Abstract
DNA double-strand breaks (DSBs) are highly cytotoxic lesions that can lead to chromosome rearrangements, genomic instability and cell death. Consequently, cells have evolved multiple mechanisms to efficiently repair DSBs to preserve genomic integrity. We have developed a DSB repair assay system, designated CDDR (CRISPR–Cas9-based Dual-fluorescent DSB Repair), that enables the detection and quantification of DSB repair outcomes in mammalian cells with high precision. CDDR is based on the introduction and subsequent resolution of one or two DSB(s) in an intrachromosomal fluorescent reporter following the expression of Cas9 and sgRNAs targeting the reporter. CDDR can discriminate between high-fidelity (HF) and error-prone non-homologous end-joining (NHEJ), as well as between proximal and distal NHEJ repair. Furthermore, CDDR can detect homology-directed repair (HDR) with high sensitivity. Using CDDR, we found HF-NHEJ to be strictly dependent on DNA Ligase IV, XRCC4 and XLF, members of the canonical branch of NHEJ pathway (c-NHEJ). Loss of these genes also stimulated HDR, and promoted error-prone distal end-joining. Deletion of the DNA repair kinase ATM, on the other hand, stimulated HF-NHEJ and suppressed HDR. These findings demonstrate the utility of CDDR in characterizing the effect of repair factors and in elucidating the balance between competing DSB repair pathways.
INTRODUCTION
DNA double-strand breaks (DSBs) are the most deleterious form of DNA damage and can lead to chromosomal translocations, genomic instability and cell death. Many of the currently available anti-cancer therapies including radiotherapy, topoisomerase inhibitors and replication inhibitors, rely on their ability to induce DSBs to effectively eliminate cancer cells. Thus, elucidating the mechanisms underlying DSB repair not only enhances our understanding of cancer etiology and the factors that affect the sensitivity of tumors to radio- and chemotherapies, but also helps identify novel molecular targets for therapeutic intervention.
Cells have evolved highly conserved mechanisms and distinct pathways to resolve DSBs. In mammalian cells, DSBs are predominantly repaired by non-homologous end-joining (NHEJ) and homology-directed repair (HDR). HDR faithfully repairs DSBs using extensive sequence homology between a pair of homologous duplex DNA molecules (1,2). This restricts HDR activity to cells encountering DSBs in S and G2 phases of the cell cycle, when a sister chromatid is available for templated repair. By contrast, NHEJ operates throughout the cell cycle and is generally considered to be error-prone, often resulting in small insertions and deletions (indels) (2,3). Repair of DSBs via NHEJ encompasses two major sub-pathways: canonical/classical NHEJ (cNHEJ), and non-canonical, alternative end-joining (alt-EJ). The c-NHEJ repair branch is dependent on the activity of the DNA-PK holoenzyme, among other DSB repair proteins including DNA Ligase IV, XRCC4 and XLF. This repair pathway involves minimal end-processing to ligate DSBs in a manner that is largely independent of sequence homology (2,3). Alt-EJ, on the other hand, functions in the absence of cNHEJ proteins and requires 5′ to 3′ end-resection, mediated by the MRN complex (MRE11, RAD50 and NBS1) and CtIP. Other repair factors implicated in alt-EJ include PARP1 and DNA Ligase I or III (1,2). Alt-EJ often involves a synthesis-dependent mechanism that requires the activity of DNA polymerase theta (Pol θ; also known as POLQ), and is directed by short tracts of sequence homology (microhomology or MH) flanking the DSBs to repair broken ends, resulting in MH-flanked larger deletions or templated insertions (1,2). As such, this type of alt-EJ repair has generally been referred to as microhomology-mediated end-joining (MMEJ) or theta-mediated end-joining (TMEJ) (1,2).
Several cell-based reporter assays have been developed to measure DSB repair activity in mammalian cells, and these have proven valuable in ascertaining the role of some DNA repair proteins in a number of mechanistically distinct repair pathways (4–30). Initial assays were based on the capacity of a cell or cell extracts to rejoin the ends of linearized plasmids, followed by quantitative measurement of the repaired plasmids by PCR or by flow cytometry if the plasmid circularization generates a cDNA coding for a fluorescent protein (4,5). These assays have been supplanted by chromosomally-integrated reporter systems that recapitulate genomic features that are lacking in plasmid-based assays (e.g. nucleosome packaging, epigenetic modifications, etc.) (6–30). The majority of these intra-chromosomal reporter assays are based on the introduction of DSBs through the expression of an endonuclease (e.g. I-SceI or Cas9) targeting specific sites within the reporter (6–30). These reporters typically encode a fluorescent protein that is either disrupted or repaired following the induction of a single or two DSB(s) at an integrated I-SceI recognition sequence, or at a site complementary to a single guide RNA (sgRNA) that guides Cas9 to the target sequence. Following the expression of I-SceI or Cas9/sgRNA, various DSB repair activities can be quantitatively measured through the gain or loss of fluorescent signals by flow cytometry. These repair activities, however, are often measured at low frequencies, in part due to poor transfection or endonuclease cutting efficiencies, and/or suboptimal reporter designs. Further limitations include variability in transfection efficiency and the requirement for normalization with additional control plasmids. These limitations impede the accurate assessment of the role of certain proteins in resolving DSBs, and the impact of particular treatments (e.g. si-RNA transfection or pharmacological inhibition) on DSB repair. Importantly, the majority of I-SceI- and Cas9-based assays exhibit significant bias towards mutagenic repair because the faithful repair of targeted DSBs regenerates the endonuclease recognition site, rendering it susceptible to multiple cleavage-repair cycles (15,17,26,31). Furthermore, most DSB repair reporters have been established in few cell lines and require significant effort for clonal amplification and validation, limiting their use by the broader research community.
In this study, we sought to develop a reporter assay system for detecting and measuring DSB repair activity that overcomes several of the limitations described above, and further enables the examination of the role of DSB repair proteins or the impact of certain treatments on DSB repair with higher precision and sensitivity. Our novel DSB repair assay system reports on the repair of Cas9-induced DSBs with high efficiency and reproducibility, and without the need for normalization steps. Using isogenic U2OS cells with deletions in key genes implicated in the repair of DSBs, we demonstrate the utility of our reporter system in elucidating the contribution of important repair proteins to the fidelity of DSB repair by NHEJ, and the utilization of distal versus proximal DSB ends in NHEJ, as well as the repair of DSBs via HDR.
MATERIALS AND METHODS
Cell culture
Human osteosarcoma U2OS cells and human embryonic kidney HEK 293T cells were acquired from ATCC, and maintained in high-glucose DMEM supplemented with 10% FBS and 1% penicillin/streptomycin (100 U/ml penicillin, 100 mg/ml streptomycin). The cell lines were validated by short tandem repeat profiling (STR) and tested negative for mycoplasma contamination using the PlasmoTest mycoplasma detection kit (Invivogen).
Cell lysis and immunoblotting
For immunoblotting, cells were lysed using RIPA buffer (50 mM Tris (pH 8.0), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with 1X Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific). Protein lysates were quantified via Bradford Assay using Protein Assay Dye Reagent (Bio-Rad) and bovine albumin for standard curve preparation. Equal amounts of protein lysate were separated on 7–12% polyacrylamide gels via electrophoresis and transferred to nitrocellulose membranes (GE Life Sciences). Membranes were blocked in 5% milk or 5% BSA for 1 h at room temperature and probed with the following antibodies at a 1:1000 dilution unless indicated otherwise: ATM (Abcam, ab78), LIG4 (Sigma Aldrich, HPA001334), Tubulin (Santa Cruz, sc-53646), XLF (Bethyl Laboratories, A300-730A-M), and XRCC4 (Santa Cruz, sc-271087). Membranes were incubated with HRP-conjugated secondary anti-mouse or anti-rabbit IgG (1:5000, DAKO, Cat #P0161 and P0448, respectively) in 5% milk or 5% BSA for 1 h at room temperature. HRP signals were detected by enhanced chemiluminescence (ECL) reagent (Millipore).
Plasmid construction
The lenti-CDDR reporter construct was generated by cloning the CDDR reporter into the pCW-Cas9 expression vector (Addgene #50661). The puromycin resistance gene was replaced with a blasticidin resistance gene using standard subcloning with restriction endonucleases and the following primer pairs: Blasticidin-Fwd: 5′-AAAAAGGATccttgcgccttttccaagg-3′, and Blasticidin-Rev: 5′-aaaaaCCCGGGTTActgcagctgcaggccC-3′. The amino- and carboxyl-terminal halves of turbo GFP were PCR-amplified using the following primers: tGFP-N-Fwd: 5′-TCGCAACGGGTTTGCCGCCAGAACACAGGAGCCACCATGGAGAGCGACGAGAGCG-3′, tGFP-N-Rev: 5′-AGACTTCCTCTGCCCTCTCCAGACCCATCCCACTGCAGCACGCCGCC-3′, tGFP-C-Fwd: 5′-ggacgagctgtacaagtaaccctgaACGTGAGCTTCAGCTACCG-3, and tGFP-C-Rev: 5′-aacccagggctgccttggaaaaggcgcaagTTATTCTTCACCGGCATCTGCATCC-3′. The mCherry cDNA was PCR-amplified using the following primers: mCherry-Fwd: 5′-GGCGGCGTGCTGCAgtgggatGGGTCTGGAGAGGGCAGAGGAAGTCTGCTAAC-3′ and mCherry-Rev: 5′-CTGAAGCTCACGTtcagggttacttgtacagctcgtccatgcc-3′. The SpCas9 region of the pCW-Cas9 vector was replaced with N-terminal tGFP, mCherry, and C-terminal tGFP fragments using Gibson Assembly (NEB Cat. # E5510S). An sgRNA target site (sgRNA-A) and a T2A self-cleaving peptide sequence were placed directly upstream the mCherry cDNA, while a secondary sgRNA target site (sgRNA-B) and multiple stop codons were positioned downstream of the mCherry cDNA. To generate the BFP donor plasmid, we replaced the mCherry cDNA of the CDDR reporter construct with BFP cDNA, and removed the PAM sequence from the sgRNA target sites. Gibson Assembly was used to assemble the N-terminal tGFP, BFP, and C-terminal tGFP fragments into the pcDNA3.1 expression vector (Invitrogen). The plasmids expressing sgRNA-A (5′-GGACGGCGGCGTGCTGCAGT-3), sgRNA-B (5′-GTAGCTGAAGCTCACGTTCA-3′) or both sgRNA-A and -B were derived from the pX330 Chimeric hSpCas9 expression vector (Addgene #42230). A puromycin resistance gene was subcloned into the pX330 backbone using standard cloning techniques. All plasmids constructed in this study were verified by Sanger sequencing (Eurofins) and have been deposited to Addgene.
Lentivirus production and cell line transduction
For lentivirus production, 1.8 μg of pMD2.G (Addgene #12259), 3.2 μg of psPAX2 (Addgene #12260), and 9 μg of lenti-CDDR vector were co-transfected into 293T cells using FuGENE-6 (Promega). Supernatants containing lentiviral particles were collected 48 h post-transfection. Viral supernatant was filtered using a 0.20 μM filter, and subsequently flash-frozen in liquid nitrogen and stored at −80°C. U2OS and 293T cells were transduced with lentiviral supernatant supplemented with 10 μg/ml Polybrene for 24 h followed by selection in the presence of blasticidin (10 μg/ml) for 4 days. The lenti-CDDR reporter was transduced at low multiplicity of infection (∼0.2 MOI) to generate stable cell lines with a single copy of the chromosomally-integrated reporter. Single cells were isolated by FACS sorting for mCherry+ cells and subsequently amplified to generate individual clones of the U2OS-CDDR and 293T-CDDR cells. For experiments requiring multiple integrated copies of the CDDR reporter, we steadily increased the MOI to generate cell lines with multiple CDDR copies.
Generation of isogenic U2OS-CDDR cells with individual gene knockouts
To generate U2OS-CDDR cells deleted of various DNA repair genes, U2OS-CDDR-Clone-1 cells were transiently transfected with Cas9 and two single guide RNAs (sgRNAs) targeting the individual genes. U2OS-CDDR-Clone-1 cells were verified to contain a single copy of the CDDR reporter on chromosome 19q13.31 (determined by whole genome sequencing; Genewiz). sgRNAs targeting the various genes were designed using Benchling and/or the UCSC genome browser CRISPR track. Pairs of sgRNA were designed to target non-overlapping sequences within early exons of each gene, and cloned into the pX330 expression vector (Addgene #42230) (Supplementary Table S1). The following sgRNAs (sense strand) were used: LIG4: sg-LIG4-1 5′-CACAAACTTCACAAACTGTT-3′, sg-LIG4-2 5′-GCAATGAGACTAATTCTTCC-3′; XRCC4: sg-XRCC4–1 5′-AGTATAACTCATTTTCTACA-3′, sg-XRCC4–2 5′-TTTGTTATTACACTTACTGA-3′; XLF: sg-XLF-1 5′-TGGGCGTGGCTACAGCTTGC-3′, sg-XLF-2 5′-TGAACAGGTGGACACTAGTG-3′; ATM: sg-ATM-1 5′-TCAACTAGAACATGATAGAG-3′, sg-ATM-2 5′-GATTCGAGATCCTGAAACAA-3′. All sgRNA-containing pX330 plasmids were verified by Sanger sequencing (Eurofins) using U6-specific primers. U2OS-CDDR-Clone-1 cells (and multicopy-CDDR U2OS cells in some instances) were co-transfected with 2.0 μg of target sgRNAs along with 0.5 μg pMSCVpuro vector (Clontech) containing a puromycin selectable marker. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. 24 h post-transfection, cells were selected in the presence of puromycin (2 μg/ml) for 48 h, after which single clones were isolated via serial dilutions of the transfected pool in the absence of puromycin. All the individual clonal lines were determined to retain sensitivity to puromycin before use in DSB repair assays.
Validation of gene deletion in U2OS cells was confirmed by genotyping and by immunoblotting. Briefly, cells were lysed overnight at 55°C in lysis buffer (100 mM NaCl, 10 mM Tris–HCl pH 8, 25 mM EDTA, and 0.5% SDS) supplemented with 20 μg of proteinase K. DNA was isolated from the lysed cells using phenol chloroform/isoamyl alcohol extraction. Genotyping was performed via PCR amplification of the targeted locus with primers flanking the two predicted Cas9 cleavage sites followed by Sanger sequencing (Eurofins). The following primers were used to amplify a 300–500 bp sequence spanning the two sgRNA target sites: LIG4-F: 5′-GGCCTTCCCTCAGAAGCTCA-3′, LIG4-R: 5′-CTCCATGAGTTCCAGTGGGTG-3′; XRCC4-F: 5′-GGTGTTTGTGTAGCTGAGAGGC-3′, XRCC4-R: 5′-CCCTGAGGACTGAAGAATAGCAC-3′; XLF-F: 5′- CCAGCATGACCCTGAGGCTC-3′, XLF-R: 5′-CCTTGGGAAACTACAGGCCAGC-3′. ATM-F: 5′-ACAGACAGTGATGTGTGTTCTG-3′, ATM-R: 5′-GCCAAATTCATATGCAAGGCAT-3′. Immunoblotting of the individual clonal knockout cell lines was further used to confirm gene deletion. The regained sensitivity of the individual clones to puromycin was confirmed by treating the clones with puromycin (2 μg/ml) for 48 h, at which time no survivors were observed.
DNA DSB repair assays
U2OS or 293T cells containing single or multiple copies of the CDDR reporter, and isogenic derivatives of individual clones of U2OS cells with single CDDR copies but with deletion of the various repair genes were seeded at a density of 7.0 × 104 cells per well in 24-well tissue culture plates. Cells were transfected with 0.5 μg of control vector without sgRNA (pX330) or with 0.5 μg of the Cas9–sgRNA vector (Cas9–sgRNA-A or Cas9–sgRNA-A+B) using Lipofectamine 3000 (Invitrogen) according to the manufacturer's protocol. For HDR analysis, an additional 0.5 μg of BFP homology donor plasmid was transfected along with the Cas9–sgRNA-A vector. 16 h post-transfection, cells were transferred to 60 mm tissue culture plates and treated with puromycin (2 μg/ml) for 48 h, at which time all of the control cells (transfected with pX330 vector only) died. For determining the frequency of NHEJ and HDR events, cells with DSBs introduced into the reporter were analyzed by flow cytometry 5 days post-transfection with the Cas9–sgRNA vector, with or without the BFP homology donor plasmid. The cells were collected in 0.5 ml PBS and analyzed on the BD LSRFortessa. FlowJo software was used to determine the fraction of GFP, mCherry, or BFP positive or negative cell populations. Where indicated, cell sorting was performed on the BD Influx Cell Sorter.
Cell cycle analysis
For cell cycle analysis, U2OS-CDDR cells with single or multiple copies of the CDDR reporter were seeded at 80–90% confluency in 10 cm tissue culture plates and transfected with 3 μg of control pcDNA3.1 vector (Invitrogen) or with 3 μg of Cas9–sgRNA-A vector and 1 μg of GFP vector (Clontech) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were harvested 24 h post-transfection for flow cytometry analysis. Cells were fixed in 1% formaldehyde solution in PBS for 1 h and resuspended in 70% ethanol for 30 min. Cells were subsequently stained in propidium iodide (PI) buffer (50 μg/ml PI (Sigma Aldrich), 10 μg/ml DNase-free RNase A, and 0.05% NP40) for an additional 30 min prior to FACS acquisition on the BD LSRFortessa. FlowJo and ModFit software were used to determine the percentage of the transfected, GFP+ cells in each phase of the cell cycle.
Pharmacological inhibition
Where indicated, individual clones of U2OS or 293T cells with single copies of the CDDR reporter were treated with the ATM kinase inhibitor KU-55933 (Tocris). KU-55933 was added 4 h post-transfection of the Cas9–sgRNA vector and replenished the following day. KU-55933 was removed 48 h post-transfection.
Surveyor nuclease assays
Surveyor nuclease assays were performed according to the manufacturer's instructions (IDT Surveyor Mutation Detection Kit). The DSB sites within the integrated reporter in transfected (cut) and non-transfected (reference) cells were PCR-amplified using the following primers tGFP-Fwd: 5′-CTACCACTTCGGCACCTAC-3′, and tGFP-Rev: 5′-CTTGAAGTGCATGTGGCTG-3′. PCR amplicons from the transfected and non-transfected cells were mixed in equal amounts and hybridized through denaturation and reannealing to form hetero- and homo-duplexes. Hybridized DNA (400 ng) was digested with Surveyor Nuclease (1 μl) in a 40 μl volume reaction supplemented with 0.15 M MgCl2 Solution (4 μl) and Surveyor Enhancer (1 μl) at 42°C for 60 min. As a negative control, PCR amplicons from the non-transfected cells were treated with Surveyor nuclease in parallel. Following digestion, stop solution was added to the reaction and the digestion products were analyzed using agarose gel electrophoresis. The presence of cleaved DNA fragments is indicative of heteroduplex formation resulting from DNA mismatches (32).
End-utilization analysis
For end-utilization analysis, genomic DNA was extracted from U2OS-CDDR-Clone-1 cells (or isogenic derivatives deleted of the indicated genes) following the induction of two DSBs at the sgRNA-A and sgRNA-B targeted sites. The cleaved and repaired locus was PCR-amplified using the tGFP primers (listed above) that anneal to regions flanking the mCherry cDNA sequence within the CDDR reporter. This PCR yielded two amplicons corresponding to the amount of proximal- and distal- NHEJ. The percentage of distal end-utilization was calculated as the band intensity derived from the distal EJ amplicon, divided by the total band intensity (the sum of the band intensity of proximal and distal EJ amplicons). Band intensity was quantified using Image Lab software (Bio-Rad).
Deep sequencing of DSB repair junctions
Genomic DNA from U2OS- or 293T-CDDR cells was isolated a minimum of 5 days following transfection with the Cas9-sgRNA vector. PCR amplicons spanning the cleaved and repaired end-joining junctions were generated using the following primers tGFP-Fwd: 5′-CTACCACTTCGGCACCTAC-3′, and tGFP-Rev: 5′-CTTGAAGTGCATGTGGCTG-3′. Next-Generation Sequencing (NGS) libraries were prepared using nested PCR as previously described (33), with first-round primers consisting of universal Illumina sequences and a priming site designed to amplify a ∼300 bp region surrounding the predicted Cas9 cleavage sites, as well as the fusion junction site formed via end-joining of the two distal DSBs (Supplementary Table S2). To amplify the DSB site that was cleaved in the presence of the BFP homology donor plasmid, we designed first-round primers that completely anneal to both the mCherry and BFP cDNA, which is possible due to high sequence similarity among the two cDNA. The custom first-round forward primers include a 1–10 nt staggered region following the Illumina adapter sequence and preceding the primer binding site in order to introduce sequencing diversity and improve sequencing quality. Second-round universal primers with unique barcodes were used to amplify the first-round PCR product (Supplementary Table S2). Qubit and qPCR measurements were used to determine library quality and concentration prior to the NGS run setup. Paired-end sequencing was performed on the MiSeq NGS Platform at the University of Virginia Sequencing Core using the 500-cycle MiSeq Kit V2 (Illumina); Read 1 = 250 cycles, Read 2 = 250 cycles, Index 1 = 8 cycles, Index 2 = 8 cycles. The high-throughput sequencing (HTS) raw data were deposited at the NCBI Sequence Read Archive (SRA) database (PRJNA628028).
Sequencing analysis of DSB repair junctions
Sequencing reads were trimmed and quality filtered using Cutadapt v2 (34). Analysis of the indels at the predicted Cas9 cleavage sites and fusion junction site was performed using CRISPResso2 to characterize the frequency and size of insertions, deletions and substitutions (35).
Statistical analysis
For DNA repair quantification by flow cytometry and for PCR end-utilization analysis, at least three independent experiments were performed for each data set, and expressed as the mean ± standard deviation (s.d.). Statistical significance was determined by two-tailed Student's t-test and P < 0.05 was considered significant. *P < 0.05, **P < 0.01, ***P < 0.001. For NGS analyses, statistical differences among samples were determined using chi-squared test with Yates’ continuity correction. Bonferroni correction was applied to all comparisons. For indel analyses, additional stringency of >1.5-fold change was applied to determine significance (P < 0.0001 and >1.5-fold change).
RESULTS
Construction and characterization of the CDDR DSB repair reporter assay system
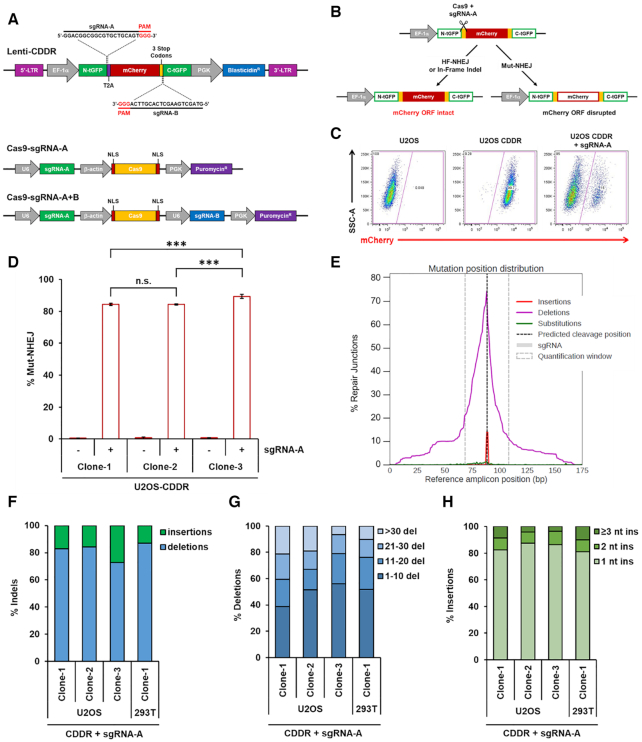
We sought to develop a non-biased assay system to measure the capacity of cells to repair DSBs with high efficiency in mammalian cells. To this end, we have engineered a dual-fluorescent DSB repair assay based on the introduction and subsequent repair of one or two DSB(s) within a chromosomally-integrated fluorescent reporter following the expression of SpCas9 endonuclease and single guide RNAs (sgRNAs) targeting the reporter. The lentiviral reporter construct (Figure 1A), dubbed ‘CDDR’ for CRISPR–Cas9-based Dual-fluorescent DSB Repair, constitutively expresses mCherry and contains a blasticidin resistance gene for selection in mammalian cells. The mCherry cDNA is flanked by two halves of TurboGFP cDNA, with the N-terminal half (N-tGFP) in frame with mCherry, but separated by a T2A self-cleaving peptide coding sequence to ensure the generation of functional mCherry (36). The T2A peptide coding sequence is preceded by a 20 nt sequence complementary to an sgRNA (sgRNA-A) that guides the Cas9 to induce a DSB between the N-tGFP and mCherry sequence. Multiple stop codons were placed downstream of the mCherry cDNA, followed by a 20 nt sequence complementary to a second sgRNA (sgRNA-B) that guides the Cas9 to induce a second DSB immediately upstream of the C-terminal half of TurboGFP (C-tGFP). The CDDR reporter is designed such that a single DSB is introduced at the sgRNA-A target site following the transient transfection of cells with a plasmid expressing both Cas9 and sgRNA-A (Figure 1A). Alternatively, two DSBs are introduced at both the sgRNA-A and -B target sites by transiently transfecting cells containing the reporter with a plasmid expressing Cas9 and both sgRNA-A and -B (Figure 1A). The Cas9-expressing plasmids contain a puromycin resistance gene for the selection of transfected cells, thus ensuring that downstream analyses will exclude cells that do not express Cas9/sgRNA(s), and further obviating the need for normalization of transfection efficiency.
Figure 1.
Detection and measurement of DSB repair using the CDDR reporter assay. (A) Schematic of the lenti-CDDR reporter (top) and Cas9–sgRNA-expressing plasmids (bottom). (B) Diagram of the phenotypic outcomes following the induction of single DSBs and subsequent repair within the CDDR reporter. (C) Representative FACS profiles of normal U2OS cells and U2OS-CDDR cells ± transfection with the Cas9–sgRNA-A plasmid. (D) Percentage of cells with single DSBs repaired via mutagenic non-homologous end-joining (Mut-NHEJ) in three independent clones of U2OS-CDDR cells. Mut-NHEJ is quantified as the percentage of mCherry-ve cells measured by FACS analysis after induction of DSBs and repair within the CDDR reporter. The percentage of mCherry-ve/GFP-ve cells is depicted using white columns with a red border throughout the study. Data represent the mean of three independent experiments ± standard deviation (s.d.). Statistical significance was determined using two-tailed Student's t-test; ***P< 0.001, n.s.: not significant. (E) Distribution of deep sequencing reads with insertions (red), deletions (purple), and substitutions (green) across the reference amplicon in U2OS-CDDR-Clone-1 cells. Indels are centered around the predicted sgRNA cleavage site (dotted line). Plot was generated with CRISPResso2 (35). (F) Frequency of deep sequencing reads categorized as insertions or deletions among total indels at the repair junctions in U2OS- and 293T-CDDR cells following the induction of single DSBs within the CDDR reporter. (G, H) Size distribution among deletions (G) and insertions (H) at the repair junctions in U2OS- and 293T-CDDR cells following the induction of single DSBs within the CDDR reporter.
The repair of a single DSB at the sgRNA-A target site may proceed through high-fidelity NHEJ (HF-NHEJ) or HDR, causing cells to retain mCherry expression (Figure 1B and C). By contrast, error-prone, mutagenic NHEJ (Mut-NHEJ), results in the loss of mCherry expression following disruption of the mCherry open-reading frame (ORF) (Figure 1B and C). However, due to reiterative cutting by the Cas9 nuclease following the repair of single DSBs by HF-NHEJ, few cells undergoing mutagenic NHEJ repair will contain small in-frame indels and, consequently, will retain mCherry expression, limiting the ability to determine the fidelity of repair at these proximal EJ junctions with high precision. Therefore, the appearance of some mCherry+ cells following the reiterative induction of single DSBs within the CDDR reporter may not represent ‘true’ proximal HF-NHEJ repair events, but instead represent repair events that generate small in-frame indels mediated by the cNHEJ pathway (discussed below).
The introduction of two synchronous DSBs at the sgRNA-A and -B target sites within the CDDR reporter overcomes the limitation of reiterative cutting, as the end-joining between the two distal DSBs destroys both sgRNA target sites and the protospacer adjacent motif (PAM) sequences recognizable by Cas9 (Figure 1A). The precise (high-fidelity) end-joining of the two distal DSBs at the predicted Cas9 cleavage sites results in the loss of the entire mCherry fragment (∼800 bp) and the fusion of the separated TurboGFP halves, generating mCherry-negative/GFP-positive (mCherry-ve/GFP+) cells. In cells containing a single copy of the CDDR reporter, this population represents distal HF-NHEJ repair events (Supplementary Figure S1A). On the other hand, if repair at these two distal ends proceeds through error-prone, mutagenic NHEJ, this not only eliminates the mCherry coding sequence, but will also generate indels that interfere with GFP expression, resulting in mCherry-ve/GFP-ve cells, which we refer to as Mut-NHEJ repair events. In both cases, repair by distal-NHEJ is not subject to the multiple cleavage-repair cycles associated with single DSBs (proximal-NHEJ). Thus, depending on whether a single or two distal DSB(s) are introduced within the reporter, it is possible to measure both proximal- and distal-NHEJ, and to report on the fidelity of NHEJ (HF- versus Mut-NHEJ).
To examine the functionality of the reporter system, we first transduced human osteosarcoma U2OS cells with the CDDR lentiviral reporter at low multiplicity of infection (MOI) to ensure that a single CDDR copy is randomly integrated into the cells (U2OS-CDDR). To determine the efficiency of introducing a DSB at the individual sgRNA target sites within the integrated reporter, we transiently transfected a pool of FACS-sorted mCherry+ U2OS-CDDR cells with a plasmid co-expressing Cas9 and either sgRNA-A or sgRNA-B. Following puromycin selection of the transfected cells (48 hours), we isolated DNA and assessed the editing efficiency at these sites using Surveyor nuclease assays (32). Briefly, we PCR-amplified the sequences flanking the DSB sites within the integrated reporter in control U2OS cells and in cells transfected with the Cas9-sgRNA plasmid. PCR amplicons from the transfected and non-transfected cells were mixed in equal amounts and hybridized through denaturation and reannealing to form hetero- and homo-duplexes. The mixture of annealed hetero- and homoduplexes was treated with Surveyor nuclease, which cleaves 3′ of mismatched sites (including base substitutions and indels) in both DNA strands (32). As a negative control, PCR amplicons from the non-transfected cells were treated in parallel with Surveyor nuclease. Following treatment, the digestion products were analyzed using agarose gel electrophoresis. We observed cleaved DNA fragments in the cells that were transfected with the Cas9-sgRNA plasmid, which is indicative of heteroduplex formation resulting from DNA mismatches (Supplementary Figure S1B). This analysis showed an editing efficiency of greater than 50% at both cleavage sites. Similar results were obtained in 293T cells containing a single copy of the integrated CDDR reporter (data not shown).
Using FACS sorting, we isolated and amplified individual clones of mCherry+ U2OS cells containing a single-copy of the CDDR reporter for further analysis. Transient transfection of three individual clones of U2OS-CDDR cells with Cas9 and sgRNA-A resulted in the loss of mCherry signal in approximately 85–90% of the cells in each of these clones, indicating a strong tendency towards mutagenic repair (Figure 1C and D). A slightly lower percentage of mCherry-ve cells (63–75%) was detected in individual clones of 293T cells containing a single-copy of the CDDR reporter (Supplementary Figure S1C and D). The bias towards mutagenic repair in cells with single DSBs was not unexpected, because accurate repair of these sites reconstitutes the target sequence, resulting in the reiterative cleavage of the reporter by the Cas9–sgRNA complex (17,26,31).
To confirm the results obtained by FACS, we performed DNA deep sequencing of the PCR-amplified repair junctions at the sgRNA-A target site, both in U2OS and 293T cells, and analyzed the frequency of indels using CRISPResso2 (35). This analysis revealed that almost all of the repair junctions in the analyzed clones in U2OS contained indels upon Cas9/sgRNA expression (data not shown), indicating, as described above, that the majority of repaired single DSBs in mCherry+ cells do not represent ‘true’ HF-NHEJ repair events, but reflect cells with small in-frame indels. Further analysis of the repair junctions indicated that most of the resulting indels (∼75–90%) were deletions centered around the predicted Cas9 cleavage site (Figure 1E and F and Supplementary Figure S1E–G). Of these deletions, the majority (∼60–80%) were <20 nt in length. (Figure 1G). The remaining indels were mainly small templated insertions, with over 80% consisting of a single nucleotide (Figure 1H). The rate of substitutions in each sample was relatively low (<1.5%), and is largely attributable to sequencing errors, given that substitutions were detected at the same frequency in amplicons from cells that were not transfected with Cas9/sgRNA, and also were not confined to the predicted Cas9 cleavage site (Figure 1E and data not shown). Notably, despite having identical sgRNA-A target sites, the frequency and size distribution of indels at the repaired DSBs varied among the individual clones analyzed, demonstrating that the genomic context within which a given sequence is cleaved influences the repair outcomes (Figure 1D, F–H and Supplementary Figure S1C).
We also examined the frequency of in-frame indels versus indels resulting in frameshifts in U2OS and 293T cells. We found that 26–32% of the indels observed in U2OS cells were in-frame, which is approximating the value expected if repair outcomes were to follow a normal distribution (Supplementary Figure S2A). In 293T-CDDR cells surviving single DSBs, the frequency of in-frame mutations decreased from 30% in the unsorted pool to 16% in the sorted mCherry-ve population (Supplementary Figure S2B). This indicates that a sizeable percentage of the mCherry+ cells in the unsorted pool contain in-frame indels that permitted mCherry expression. Thus, in both 293T and U2OS cells, mCherry-positivity following the induction of single DSBs largely represents cells with small in-frame indels. On the other hand, mCherry-negativity is strictly associated with Mut-NHEJ, and sequencing analysis of the repair junctions revealed a significant increase in modified sequences in FACS-sorted mCherry-ve cells relative to unsorted cells (from 89% to 97%), which supports equating mCherry-negativity with Mut-NHEJ (Supplementary Figure S2C). Moreover, with the exception of the appearance of larger (>30 bp) deletions in the sorted mCherry-ve cells relative to the unsorted cells, both the frequency and the size distribution of insertions and deletions were similar in these two cell population, indicating that sorting is generally not necessary for indel analysis (Supplementary Figure S2D–F). These results also indicate that the loss of mCherry signal is the result of targeted genomic editing, and not a consequence of potential interference with mCherry transcription.
To test whether DSB repair outcomes are affected by the number of breaks induced within the cells, we generated U2OS cells with increasing copy number of the CDDR reporter by transducing the cells with increasing titer of the CDDR lentivirus. Transient transfection of these multicopy-CDDR U2OS cells with the Cas9–sgRNA-A plasmid resulted in a copy number-dependent decrease of Mut-NHEJ with a corresponding increase in cells retaining mCherry expression (Supplementary Figure S3A). These results were confirmed in four independent clones of U2OS with multiple copies of the CDDR reporter (Supplementary Figure S3B). Furthermore, deep sequencing of the repair junctions in one of these multicopy-CDDR clones (Clone-1) revealed that the size and distribution of indels were similar to those observed in independent clones of single-copy U2OS-CDDR cells, with the exception of a slight increase in the rate of insertions relative to deletions (Supplementary Figure S3C–F). The notable decrease in Mut-NHEJ repair in cells containing multiple copies of the CDDR reporter may reflect enhanced repair via the HDR pathway, with the multiple copies serving as templates for HDR or may indicate a lack of Cas9 cleavage at the sgRNA-A target site within one or more copies of the CDDR reporter. Alternatively, this could reflect an actual increase in repair fidelity (HF-NHEJ or HDR), potentially through enhanced checkpoint activation and/or activated DNA repair proteins. Consistent with the latter possibility, we found that DSB induction in U2OS cells with multiple CDDR copies resulted in a twofold increase in the percentage of cells with G2/M DNA content compared to cells with a single copy (Supplementary Figure S3G). This suggests that in cells with multiple DSBs, persistent unrepaired DNA damage may lead to increased checkpoint activation, with more cells arresting in G2 phase.
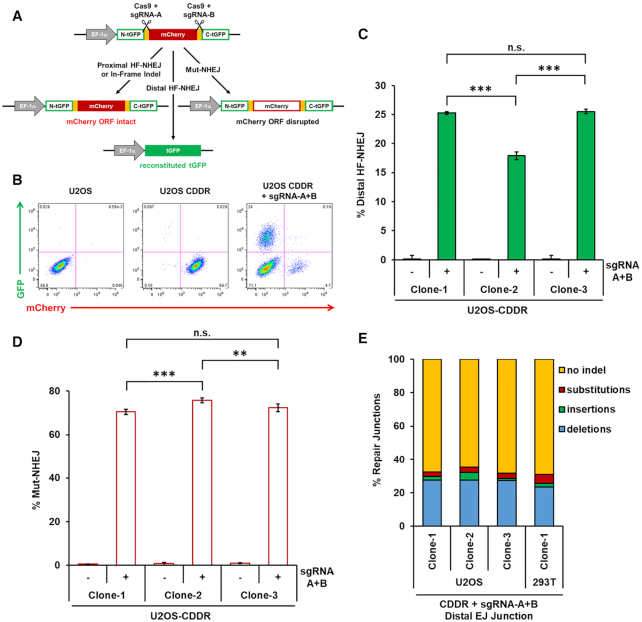
CDDR enables the accurate reporting of distal NHEJ repair events in mammalian cells
To investigate the ability of the CDDR repair assay system to measure the fidelity of NHEJ repair without the mutagenic bias imposed by the reiterative cutting of single DSBs, we transfected individual clones of U2OS and 293T cells containing single copies of the CDDR reporter with a plasmid expressing Cas9, and both sgRNA-A and sgRNA-B. HF-NHEJ between the two distal DSBs is predicted to result in the loss of mCherry signal concurrent with a gain of GFP expression (mCherry-ve/GFP+) (Figure 2A and B). On the other hand, inaccurate repair (Mut-NHEJ) of the DSBs will generate indels with the consequent loss of mCherry signal but without gaining GFP expression (mCherry-ve/GFP-ve) (Figure 2A and B). In practice, we found that the induction of two DSBs within the CDDR reporter in single-copy U2OS-CDDR cells resulted in 18–25% of the cells expressing GFP and losing mCherry expression, demonstrating repair by distal HF-NHEJ (Figure 2B and C). The variability in the frequency of cells undergoing distal HF-NHEJ among the individual clones of U2OS cells with different integration sites of the CDDR reporter supports the conclusion that the genomic context within which a DSB is introduced, in addition to the immediate flanking sequences, can influence repair outcomes (37–40). The majority of cells (70–76%) were negative for both mCherry and GFP expression (mCherry-ve/GFP-ve), representing Mut-NHEJ repair of both proximal and distal end-joining events (Figure 2D). The remaining 2–6% of the cells that retained mCherry expression (mCherry+/GFP-ve) represent cells that were either repaired via proximal HF-NHEJ, or those repaired via proximal NHEJ, but contained small in-frame indels that do not interfere with mCherry expression (Supplementary Figure S4A). A slightly larger proportion of single-copy 293T-CDDR cells (25–41%) underwent distal HF-NHEJ (mCherry-ve/GFP+ cells) (Supplementary Figure S4B). The ∼20–40% GFP-positivity observed in U2OS and 293T cells following repair of DSBs reveals robust distal HF-NHEJ activity in mammalian cells, and reflects only the cells in which the two DSBs are processed simultaneously.
Figure 2.
Measuring high-fidelity (HF) and mutagenic (Mut) NHEJ at proximal and distal DSBs using the CDDR reporter. (A) Diagram of the phenotypic outcomes following the induction of two DSBs and subsequent repair within the CDDR reporter. (B) Representative FACS profiles of normal U2OS cells and U2OS-CDDR cells ± transfection with the Cas9–sgRNA-A+B plasmid. (C) Percentage of cells with DSBs repaired via distal high-fidelity NHEJ (HF-NHEJ) in three independent clones of U2OS-CDDR cells. Distal HF-NHEJ is quantified as the percentage of mCherry-ve/GFP+ cells measured by FACS after induction of two DSBs and subsequent repair within the CDDR reporter. The percentage of GFP+ cells is depicted using green columns throughout the study. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; ***P< 0.001, n.s.: not significant. (D) Percentage of cells with DSBs repaired via Mut-NHEJ in three independent clones of U2OS-CDDR cells. Mut-NHEJ is quantified as the percentage of mCherry-ve/GFP-ve cells after induction of two DSBs and subsequent repair within the CDDR reporter. The percentage of mCherry-ve/GFP-ve cells is depicted using white columns with a red border throughout the study. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; **P< 0.01, ***P< 0.001, n.s.: not significant. (E) Frequency of deep sequencing reads categorized as insertions, deletions, substitutions, or no indel at the repair junctions formed by the end-joining of the two distal DSBs (distal EJ junctions) within the CDDR reporter in U2OS- and 293T-CDDR cells.
To further characterize the resulting repair products in cells with two DSBs, we PCR-amplified the repair junctions within the CDDR reporter using primers flanking the mCherry cDNA (Supplementary Figure S4C). As predicted, this yielded two major bands on an agarose gel: a slow-migrating band corresponding to the independent repair of each DSB site (proximal NHEJ), and a fast-migrating band corresponding to the end-joining of the two distal DSBs (distal NHEJ) (Supplementary Figure S4D). We performed nested PCR to amplify and barcode the repair junctions at both sgRNA cleavage sites, as well as the junctions generated by the fusion of the two distal DSBs. Analysis of the proximally-ligated junctions at the sgRNA-A target site revealed indel frequencies similar to those observed following the repair of a single DSB at this site (Junction A) (Supplementary Figure S5A-D). However, unlike the indel distribution at the sgRNA-A target site, which consisted of primarily deletions (70–87%), the sgRNA-B target site contained mostly 1–2 nt insertions (67–79%; Junction B) (Supplementary Figure S5E-H). Consequently, the sgRNA-B target site had a much greater percentage of indels resulting in frameshifts compared the sgRNA-A target site (Supplementary Figure S5B and F). The striking contrast in indel distribution between the two Cas9 cleavage sites was not unexpected given that editing outcomes are dependent upon the DNA sequence at the target site (37–40). Sequence analysis of the fusion junctions (distal end-joining) indicated that approximately 70% of these junctions in U2OS or 293T cells lacked indels (other than the deletion of the mCherry sequence) (Figure 1E). Thus, in cells wherein the two distal DSBs were ligated, roughly 70% of total distal end-joining is high-fidelity (distal HF-NHEJ). Of the fusion junctions that were repaired via Mut-NHEJ, the majority of indels (86–96%) consisted of deletions that were mostly <20 nt in length, with insertions being relatively infrequent (Supplementary Figure S5I-L).
Enriching for GFP+ cells by FACS, followed by deep sequencing analysis of the distal repair junctions revealed an anticipated decrease in indels, from 26% in the unsorted pool, to less than 9% in GFP-sorted 293T cells (Supplementary Figure S6A). The presence of a small proportion of indels remaining in the sorted population was not a consequence of small in-frame indels that permitted expression of GFP (Supplementary Figure S6B), but likely reflects limitations in sorting efficiency due to inherent variance within FACS distributions that can only be reduced by numerous rounds of sorting (41). Despite these limitations, the significant reduction of indels in the population enriched for GFP+ cells supports equating GFP positivity with high-fidelity distal NHEJ events.
In a heterogenous pool or individual clones of U2OS cells containing multiple copies of the CDDR reporter, we observed a larger population of GFP+/mCherry-ve cells (40–62%) (Supplementary Figure S7A-C). An additional 7–14% of cells expressed both GFP and mCherry (GFP+/mCherry+), and these represent cells in which at least one set of DSBs is repaired by distal HF-NHEJ, with mCherry expressed from another CDDR copy that is either repaired via HDR, HF-NHEJ, or Mut-NHEJ but with small in-frame indels that do not interfere with mCherry expression (Supplementary Figure S7A–C). The apparent increase in GFP-positivity (distal HF-NHEJ) in cells with multiple copies of the CDDR reporter may reflect the sufficiency of a single HF-NHEJ event in a given cell to result in GFP expression even if all other DSBs within the cell are repaired via Mut-NHEJ, but may also be due to enhanced checkpoint activation that favors HF-NHEJ or HDR repair (Supplementary Figure S3G).
Overall, these findings demonstrate that the CDDR reporter has the ability to quantify NHEJ repair events that occur at proximal or distal DSBs and can distinguish between HF- and Mut-NHEJ with high sensitivity and efficiency, and without the need for normalization.
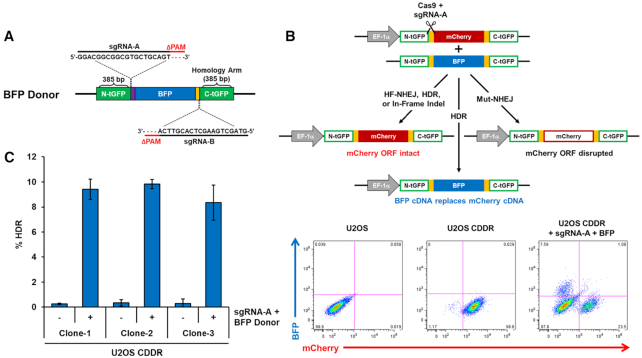
Efficient and robust detection of HDR activity using the CDDR reporter system in mammalian cells
To test whether the CDDR reporter could be used for detecting HDR events, we designed a donor plasmid that contains a promoter-less blue fluorescent protein (BFP) coding sequence flanked by two homology arms to the tGFP halves (∼400 bp in length), but with deletions of the PAM sequences at both sgRNA target sites to prevent their cleavage by Cas9 after recombination (Figure 3A). Repair of a single DSB within the reporter via HDR using the donor plasmid as a homologous template is expected to replace the mCherry cDNA with BFP cDNA (Figure 3B). This will generate BFP+/mCherry-ve cells without subjecting the recombined locus to further cutting by Cas9. We co-transfected U2OS- or 293T-CDDR cells with the Cas9–sgRNA-A plasmid along with the BFP homology donor plasmid described above. Following selection with puromycin, we found that 8–10% of U2OS cells and 1.5–3% of 293T cells were BFP+/mCherry-ve (Figure 3B, C and Supplementary Figure S8A). The reason for the lower HDR activity in 293T compared to U2OS cells is not entirely clear, but may reflect certain genetic or epigenetic differences between the two cell lines. No BFP+ cells were detected when cells were transfected with only the BFP donor plasmid (i.e. in the absence of Cas9/sgRNA; data not shown), demonstrating that BFP production is a result of HDR activity at the CDDR locus, and not due to random integration of the BFP donor sequence within the genome. This was further confirmed by deep sequencing of the recombination site from BFP-sorted cells, which demonstrated the precise replacement of the BFP cDNA (with the deleted PAM sequence) in place of the mCherry cDNA sequence (data not shown). Therefore, BFP-positivity can be used as a surrogate measurement of HDR activity in mammalian cells using the CDDR reporter system.
Figure 3.
The CDDR reporter enables the detection and quantification of HDR events with high sensitivity. (A) Schematic of the BFP donor plasmid used for homology-directed repair. (B) Top: diagram of the phenotypic outcomes following the induction of single DSBs in the presence of a BFP cDNA donor template. Bottom: Representative FACS profiles of normal U2OS cells and U2OS-CDDR cells ± co-transfection with Cas9–sgRNA-A and the BFP homology donor plasmid. (C) Percentage of cells with single DSBs repaired via HDR in three independent clones of U2OS-CDDR cells. HDR is quantified as the percentage of BFP+/mCherry-ve cells measured by FACS analysis after DSB induction and subsequent repair in the presence of a BFP cDNA donor template. The percentage of BFP+ cells is depicted using blue columns throughout the study. Data represent the mean of three independent experiments ± s.d.
These HDR events (BFP positivity) are detected at a higher frequency than those detected by other established HDR reporters (10,13,14,16,23–25). Yet, they represent only a fraction of the total HDR in these cells, since HDR activity can also repair the DSB without recombination of the entire BFP cDNA. In this scenario, the mCherry cDNA is restored using short homologous sequences present on the BFP donor template that are adjacent to the DSB site, but will contain a 3 bp deletion of the PAM sequence (which is only present on the BFP donor template). This is possible due to the high sequence similarity between the two cDNA, which is not uncommon among fluorescent proteins (42). Because the PAM sequence for SpCas9 is only three nucleotides, its removal following HDR repair leaves the mCherry cDNA sequence in-frame, resulting in mCherry+ cells. Consistently, we observed an increase in mCherry-positivity in cells with single DSBs repaired in the presence of the HDR donor plasmid than in cells repairing these DSBs in the absence of the donor plasmid (Supplementary Figure S8B). This was further corroborated by deep sequencing analysis of the repair junctions, revealing that ∼40% of the sequencing reads that aligned to the CDDR reporter (mCherry cDNA sequence) were lacking the 3 bp PAM motif, but otherwise absent of indels (Supplementary Figure S8D). In 293T cells, the frequency of sequencing reads deleted of the PAM motif (26%) was considerably less than the 40% of reads observed in U2OS cells (Supplementary Figure S8C). This is consistent with the smaller percentage of BFP-positivity in 293T cells detected via FACS, and reflects reduced HDR activity in 293T relative to U2OS cells.
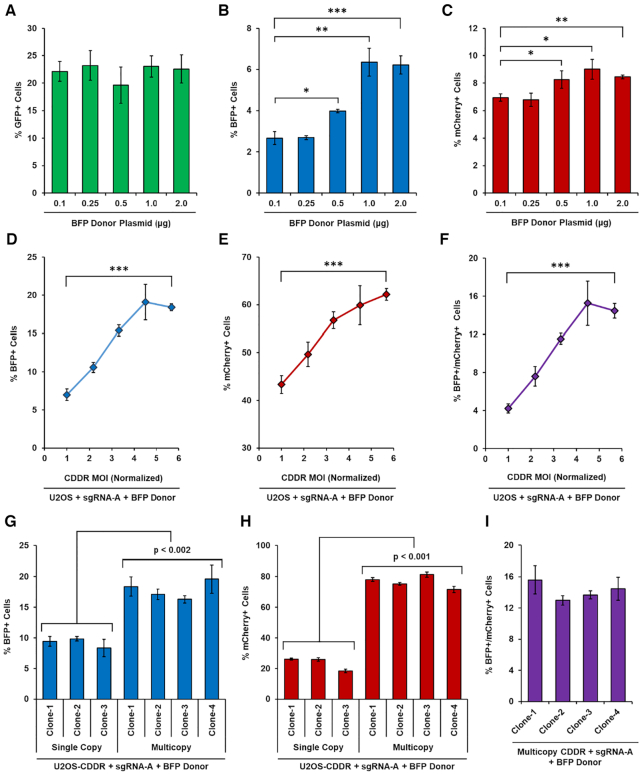
To determine whether NHEJ and HDR events can be measured simultaneously without the complication of reiterative cleavage events, we co-transfected single-copy U2OS-CDDR cells with the Cas9–sgRNA-A+B plasmid and with increasing amounts of the BFP donor plasmid. While the percentage of GFP+ cells remained relatively constant (20–23%), the percentage of BFP+ cells rose steadily with increasing amounts of BFP donor plasmid, as did the percentage of mCherry+ cells (Figure 4A–C). This implies that repair of DSBs through HDR is not necessarily at the expense of HF-NHEJ. Rather, the increase in HDR activity was accompanied by reduced Mut-NHEJ (Supplementary Figure S9A). One interpretation for this observation is that in the presence of homologous donor DNA, resected DSB ends have a greater likelihood of being repaired via HDR rather than mutagenic repair pathways (i.e. alt-EJ and SSA). However, non-resected DSB ends remain committed to repair via HF-NHEJ (predominantly through c-NHEJ) irrespective of homologous donor template concentration.
Figure 4.
Increasing HDR activity in cells using the CDDR reporter system. (A–C) Percentage of GFP+ (A), BFP+ (B) or mCherry+ (C) cells following the induction of two DSBs within the CDDR reporter after co-transfection with Cas9–sgRNA-A+B along with increasing amounts of BFP donor plasmid in U2OS-CDDR-Clone-1 cells. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; *P< 0.05, **P< 0.01, ***P< 0.001. (D–F) Percentage of cells with single DSBs repaired via HDR exclusively (% BFP+ cells; D), or by proximal HF-NHEJ or HDR (% mCherry+ cells; E), or by at least one HDR event (BFP+/mCherry+ cells; F) following DSB induction and subsequent repair in the presence of a BFP cDNA donor template in U2OS cells transduced with increasing MOI of lenti-CDDR. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; ***P< 0.001. (G, H) Percentage of BFP+ (G) or mCherry+ (H) cells following DSB induction and subsequent repair in the presence of a BFP cDNA donor template in independent clones of U2OS cells containing single or multiple copies of the CDDR reporter. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-tests; P< 0.002 for all comparisons between single and multicopy CDDR clones. (I) Percentage of BFP+/mCherry+ cells following DSB induction and subsequent repair in the presence of a BFP cDNA donor template in four independent clones of U2OS cells containing multiple copies of the CDDR reporter. Data represent the mean of three independent experiments ± s.d.
Increasing the copy number of the CDDR reporter in U2OS cells also resulted in a copy number-dependent increase in BFP+ cells, as well as an increase in cells retaining mCherry expression or expressing both BFP and mCherry (Figure 4D-I), and was accompanied by a decrease in Mut-NHEJ (Supplementary Figure S9B). The increase in both BFP and mCherry expression in cells with multiple copies of the CDDR reporter could reflect the sufficiency of a single recombination event in a given cell to result in BFP expression (or mCherry expression due to recombination only at the proximal PAM sequence), and/or increased HDR activity due to enhanced checkpoint activation (Supplementary Figure S3G). Collectively, these results show that HDR activity can be enhanced in a dose-dependent manner, either through increasing the CDDR copy number, or by increasing the concentration of homologous donor plasmid. In addition, our findings demonstrate that the CDDR reporter system can be used to measure the balance between multiple competing DSB repair pathways in mammalian cells.
DNA Ligase IV, XRCC4 and XLF are essential for HF-NHEJ and suppress distal end-utilization
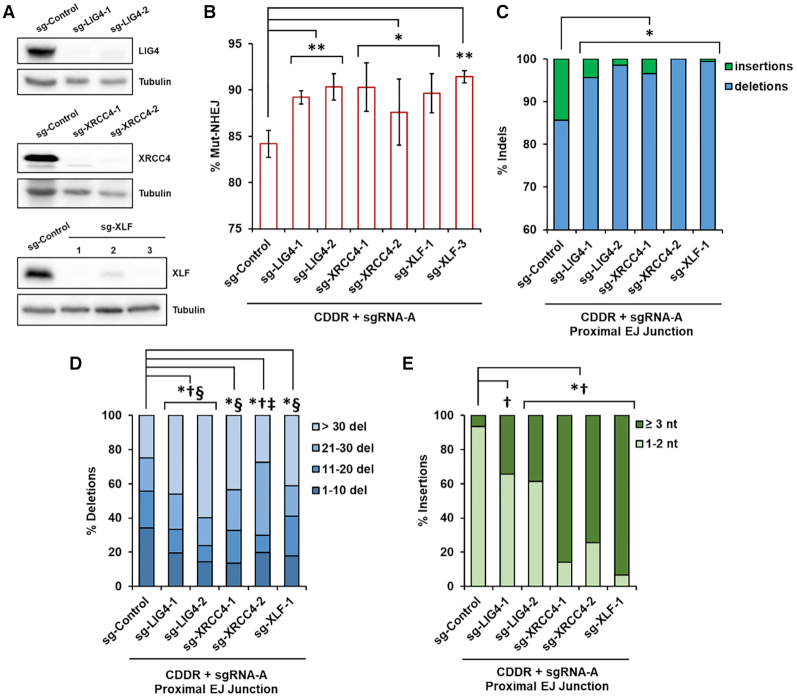
Using the CDDR reporter assay system, we sought to evaluate the contribution of key repair proteins to resolving DSBs. We deleted several DSB repair genes using CRISPR–Cas9 in U2OS cells containing a single copy of the CDDR reporter located in chromosome 19 at q13.31 (U2OS-CDDR-Clone-1) (Supplementary Figure S10A). We first examined genes involved in the repair of DSBs via the canonical branch of NHEJ, including LIG4 (encoding DNA Ligase IV), XRCC4, and XLF. These genes encode core components of the cNHEJ pathway that mediate the ligation of minimally processed DSB ends (2,3). The deletion of these genes in multiple clones was achieved by transiently transfecting U2OS-CDDR-Clone-1 cells with plasmids expressing Cas9 and two sgRNAs targeting early exons of each gene, and was confirmed by genotyping and immunoblotting (Figure 5A and Supplementary Table S1).
Figure 5.
The cNHEJ proteins DNA Ligase IV, XRCC4, and XLF suppress mutagenic NHEJ and deletion mutagenesis in the repair of DSBs. (A) Immunoblots of protein lysates from independent clones of U2OS-CDDR-Clone-1 cells deleted of LIG4, XRCC4 or XLF. (B) Percentage of cells with single DSBs repaired via Mut-NHEJ (% mCherry-ve cells) in the indicated cell lines. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; *P< 0.05, **P< 0.01. (C) Frequency of deep sequencing reads categorized as insertions or deletions among total indels at repair junctions following the induction of single DSBs within the CDDR reporter. Statistical significance was assessed using chi-squared tests with Yates' continuity correction. Bonferroni correction and an additional stringency of >1.5-fold change was applied to all comparisons; P< 0.0001 and >1.5-fold change: *insertions. (D) Size distribution among deletions at repair junctions following the induction of single DSBs within the CDDR reporter. Statistical significance was assessed using chi-squared tests with Yates' continuity correction and Bonferroni correction; P< 0.0001 and >1.5-fold change: *1–10 del, †11–20 del, ‡21–30 del, §>30 del. (E) Frequency of insertions of either 1–2 nt or ≥3 nt at repair junctions following the induction of single DSBs within the CDDR reporter. Statistical significance was assessed using chi-squared tests with Yates' continuity correction and Bonferroni correction; P< 0.0001 and >1.5-fold change: *1–2 nt ins, †≥ 3 nt ins.
We transfected control U2OS-CDDR-Clone-1 or independent clonal derivatives of these cells (sg-LIG4, sg-XRCC4, sg-XLF) with the Cas9–sgRNA-A plasmid, and analyzed the repair of DSBs by flow cytometry. From this analysis, we found that deletion of LIG4, XRCC4 or XLF significantly increased the percentage of mCherry-ve cells (Figure 5B). Although sequencing analysis indicates that the majority of repair junctions contained indels in both the control U2OS-CDDR-Clone-1 cells and in the cNHEJ-deficient cell lines as expected, we observed a significant increase in deletions (particularly deletions > 30 nt), with a corresponding decrease in the frequency of insertions (from 14% to <5%) in the cNHEJ-deficient cell lines, which is indicative of elevated repair by mutagenic alt-EJ (Figure 5C and D). Furthermore, although the total frequency of insertions was reduced in the c-NHEJ-deficient cells, the fraction of insertions ≥3 nt in this cell population increased significantly relative to control parental cells (Figure 5E), consistent with previous reports (43). These findings indicate that DNA Ligase IV, XRCC4 and XLF are not only necessary for suppressing deletion mutagenesis, but also, and perhaps surprisingly, for promoting small templated insertion mutagenesis. Interestingly, the indels observed in cells deficient of XRCC4 and XLF, but not of LIG4, were enriched for in-frame indels (Supplementary Figure S10B). The majority of these were 12 bp or larger (Supplementary Figure S10C), which explains the increase in mCherry-negativity in these cells (Figure 5B). Although we did not observe an increase of in-frame indels in the LIG4-deficient cells, the deletions in these cells were nonetheless significantly larger than those observed in control parental cells (Figure 5D and Supplementary Figure S10B and C). Because these larger deletions are also enriched for microhomology use (data not shown), we conclude that even in the context of reiterative cutting at the proximal junctions within the CDDR reporter, a reduction in the mCherry+ cell population in cells deficient in some DNA repair factors (as is the case here for LIG4, XRCC4 and XLF) may be used as a quantitative measure for the shunting of repair from c-NHEJ to alt-EJ.
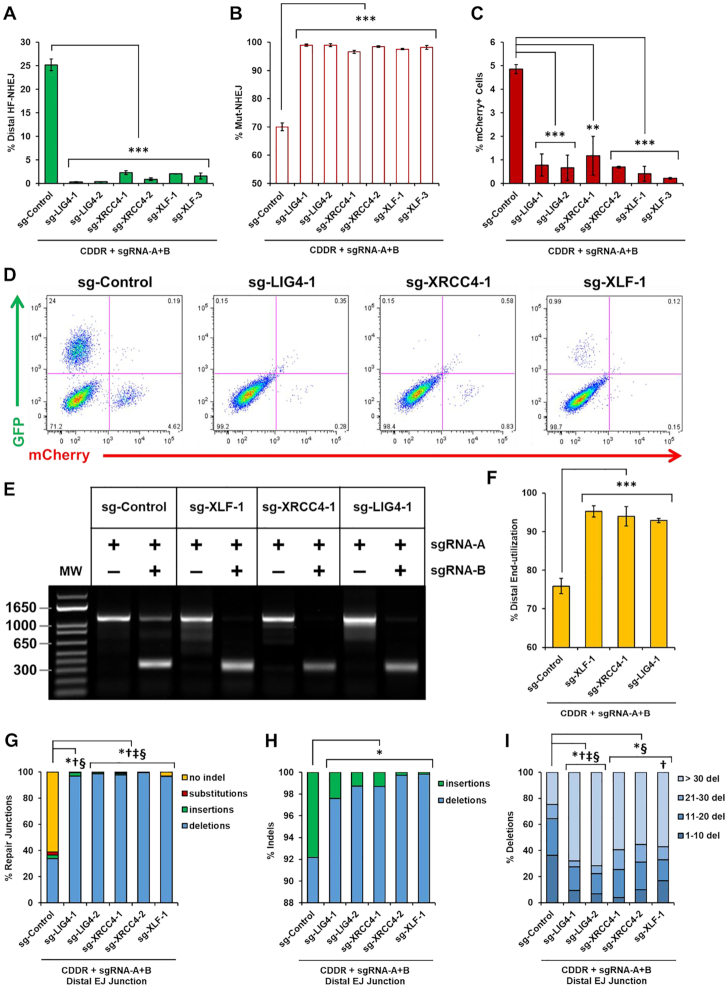
We next analyzed the impact of LIG4, XRCC4 and XLF on the repair of two distal DSBs following the expression of Cas9 and sgRNA-A+B. Along with an increase in proximal Mut-NHEJ, the deletion of any of these genes abolished distal HF-NHEJ, with nearly all of the cells losing mCherry expression (GFP-ve/mCherry-ve) (Figure 6A–D). Notably, distal HF-NHEJ was not affected in sg-XLF-2 cells that retained residual XLF expression compared to the parental U2OS-CDDR-Clone-1 cells (Figure 5A and data not shown), highlighting the necessity of complete genetic ablation, at least for some genes, to detect an impact on the repair of DSBs. This also suggests that partial depletion of some repair proteins (e.g. by siRNA or shRNA silencing) may be insufficient for revealing their role in DSB repair. The lack of GFP production in cells deleted of LIG4, XRCC4 or XLF could result from increased mutagenic repair, and/or a decrease in distal end-utilization. If proximal end-joining of each DSB is favored over the ligation of the two distal DSBs, this would inevitably lead to a reduction in GFP signal. To investigate this possibility, we determined the relative amount of distal end-utilization by PCR amplification of the repair junctions and measured the band intensity of the PCR products representing either proximal (slow-migrating band) or distal (fast-migrating band) end-joining (Supplementary Figure S4C). This analysis showed an increase in distal end-utilization from ∼75% in control cells to ∼95% in cells deficient of LIG4, XRCC4 and XLF (Figure 6E and F). Therefore, the absence of GFP signal in these cells is not due to reduced end-utilization, but reflects enhanced mutagenic repair at the distal EJ junctions. These results were corroborated by deep sequencing analysis of the distal EJ junctions, which showed significantly elevated mutagenic repair, with indels increasing from ∼40% to nearly 100% in cells deleted of LIG4, XRCC4 or XLF (Figure 6G). These indels, similar to those detected at proximal EJ junctions, were enriched for larger deletions (>30 nt) with a significant reduction of insertions (Figure 6H and I), and with increased microhomology use (data not shown). Likewise, these repair products were enriched for larger in-frame indels (≥12 bp) (Supplementary Figure S10D and E). When the two DSBs were repaired independently (generating PCR amplicons corresponding to the slower migrating bands), we observed a similar increase in larger deletions, and reduction of insertions at both of the proximal EJ sites (Supplementary Figure S10F and G). Even at the sgRNA-B target site, where insertions are far more prevalent, the percentage of insertions decreased from 75% to <22% in cNHEJ-deficient cells (Supplementary Figure S10G). Collectively, our findings show that DNA Ligase IV, XRCC4 and XLF are essential for the fidelity of NHEJ repair, promote insertion mutagenesis, and suppress the end-joining of distal DSBs. This also supports the role of these proteins in the stabilization of proximal DSB ends through end-bridging during short-range synapsis (44–46).
Figure 6.
DNA Ligase IV, XRCC4, and XLF are essential for high-fidelity NHEJ and suppress distal end-utilization. (A, B) Percentage of cells with DSBs repaired via distal HF-NHEJ (% GFP+ cells; A), or Mut-NHEJ (% mCherry-ve/GFP-ve cells; B) following the induction of two DSBs within the CDDR reporter in U2OS-CDDR-Clone-1 cells and clonal derivatives deleted of LIG4, XRCC4 or XLF. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; ***P< 0.001. (C) Percentage of mCherry+ cells following the induction of two DSBs within the CDDR reporter in U2OS-CDDR-Clone-1 cells and clonal derivatives deleted of LIG4, XRCC4 or XLF. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; **P< 0.01, ***P< 0.001. (D) Representative FACS profiles of U2OS-CDDR-Clone-1 cells and clonal derivates deleted of LIG4, XRCC4 or XLF, following the induction of two DSBs within the CDDR reporter and subsequent repair. (E) Representative agarose gel displaying PCR amplification of the CDDR reporter, using primers flanking the mCherry cDNA, in U2OS-CDDR-Clone-1 cells and in clonal derivatives deleted of XLF, XRCC4 and LIG4, following transfection with the Cas9–sgRNA-A or Cas9–sgRNA-A+B plasmids. (F) Frequency of distal end-utilization, calculated by dividing the band image intensity of the distal EJ amplicon (lower band) by the sum of the band image intensity of the proximal and distal EJ amplicons in the cells transfected with Cas9–sgRNA-A+B. Band image intensity was quantified using Image Lab software (Bio-Rad). Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; ***P< 0.001. (G) Frequency of deep sequencing reads categorized as insertions, deletions, substitutions, or no indel at distal EJ junctions following the induction of two DSBs within the CDDR reporter in the indicated cell lines. Statistical significance was assessed using chi-squared tests with Yates' continuity correction and Bonferroni correction; P< 0.0001 and >1.5-fold change: *no indel, †substitutions, ‡insertions, §deletions. (H) Frequency of deep sequencing reads categorized as insertions or deletions among total indels at distal EJ junctions following the induction of two DSBs within the CDDR reporter in the indicated cell lines. Statistical significance was assessed using chi-squared tests with Yates' continuity correction. Bonferroni correction and an additional stringency of >1.5-fold change was applied to all comparisons; P< 0.0001 and >1.5-fold change: *insertions. (I) Size distribution among deletions at distal EJ junctions following the induction of two DSBs within the CDDR reporter in the indicated cell lines. Statistical significance was assessed using chi-squared tests with Yates' continuity correction and Bonferroni correction; P<0.0001 and >1.5-fold change: *1–10 del, †11–20 del, ‡21–30 del, §>30 del.
Loss of ATM promotes HF-NHEJ and distal end-utilization
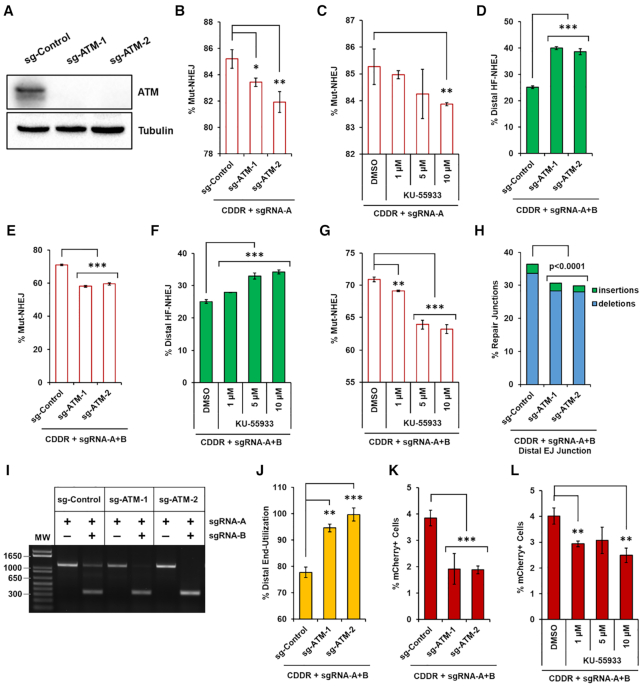
We next sought to validate our reporter system in the context of ATM deletion, because this protein, unlike DNA Ligase IV, XRCC4 and XLF, promotes resection-dependent repair (47,48). ATM is a multifunctional kinase that phosphorylates hundreds of substrates to coordinate the cellular response to DNA damage (47,49). At DSBs, ATM phosphorylates all members of the MRE11-RAD50-NBS1 (MRN) complex to initiate end-resection, generating 3′ single-stranded DNA (ssDNA) overhangs (48). End-resection is an important determinant of repair pathway choice and when resection is blocked, repair via cNHEJ is favored (1,2). To investigate the contribution of ATM activity to NHEJ repair using the CDDR reporter system, we deleted ATM in isogenic U2OS-CDDR-Clone-1 cells through transient transfection of Cas9 and two sgRNAs targeting exons 2 and 3 of ATM, and verified its deletion in individual clones (Figure 7A). In cells encountering single DSBs within the CDDR reporter, we found that the deletion of ATM led to a statistically significant reduction in the percentage of mCherry-ve cells after DSB repair (Figure 7B). Likewise, pharmacological inhibition of ATM activity with the ATM-specific inhibitor KU-55933 (50) also led to a reduction in mCherry-ve cells (Figure 7C). This apparent reduction in mCherry negativity in ATM-deficient cells is indicative of a shift in repair from resection-dependent repair (mutagenic alt-EJ) to cNHEJ, because indels in the ATM-deficient cells were enriched for smaller deletions (Supplementary Figure S11A), and a greater percentage of smaller in-frame deletions (Supplementary Figure S11B and C).
Figure 7.
Loss of ATM promotes high-fidelity NHEJ and distal end-utilization. (A) Immunoblots of protein lysates from U2OS-CDDR-Clone-1 cells and clonal derivatives deleted of ATM. (B) Percentage of cells with single DSBs repaired via Mut-NHEJ (% mCherry-ve cells) following DSB induction in the indicated cell lines. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; *P<0.05, **P< 0.01. (C) Percentage of cells with single DSBs repaired via Mut-NHEJ (% mCherry-ve cells) following DSB induction in U2OS-CDDR-Clone-1 cells treated with DMSO or the indicated doses of ATM-specific inhibitor KU-55933. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; **P< 0.01. (D, E) Percentage of cells with DSBs repaired via distal HF-NHEJ (% GFP+ cells; D), or Mut-NHEJ (% mCherry-ve/GFP-ve cells; E) following the induction of two DSBs within the CDDR reporter in U2OS-CDDR-Clone-1 cells and clonal derivatives deleted of ATM. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; ***P< 0.001. (F, G) Percentage of cells with DSBs repaired via distal HF-NHEJ (% GFP+ cells; F) or Mut-NHEJ (% mCherry-ve/GFP-ve cells; G) following the induction of two DSBs within the CDDR reporter in U2OS-CDDR-Clone-1 cells treated with DMSO or the indicated doses of ATM-specific inhibitor KU-55933. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; **P< 0.01, ***P< 0.001. (H) Frequency of deep sequencing reads categorized as insertions or deletions at distal EJ junctions following the induction of two DSBs within the CDDR reporter in U2OS-CDDR-Clone-1 cells and clonal derivatives deleted of ATM. Statistical significance was assessed using chi-squared tests with Yates' continuity correction and Bonferroni correction. (I) Representative agarose gel displaying PCR amplification of the CDDR reporter in U2OS-CDDR-Clone-1 cells and clonal derivatives deleted of ATM and transfected with Cas9–sgRNA-A or Cas9–sgRNA-A+B. (J) Frequency of distal end-utilization following the induction of two DSBs within the CDDR reporter in the indicated cell lines. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; **P< 0.01, ***P< 0.001. (K) Percentage of mCherry+ cells following the induction of two DSBs within the CDDR reporter in U2OS-CDDR-Clone-1 cells and clonal derivatives deleted of ATM. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; ***P< 0.001. (L) Percentage of mCherry+ cells following the induction of two DSBs within the CDDR reporter in U2OS-CDDR-Clone-1 cells treated with DMSO or the indicated doses of ATM-specific inhibitor KU-55933. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; **P< 0.01.
Following the induction of two DSBs within the CDDR reporter, we observed a significant increase in distal HF-NHEJ in cells deleted of ATM (Figure 7D and E). Similar results were found in U2OS-CDDR-Clone-1 cells treated with KU-55933 (Figure 7F and G). The stimulatory effect of ATM inhibition on distal HF-NHEJ was also observed in 29T-CDDR-Clone-1 cells (Supplementary Figure S12A and B), and in U2OS cells containing multiple copies of the CDDR reporter (Supplementary Figure S12C and D). Sequencing analysis of the distal repair junctions in U2OS-CDDR-Clone-1 cells deleted of ATM showed a small but reproducible reduction in indel frequency (Figure 7H), potentially reflecting enhanced distal end-utilization (distal NHEJ) at the expense of proximal ligation in ATM-deficient cells (51). Indeed, PCR amplification of the distal and proximal EJ junctions revealed that the deletion of ATM promotes distal end-utilization (Figure 7I and J). Thus, the increase in distal HF-NHEJ over proximal HF-NHEJ observed in ATM−/− cells is also a consequence of enhanced distal end-utilization. Thus, it was not surprising that in ATM-deficient cells we observed reduced mCherry+ cells when two DSBs were introduced within the reporter (Figure 7K and L). When analyzing the size of indels at the distal repair junctions, we did not observe a consistent increase in smaller deletions (Supplementary Figure S11D) or smaller in-frame indels among the ATM-deficient cells (Supplementary Figures S11E and F), presumably due to the increase in HF-distal end-joining observed in these cells.
DNA Ligase IV, XRCC4 and XLF suppress HDR, whereas ATM promotes HDR
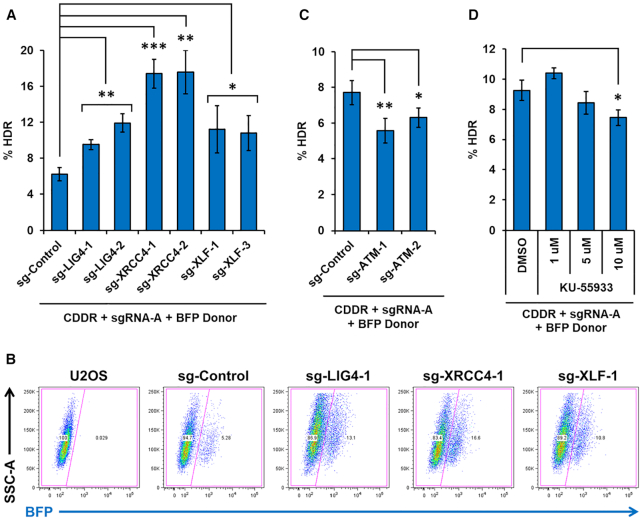
To validate the ability of the CDDR reporter assay system to determine the impact of various repair proteins on HDR activity, we examined the role of LIG4, XRCC4 and XLF on the repair of DSBs in the presence of a BFP homology donor plasmid. Numerous studies have shown that genetic deficiency or inhibition of DNA ligase IV increases HDR activity (52–59). Consistent with these reports, we observed significantly elevated HDR activity in LIG4−/− cells (Figure 8A and B). We observed a similar increase in HDR activity in cells deleted of XRCC4 or XLF (Figure 8A and B). Intriguingly, we found that although the loss of any of these proteins completely abolished HF-NHEJ, the loss of XRCC4 exerted the most stimulatory impact on HDR (∼3-fold increase). This suggests that XRCC4 may play a role in suppressing HDR beyond stimulating LIG4 activity. Collectively, our findings indicate that LIG4, XRCC4 and XLF all play an active role in suppressing HDR, which likely involves the stabilization of the DSB and/or suppression of hyper-resection by these factors.
Figure 8.
DNA Ligase IV, XRCC4, and XLF suppress HDR, whereas ATM promotes HDR. (A) Percentage of cells with single DSBs repaired via HDR (% BFP+ cells) following DSB induction and subsequent repair in the presence of a BFP cDNA donor template in U2OS-CDDR-Clone-1 cells and clonal derivatives deleted of LIG4, XRCC4, or XLF. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; *P< 0.05, **P< 0.01, ***P< 0.001. (B) Representative FACS profiles of U2OS cells, U2OS-CDDR-Clone-1 cells, and clonal derivates of the latter deleted of LIG4, XRCC4, or XLF, following the induction of single DSBs and subsequent repair in the presence of a BFP cDNA donor template. (C) Percentage of cells with single DSBs repaired via HDR (% BFP+ cells) in U2OS-CDDR-Clone-1 cells and clonal derivatives deleted of ATM. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; *P< 0.05, **P< 0.01. (D) Percentage of cells with single DSBs repaired via HDR (% BFP+ cells) in U2OS-CDDR-Clone-1 cells treated with DMSO or increasing doses of ATM-specific inhibitor KU-55933. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; *P< 0.05.
Next, we sought to examine the role of ATM on the repair of DSBs by HDR. Beyond its role in promoting end-resection via MRN phosphorylation, ATM also stimulates HDR through the phosphorylation of CtIP and BRCA1 (60–62). However, HDR can occur in the absence of ATM, suggesting that ATM may not be essential for HDR activity (47). Using the CDDR system, we found that deletion of ATM suppressed HDR, but only subtly (Figure 8C). Likewise, pharmacological inhibition of ATM activity in U2OS-CDDR-Clone-1 cells led to a slight reduction in HDR (Figure 8D). These findings are consistent with the notion that although ATM supports HDR, it is clearly not essential for this repair activity (1).
DISCUSSION
Elucidating the mechanisms and pathways regulating DSB repair increases our understanding of cancer etiology and helps identify novel molecular targets for cancer therapeutics. This can be achieved through the detection and quantification of repair activity at DSBs with high precision. In this study, we have developed a new DSB repair assay system based on the introduction and subsequent resolution of one or two Cas9-mediated DSB(s) in a chromosomally-integrated fluorescent reporter. Quantitative flow cytometry reports on the repair of these DSBs by NHEJ or by HDR with high accuracy and reproducibility, and without the need for cell sorting or normalization for transfection efficiency. Deep sequencing analysis of the repair junctions corroborate the results obtained by flow cytometry and provide comprehensive analyses of the repair products generated by the various repair pathways. Importantly, the CDDR system is a multifunctional assay that reports on high-fidelity and mutagenic repair via the NHEJ pathway, and can further discriminate between proximal and distal NHEJ using a single reporter cassette. In addition, CDDR can be used in the presence of a BFP donor plasmid to detect HDR with greater sensitivity than previously developed HDR assays (10,13,14,16,23–25).
Although Cas9 primarily induces blunt-ended DSBs, many pathological DSBs are incompatible and require additional end-processing to generate ligatable ends. We thus recognize that Cas9-based reporter systems, including CDDR, are limited in the study of proteins involved in end-resection and end-processing during cNHEJ, such as the nuclease Artemis or the DNA polymerases Pol μ and Pol λ. Because blunt ends are preferentially repaired without end-resection, and are more likely to be precisely ligated than incompatible ends, it is not altogether surprising that the accurate end-joining of distal DSBs is robust, as demonstrated in the present study and in previous reports (21,22,63). By contrast, I-SceI-induced DSBs leave behind sticky 3′-overhangs of 4 bp in length (64), and I-SceI-based reporters designed with two I-SceI sites in opposite orientation produce non-complementary overhangs that require end-processing for ligation (3,26). This is an important distinction between reporters using Cas9 and oppositely oriented I-SceI target sites, and limits the comparison of results obtained using different endonuclease reporter systems. However, previous studies have demonstrated comparable levels of accurate NHEJ between two distal DSBs using Cas9 and same-oriented I-SceI target sites, at 70% and 80% respectively (63). This indicates that distal HF-NHEJ is robust when ends are readily ligatable, irrespective of the endonuclease used to produce these breaks.
Similar to other DSB repair assays, the CDDR system relies on the activity of endonucleases for inducing DSBs. This poses a challenge when analyzing DSB repair activity in these contexts, as these endonucleases may create physical barriers that interfere with the normal processing of spontaneously generated DSBs or DSBs induced through non-enzymatic activity such as ionizing radiation. Consistent with this notion, Clarke et al. has recently demonstrated that Cas9 remains stably bound to these breaks before it is displaced by repair factors (65). One such factor is RNA polymerase II (Pol II), which has the capacity to displace the Cas9 protein in an orientation-dependent fashion (65). This suggests that Cas9-induced DSBs encountered at transcriptionally active sites may be processed differently than those in inter-genic or transcriptionally silent chromosomal regions. The inability of KU proteins to displace Cas9 from DSBs further suggests that certain repair proteins may not have easy access to repair these lesions and may require additional proteins for repairing Cas9-inducible DSBs, and therefore bias against repair via the cNHEJ pathway (65). Similar complications may exist for the repair of DSBs induced by other nucleases (e.g. I-SceI or HO endonuclease) and caution is warranted when analyzing repair outcomes in the context of these ‘artificial’ breaks. Thus, while the CDDR and other DSB repair assays may be very useful for the rapid identification and quantification of repair outcomes following the perturbations of certain repair factors, it is important that these factors be analyzed in the context of DSBs that are induced in the absence of these potential physical barriers.
Several of the currently available NHEJ reporter assays exhibit significant bias toward mutagenic repair at single DSBs because the faithful repair of these breaks regenerates the endonuclease recognition site, rendering it susceptible to multiple cleavage-repair cycles (15,17,26,31). This bias is also manifested in the CDDR assay system when single DBSs are introduced within the reporter. Future efforts will focus on alleviating this limitation possibly through establishing temporal control over Cas9 activity. This may require the conditional expression of an active but degradable form of Cas9 that may permit investigating DSB repair with limited cleavage-repair cycles (66–69). The ability to control the activation and degradation of Cas9 would also enable the generation of a stable lines containing both the CDDR reporter cassette, and the Cas9/sgRNA construct, without the need for transfection and subsequent antibiotic selection, thus permitting faster analysis of repair activity.
An important advantage of the CDDR repair assay system is its ability to overcome the limitation of reiterative break-repair cycles through the induction of two distal DSBs within the reporter. The repair of these breaks eliminates sites recognizable by Cas9, and thus reports on single cleavage-repair events. Furthermore, the use of a 2-cut system allows for simultaneous monitoring of end-joining at proximal and distal DSBs, with important applications for studying chromosomal rearrangements caused by incorrect end-joining between distal DSBs. The results reported herein indicate that the repair of blunt-ended Cas9-induced DSBs via distal NHEJ is largely error-free, with ∼70% of cells rejoining the distal ends without any modification. The remaining 30% of cells repair DSBs via Mut-NHEJ, resulting in small indels that vary in their distribution (deletions vs. insertions) based on the sequence flanking the DSB site, as well as the genomic context within which these sequences are found. Our findings are consistent with previous studies supporting the intrinsic precision of NHEJ (21,22,63,70), particularly when ends require minimal processing and are readily ligatable. It would be interesting to test whether similar repair outcomes are obtained for DSBs with compatible staggered ends; for example, by replacing the Cas9 endonuclease in our assays with the Cpf1 endonuclease, which induces DSBs with staggered ends (71).
Bhargava et al. recently reported 54% GFP positivity (normalized to transfection efficiency), reflecting no-indel distal-NHEJ in U2OS cells following the repair of two Cas9-induced DSBs separated by a 46 nt spacer within an intrachromosomal split GFP reporter (EJ7-GFP) (22). Our results however, show that although the rate of distal NHEJ is robust, it generally does not exceed 25% in U2OS-CDDR cells and 40% in 293T-CDDR cells. The higher frequency of basal HF-distal EJ reported by Bhargava et al. could result from: (a) enhanced distal HF-NHEJ at the FRT site harboring the EJ7-GFP reporter; (b) enhanced distal ligation at DSBs that are separated by shorter distances; and/or (c) the presence of more than a single copy of the EJ7-GFP reporter in these cells. Consistent with the latter possibility, we found that increasing the copy number of the CDDR reporter in U2OS resulted in 40–62% GFP positivity.
Through the use of isogenic cell lines with deletions of key DSB repair proteins, we found that HF-NHEJ activity is strictly dependent on the core cNHEJ factors DNA Ligase IV, XRCC4 and XLF. In their absence, the repair of DSBs is shunted towards resection-dependent repair pathways, such as HDR and alt-EJ. Alt-EJ functions in the absence of cNHEJ proteins and utilizes short stretches of sequence homology to repair broken ends, resulting in larger deletions (72). Accordingly, we observed a significant increase in larger deletions, particularly deletions > 30 nt, at both proximal and distal EJ repair junctions in c-NHEJ-deficient cells, as well as an increase in microhomology use at these deletion junctions (data not shown), implicating elevated repair via MMEJ. Likewise, repair of single DSBs in the presence of a BFP donor plasmid led to a significant increase in HDR activity in these cells. Unlike XRCC4, XLF or LIG4, however, the loss of ATM stimulated HF-NHEJ and suppressed HDR, indicating shunting from resection-dependent repair towards cNHEJ. The impact of deleting or inhibiting the activity of various repair factors presented in this study has been summarized in Supplementary Figure S13.
Interestingly, cells lacking DNA Ligase IV, XRCC4 or XLF not only lost HF-NHEJ, but also exhibited enhanced distal end-utilization at the expense of proximal end-utilization. This implies these proteins are not only important for rejoining DSB ends with high-fidelity, but also for stabilizing the DSB junctions and guarding against the loss of genetic material between two nearby DSBs. Consistent with this notion, previous studies have reported an increase in chromosomal translocations in mammalian cells deleted of XRCC4 or DNA Ligase IV (73–75).
The stabilization of broken ends at DSBs is not limited to the cNHEJ factors described above, but also requires ATM, as deletion of ATM was also found to enhance distal end-utilization over proximal end-utilization. The increase in distal-end utilization observed in cells deleted of ATM, as well as cells deleted of DNA Ligase IV, XRCC4, or XLF, could be due to increased break persistence, and/or decreased stability of the proximal ends (i.e. defective end-tethering) (51). The longer a break persists, the greater the likelihood of both DSBs occurring simultaneously, leading to enhanced distal end-joining. This model is consistent with previous reports showing DSB persistence in cells deficient of ATM and DNA Ligase IV (76–79). Another potential mechanism by which cNHEJ factors and ATM could limit distal end-utilization is through faithful end-tethering of DSBs during repair (51). Deficiency of these repair factors could destabilize the broken ends, leading to a loss of genetic material between the DSBs. Given that ATM phosphorylates hundreds of substrates in the DNA damage response, it is conceivable that some of these substrates are important for DSB end-stabilization and support faithful end-tethering during repair (47,51). Overall, these findings support the notion that multiple factors from various molecular pathways stabilize the broken ends of DSBs to maintain genomic integrity (21,22,44–46,73–75).
Collectively, the findings reported in this study highlight the utility of CDDR reporter system in measuring DSB repair outcomes in mammalian cells. Because these assays do not require normalization steps, and are insensitive to variability in transfection efficiency, they greatly facilitate the analysis of various proteins on their contribution to the repair of DSBs. When combined with genetic or pharmacological manipulation of various repair factors, our findings reveal distinct new features for a subset of DSB repair proteins that may be critical for the predisposition of cells deficient in these repair factors to genomic instability, cancer development, and sensitivity to DSB-inducing anticancer therapies. Furthermore, the CDDR reporter system can be readily applied to study the manipulation of experimental variables to enhance targeted genome editing using the CRISPR–Cas9 system, with applications for improving precise gene deletion or HDR-mediated incorporation of exogenous DNA fragments.
DATA AVAILABILITY
The high-throughput sequencing (HTS) raw data associated with this manuscript were deposited at the NCBI Sequence Read Archive (SRA) database (PRJNA628028). Plasmids constructed in this study are available through Addgene with the following ID numbers: lenti-CDDR (149355), Cas9-sgRNA-A (149369), Cas9-sgRNA-A+B (149370), BFP Homology Donor (149372). Additional materials reported in this study are available from the corresponding author upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the University of Virginia Genome Analysis & Technology Core (GATC) and Flow Cytometry Core for providing technical assistance.
Contributor Information
Rebeka Eki, Department of Radiation Oncology, University of Virginia, Charlottesville, VA 22908, USA; Department of Biochemistry and Molecular Genetics, University of Virginia, Charlottesville, VA 22908, USA; Center for Cell Signaling, University of Virginia, Charlottesville, VA 22908, USA.
Jane She, Department of Radiation Oncology, University of Virginia, Charlottesville, VA 22908, USA.
Mahmut Parlak, Department of Radiation Oncology, University of Virginia, Charlottesville, VA 22908, USA.
Mouadh Benamar, Department of Radiation Oncology, University of Virginia, Charlottesville, VA 22908, USA; Department of Biochemistry and Molecular Genetics, University of Virginia, Charlottesville, VA 22908, USA; Center for Cell Signaling, University of Virginia, Charlottesville, VA 22908, USA.
Kang-Ping Du, Department of Radiation Oncology, University of Virginia, Charlottesville, VA 22908, USA.
Pankaj Kumar, Department of Biochemistry and Molecular Genetics, University of Virginia, Charlottesville, VA 22908, USA.
Tarek Abbas, Department of Radiation Oncology, University of Virginia, Charlottesville, VA 22908, USA; Department of Biochemistry and Molecular Genetics, University of Virginia, Charlottesville, VA 22908, USA; Center for Cell Signaling, University of Virginia, Charlottesville, VA 22908, USA; Cancer Center, University of Virginia, Charlottesville, VA 22908, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of General Medical Sciences [R01GM135376 to T.A.]; National Institutes of Health [T32 CA009109 to R.E.]. Funding for open access charge: National Institute of General Medical Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ceccaldi R., Rondinelli B., D’andrea A.D.. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016; 26:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scully R., Panday A., Elango R., Willis N.A.. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019; 20:698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R.. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017; 18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Verkaik N.S., Esveldt-van Lange R.E.E., van Heemst D., Brüggenwirth H.T., Hoeijmakers J.H.J., Zdzienicka M.Z., van Gent D.C.. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol. 2002; 32:701–709. [DOI] [PubMed] [Google Scholar]
- 5. Pastwa E., Somiari R.I., Malinowski M., Somiari S.B., Winters T.A.. In vitro non-homologous DNA end joining assays-The 20th anniversary. Int. J. Biochem. Cell Biol. 2009; 41:1254–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mansour W.Y., Schumacher S., Rosskopf R., Rhein T., Schmidt-Petersen F., Gatzemeier F., Haag F., Borgmann K., Willers H., Dahm-Daphi J.. Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res. 2008; 36:4088–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mansour W.Y., Rhein T., Dahm-Daphi J.. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res. 2010; 38:6065–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mao Z., Seluanov A., Jiang Y., Gorbunova V.. TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:13068–13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mao Z., Bozzella M., Seluanov A., Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst). 2008; 7:1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seluanov A., Mao Z., Gorbunova V.. Analysis of DNA double-strand break (DSB) repair in mammalian cells. J. Vis. Exp. 2010; 8:2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Y., Zhang H., Xu Z., Tang H., Geng A., Cai B., Su T., Shi J., Jiang C., Tian X. et al.. A PARP1-BRG1-SIRT1 axis promotes HR repair by reducing nucleosome density at DNA damage sites. Nucleic Acids Res. 2019; 47:8563–8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xie A., Kwok A., Scully R.. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 2009; 16:814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coleman K.A., Greenberg R.A.. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J. Biol. Chem. 2011; 286:13669–13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Certo M.T., Ryu B.Y., Annis J.E., Garibov M., Jarjour J., Rawlings D.J., Scharenberg A.M.. Tracking genome engineering outcome at individual DNA breakpoints. Nat. Methods. 2011; 8:671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bindra R.S., Goglia A.G., Jasin M., Powell S.N.. Development of an assay to measure mutagenic non-homologous end-joining repair activity in mammalian cells. Nucleic Acids Res. 2013; 41:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pierce A.J., Johnson R.D., Thompson L.H., Jasin M.. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999; 13:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gomez-Cabello D., Jimeno S., Fernández-Ávila M.J., Huertas P.. New tools to study DNA double-strand break repair pathway choice. PLoS One. 2013; 8:e77206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kuhar R., Gwiazda K.S., Humbert O., Mandt T., Pangallo J., Brault M., Khan I., Maizels N., Rawlings D.J., Scharenberg A.M. et al.. Novel fluorescent genome editing reporters for monitoring DNA repair pathway utilization at endonuclease-induced breaks. Nucleic Acids Res. 2016; 42:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kostyrko K., Mermod N.. Assays for DNA double-strand break repair by microhomology-based end-joining repair mechanisms. Nucleic Acids Res. 2016; 44:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gunn A., Stark J.M.. I-SceI-Based Assays to Examine Distinct Repair Outcomes of Mammalian Chromosomal Double Strand Breaks. 2012; 920:Totowa: Humana Press; 379–391. [DOI] [PubMed] [Google Scholar]
- 21. Bhargava R., Carson C.R., Lee G., Stark J.M.. Contribution of canonical nonhomologous end joining to chromosomal rearrangements is enhanced by ATM kinase deficiency. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhargava R., Sandhu M., Muk S., Lee G., Vaidehi N., Stark J.M.. C-NHEJ without indels is robust and requires synergistic function of distinct XLF domains. Nat. Commun. 2018; 9:2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arnoult N., Correia A., Ma J., Merlo A., Garcia-Gomez S., Maric M., Tognetti M., Benner C.W., Boulton S.J., Saghatelian A. et al.. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature. 2017; 549:548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pierce A.J., Hu P., Han M., Ellis N., Jasin M.. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001; 15:3237–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stark J.M., Pierce A.J., Oh J., Pastink A., Jasin M.. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004; 24:9305–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brandsma I., Gent D.C.. Pathway choice in DNA double strand break repair: observations of a balancing act. Genome Integr. 2012; 3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guirouilh-Barbat J., Huck S., Bertrand P., Pirzio L., Desmaze C., Sabatier L., Lopez B.S.. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004; 14:611–623. [DOI] [PubMed] [Google Scholar]
- 28. Guirouilh-Barbat J., Rass E., Plo I., Bertrand P., Lopez B.S.. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:20902–20907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rass E., Grabarz A., Plo I., Gautier J., Bertrand P., Lopez B.S.. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009; 16:819–824. [DOI] [PubMed] [Google Scholar]
- 30. Schulte-Uentrop L., El-Awady R.A., Schliecker L., Willers H., Dahm-Daphi J.. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res. 2008; 36:2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bennardo N., Gunn A., Cheng A., Hasty P., Stark J.M.. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet. 2009; 5:e1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., Rebar E.J.. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010; 649:247–256. [DOI] [PubMed] [Google Scholar]
- 33. Joung J., Konermann S., Gootenberg J.S., Abudayyeh O.O., Platt R.J., Brigham M.D., Sanjana N.E., Zhang F.. Genome-scale CRISPR–Cas9 knockout and transcriptional activation screening. Nat. Protoc. 2017; 12:828–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10–12. [Google Scholar]
- 35. Clement K., Rees H., Canver M.C., Gehrke J.M., Farouni R., Hsu J.Y., Cole M.A., Liu D.R., Joung J.K., Bauer D.E. et al.. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019; 37:224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu Z., Chen O., Wall J.B.J., Zheng M., Zhou Y., Wang L., Ruth Vaseghi H., Qian L., Liu J.. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 2017; 7:2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bae S., Kweon J., Kim H.S., Kim J.S.. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods. 2014; 11:705–706. [DOI] [PubMed] [Google Scholar]
- 38. van Overbeek M., Capurso D., Carter M.M.M., Thompson M.S.S., Frias E., Russ C., Reece-Hoyes J.S.S., Nye C., Gradia S., Vidal B. et al.. DNA Repair Profiling Reveals Nonrandom Outcomes at Cas9-Mediated Breaks. Mol. Cell. 2016; 63:633–646. [DOI] [PubMed] [Google Scholar]
- 39. Lemos B. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E2040–E2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shou J., Li J., Liu Y., Wu Q.. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol. Cell. 2018; 71:498–509. [DOI] [PubMed] [Google Scholar]
- 41. Yu J.S., Pertusi D.A., Adeniran A.V., Tyo K.E.J.J., Wren J.. CellSort: a support vector machine tool for optimizing fluorescence-activated cell sorting and reducing experimental effort. Bioinformatics. 2017; 33:909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chudakov D.M., Matz M.V., Lukyanov S., Lukyanov K.A.. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 2010; 90:1103–1163. [DOI] [PubMed] [Google Scholar]
- 43. Wyatt D.W., Feng W., Conlin M.P., Yousefzadeh M.J., Roberts S.A., Mieczkowski P., Wood R.D., Gupta G.P., Ramsden D.A.. Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol. Cell. 2016; 63:662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brouwer I., Sitters G., Candelli A., Heerema S.J., Heller I., Melo De A.J., Zhang H., Normanno D., Modesti M., Peterman E.J.G. et al.. Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature. 2016; 535:566–569. [DOI] [PubMed] [Google Scholar]
- 45. Roy S., de Melo A.J., Xu Y., Tadi S.K., Négrel A., Hendrickson E., Modesti M., Meek K.. XRCC4/XLF interaction is variably required for DNA repair and is not required for ligase IV stimulation. Mol. Cell. Biol. 2015; 35:3017–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Graham T.G.W., Walter J.C., Loparo J.J.. Two-stage synapsis of DNA ends during non-homologous end joining. Mol. Cell. 2016; 61:850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blackford A.N., Jackson S.P.. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell. 2017; 66:801–817. [DOI] [PubMed] [Google Scholar]
- 48. Lavin M.F., Kozlov S., Gatei M., Kijas A.W.. ATM-dependent phosphorylation of all three members of the MRN complex: from sensor to adaptor. Biomolecules. 2015; 5:2877–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y. et al.. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007; 316:1160–1166. [DOI] [PubMed] [Google Scholar]
- 50. Hickson I., Zhao Y., Richardson C.J., Green S.J., Martin N.M.B., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C.M.. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004; 64:9152–9159. [DOI] [PubMed] [Google Scholar]
- 51. Bennardo N., Stark J.M.. ATM limits incorrect end utilization during non-homologous end joining of multiple chromosome breaks. PLoS Genet. 2010; 6:e1001194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bozas A., Beumer K.J., Trautman J.K., Carroll D.. Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila. Genetics. 2009; 182:641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chu V.V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R.. Increasing the efficiency of homology-directed repair for CRISPR–Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015; 33:543–548. [DOI] [PubMed] [Google Scholar]
- 54. Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L.. Increasing the efficiency of precise genome editing with CRISPR–Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015; 33:538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pinder J., Salsman J., Dellaire G.. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015; 43:9379–9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Robert F., Barbeau M., Éthier S., Dostie J., Pelletier J.. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med. 2015; 7:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Singh P., Schimenti J.C., Bolcun-Filas E.. A mouse geneticist's practical guide to CRISPR applications. Genetics. 2015; 199:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hu Z., Shi Z., Guo X., Jiang B., Wang G., Luo D., Chen Y., Zhu Y.S.. Ligase IV inhibitor SCR7 enhances gene editing directed by CRISPR–Cas9 and ssODN in human cancer cells. Cell Biosci. 2018; 8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang J.-P.J.P., Li X.L., Li G.H., Chen W., Arakaki C., Botimer G.D., Baylink D., Zhang L., Wen W., Fu Y.W. et al.. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017; 18:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang H., Shi L.Z., Wong C.C.L., Han X., Hwang P.Y.H., Truong L.N., Zhu Q., Shao Z., Chen D.J., Berns M.W. et al.. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013; 9:e1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chanut P., Britton S., Coates J., Jackson S.P., Calsou P.. Coordinated nuclease activities counteract Ku at single-ended DNA double-strand breaks. Nat. Commun. 2016; 7:12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li S., Ting N.S.Y., Zheng L., Chen P.L., Ziv Y., Shiloh Y., Lee E.Y.H.P., Lee W.H.. Functional link of BRCA1 and ataxia telangiectasia gone product in DNA damage response. Nature. 2000; 406:210–215. [DOI] [PubMed] [Google Scholar]
- 63. Guo T., Feng Y.L., Xiao J.J., Liu Q., Sun X.N., Xiang J.F., Kong N., Liu S.C., Chen G.Q., Wang Y. et al.. Harnessing accurate non-homologous end joining for efficient precise deletion in CRISPR/Cas9-mediated genome editing. Genome Biol. 2018; 19:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Niu Y., Tenney K., Li H., Gimble F.S.. Engineering variants of the I-SceI homing endonuclease with strand-specific and site-specific DNA-nicking activity. J. Mol. Biol. 2008; 382:188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Clarke R., Heler R., MacDougall M.S., Yeo N.C., Chavez A., Regan M., Hanakahi L., Church G.M., Marraffini L.A., Merrill B.J.. Enhanced bacterial immunity and mammalian genome editing via RNA-polymerase-mediated dislodging of Cas9 from double-strand DNA breaks. Mol. Cell. 2018; 71:42–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gangopadhyay S.A., Cox K.J., Manna D., Lim D., Maji B., Zhou Q., Choudhary A.. Precision Control of CRISPR–Cas9 Using Small Molecules and Light. Biochemistry. 2019; 58:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kleinjan D.A., Wardrope C., Nga Sou S., Rosser S.J.. Drug-tunable multidimensional synthetic gene control using inducible degron-tagged dCas9 effectors. Nat. Commun. 2017; 8:1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Davis K.M., Pattanayak V., Thompson D.B., Zuris J.A., Liu D.R.. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015; 11:316–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Senturk S., Shirole N.H., Nowak D.G., Corbo V., Pal D., Vaughan A., Tuveson D.A., Trotman L.C., Kinney J.B., Sordella R.. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nat. Commun. 2017; 8:14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bétermier M., Bertrand P., Lopez B.S.. Is non-homologous end-joining really an inherently error-prone process. PLoS Genet. 2014; 10:e1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., Van Der Oost J., Regev A. et al.. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bennardo N., Cheng A., Huang N., Stark J.M.. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008; 4:e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Simsek D., Jasin M.. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat. Struct. Mol. Biol. 2010; 17:410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Soni A., Siemann M., Grabos M., Murmann T., Pantelias G.E., Iliakis G.. Requirement for Parp-1 and DNA Ligases 1 or 3 but not of Xrcc1 in chromosomal translocation formation by backup end joining. Nucleic Acids Res. 2014; 42:6380–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Soni A., Siemann M., Pantelias G.E., Iliakis G.. Marked contribution of alternative end-joining to chromosome-translocation-formation by stochastically induced DNA double-strand-breaks in G2-phase human cells. Mutat. Res. - Genet. Toxicol. Environ. Mutagen. 2015; 793:2–8. [DOI] [PubMed] [Google Scholar]
- 76. Callén E., Jankovic M., Difilippantonio S., Daniel J.A.J.J.A., Chen H.-T.H.T., Celeste A., Pellegrini M., McBride K., Wangsa D., Bredemeyer A.L. et al.. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007; 130:63–75. [DOI] [PubMed] [Google Scholar]
- 77. Callén E., Jankovic M., Wong N., Zha S., Chen H.. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell. 2009; 34:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bredemeyer A.A.L., Sharma G.G.G., Huang C.Y.C.-Y., Helmink B.B.A.B., Walker L.L.M., Khor K.C., Nuskey B., Sullivan K.E., Pandita T.K., Bassing C.H. et al.. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006; 442:466–470. [DOI] [PubMed] [Google Scholar]
- 79. Kuhne M., Riballo E., Rief N., Rothkamm K., Jeggo P.A.P., Kühne M., Riballo E., Rief N., Rothkamm K., Jeggo P.A.P. et al.. A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res. 2004; 64:500–508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The high-throughput sequencing (HTS) raw data associated with this manuscript were deposited at the NCBI Sequence Read Archive (SRA) database (PRJNA628028). Plasmids constructed in this study are available through Addgene with the following ID numbers: lenti-CDDR (149355), Cas9-sgRNA-A (149369), Cas9-sgRNA-A+B (149370), BFP Homology Donor (149372). Additional materials reported in this study are available from the corresponding author upon request.