Abstract
LSH, a SNF2 family DNA helicase, is a key regulator of DNA methylation in mammals. How LSH facilitates DNA methylation is not well defined. While previous studies with mouse embryonic stem cells (mESc) and fibroblasts (MEFs) derived from Lsh knockout mice have revealed a role of Lsh in de novo DNA methylation by Dnmt3a/3b, here we report that LSH contributes to DNA methylation in various cell lines primarily by promoting DNA methylation by DNMT1. We show that loss of LSH has a much bigger effect in DNA methylation than loss of DNMT3A and DNMT3B. Mechanistically, we demonstrate that LSH interacts with UHRF1 but not DNMT1 and facilitates UHRF1 chromatin association and UHRF1-catalyzed histone H3 ubiquitination in an ATPase activity-dependent manner, which in turn promotes DNMT1 recruitment to replication fork and DNA methylation. Notably, UHRF1 also enhances LSH association with the replication fork. Thus, our study identifies LSH as an essential factor for DNA methylation by DNMT1 and provides novel insight into how a feed-forward loop between LSH and UHRF1 facilitates DNMT1-mediated maintenance of DNA methylation in chromatin.
INTRODUCTION
DNA methylation in cytosine is a conserved epigenetic modification essential for embryonic development and cell differentiation in mammals (1–4). While all three active DNA methyltransferases, namely DNMT3A, DNMT3B and DNMT1 act cooperatively to set up patterns of DNA methylation during embryonic development, DNMT1 is generally considered as the primary enzyme responsible for maintenance of DNA methylation patterns in somatic cells (5–7). Consistent with a role in maintaining patterns of DNA methylation upon DNA replication, DNMT1 is recruited to DNA replication fork and preferentially converts hemi-methylated CpGs generated during DNA replication to fully methylated sites. Studies over the last decade or so have established UHRF1 as an essential accessory factor required for targeting DNMT1 to replication fork (8,9).
UHRF1 is a multi-functional domain protein. It binds to newly replicated DNA in S phase by specific recognition of hemi-methylated CpG and histone tails (10–17). Multiple lines of evidence support that UHRF1 in turn ubiquitinates histone H3 and recruits DNMT1 to replication fork at least in part through an interaction between ubiquitinated H3 and DNMT1 (18,19). The binding of ubiquitinated H3 also stimulates DNMT1 enzymatic activity (20). Although early studies suggest that DNA maintenance methylation is a rapid process and thus may occur before chromatin assembly, the dependence of DNMT1 recruitment and activation on ubiquitinated histones, together with recent findings that there is a global delay in nascent strand DNA methylation (21), indicates that DNA methylation by DNMT1 takes place at least in part in chromatin. In its default state, chromatin structure limits DNA methylation by DNMT1. How UHRF1/DNMT1 gains access to chromatin in S phase is presently unknown.
ATPase chromatin remodeling proteins can mobilize and restructure nucleosomes through the energy of ATP hydrolysis and thus promote chromatin DNA accessibility (22,23). LSH (also known as Hells, PASG and SMARCA6) and its plant homolog DDM are the SNF2 family DNA helicases that play a critical role in global DNA methylation both in mammals and plants (24,25). Targeted deletion of Lsh in mice results in perinatal lethality and substantial loss of DNA methylation throughout the genome (24,26–28). Consistent with its helicase activity, LSH has been shown to promote DNA methylation depending on its ATPase activity (29,30). LSH has also been shown to interact with DNMT3A and DNMT3B but not DNMT1 and promote de novo but not the maintenance of pre-existed DNA methylation in an episomal DNA based assay (26,31). Thus, the current prevailing view is that LSH contributes to DNA methylation in mammals by promoting de novo methylation by DNMT3A/3B. However, we noted that nearly all these mechanistic studies were carried out by using Lsh−/− embryonic stem (ES) cells or embryonic fibroblast (MEF) cells derived from Lsh knockout mice, whose DNA methylation pattern has gone through drastic demethylation and remethylation reprogramming in early embryonic development and the remethylation process is in fact determined by a coordinated function of all three DNA methyltransferases. It remains to be vigorously tested if and how LSH is involved in DNA maintenance methylation by UHRF1/DNMT1 axis.
In this study, we have generated multiple LSH-null cell lines by using CRISPR/Cas9 technology. We show that loss of LSH has a much bigger effect in DNA methylation than that of both DNMT3A and DNMT3B, suggesting that LSH also plays a role in DNA methylation by DNMT1. Although LSH does not interact with DNMT1, we show that it interacts with UHRF1 and enhances UHRF1 chromatin association and activity for H3 ubiquitination, and consequently promotes DNMT1 recruitment in the S phase of cell cycle. Interestingly, we find that UHRF1 is also required for efficient targeting of LSH to replication fork. Our study thus not only reveals a critical role of LSH in DNA methylation by DNMT1 but also provides novel insight into the underlying mechanisms.
MATERIALS AND METHODS
Cell culture
Human embryonic kidney 293T cell line (HEK293T), human cervical cancer cell line (HeLa) and mouse embryo fibroblast cell line (NIH3T3) were maintained in DMEM medium (Gibco) containing 10% fetal calf serum (Gemini), 100 U/ml penicillin and 100 μg/ml streptomycin (Millipore). Human HCT116 colon cancer cells were maintained in McCoy's 5A medium (Gibco) containing 10% fetal calf serum (Gemini), 100 U/ml penicillin, and 100 μg/ml streptomycin (Millipore). The cultured cells were maintained at 37°C in a humidified incubator with 5% CO2.
Knockdown by shRNA
For knockdown of target genes by shRNAs, shRNA lentiviral particles were packaged and transduced into the indicated cells according to the manufacturer's guidelines.
The sequences of UHRF1 shRNA were listed below:
shUHRF1-#1, 5′- GCCTTTGAT TCGTTCCTTCTT-3′;
shUHRF1-#2, 5′- ATGTGGGATGAGACGGAATTG-3′.
The sequences of LSH shRNA were listed below:
shLSH-#1, 5′- GATCAAGAGAGAAGGTCATTA-3′;
shLSH-#2, 5′- GAACAAAGAAGTATCCATATT -3′.
Generation of LSH KO, DNMT3A/3B DKO and DNMT3A/3B/LSH TKO cells by the CRISPR–Cas9
The LSH KO, DNMT3A/DNMT3B DKO and LSH/ DNMT3A /DNMT3B TKO cell lines were obtained by the CRISPR–Cas9 system essentially as described (32) with the following guide RNAs:
CAGTTAGGAAGTGTAGACAA for LSH;
GCTACCACGCCTGAGCCCGT for DNMT3A;
AGACTCGATCCTCGTC AACG for DNMT3B.
The same guide RNA sequence was used for knockout of human LSH gene in HeLa, HCT116 and MCF7 cells and mouse Lsh gene in NIH3T3 cells since the targeting sequence selected is identical between human LSH and mouse Lsh genes.
Generation of LSH-KO HeLa cell lines re-expressing wild-type LSH and mutants
The LSH-KO HeLa cell lines were transfected with constructs encoding wild-type and LSH mutants as indicated. The stable transfected cells were obtained by selection in culture medium with 50 μg/ml hygromycin (Sigma) for about 2 weeks.
Western blot analysis
Cells were directly lysed by 1 × SDS loading buffer (62.5 mM Tris–HCl, pH 6.8; 2% w/v SDS; 10% glycerol; 1% v/v β-mercaptoethanol; 0.01% w/v bromophenol blue). Lysates were boiled at 95°C for 15 min and then subjected to gel separation by SDS-PAGE. Proteins were transferred to the nitrocellulose membrane (GE Healthcare Life Science), and the membranes were blocked with 8% milk for 1 h at room temperature. After overnight incubation at 4°C with primary antibodies, the membranes were washed three times with PBST buffer (0.1% Tween-20 in 1 × PBS), followed by incubation with Alexa Fluor® 680 goat anti-rabbit or Alexa Fluor® 790 goat anti-mouse antibody (Jackson ImmunoResearch, dilution: 1:10 000) for 1 h at room temperature. The membranes were visualized by the Odyssey CLx Imaging System (LI-COR Bioscience). Quantification was performed by using ImageJ software and the value of UHRF1 or ub-H3 in the control sample was set as 1.
Immunofluorescence staining assay
Cells grown on slides were washed with 1× PBS and fixed with 4% paraformaldehyde for 20 min at 4°C. After fixation, the cells were permeabilized with 1% Triton X-100 in 1× PBS for 20 min at 4°C and then the cells were blocked with 5% bovine serum albumin (BSA) (Sigma) in 1× PBS for 1 hr at 37°C and incubated with primary antibodies overnight at 4°C. After washing three times with 1 × PBS, the cells were incubated with Alexa Fluor® 594 goat anti-rabbit IgG (Jackson ImmunoResearch, 111-585-003, 1:500) and Alexa Fluor® 488 goat anti-mouse IgG (Jackson ImmunoResearch, 115-545-003, 1:500) at 37°C for 1 h. Finally, the nuclei were stained by Hoechst 33342 (Sigma). After washing three times with 1× PBS, the images were acquired with a Leica SP8 confocal microscope.
Co-immunoprecipitation assay
For co-immunoprecipitation of exogenous proteins, the indicated plasmid(s) were transfected into HEK 293T or HCT116 cells. The cells were collected 48 hr after transfection and lysed in IP Lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 1 mM EDTA, 8% glycerol, 1× protease inhibitor cocktail (MCE), and 1 mM DTT (Amresco)). The lysates were cleared by centrifugation at 12 000 rpm for 20 min at 4°C. The supernatant was directly incubated with anti-Flag M2-affinity beads (Biotool) or antibodies as indicated for 3 h at 4°C. For co-immunoprecipitation of endogenous proteins, antibodies were added at a concentration of 1 μg/mg of lysates and incubated overnight at 4°C, followed by antibody-protein complex capture with Protein G/ Protein A Sepharose beads (Santa Cruz). After extensive washing, complexes were boiled in 1× SDS loading buffer, separated by SDS-PAGE, and analyzed by western blotting.
In vitro pulldown assay
To express UHRF1 in bacteria, PCR products encoding UHRF1 were ligated into pGEX4T-1 vector. GST-UHRF1 fusion protein was expressed and purified from E. coli. To purify FLAG-tagged LSH protein from mammalian cells, the 293T cells were transfected with plasmid encoding FLAG-LSH for 48 h. The cells were collected and lysed in high salt Lysis buffer (25 mM Tris–HCl, pH 8.0, 500 mM NaCl, 1% Triton X-100, 2 mM EDTA, 1× protease inhibitor cocktail, 1 mM DTT). FLAG-LSH protein was then captured with anti-FLAG M2-affinity beads and eluted with FLAG-peptide elution buffer (100 μg/ml FLAG-peptides, 50 mM Tris-HCl, pH 8.0, 10% glycerol, 1 mM EDTA, 1× protease inhibitor cocktail, 1 mM DTT). For pulldown assay to test in vitro binding of UHRF1 and LSH, 1 μg of purified FLAG-LSH and 2μg of bead-bound GST-UHRF1 beads were incubated in pulldown binding buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0. 1% Triton X-100, 1 mM EDTA, 8% glycerol, 1× protease inhibitor cocktail (MCE), and 1 mM DTT (Amresco)) for 6 h at 4°C. The resulting beads were washed three times with ice-cold pulldown wash buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0. 1% Triton X-100, 1 mM EDTA, 1× protease inhibitor cocktail (MCE) and 1 mM DTT (Amresco)). After extensive washing, complexes were boiled in 1 × SDS loading buffer and analyzed by western blotting and Coomassie blue staining.
Histone acid extraction
Histone acid extraction was performed as described (18).
In vitro ubiquitination assay
Recombinant polynucleosomes (H3.1) were obtained commercially (Active Motif, 31466). Human UHRF1 was cloned into pGEX4T1 plasmid as N-terminal GST-tagged fusion protein. GST-UHRF1 protein was expressed in E. coli BL21 cells and purified by GST affinity column and full-length UHRF1 proteins were obtained after thrombin cleavage. Recombinant UHRF1 proteins were concentrated using the Amicon Ultra 50K Centrifuge Filter Devices (Millipore). Ubiquitination assays were performed in 10 μl reactions containing 50 nM His6-UBE1 (Boston Biochem, E-304), 500 nM E2 GST-UBE2D1 (UbcH5a, Boston Biochem, E2-615), 1 μg ubiquitin, 0.5 μg UHRF1, 0.5 μg FLAG-LSH, 1 μg polynucleosomes, 50 mM Tris–HCl pH 7.5, 5 mM MgCl2 and 2 mM ATP. Assays were performed at 37°C for the indicated time. Reactions were stopped by adding 10 μl 2× SDS loading buffer and resolved by SDS-PAGE followed by western blotting.
Total RNA extraction and quantitative RT-PCR
Total RNA was extracted from cells using RNAiso Plus Reagent (Takara) according to the manufacturer's instructions. For RT-qPCR analysis, 2 μg total RNA were reverse-transcribed with TransScriptR One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech Co., Ltd). Gene expression levels were determined using qPCR with the CFX96 Real-Time System (Bio-Rad) using TransStart Green qPCR SuperMix (TransGen Biotech Co., Ltd) and normalized to GAPDH expression. The data represent mean ± STD for three repeats. Primers for RT-PCR are listed below.
5′-TCACATGGTAAGCGGGATGTC-3′ (forward)
5′-CGCACTATTGGCCACACATTC-3′ (reverse) for ERV #1;
5′- AGCAGGTCAGGTGCCTGTAACATT-3′ (forward)
5′-TGGTGCCGTAGGATTAAGTCTCCT-3′ (reverse) for ERV #2;
5′-GGCCA TCAGAGTCTAAACCACG-3′ (forward)
5′-CTGACTTTCTGGGGGTGGCCG-3′ (reverse) for ERV #3;
5′-CAACATAGTGAAACCCCGTCTCT-3′ (forward)
5′-GCCTCAGCCTCCCGAGTAG-3′ (reverse) for Alu #1;
5′-CATGGTGAAACC CCGTCTCTA-3′ (forward)
5′-GCCTCAGCCTCCCGAGTAG-3′ (reverse) for Alu #2;
5′-TAACCAATACAGAGAAGTGC-3′ (forward)
5′-GATAATATCCT GCAGAGTGT-3′ (reverse) for Line1.
HPLC analysis of 5mC
To prepare genomic DNA, cells were resuspended with cell lysis buffer (10 mM Tris–HCl pH 7.5, 10 mM EDTA, 10 mM NaCl, 0.5% sarcosyl, 0.1 mg/ml RNase (CWBIO)) and incubated at 37°C overnight. Then, protease K was added to a final concentration of 0.2 mg/ml and incubated at 65°C for 24 h. Genomic DNA was then extracted by phenol/chloroform and ethanol precipitated. DNA samples were dissolved in ddH2O. To hydrolyze the genomic DNA, 25 μg of denatured genomic DNA were incubated overnight in 133 μ; of the hydrolysis solution (40 mM NaAc pH 5.3, 1 mM ZnSO4, 1.5 U/ml nuclease P1 (Wako)) at 37°C overnight and 20 U CIP (NEB) and 15 μl CutSmart buffer (NEB) was then added and incubated for additional 4 h at 37°C. 50 μl of hydrolyzates were then analyzed using an HPLC system (Agilent Technologies, 1100 Series) equipped with an Agilent Eclipse XDB-C18 column (5 μm, 4.6 × 250 mm, Agilent Technologies Inc.). The levels of dC and 5mdC were detected by UV-detector at 280 nm wavelength.
Dot-blot analysis of 5hmC
Genomic DNA was denatured by heating at 95°C, immediately cooled on ice and loaded on nitrocellulose membrane (GE Healthcare Life Science). After UV cross-linking, the membrane was blocked with 8% milk for 1 h at room temperature. After overnight incubation at 4°C with 5hmC antibody (Active Motif, 39769), the membranes were washed three times with PBST buffer (0.1% Tween-20 in 1 × PBS), followed by incubation with Alexa Fluor® 680 goat anti-rabbit antibody (Jackson ImmunoResearch, dilution: 1:10 000) for 1 h at room temperature. The membranes were visualized by the Odyssey CLx Imaging System (LI-COR Bioscience).
Bisulfite DNA sequencing analysis
Bisulfite conversion was performed using the EZ DNA Methylation-GoldTM Kit (ZYMO Research) according to the Instruction manual. Bisulfite converted DNA was used in PCR amplification by TaKaRa Ex Taq HS (Takara). The PCR products were purified by gel extraction for Illumina® (ND102-0102) high-throughput sequencing. The primers for PCR amplification are listed below.
5′-GAGATTATATTTTATATTTGGTTTAGAGGG-3′ (forward)
5′-AACTATAATAAACTCCACCCAATTC-3′ (reverse) for human LINE-1;
5′-ATATTAAGGGAATTTAGAGGTTGG-3′ (forward)
5′-CCCCTACACACCTATAAATATTTC-3′ (reverse) for human HERV-K.
RRBS data analysis
RRBS analysis is essentially as described (33). RRBS reads were mapped to human genome hg38 by bismark (v0.19.0) (34). Only unique mapped reads were kept and CpGs with ≥5 reads mapped were used for further analysis. The mean profiles around genes were generated by deepTools (35). The genomic locations of down regulated (diff > 30) CpGs and target repeats (with the enrichment score) were annotated by annotatePeak.pl from HOMER package (36). Repeat annotations were carried out by RepeatMasker (37) for human were downloaded from UCSC. The CpG island annotations for hg38 were also downloaded from UCSC. Correlation of CpG methylation changes and gene expression were carried out for gene's transcription start site upstream 2 kb and downstream 2 kb, requiring the more than five CpGs in the TSS region, and the change of methylation is mean of the CpGs in the TSS region.
RNA-seq data analysis
Duplicates of RNA samples from control and LSH KO cells were carried out for RNA-seq. Paired-end RNA-seq reads for replicates of parental HeLa and LSH-KO cells were mapped to hg38 by STAR (v2.4.0d) (38) and then quantified by Cuffdiff (v2.2.1) (39). Differentially expressed genes (DEGs) were required P-value ≤1e–2 and fold change ≥2 as reported by Cuffdiff. The enriched GO terms for the DEGs were called by findGO.pl in HOMER package.
EdU staining assay
The EdU staining assay was performed according to the RiBoBio (C10310) Cell-Light™ EdU Fluorescent Detection Kit with a slight modification. In brief, cells grown on 48 wells were labeled with 20 μM EdU (5-ethynyl-2’-deoxyuridine) for ∼30 min at 37°C, washed with 1× PBS twice and fixed with 4% paraformaldehyde for 20 min at 4°C before neutralization with glycine (2 mg/ml). The cells were then permeabilized, blocked and incubated with antibodies as described above. Finally, the EdU incorporated into DNA was stained in Apollo reaction buffer at 37°C for 30 min. Slides were washed by methanol once and PBS twice and then for fluorescent detection with a Leica SP8 confocal microscope.
Modified eSPAN assay
To assay the association of UHRF1 or LSH with newly replicated DNA, the cells were grown in medium with addition of BrdU under experimental conditions for 30 min. For the first step ChIP, the cells were crosslinked with 1% formaldehyde for 15 min before neutralization with 0.125 M glycine. The cells were lysed by sonication in ChIP Lysis buffer (25 mM Tris–HCl pH 8.0, 0.1% SDS, 1 mM EDTA, 1× protease inhibitor cocktail (MCE)), and soluble cell extracts were recovered after centrifugation at 12 000 rpm and 4°C for 20 min. The indicated antibodies were added, followed by overnight incubation at 4°C on a rotator. Chromatin-antibody complexes were isolated with 20 μl of Protein-A Sepharose beads (Santa Cruz) blocked by sperm DNA and bovine serum albumin. After extensive washing, protein/DNA complexes were eluted from the beads in Elution buffer (50 mM Tris–HCl pH 8.0, 1% SDS, 10 mM EDTA) at 65°C. Immunoprecipitated DNA was purified by phenol/chloroform extraction and dissolved in TE buffer. The newly replicated, BrdU-containing DNA was then isolated by BrdU immunoprecipitation (BrdU-IP) (40) and analyzed by real-time quantitative PCR. The primers used were listed below.
5′- AAGGTCAATGGCAGAAAAGAA-3′ (forward)
5′- CAACGAAGGCCACAA GATGTC-3′ (reverse) for α-sat-#1;
5′-TCATTCCCACAAACTGCGTTG-3′ (forward)
5′-TCCAACGAAGGCCACAAGA-3′ (reverse) for α-sat-#2;
5′-CAGCCTGGGTGATAGAGCAAG-3′ (forward)
5′-AGAGAAAGAGGAAACACAAGGAGC-3′ (reverse) for Alu;
5′-AGAGGAAGGAATGCCTCTTGCAGT-3′ (forward)
5′-TTACAAAGCAGTATTGCTGCCCGC-3′ (reverse) for ERV;
5′-AGCCTAACTGGGAGGCACCC-3′ (forward)
5′-GATGATGGTGATGTACAGATGGG-3′ (reverse) for Line1.
Biochemical fractionation of chromatin-associated proteins
To assay the association of protein of interest with chromatin in S phase of cells, the cells were synchronized to S phase by aphidicolin treatment followed by release into fresh medium for 2 h. Fractionation of cytosol, nuclear and chromatin fractions were performed essentially as described (41).
Statistical analysis
All quantified data represent mean ± STD for repeats, as referenced in figure legend. HPLC/RT-PCR/Cell Growth Curve analyses were performed at least in three biological replicates (n = 3). P-values <0.05 was considered significant. ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05.
RESULTS
Knockout of LSH results in substantial loss of DNA methylation in multiple cell lines
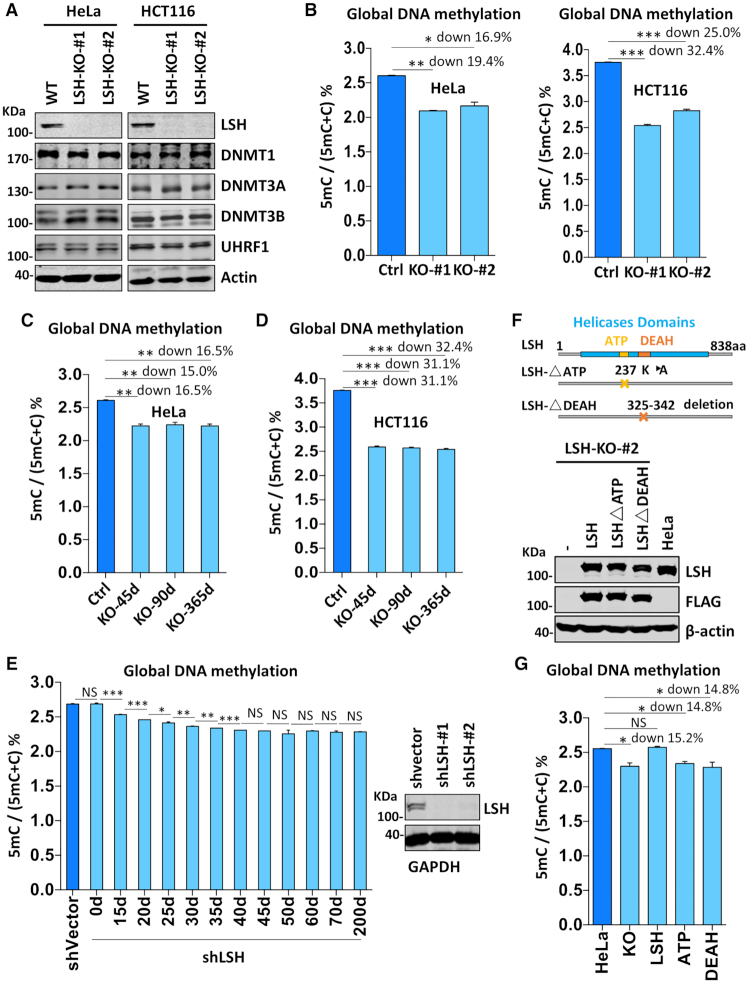
To investigate the role of LSH in DNA methylation, we generated LSH knockout (KO) cell lines from HeLa, HCT116, MCF7 and NIH3T3 cells as outlined in Supplemental Figure S1A and S1B. As the selected targeting sequence in human LSH and mouse Lsh genes is identical, the same guide RNA construct was used for knockout of LSH/Lsh in both human and mouse cell lines. The successful disruption of LSH alleles was confirmed by genomic DNA sequencing, as evident by short insertion and/or deletion of LSH sequences around the targeting region (Supplemental Figure S1C). We further validated the knockout status by western blotting analysis using two different LSH-specific antibodies, showing the lack of LSH protein in these selected knockout cell clones (Supplemental Figure S1D). Multiple LSH KO clones were generated for each cell line and all LSH KO cells showed a reduced proliferation (representative results for HeLa and HCT116 in Supplemental Figure S2).
To analyze the effect of loss of LSH on DNA methylation, we first analyzed if loss of LSH affects the expression of proteins involved in DNA methylation in HeLa and HCT116 cells. As shown in Figure 1A, loss of LSH did not significantly affect the levels of DNMT1, DNMT3A, DNMT3B and UHRF1 proteins. We then prepared genomic DNA from control and two individual KO clones for each cell type and quantitatively determined the level of DNA methylation (5-mC) by High-Performance Liquid Chromatography (HPLC). As shown in Figure 1B, LSH knockout resulted in a substantial reduction of global DNA methylation in both HeLa (an average of 18.2% reduction) and HCT116 cells (an average of 28.7% reduction). Similarly, we found that LSH knockout also led to substantial reduction of global DNA methylation in MCF7 (an average of 13.1% reduction) and NIH3T3 cells (an average of 21.2% reduction) (Supplemental Figure S1E). Consistent with the marked reduction of global DNA methylation measured by HPLC, bisulfite DNA sequencing analysis revealed strong reduction of DNA methylation in LINE-1 repetitive sequences in LSH-KO HeLa cells (Supplemental Figure S3). On the other hand, loss of LSH had not significantly effect on DNA methylation of HERV-K sequences, suggesting that LSH is not equally required for DNA methylation in all sites.
Figure 1.
LSH is required for DNA methylation in various cells. (A) Western blots showing that knockout of LSH did not affect the levels of DNMT1, DNMT3A, DNMT3B and UHRF1 proteins in HeLa and HCT116 cells. (B) Quantification of the levels of 5mC in genomic DNA derived from control and LSH-KO cells by HPLC. The levels of 5mC were shown as 5mC/(5mC + C)%. *P < 0.05; **P< 0, 005, ***P< 0. 0005, n = 3, error bar represents SEM in all 5mC measurement. (C) The levels of 5mC in genomic DNA derived from HeLa LSH-KO#2 cells consecutively cultured for various days determined by HPLC. The culture was regularly splited every three days. ** P< 0.005, n = 3, error bar represents SEM in all 5mC measurement. (D) The levels of 5mC in genomic DNA derived from HCT116 LSH-KO#1 cells consecutively cultured for various days determined by HPLC. ***P< 0. 0005, n = 3, error bar represents SEM in all 5mC measurement. (E) The levels of 5mC in genomic DNA derived from LSH-knockdown HeLa cells cultured for various times determined by HPLC. *P < 0.05; ** P< 0.005, ***P< 0. 0005, n = 3, error bar represents SEM in all 5mC measurement. (F) Western blots showing re-expression of wild-type and mutant LSH in stable culture derived from HeLa LSH-KO#2. Also shown at top are schematic drawings of wild-type and mutant LSH. (G) DNA methylation measurement by HPLC showing re-expression of wild-type but not mutant LSH restored DNA methylation in HeLa LSH-KO#2 cells. *P < 0.05, n = 3, error bar represents SEM in all 5mC measurement.
Notably, the reduced level of DNA methylation in LSH KO cells could be stably maintained upon a long-term consecutive culture (Figure 1C and D). This observation was confirmed by using shRNA-mediated LSH knockdown cells (Figure 1E). Together, these data indicate that LSH is generally required for maintaining global DNA methylation in mammalian cells. However, a reduced but new homeostasis in DNA methylation can be reached in the absence of LSH.
To ensure that the observed reduction in global DNA methylation is indeed due to loss of LSH, we reintroduced wild-type and mutant LSH into a LSH-KO HeLa cell line as detailed in Materials and Methods (Figure 1F). Subsequent DNA methylation analysis demonstrated that re-expression of wild-type LSH was able to restore the DNA methylation to the level in control HeLa cells (Figure 1G). However, re-expression of either an ATPase-deficient mutant or a mutant with deletion of DEAH domain failed to rescue DNA methylation defect (Figure 1G). Thus, consistent with previous reports observed in Lsh-KO mES cells (29,31), chromatin-remodeling activity is required for LSH to promote DNA methylation in cultured cells.
LSH primarily promotes DNA methylation by DNMT1
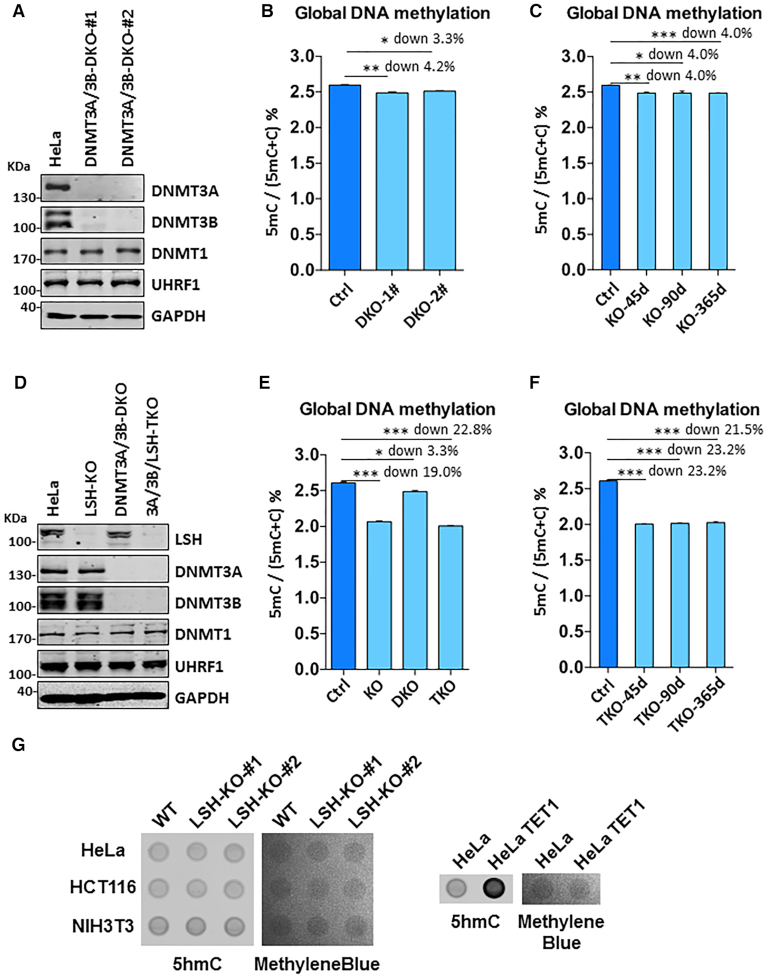
Having demonstrated that loss of LSH leads to substantial reduction of DNA methylation, we next tested if LSH contributes to DNA methylation by promoting de novo methylation by DNMT3A/DNMT3B and/or maintenance methylation by DNMT1. To this end, we first generated DNMT3A/DNMT3B double knockout (DKO) HeLa cells by CRISPR/Cas9 (Supplemental Figure S4A). The disruption of both DNMT3A and DNMT3B genes and lack of DNMT3A and DNMT3B proteins in two independent clones were confirmed by DNA sequencing (Supplemental Figure S4B) and Western blot analysis (Figure 2A), respectively. Consistent with our recent observation (42), we found that double knockout of DNMT3A and DNMT3B in HeLa cells only led to ∼4% reduction of DNA methylation (Figure 2B), substantially less than what was observed in LSH-KO HeLa cells (down by 18.2%). Like LSH-KO cells, DNMT3A/3B-DKO cells could stably maintain a reduced level of DNA methylation upon a long-term culture (Figure 2C). In this regard, minor reduction of DNA methylation was also observed in DNMT3A-KO, DNMT3B-KO and DNMT3A/3B-DKO HCT116 cells (43). Thus, DNMT3A and DNMT3B contribute only modestly to the global levels of DNA methylation in HeLa and HCT116 cells, revealing that LSH regulates DNA methylation far beyond its potential effect on de novo methylation by DNMT3A/3B.
Figure 2.
LSH regulates DNA methylation extending beyond its potential effect on DNA methylation by DNMT3A and DNMT3B. (A) Western blots showing knockout of both DNMT3A and DNMT3B did not affect the expression of DNMT1 and UHRF1 in HeLa cells. (B) Quantification of 5mC levels in genomic DNA derived from control and DNMT3A/3B DKO cells by HPLC. *P < 0.05, ** P< 0.005, n = 3, error bar represents SEM in all 5mC measurement. (C) Quantitative measurement of 5mC levels in genomic DNA derived from DNMT3A/3B DKO cells cultured for various days. *P < 0.05, ** P< 0.005, *** P< 0. 0005, n = 3, error bar represents SEM in all 5mC measurement. (D) Western blots validating the lack of LSH, DNMT3A/3B, and DNMT3A/3B/LSH in LSH-KO, DNMT3A/3B-DKO and DNMT3A/3B/LSH-TKO cells, respectively. (E) Quantitative measurement of 5mC levels in genomic DNA derived from LSH-KO, DNMT3A/3B-DKO and DNMT3A/3B/LSH-TKO cells. *P < 0.05, *** P< 0.0005, n = 3, error bar represents SEM in all 5mC measurement. (F) Quantitative measurement of the levels of 5mC in DNMT3A/3B/LSH-TKO cells cultured for various days. *** P< 0.0005, n = 3, error bar represents SEM in all 5mC measurement. (G) Dot-blot analysis of 5hmC in control and LSH-KO cells. Genomic DNA was prepared from each cell line and analyzed by dot-blot using anti-5hmC-specific antibody.
To test that LSH indeed contributes to DNA methylation by DNMT1, we further knocked out LSH in the DNMT3A/3B-DKO HeLa cells. The resulting DNMT3A/3B/LSH-TKO cells were obtained and verified by DNA sequencing (Supplemental Figure S4C) and western blot analysis (Figure 2D), respectively. Subsequent quantitative DNA methylation analysis by HPLC showed that knockout of LSH in DNMT3A/3B-DKO HeLa cells led to a marked reduction of DNA methylation (down by 22.8% versus control, compared to down by 19.0% in LSH-KO and 3.3% in DNMT3A/3B-DKO) (Figure 2E). Again, this markedly reduced DNA methylation level could be stably maintained in TKO cells in culture (Figure 2F). As triple knockout of DNMT3A/3B/LSH led to a reduction of DNA methylation that is nearly the sum of DNA methylation reduction in DNMT3A/3B TKO cells and LSH KO cells, LSH and DNMT3A/3B appear to independently and additively control global DNA methylation. Given that knockout of both DNMT3A/3B in HeLa and HCT116 cells only led to mild reduction of DNA methylation, whereas loss of LSH inevitably resulted in much more severe reduction of global DNA methylation in all cell lines we tested, our data thus suggest that LSH promotes DNA methylation in these cells primarily through enhancing DNA methylation by DNMT1. Consistent with this idea as well as previous observation in HCT116 cells, we found that knockout of DNMT1 in HeLa cells essentially abolished DNA methylation as revealed by immunostaining using anti-5mC antibody (Supplemental Figure S5). Unlike the case of LSH, we could not generate DNMT1-KO HeLa cell line, presumably due to cell death instigated by DNMT1 knockout, as observed in DNMT1-KO HCT116 cells. Thus, DNMT1 is responsible for bulk DNA methylation in these cultured cells and required for cell viability, as expected.
In theory, reduced DNA methylation in LSH-KO cells could be due to increased DNA demethylation via TET family proteins. If this were the case, we would expect to see increased level of 5-hydroxylmethyl-C (5hmC). However, by dot-blot analysis using an anti-5hmC specific antibody, we failed to detect any significant increase of 5hmC in genomic DNA prepared from LSH-KO HeLa, HCT116 and NIH3T3 cells (Figure 2G, left panel). However, the level of 5hmC was drastically increased in genomic DNA prepared from HeLa cells ectopically expressing TET1 proteins (Figure 2G, right panel), thus validating our dot-blot assay for 5hmC. As no increased 5hmC was detected in all LSH-KO cell lines, we exclude increased DNA demethylation as the potential explanation for the substantial reduction of DNA methylation in LSH-KO cells.
Taken together, our data suggest that LSH contributes to global DNA methylation primarily by promoting DNA methylation by DNMT1.
Loss of LSH broadly affects DNA methylation
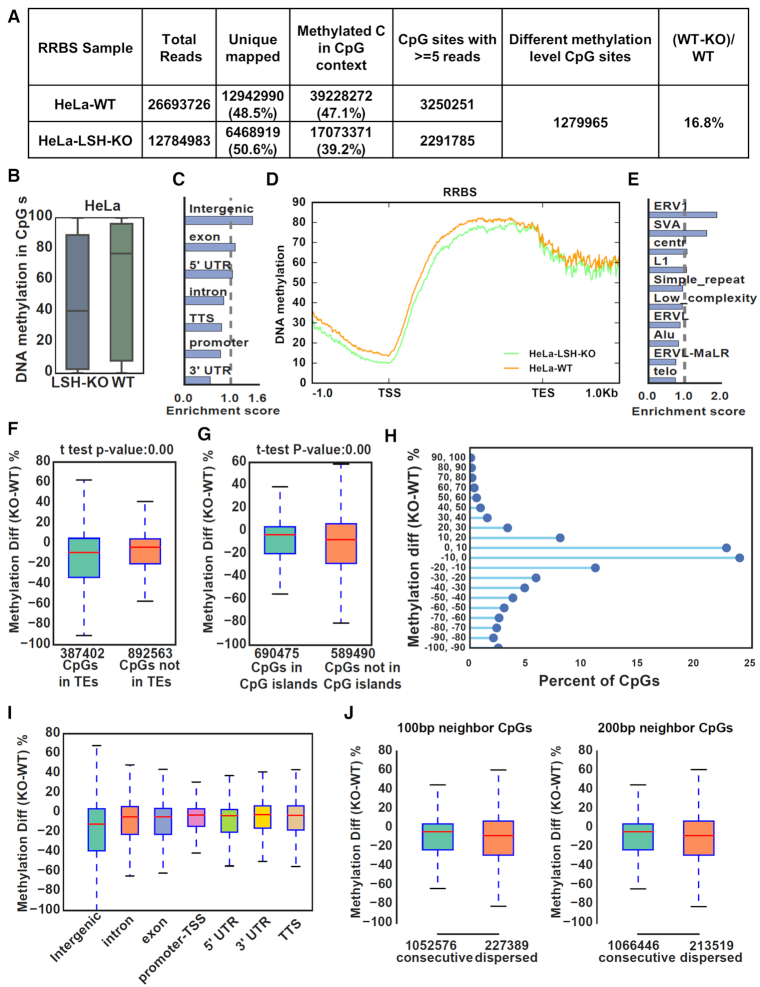
To elucidate further the role of LSH in DNA methylation, we analyzed the DNA methylation landscape in control HeLa and HeLa LSH-KO#2, and control HCT116 and HCT116 LSH-KO#2 cells by reduced representative bisulfite sequencing (RRBS) analysis (33). Although two sets of data differ in the levels of DNA methylation reduction, the general features are the same and thus we present the data from HeLa cells, whereas the data for HCT116 cells were shown in Supplemental Figure S6. As summarized in Figure 3A, while 47.1% uniquely sequenced CpG sites were methylated in control HeLa cells, it dropped to 39.2% in LSH-KO cells, representing a 16.8% reduction in DNA methylation, a number very close to 16.9% reduction measured by HPLC (Figure 1B). By defining differentially methylated CpGs at false discovery rate (FDR) <0.05, we identified 1.28 million differentially methylated CpG sites that were covered with at least 5 reads for both control and LSH-KO samples (Figure 3A). When each CpG site was plotted according to their methylation rate, loss of LSH clearly led to a substantial reduction of DNA methylation (Figure 3B). When differentially methylated CpG sites were plotted according to their distribution in genomic elements, reduced methylation is relatively enriched in intergenic than genic regions (Figure 3C). Within the genic region, differentially methylated CpG sites were under-represented in the promoter and 3′ UTR (Figure 3C and D), consistent with a lower DNA methylation in TSS and 3′ UTR. Among the transposable elements (TEs), differentially methylated CpGs were relatively enriched in ERV1 and SVA (Figure 3E). When CpG sites were divided into sites in TEs and not in TEs, a slight enrichment for differentially methylated sites in TEs was observed (Figure 3F). In contrast, differentially methylated CpGs were less enriched in CpG islands versus non-CpG islands (Figure 3G), suggesting that LSH is prone to promote maintenance of DNA methylation in non-CpG island regions. Similar changes in DNA methylation were observed for LSH-KO HCT116 cells (Supplemental Figure S6). These results indicate that, although loss of LSH has a slightly severe effect on DNA methylation in the heterochromatic regions enriched of transposons and repeats, it down-regulates genome-wide DNA methylation. These results are reminiscent of DNA methylation changes in Lsh−/− mES and MEF cells (27,28), although to a lesser extent.
Figure 3.
Genome-wide DNA methylation analysis reveals a widespread role of LSH in DNA methylation. (A) Summary of RRBS data for control HeLa and LSH KO cells. Only uniquely mapped reads were kept and CpGs with ≥ 5 reads mapped were used for further analysis. (B) Global distribution and median levels of CpG methylation in control HeLa and LSH KO cells. (C) Genomic distribution of CpGs with highly reduced methylation in LSH-KO HeLa cells (WT-KO ≥ 30%, 276 513 CpGs). (D) Average CpG methylation levels for all genes and flanking 1kb regions in control HeLa and LSH-KO cells. TSS, transcription start site; TES, transcription end site. (E) Distribution of CpGs with highly reduced methylation (WT-KO ≥ 30%) that were mapped to transposable elements. (F) Comparison of methylation differences of CpG sites located within and not in transposable elements regions between control and LSH-KO cells. (G) Comparison of methylation differences of CpG sites in and not in CpG islands between control and LSH-KO cells. (H) All CpG sites were plotted according to the difference in levels of CpG methylation between control and LSH-KO cells. The Y axes represents distribution of CpG sites with DNA methylation in KO cells that were reduced from 0 to 100% or increased from 0 to 100% as compared to control. X axes represents the percentage of CpG sites in each category with a total of 100%. (I) CpG methylation changes between control and LSH-KO cells at different genomic regions. (J) CpG methylation changes at consecutive CpG sites and dispersed sites. Consecutive CpGs were defined as more than one CpG located in 100 bp (left) and 200 bp (right).
It is noteworthy that, although loss of LSH led to a marked reduction of global DNA methylation, a small fraction of differentially methylated CpGs actually exhibited an increased DNA methylation (Figure 3H). The CpG sites with increased DNA methylation were distributed in all elements (Figure 3I). A recent study demonstrates that DDM1 and LSH allow methylation of DNA wrapped in nucleosomes and inactivation of DDM1/LSH biases DNA methylation toward nucleosome-free linker DNA (30). We suggest the observed gain of methylation could reflect this methylation redistribution in nucleosome-free linker DNA in the absence of LSH.
As DNA maintenance methylation by DNMT1 is influenced by CpG density, we also analyzed if CpG density affects the degree of DNA methylation reduction. As shown in Figure 3J, loss of DNA methylation is more severe in dispersed CpG sites. This result also supports a role of LSH in promoting DNA maintenance methylation by DNMT1.
Reduction in DNA methylation correlates better with up-regulation of transcription
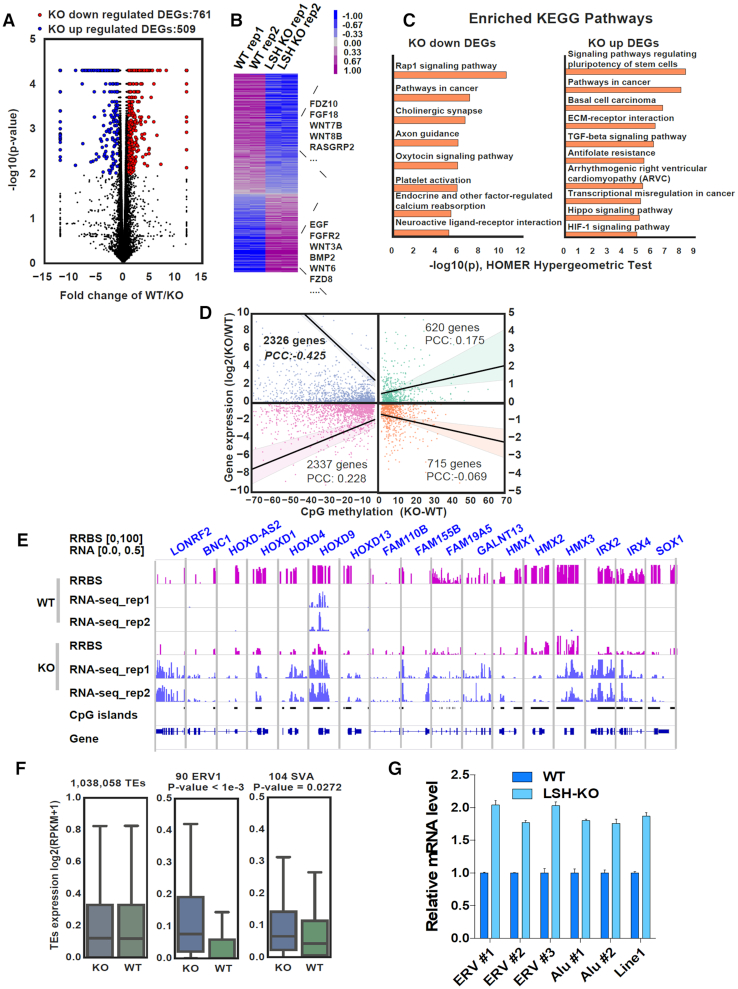
Previous studies indicate that there is no clear correlation between changes in DNA methylation and gene expression in Lsh−/− ES and MEF cells (27), possibly due to complex epigenetic compensation aroused during embryonic development upon loss of Lsh. Knockout of LSH in cultured cells is unlikely to evoke complex epigenetic compensation and thus provides a model to investigate how loss of LSH and its associated reduction of DNA methylation affects gene expression. To this end, we carried out RNA-seq analysis in control and LSH-KO HeLa cells. Using biological duplicates, 761 genes were identified as down-regulated and 609 genes as up-regulated upon loss of LSH in LSH-KO cells (Figure 4A and B). Gene ontology analysis revealed that down-regulated genes were enriched in pathways in cancer and neurogenesis, whereas up-regulated genes were skewed toward pluripotency of stem cells and pathways in cancer (Figure 4C). We also analyzed the correlation between changes in DNA methylation and gene expression. As shown in Figure 4D, reduced DNA methylation in promoter showed a better correlation with up-regulated expression (PCC –0. 425) than with down-regulated gene expression (PCC 0. 228). For examples, significant reduction of DNA methylation were observed at the CpG islands of multiple HOXD and FAM genes in LSH-KO cells, which were associated with elevated levels of corresponding transcripts (Figure 4E). Also consistent with a severe reduction of DNA methylation in ERV1 and SVA elements, RNA-seq data revealed increased transcripts for both TE elements, although no significant difference was observed if all TEs were taken in count (Figure 4F). We further confirmed by quantitative RT-PCR analysis that loss of LSH led to increased levels of ERVs, Alu and Line-1 transcripts (Figure 4G). This analysis supports a role of DNA methylation in regulation of gene expression and repression of transposons.
Figure 4.
Reduced promoter DNA methylation correlates with increased transcripts. (A) Volcano plot for differentially expressed genes between control and LSH KO HeLa cells. (B) Heatmap visualization of differentially expressed genes. (C) Enriched GO terms for the LSH-KO up- and down-regulated genes. (D) Correlation of DNA methylation changes at promoter and gene expression levels. Changes of DNA methylation represent the mean difference of CpGs in the TSS region. (E) Examples of genes with significantly decreased DNA methylation in CpG islands and up-regulated RNA transcripts in LSH-KO HeLa cells. (F) Expression changes at transposable regions for all TEs (left), ERV1 family (middle) and SVA family (right). Repeats of ERV1 and SVA families were selected as they had significant decrease of mean CpG methylations (KO-WT ≥ 30%). (G) qRT-PCR analysis of specific transposable elements. *** P< 0.0005.
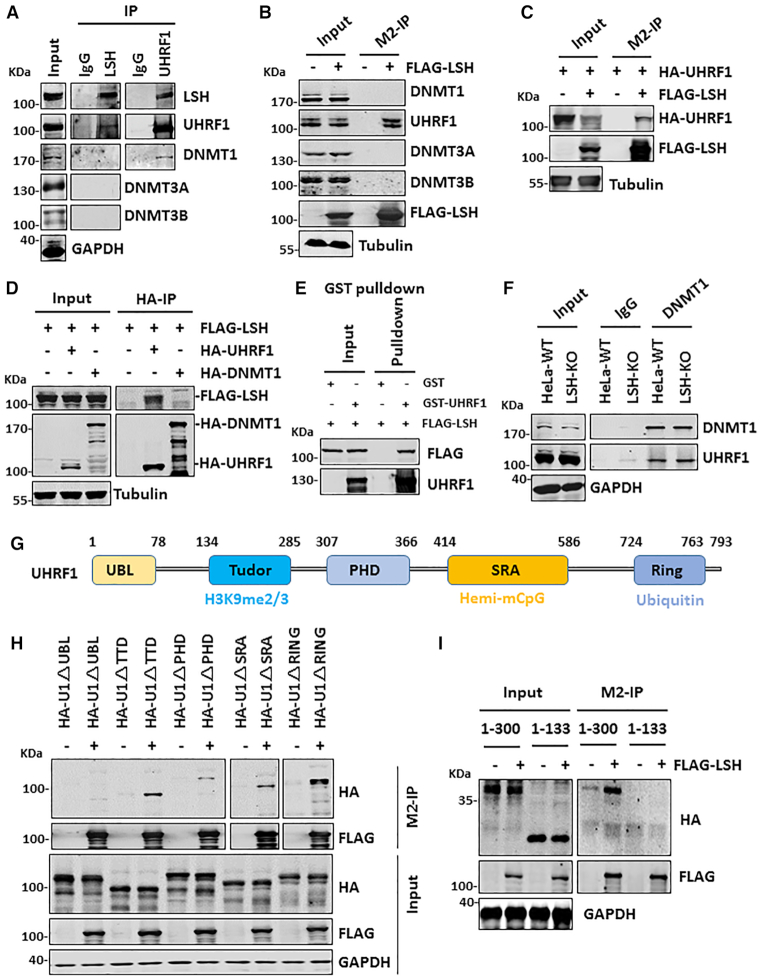
LSH interacts with UHRF1
We next investigated the underlying mechanism by which LSH facilitates DNA methylation by DNMT1. To this end, we first analyzed the interaction of LSH with all three DNA methyltransferases and UHRF1 in HeLa cells by co-immunoprecipitation (co-IP) assay. Although LSH was reported to interact with DNMT3A and DNMT3B (26) or DNMT3B (44) but not DNMT1, we failed to detect the presence of all three DNMTs when LSH was immunoprecipitated from HeLa cellular extracts (Figure 5A). However, UHRF1 was readily detected in IP of LSH (Figure 5A). Furthermore, LSH was also readily detected in IP of UHRF1 (Figure 5A). While DNMT1 was not detected in IP of LSH, it co-immunoprecipitated with UHRF1, a result consistent with previous publications (13,45). In support of a specific interaction between LSH and UHRF1, we observed that ectopically expressed FLAG-tagged LSH co-immunoprecipitated with endogenous UHRF1 but not DNMT1, DNMT3A and DNMT3B (Figure 5B). We further validated the protein-protein interaction between UHRF1 and LSH by reciprocal IP of ectopically expressed proteins (Figure 5C and D). Note that, although we failed to detect an interaction between LSH and endogenous DNMT3A/3B under our experimental conditions, we did observe co-IP between ectopically expressed LSH and DNMT3A/3B, but not with DNMT1 (Supplemental Figure S7A), in line with previous publications (26,44), suggesting that LSH could interact weakly with DNMT3A and DNMT3B. Co-IP experiments with HCT116 cellular extracts confirmed that endogenous LSH interacted with UHRF1 but not DNMT1 (Supplemental Figure S7B, upper panel). Co-IP with ectopically expressed FLAG-LSH in HCT116 further confirmed a robust interaction between LSH and UHRF1 (Supplemental Figure S7B, lower panel). A weak DNMT1 signal was also detected, but this could be due to the interaction between UHRF1 and DNMT1. By using purified recombinant FLAG-tagged LSH and GST-UHRF1 (Supplemental Figure S7C), we observed efficient pulldown of FLAG-LSH by GST-UHRF1, supporting the direct protein-protein interaction between LSH and UHRF1 (Figure 5E). By using cellular extracts derived from LSH-KO HeLa cells, we found loss of LSH did not affect the interaction between UHRF1 and DNMT1 (Figure 5F). Taken together, our data reveal a specific interaction between LSH and UHRF1 and no interaction between LSH and DNMT1, suggesting that LSH is likely to promote DNA methylation by DNMT1 through its interaction with UHRF1.
Figure 5.
LSH specifically interacts with UHRF1. (A) Western blots showing reciprocal co-IP of endogenous LSH and UHRF1 in HeLa cell extracts. Note DNMT3A, DNMT3B and DNMT1 were not detected in IP of LSH, whereas DNMT1 was detected in IP of UHRF1. (B) Western blots showing ectopically expressed FLAG-LSH co-immunoprecipitated with endogenous UHRF1 but not DNMT3A, DNMT3B and DNMT1. (C) Western blots showing co-IP of ectopically expressed HA-UHRF1 and FLAG-LSH. (D) Western blots showing co-IP of ectopically expressed FLAG-LSH with HA-UHRF1 but not HA-DNMT1. (E) In vitro GST pulldown assay showing purified FLAG-LSH bound to recombinant GST-UHRF1 but not control GST proteins. (F) Western blots showing knockout of LSH did not affect the interaction between UHRF1 and DNMT1. (G) Schematic illustration of UHRF1 structural and functional domains. (H) Western blots showing deletion of UBL domain impaired interaction between UHRF1 and LSH. (I) Western blots showing the UBL domain is not sufficient for interaction with LSH.
We next determined the region in UHRF1 that is responsible for interaction with LSH. Using a series of mutants with deletion of each individual structural/functional domain (Figure 5G), we found that deletion of the N-terminal UBL domain, but not other regions, impaired the interaction between UHRF1 and LSH (Figure 5H). This region has recently been shown to be important for UHRF1’s function in DNA methylation (46,47). However, the UBL domain itself is not sufficient for binding, as the interaction was observed for the UHRF1 fragment containing amino acids 1–300 but not 1–133 (Figure 5I). Using a series of C-terminal truncated UHRF1 mutants (45), we confirmed the UHRF1 1–300 fragment is sufficient for interaction with LSH (Supplemental Figure S7D).
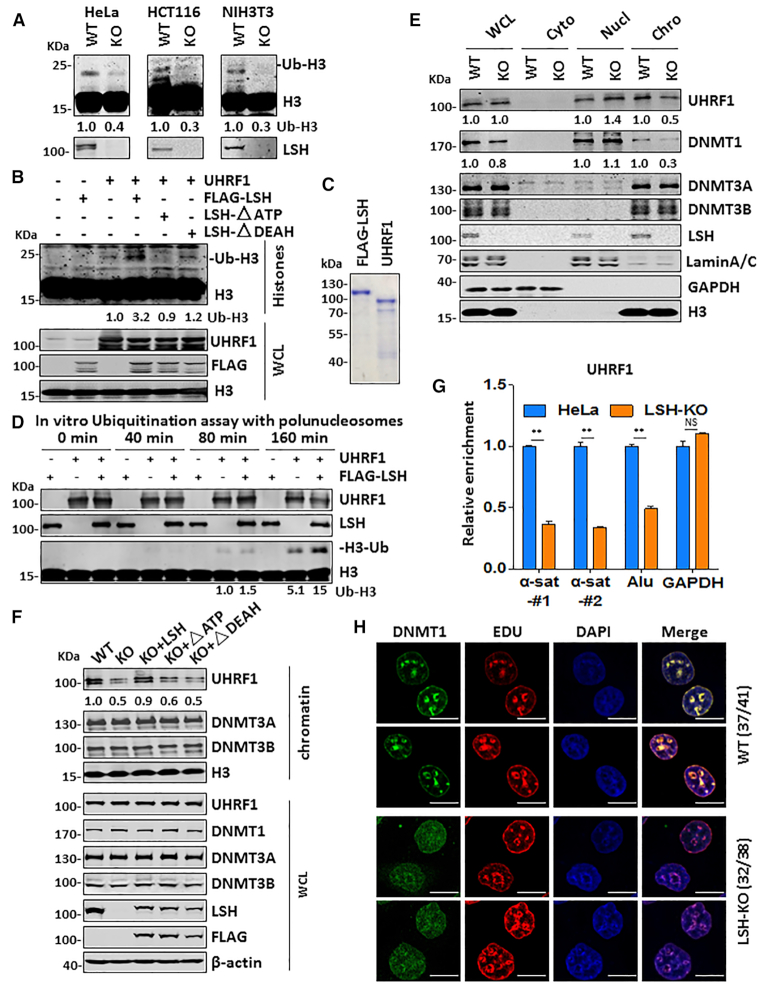
LSH enhances histone H3 ubiquitination by UHRF1
Recent studies have led to a working model that in S phase of cell cycle UHRF1-catalyzed histone H3 ubiquitination is critical for recruitment of DNMT1 to replication foci and subsequent DNA maintenance methylation (18–20). To elucidate how loss of LSH impairs DNA methylation by DNMT1, we examined if LSH regulates histone H3 ubiquitination by UHRF1. To this end, we synchronized both control and LSH-KO HeLa, HCT116 and NIH3T3 cells to S phase by aphidicolin treatment followed by releasing cells into fresh medium for 2 h. Subsequent cell cycle analysis by FACS indicated that this treatment synchronized more than 80% cells to the S phase, whereas in the control untreated cells the S phase cells ranged from 22.41% in HeLa to 6.47% in NIH3T3 cells (Supplemental Figure S8). We then examined the levels of ubiquitinated H3 by western blotting analysis. As shown in Figure 6A, a marked reduction of ubiquitinated H3 was observed for all LSH-KO cells when compared to that in control cells, suggesting that LSH is required for effective H3 ubiquitination by UHRF1 in S phase. In support of this idea, we observed that co-expression of LSH markedly enhanced H3 ubiquitination by UHRF1 (Figure 6B). Furthermore, LSH enhanced UHRF1’s H3 ubiquitination activity in an ATPase-dependent manner (Figure 6B), suggesting that LSH enhances H3 ubiquitination by UHRF1 through its chromatin-remodeling activity.
Figure 6.
LSH promotes histone H3 ubiquitination, UHRF1 association with chromatin and DNMT1 recruitment in S phase of cell cycle. (A) Western blots showing significantly reduced H3 ubiquitination in LSH-KO HeLa, HCT116 and NIH3T3 cells synchronized to S phase of cell cycle. (B) Western blots showing co-expression of wild-type but not LSH mutants enhanced UHRF1-catalyzed H3 ubiquitination. Image J was used for quantitation of western blots. (C) Coomassie blue staining showing FLAG-LSH expressed and purified from HEK293T cells and full-length UHRF1 from E.coli. GST-UHRF1 was expressed and purified from E.coli by using glutathione agarose beads, and UHRF1 was then eluted from beads by thrombin cleavage. (D) In vitro ubiquitination assay showing LSH enhanced histone H3 ubiquitination by UHRF1. Recombinant polynucleosomes were used for substrate. Reactions were carried out for various times, with addition of E1 (6xHis-UBE1), E2 (GST-UBCH5a) and Ub, together with UHRF1 and or FLAG-LSH as indicated. (E) Cellular fractionation followed by western blot analysis showing LSH knockout reduced UHRF1 and DNMT1 chromatin association in S phase. The cells were synchronized to S phase by aphidicolin treatment followed by culture in fresh medium for 2 h. (F) Western blots showing re-expression of wild-type but not LSH mutants restored UHRF1 and DNMT1 chromatin association in S phase. The cells were synchronized to S phase as above. (G) Modified eSPAN assay showing reduced association of UHRF1 with newly replicated repetitive sequences. ** P< 0.005. (H) IF analysis showing reduced DNMT1 enrichment in EdU labeled pericentromeric heterochromatin foci in LSH-KO HeLa cells. Note these cells were within mid-later S phase according to EdU labeling of pericentromeric heterochromatin. Scale bar, 10 μm.
We next tested if LSH promotes H3 ubiquitination by UHRF1 in chromatin in vitro. To this end, we ectopically expressed FLAG-LSH in HEK293T cells and purified FLAG-LSH by using anti-FLAG M2 agarose beads (Figure 6C). We also prepared recombinant full-length UHRF1 from E.coli (Figure 6C). We then carried out in vitro ubiquitination reactions with recombinant polynucleosomes as substrate. As shown in a representative time-course experiment in Figure 6D, addition of LSH enhanced H3 ubiquitination by UHRF1. Thus, our in vitro and cell-based experiments demonstrated for the first time that LSH promotes histone H3 ubiquitination by UHRF1.
LSH enhances UHRF1 and DNMT1 chromatin association in S phase of cell cycle
To understand how loss of LSH impairs UHRF1-catalyzed H3 ubiquitination, we examined whether LSH regulates UHRF1 chromatin association in S phase of cell cycle. As shown in Figure 6E, biochemical fractionation revealed that loss of LSH markedly reduced the level of chromatin-associated UHRF1 in S phase of cell cycle. Loss of LSH did not affect the overall level of UHRF1 (Figure 6E and Figure 1A), excluding the possibility that LSH affects H3 ubiquitination by control the level of UHRF1. Moreover, we observed that loss of LSH also reduced the level of chromatin-associated DNMT1 (Figure 6E). However, loss of LSH had no effect on the association of DNMT3A and DNMT3B with chromatin (Figure 6E). The same results were observed when control and LSH-KO HCT116 cells were analyzed (Supplemental Figure S9A). Thus, loss of LSH impairs UHRF1 binding of chromatin and subsequent DNMT1 recruitment in S phase.
To ascertain the impaired chromatin association of UHRF1 is due to loss of LSH, we examined UHRF1 chromatin association in LSH-KO HeLa cells that were complemented with either wild-type or mutant LSH. As shown in Figure 6F, re-expression of wild-type but not ATPase-deficient or DEAH domain deletion mutant restored chromatin association of UHRF1 in S phase. Thus, LSH and its chromatin remodeler activity is required for efficient access of UHRF1 to chromatin in S phase.
To directly demonstrate that LSH indeed enhances UHRF1 binding of newly replicated DNA, we used a modified eSPAN (enrichment and sequencing of protein-associated nascent DNA) assay (48), in which unsynchronized cells were cultured in the presence of nucleotide analog BrdU for 30 min to label the newly replicated DNA. The cells were then treated with formaldehyde to cross-link chromatin-associated proteins with DNA, and after sonication, the UHRF1-assocaited chromatin fragments were immunoprecipitated with anti-UHRF1 antibody. The UHRF1 associated, BrdU-labeled nascent DNA was then recovered by using anti-BrdU antibody and quantified by qPCR analysis. As shown in Figure 6G, we consistently observed that loss of LSH resulted in reduced association of UHRF1 with pericentromeric heterochromatin α-satellite sequences and Alu repetitive sequences, both of which are known to be hypermethylated, but not the unmethylated GAPDH locus. We thus conclude that loss of LSH impairs UHRF1 association with replication fork containing hemi-methylated DNA.
During mid to late S phase, both UHRF1 and DNMT1 are characteristically enriched at pericentromeric heterochromatin regions and can be visualized as clear foci in immunostaining (13,45,49,50). We thus briefly labeled S phase cells by nucleotide analog EdU (5-ethynyl-2’-deoxyuridine) and carried out double immunostaining for EdU and DNMT1 in both control and LSH-KO cells. While in control cells clear DNMT1 foci were observed and found to co-localize with EdU-labeled spots (late replicated pericentromeric heterochromatin regions), the DNMT1 foci were much weaker in LSH-KO cells (Figure 6H), consistent with a reduced DNMT1 chromatin association in LSH-KO cells as revealed by chromatin fractionation (Figure 6E) and eSPAN assay (Figure 6G). Similar experiments for UHRF1 were not successful, due to a lack of specific UHRF1 antibody suitable for detection of endogenous UHRF1 by immunostaining (data not shown). By using GFP-tagged UHRF1, we did observe co-localization of GFP-UHRF1 and LSH with bright DAPI spots during S phase of cell cycle (Supplemental Figure S9B) and that loss of LSH substantially reduced the percentage of S phase cells with bright GFP-UHRF1 foci (Supplemental Figure S9C, D). Altogether, these results provide compelling evidence that LSH is required for efficient chromatin association of UHRF1 and subsequent DNMT1 recruitment in S phase of cell cycle.
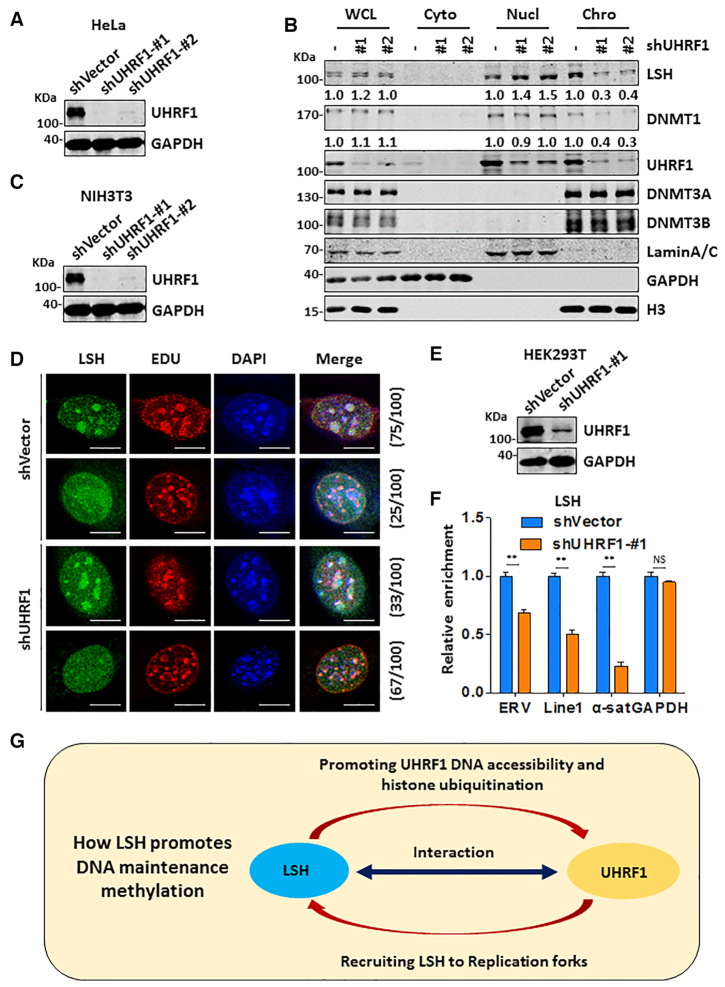
UHRF1 has a role in targeting LSH to replication fork
Using ectopically expressed LSH, a previous study observed colocalization of LSH with Dnmt1 in pericentromeric regions only in late S phase NIH3T3 and MEF cells (51), providing evidence that LSH is present in DNA replication fork. However, how LSH is recruited to pericentromeric regions in late S phase is not known. Our finding that UHRF1 interacts with LSH raises the possibility whether UHRF1 plays a role in recruiting LSH to DNA replication fork. To test this idea, we first established experimental condition to knock down UHRF1 in HeLa cells by two distinct shRNAs (Figure 7A). We then synchronized control and shUHRF1-treated cells to S phase by aphidicolin treatment followed by releasing cells into fresh culture medium for 2 h. Subsequent cellular fractionation analysis revealed that knockdown of UHRF1 reduced the level of chromatin-associated LSH and a concomitant increase of LSH in soluble nuclear fraction (Figure 7B). Note that knockdown of UHRF1 also reduced the level of chromatin-associated DNMT1, but had no effect on binding of chromatin by DNMT3A and DNMT3B (Figure 7B). This result provides first evidence that UHRF1 is required for LSH binding to chromatin in S phase of cell cycle.
Figure 7.
UHRF1 enhances LSH chromatin association during S phase of cell cycle. (A) Western blots showing efficient knockdown of UHRF1 by two specific shRNAs in HeLa cells. (B) Cellular fractionation followed by western blot analysis showing UHRF1 knockdown in HeLa cells impaired LSH chromatin association in S phase. (C) Western blots showing efficient knockdown of UHRF1 in NIH3T3 cells by two specific shRNAs. (D) IF analysis showing knockdown of UHRF1 substantially reduced mid-to-late S phase cells with EdU-colocalized LSH foci. Scale bar, 10 μm. (E) Western blots showing efficient knockdown of UHRF1 in HEK293T cells by a UHRF1-specific shRNA. (F) Modified eSPAN assay showing reduced association of LSH with newly replicated repetitive sequences upon knockdown of UHRF1 in HEK293T cells. ** P< 0.005. (G) Working model illustrating how LSH and UHRF1 work together to promote DNA methylation by DNMT1.
To further test the role of UHRF1 in LSH recruitment, we also knocked down UHRF1 in NIH3T3 cells by two distinct shRNAs (Figure 7C) and examined LSH pericentromeric heterochromatin association by immunostaining. To mark cells in S phase, we pulse-labeled NIH3T3 cells with EdU. Consistent with the previous observation (51), we observed that, in the majority of late S phase control cells with characteristic large, DAPI-colocalized EdU foci, there were LSH foci that showed colocalization with both EdU and DAPI foci (75%). However, knockdown of UHRF1 substantially reduced the number of cells with colocalized LSH and EDU foci (Figure 7D) (33%), in agreement with a role of UHRF1 in recruiting LSH to replication forks.
To test directly a role of UHRF1 in targeting LSH to replicating DNA, we resorted again using our modified eSPAN assay as described above to compare the association of LSH with nascent DNA in control and UHRF1-knockdown HEK293T cells (Figure 7E). As shown in Figure 7F, knockdown of UHRF1 variably reduced the association of LSH with replicating ERV, LINE1 and α-satellite, but not GAPDH. Taken together, these results indicate that UHRF1 is also required for efficient association of LSH with replicating heterochromatin.
DISCUSSION
Besides three DNMTs, LSH and UHRF1 are well recognized for their crucial roles in DNA methylation (10,13,24,25,27). Lsh knockout mice die shortly after birth and its genomic DNA is severely hypomethylated in repetitive elements and single copy genes (24). Similarly, DNA hypomethylation in Lsh−/− ES and MEF cells also occur genome-wide (26–28,52). By using an episomal DNA-based assay, Zhu et al. reported that the acquisition of DNA methylation, but not the maintenance of pre-methylated episomes, is impaired in Lsh−/− MEF cells. LSH was found to interact with both DNMT3A and DNMT3B but not DNMT1, leading to the conclusion that LSH is directly involved in the control of de novo methylation by DNMT3A/3B but not maintenance methylation by DNMT1 (26). Termanis et al. reported that re-expression of LSH was able to reestablish DNA methylation and silencing of misregulated genes in Lsh−/− MEF cells, possibly through an interaction with DNMT3B (31). Thus, the current view is that LSH is crucial for DNA methylation due to its role in promoting de novo DNA methylation by DNMT3A/3B. In contrast to this prevailing view, our data indicate that LSH facilitates DNA methylation primarily through enhancing methylation by DNMT1. This is evident as a much severe loss of DNA methylation was observed in LSH KO cells than that in DNMT3A/3B DKO cells (Figure 2) and loss of LSH did not elevate DNA demethylation via TET family proteins (Figure 2G and data not shown). A minor contribution of DNMT3A/3B to global DNA methylation in cultured cells is unlikely unique to HeLa cells, as it is also observed in HCT116 cells (43). A critical role for LSH in DNA methylation by DNMT1 can be explained by a previously unrecognized molecular link between LSH and UHRF1. While we confirmed a lack of interaction between LSH and DNMT1, we found that LSH interacts strongly and directly with UHRF1 (Figure 5). We believe this interaction is functionally relevant, because LSH promotes UHRF1 chromatin association and H3 ubiquitination in S phase as well as in vitro (Figure 6). The interaction between UHRF1 and LSH also provides an explanation of their mutual stimulation on chromatin association in S phase and why LSH is enriched in pericentromeric heterochromatins in late S phase (51). Altogether, our study reveals for the first time a role of and mechanism by which LSH facilitates DNA methylation through the UHRF1/DNMT1 axis.
While our study has provided clear evidence that LSH has a role in promoting DNA methylation by DNMT1, our study does not exclude the possibility that LSH can also facilitate methylation by DNMT3A/3B. In this regard, we noticed that reduction of DNA methylation in Lsh−/− embryos and Lsh−/− MEFs is more extensive than what we have observed in all four types of LSH-KO cells (24,27,28,53). As early embryonic development undergoes an extensive DNA demethylation process (54–56), the more broader DNA hypomethylation in Lsh−/− embryos and MEFs likely reflects an impaired re-establishment of DNA methylation that is dependent on a coordinated function of all thee active DNMTs. Nevertheless, to define precisely the role of LSH in de novo methylation by DNMT3A/3B, one has to resort to DNMT1 knockout cells. As DNMT1 knockout is lethal in cells such as HCT116 (57) and HeLa (data not shown), it is of interest in the future to examine if Lsh indeed promotes DNA methylation by Dnmt3a and Dnmt3b in Dnmt1−/− mouse ES cells, which are viable (58,59).
Immunodeficiency, centromeric instability, and facial anomalies (ICF) syndrome is a rare autosomal recessive disorder characterized by reduced immunoglobulin levels in the serum and recurrent infection. Mutations in four genes, namely DNMT3B, ZBTB24, CDCA7 and LSH, have been causally linked to ICF syndrome (60), providing a strong evidence for the functional link between LSH and DNMT3B. Interestingly, by analyzing and comparing methylomes of ICF patients from all four genotypes, Velasco et al. found that DNMT3B mutations affect distinct genomic context of DNA methylation from the other three gene mutations. For example, they found preferential hypomethylation of CpG islands in ICF1 (DNMT3B mutation) patients, whereas perturbations of DNA methylation at genomic loci with heterochromatin hallmarks are observed in ICF2 (ZBTB24), ICF3 (CDCA7) and ICF4 (LSH) patients. They also showed that DNA hypomethylation is much less extensive in ICF1 (DNMT3B mutation) than that in ICF2, ICF3 and ICF4. This is reminiscent of our data that loss of DNMT3B has a much weak effect on DNA methylation than loss of LSH. Furthermore, a recent study has revealed a role of CDCA7 and LSH in classical nonhomologous end joining (C-NHEJ) and conclude that the defect in C-NHEJ may account for some common features of ICF cells, including centromeric instability, abnormal chromosome segregation and apoptosis (61). Although less prominent, cells with mutations in DNMT3B and ZBTB24 also showed similar defects. Thus, ICF syndrome may be due to defect in DNA repair, not necessarily DNA methylation.
Our finding that reduced levels of DNA methylation can be stably maintained in DNMT3A/3B DKO and DNMT3A/3B/LSH TKO cells is in support of a minor de novo activity for DNMT1 (62–64). Given the existence of active demethylation by TET family proteins and <100% maintenance efficiency (56,62,65), one would expect to see progressive reduction of DNA methylation if DNMT1 has no de novo activity. Consistent with this idea, a recent study provides compelling evidence for DNMT1 de novo activity during oogenesis and in somatic cells (66).
Our RRBS analysis revealed that, although loss of LSH led to substantial reduction of global DNA methylation, a fraction of CpG sites was actually found to gain in DNA methylation. Previous in vitro studies demonstrated that DNMT1 lacks the ability to methylate nucleosomal CpG sites (67,68). In accord with the recent report that DDM1 and LSH are required for DNA methylation in wrapped nucleosomes through nucleosome reconfiguration and/or sliding and that DDM1/LSH may reflect the evolution divergence in nucleosomal or linker DNA methylation (30), we propose that the sites with gain of DNA methylation may localize in the linker regions and reflect skewed methylation toward nucleosomal linker regions in the absence of LSH. This shift in DNA methylation may also contribute to the observed new homeostasis of DNA methylation in each type of LSH-KO cells. Future work is needed to validate this predication.
Although DNA methylation is well accepted as a mechanism for transcription repression and genome integrity, previous studies with Lsh−/− mouse ES and MEF cells showed that global loss of DNA methylation correlates with aberrant expression of a subset of repetitive sequences but has little effect on unique gene expression (27). This is likely due to epigenetic compensation evoked during embryonic development. We surmised that epigenetic compensation is less likely to occur when LSH is directly disrupted in cultured cells. Consistent with this idea, we observed that loss of LSH in HeLa cells has a broad effect on steady state expression levels of repetitive sequences as well as unique genes (Figure 4). Furthermore, loss of DNA methylation in promoter correlates better with up-regulated than down-regulated transcripts, which is not observed in Lsh−/− MEF cells. Nevertheless, the loss of LSH on gene expression is likely complicated, as LSH has been shown to affect not only DNA methylation but also histone modification and participate in transcription, DNA repairs and possibly other processes (44,69–71).
Our finding that LSH promotes UHRF1 chromatin accessibility also provides a solution to an emerging issue in DNA maintenance methylation by DNMT1. Although DNA maintenance methylation is long believed to be a rapid process coupled with DNA replication, the findings that histone tail and ubiquitination play critical roles in targeting UHRF1 and DNMT1 to DNA replication fork all suggest that DNMT1 is likely to carry out DNA methylation at least in part in the context of chromatin (18–20,49,72). In further support of this idea, recent studies have revealed a global delay for a subset of CpGs in nascent strand DNA methylation (21,73). As maintenance methylation by DNMT1 is initiated by binding of UHRF1 to hemi-methylated CpGs, how UHRF1 gains access to hemi-methylated CpGs in chromatin is previously unknown. Our study demonstrates that LSH promotes UHRF1 association with chromatin and newly replicated DNA in S phase in an ATPase-dependent manner (Figure 6), thus identifying LSH as a crucial chromatin remodeler that promotes UHRF1 access to chromatin, most likely through nucleosome reconfiguration and/or sliding. Consistent with this idea, we found that LSH also promotes UHRF1-dependent H3 ubiquitination and DNMT1 recruitment. Altogether, we propose that LSH and UHRF1 work cooperatively to promote DNA methylation by DNMT1 (Figure 7G). Through protein-protein interaction between LSH and UHRF1, LSH and UHRF1 mutually facilitate each other's association with replication fork. This enhances H3 ubiquitination by UHRF1 and subsequent recruitment and DNA methylation by DNMT1.
DATA AVAILABILITY
The RRBS data and RNA-seq data were submitted to the GEO repository under accession number GEO: GSE136931.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Cheng-Ming Chiang (University of Texas Southwestern Medical Center) for critical reading of the manuscript. We thank members of Wong's laboratory for valuable discussion. We are very grateful to Dr Kathrin Muegge (National Cancer Institute/NIH) for kindly providing LSH and mutant constructs.
Authors contributions: M.H. generated all LSH-KO, DNMT3A/3B-DKO and DNMT3A/3B/LSH-TKO cells and M.H. and J.L. performed DNA methylation analysis and fractionation and cellular assays. J.L. also carried out in vitro ubiquitination assay. M.H., W.L., Q.Z. and J.L. carried out interaction analysis. Y.C. and J.J.H. performed RRBS and RNA-seq data analysis. H.Z. and Q.W. carried out RRBS sequencing. J.F. and J.W. supervised the project and M.H. and J.W. wrote the manuscript.
Contributor Information
Mengmeng Han, Shanghai Key Laboratory of Regulatory Biology, Fengxian District Central Hospital-ECNU Joint Center of Translational Medicine, Institute of Biomedical Sciences and School of Life Sciences, East China Normal University, Shanghai 200241, China.
Jialun Li, Shanghai Key Laboratory of Regulatory Biology, Fengxian District Central Hospital-ECNU Joint Center of Translational Medicine, Institute of Biomedical Sciences and School of Life Sciences, East China Normal University, Shanghai 200241, China; Joint Center for Translational Medicine, Fengxian District Central Hospital, 6600th Nanfeng Road, Fengxian District, Shanghai 201499, China.
Yaqiang Cao, CAS Key Laboratory of Computational Biology, CAS-MPG Partner Institute for Computational Biology, Shanghai Institutes of Nutrition and Health, Shanghai Institutes for Biological Sciences, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Room122, 320 Yue Yang Road, Shanghai 200031, China.
Yuanyong Huang, Shanghai Key Laboratory of Regulatory Biology, Fengxian District Central Hospital-ECNU Joint Center of Translational Medicine, Institute of Biomedical Sciences and School of Life Sciences, East China Normal University, Shanghai 200241, China.
Wen Li, Shanghai Key Laboratory of Regulatory Biology, Fengxian District Central Hospital-ECNU Joint Center of Translational Medicine, Institute of Biomedical Sciences and School of Life Sciences, East China Normal University, Shanghai 200241, China.
Haijun Zhu, NHC Key Laboratory of Reproduction Regulation (Shanghai Institute of Planned Parenthood Research), Fudan University, Shanghai 200032, China.
Qian Zhao, Shanghai Key Laboratory of Regulatory Biology, Fengxian District Central Hospital-ECNU Joint Center of Translational Medicine, Institute of Biomedical Sciences and School of Life Sciences, East China Normal University, Shanghai 200241, China.
Jing-Dong Jackie Han, CAS Key Laboratory of Computational Biology, CAS-MPG Partner Institute for Computational Biology, Shanghai Institutes of Nutrition and Health, Shanghai Institutes for Biological Sciences, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Room122, 320 Yue Yang Road, Shanghai 200031, China.
Qihan Wu, NHC Key Laboratory of Reproduction Regulation (Shanghai Institute of Planned Parenthood Research), Fudan University, Shanghai 200032, China.
Jiwen Li, Shanghai Key Laboratory of Regulatory Biology, Fengxian District Central Hospital-ECNU Joint Center of Translational Medicine, Institute of Biomedical Sciences and School of Life Sciences, East China Normal University, Shanghai 200241, China.
Jing Feng, Joint Center for Translational Medicine, Fengxian District Central Hospital, 6600th Nanfeng Road, Fengxian District, Shanghai 201499, China; Department of Laboratory Medicine & Central Laboratory, Southern Medical University Affiliated Fengxian Hospital, Shanghai 201499, China.
Jiemin Wong, Shanghai Key Laboratory of Regulatory Biology, Fengxian District Central Hospital-ECNU Joint Center of Translational Medicine, Institute of Biomedical Sciences and School of Life Sciences, East China Normal University, Shanghai 200241, China; Joint Center for Translational Medicine, Fengxian District Central Hospital, 6600th Nanfeng Road, Fengxian District, Shanghai 201499, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China [2017YFA054201] and National Natural Science Foundation of China [31730048, 81530078 to J.W., 31900453 to J.L., 81672624 to J.F.];. Funding for open access charge: Ministry of Science and Technology of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Li E., Zhang Y.. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014; 6:a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012; 13:484–492. [DOI] [PubMed] [Google Scholar]
- 3. Smith Z.D., Meissner A.. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013; 14:204–220. [DOI] [PubMed] [Google Scholar]
- 4. Schubeler D. Function and information content of DNA methylation. Nature. 2015; 517:321–326. [DOI] [PubMed] [Google Scholar]
- 5. Jones P.A., Liang G.. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 2009; 10:805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gowher H., Jeltsch A.. Mammalian DNA methyltransferases: new discoveries and open questions. Biochem. Soc. Trans. 2018; 46:1191–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edwards J.R., Yarychkivska O., Boulard M., Bestor T.H.. DNA methylation and DNA methyltransferases. Epigenet. Chromatin. 2017; 10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xie S., Qian C.. The growing complexity of UHRF1-mediated maintenance DNA methylation. Genes (Basel). 2018; 9:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bronner C., Alhosin M., Hamiche A., Mousli M.. Coordinated dialogue between UHRF1 and DNMT1 to ensure faithful inheritance of methylated DNA patterns. Genes (Basel). 2019; 10:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bostick M., Kim J.K., Esteve P.O., Clark A., Pradhan S., Jacobsen S.E.. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007; 317:1760–1764. [DOI] [PubMed] [Google Scholar]
- 11. Hashimoto H., Horton J.R., Zhang X., Bostick M., Jacobsen S.E., Cheng X.. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008; 455:826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karagianni P., Amazit L., Qin J., Wong J.. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell. Biol. 2008; 28:705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K. et al.. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007; 450:908–912. [DOI] [PubMed] [Google Scholar]
- 14. Arita K., Ariyoshi M., Tochio H., Nakamura Y., Shirakawa M.. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008; 455:818–821. [DOI] [PubMed] [Google Scholar]
- 15. Avvakumov G.V., Walker J.R., Xue S., Li Y., Duan S., Bronner C., Arrowsmith C.H., Dhe-Paganon S.. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008; 455:822–825. [DOI] [PubMed] [Google Scholar]
- 16. Rottach A., Frauer C., Pichler G., Bonapace I.M., Spada F., Leonhardt H.. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res. 2010; 38:1796–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nady N., Lemak A., Walker J.R., Avvakumov G.V., Kareta M.S., Achour M., Xue S., Duan S., Allali-Hassani A., Zuo X. et al.. Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J. Biol. Chem. 2011; 286:24300–24311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nishiyama A., Yamaguchi L., Sharif J., Johmura Y., Kawamura T., Nakanishi K., Shimamura S., Arita K., Kodama T., Ishikawa F. et al.. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013; 502:249–253. [DOI] [PubMed] [Google Scholar]
- 19. Qin W., Wolf P., Liu N., Link S., Smets M., La Mastra F., Forne I., Pichler G., Horl D., Fellinger K. et al.. DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res. 2015; 25:911–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ishiyama S., Nishiyama A., Saeki Y., Moritsugu K., Morimoto D., Yamaguchi L., Arai N., Matsumura R., Kawakami T., Mishima Y. et al.. Structure of the Dnmt1 reader module complexed with a unique two-mono-ubiquitin mark on histone H3 reveals the basis for DNA methylation maintenance. Mol. Cell. 2017; 68:350–360. [DOI] [PubMed] [Google Scholar]
- 21. Charlton J., Downing T.L., Smith Z.D., Gu H.C., Clement K., Pop R., Akopian V., Klages S., Santos D.P., Tsankov A.M. et al.. Global delay in nascent strand DNA methylation. Nat. Struct. Mol. Biol. 2018; 25:327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Narlikar G.J., Sundaramoorthy R., Owen-Hughes T.. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013; 154:490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou C.Y., Johnson S.L., Gamarra N.I., Narlikar G.J.. Mechanisms of ATP-dependent chromatin remodeling motors. Annu. Rev. Biophys. 2016; 45:153–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dennis K., Fan T., Geiman T., Yan Q., Muegge K.. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 2001; 15:2940–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zemach A., Kim M.Y., Hsieh P.H., Coleman-Derr D., Eshed-Williams L., Thao K., Harmer S.L., Zilberman D.. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013; 153:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu H., Geiman T.M., Xi S., Jiang Q., Schmidtmann A., Chen T., Li E., Muegge K.. Lsh is involved in de novo methylation of DNA. EMBO J. 2006; 25:335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu W., McIntosh C., Lister R., Zhu I., Han Y., Ren J., Landsman D., Lee E., Briones V., Terashima M. et al.. Genome-wide DNA methylation patterns in LSH mutant reveals de-repression of repeat elements and redundant epigenetic silencing pathways. Genome Res. 2014; 24:1613–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Myant K., Termanis A., Sundaram A.Y., Boe T., Li C., Merusi C., Burrage J., de Las Heras J.I., Stancheva I.. LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res. 2011; 21:83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ren J., Briones V., Barbour S., Yu W., Han Y., Terashima M., Muegge K.. The ATP binding site of the chromatin remodeling homolog Lsh is required for nucleosome density and de novo DNA methylation at repeat sequences. Nucleic Acids Res. 2015; 43:1444–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lyons D.B., Zilberman D.. DDM1 and Lsh remodelers allow methylation of DNA wrapped in nucleosomes. Elife. 2017; 6:e30674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Termanis A., Torrea N., Culley J., Kerr A., Ramsahoye B., Stancheva I.. The SNF2 family ATPase LSH promotes cell-autonomous de novo DNA methylation in somatic cells. Nucleic Acids Res. 2016; 44:7592–7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meissner A., Gnirke A., Bell G.W., Ramsahoye B., Lander E.S., Jaenisch R.. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005; 33:5868–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krueger F., Andrews S.R.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T.. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014; 42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tempel S. Using and understanding repeat masker. Methods Mol. Biol. 2012; 859:29–51. [DOI] [PubMed] [Google Scholar]
- 38. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Viggiani C.J., Knott S.R., Aparicio O.M.. Genome-wide analysis of DNA synthesis by BrdU immunoprecipitation on tiling microarrays (BrdU-IP-chip) in Saccharomyces cerevisiae. Cold Spring Harb. Protoc. 2010; 2010:pdb prot5385. [DOI] [PubMed] [Google Scholar]
- 41. Jeong S., Liang G., Sharma S., Lin J.C., Choi S.H., Han H., Yoo C.B., Egger G., Yang A.S., Jones P.A.. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol. Cell. Biol. 2009; 29:5366–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang H., Gao Q., Tan S., You J., Lyu C., Zhang Y., Han M., Chen Z., Li J., Wang H. et al.. SET8 prevents excessive DNA methylation by methylation-mediated degradation of UHRF1 and DNMT1. Nucleic Acids Res. 2019; 47:9053–9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cai Y., Tsai H.C., Yen R.C., Zhang Y.W., Kong X., Wang W., Xia L., Baylin S.B.. Critical threshold levels of DNA methyltransferase 1 are required to maintain DNA methylation across the genome in human cancer cells. Genome Res. 2017; 27:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Myant K., Stancheva I.. LSH cooperates with DNA methyltransferases to repress transcription. Mol. Cell. Biol. 2008; 28:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang J., Gao Q., Li P., Liu X., Jia Y., Wu W., Li J., Dong S., Koseki H., Wong J.. S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance. Cell Res. 2011; 21:1723–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. DaRosa P.A., Harrison J.S., Zelter A., Davis T.N., Brzovic P., Kuhlman B., Klevit R.E.. A bifunctional role for the UHRF1 UBL domain in the control of Hemi-methylated DNA-dependent histone ubiquitylation. Mol. Cell. 2018; 72:753–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Foster B.M., Stolz P., Mulholland C.B., Montoya A., Kramer H., Bultmann S., Bartke T.. Critical role of the UBL domain in stimulating the E3 ubiquitin ligase activity of UHRF1 toward chromatin. Mol. Cell. 2018; 72:739–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu C., Gan H., Han J., Zhou Z.X., Jia S., Chabes A., Farrugia G., Ordog T., Zhang Z.. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol. Cell. 2014; 56:551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu X., Gao Q., Li P., Zhao Q., Zhang J., Li J., Koseki H., Wong J.. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun. 2013; 4:1563. [DOI] [PubMed] [Google Scholar]
- 50. Leonhardt H., Page A.W., Weier H.U., Bestor T.H.. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992; 71:865–873. [DOI] [PubMed] [Google Scholar]
- 51. Yan Q., Cho E., Lockett S., Muegge K.. Association of Lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin. Mol. Cell. Biol. 2003; 23:8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tao Y., Xi S., Shan J., Maunakea A., Che A., Briones V., Lee E.Y., Geiman T., Huang J., Stephens R. et al.. Lsh, chromatin remodeling family member, modulates genome-wide cytosine methylation patterns at nonrepeat sequences. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:5626–5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu W., Briones V., Lister R., McIntosh C., Han Y., Lee E.Y., Ren J., Terashima M., Leighty R.M., Ecker J.R. et al.. CG hypomethylation in Lsh-/- mouse embryonic fibroblasts is associated with de novo H3K4me1 formation and altered cellular plasticity. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:5890–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Smith Z.D., Chan M.M., Humm K.C., Karnik R., Mekhoubad S., Regev A., Eggan K., Meissner A.. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014; 511:611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang L., Zhang J., Duan J., Gao X., Zhu W., Lu X., Yang L., Zhang J., Li G., Ci W. et al.. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014; 157:979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu X., Zhang Y.. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 2017; 18:517–534. [DOI] [PubMed] [Google Scholar]
- 57. Chen T., Hevi S., Gay F., Tsujimoto N., He T., Zhang B., Ueda Y., Li E.. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 2007; 39:391–396. [DOI] [PubMed] [Google Scholar]
- 58. Li E., Bestor T.H., Jaenisch R.. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992; 69:915–926. [DOI] [PubMed] [Google Scholar]
- 59. Tsumura A., Hayakawa T., Kumaki Y., Takebayashi S., Sakaue M., Matsuoka C., Shimotohno K., Ishikawa F., Li E., Ueda H.R. et al.. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006; 11:805–814. [DOI] [PubMed] [Google Scholar]
- 60. Velasco G., Grillo G., Touleimat N., Ferry L., Ivkovic I., Ribierre F., Deleuze J.F., Chantalat S., Picard C., Francastel C.. Comparative methylome analysis of ICF patients identifies heterochromatin loci that require ZBTB24, CDCA7 and HELLS for their methylated state. Hum. Mol. Genet. 2018; 27:2409–2424. [DOI] [PubMed] [Google Scholar]
- 61. Unoki M., Funabiki H., Velasco G., Francastel C., Sasaki H.. CDCA7 and HELLS mutations undermine nonhomologous end joining in centromeric instability syndrome. J. Clin. Invest. 2019; 129:78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Arand J., Spieler D., Karius T., Branco M.R., Meilinger D., Meissner A., Jenuwein T., Xu G., Leonhardt H., Wolf V. et al.. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLos Genet. 2012; 8:e1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gowher H., Stockdale C.J., Goyal R., Ferreira H., Owen-Hughes T., Jeltsch A.. De novo methylation of nucleosomal DNA by the mammalian Dnmt1 and Dnmt3A DNA methyltransferases. Biochemistry. 2005; 44:9899–9904. [DOI] [PubMed] [Google Scholar]
- 64. Song J., Rechkoblit O., Bestor T.H., Patel D.J.. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science. 2011; 331:1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xu G.L., Wong J.M.. Oxidative DNA demethylation mediated by Tet enzymes. Natl. Sci. Rev. 2015; 2:318–328. [Google Scholar]
- 66. Li Y., Zhang Z., Chen J., Liu W., Lai W., Liu B., Li X., Liu L., Xu S., Dong Q. et al.. Stella safeguards the oocyte methylome by preventing de novo methylation mediated by DNMT1. Nature. 2018; 564:136–140. [DOI] [PubMed] [Google Scholar]
- 67. Robertson A.K., Geiman T.M., Sankpal U.T., Hager G.L., Robertson K.D.. Effects of chromatin structure on the enzymatic and DNA binding functions of DNA methyltransferases DNMT1 and Dnmt3a in vitro. Biochem. Biophys. Res. Commun. 2004; 322:110–118. [DOI] [PubMed] [Google Scholar]
- 68. Schrader A., Gross T., Thalhammer V., Langst G.. Characterization of Dnmt1 binding and DNA methylation on nucleosomes and nucleosomal arrays. PLoS One. 2015; 10:e0140076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yan Q., Huang J., Fan T., Zhu H., Muegge K.. Lsh, a modulator of CpG methylation, is crucial for normal histone methylation. EMBO J. 2003; 22:5154–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huang J., Fan T., Yan Q., Zhu H., Fox S., Issaq H.J., Best L., Gangi L., Munroe D., Muegge K.. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res. 2004; 32:5019–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Burrage J., Termanis A., Geissner A., Myant K., Gordon K., Stancheva I.. The SNF2 family ATPase LSH promotes phosphorylation of H2AX and efficient repair of DNA double-strand breaks in mammalian cells. J. Cell Sci. 2012; 125:5524–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rothbart S.B., Dickson B.M., Ong M.S., Krajewski K., Houliston S., Kireev D.B., Arrowsmith C.H., Strahl B.D.. Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 2013; 27:1288–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Xu C., Corces V.G.. Nascent DNA methylome mapping reveals inheritance of hemimethylation at CTCF/cohesin sites. Science. 2018; 359:1166–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RRBS data and RNA-seq data were submitted to the GEO repository under accession number GEO: GSE136931.