle progression, E2F1 is ubiquitinated by several E3 ligases leading to its degradation during cellular transition from S into G2 and from G2 into the M-phase, respectively. Thus, SCFskp2 is known to ubiquitinate E2F1 in the late S-phase (53) and the anaphase-promoting complex/cyclosome (APC/c) targets E2F1 during the early M-phase (prometaphase), either via the adaptor protein Cdh1 (54) or Cdc20 (55). Upon B[a]P exposure, enhanced ubiquitination and increased overall proteasomal activity was observed. Moreover B[a]P-induced loss of E2F1 was abrogated using a proteasomal or a SCFskp2 inhibitor. Inhibition of the APC/c complex had no impact on E2F1 degradation even at high concentrations. These data indicate that within the G2-phase, the abrogation of the E2F1 signalling and subsequent downregulation of DNA repair genes is caused by proteasomal degradation of E2F1.
B[a]P abrogates E2F1 signalling in the G1-phase by activation of the DREAM complex
Since B[a]P treatment caused DNA repair gene repression in G1 and G2, additional mechanisms must prohibit synthesis of E2F1 in G1 cells. Interestingly, p53 has been reported to be indirectly associated with transcriptional repression through activation of p21 and recruitment of the E2F4-associated repressor complex DREAM (56). More precisely, p21 inhibits CDK1 and CDK2 activity, thereby reducing phosphorylation of the Rb proteins. This leads to activation of p130-E2F4 and inactivation of Rb-E2F1, which inhibits cell cycle progression (45,46). Associated with its role as CDK inhibitor, p21 is responsible for the formation of the repressive DREAM complex (57). The core of the DREAM complex is formed by the MuvB complex, which itself consists of the MuvB-related proteins LIN9, LIN37, LIN52, LIN54 and RBBP4 (58). Throughout the cell cycle, several proteins bind to the MuvB core to increase or decrease transcriptional activity. Thus, B-MYB and FOXM1 binding can enhance the transcriptional activator function of the MuvB complex. In contrast, binding of p130 in its hypo-phosphorylated form together with E2F4 and DP1 turns the MuvB complex into the repressive DREAM complex. DREAM can bind to E2F consensus sequence in the DNA, which is supported by CHR-like element (CLE) in a distance of 4 bp from the E2F binding site, via the linked E2F4, thereby shielding the promoter from activating E2Fs (57,59).
E2F1, which is transcriptionally activated in G1, also represents a target of the DREAM complex. Upon B[a]P exposure, a strong transcriptional repression of E2F1 was associated with activation of the DREAM complex, as detected by hypo-phosphorylation of p130. Moreover, pharmacological inhibition and knockdown of p21 abrogated the transcriptional repression of E2F1, MSH2, MSH6, EXO1 and RAD51, suggesting that these repair genes are also targets of the DREAM complex, which is activated upon genotoxic stress. Indeed, ChIP experiments using E2F4 antibodies showed an increased binding after B[a]P exposure. This is in line with data revealing MSH2 and MSH6 as E2F4 targets (34,60). Besides MSH2 and MSH6, additional DNA repair factors have been described to be targeted by the DREAM complex, including the members of the Fanconi Anemia pathway (61,62). Indeed, also these genes were transcriptionally repressed upon B[a]P exposure (Supplementary Figure S5C), further indicating the importance of the DREAM complex in regulating DNA repair upon genotoxic stress.
DNA repair gene repression is an early event in DNA-damage induced senescence
Since the activity of the E2F1 pathway is strongly associated with cell cycle progression and distribution, one might speculate that the observed downregulation of DNA repair is a side-effect of proliferation inhibition and cell cycle redistribution. Several findings, however, contradict this interpretation. (i) At early (Supplementary Figure S6A) and late (Figures 4C, 7B) time points, a uniform decrease of cells in G1 and increase in the G2-phase was observed. Since MSH2 and RAD51 are reported to be higher expressed in S/G2 than in the G1-phase (63), the repression of these factors following genotoxin treatment is very unlikely to be caused by mere cell cycle redistribution. In this case, an increase in expression would be expected due to the lower G1 and increased G2 cell population. (ii) Analysing gene expression in untreated G1 and G2 cells showed only for MSH2 a higher expression in G2 and for E2F1 in G1 (Supplementary Figure S6B). Upon B[a]P exposure the expression of EXO1, MSH6, RAD51 and E2F1 was reduced in G1 and G2 to a similar level. Only for MSH2 a stronger repression was observed in G2 compared to G1, which was due to its higher basal expression in the G2-phase. Thus, our results show that repression of the repair factors occurs in G1 and G2. (iii) Cellular quiescence is not sufficient to silence MSH2, MSH6 or RAD51. Accordingly, non-proliferative, confluent MCF7 cells neither showed E2F1 degradation nor reduced pp130 expression. Furthermore, neither induction of p21 nor repression of MSH2, MSH6 or RAD51 was observed in confluent (contact inhibited) MCF7 cells. This indicates that the repression of DNA repair only occurs in cells upon genotoxic stress and not upon non-genotoxic stress-induced cell cycle arrest.
We further provide evidence that the repression of DNA repair is a consequence of genotoxin-induced senescence (for review on senescence see (64–67)). The p21-triggered, irreversible cell cycle arrest is essentially involved in blocking E2F1 signalling through induction of E2F1 degradation in G2 and activation of the DREAM complex in G1 and thereby represses MMR and HR. Similar results were obtained in glioma cells, in which only p53/p21 proficient cell lines were able to induce senescence and responded with repression of MSH2, MSH6, EXO1 and RAD51 upon treatment with TMZ (33). p53/p21 deficient cell lines showed neither senescence nor DNA repair repression, although they were strongly arrested in G2 following genotoxin treatment (33). Finally, separation of C12FDG positive and negative cells indicates, that upon B[a]P exposure, repression of DNA repair is much stronger in later phases of senescence than in cells that have not reached this state yet, although they show a similar cell cycle distribution.
Additional alterations in DNA repair gene expression
Besides B[a]P, also the active metabolite BPDE was able to induce senescence and repression of MSH2, MSH6, EXO1 and RAD51, which was observed in MCF7 and VH10tert cells (Supplementary Figure S3A/B) as well as primary bronchial epithelial cells (Supplementary Figure S7). Importantly, ionizing radiation also caused cellular senescence, which was preceded by repression of these repair factors (Supplementary Figure S8), indicating that this mechanism is independent of cell type and genotoxic stimuli. We should note that upon B[a]P and IR multiple crosslink repair-regulating FANC-components, which also represent DREAM targets, were also dysregulated (Supplementary Figures S5C, 8B). Interestingly, additional analyses showed clear differences between the different DNA repair pathways (Supplementary Figures S9–11). While the expression of NER factors was not dysregulated, additional MMR factors like MLH1, PMS1 and PMS2 were found to be strongly repressed. Concerning BER factors, among 17 tested components only ligase I and polymerase beta (PolB) showed strong repression upon treatment with B[a]P and IR. Furthermore, the different DNA glycosylases were not affected. Similar results were also observed for NHEJ, where only repression of Ku70 was observed. In the case of HR, additional genes like RAD51C, RAD54B and XRCC2 were strongly repressed beside RAD51. Overall, our results show that specifically ICL repair, HR and MMR are dysregulated during B[a]P-induced senescence.
Biological consequence of DNA repair repression in senescent cells
It is conceivable that the observed strong alterations in DNA repair activity influence the fate of senescent cells. Genotoxin-induced senescence is characterized by sustained activation of the DDR, implicating the presence of unrepaired DNA lesions (which are presumably DSBs). Upon B[a]P/BPDE treatment, a large fraction of cells is arrested in G2, in which HR acts as a main pathway for the repair of DSBs. Therefore, we speculate that downregulation of HR prevents the repair of critical DSBs, which causes a sustained trigger activating the DDR, thus maintaining the senescent state. Of note, B[a]P is highly carcinogenic (4,5). Carcinogenesis requires proliferation of exposed cells and expansion of initiated cells. Therefore, it is reasonable to conclude that induction of senescence protects from carcinogenesis. However, it has been shown that cells can escape senescence (68–70). In this case, the limited repair during the senescent phase may increase chromosomal instability (by deficient HR) and mutation frequency (by deficient MMR). Therefore, the cells evading or escaping senescence might be prone to undergo carcinogenesis at a later stage. To analyse the impact of compromised DNA repair on senescence maintenance and to analyse the impact of senescence-evading or senescence-escaping cells on carcinogenesis will be an important task for the future.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Birgit Rasenberger for excellent technical assistance. We acknowledge the Z-project (Project-ID 393547839 – SFB 1361, subproject Z01) for technical support concerning the detection of DNA repair activity. Support by IMB’s Flow Cytometry Core Facility is gratefully acknowledged.
Contributor Information
Sebastian Allmann, Institute of Toxicology, University Medical Center, Johannes Gutenberg University of Mainz, Obere Zahlbacher Str. 67, D-55131 Mainz, Germany.
Laura Mayer, Institute of Toxicology, University Medical Center, Johannes Gutenberg University of Mainz, Obere Zahlbacher Str. 67, D-55131 Mainz, Germany.
Jessika Olma, Institute of Toxicology, University Medical Center, Johannes Gutenberg University of Mainz, Obere Zahlbacher Str. 67, D-55131 Mainz, Germany.
Bernd Kaina, Institute of Toxicology, University Medical Center, Johannes Gutenberg University of Mainz, Obere Zahlbacher Str. 67, D-55131 Mainz, Germany.
Thomas G Hofmann, Institute of Toxicology, University Medical Center, Johannes Gutenberg University of Mainz, Obere Zahlbacher Str. 67, D-55131 Mainz, Germany.
Maja T Tomicic, Institute of Toxicology, University Medical Center, Johannes Gutenberg University of Mainz, Obere Zahlbacher Str. 67, D-55131 Mainz, Germany.
Markus Christmann, Institute of Toxicology, University Medical Center, Johannes Gutenberg University of Mainz, Obere Zahlbacher Str. 67, D-55131 Mainz, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) [393547839 – SFB 1361, subproject 05 and DFG CH 665/5-1 to M.C.]; Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) [393547839 – SFB 1361, subproject 19 to T.G.H.]; Intramural Research Funding of the University Medical Center Mainz (to M.T.T.). Funding for open access charge: Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) [393547839 – SFB 1361, central grant].
Conflict of interest statement. None declared.
REFERENCES
- 1. Christmann M., Kaina B.. Transcriptional regulation of human DNA repair genes following genotoxic stress: trigger mechanisms, inducible responses and genotoxic adaptation. Nucleic Acids Res. 2013; 41:8403–8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Christmann M., Boisseau C., Kitzinger R., Berac C., Allmann S., Sommer T., Aasland D., Kaina B., Tomicic M.T.. Adaptive upregulation of DNA repair genes following benzo(a)pyrene diol epoxide protects against cell death at the expense of mutations. Nucleic Acids Res. 2016; 44:10727–10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hattemer-Frey H.A., Travis C.C.. Benzo-a-pyrene: environmental partitioning and human exposure. Toxicol. Ind. Health. 1991; 7:141–157. [DOI] [PubMed] [Google Scholar]
- 4. Baan R., Grosse Y., Straif K., Secretan B., El Ghissassi F., Bouvard V., Benbrahim-Tallaa L., Guha N., Freeman C., Galichet L. et al.. A review of human carcinogens–Part F: chemical agents and related occupations. Lancet. Oncol. 2009; 10:1143–1144. [DOI] [PubMed] [Google Scholar]
- 5. Shimada T., Fujii-Kuriyama Y.. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004; 95:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Borgen A., Darvey H., Castagnoli N., Crocker T.T., Rasmussen R.E., Wang I.Y.. Metabolic conversion of benzo(a)pyrene by Syrian hamster liver microsomes and binding of metabolites to deoxyribonucleic acid. J. Med. Chem. 1973; 16:502–506. [DOI] [PubMed] [Google Scholar]
- 7. Huberman E., Sachs L., Yang S.K., Gelboin V.. Identification of mutagenic metabolites of benzo(a)pyrene in mammalian cells. PNAS. 1976; 73:607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Newbold R.F., Brookes P.. Exceptional mutagenicity of a benzo(a)pyrene diol epoxide in cultured mammalian cells. Nature. 1976; 261:52–54. [DOI] [PubMed] [Google Scholar]
- 9. Slaga T.J., Viaje A., Betty D.L., Brachen W., Buty S.G., Scribner J.D.. Skin tumor initiating ability of benzo(a)pyrene 4,5- 7,5- and 7,8-diol-9,10-epoxides and 7,8-diol. Cancer Lett. 1976; 2:115–121. [DOI] [PubMed] [Google Scholar]
- 10. Kim J.H., Stansbury K.H., Walker N.J., Trush M.A., Strickland P.T., Sutter T.R.. Metabolism of benzo[a]pyrene and benzo[a]pyrene-7,8-diol by human cytochrome P450 1B1. Carcinogenesis. 1998; 19:1847–1853. [DOI] [PubMed] [Google Scholar]
- 11. Yang S.K., McCourt D.W., Leutz J.C., Gelboin H.V.. Benzo[a]pyrene diol epoxides: mechanism of enzymatic formation and optically active intermediates. Science. 1977; 196:1199–1201. [DOI] [PubMed] [Google Scholar]
- 12. Phillips D.H., Hewer A., Seidel A., Steinbrecher T., Schrode R., Oesch F., Glatt H.. Relationship between mutagenicity and DNA adduct formation in mammalian cells for fjord- and bay-region diol-epoxides of polycyclic aromatic hydrocarbons. Chem. Biol. Interact. 1991; 80:177–186. [DOI] [PubMed] [Google Scholar]
- 13. Hockley S.L., Arlt V.M., Brewer D., Giddings I., Phillips D.H.. Time- and concentration-dependent changes in gene expression induced by benzo(a)pyrene in two human cell lines, MCF-7 and HepG2. BMC Genomics. 2006; 7:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Osborne M.R., Beland F.A., Harvey R.G., Brookes P.. The reaction of (+/-)-7alpha, 8beta-dihydroxy-9beta, 10beta-epoxy-7,8,9,10-tetrahydrobenzo(a)pyrene with DNA. Int. J. Cancer. 1976; 18:362–368. [DOI] [PubMed] [Google Scholar]
- 15. Hess M.T., Gunz D., Luneva N., Geacintov N.E., Naegeli H.. Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide-guanine adducts by human nucleotide excision repair enzymes. Mol. Cell. Biol. 1997; 17:7069–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dreij K., Seidel A., Jernstrom B.. Differential removal of DNA adducts derived from anti-diol epoxides of dibenzo[a,l]pyrene and benzo[a]pyrene in human cells. Chem. Res. Toxicol. 2005; 18:655–664. [DOI] [PubMed] [Google Scholar]
- 17. Gunz D., Hess M.T., Naegeli H.. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 1996; 271:25089–25098. [DOI] [PubMed] [Google Scholar]
- 18. Hoeijmakers J.H. DNA damage, aging, and cancer. N. Engl. J. Med. 2009; 361:1475–1485. [DOI] [PubMed] [Google Scholar]
- 19. Kress J.M., Dio L.D., Heck L., Pulliero A., Izzotti A., Laarmann K., Fritz G., Kaina B.. Human primary endothelial cells are impaired in nucleotide excision repair and sensitive to benzo[a]pyrene compared with smooth muscle cells and pericytes. Sci. Rep. 2019; 9:13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rechkoblit O., Zhang Y., Guo D., Wang Z., Amin S., Krzeminsky J., Louneva N., Geacintov N.E.. trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J. Biol. Chem. 2002; 277:30488–30494. [DOI] [PubMed] [Google Scholar]
- 21. Suzuki N., Ohashi E., Kolbanovskiy A., Geacintov N.E., Grollman A.P., Ohmori H., Shibutani S.. Translesion synthesis by human DNA polymerase kappa on a DNA template containing a single stereoisomer of dG-(+)- or dG-(-)-anti-N(2)-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene). Biochemistry. 2002; 41:6100–6106. [DOI] [PubMed] [Google Scholar]
- 22. Zhang Y., Yuan F., Wu X., Wang M., Rechkoblit O., Taylor J.S., Geacintov N.E., Wang Z.. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000; 28:4138–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y., Yuan F., Wu X., Rechkoblit O., Taylor J.S., Geacintov N.E., Wang Z.. Error-prone lesion bypass by human DNA polymerase eta. Nucleic Acids Res. 2000; 28:4717–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chiapperino D., Kroth H., Kramarczuk I.H., Sayer J.M., Masutani C., Hanaoka F., Jerina D.M., Cheh A.M.. Preferential misincorporation of purine nucleotides by human DNA polymerase eta opposite benzo[a]pyrene 7,8-diol 9,10-epoxide deoxyguanosine adducts. J. Biol. Chem. 2002; 277:11765–11771. [DOI] [PubMed] [Google Scholar]
- 25. Hockley S.L., Arlt V.M., Brewer D., Te Poele R., Workman P., Giddings I., Phillips D.H.. AHR- and DNA-damage-mediated gene expression responses induced by Benzo(a)pyrene in human cell lines. Chem. Res. Toxicol. 2007; 20:1797–1810. [DOI] [PubMed] [Google Scholar]
- 26. Hockley S.L., Arlt V.M., Jahnke G., Hartwig A., Giddings I., Phillips D.H.. Identification through microarray gene expression analysis of cellular responses to benzo(a)pyrene and its diol-epoxide that are dependent or independent of p53. Carcinogenesis. 2007; 29:202–210. [DOI] [PubMed] [Google Scholar]
- 27. Wang A., Gu J., Judson-Kremer K., Powell K.L., Mistry H., Simhambhatla P., Aldaz C.M., Gaddis S., MacLeod M.C.. Response of human mammary epithelial cells to DNA damage induced by BPDE: involvement of novel regulatory pathways. Carcinogenesis. 2003; 24:225–234. [DOI] [PubMed] [Google Scholar]
- 28. Mahadevan B., Keshava C., Musafia-Jeknic T., Pecaj A., Weston A., Baird W.M.. Altered gene expression patterns in MCF-7 cells induced by the urban dust particulate complex mixture standard reference material 1649a. Cancer Res. 2005; 65:1251–1258. [DOI] [PubMed] [Google Scholar]
- 29. Christmann M., Tomicic M.T., Roos W.P., Kaina B.. Mechanisms of human DNA repair: an update. Toxicology. 2003; 193:3–34. [DOI] [PubMed] [Google Scholar]
- 30. Tomicic M.T., Steigerwald C., Rasenberger B., Brozovic A., Christmann M.. Functional mismatch repair and inactive p53 drive sensitization of colorectal cancer cells to irinotecan via the IAP antagonist BV6. Arch. Toxicol. 2019; 93:2265–2277. [DOI] [PubMed] [Google Scholar]
- 31. Platt K., Oesch F.. Efficient synthesis of non-K-region trans-dihydro diols of polycyclic aromatic hydrocarbons from o-quinones and catechols. J. Org. Chem. 1983; 48:265–268. [Google Scholar]
- 32. Yagi H., Thakker D.R., Hernandez O., Koreeda M., Jerina D.M.. Synthesis and reactions of the highly mutagenic 7,8-diol 9,10-epoxides of the carcinogen benzo[a]pyrene. J. Am. Chem. Soc. 1977; 99:1604–1611. [DOI] [PubMed] [Google Scholar]
- 33. Aasland D., Gotzinger L., Hauck L., Berte N., Meyer J., Effenberger M., Schneider S., Reuber E.E., Roos W.P., Tomicic M.T et al.. Temozolomide induces senescence and repression of DNA repair pathways in glioblastoma cells via activation of ATR-CHK1, p21, and NF-kappaB. Cancer Res. 2019; 79:99–113. [DOI] [PubMed] [Google Scholar]
- 34. Fischer M., Grossmann P., Padi M., DeCaprio J.A.. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 2016; 44:6070–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dosch J., Christmann M., Kaina B.. Mismatch G-T binding activity and MSH2 expression is quantitatively related to sensitivity of cells to methylating agents. Carcinogenesis. 1998; 19:567–573. [DOI] [PubMed] [Google Scholar]
- 36. Christmann M., Kaina B.. Nuclear translocation of mismatch repair proteins MSH2 and MSH6 as a response of cells to alkylating agents. J. Biol. Chem. 2000; 275:36256–36262. [DOI] [PubMed] [Google Scholar]
- 37. Christmann M., Tomicic M.T., Kaina B.. Phosphorylation of mismatch repair proteins MSH2 and MSH6 affecting MutS{alpha} mismatch-binding activity. Nucl. Acids. Res. 2002; 30:1959–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Christmann M., Diesler K., Majhen D., Steigerwald C., Berte N., Freund H., Stojanovic N., Kaina B., Osmak M., Ambriovic-Ristov A. et al.. Integrin alphaVbeta3 silencing sensitizes malignant glioma cells to temozolomide by suppression of homologous recombination repair. Oncotarget. 2017; 8:27754–27771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takahashi Y., Rayman J.B., Dynlacht B.D.. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000; 14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 40. Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D.. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002; 16:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cam H., Balciunaite E., Blais A., Spektor A., Scarpulla R.C., Young R., Kluger Y., Dynlacht B.D.. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell. 2004; 16:399–411. [DOI] [PubMed] [Google Scholar]
- 42. Roos W.P., Christmann M., Fraser S.T., Kaina B.. Mouse embryonic stem cells are hypersensitive to apoptosis triggered by the DNA damage O(6)-methylguanine due to high E2F1 regulated mismatch repair. Cell Death Differ. 2007; 14:1422–1432. [DOI] [PubMed] [Google Scholar]
- 43. Janicke R.U., Sprengart M.L., Wati M.R., Porter A.G.. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J. Biol. Chem. 1998; 273:9357–9360. [DOI] [PubMed] [Google Scholar]
- 44. Dubrez L. Regulation of E2F1 transcription factor by ubiquitin conjugation. Int. J. Mol. Sci. 2017; 18:2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weinberg R.A. The retinoblastoma protein and cell cycle control. Cell. 1995; 81:323–330. [DOI] [PubMed] [Google Scholar]
- 46. Dimova D.K., Dyson N.J.. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005; 24:2810–2826. [DOI] [PubMed] [Google Scholar]
- 47. el-Deiry W.S., Kern S.E., Pietenpol J.A., Kinzler K.W., Vogelstein B.. Definition of a consensus binding site for p53. Nat. Genet. 1992; 1:45–49. [DOI] [PubMed] [Google Scholar]
- 48. Chen Y., Huang C., Bai C., Gao H., Ma R., Liu X., Dong Q.. Benzo[alpha]pyrene repressed DNA mismatch repair in human breast cancer cells. Toxicology. 2013; 304:167–172. [DOI] [PubMed] [Google Scholar]
- 49. Polager S., Kalma Y., Berkovich E., Ginsberg D.. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene. 2002; 21:437–446. [DOI] [PubMed] [Google Scholar]
- 50. Lin W.C., Lin F.T., Nevins J.R.. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001; 15:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- 51. Blattner C., Sparks A., Lane D.. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol. Cell. Biol. 1999; 19:3704–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pediconi N., Ianari A., Costanzo A., Belloni L., Gallo R., Cimino L., Porcellini A., Screpanti I., Balsano C., Alesse E. et al.. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat. Cell Biol. 2003; 5:552–558. [DOI] [PubMed] [Google Scholar]
- 53. Marti A., Wirbelauer C., Scheffner M., Krek W.. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1999; 1:14–19. [DOI] [PubMed] [Google Scholar]
- 54. Budhavarapu V.N., White E.D., Mahanic C.S., Chen L., Lin F.T., Lin W.C.. Regulation of E2F1 by APC/C Cdh1 via K11 linkage-specific ubiquitin chain formation. Cell Cycle. 2012; 11:2030–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Peart M.J., Poyurovsky M.V., Kass E.M., Urist M., Verschuren E.W., Summers M.K., Jackson P.K., Prives C.. APC/C(Cdc20) targets E2F1 for degradation in prometaphase. Cell Cycle. 2010; 9:3956–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Benson E.K., Mungamuri S.K., Attie O., Kracikova M., Sachidanandam R., Manfredi J.J., Aaronson S.A.. p53-dependent gene repression through p21 is mediated by recruitment of E2F4 repression complexes. Oncogene. 2014; 33:3959–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Litovchick L., Sadasivam S., Florens L., Zhu X., Swanson S.K., Velmurugan S., Chen R., Washburn M.P., Liu X.S., DeCaprio J.A.. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell. 2007; 26:539–551. [DOI] [PubMed] [Google Scholar]
- 58. Schmit F., Korenjak M., Mannefeld M., Schmitt K., Franke C., von Eyss B., Gagrica S., Hanel F., Brehm A., Gaubatz S.. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007; 6:1903–1913. [DOI] [PubMed] [Google Scholar]
- 59. Muller G.A., Stangner K., Schmitt T., Wintsche A., Engeland K.. Timing of transcription during the cell cycle: protein complexes binding to E2F, E2F/CLE, CDE/CHR, or CHR promoter elements define early and late cell cycle gene expression. Oncotarget. 2017; 8:97736–97748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fischer M., Quaas M., Steiner L., Engeland K.. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res. 2016; 44:164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fischer M., Steiner L., Engeland K.. The transcription factor p53: not a repressor, solely an activator. Cell Cycle. 2014; 13:3037–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Engeland K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018; 25:114–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mjelle R., Hegre S.A., Aas P.A., Slupphaug G., Drablos F., Saetrom P., Krokan H.E.. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair (Amst.). 2015; 30:53–67. [DOI] [PubMed] [Google Scholar]
- 64. d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer. 2008; 8:512–522. [DOI] [PubMed] [Google Scholar]
- 65. Muller M. Cellular senescence: molecular mechanisms, in vivo significance, and redox considerations. Antioxid. Redox Signal. 2009; 11:59–98. [DOI] [PubMed] [Google Scholar]
- 66. Fridman A.L., Tainsky M.A.. Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene. 2008; 27:5975–5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cichowski K., Hahn W.C.. Unexpected pieces to the senescence puzzle. Cell. 2008; 133:958–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Beausejour C.M., Krtolica A., Galimi F., Narita M., Lowe S.W., Yaswen P., Campisi J.. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003; 22:4212–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Michishita E., Nakabayashi K., Ogino H., Suzuki T., Fujii M., Ayusawa D.. DNA topoisomerase inhibitors induce reversible senescence in normal human fibroblasts. Biochem. Biophys. Res. Commun. 1998; 253:667–671. [DOI] [PubMed] [Google Scholar]
- 70. Chitikova Z.V., Gordeev S.A., Bykova T.V., Zubova S.G., Pospelov V.A., Pospelova T.V.. Sustained activation of DNA damage response in irradiated apoptosis-resistant cells induces reversible senescence associated with mTOR downregulation and expression of stem cell markers. Cell Cycle. 2014; 13:1424–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.