Abstract
Eukaryotic cells compartmentalize their internal milieu in order to achieve specific reactions in time and space. This organization in distinct compartments is essential to allow subcellular processing of regulatory signals and generate specific cellular responses. In the nucleus, genetic information is packaged in the form of chromatin, an organized and repeated nucleoprotein structure that is a source of epigenetic information. In addition, cells organize the distribution of macromolecules via various membrane-less nuclear organelles, which have gathered considerable attention in the last few years. The macromolecular multiprotein complexes known as Promyelocytic Leukemia Nuclear Bodies (PML NBs) are an archetype for nuclear membrane-less organelles. Chromatin interactions with nuclear bodies are important to regulate genome function. In this review, we will focus on the dynamic interplay between PML NBs and chromatin. We report how the structure and formation of PML NBs, which may involve phase separation mechanisms, might impact their functions in the regulation of chromatin dynamics. In particular, we will discuss how PML NBs participate in the chromatinization of viral genomes, as well as in the control of specific cellular chromatin assembly pathways which govern physiological mechanisms such as senescence or telomere maintenance.
INTRODUCTION
Eukaryotic cells package ∼2 m of DNA into a nucleus of a few micrometers diameter together with all the biological macromolecules required to organize, replicate, and interpret this genetic information. Mechanisms have thus evolved to organize this crowded environment. Our genetic material is packaged in a complex nucleoprotein structure called chromatin, whose basic unit, the nucleosome is composed of an octamer of histones comprising two copies of each core histone H2A, H2B, H3 and H4, around which is wrapped 147 bp of DNA (1). Targeted deposition of histone variants, or addition of specific post-translational modifications to histones and DNA provide a large repertoire of epigenetic information that can modulate chromatin accessibility and gene expression, and thus regulate cell identity (2). On the other hand, spatial and temporal distribution of macromolecules is organized through membrane-bound and membrane-less organelles which participate in the compartmentalization of biochemical reactions in the nucleus. Liquid–liquid phase separation (LLPS) has recently emerged as a new biophysical paradigm providing a mechanistical basis for membrane-less organelles assembly in a spontaneous manner (3–8). Upon specific biophysical conditions (pH, temperature, concentration, nature of the macromolecule, etc.), biological macromolecules concentrate in phase-separated liquid-like droplets, which coexist with a dilute phase, like oil drops in water. This process is energetically favorable and allows the formation of membrane-less compartments, called biomolecular condensates (3, 4).
Promyelocytic leukemia (PML) nuclear bodies (NBs) (also known as ND10) are an archetype of membrane-less organelles, that concentrate proteins at discrete sites within the nucleoplasm (9,10). They form a sphere of ∼0.1–1 μm in diameter and are present in the majority of mammalian cell nuclei (9). PML NBs were discovered through their disorganization in acute promyelocytic leukemia (APL). The PML gene was identified at the breakpoint of a common translocation t(15,17) resulting in a fusion protein with retinoic acid receptor alpha (RARα) that drives APL (11). The tumor-suppressor PML protein is the main organizer of PML NBs and forms a shell surrounding an inner core of dozens of proteins (12). PML NBs have been implicated in a wide range of biological processes such as senescence, antiviral defense or stemness. They may act both to concentrate components to facilitate biochemical reactions such as sumoylation or as storage compartments regulating protein availability in the nucleus.
In this review, we will discuss how the biophysical process of LLPS may participate in the multi-step biogenesis of PML NBs (3,6). We will consider the interplay between PML NBs and the regulation of chromatin dynamics in light of this new paradigm of phase separation and explore its various functional implications. We will provide some perspective on how the partitioning of various chromatin-related factors in the PML NBs might provide a means to fine-tune gene expression and chromatin plasticity.
STRUCTURE AND FORMATION OF PML NBs
Structure of PML and PML NBs
The structure of PML NBs has been extensively studied using both light and electron microscopy. There are typically 5–30 PML nuclear bodies per nucleus depending on the cell type, cell-cycle phase or physiological state (13). In immunofluorescence, they appear as nuclear dot-shaped spherical structures that reside in the interchromatin nuclear space (14). By electron or super-resolution light microscopy, it was observed that PML protein is concentrated in a ≈100 nm thick shell at the periphery of nuclear bodies, surrounding an inner core filled with dozens of factors (15,16). More than 170 proteins have been found to reside either constitutively or transiently in PML NBs (12). Among them, the nuclear antigen Sp100 was the first characterized protein to localize in these nuclear bodies (17) and is found together with PML in the periphery (16).
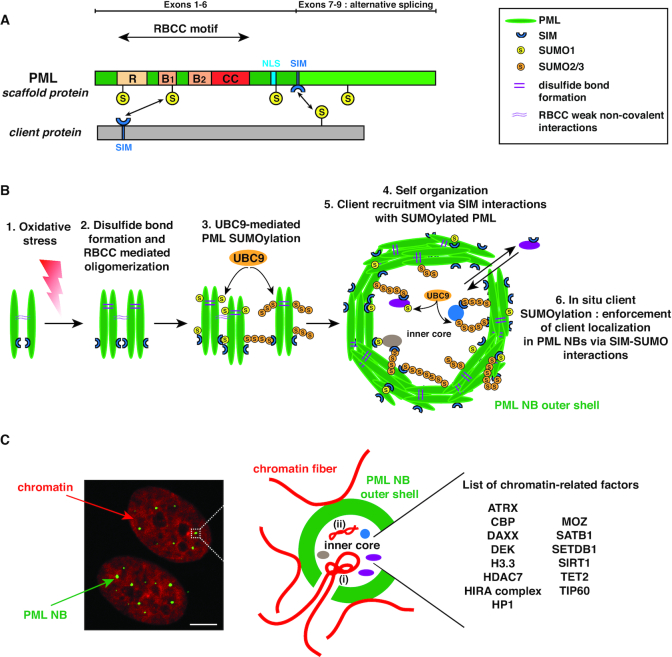
The PML protein (also known as TRIM19) is an essential component of PML NBs (Figure 1A). PML belongs to the family of tripartite motif (TRIM)-containing proteins characterized by a conserved RBCC motif consisting of a RING finger domain (R) followed by two cysteine-histidine-rich B-box domains (B) and an alpha-helical coiled-coil domain (CC). While alternative splicing of C-terminal exons generates seven different isoforms of PML, they all contain the conserved RBCC motif in their N-terminal part (13). All PML isoforms, except PML-VII, show a predominantly nuclear pattern due to the conservation of the nuclear localization signal (NLS) present in exon 6, but may also display specific functions that will not be discussed in the present study (for review (18)). PML protein sustains multiple post-translational modifications including SUMOylation, the covalent attachment of a small ubiquitin-like modifier (SUMO) protein to a protein-specific lysine residue (19). PML main SUMOylation sites are lysines K65, K160 and K490 (20), although SUMOylation of other lysines, like K616 has also been reported (21). PML also contains a SUMO interacting motif (SIM) at position aa 556–562 enabling it to interact with SUMOylated proteins (22) (Figure 1A). PML’s branched SUMO chains and SIM motifs may provide a ‘molecular glue’ to stabilize proteins within PML NBs (see below).
Figure 1.
Structure of PML and organization of PML NBs. (A) Structure of the PML protein scaffold. All PML isoforms (I–VII), ranging from 882aa (PML-I) to 435aa (PML-VII), possess a conserved RBCC/TRIM motif in their N-terminal part. The different C-terminal parts of PML-I to VI are generated through alternative splicing of the 3′ exons 7 to 9 of the unique PML gene, while PML-VII only possesses exons (1–4 and 7b). SUMO modification sites (S) are indicated at lysine positions K65, K160, K490 and K616. The NLS (Nuclear Localisation Sequence) and the SIM (SUMO Interacting Motif) are indicated. NB: PML structure is not to scale. (B) Formation of canonical PML NBs. (1) Oxidative stress triggers PML cross-linking by disulfide bond formation. (2) Together with RBCC weak non-covalent interactions, this triggers oligomerization of non SUMOylated PML proteins. (3) UBC9-mediated (poly-)SUMOylation of PML then allows multiple SUMO-SIM interactions, (4) which stabilize the formation of the self-organized matrix-associated outer shell, possibly involving liquid-liquid phase separation mechanisms. Of note SUMO1 modification (yellow) is mostly present in the PML NB outer shell, while the poly-SUMO2/3 chains (orange) present in the shell also protrude to variable degrees in the interior of the PML NB. (5) Client proteins are recruited in the outer shell (eg Sp100 not shown) as well as in the inner core through specific interactions of their SIM with the SUMOylated-PML scaffold. (6) UBC9 SUMOylation of client proteins then enforces their sequestration in PML NBs. Turnover of client proteins is relatively rapid ranging from seconds to a few minutes. (C) PML NBs are interspersed in the chromatin. (left) Immunofluorescence analysis of human primary BJ fibroblasts stained by PML (green) and DAPI (red). Scale bar is 10μm. (right) Scheme showing PML NBs (green) surrounded by chromatin loops (red). Cellular loci, such as telomeres, can localize partly within PML NBs in specific cases (i) (see main text). Chromatin-related factors (histone modifiers, histone readers and histone chaperones) as well as viral genomes (ii) localize inside PML NBs.
Formation of PML NBs and LLPS
While early models put forward an interaction between PML-conjugated SUMO and PML-SIM to nucleate NBs (22–24), recent analyses brought new insights on the multi-step formation of PML NBs and the recruitment of their protein constituents possibly through phase separation mechanisms.
PML, which is the main organizer of PML NBs, is a multivalent protein with multiple modular domains and interaction motifs, a key feature that can enable polymerization-driven liquid-liquid phase separation. PML is essential for the structural integrity of PML NBs (25) and is therefore referred to as a scaffold protein. Other proteins that permanently or transiently reside in PML NBs are called client proteins (6). The first phase of PML NB formation relies on intermolecular covalent disulfide linkage of oxidized PML monomers, as well as non-covalent interactions between the RBCC domains to drive assembly of PML multimers forming the primary nuclear body outer shell (24,26,27). These PML multimers are absent in MEF Pml–/– cells reconstituted with a PML mutated in the RBCC domain, emphasizing the importance of this domain for nuclear body formation (27). Recent crystallographic studies of PML RING and B1 domains put forward a cooperative mechanism in which a RING tetramerization step is followed by B1 polymerization of PML to allow macromolecular scaffolding of PML (28,29). On the contrary, PML SUMO-mutants (e.g. PML 3KR), or devoid of their SIM (PMLΔSIM) allow the formation of PML multimers and the formation of spherical PML NBs exactly like the WT structures when introduced into MEFs Pml–/– (27). This underscores the non-essential role of the PML SUMO-SIM interactions for the initial steps of PML NB formation. Of note, studies also point out the importance of C-terminal regions of specific PML isoforms in the oligomerization process of PML (30,31). The next step involves the recruitment of UBC9, the only SUMO E2-conjugating enzyme known so far, which is dependent on the RBCC oligomerization of PML (28). UBC9-mediated SUMOylation of PML then regulates and enforces PML-PML interactions via intermolecular SUMO-SIM interactions consistent with their importance to form mature PML NBs (22–24,27) (Figure 1B). In addition, SUMOylated PML drives the multivalent recruitement of inner core client proteins through their SIM, to form mature PML NBs (see below).
Membrane-less organelles biogenesis has recently been revisited through the prism of LLPS which stipulates that above a concentration threshold some proteins may phase separate and form liquid-like droplets with a distinct composition from the surrounding environment (3,8,32). Beautiful in vitro experiments demonstrated that mixes of polySUMO-polySIM polymers allow droplet formation that can recruit SUMO/SIM clients depending on the number of free sites remaining on the polymer (6). When transfected into cells, these polymers trigger the formation of condensates, that can be induced specifically at telomeres, and regulate partitioning of SUMO/SIM-containing clients (6,33,34). While PML is a multivalent protein with several identified SUMO/SIM interaction modules that could contribute to a possible phase separation of PML NBs, phase transition properties of the PML protein itself have yet to be demonstrated. In addition, this SUMO/SIM condensation process is not sufficient alone to explain the specific architecture of PML NBs, which exhibit a spherical shell formed by the oligomerized PML protein surrounding an inner core of client proteins (35). This dual phase architecture is rather unique among membrane-less organelles, and it remains to be determined to what extent the shell and the inner core present different solid-like versus liquid-like biophysical properties. Yet, the existence of multiphase biomolecular condensates, formed by LLPS such as the nucleolus (3,36) does not exclude the possible contribution of LLPS to PML NBs biogenesis. In particular, one hallmark feature of the LLPS model, ie, the concentration buffering/dependence, is validated by PML NBs (3,8,37). Size of the PML NBs scales up when increasing the concentration of PML as observed upon IFN-I treatment or senescence entry (38–42). On the contrary PML NBs are dissolved when artifically expanding the volume of the nucleus (43). In addition, PML NBs exhibit many other properties that meet the criteria defining LLPS-based structures, including a spherical shape, fusion/fission events in physiological or stress conditions, or high molecular mobility of internal components (Table 1). Nevertheless, many of this evidence remains qualitative and only provides indirect evidence for LLPS in vivo (32).
Table 1.
Summary of LLPS criteria that are matched or not by canonical PML NBs. In this table, we put forward the experimental evidence that sustains or not the involvement of LLPS in biogenesis of canonical PML NBs. Criteria listed here have been chosen based on the following reviews (4,8,32) and may not be all necessary/sufficient to prove LLPS. n.d. : non determined
| LLPS criterion | Criterion met by PML NBs | Experimental evidence | References |
|---|---|---|---|
| Spherical shape (roundness): liquid droplets have a spherical shape driven by surface tension | Yes | Super-resolution microscopy or transmission electron microscopy of PML NBs show sphericity of these nuclear bodies | (15,16,27,35) |
| Fusion/fission: like oil droplets in water, biomolecular condensates have the ability to fuse or drip | Yes | Time-lapse observations of PML NBs confirms their ability to undergo fusion/fission events during DNA replication or upon various stress conditions such as DNA damage, heat shock or physical pressure | (15,171–172) |
| Molecular mobility a: liquid condensates are characterized by a high mobility of proteins within them which is essentially depending on diffusion | Partially | FRAP experiments underlined fast recovery times for client proteins such as DAXX, CBP or BLM in the range of seconds, while PML isoforms exhibit slightly slower recovery times in the range of a few minutes compatible with the liquid-like nature of PML NBs. However, long recovery rates have been observed for specific isoforms such as PML V which may contribute to the structural integrity of nuclear bodies and could act as a stable scaffold for the recruitment of faster-exchanging molecules such as DAXX or CBP | (14–15,115) |
| Concentration buffering/dependence: LLPS is a function of concentration: past the critical concentration required for droplet formation, production of more protein increases droplet size but does not change concentration in either phase | Yes | Increase in PML intracellular concentration, as observed upon IFN-I treatment or senescence entry, results in an increased PML NBs size, while a decrease in PML protein concentration dissolves PML NBs in vivo | (38–43) |
| Interfacial boundary b: phase-separated proteins should preferentially move within the droplet. Presence of a phase boundary should reduce diffusion across the boundary | Partially | Diffusion coefficient for NLS-GFP was determined in nucleoplasm or in PML NBs by FCS. This demonstrated a 3-fold reduction in the diffusion coefficient inside the PML NBs as well as reduced exchanges of NLS-GFP between PML NBs and the nucleoplasm | (16,115) |
| Undergoes LLPS in vitro/in vivo | Partially | Not demonstrated for the PML protein itself. Yet, polySUMO-polySIM polymers form droplets in vitro and in vivo and recruit SUMO/SIM containing protein clients in vitro and in vivo | (6,33–34) |
| Temperature/ion strenght/pH dependance: measure of droplet formation in vivo should show dependance on temperature, ion concentration or pH | n.d. | n.d. | - |
| Sensitivity to 1,6-hexanediol: this chemical compound perturbs weak hydrophobic interactions that are involved in LLPS. Yet, sensitivity to 1,6 hexanediol is neither necessary nor sufficient to demonstate that a structure is formed by LLPS | n.d. | n.d. | - |
| Optodroplet assay: investigate whether expression of a fusion protein (protein of interest fused to a photolyase domain that can self-associate upon blue light) facilitates droplet formation in vivo upon blue light stimulation. Results should be interpreted with caution since these experiments rely on an artificial fusion protein system and should thus be combined with other experiments to prove LLPS in vivo | n.d. | n.d. | - |
aMolecular mobility is traditionally measured by Fluorescence Recovery After Photobleaching (FRAP). However, it should be noted that the use of recovery time as a marker of LLPS is insufficient per se since rapid recovery can result from a variety of mechanisms (32). One critical point is to demonstrate that the recovery rate is truely dominated by diffusion (rather than binding), which can be assessed by performing FRAP with various sizes of the bleach spot (32), which has not been performed yet in PML NBs.
bDiffusion across the boundary can be measured by fluorescence correlation spectroscopy (FCS) or single-molecule tracking (SMT). Alternatively, FRAP performed on half of the condensate, as performed in (45) provides an original and quantitative measure for the presence of a impermeable boundary, which could potentially be applied to PML NBs.
It is also important to discriminate between true LLPS and alternative mechanisms that could concentrate factors in a given place. In particular, it was recently shown that the transient non-specific binding of RNA polymerase II to the naked DNA of the herpes simplex virus 1 (HSV-1) genome during the lytic phase, leads to a viral DNA-mediated nuclear compartmentalization of replication foci through a mechanism distinct from LLPS (44), and rather driven by polymer-polymer phase separation (PPPS) (for review (37)). This chromatin bridging mechanism also explains the formation of heterochromatin foci that behave as collapsed chromatin globules (45), despite the fact that Heterochromatin Protein 1 (HP1) can undergo LLPS (46,47). As described above, several lines of evidence rather support a contribution of LLPS mechanisms for PML NBs biogenesis, independently of the chromatin polymer (Table 1). Yet, presence of DNA nucleation sites may help to recruit and concentrate PML proteins to reach the saturation concentration required for PML NBs droplet formation. PML NBs can be formed de novo at telomeric DNA and subsequently detach from them, suggesting that a nucleation site could mediate the formation of a subset of PML NBs (48). In addition, forced tethering of PML proteins to chromatin by the LacO/LacI or dCas9 systems induces PML NB formation at the targeted locus, suggesting that chromatin-bound PML proteins could be seeds for PML NB formation at specific loci by reaching the saturation concentration for condensate formation (45,49–51).
Many membrane-less organelles, such as nucleolus or Cajal bodies, are condensates that contain, in addition to proteins, RNA molecules which are critical for LLPS. PML NBs are found in regions of high transcriptional activity (14, 52) and early studies showed that nascent RNA could be found inside PML NBs in normal conditions (53) or upon interferon (IFN) α or IFNγ stimulation (54,55). In addition, it was recently shown that the long non-coding RNA (lncRNA) telomeric repeat-containing RNA (TERRA) is found within PML NBs of cells that activate a specific telomere maintenance pathway (56) (see below). Yet, it remains controversial to what extent bulk RNAs physically localize within PML NBs. Other publications showed that nascent RNAs are not enriched in PML NBs (57) but rather accumulate in their vicinity together with highly acetylated blocks of chromatin (14). In addition, brief transcriptional inhibition does not dramatically impact PML NBs structure (54,58), in contrast to nucleoli (59,60), and RNAs are not required per se for PML NBs biogenesis (6).
PML NBs thus appear as membrane-less organelles formed through a multi-step process that initially involves PML polymerization-driven shell formation followed by multivalent SUMO–SIM interactions of the PML scaffold and partners that could regulate liquid–liquid phase separation of PML NBs and their composition (see below), without any contributing RNA. We refer hereafter to these nuclear bodies as canonical PML NBs.
Alternative PML-containing structures
Remarkably, a continuum of PML-containing structures has been observed, in which the liquid-like properties of canonical PML NBs seem to be lost. After entry into mitosis, PML NBs undergo dramatic rearrangements (61), and partition in distinct larger aggregates of PML proteins called MAPPs (Mitotic Accumulation of PML Protein) (62) (for review (63)). MAPPs neither undergo rapid exchange of the PML protein, nor fusion/fission processes (62), that are essential criteria for LLPS (Table 1). PML undergoes an extensive de-SUMOylation preventing it from recruiting SIM-containing proteins such as the regular components of PML NBs, Sp100 or Death Domain-Associated Protein (DAXX) (61,62) (see below). In early G1, PML NBs, in the form of the so-called CyPNs (Cytoplasmic assemblies of PML and Nucleoporins), become associated with karyopherin KPNB1 which recruits FG-repeat-containing nucleoporins. The latter have the ability to form an hydrogel that encapsulates PML aggregates, which could facilitate their re-solubilization and nuclear import (64,65). Sequential recruitment of PML NBs components then occurs allowing reformation of mature canonical PML NBs (62,66). Other atypical PML-containing structures associated with nuclear lipid droplets were observed in cells after fatty acid stress (67,68). These so-called lipid-associated PML structures (LAPS) differ from canonical PML NBs as they lack SUMO1, Sp100 and DAXX proteins (68). PML only occupies part of the surface of the nuclear lipid droplets and is not required for their formation but is necessary for their functional maturation (68). In human embryonic stem cells (hESCs), PML-containing structures, also devoid of SUMO1, Sp100 or DAXX, show particular morphological types in the forms of long-linear rods or rosettes (69), which may be used as an indicator of the pluripotent state of the cells (70). Finally, our data characterizing PML NBs in sensory neurons within human trigeminal ganglia show the presence of large aggregates of PML, lacking SUMO1, Sp100 and DAXX (71) (and unpublished). Thus, self-association of the PML protein scaffold allows to form various alternative PML-containing structures that do not exhibit LLPS properties. We hypothesize that the presence of SUMO1, which can undergo LLPS in vitro (6), is key to promote formation of canonical PML NBs possibly via triggering LLPS and thus partitioning of regular client proteins such as DAXX or Sp100 in PML NBs.
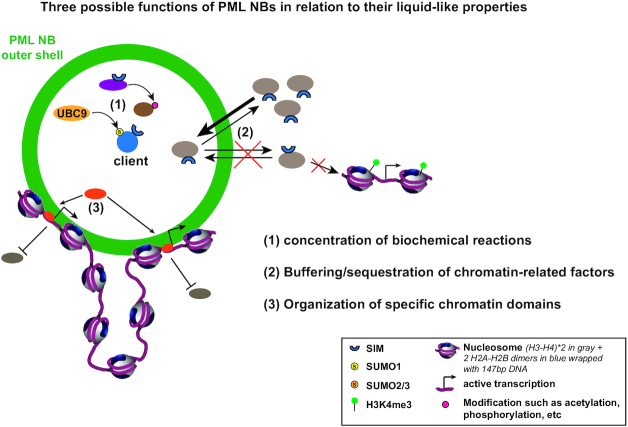
While canonical PML NBs appear as discrete foci interspersed between chromatin (Figure 1C), we will now focus on understanding the physical and functional connection of PML NBs with chromatin and investigate how the compartmentalization activity of PML NBs through phase separation mechanisms provides multiple strategies to regulate chromatin-related factors partitioning and chromatin dynamics. In particular, we can envisage three main non-exclusive processes that will be discussed: (1) PML NBs may be hotspots for modifications such as SUMOylation, potentially modulating the activity of chromatin-related factors; (2) PML NBs may store/sequestrate chromatin-related factors and control their dynamic release thereby fine-tuning the nucleoplasmic pool of a given factor and (3) PML NBs may help targeting chromatin-related factors to specific chromatin-associated regions by compartmentalizing them (Figure 2).
Figure 2.
Three possible functions of PML NBs in relation to their liquid-like properties. Liquid properties are advantageous for the cells by providing the ability of fast and easy rearrangements of macromolecules. Yet, the separation of the “liquid" nucleoplasm in several membrane-less condensates including PML NBs is essential to allow the formation of small reaction volumes with a different composition from the outside. Description of PML NBs as biomolecular condensates can illuminate the understanding of their function. We can envisage three important functions which may explain their roles in chromatin dynamics: (1) PML NBs may concentrate biochemical reactions. The biochemical environment within phase-separated PML NBs is different from the nucleoplasm and could serve to regulate (i) the kinetics of enzymatic reactions or (ii) the specificity of the modifications catalyzed. This is consistent with the described role of PML NBs as sumoylation hotspots, but could also apply for other modifications such as phosphorylation, acetylation, ubiquitination, or protein degradation. An example of the SUMOylation of a given client by UBC9 or of another client modification by a specific enzyme is shown. (2) PML NBs may buffer/sequester proteins via liquid-liquid phase separation of these client proteins. Increase in PML/client concentration may trigger accumulation of a given protein in PML NBs as a means to buffer the amount of the free protein in the nucleoplasm (as observed early for CBP for example). In addition, protein sequestration in PML NBs might affect their known activity as observed for DAXX. (3) PML NBs may help to organize specific nuclear domains, such as chromatin domains. PML NBs are interspersed in the active chromatin compartment and could potentially help to organize this compartment by pulling together genomic loci with similar transcriptional regulation. Of note, these three functions are not mutually exclusive and may serve altogether to regulate chromatin dynamics. Concentration of various factors in PML NBs together with specific genomic loci may help to catalyse specific reactions at given loci, as in the case of the ALT pathway for example (see Figure 3).
A CONNECTION OF PML NBs WITH CHROMATIN
PML NBs contain multiple chromatin associated proteins, including histones and histone chaperones
The idea of a role for PML NBs in the regulation of chromatin dynamics emerged via the identification of numerous chromatin-modifying factors within PML NBs, such as the CREB-Binding Protein (CBP), an histone acetyltransferase (HAT) involved in transcriptional regulation (53,72,73), or HP1 (74–77). HP1 is a key protein involved in heterochromatin formation, which interacts with Sp100, a constitutive component of PML NBs, and which localizes within these bodies in interphase as well as in senescent cells (74–78), suggesting very early on a connection of PML NBs with chromatin dynamics. Together with HP1, DAXX was identified as a constitutive PML NBs component (25,77) but it was not until its identification as a histone chaperone that the connection with chromatin dynamics was made (79). DAXX associates with the chromatin remodeler ATRX to form an H3.3-specific histone chaperone essential for H3.3 deposition at heterochromatin loci (79–81), and is required for ATRX localization in PML NBs (82–85) (Table 2).
Table 2.
List of histone chaperones, histone modifiers or histone readers localizing within PML NBs. Only proteins with known localization in PML NBs are listed, those that interact with PML, but whose localization in PML NBs has not yet been proven, have been omitted. Presence of validated SUMOylation sites or SIM motifs is indicated, putative sites/motifs identified by bioinformatic analysis or in SUMO screens are not shown. Positions refers to human proteins unless stated otherwise. While HP1 has been shown to be SUMOylated (236), it remains to be determined whether this SUMOylation controls its localization in PML NBs. The function related to the localization in PML NBs is also depicted. n.d. : non determined. hMSCs : human mesenchymal stem cells. MARs : Matrix attachment regions
| Protein | Protein function | SUMO | SIM | Recruitment | Function related to localization in PML NBs | References for PML NB localization | References for SUMO/SIM |
|---|---|---|---|---|---|---|---|
| ATRX | H3.3 histone chaperone | n.d. | n.d. | Constitutive, DAXX-dependent | Heterochromatin establishment | (82–83) | - |
| CBP | Histone acetyltransferase | n.d. | n.d. | Constitutive | Transcriptional regulation via p53 acetylation | (53,72–73) | - |
| DAXX | H3.3 histone chaperone | Multiple lysine residues | SIM1 IIVL (aa 7-10) and SIM2 IIVLSDSD (aa 733–740) | Constitutive | Transcriptional regulation, heterochromatin establishment, H3.3 recruitment in PML NBs, H3.3-dependent chromatin assembly | (25,61) | (118) |
| DEK | H3.3 histone chaperone | n.d. | AKRE (aa 260–263) (not validated by mutation) | Constitutive (hMSCs) | Maintenance of an H3.3 soluble pool available for recruitment in PML NBs | (101) | (101) |
| H3.3 | Histone H3 variant found in transcriptionally active regions and specific heterochromatic regions | n.d. | n.d. | Constitutive as well in senescence, DAXX-dependent | H3.3 soluble pool available for triage between histone chaperones | (84–85) | - |
| HDAC7 | Class IIA histone deacetylase | n.d. | n.d. | Constitutive in a subset of PML NBs, increased upon TNF-α | Transcriptional regulation (sequestration in PML NBs to relieve gene repression) | (108) | - |
| HIRA complex | H3.3 histone chaperone complex composed of HIRA, UBN1, CABIN1 and transiently ASF1A | n.d. | n.d. | Stress-induced (senescence, IFN, viral infection) | H3.3-dependent chromatin assembly in transcriptionally active regions, sequestration mechanism ? | (78,95–100) | - |
| HP1 | Heterochromatin protein 1 | K84 + alternative usage of various lysines residues | n.d. | Constitutive as well as in senescence | Heterochromatin establishment, in particular at cell-cycle genes during senescence | (75–78) | (236) |
| MOZ (KAT6A) | Histone acetyltransferase | n.d. | n.d. | Stress-induced (DNA damage, senescence) | Transcriptional regulation via p53 acetylation | (112) | - |
| SATB1 | Chromatin organizer by anchoring of MARs to the nuclear matrix, transcriptional regulator | K744 | n.d. | Constitutive in a subset of PML NBs, SUMO-dependent | Transcriptional regulation in immune cells, regulation of SATB1 levels by caspase-induced cleavage | (143,182) | (182) |
| SETDB1 | Histone H3K9 trimethyltransferase | n.d. | IIEI (aa 125–129) | Constitutive | H3K9me3 heterochromatin establishment at specific loci (such as Id2 gene) + maintenance of PML NBs | (102) | (122) |
| SIRT1 | NAD-dependent histone deacetylase | n.d. | n.d. | Stress-induced (PML-IV overexpression, senescence) | Deacetylation of p53 leading to repression of p53-mediated transactivation | (109) | - |
| TET2 | Oxidation of 5mC to promote DNA demethylation | n.d. | n.d. | Chemotherapy-induced, dependent on PML C-Terminus | Chemotherapy-induced demethylation of specific genes | (113) | - |
| TIP60 (KAT5) | Histone acetyltransferase | K430 and K451 | n.d. | UV-induced, SUMO-dependent, PML3-dependent | UV-induced DNA damage response (p53 recruitment in PML NBs and stabilization), SUMOylation promotes HAT activity, regulation of KAT5A stability | (110–111) | (110) |
Histone chaperones are dedicated proteins, which associate with non-nucleosomal histones and escort them throughout their cellular life in processes ranging from nuclear import, storage, assembly/disassembly onto chromatin during several DNA metabolic processes (86). Histone chaperones can be distinguished on the basis of their histone binding selectivity with a preference for H2A–H2B or H3–H4 histones and with additional selectivity towards specific histone variants. The replicative histone variants represent the bulk of histones and are expressed in S-phase, while replacement variants are expressed constitutively at lower levels. Among the H3-H4 histone chaperones, the CAF-1 complex is involved in the specific deposition of the H3.1 replicative histone variant in a DNA-synthesis dependent manner, while HIRA and DAXX-ATRX are H3.3 specific histone chaperones complexes implicated in H3.3–H4 deposition in a DNA-synthesis independent manner (for review (86)). H3.3 deposition was initially identified as characteristic of euchromatic transcriptionally active regions with high histone turnover (87–89). HIRA interacts with RNA polymerase II (90), specific transcription factors (TFs) (91) or replication protein A (RPA) found in nucleosome-free regions (92), thus mediating H3.3 deposition at active regulatory elements such as enhancers, promoters or gene bodies (80,92). Although unexpected, H3.3 was later found enriched in heterochromatin loci such as telomeres or pericentromeric chromatin where it is deposited by the DAXX-ATRX complex (79–81). This reflects the double face of H3.3 histone variant in gene regulation, which is context-dependent (for review (86,93,94)).
Interestingly, PML NBs seem to have a strong connection with the H3.3 chromatin assembly pathway. In addition to the constitutive localization of the DAXX–ATRX complex in PML NBs, the HIRA H3.3-specific histone chaperone complex, composed of HIRA, UBN1, CABIN-1 and transiently ASF1A, also localizes in PML NBs upon senescence entry (78,95–97) and upon viral infection-associated type I interferon (IFN-I) signaling (98–100) (see below). Furthermore, soluble newly-synthesized H3.3-H4 histone dimers are brought to PML NBs in a DAXX-dependent manner before deposition onto chromatin (84,85), thus suggesting that PML NBs may be important regulatory sites for the sorting of H3.3 among various histone chaperones complexes and further incorporation of H3.3 onto chromatin. Of note, another H3.3 specific chaperone called DEK also localizes in PML NBs in stem cells, and may participate in the maintenance of an H3.3 soluble pool available for association with other chaperones in PML NBs (101) (Table 2 and see below).
Finally, SETDB1, an histone H3 lysine 9 (H3K9) specific methyltransferase also localizes constitutively in PML NBs, which may be related to the transcriptional repression of specific genes (102). Given the connection of SETDB1 with DAXX–ATRX in the heterochromatinization of retroelements in mouse ES cells (103,104), further studies will be required to determine the function of the SETDB1 pool in PML NBs in regards to its function in heterochromatin maintenance. Other H3K9 methyltransferases, such as SUV39H1 and G9a, or EZH2, an H3K27 methyltransferase member of the Polycomb Repressive Complex 2 (PRC2), associate with PML but it remains to be shown whether they actually localize in PML NBs (105–107). Interestingly, other histone modifiers, such as TIP60 (KAT5) and MOZ (KAT6A) histone acetyltransferases, histone deacetylase 7 (HDAC7), or SIRT1, partition in PML NBs to regulate chromatin dynamics and transcriptional regulation (108–112). Finally, PML NBs are also associated with DNA demethylation activities through the recruitment of the ten-eleven translocation dioxygenase 2 enzyme (TET2) in response to chemotherapy and exclusion of DNA methyltransferase 3A (DNMT3A) (113,114) (Table 2).
Regulation of the composition of PML NBs in chromatin-related factors
How do cells regulate the composition of PML NBs and what molecular mechanisms govern the recruitment of the chromatin-associated factors in PML NBs? Indeed, while PML NBs are macroscopically stable and persist for hours/days, photobleaching recovery experiments showed that they are highly dynamic at the molecular level, turning over their contents on time-scales from seconds to minutes (15,115). In light of the LLPS paradigm, we will now explore how the partitioning of various histone chaperones in the membrane-less PML NBs is regulated. The number of interaction modules (valency) and their affinity are key parameters controlling phase separation and could thus enable compositional control of PML NBs (6). In particular, changes in concentration or specific post-translational modifications (e.g. SUMOylation) of the PML scaffold or of a client protein, modify the valency of free sites available, and thus the affinity between interacting modules regulating PML NB composition (6). SUMOylation of PML is not required for PML NB formation but is essential for the recruitment of partners containing one or several SIM motifs (23–25,27,116,117). Indeed, DAXX possesses 1 SIM motif, I733IVLSDSD740, at its C-terminus which is both critical for its localisation in PML NBs as shown by deletion experiments, and sufficient for the localization of a GFP-DAXXSIM fusion protein in PML NBs (27,118). Interestingly, DAXX can also be SUMOylated (118–121), but the forced fusion of SUMO1 or SUMO2 with DAXX is insufficient to rescue the DAXXΔSIM localization in PML condensates. Hence, in normal cell conditions, the presence of a SIM with affinity for SUMOylated PML is necessary and sufficient for a constitutive localization of DAXX in PML NBs. SUMOylation of DAXX by UBC9 present in the PML NBs then enforces its sequestration within the condensates by intermolecular interactions with PML SIM (27).
Interestingly, bioinformatics analysis of chromatin associated factors such as DEK or SETDB1, which can localize in PML NBs, identified putative SIM in these proteins with SETDB1’s SIM being crucial for its interaction with SUMOylated proteins (101,122,123). It is tempting to speculate that these SIM motifs could be implicated in their recruitment to PML NBs, yet further studies will need to confirm the exact sequence requirements. Overexpression of the PML protein itself or increase in PML SUMOylation (eg following IFN treatment (38–40)) increases the number of available SUMO groups on PML (multivalency), which can trigger the switch-like recruitment of client proteins such as CBP (6,124). On the contrary, ectopic overexpression of client proteins, such as HIRA, UBN1 or CBP, leads to their recruitment to PML NBs, which may result from an increased valency of the client upon higher concentration and thus suggests a buffering mechanism for excess nucleoplasmic protein (95,124,125) (as represented in Figure 2 item (2)). Thus, caution should be taken when concluding on the localization of a given protein only based on the overexpression of an ectopic form. SETDB1 as well as HIRA also possess putative SUMO sites and have experimentally been found in screens for SUMOylated proteins (119–121,126). Whether these sites are essential to enforce their localization in PML NBs or regulate their turnover as observed for Sp100 (115) remains to be tested.
Changes in affinity may also be regulated by post-translational modifications of the scaffold (PML) or of the client proteins. Phosphorylation adjacent to the SIM motifs, as observed for the phosphoSIMs of PML and DAXX, leads to an increased affinity towards SUMO1 via interaction with specific SUMO1 lysine residues (127–130). On the contrary, acetylation of SUMO1 decreases the affinity for SIM, as observed for DAXX which then looses its localization in PML NBs (131), and thus participates in the regulation of client partitioning into biomolecular condensates. Of note, acetylation of SUMO1 at key lysine residues alters binding to the phosphoSIMs of PML or DAXX showing the structural plasticity of SUMO-SIM interactions that can be controlled by residue-specific post-translational modifications (132). Phosphorylation of HIRA by glycogen synthase kinase 3β (GSK-3β) has been proposed to regulate its localization in PML NBs upon senescence (133), but does not seem to play a role in IFN-mediated relocalization of HIRA in PML NBs (100). Recently, a large RNAi screen also identified Homeodomain-Interacting Protein Kinases (HIPK1 & 2) as important regulators of PML NBs composition. Overexpression of these proteins led to a decreased accumulation of Sp100 in PML NBs, but not of PML itself. This suggests a role of HIPK1 & 2 in controlling the condensation of proteins in PML NBs by phosphorylation (134). The use of kinase-dead enzymes should rule out a possible titration effect where HIPK1 & 2 overexpression could saturate SUMO sites via binding of their SIM (135), hence displacing Sp100.
Thus, PML NBs contain multiple chromatin-associated factors whose localization is regulated by a switch-like partionning between the diffuse and the condensed phase controlled by the multivalency of PML and of the client protein itself. We will now discuss the possible physical contacts of PML NBs with chromatin.
A physical connection of PML NBs with chromatin
The use of an analytic electron microscopic method called Electron Spectroscopic Imaging (ESI) was instrumental in precisely determining the nucleic acid-based regions and protein-based regions within and around PML NBs (14). Boisvert and colleagues demonstrated that the PML NB core is a protein-based structure and that PML NBs are devoid of nucleic acids in normal conditions. Yet, nascent RNA, as well as highly acetylated blocks of chromatin were found to accumulate in the vicinity of PML NBs suggesting an association of these nuclear bodies with transcriptionally active chromatin (14) (see below). Further ESI studies demonstrated that the protein cores of PML NBs are surrounded by chromatin fibers and make direct physical contact with them, allowing the positional stability of PML NBs (58). PML NBs are also found adjacent to replication foci labelled by BrdU in middle-late S-phase cells (57).
While PML NBs physically contact chromatin, additional studies explored their associations with specific regions of the genome. The use of immuno-DNA FISH to combine immunolocalization of PML NBs with localization of specific genomic loci provided convincing data to demonstrate the specific association of PML NBs with cellular chromosomal loci. Using this approach, Shiels et al. demonstrated for the first time a non-random association of a specific locus, the major histocompatibility complex (MHC) on chromosome 6, with PML NBs in human primary fibroblasts (136), which was consistent with the role of PML in the upregulation of MHC specific genes (137). The association of this gene-rich locus with PML NBs was neither dependent on transcription nor on cell cycle phase, and could be observed when the locus was placed on chromosome 18. Further immuno-DNA FISH studies showed a specific association of PML NBs with the TP53 gene locus, but not BCL2 in jurkat cells (138) as well as a more general association of PML NBs with regions of high transcriptional activity (52). Of note, the association of PML NBs with specific loci might be cell-cycle specific since association of the histone gene cluster was increased in S-phase when canonical histones genes are transcribed (52). Similarly, it was found that PML NBs are preferentially juxtaposed to centromeres during G2-phase (77). Juxtaposition of genomic loci to PML NBs may be a means to regulate specific gene expression (see below). PML NBs show significant association with the Oct3/4 locus in ESCs, with a decrease upon differentiation in Neural Precursor Cells (NPCs), correlating with the decrease in Oct3/4 expression (139). In addition, Salsman et al. showed that PML NBs are not only juxtaposed to the DDTI4 gene locus, but they are also closely associated with the DDTI4 RNA transcriptional foci, as shown by immuno-RNA FISH, and consistent with the decreased expression of DDIT4 upon PML loss (140).
Interestingly, IFNγ increases the spatial proximity between PML NBs and the MHC class II gene cluster and PML is required for the IFNγ-induced MHC class II gene transcription (141). In particular, the association of a gene from this locus, the DRA gene, with PML NBs is maintained after IFNγ shut-off and is required to keep a prolonged permissive chromatin state on the DRA promoter (142). This underlines the importance of the PML NBs spatial proximity with specific loci to mediate epigenetic memory of a stimulus through cell divisions to increase responsiveness of gene expression to future activation signals (142). The connection of PML with the MHC locus was further substantiated by genomic studies using ChIP showing that PML is directly associated with specific regions within the MHC class-I locus (143). Together with special AT-rich sequence binding protein 1 (SATB1), PML is involved in the chromatin-loop organization of the MHC class-I locus and regulates a distinct set of genes within this locus upon IFNγ treatment (143). Recent ChiP-Seq analysis of PML binding regions in MEFs also found PML enriched at heterochromatin gene-poor loci called PML-associated domains (PADs) (144). However, even if ChIP experiments overcome the a priori assumptions for selecting a genomic locus, ChIP cannot distinguish between the nucleoplasmic pool of PML and PML that is located within PML NBs. In particular, the recent ChIP-Seq analysis against PML in MEFs illustrates that most of the loci immunoprecipitated with PML do not localize in PML NBs and PML association at these loci is required to preserve their H3K9me3 heterochromatic state (144). Immuno-FISH therefore remains an indispensable control to assess whether the association of PML with specific chromatin loci happens through PML NBs.
To overcome limitations of immuno-FISH and ChIP, the Bazett-Jones’ team developped a method called immuno-TRAP which allows the deposition of biotin onto DNA in close proximity to PML NBs. DNA can then be purified with streptavidin agarose beads and analyzed in a unbiased manner (145). Using various FOSMIDS, the authors confirmed an interaction of PML NBs with TP53 locus and uncover an association with the PML locus itself. The associations observed were cell-type specific and dependent on the cell's physiological state since IFNα treatment modified the loci association with PML NBs (145). Importantly, the use of an engineered APEX2 peroxidase fused to PML in mouse ESCs to mediate chromatin labeling and purification combined with deep-sequencing (ALaP-Seq) allowed the identification of chromatin regions proximal to PML NBs (114). The authors confirmed the association of PML NBs with regulatory regions of active genes in a genome-wide manner, as well as identified novel hotspots regions, such as the short arm of the Y chromosome, frequently associated with PML NBs (114). The use of a PML RING domain mutant, that is diffuse in the nucleoplasm, only gave very few peaks demonstrating that the majority of the ALaP-Seq peaks truely reflect chromatin interactions with PML NBs, but not with the nucleoplasmic pool of PML.
In parallel to the connection of PML NBs with cellular loci, it was early demonstrated that the genomes of specific viruses (HSV-1, Simian Virus 40 (SV40) and adenovirus) were also juxtaposed to PML NBs during the early stages of lytic virus infection (146,147). A role of PML NBs as potential docking sites for viruses favoring their replication and/or transcription was then confirmed for HSV-1 (148,149), human papilloma virus 11 (150), Epstein-Barr Virus (151), and bovine papillomavirus (152), suggesting that PML NBs could facilitate the infectious process under certain circumstances. In contrast, the presence of a foreign plasmid transgene or of latent human immunodeficiency virus (HIV) proviruses next to PML NBs was associated with transcriptional silencing (107,153). These examples suggest that cells could handle foreign genomes of viral origin in a non-random fashion, and that PML NBs are likely to show a Janus activity depending on the virus and the stage of the viral infection (see below).
PML NBs can contain DNA/chromatin in specific cases
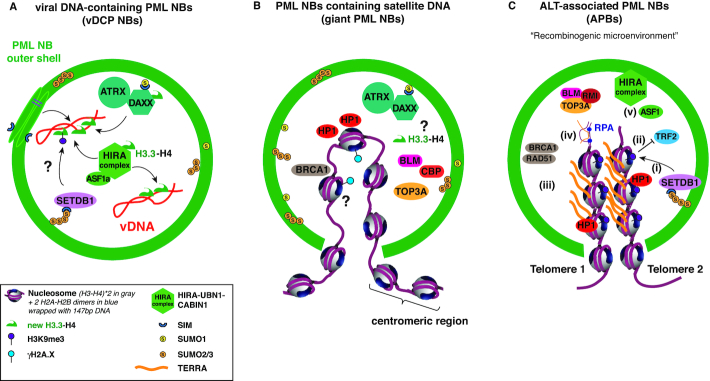
Specific cellular or viral loci can be juxtaposed to PML NBs. However, in certain cases, particular DNAs are located inside the nuclear bodies. The first example comes from telomerase-negative tumors and tumor-derived human cell lines that have been shown to maintain their telomeres length by a mechanism called alternative lengthening of telomeres (ALT) (154–156). ALT cells and tumors contain specific structures of PML NBs referred to as ALT-associated PML nuclear Bodies (APBs). High resolution microscopy images show that these APBs contain telomeric DNA in the interior of the structure, in addition to the PML protein and its partners (16,157,158) (Figure 3). In another specific pathology, the Immunodeficiency, Centromeric region instability, and Facial anomalies syndrome (ICF), enlarged ring-like PML structures, namely giant PML NBs, were observed in ICF G2 nuclei (159). ICF is a rare autosomal recessive disorder associated with mutations in the DNA-methyltransferase DNMT3B gene causing the hypomethylation and decondensation of the heterochromatic structure of satellite DNA mostly in pericentromeric regions of chromosomes 1 (1qh), 16 (16qh) and 9 (9qh) (160–162). Giant PML NBs contain the undercondensed 1qh or 16qh heterochromatin in the inner core with PML forming the outer shell (159) (Figure 3). Other PML NBs constituents, such as HP1, DAXX, ATRX, SP100, SUMO1, CBP and the DNA repair-associated factors BLM, TOP3A and BRCA1 were also found inside the structure adopting a specific multilayered organization (159).
Figure 3.
PML NBs directly regulate chromatin dynamics of DNA sequences found in the condensate. (A) Viral DNA-containing PML NBs (vDCP NBs) are specific PML NBs encasing the HSV-1 latent viral genome. Both H3.3 histone chaperone complexes (DAXX-ATRX and HIRA complexes) are found in these structures together with H3.3–H4. These complexes are essential for the H3.3 chromatinization of the virus, together with PML. H3.3 is decorated with the heterochromatin mark H3K9me3, which could be deposited by SETDB1 (question mark), a known client protein localizing constitutively in PML NBs. (B) PML NBs containing satellite DNA are found in the ICF syndrome in the form of giant PML NBs. These structures contain proteins organized in ordered concentric layers around the satellite DNA core, in the following order from the center: HP1 proteins, DAXX–ATRX complex, CBP/BLM/TOP3A, surrounded by a sphere of SUMO1/SP100 and then PML protein (concentric layers not shown). While the heterochromatin nature of the satellite DNA is atypical with absence of the constitutive H3K9me3 mark despite HP1 presence, the presence of γH2A.X in some giant PML NBs (25%) nevertheless suggests that satellite DNA is associated with chromatin inside PML NBs. Of note, normal PML NBs can also contain satellite DNA in G2 phase. PML NBs-containing satellite DNA may help remodelling and maintenance of the heterochromatin structure present at late-replicating satellite DNA. (C) ALT-associated PML NBs (APBs) are a hallmark of the ALT pathway. Here we only focus on the chromatin dynamics in APBs, and neither display the numerous repair factors present in APBs nor the mechanisms involved in ALT. Telomeric DNA localizes within PML NBs together with specific chromatin-related factors such as SETDB1, ASF1, or HIRA. Recent data suggest that telomeric DNA repeats are more compact, with higher levels of H3K9me3 deposited by SETDB1 (i), and bound less TRF2 in ABPs than regular telomeres (ii), which would cause telomeric deprotection and promote telomeric recombination. Increase in TERRA transcription (orange lines) is also observed (iii) and fuels the ALT process by increasing RNA:DNA hybrids (iv) and thus replicative stress. Depletion of the histone chaperone Asf1 promotes histone management dysfunction during telomeric replication and is sufficient to trigger ALT (v).
In the case of viral infection with HSV-1, a dsDNA virus, the latent viral genome does not integrate in the host genome, and remains as a chromatinized episomal form in the nucleus of infected cells. We and others have shown by confocal microscopy that positioning of the latent HSV-1 genomes is not random and instead, the viral genome is encased in PML NBs forming structures called viral DNA-containing PML NBs (vDCP-NBs) or ND10-like (71,163–165). vDCP NBs contain, just like APBs and giant PML NBs, most of the PML partners, including the DAXX–ATRX complex, as well as all members of the H3.3 histone chaperone HIRA complex (98–100) (Figure 3). Interestingly, a physical and functional association of the genome of an RNA virus, the hepatitis delta virus was observed with PML NBs. The particular antigenomic RNA co-localizes with PML NBs but contrarily to the APBs, giant PML NBs and vDCP NBs, resides at the edge of a rim-like structure that shows in the inside the presence of the PML, SP100, DAXX and SUMO-1 proteins (166). This peculiar association plays a role in viral RNA synthesis mediated by host RNA polymerase II (167), but has not been studied further and remains so far the only example of a viral RNA product closely associated with PML NBs. Finally, an exogenous cytomegalovirus promoter-containing transgene array is found at the center of PML NBs, with PML and SUMOs forming a ring structure around it, as observed by confocal microscopy, supporting the evidence of DNA in the interior of PML NBs (168,169). Altogether these specific examples show that PML NBs have a strong physical connection with specific genomic loci and can entrap particular DNAs, supporting an important role in regulating DNA-metabolic processes (see below).
Regulation of the physical connection of PML NBs with chromatin loci
PML NBs are very dynamic entities whose number and size varies depending on the cell cycle and on various stimuli (61,170). During interphase, nucleoplasm is separated from the cytoplasm by the nuclear envelope forming a selective barrier. PML NBs exhibit apparent stability in the nucleus of unperturbed healthy cells. Yet they are actively remodeled during S-phase due to chromatin topological changes without major changes in PML protein levels or biochemical alterations. In S-phase, chromatin that undergoes replication retracts from PML NBs and actively pulls the PML NBs apart causing their fragmentation in smaller PML NBs by a fission mechanism (171). High rates of PML protein exchange between the nucleoplasmic pool and the PML NBs then ensures that the nascent PML microbodies increase in size by G2. Of note, fusion events can also contribute to the regulation of PML NBs size in S-phase (171).
Reduction of the physical contacts between PML NBs and chromatin can also be induced by specific stresses, such as heat shock, transcriptional repression, apoptosis induction, DNA damage or oxidative stress (58,114,172,173). This triggers the formation of newly formed microbodies by fission as well as in their increased mobility underscoring the importance of chromatin interactions for the structural and morphological integrity as well as the dynamics of PML NBs (58,174). Increase in PML NBs number may also be linked to chromatin decondensation mediated by an ATM-KAP1 axis during DNA damage, or as observed upon HDAC inhibition (172,173). Biomolecular condensates can fuse, coalesce and drip, which are typical properties of liquid assemblies (4). Fission and fusion events of PML NBs observed across the cell cycle or following various stresses thus appear as a convincing feature that would sustain the hypothesis of a liquid-like behavior for these nuclear bodies. Changes in the amount of PML NBs contacts with chromatin across the cell-cycle or following various stresses can thus provide many regulation opportunities for the cells that will need to be explored further.
PML NBs ARE IMPORTANT FOR THE CHROMATINIZATION OF VIRAL GENOMES
The discovery of the association of PML NBs with the genomes of several viruses suggests that PML NBs and their chromatin-related factors mediate their antiviral activity partly through this physical interaction. Remarkably, viruses have evolved several strategies to counteract these antiviral effects by encoding specific anti-PML NBs viral proteins. This is the case for HSV-1 infected cell protein 0 (ICP0), which induces the proteasomal-dependent degradation of SUMOylated forms of PML, leading to PML NBs disappearance. Other viruses directly target the SUMO modification of PML (by preventing it or removing it), thus altering the multivalent potential of PML and the regulation of their protein composition by phase separation (for reviews (175–177).
The packaging of viral DNA with cellular histones carrying specific post-translational modifications allows for a transcriptional control of viral expression (for review (178)). As mentioned above, latent HIV provirus juxtaposed to PML NBs is transcriptionally silent, while reactivation of the virus correlates with displacement of the provirus away from the nuclear bodies (107). Interestingly, the transcriptional repression activity of PML requires its binding to the latent chromatinized provirus, which allows the recruitment of the G9a methyltransferase responsible for H3K9 dimethylation (H3K9me2) on the provirus promoter. Knockdown of PML results in a decrease of provirus-bound G9a, a loss of H3K9me3 heterochromatin marks on the silent viral promoter and a concomitant gain in the transcriptionally-prone H3K4me3 marks (107). Another compelling example of the involvement of PML NBs in the chromatinization of a viral genome came from our lab with regards to the chromatinization of the nucleosome-free HSV-1 genome entering the nucleus of the infected cells, prior to the establishment of latency (99). PML NBs encase the latent viral genome together with the H3.3 chaperone complexes DAXX-ATRX and HIRA, thus allowing the concentration of histone chaperones together with the viral genome in a condensed phase (99) (Figures 2 and 3A). The two H3.3 histone chaperone complexes likely play redundant role in the chromatinization of latent HSV-1 genomes with H3.3-containing nucleosomes, together with the PML protein itself (99). Interestingly, H3.3 is modified with the repressive heterochromatin mark H3K9me3 on the HSV-1 latent genome and destabilization of PML NBs containing the viral genome by ICP0 results in the recovery of a lytic transcriptional program (99), underscoring the repressive function of the PML NB-H3.3 axis in virus latency (Figure 3A). However, HIRA mediated-deposition of H3.3 on HSV-1 genomes during the lytic phase is required for viral RNAs transcription (179), consistent with the double face of H3.3 which can thus be deposited in actively transcribed regions depending on the infectious context. Of note, the switch-like partitioning of HIRA in the PML-NBs following infection by HSV-1 (98–100), correlates with the transcriptional upregulation of host genes, including interferon stimulated genes (ISGs), and contributes to the intrinsic and innate immune defenses against HSV-1 infection (98,100). Therefore, the PML NBs antiviral activity could act both directly through the chromatinization-associated transcriptional regulation of the viral genomes, and indirectly through the regulation of ISGs. PML NBs thus play an essential role in the regulation of viral chromatinization with specific histones variants/marks and are essential for the epigenetic control of viral expression.
PML NBs PARTICIPATE IN THE REGULATION OF CELLULAR CHROMATIN DYNAMICS
PML NBs and the dynamics of chromatin during transcriptional regulation
PML NBs are present in regions of high transcriptional activity (14,52). The recent ALaP-Seq analysis of chromatin regions proximal to PML NBs confirms that PML NBs associate primarily with regulatory regions of active genes in a genome-wide manner (114). In addition to their role in the transcriptional regulation of viral genes, they also regulate transcription of cellular loci (for review (180)). While the PML protein itself can act as a transcriptional co-activator or co-repressor, we focus here specifically on the interplay between PML NBs and transcriptional regulation through the prism of chromatin dynamics and in light of their liquid-like properties (Figure 4).
Figure 4.
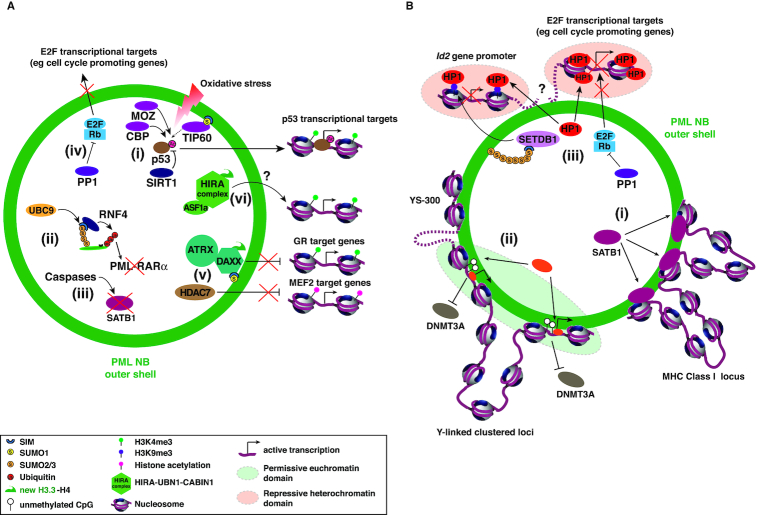
Role of PML NBs in transcriptional regulation. PML NBs has a dual effect on gene expression both facilitating or repressing expression of specific genes. (A) PML NBs regulates transcription through specific modifications of transcription factors or by modulating the availability of transcription factors or chromatin-related factors. (i) Upon Ras-induced senescence entry, p53 localizes in PML NBs which promotes its phosphorylation on serine 15 (not shown) as well as its acetylation on lysine 382 by CBP or MOZ, which may be counteracted by SIRT1. These PML-dependent modifications are required for p53 transactivation activity. TIP60 SUMO-dependent relocalization in PML NBs upon UV damage may also participate in p53 recruitment and stabilization (dashed arrow), thus favoring its transactivation activity. Oxidative stress can also trigger PML-dependent p53 activation conveying the ROS response (237). (ii) PML NBs can regulate proteins levels by SUMO-dependent poly-ubiquitination by RNF4 and subsequent proteasome-mediated degradation as observed for PML-RARα, or (iii) by caspase degradation as observed for SATB1. (iv) In senescence, PML NBs concentrate Protein Phosphatase 1 alpha (PP1α) together with Rb preventing its CDK-dependent phosphorylation and thus inhibiting E2F which remains sequestered in PML NBs and cannot activate cell-cycle promoting genes. (v) The DAXX histone chaperone brings new H3.3-H4 dimers within PML NBs but may then be sequestered preventing the transcriptional repression of its target genes such as Glucocorticoid receptor (GR) target genes. HDAC7 may also be sequestered to prevent repression of MEF2 target genes. (vi) The role for HIRA complex localization in PML NBs remains more enigmatic (question mark). (B) PML NBs could also participate in establishing chromatin domains that are either permissive or refractory to transcription. (i) Interaction between SATB1 and PML is essential to establish a specific chromatin-loop structure at the MHC class I locus and may serve to regulate transcriptional activity of genes within this locus. (ii) PML NBs can also provide a transcriptionally-permissive chromatin environment to neighboring loci (dashed green circle). In particular, binding to the short arm of the Y-chromosome (region YS300) to PML NBs allows anchoring of specific Y-linked gene promoters that are located away from this region (dashed line). PML NBs allow the maintenance of their transcriptional activity by excluding DNMT3A and preventing DNA methylation on these proximal promoters. Specific transcription factors or chromatin-related factors located in PML NBs (orange factor) could also contribute to gene expression in these chromatin domains. (iii) On the contrary, PML NBs may help to concentrate HP1 proteins on specific loci, possibly through phase separation of heterochromatin (dashed red circle), to promote repression of genes such as E2F target genes. SETDB1 may also participate in creating a repressing heterochromatin environment by depositing H3K9me3 on gene promoters such as for the Id2 gene. However, it remains to be determined whether these repressed loci are found in vicinity of PML NBs (question mark).
First, PML NBs can regulate transcriptional activity through specific modifications of transcriptional factors as examplified for p53 acetylation and phosphorylation in senescence (see below) (Figure 4Ai). PML NBs are also known SUMOylation hotspots through concentration of the SUMO E2-conjugating enzyme UBC9 (12,27). SUMOylation could serve to regulate client activity as observed for TIP60-induced HAT activity upon UV damage (110) (Figure 4Ai). Poly-SUMOylation may initiate polyubiquitination by the SUMO-targeted Ubiquitin ligase (STUbl) RNF4 and subsequent proteasome-mediated degration, as observed for the degradation of the PML-RARα fusion protein (116,181) (Figure 4Aii). SUMOylation may also trigger caspase-dependent degradation of proteins within PML NBs as observed for SATB1, impairing its role in chromatin loop organization and transcriptional regulation (143,182) (Figure 4Aiii). However, the abundance of other chromatin related factors, such as HIRA, does not change upon relocalization in PML NBs (98,100) and it remains to be investigated whether SUMOylation could serve to regulate histone chaperone activity.
Second, PML NBs can regulate transcription by modulating the availability of chromatin associated factors within PML NBs. E2Fs transcription factors may be sequestered by pRb within PML NBs upon senescence preventing E2F target genes transcription (see below) (Figure 4Aiv). On the contrary, sequestration of the histone deacetylase HDAC7, a potent transcriptional repressor, in PML NBs could participate in upregulation of MEF2 target genes (108) (Figure 4Av). Dynamic changes in histone chaperone localization might also be a means to fine-tune gene expression. The H3.3 histone chaperone DAXX acts as a potent transcriptional repressor and is a well-studied PML NBs component. Sequestration of DAXX in PML NBs releases transcriptional repression on reporter genes or specific cellular genes, such as glucocorticoid receptor target genes, whereas disruption of DAXX localization in PML NBs by ICP0 or expression of SUMOylation-defective PML mutant fails to relieve DAXX-mediated transcriptional repression (183–185) (Figure 4Av). HIRA localizes in PML NBs upon specific stresses, such as IFN treatment, without any global change in the amount of HIRA RNA or protein levels (98,100). This anticipates a drop of HIRA concentration in the rest of the nucleoplasm through a sequestration mechanism in PML NBs. We can hypothesize that the depletion of HIRA from genomic loci could have a global impact on H3.3 dynamics at specific genes located at a distance from PML NBs, but this titration effect remains to be investigated (Figure 4Avi).
Third, PML NBs could participate in establishing chromatin domains that are either permissive or refractory to transcription (Figure 4B). A recent paradigm shift in the field of transcriptional regulation has put forward a phase separation model for transcriptional control, in which multi-molecular assemblies would form by phase separation bridging enhancers and promoters allowing gene activation (186). As biomolecular condensates contacting specific chromatin loci, PML NBs could participate in forming specific transcriptional conditions on genomic loci. Using a novel CasDrop technology, a Crispr–Cas9-based optogenetic technology allowing local concentration of droplets at specific genomic loci, Shin et al. recently showed that condensates form preferentially in low-density chromatin regions (like PML NBs) and are able to mechanically pull together targeted genomic loci (187). Although CasDrop is an artificial system with the tethering of specific proteins able to phase separate onto genomic loci, the mechanical pulling of distal genomic loci may indeed occur in vivo for PML NBs. In particular, at the MHC locus, PML NBs might regulate transcription of specific genes through the formation of SATB1-associated specific chromatin loops, bringing closer some distal genes in the locus (143) (Figure 4Bi). In addition, using ALaP-Seq Kurihara et al. recently showed that anchoring of PML NBs with the short arm of the Y chromosome (YS300) promotes the association of nearby Y-linked genes with PML NBs, which is required to maintain their expression through DNMT3A exclusion (114) (Figure 4Bii). Deletion of most of the YS300 sequence results in dissociation of Y-linked genes from the PML NBs and their downregulation. More generally, the authors showed that PML NBs associate significantly with regulatory regions, such as enhancers or promoters, that are located in an open chromatin environment (as confirmed by the ATAC-Seq, H3K27ac and H3K4me3 epigenetic signature), and correlates with the expression levels of the associated genes. PML NBs may thus play a novel role in the 3D organization of chromatin by providing a specific nuclear space protected from DNMT3A action and therefore create a transcriptionally-permissive chromatin environment with hypomethylated gene promoters (Figure 4Bii).
Alternatively, PML NBs might also help to organize repressive domains. As mentionned above, SETDB1 constitutively localizes within PML NBs and structural integrity of these nuclear bodies is essential for SETDB1 targeting to the Id2 gene (102). SETDB1 deposits H3K9me3 mark on the Id2 gene promoter, allowing its transcritional repression (102) (Figure 4Biii). In this case, concentration of SETDB1 in PML NBs together with HP1α, which is known to allow phase-separation formation of heterochromatin together with H3K9me3 (46,47,188) may help to organize a chromatin repressive structure around the Id2 gene promoter (Figure 4Biii). Interestingly, the establishment of specific chromatin domains is not necessarily mutually exclusive with the buffering/sequestration role of PML NBs. Specific targeting of genomic loci to PML NBs could provide a way to regulate them by binding of a given client protein, while other loci remaining away from PML NBs could show a depletion of this same protein.
While some genomic loci localize within PML NBs or are juxtaposed to PML NBs, we could wonder about the order of events between PML NBs biogenesis and specific genomic loci targeting. Indeed, transcription factors and chromatin-related factors could help bring genomic loci in close proximity or even within PML NBs through interaction with chromatin, after the formation of PML NBs. Another alternative model suggests that transcription factors or other chromatin-bound proteins could first bind to genomic loci and then recruit PML. This could help the nucleation and possible phase separation of PML NBs at a given locus, aided by the fusion with pre-existing PML NBs to create a transcriptional specific environment. In the case of HSV-1 infection, it is interesting to note that chromatin-related factors, such as DAXX, are indeed recruited to HSV-1 genomes very early after infection, before the detection of PML (99,189). PML would then bind the viral genome and fuse with a pre-existing nearby PML NB (189).
Presence of numerous PML NBs client proteins that are associated with heterochromatin formation (e.g. DAXX, ATRX, HP1, SETDB1) hinted at the probable implication of PML NBs in heterochromatin dynamics. In the next sections, we will develop the various roles of PML NBs in heterochromatin formation and maintenance in specific cell states, such as senescence, as well as in specific regions, such as pericentromeres or telomeres.
PML NBs and the regulation of chromatin dynamics in senescent cells
Cell senescence is defined by a permanent arrest of the cell cycle that can be induced by various stresses, such as telomeres attrition (replicative senescence), oncogene activation or genotoxic insults. Senescence is therefore considered as a defense mechanism against tumoral transformation (190). Chromatin of senescent cells undergoes massive reorganization, with the condensation of each chromosome to form Senescence-Associated Heterochromatin Foci (SAHF) (191–193), which are enriched in heterochromatic markers such as H3K9me3, HP1 and histone macroH2A (for review (194)). This organization may contribute to the maintenance of a specific senescent gene expression profile, with the down-regulation of cell cycle promoting E2F target genes and the upregulation of genes coding for factors of the senescence-associated secretory phenotype (SASP), reinforcing the senescent state (194). The first evidence linking PML NBs with senescence came from the observation that PML NBs dramatically increase in size and number upon senescence entry (41,42). PML depleted cells are impaired in their ability to undergo senescence and conversely PML overexpression triggers senescence entry in a p53 and pRb/E2F pathways-dependent manner (41,42,195). SAHF formation is also tightly related to PML NBs, eventhough SAHF per se are not found inside PML NBs (78). Indeed, expression of the dominant negative PML-RARα fusion protein, which impairs PML NBs formation, eliminates SAHF formation in cells induced in senescence (196).
All three functions of PML NBs described in Figure 2 could be at play during senescence. First, these nuclear bodies could act as specific biochemical reactors to ensure p53 phosphorylation/acetylation and pRb hypophosphorylation, by concentrating them with modifying enzymes such as CBP, MOZ, TIP60 and Protein Phosphatase 1 alpha (PP1α), respectively (41,42,112,197) (Figure 4Ai and 4Aiv). Of note, SIRT1 localization in PML NBs upon senescence induction may counteract CBP-mediated p53 acetylation and thus repress p53 target genes (109) (Figure 4Ai). Second, PML NBs may also serve to sequestrate various client proteins such as the abundant E2Fs activators (E2F1-3), which localize in PML NBs in a pRb-dependent manner in oncogene-induced senescent cells. PML NBs prevent the CDK-dependent phosphorylation of pRb by concentrating it with PP1α, leading to E2Fs sequestration and obstruction of their cell-cycle promoting activity (197) (Figure 4Aiv). Moreover, all HP1 isoforms transiently accumulate within PML NBs in the early stages of senescence, before their stable incorporation into SAHF (78,196). This may be linked to a potential role of PML NBs in targeting HP1 to defined juxtaposed chromatin regions. In particular, in senescent cells, PML associates with promoters of E2F target genes and is required for their H3K9me3-mediated heterochromatinization in a pRb-dependent manner (Figure 4Biii). This silencing in turn is required to prevent DNA replication and cell cycle progression (197,198). As discussed above for the Id2 gene, PML NBs could serve as concentration sites for HP1, enabling heterochromatin formation by liquid–liquid phase separation at E2F target genes. Finally, upon senescence entry, members of the HIRA chaperone complex relocalize within H3.3-containing PML NBs (78,95,97) and this specific accumulation is required for proper SAHF formation (78,196). However, since no enrichment of H3.3 is observed in SAHF structures (85), the exact role of HIRA complex in SAHF formation is still unclear and HIRA could play an indirect role in senescence via keeping the senescent expressed genes active (199).
PML NBs and pericentromeric heterochromatin dynamics
First evidence of PML NBs association with pericentromeric chromatin arose from observation in cells derived from patients with the ICF syndrome (159,200) (see above). In those cells, decondensed pericentromeric regions are found within giant PML NBs together with DAXX, ATRX and HP1 (159,200). A subsequent study on normal cells also showed the localization of human pericentromeric DNA repeats within a subset of PML NBs in the G2 phase of the cell cycle, suggesting a general role of PML NBs in re-establishing condensed chromatin on these late-replicated regions (159). However, it remains to be shown if chromatin assembly can happen within PML NBs at these regions, since heterochromatin modifications, such as H3K9me3 or H4K20me3, are absent (Figure 3B). Interestingly, DAXX, which localizes in PML NBs, is required for H3.3 incorporation in the pericentromeric satellites repeats, as well as the PML protein itself (79,85). By modulating DAXX levels, it was shown that H3.3 deposition in pericentromeric regions is linked to the transcription of the repeats (79,85). This might promote further HP1 recruitment and heterochromatin condensation via a ncRNA-mediated mechanism, as observed during early development or in fission yeast (201–203). Of note, a recent study in MEF cells challenges the idea of a PML NB-dependent H3.3 incorporation in pericentromeric regions. PML depleted cells show an accumulation of H3.3 at pericentromeric repeats suggesting that the PML protein prevents H3.3 incorporation at these sites (204). Consistently, depletion of PML increases H3.3 incorporation at a repetitive heterochromatin transgene array which localizes in PML NBs in S phase (169). Explaining such discrepancies will require more investigations in a cell-cycle controlled manner in order to better characterize the roles of PML NBs in pericentromeric chromatin dynamics, and in particular the H3.3 chromatin assembly pathway.
PML NBs and heterochromatin dynamics at telomeres
PML NBs association with telomeres was first suggested by the appearance of APBs in ALT tumors cells, where telomeres repeats are found within PML NBs (157) (see above). ALT is a telomerase-independent, but recombination-dependent, process involving replication stress as well as break-induced replication (BIR) mechanism in APBs to extend telomeres. APBs indeed contain numerous proteins involved in homologous recombination, such as Rad51, breast cancer susceptibility protein 1 (BRCA1), or RPA. While this review does not intend to provide an extensive description of the APBs structure and functions (for review (205,206)), we will nevertheless give a few highlights on the recent connections between PML NBs, chromatin dynamics and the ALT process. PML NBs may act to concentrate specific DNA repair and recombination factors, as well as chromatin modifiers, in a condensate organized around telomeric chromatin (Figures 2 and 3C). Interestingly, a series of recent studies provides evidence for this model. Formation of APB-like condensates by using a scaffold made of poly(SUMO)–poly(SIM) motifs tethered at telomeres or by using a chemically induced protein dimerization approach to tether SIM at telomeres, induced telomere clustering within the artificially engineered APBs in vivo (33,34). Overexpression of the helicase BLM triggered ALT-like phenotypes in presence of these reconstituted APB-like condensates suggesting that concentration of specific DNA repair factors, together with the clustering of telomeres to provide repair templates, are required for induction of the ALT phenotype (33). Stochiometry of the SUMO-SIM interactions controls recruitment of BLM or of the full BLM–TOP3A–RMI (BTR) complex at telomeric ends in APBs (33,207). The BTR complex is essential for telomere lengthening and its artificial tethering to telomeres can bypass the absence of PML to induce ALT (207). Moreover, by applying an assay to visualize telomeric DNA synthesis in human cells, it was shown that ALT DNA synthesis occurs exclusively in APBs and is dependent on PML (208), consistent with the essential role of PML NBs for ALT (209). Thus PML NBs cluster telomeric ends in a condensate together with all required factors to provide a ‘recombinogenic microenvironment’ in order to promote ALT.
APBs are also tightly linked to heterochromatin establishment/dynamics at telomeres. A strong correlation between ATRX/DAXX inactivation and the ALT phenotype has been observed in specific tumors, although loss of these chaperones is not sufficient to trigger ALT immediately, but may induce ALT via a gradual telomeric DNA replication dysfunction (210–213). Interestingly, depletion of the histone chaperone Asf1 enhances replicative stress posed at telomeres and provokes ALT appearance (214). The histone chaperone HIRA is also present in APBs of ALT cells and is required for the localization of HP1 in APBs (96). Since HIRA is in complex with UBN1, which is known to associate with an H3K9 histone methyltransferase activity (95), it could help the H3K9 trimethylation of telomeric DNA and ensure HP1 binding to telomeres in APBs. In this regard, it is interesting to note that SETDB1 is required for the H3K9me3 modification of telomeric chromatin in both mouse ES cells and human ALT cells (215). In contrast to the prevailing view of heterochromatin inhibiting the recombination process at telomeres in ALT cells (216), it was shown that increase in H3K9me3-heterochromatin mediated by SETDB1 at telomeres promotes ALT features and telomeric transcription of TERRA (215), which localizes in PML NBs (56) (Figure 3C). On the contrary, upon SETDB1 knockdown, decrease of H3K9me3 diminishes the number of APBs, as well as telomeric transcription and abrogates ALT (215). These results are consistent with a previous study showing that the knockdown of factors involved in heterochromatin formation, such as the histone H4K20 methyltransferase SUV4-20H2 (KMT5C), HP1γ or the histone H3K4 demethylase LSD1, reduces the number of APBs (209). The local compaction of chromatin could prevent TRF2 binding (209), a protein part of the shelterin complex protecting telomeres, thus facilitating recombination. Increased heterochromatin features at telomeric DNA could also facilitate phase transitions, as observed for regular heterochromatin (46,47,188), to form the APB recombinogenic microenvironment. Interestingly, telomeric RNA present in ABPs generates RNA:DNA telomeric hybrids, which could promote replication stress and structures prone to engage into recombination (56) (Figure 3C). Reduction of TERRA transcription by a decrease of heterochromatin composition of telomeres within APBs or decrease in the amount of RNA:DNA hybrids by RNAseH1 or FANCM can impair the ALT process (56,217).
In addition to their role in the ALT pathway, further studies showed that PML NBs are also modulating chromatin structure at telomeres in non-neoplastic cells. Indeed, in mouse ES cells, FISH experiments showed that telomeres colocalize with PML NBs together with DAXX, ATRX and the histone variant H3.3, prominently in S phase (218). This is consistent with the known function of DAXX-ATRX complex for the incorporation of H3.3 at telomeric repeats in ES cells, associated with TERRA repression (80,219). ALaP-Seq survey showed that telomeric sequences were significantly enriched for PML, confirming telomeres association with PML NBs in mouse ESCs (114). PML depletion leads to a reduction of ATRX-dependent H3.3 incorporation at telomeres driving the appearance of damage in the form of telomere-induced dysfunctional foci (TIFs) (218). Interestingly, telomere-associated PML NBs are linked to the pluripotency and their near absence in somatic cell types coincides with a switch in the epigenetic signature of telomeric chromatin towards a more compact structure (218). Indeed, telomeres of somatic cells show a decrease of MNase accessibility, as well as higher levels of repressive chromatin marks including H3K9me3 and H4K20me3 (220), which can be reproduced in the absence of PML in ES cells (218). Therefore, PML NBs constitute an essential platform for the regulation of telomeric chromatin and seem important to maintain a ‘less heterochromatic’ structure of pluripotent cells telomeres, which could favor telomere-elongating activities by telomerase (highly expressed in human and mouse ES cells) and the capacity for continual telomere-renewal important for ES cells pluripotency (220).
Although telomere-associated PML NBs are mostly lost with differentiation, colocalization of PML NBs with telomeres was observed in cells with shortened or damaged telomeres (221). This suggests that PML NBs may participate in telomeres surveillance in somatic cells (221), which could be linked to their function in DNA repair (for review (222)) as well as in telomeric maintenance in ALT cells. While PML NBs no longer seem associated with telomeres in differentiated cells, it was recently shown by ChIP-Seq that PML protein prevents H3.3 incorporation at telomeres in MEFs and is required for maintenance of an H4K20me3-rich heterochromatin structure at the opposite of its role in embryonic cells (204). This ‘guardian’ function of PML in somatic cells is likely independent of PML NBs given that most PML immunoprecipitated comes from the nucleoplasmic pool (144) and will require further investigations.
CONCLUSION AND FUTURE PERSPECTIVES
Phase separation as an organizing principle in biology allowed us to revisit the functional roles of PML NBs in chromatin dynamics under a new light. As discussed throughout the review, PML NBs exhibit remarkable plasticity, both in terms of composition, dynamic changes during the cell cycle and roles related to chromatin dynamics, which apply in different physiological contexts or at different times. The sensitivity of the phase separation mechanism to environmental changes makes PML NBs ideal nuclear bodies to process external stimuli towards specific chromatin responses.
PML NBs formation can be efficiently induced by known therapeutic drugs, such as Arsenic trioxide or IFN, thereby opening interesting therapeutic avenues. Drug-induced PML NB formation could favor senescence entry through p53 activation and HIRA localization in PML NBs, as well as promote specific client degradation of undesired proteins. In addition, the tight relationship between PML NBs and telomeric elongation in ALT cells is also of particular interest to target specific cancer cells. Targeting the replicative stress response of ALT cells by ATR inhibition or FANCM+BLM/BRCA1 depletion can prove useful to induce specific lethality of these cells (223–225). Others inhibitors against one or several of the DNA repair/recombination factors (TOP3A, POLD3, POLD4 etc.), or against the protein TSYPL5, a recently identified protein critical for ALT positive cells viability (226), could potentially provide additional therapeutic strategies for treating ALT tumors. Physiological relevance of this approach was confirmed in colorectal cancer (CRC) tissues with short telomeres, correlating with increased PML levels, APB formation and proliferation rate (227). Inhibition of ATR in patient-derived colorectal cancer organoids reduced APB formation, shortened telomeres and reduced proliferation (227).
Important questions remain opened for future studies on PML NBs and chromatin dynamics. In particular, the following challenges should be addressed in the next years to help decipher the many facets of PML NBs:
What is the role of nucleoplasmic PML versus PML NBs? It will be important to distinguish the functions of the free PML protein, versus the one that is present in PML NBs. The use of APEX2-mediated chromatin labeling and purification in conjunction with PML WT or the nucleoplasmic diffused PML RING domain mutant has allowed to identify chromatin-PML NBs associations in a genome-wide fashion (114). This technique (or others related techniques such as BioID (228)) could be applied to investigate the PML NB proteome, and thus provide a more comprehensive picture of PML NBs functions.
How heterogenous are PML NBs? To reconcile the divergent roles of PML NBs in both transcriptional activation and transcriptional repression, it can be envisaged that PML NBs could have a different composition relative to PML isoforms or client proteins, or a different localization by being targeted to various genomic loci throughout the nucleus. Alternatively, PML NBs could be subdivided in several functional subdomains with varying PML isoforms or client proteins concentrations within the subparts of the same nuclear body. Interestingly, subcompartmentalization of phase-separated condensates has recently been observed with co-existing liquid phases within a condensate regulated by different phosphorylation patterns. This subcompartmentalization allows the spatial coupling of translation regulation with mRNA deadenylation rates (229). Whether changing composition of PML NBs does actually matter for the regulation of chromatin dynamics should be explored in the future.
Are PML NBs implicated in chromatin dynamics during DNA repair? PML NBs accumulate various proteins involved in DNA repair pathways and their role in the maintenance of genome integrity has been extensively studied (for review (222)). Whether they actually regulate histone disassembly/assembly processes that are known to occur during DNA repair remains unknown. Given the accumulation of various H3.3 histone chaperones in PML NBs, and their connection with DNA repair (230), as well as the localization of damaged foci juxtaposed to PML NBs (172,222,231,232), it is tempting to speculate that they could also potentially regulate chromatin dynamics at DNA repair foci, from within PML NBs.
What is the actual role of PML NBs in diseases with strong inflammatory signature? Idiopathic inflammatory myopathies are characterized by muscle impairment associated with a strong IFN-signature (233). Since PML NBs number and size are controlled by IFN (38–40), we could speculate that their role in transcriptional regulation and chromatin dynamics could participate in the phenotypical alterations observed in these myopathies. Moreover, the recent SARS-CoV-2 pandemic has underlined the importance of IFN-I to activate ISGs and mediate antiviral reponse. Defects in IFNα/β production in patients correlated with the severity of the disease (234,235), but whether this depends on PML NBs functions remains to be investigated.
To conclude, many challenges still lie ahead to understand the complex functions of these fascinating nuclear bodies.
ACKNOWLEDGEMENTS
We apologize to the authors whose work has not been cited due to length restrictions. We thank all our collaborators who contributed to the studies from our laboratory in connection with the present review. We thank Olivier Binda and Faouzi Baklouti for their comments on the manuscript and Fabian Erdel for helpful discussion on LLPS.
Contributor Information
Armelle Corpet, Univ Lyon, Université Claude Bernard Lyon 1, CNRS UMR 5310, INSERM U 1217, LabEx DEVweCAN, Institut NeuroMyoGène (INMG), team Chromatin Dynamics, Nuclear Domains, Virus F-69008, Lyon, France.
Constance Kleijwegt, Univ Lyon, Université Claude Bernard Lyon 1, CNRS UMR 5310, INSERM U 1217, LabEx DEVweCAN, Institut NeuroMyoGène (INMG), team Chromatin Dynamics, Nuclear Domains, Virus F-69008, Lyon, France.
Simon Roubille, Univ Lyon, Université Claude Bernard Lyon 1, CNRS UMR 5310, INSERM U 1217, LabEx DEVweCAN, Institut NeuroMyoGène (INMG), team Chromatin Dynamics, Nuclear Domains, Virus F-69008, Lyon, France.
Franceline Juillard, Univ Lyon, Université Claude Bernard Lyon 1, CNRS UMR 5310, INSERM U 1217, LabEx DEVweCAN, Institut NeuroMyoGène (INMG), team Chromatin Dynamics, Nuclear Domains, Virus F-69008, Lyon, France.
Karine Jacquet, Univ Lyon, Université Claude Bernard Lyon 1, CNRS UMR 5310, INSERM U 1217, LabEx DEVweCAN, Institut NeuroMyoGène (INMG), team Chromatin Dynamics, Nuclear Domains, Virus F-69008, Lyon, France.
Pascale Texier, Univ Lyon, Université Claude Bernard Lyon 1, CNRS UMR 5310, INSERM U 1217, LabEx DEVweCAN, Institut NeuroMyoGène (INMG), team Chromatin Dynamics, Nuclear Domains, Virus F-69008, Lyon, France.
Patrick Lomonte, Univ Lyon, Université Claude Bernard Lyon 1, CNRS UMR 5310, INSERM U 1217, LabEx DEVweCAN, Institut NeuroMyoGène (INMG), team Chromatin Dynamics, Nuclear Domains, Virus F-69008, Lyon, France.
FUNDING
P.L. laboratory is funded by grants from the Centre National de la Recherche Scientifique (CNRS) (http://www.cnrs.fr), Institut National de la Santé et de la Recherche Médicale (INSERM) (https://www.inserm.fr), University Claude Bernard Lyon 1 (https://www.univ-lyon1.fr), French National Agency for Research-ANR [CENTROLAT ANR-05-MIIM-008-01/02; VIRUCEPTION ANR-13-BSV3-0001-01; EPIPRO ANR-18-CE15-0014-01 http://www.agence-nationale-recherche.fr]; LabEX DEVweCAN and DEV2CAN [ANR-10-LABX-61, http://www.agence-nationale-recherche.fr]; Comité départemental du Rhône de La Ligue contre le cancer and the FINOVI foundation [142690]; P.L. is a CNRS Research Director and AC is assistant professor in the University Claude Bernard Lyon 1. Funding for open access charge: LabEX.
Conflict of interest statement. None declared.
REFERENCES
- 1. Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997; 389:251–260. [DOI] [PubMed] [Google Scholar]
- 2. Allis C.D., Jenuwein T.. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016; 17:487–500. [DOI] [PubMed] [Google Scholar]
- 3. Banani S.F., Lee H.O., Hyman A.A., Rosen M.K.. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell. Biol. 2017; 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alberti S., Gladfelter A., Mittag T.. Considerations and challengesin studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019; 176:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mir M., Bickmore W., Furlong E.E.M., Narlikar G.. Chromatin topology, condensates and gene regulation: shifting paradigms or just a phase. Development. 2019; 146:dev182766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., Rosen M.K.. Compositional control of phase-separated cellular bodies. Cell. 2016; 166:651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hyman A.A., Weber C.A., Jülicher F.. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014; 30:39–58. [DOI] [PubMed] [Google Scholar]
- 8. A P., Weber S.C.. Evidence for and against liquid-liquid phase separation in the nucleus. ncRNA. 2019; 5:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lallemand-Breitenbach V., de Thé H.. ScienceDirectPML nuclear bodies: from architecture to function. Curr. Opin. Cell Biol. 2018; 52:154–161. [DOI] [PubMed] [Google Scholar]
- 10. Li Y., Ma X., Wu W., Chen Z., Meng G.. PML nuclear body biogenesis, carcinogenesis, and targeted therapy. Trends Cancer. 2020; doi:10.1016/j.trecan.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 11. de Thé H., Le Bras M., Lallemand-Breitenbach V.. The cell biology of disease: acute promyelocytic leukemia, arsenic, and PML bodies. J. Cell Biol. 2012; 198:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Damme E., Laukens K., Dang T.H., Van Ostade X.. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int. J. Biol. Sci. 2010; 6:51–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bernardi R., Pandolfi P.P.. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007; 8:1006–1016. [DOI] [PubMed] [Google Scholar]
- 14. Boisvert F.-M., Hendzel M.J., Bazett-Jones D.P.. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 2000; 148:283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoischen C., Monajembashi S., Weisshart K., Hemmerich P.. Multimodal light microscopy approaches to reveal structural and functional properties of promyelocytic leukemia nuclear bodies. Front. Oncol. 2018; 8:3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lang M., Jegou T., Chung I., Richter K., Munch S., Udvarhelyi A., Cremer C., Hemmerich P., Engelhardt J., Hell S.W. et al.. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J. Cell Sci. 2010; 123:392–400. [DOI] [PubMed] [Google Scholar]
- 17. Szostecki C., Guldner H.H., Netter H.J., Will H.. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J. Immunol. 1990; 145:4338–4347. [PubMed] [Google Scholar]
- 18. Nisole S., Maroui M.A., Mascle X.H., Aubry M., Chelbi-Alix M.K.. Differential roles of PML isoforms. Front. Oncol. 2013; 3:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seeler J.-S., Dejean A.. SUMO and the robustness of cancer. Nat. Rev. Cancer. 2017; 17:184–197. [DOI] [PubMed] [Google Scholar]
- 20. Kamitani T., Kito K., Nguyen H.P., Wada H., Fukuda-Kamitani T., Yeh E.T.. Identification of three major sentrinization sites in PML. J. Biol. Chem. 1998; 273:26675–26682. [DOI] [PubMed] [Google Scholar]
- 21. Cuchet-Lourenço D., Boutell C., Lukashchuk V., Grant K., Sykes A., Murray J., Orr A., Everett R.D.. SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog. 2011; 7:e1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shen T.H., Lin H.-K., Scaglioni P.P., Yung T.M., Pandolfi P.P.. The mechanisms of PML-nuclear body formation. Mol. Cell. 2006; 24:331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhong S., Müller S., Ronchetti S., Freemont P.S., Dejean A., Pandolfi P.P.. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000; 95:2748–2752. [PubMed] [Google Scholar]
- 24. Lallemand-Breitenbach V., Zhu J., Puvion F., Koken M., Honoré N., Doubeikovsky A., Duprez E., Pandolfi P.P., Puvion E., Freemont P. et al.. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 2001; 193:1361–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ishov A.M., Sotnikov A.G., Negorev D., Vladimirova O.V., Neff N., Kamitani T., Yeh E.T., Strauss J.F., Maul G.G.. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999; 147:221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jeanne M., Lallemand-Breitenbach V., Ferhi O., Koken M., Le Bras M., Duffort S., Peres L., Berthier C., Soilihi H., Raught B. et al.. PML/RARA oxidation and arsenic binding initiate the antileukemia response of As. Cancer Cell. 2010; 18:88–98. [DOI] [PubMed] [Google Scholar]
- 27. Sahin U., Ferhi O., Jeanne M., Benhenda S., Berthier C., Jollivet F., Niwa-Kawakita M., Faklaris O., Setterblad N., de The H. et al.. Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J. Cell Biol. 2014; 204:931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang P., Benhenda S., Wu H., Lallemand-Breitenbach V., Zhen T., Jollivet F., Peres L., Li Y., Chen S.-J., Chen Z. et al.. RING tetramerization is required for nuclear body biogenesis and PML sumoylation. Nat. Commun. 2018; 1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Y., Ma X., Chen Z., Wu H., Wang P., Wu W., Cheng N., Zeng L., Zhang H., Cai X. et al.. B1 oligomerization regulates PML nuclear body biogenesis and leukemogenesis. Nat. Commun. 2019; 10:3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Geng Y., Monajembashi S., Shao A., Cui D., He W., Chen Z., Hemmerich P., Tang J.. Contribution of the C-terminal regions of promyelocytic leukemia protein (PML) isoforms II and V to PML nuclear body formation. J. Biol. Chem. 2012; 287:30729–30742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li C., Peng Q., Wan X., Sun H., Tang J.. C-terminal motifs in promyelocytic leukemia protein isoforms critically regulate PML nuclear body formation. J. Cell Sci. 2017; 130:3496–3506. [DOI] [PubMed] [Google Scholar]
- 32. McSwiggen D.T., Mir M., Darzacq X., Tjian R.. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev. 2019; 33:1619–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Min J., Wright W.E., Shay J.W.. Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 2019; 33:814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang H., Zhao R., Tones J., Liu M., Dilley R.L., Chenoweth D.M., Greenberg R.A., Lampson M.A.. Nuclear body phase separation drives telomere clustering in ALT cancer cells. Mol. Biol. Cell. 2020; 31:2048–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lallemand-Breitenbach V., de The H.. PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010; 2:a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M., Kriwacki R.W., Pappu R.V., Brangwynne C.P.. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016; 165:1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Erdel F., Rippe K.. Formation of chromatin subcompartments by phase separation. Biophys. J. 2018; 114:2262–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lavau C., Marchio A., Fagioli M., Jansen J., Falini B., Lebon P., Grosveld F., Pandolfi P.P., Pelicci P.G., Dejean A.. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene. 1995; 11:871–876. [PubMed] [Google Scholar]
- 39. Stadler M., Chelbi-Alix M.K., Koken M.H., Venturini L., Lee C., Saïb A., Quignon F., Pelicano L., Guillemin M.C., Schindler C.. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene. 1995; 11:2565–2573. [PubMed] [Google Scholar]
- 40. Chelbi-Alix M.K., Pelicano L., Quignon F., Koken M.H., Venturini L., Stadler M., Pavlovic J., Degos L., de The H.. Induction of the PML protein by interferons in normal and APL cells. Leukemia. 1995; 9:2027–2033. [PubMed] [Google Scholar]
- 41. Ferbeyre G., de Stanchina E., Querido E., Baptiste N., Prives C., Lowe S.W.. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 2000; 14:2015–2027. [PMC free article] [PubMed] [Google Scholar]
- 42. Pearson M., Carbone R., Sebastiani C., Cioce M., Fagioli M., Saito S., Higashimoto Y., Appella E., Minucci S., Pandolfi P.P. et al.. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000; 406:207–210. [DOI] [PubMed] [Google Scholar]
- 43. Hancock R. A role for macromolecular crowding effects in the assembly and function of compartments in the nucleus. J. Struct. Biol. 2004; 146:281–290. [DOI] [PubMed] [Google Scholar]
- 44. McSwiggen D.T., Hansen A.S., Teves S.S., Marie-Nelly H., Hao Y., Heckert A.B., Umemoto K.K., Dugast-Darzacq C., Tjian R., Darzacq X.. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. Elife. 2019; 8:e47098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Erdel F., Rademacher A., Vlijm R., Tünnermann J., Frank L., Weinmann R., Schweigert E., Yserentant K., Hummert J., Bauer C. et al.. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol. Cell. 2020; 78:236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., Karpen G.H.. Phase separation drives heterochromatin domainformation. Nature. 2017; 547:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L., Agard D.A., Redding S., Narlikar G.J.. Liquid droplet formation by HP1. Nature. 2017; 547:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brouwer A.K., Schimmel J., Wiegant J.C.A.G., Vertegaal A.C.O., Tanke H.J., Dirks R.W.. Telomeric DNA mediates de novo PML body formation. Mol. Biol. Cell. 2009; 20:4804–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kaiser T.E., Intine R.V., Dundr M.. De novo formation of a subnuclear body. Science. 2008; 322:1713–1717. [DOI] [PubMed] [Google Scholar]
- 50. Chung I., Leonhardt H., Rippe K.. De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation. J. Cell Sci. 2011; 124:3603–3618. [DOI] [PubMed] [Google Scholar]
- 51. Wang H., Xu X., Nguyen C.M., Liu Y., Gao Y., Lin X., Daley T., Kipniss N.H., La Russa M., Qi L.S.. CRISPR-mediated programmable 3D genome positioning and nuclear organization. Cell. 2018; 175:1405–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang J. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 2004; 164:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. LaMorte V.J., Dyck J.A., Ochs R.L., Evans R.M.. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc. Natl Acad. Sci. U.S.A. 1998; 95:4991–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fuchsová B., Novák P., Kafková J., Hozák P.. Nuclear DNA helicase II is recruited to IFN-α–activated transcription sites at PML nuclear bodies. J. Cell Biol. 2002; 158:463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kiesslich A., Mikecz von A., Hemmerich P.. Cell cycle-dependent association of PML bodies with sites of active transcription in nuclei of mammalian cells. J. Struct. Biol. 2002; 140:167–179. [DOI] [PubMed] [Google Scholar]
- 56. Arora R., Lee Y., Wischnewski H., Brun C.M., Schwarz T., Azzalin C.M.. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014; 5:5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grande M.A., van der Kraan I., van Steensel B., Schul W., de The H., van der Voort H.T., de Jong L., van Driel R.. PML-containing nuclear bodies: their spatial distribution in relation to other nuclear components. J. Cell. Biochem. 1996; 63:280–291. [DOI] [PubMed] [Google Scholar]
- 58. Eskiw C.H., Dellaire G., Bazett-Jones D.P.. Chromatin contributes to structural integrity of promyelocytic leukemia bodies through a SUMO-1-independent mechanism. J. Biol. Chem. 2004; 279:9577–9585. [DOI] [PubMed] [Google Scholar]
- 59. Berry J., Weber S.C., Vaidya N., Haataja M., Brangwynne C.P.. RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci U.S.A. 2015; 112:E5237–E5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Falahati H., Pelham-Webb B., Blythe S., Wieschaus E.. Nucleation by rRNA dictates the precision of nucleolus assembly. Curr. Biol. 2016; 26:277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Everett R.D., Lomonte P., Sternsdorf T., van Driel R., Orr A.. Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 1999; 112:4581–4588. [DOI] [PubMed] [Google Scholar]
- 62. Dellaire G. Mitotic accumulations of PML protein contribute to the re-establishment of PML nuclear bodies in G1. J. Cell Sci. 2006; 119:1034–1042. [DOI] [PubMed] [Google Scholar]
- 63. Lång A., Lång E., Bøe S.O.. PML bodies in mitosis. Cells. 2019; 8:893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jul-Larsen A., Grudic A., Bjerkvig R., Ove Boe S.. Cell-cycle regulation and dynamics of cytoplasmic compartments containing the promyelocytic leukemia protein and nucleoporins. J. Cell Sci. 2009; 122:1201–1210. [DOI] [PubMed] [Google Scholar]
- 65. Lång A., Eriksson J., Schink K.O., Lång E., Blicher P., Połeć A., Brech A., Dalhus B., Bøe S.O.. Visualization of PML nuclear import complexes reveals FG-repeat nucleoporins at cargo retrieval sites. nucleus. 2017; 8:404–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen Y.-C.M., Kappel C., Beaudouin J., Eils R., Spector D.L.. Live cell dynamics of promyelocytic leukemia nuclear bodies upon entry into and exit from mitosis. Mol. Biol. Cell. 2008; 19:3147–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ohsaki Y., Kawai T., Yoshikawa Y., Cheng J., Jokitalo E., Fujimoto T.. PML isoform II plays a critical role in nuclear lipid droplet formation. J. Cell Biol. 2016; 212:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee J., Salsman J., Foster J., Dellaire G., Ridgway N.D.. Lipid-associated PML structures assemble nuclear lipid droplets containing CCTα and Lipin1. Life Sci. Alliance. 2020; 3:e202000751-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Butler J.T., Hall L.L., Smith K.P., Lawrence J.B.. Changing nuclear landscape and unique PML structures during early epigenetic transitions of human embryonic stem cells. J. Cell. Biochem. 2009; 107:609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tokunaga K., Saitoh N., Goldberg I.G., Sakamoto C., Yasuda Y., Yoshida Y., Yamanaka S., Nakao M.. Computational image analysis of colony and nuclear morphology to evaluate human induced pluripotent stem cells. Sci. Rep. 2014; 4:704–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Maroui M.A., Calle A., Cohen C., Streichenberger N., Texier P., Takissian J., Rousseau A., Poccardi N., Welsch J., Corpet A. et al.. Latency entry of herpes simplex virus 1 is determined by the interaction of its genome with the nuclear environment. PLoS Pathog. 2016; 12:e1005834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Doucas V., Tini M., Egan D.A., Evans R.M.. Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling. Proc. Natl Acad. Sci. U.S.A. 1999; 96:2627–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mikecz von A., Zhang S., Montminy M., Tan E.M., Hemmerich P.. CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol. 2000; 150:265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lehming N., Le Saux A., Schüller J., Ptashne M.. Chromatin components as part of a putative transcriptional repressing complex. Proc. Natl Acad. Sci. U..S.A. 1998; 95:7322–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Seeler J.S., Marchio A., Sitterlin D., Transy C., Dejean A.. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl Acad. Sci. U.S.A. 1998; 95:7316–7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hayakawa T. Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J. Cell Sci. 2003; 116:3327–3338. [DOI] [PubMed] [Google Scholar]
- 77. Everett R.D., Earnshaw W.C., Pluta A.F., Sternsdorf T., Ainsztein A.M., Carmena M., Ruchaud S., Hsu W.L., Orr A.. A dynamic connection between centromeres and ND10 proteins. J. Cell Sci. 1999; 112:3443–3454. [DOI] [PubMed] [Google Scholar]
- 78. Zhang R., Poustovoitov M.V., Ye X., Santos H.A., Chen W., Daganzo S.M., Erzberger J.P., Serebriiskii I.G., Canutescu A.A., Dunbrack R.L. et al.. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell. 2005; 8:19–30. [DOI] [PubMed] [Google Scholar]
- 79. Drane P., Ouararhni K., Depaux A., Shuaib M., Hamiche A.. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010; 24:1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Goldberg A.D., Banaszynski L.A., Noh K.-M., Lewis P.W., Elsaesser S.J., Stadler S., Dewell S., Law M., Guo X., Li X. et al.. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010; 140:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lewis P.W., Elsaesser S.J., Noh K.-M., Stadler S.C., Allis C.D.. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:14075–14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xue Y., Gibbons R., Yan Z., Yang D., McDowell T.L., Sechi S., Qin J., Zhou S., Higgs D., Wang W.. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl Acad. Sci. U.S.A. 2003; 100:10635–10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ishov A.M. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J. Cell Sci. 2004; 117:3807–3820. [DOI] [PubMed] [Google Scholar]
- 84. Delbarre E., Ivanauskiene K., Küntziger T., Collas P.. DAXX-dependent supply of soluble (H3.3-H4) dimers to PML bodies pending deposition into chromatin. Genome Res. 2013; 23:440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Corpet A., Olbrich T., Gwerder M., Fink D., Stucki M.. Dynamics of histone H3.3 deposition in proliferating and senescent cells reveals a DAXX-dependent targeting to PML-NBs important for pericentromeric heterochromatin organization. Cell Cycle. 2014; 113:E3213–E3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gurard-Levin Z.A., Quivy J.-P., Almouzni G.. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 2014; 83:487–517. [DOI] [PubMed] [Google Scholar]
- 87. Ahmad K., Henikoff S.. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002; 9:1191–1200. [DOI] [PubMed] [Google Scholar]
- 88. Mito Y., Henikoff J.G., Henikoff S.. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005; 37:1090–1097. [DOI] [PubMed] [Google Scholar]
- 89. Deaton A.M., Gómez-Rodríguez M., Mieczkowski J., Tolstorukov M.Y., Kundu S., Sadreyev R.I., Jansen L.E., Kingston R.E.. Enhancer regions show high histone H3.3 turnover that changes during differentiation. Elife. 2016; 5:e15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ray-Gallet D., Woolfe A., Vassias I., Pellentz C., Lacoste N., Puri A., Schultz D.C., Pchelintsev N.A., Adams P.D., Jansen L.E.T. et al.. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell. 2011; 44:928–941. [DOI] [PubMed] [Google Scholar]
- 91. Soni S., Pchelintsev N., Adams P.D., Bieker J.J.. Transcription factor EKLF (KLF1) recruitment of the histone chaperone HIRA is essential for β-globin gene expression. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:13337–13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhang H., Gan H., Wang Z., Lee J.-H., Zhou H., Ordog T., Wold M.S., Ljungman M., Zhang Z.. RPA Interacts with HIRA and Regulates H3.3 Deposition at Gene Regulatory Elements in Mammalian Cells. Mol. Cell. 2017; 65:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Szenker E., Ray-Gallet D., Almouzni G.. The double face of the histone variant H3.3. Cell Res. 2011; 21:421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Martire S., Banaszynski L.A.. The roles of histone variants in fine-tuning chromatin organization and function. Nat. Rev. Mol. Cell Biol. 2020; 21:522–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Banumathy G., Somaiah N., Zhang R., Tang Y., Hoffmann J., Andrake M., Ceulemans H., Schultz D., Marmorstein R., Adams P.D.. Human UBN1 is an ortholog of yeast Hpc2p and Has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells. Mol. Cell. Biol. 2009; 29:758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jiang W.-Q., Nguyen A., Cao Y., Chang A.C.-M., Reddel R.R.. HP1-mediated formation of alternative lengthening of telomeres-associated PML bodies requires HIRA but not ASF1a. PLoS One. 2011; 6:e17036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Rai T.S., Puri A., McBryan T., Hoffman J., Tang Y., Pchelintsev N.A., van Tuyn J., Marmorstein R., Schultz D.C., Adams P.D.. Human CABIN1 Is a Functional Member of the Human HIRA/UBN1/ASF1a Histone H3.3 Chaperone Complex. Mol. Cell. Biol. 2011; 31:4107–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Rai T.S., Glass M., Cole J.J., Rather M.I., Marsden M., Neilson M., Brock C., Humphreys I.R., Everett R.D., Adams P.D.. Histone chaperone HIRA deposits histone H3.3 onto foreign viral DNA and contributes to anti-viral intrinsic immunity. Nucleic Acids Res. 2017; 45:11673–11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cohen C., Corpet A., Roubille S., Maroui M.A., Poccardi N., Rousseau A., Kleijwegt C., Binda O., Texier P., Sawtell N. et al.. Promyelocytic leukemia (PML) nuclear bodies (NBs) induce latent/quiescent HSV-1 genomes chromatinization through a PML NB/Histone H3.3/H3.3 Chaperone Axis. PLoS Pathog. 2018; 14:e1007313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. McFarlane S., Orr A., Roberts A.P.E., Conn K.L., Iliev V., Loney C., da Silva Filipe A., Smollett K., Gu Q., Robertson N. et al.. The histone chaperone HIRA promotes the induction of host innate immune defences in response to HSV-1 infection. PLoS Pathog. 2019; 15:e1007667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ivanauskiene K., Delbarre E., McGhie J.D., Kuntziger T., Wong L.H., Collas P.. The PML-associated protein DEK regulates the balance of H3.3 loading on chromatin and is important for telomere integrity. Genome Res. 2014; 24:1584–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Cho S., Park J.S., Kang Y.-K.. Dual functions of histone-lysine N-methyltransferase Setdb1 protein at promyelocytic leukemia-nuclear body (PML-NB): maintaining PML-NB structure and regulating the expression of its associated genes. J. Biol. Chem. 2011; 286:41115–41124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Matsui T., Leung D., Miyashita H., Maksakova I.A., Miyachi H., Kimura H., Tachibana M., Lorincz M.C., Shinkai Y.. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010; 464:927–931. [DOI] [PubMed] [Google Scholar]
- 104. Elsässer S.J., Noh K.-M., Diaz N., Allis C.D., Banaszynski L.A.. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015; 522:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Carbone R., Botrugno O.A., Ronzoni S., Insinga A., Di Croce L., Pelicci P.G., Minucci S.. Recruitment of the histone methyltransferase SUV39H1 and its role in the oncogenic properties of the leukemia-associated PML-retinoic acid receptor fusion protein. Mol. Cell. Biol. 2006; 26:1288–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Villa R., Pasini D., Gutierrez A., Morey L., Occhionorelli M., Viré E., Nomdedeu J.F., Jenuwein T., Pelicci P.G., Minucci S. et al.. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007; 11:513–525. [DOI] [PubMed] [Google Scholar]
- 107. Lusic M., Marini B., Ali H., Lucic B., Luzzati R., Giacca M.. Proximity to PML nuclear bodies regulates HIV-1 latency in CD4+ T cells. Cell Host Microbe. 2013; 13:665–677. [DOI] [PubMed] [Google Scholar]
- 108. Gao C., Cheng X., Lam M., Liu Y., Liu Q., Chang K.-S., Kao H.-Y.. Signal-dependent regulation of transcription by histone deacetylase 7 involves recruitment to promyelocytic leukemia protein nuclear bodies. Mol. Biol. Cell. 2008; 19:3020–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Langley E., Pearson M., Faretta M., Bauer U.-M., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T.. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002; 21:2383–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cheng Z., Ke Y., Ding X., Wang F., Wang H., Ahmed K., Liu Z., Xu Y., Aikhionbare F., Yan H. et al.. Functional characterization of TIP60 sumoylation in UV-irradiated DNA damage response. Oncogene. 2007; 27:931–941. [DOI] [PubMed] [Google Scholar]
- 111. Wu Q., Hu H., Lan J., Emenari C., Wang Z., Chang K.-S., Huang H., Yao X.. PML3 orchestrates the nuclear dynamics and function of TIP60. J. Biol. Chem. 2009; 284:8747–8759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Rokudai S., Laptenko O., Arnal S.M., Taya Y., Kitabayashi I., Prives C.. MOZ increases p53 acetylation and premature senescence through its complex formation with PML. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:3895–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Song C., Wang L., Wu X., Wang K., Xie D., Xiao Q., Li S., Jiang K., Liao L., Yates J.R. et al.. PML Recruits TET2 to Regulate DNA Modification and Cell Proliferation in Response to Chemotherapeutic Agent. Cancer Res. 2018; 78:2475–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kurihara M., Kato K., Sanbo C., Shigenobu S., Ohkawa Y., Fuchigami T., Miyanari Y.. Genomic profiling by ALaP-Seq reveals transcriptional regulation by PML bodies through DNMT3A exclusion. Mol. Cell. 2020; 78:493–505. [DOI] [PubMed] [Google Scholar]
- 115. Weidtkamp-Peters S., Lenser T., Negorev D., Gerstner N., Hofmann T.G., Schwanitz G., Hoischen C., Maul G., Dittrich P., Hemmerich P.. Dynamics of component exchange at PML nuclear bodies. J. Cell Sci. 2008; 121:2731–2743. [DOI] [PubMed] [Google Scholar]
- 116. Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de Thé H.. Arsenic degrades PML or PML–RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 2008; 10:547–555. [DOI] [PubMed] [Google Scholar]
- 117. Zhu J., Zhou J., Peres L., Riaucoux F., Honoré N., Kogan S., de Thé H.. A sumoylation site in PML/RARA is essential for leukemic transformation. Cancer Cell. 2005; 7:143–153. [DOI] [PubMed] [Google Scholar]
- 118. Lin D.-Y., Huang Y.-S., Jeng J.-C., Kuo H.-Y., Chang C.-C., Chao T.-T., Ho C.-C., Chen Y.-C., Lin T.-P., Fang H.-I. et al.. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell. 2006; 24:341–354. [DOI] [PubMed] [Google Scholar]
- 119. Lamoliatte F., Caron D., Durette C., Mahrouche L., Maroui M.A., Caron-Lizotte O., Bonneil E., Chelbi-Alix M.K., Thibault P.. Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat. Commun. 2014; 5:5409. [DOI] [PubMed] [Google Scholar]
- 120. Hendriks I.A., D'Souza R.C.J., Yang B., Vries M.V.-D., Mann M., Vertegaal A.C.O.. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014; 21:927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Schimmel J., Eifler K., Sigurðsson J.O., Cuijpers S.A.G., Hendriks I.A., Vries M.V.-D., Kelstrup C.D., Francavilla C., Medema R.H., Olsen J.V. et al.. Uncovering SUMOylation dynamicsduring cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol. Cell. 2014; 53:1053–1066. [DOI] [PubMed] [Google Scholar]
- 122. Ivanov A.V., Peng H., Yurchenko V., Yap K.L., Negorev D.G., Schultz D.C., Psulkowski E., Fredericks W.J., White D.E., Maul G.G. et al.. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell. 2007; 28:823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ninova M., Chen Y.-C.A., Godneeva B., Rogers A.K., Luo Y., Fejes Tóth K., Aravin A.A.. Su(var)2-10 and the SUMO pathway link piRNA-guided target recognition to chromatin silencing. Mol. Cell. 2020; 77:556–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Boisvert F.M., Kruhlak M.J., Box A.K., Hendzel M.J., Bazett-Jones D.P.. The transcription coactivator CBP is a dynamic component of the promyelocytic leukemia nuclear body. J. Cell Biol. 2001; 152:1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Tang Y., Puri A., Ricketts M.D., Rai T.S., Hoffmann J., Hoi E., Adams P.D., Schultz D.C., Marmorstein R.. Identification of an ubinuclein 1 region required for stability and function of the human HIRA/UBN1/CABIN1/ASF1a histone H3.3 chaperone complex. Biochemistry. 2012; 51:2366–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Tammsalu T., Matic I., Jaffray E.G., Ibrahim A.F.M., Tatham M.H., Hay R.T.. Proteome-wide identification of SUMO2 modification sites. Sci. Signal. 2014; 7:rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Chang C.-C., Naik M.T., Huang Y.-S., Jeng J.-C., Liao P.-H., Kuo H.-Y., Ho C.-C., Hsieh Y.-L., Lin C.-H., Huang N.-J. et al.. Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol. Cell. 2011; 42:62–74. [DOI] [PubMed] [Google Scholar]
- 128. Percherancier Y., Germain-Desprez D., Galisson F., Mascle X.H., Dianoux L., Estephan P., Chelbi-Alix M.K., Aubry M.. Role of SUMO in RNF4-mediated promyelocytic leukemia protein (PML) degradation: sumoylation of PML and phospho-switch control of its SUMO binding domain dissected in living cells. J. Biol. Chem. 2009; 284:16595–16608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cappadocia L., Mascle X.H., Bourdeau V., Tremblay-Belzile S., Chaker-Margot M., Lussier-Price M., Wada J., Sakaguchi K., Aubry M., Ferbeyre G. et al.. Structural and functional characterizationof the phosphorylation-dependent interaction between PML and SUMO1. Structure/Folding Des. 2015; 23:126–138. [DOI] [PubMed] [Google Scholar]
- 130. Stehmeier P., Müller S.. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol. Cell. 2009; 33:400–409. [DOI] [PubMed] [Google Scholar]
- 131. Ullmann R., Chien C.D., Avantaggiati M.L., Müller S.. An acetylation switch regulates SUMO-dependent protein interaction networks. Mol. Cell. 2012; 46:759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Mascle X.H., Gagnon C., Wahba H.M., Lussier-Price M., Cappadocia L., Sakaguchi K., Omichinski J.G.. Acetylation of SUMO1 alters interactions with the SIMs of PML and Daxx in a protein-specific manner. Structure. 2020; 28:157–168. [DOI] [PubMed] [Google Scholar]
- 133. Ye X., Zerlanko B., Kennedy A., Banumathy G., Zhang R., Adams P.D.. Downregulation of Wnt signaling is a trigger for formation of facultative heterochromatin and onset of cell senescence in primary human cells. Mol. Cell. 2007; 27:183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Berchtold D., Battich N., Pelkmans L.. A systems-level study reveals regulators of membrane-less organelles in human cells. Mol. Cell. 2018; 72:1035–1049. [DOI] [PubMed] [Google Scholar]
- 135. la Vega de L., Fröbius K., Moreno R., Calzado M.A., Geng H., Schmitz M.L.. Control of nuclear HIPK2 localization and function by a SUMO interaction motif. BBA - Mol. Cell Res. 2011; 1813:283–297. [DOI] [PubMed] [Google Scholar]
- 136. Shiels C., Islam S.A., Vatcheva R., Sasieni P., Sternberg M.J., Freemont P.S., Sheer D.. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J. Cell Sci. 2001; 114:3705–3716. [DOI] [PubMed] [Google Scholar]
- 137. Zheng P., Guo Y., Niu Q., Levy D.E., Dyck J.A., Lu S., Sheiman L.A., Liu Y.. Proto-oncogene PML controls genes devoted to MHC class I antigen presentation. Nature. 1998; 396:373–376. [DOI] [PubMed] [Google Scholar]
- 138. Sun Y., Durrin L.K., Krontiris T.G.. Specific interaction of PML bodies with the TP53 locus in Jurkat interphase nuclei. Genomics. 2003; 82:250–252. [DOI] [PubMed] [Google Scholar]
- 139. Aoto T., Saitoh N., Ichimura T., Niwa H., Nakao M.. Nuclear and chromatin reorganization in the MHC-Oct3/4 locus at developmental phases of embryonic stem cell differentiation. Dev. Biol. 2006; 298:354–367. [DOI] [PubMed] [Google Scholar]
- 140. Salsman J., Stathakis A., Parker E., Chung D., Anthes L.E., Koskowich K.L., Lahsaee S., Gaston D., Kukurba K.R., Smith K.S. et al.. PML nuclear bodies contribute to the basal expression of the mTOR inhibitor DDIT4. Sci. Rep. 2017; 7:45038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ulbricht T., Alzrigat M., Horch A., Reuter N., Mikecz von A., Steimle V., Schmitt E., Krämer O.H., Stamminger T., Hemmerich P.. PML promotes MHC class II gene expression by stabilizing the class II transactivator. J. Cell Biol. 2012; 199:49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Gialitakis M., Arampatzi P., Makatounakis T., Papamatheakis J.. Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol. Cell. Biol. 2010; 30:2046–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Kumar P.P., Bischof O., Purbey P.K., Notani D., Urlaub H., Dejean A., Galande S.. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 2007; 9:45–56. [DOI] [PubMed] [Google Scholar]
- 144. Delbarre E., Ivanauskiene K., Spirkoski J., Shah A., Vekterud K., Moskaug J.Ø., Bøe S.O., Wong L.H., Küntziger T., Collas P.. PML protein organizes heterochromatin domains where it regulates histone H3.3 deposition by ATRX/DAXX. Genome Res. 2017; 27:913–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Ching R.W., Ahmed K., Boutros P.C., Penn L.Z., Bazett-Jones D.P.. Identifying gene locus associations with promyelocytic leukemia nuclear bodies using immuno-TRAP. J. Cell Biol. 2013; 201:325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Ishov A.M., Maul G.G.. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 1996; 134:815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Maul G.G., Ishov A.M., Everett R.D.. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996; 217:67–75. [DOI] [PubMed] [Google Scholar]
- 148. Sourvinos G., Everett R.D.. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. EMBO J. 2002; 21:4989–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Tang Q., Li L., Ishov A.M., Revol V., Epstein A.L., Maul G.G.. Determination of minimum herpes simplex virus type 1 components necessary to localize transcriptionally active DNA to ND10. J. Virol. 2003; 77:5821–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Swindle C.S., Zou N., Van Tine B.A., Shaw G.M., Engler J.A., Chow L.T.. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 1999; 73:1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Bell P., Lieberman P.M., Maul G.G.. Lytic but not latent replication of epstein-barr virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J. Virol. 2000; 74:11800–11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Day P.M., Baker C.C., Lowy D.R., Schiller J.T.. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc. Natl Acad. Sci. U.S.A. 2004; 101:14252–14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Bishop C.L., Ramalho M., Nadkarni N., May Kong W., Higgins C.F., Krauzewicz N.. Role for centromeric heterochromatin and PML nuclear bodies in the cellular response to foreign DNA. Mol. Cell. Biol. 2006; 26:2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Bryan T.M., Englezou A., Gupta J., Bacchetti S., Reddel R.R.. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995; 14:4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Bryan T.M., Englezou A., Dalla-Pozza L., Dunham M.A., Reddel R.R.. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997; 3:1271–1274. [DOI] [PubMed] [Google Scholar]
- 156. Cesare A.J., Reddel R.R.. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 2010; 11:319–330. [DOI] [PubMed] [Google Scholar]
- 157. Yeager T.R., Neumann A.A., Englezou A., Huschtscha L.I., Noble J.R., Reddel R.R.. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999; 59:4175–4179. [PubMed] [Google Scholar]
- 158. Draskovic I., Arnoult N., Steiner V., Bacchetti S., Lomonte P., Londoño-Vallejo A.. Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:15726–15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Luciani J.J., Depetris D., Usson Y., Metzler-Guillemain C., Mignon-Ravix C., Mitchell M.J., Megarbane A., Sarda P., Sirma H., Moncla A. et al.. PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. J. Cell Sci. 2006; 119:2518–2531. [DOI] [PubMed] [Google Scholar]
- 160. Smeets D.F., Moog U., Weemaes C.M., Vaes-Peeters G., Merkx G.F., Niehof J.P., Hamers G.. ICF syndrome: a new case and review of the literature. Hum. Genet. 1994; 94:240–246. [DOI] [PubMed] [Google Scholar]
- 161. Jeanpierre M., Turleau C., Aurias A., Prieur M., Ledeist F., Fischer A., Viegas-Péquignot E.. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum. Mol. Genet. 1993; 2:731–735. [DOI] [PubMed] [Google Scholar]
- 162. Xu G.L., Bestor T.H., Bourc’his D., Hsieh C.L., Tommerup N., Bugge M., Hulten M., Qu X., Russo J.J., Viegas-Péquignot E.. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999; 402:187–191. [DOI] [PubMed] [Google Scholar]
- 163. Catez F., Picard C., Held K., Gross S., Rousseau A., Theil D., Sawtell N., Labetoulle M., Lomonte P.. HSV-1 genome subnuclear positioning and associations with host-cell PML-NBs and centromeres regulate LAT locus transcription during latency in neurons. PLoS Pathog. 2012; 8:e1002852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Everett R.D., Murray J., Orr A., Preston C.M.. Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J. Virol. 2007; 81:10991–11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Alandijany T., Roberts A.P.E., Conn K.L., Loney C., McFarlane S., Orr A., Boutell C.. Distinct temporal roles for the promyelocytic leukaemia (PML) protein in the sequential regulation of intracellular host immunity to HSV-1 infection. PLoS Pathog. 2018; 14:e1006769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Bell P., Brazas R., Ganem D., Maul G.G.. Hepatitis delta virus replication generates complexes of large hepatitis delta antigen and antigenomic RNA that affiliate with and alter nuclear domain 10. J. Virol. 2000; 74:5329–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Li Y.-J., Macnaughton T., Gao L., Lai M.M.C.. RNA-templated replication of hepatitis delta virus: genomic and antigenomic RNAs associate with different nuclear bodies. J. Virol. 2006; 80:6478–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Newhart A., Rafalska-Metcalf I.U., Yang T., Negorev D.G., Janicki S.M.. Single-cell analysis of Daxx and ATRX-dependent transcriptional repression. J. Cell Sci. 2012; 125:5489–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Shastrula P.K., Sierra I., Deng Z., Keeney F., Hayden J.E., Lieberman P.M., Janicki S.M.. PML is recruited to heterochromatin during S phase and represses DAXX-mediated histone H3.3 chromatin assembly. J. Cell Sci. 2019; 132:jcs220970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Dellaire G., Bazett-Jones D.P.. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays. 2004; 26:963–977. [DOI] [PubMed] [Google Scholar]
- 171. Dellaire G., Ching R.W., Dehghani H., Ren Y., Bazett-Jones D.P.. The number of PML nuclear bodies increases in early S phase by a fission mechanism. J. Cell Sci. 2006; 119:1026–1033. [DOI] [PubMed] [Google Scholar]
- 172. Dellaire G., Ching R.W., Ahmed K., Jalali F., Tse K.C.K., Bristow R.G., Bazett-Jones D.P.. Promyelocytic leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J. Cell Biol. 2006; 175:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Kepkay R., Attwood K.M., Ziv Y., Shiloh Y., Dellaire G.. KAP1 depletion increases PML nuclear body number in concert with ultrastructural changes in chromatin. Cell Cycle. 2011; 10:308–322. [DOI] [PubMed] [Google Scholar]
- 174. Eskiw C.H., Dellaire G., Mymryk J.S., Bazett-Jones D.P.. Size, position and dynamic behavior of PML nuclear bodies following cell stress as a paradigm for supramolecular trafficking and assembly. J. Cell Sci. 2003; 116:4455–4466. [DOI] [PubMed] [Google Scholar]
- 175. Everett R.D., Chelbi-Alix M.K.. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007; 89:819–830. [DOI] [PubMed] [Google Scholar]
- 176. Everett R.D., Boutell C., Hale B.G.. Interplay between viruses and hostsumoylation pathways. Nat Rev Micro. 2013; 11:400–411. [DOI] [PubMed] [Google Scholar]
- 177. Lomonte P. The interaction between herpes simplex virus 1 genome and promyelocytic leukemia nuclear bodies (PML-NBs) as a hallmark of the entry in latency. Microb Cell. 2016; 3:569–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Knipe D.M. Nuclear sensing of viral DNA, epigenetic regulation of herpes simplex virus infection, and innate immunity. Virology. 2015; 479–480:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Placek B.J., Huang J., Kent J.R., Dorsey J., Rice L., Fraser N.W., Berger S.L.. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J. Virol. 2009; 83:1416–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Zhong S., Salomoni P., Pandolfi P.P.. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2000; 2:E85–E90. [DOI] [PubMed] [Google Scholar]
- 181. Tatham M.H., Geoffroy M.-C., Shen L., Plechanovova A., Hattersley N., Jaffray E.G., Palvimo J.J., Hay R.T.. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008; 10:538–546. [DOI] [PubMed] [Google Scholar]
- 182. Tan J.-A.T., Sun Y., Song J., Chen Y., Krontiris T.G., Durrin L.K.. SUMO conjugation to the matrix attachment region-binding protein, special AT-rich sequence-binding protein-1 (SATB1), targets SATB1 to promyelocytic nuclear bodies where it undergoes caspase cleavage. J. Biol. Chem. 2008; 283:18124–18134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Li H., Leo C., Zhu J., Wu X., O’Neil J., Park E.J., Chen J.D.. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 2000; 20:1784–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Lin D.-Y., Lai M.-Z., Ann D.K., Shih H.-M.. Promyelocytic leukemia protein (PML) functions as a glucocorticoid receptor co-activator by sequestering Daxx to the PML oncogenic domains (PODs) to enhance its transactivation potential. J. Biol. Chem. 2003; 278:15958–15965. [DOI] [PubMed] [Google Scholar]
- 185. Newhart A., Rafalska-Metcalf I.U., Yang T., Joo L.M., Powers S.L., Kossenkov A.V., Lopez-Jones M., Singer R.H., Showe L.C., Skordalakes E. et al.. Single cell analysis of RNA-mediated histone H3.3 recruitment to a cytomegalovirus promoter-regulated transcription site. J. Biol. Chem. 2013; 288:19882–19899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A.. A phase separation model for transcriptional control. Cell. 2017; 169:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Shin Y., Chang Y.-C., Lee D.S.W., Berry J., Sanders D.W., Ronceray P., Wingreen N.S., Haataja M., Brangwynne C.P.. Liquid nuclear condensates mechanically sense and restructure the genome. Cell. 2018; 175:1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Wang L., Gao Y., Zheng X., Liu C., Dong S., Li R., Zhang G., Wei Y., Qu H., Li Y. et al.. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol. Cell. 2019; 76:646–659. [DOI] [PubMed] [Google Scholar]
- 189. Everett R.D. Dynamic response of IFI16 and promyelocytic leukemia nuclear body components to herpes simplex virus 1 infection. J. Virol. 2016; 90:167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Criscione S.W., Teo Y.V., Neretti N.. The chromatin landscape of cellular senescence. Trends Genet. 2016; 32:751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Zhang R., Chen W., Adams P.D.. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 2007; 27:2343–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Narita M., Nũnez S., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W.. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003; 113:703–716. [DOI] [PubMed] [Google Scholar]
- 193. Funayama R., Saito M., Tanobe H., Ishikawa F.. Loss of linker histone H1 in cellular senescence. J. Cell Biol. 2006; 175:869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Corpet A., Stucki M.. Chromatin maintenance and dynamics in senescence: a spotlight on SAHF formation and the epigenome of senescent cells. Chromosoma. 2014; 123:423–436. [DOI] [PubMed] [Google Scholar]
- 195. de Stanchina E., Querido E., Narita M., Davuluri R.V., Pandolfi P.P., Ferbeyre G., Lowe S.W.. PML is a direct p53 target that modulates p53 effector functions. Mol. Cell. 2004; 13:523–535. [DOI] [PubMed] [Google Scholar]
- 196. Ye X., Zerlanko B., Zhang R., Somaiah N., Lipinski M., Salomoni P., Adams P.D.. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 2007; 27:2452–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Vernier M., Bourdeau V., Gaumont-Leclerc M.-F., Moiseeva O., Begin V., Saad F., Mes-Masson A.-M., Ferbeyre G.. Regulation of E2Fs and senescence by PML nuclear bodies. Genes Dev. 2011; 25:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Talluri S., Dick F.A.. The retinoblastoma protein and PML collaborate to organize heterochromatin and silence E2F-responsive genes during senescence. Cell Cycle. 2014; 13:641–651. [DOI] [PubMed] [Google Scholar]
- 199. Rai T.S., Cole J.J., Nelson D.M., Dikovskaya D., Faller W.J., Vizioli M.G., Hewitt R.N., Anannya O., McBryan T., Manoharan I. et al.. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev. 2014; 28:2712–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Luciani J.J., Depetris D., Missirian C., Mignon-Ravix C., Metzler-Guillemain C., Megarbane A., Moncla A., Mattei M.-G.. Subcellular distribution of HP1 proteins is altered in ICF syndrome. Eur. J. Hum. Genet. 2004; 13:41–51. [DOI] [PubMed] [Google Scholar]
- 201. Santenard A., Ziegler-Birling C., Koch M., Tora L., Bannister A.J., Torres-Padilla M.-E.. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat. Cell Biol. 2010; 12:853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Nishibuchi G., Dejardin J.. The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals. 2017; 25:77–87. [DOI] [PubMed] [Google Scholar]
- 203. Probst A.V., Okamoto I., Casanova M., Marjou El, LeBaccon F., Almouzni G.. A strand-specific burst in transcriptionof pericentric satellites is required for chromocenter formation and early mouse development. Dev. Cell. 2010; 19:625–638. [DOI] [PubMed] [Google Scholar]
- 204. Spirkoski J., Shah A., Reiner A.H., Collas P., Delbarre E.. Biochemical and biophysical research communications. Biochem. Biophys. Res. Commun. 2019; 511:882–888. [DOI] [PubMed] [Google Scholar]
- 205. Chung I., Osterwald S., Deeg K.I., Rippe K.. PML body meets telomere. nucleus. 2012; 3:263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Zhang J.-M., Zou L.. Alternative lengthening of telomeres: from molecular mechanisms to therapeutic outlooks. Cell & Bioscience. 2020; 10:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Loe T.K., Li J.S.Z., Zhang Y., Azeroglu B., Boddy M.N., Denchi E.L.. Telomere length heterogeneity in ALT cells is maintained by PML-dependent localization of the BTR complex to telomeres. Genes Dev. 2020; 34:650–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Zhang J.-M., Yadav T., Ouyang J., Lan L., Zou L.. Alternative lengthening of telomeres through two distinct break-induced replication pathways. Cell Reports. 2019; 26:955–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Osterwald S., Deeg K.I., Chung I., Parisotto D., Worz S., Rohr K., Erfle H., Rippe K.. PML induces compaction, TRF2 depletion and DNA damage signaling at telomeres and promotes their alternative lengthening. J. Cell Sci. 2015; 128:1887–1900. [DOI] [PubMed] [Google Scholar]
- 210. Heaphy C.M., de Wilde R.F., Jiao Y., Klein A.P., Edil B.H., Shi C., Bettegowda C., Rodriguez F.J., Eberhart C.G., Hebbar S. et al.. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011; 333:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Lovejoy C.A., Li W., Reisenweber S., Thongthip S., Bruno J., de Lange T., De S., Petrini J.H.J., Sung P.A., Jasin M. et al.. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLos Genet. 2012; 8:e1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Ceccarelli M., Barthel F.P., Malta T.M., Sabedot T.S., Salama S.R., Murray B.A., Morozova O., Newton Y., Radenbaugh A., Pagnotta S.M. et al.. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016; 164:550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Li F., Deng Z., Zhang L., Wu C., Jin Y., Hwang I., Vladimirova O., Xu L., Yang L., Lu B. et al.. ATRX loss induces telomere dysfunction and necessitates induction of alternative lengthening of telomeres during human cell immortalization. EMBO J. 2019; 38:e96659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. O'Sullivan R.J., Arnoult N., Lackner D.H., Oganesian L., Haggblom C., Corpet A., Almouzni G., Karlseder J.. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat. Struct. Mol. Biol. 2014; 21:167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Gauchier M., Kan S., Barral A., Sauzet S., Agirre E., Bonnell E., Saksouk N., Barth T.K., Ide S., Urbach S. et al.. SETDB1-dependent heterochromatin stimulates alternative lengthening of telomeres. Sci. Adv. 2019; 5:eaav3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Episkopou H., Draskovic I., Van Beneden A., Tilman G., Mattiussi M., Gobin M., Arnoult N., Londoño-Vallejo A., Decottignies A.. Alternative lengthening of telomeres is characterized by reduced compaction of telomeric chromatin. Nucleic Acids Res. 2014; 42:4391–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Pan X., Chen Y., Biju B., Ahmed N., Kong J., Goldenberg M., Huang J., Mohan N., Klosek S., Parsa K. et al.. FANCM suppresses DNA replication stress at ALT telomeres by disrupting TERRA R-loops. Sci. Rep. 2019; 9:19110–19114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Chang F.T.M., McGhie J.D., Chan F.L., Tang M.C., Anderson M.A., Mann J.R., Andy Choo K.H., Wong L.H.. PML bodies provide an important platform for the maintenance of telomeric chromatin integrity in embryonic stem cells. Nucleic Acids Res. 2013; 41:4447–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Wong L.H., McGhie J.D., Sim M., Anderson M.A., Ahn S., Hannan R.D., George A.J., Morgan K.A., Mann J.R., Choo K.H.A.. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010; 20:351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Wong L.H., Ren H., Williams E., McGhie J., Ahn S., Sim M., Tam A., Earle E., Anderson M.A., Mann J. et al.. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009; 19:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Marchesini M., Matocci R., Tasselli L., Cambiaghi V., Orleth A., Furia L., Marinelli C., Lombardi S., Sammarelli G., Aversa F. et al.. PML is required for telomere stability in non-neoplastic human cells. Oncogene. 2016; 35:1811–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Chang H.R., Munkhjargal A., Kim M.-J., Park S.Y., Jung E., Ryu J.-H., Yang Y., Lim J.-S., Kim Y.. The functional roles of PML nuclear bodies in genome maintenance. Mut. Res./Fundam. Mol. Mech. Mutagen. 2018; 809:99–107. [DOI] [PubMed] [Google Scholar]
- 223. Pan X., Ahmed N., Kong J., Zhang D.. Breaking the end: Target the replication stress response at the ALT telomeres for cancer therapy. Mol. Cell. Oncol. 2017; 4:e1360978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Pan X., Drosopoulos W.C., Sethi L., Madireddy A., Schildkraut C.L., Zhang D.. FANCM, BRCA1, and BLM cooperatively resolve the replication stress at the ALT telomeres. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E5940–E5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Flynn R.L., Cox K.E., Jeitany M., Wakimoto H., Bryll A.R., Ganem N.J., Bersani F., Pineda J.R., Suva M.L., Benes C.H. et al.. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015; 347:273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Episkopou H., Diman A., Claude E., Viceconte N., Decottignies A.. TSPYL5 depletion induces specific death of ALT cells through USP7-dependent proteasomal degradation of POT1. Mol. Cell. 2019; 75:469–482. [DOI] [PubMed] [Google Scholar]
- 227. Gong P., Wang H., Zhang J., Fu Y., Zhu Z., Wang J., Yin Y., Wang H., Zhou Z., Yang J. et al.. Telomere maintenance-associated PML is a potential specific therapeutic target of human colorectal cancer. Transl. Oncol. 2019; 12:1164–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Roux K.J., Kim D.I., Raida M., Burke B.. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012; 196:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Kim T.H., Tsang B., Vernon R.M., Sonenberg N., Kay L.E., Forman-Kay J.D.. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science. 2019; 365:825–829. [DOI] [PubMed] [Google Scholar]
- 230. Dabin J., Fortuny A., Polo S.E.. Epigenome maintenance in response to DNA damage. Mol. Cell. 2016; 62:712–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Vancurova M., Hanzlikova H., Knoblochova L., Kosla J., Majera D., Mistrik M., Burdova K., Hodny Z., Bartek J.. PML nuclear bodies are recruited to persistent DNA damage lesions in an RNF168-53BP1 dependent manner and contribute to DNA repair. DNA Repair (Amst.). 2019; 78:114–127. [DOI] [PubMed] [Google Scholar]
- 232. Carbone R., Pearson M., Minucci S., Pelicci P.G.. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene. 2002; 21:1633–1640. [DOI] [PubMed] [Google Scholar]
- 233. Gallay L., Mouchiroud G., Chazaud B.. Interferon-signature in idiopathic inflammatory myopathies. Curr. Opin. Rheumatol. 2019; 31:634–642. [DOI] [PubMed] [Google Scholar]
- 234. Sophie Trouillet-Assant P., Sebastien Viel P.P., Alexandre Gaymard P.P., Sylvie Pons M.S., Jean-Christophe Richard M.P., Magali Perret M.S., Marine Villard P., Karen Brengel-Pesce P., Bruno Lina M.P., Mehdi Mezidi M.D. et al.. Type I IFN immunoprofiling in COVID-19 patients. J. Allergy Clin. Immunol. 2020; 146:206–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C. et al.. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020; 31:eabc6027-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Maison C., Bailly D., Roche D., de Oca R.M., Probst A.V., Vassias I., Dingli F., Lombard B., Loew D., Quivy J.-P. et al.. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat. Genet. 2011; 43:220–227. [DOI] [PubMed] [Google Scholar]
- 237. Niwa-Kawakita M., Ferhi O., Soilihi H., Le Bras M., Lallemand-Breitenbach V., de Thé H.. PML is a ROS sensor activating p53 upon oxidative stress. J. Exp. Med. 2017; 214:3197–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]