Abstract
Background
The analysis of single nucleotide polymorphism (SNPs) in drug-resistance associated genes is a commonly used strategy for the surveillance of anti-malarial drug resistance in populations of parasites. The present study was designed and performed to provide genetic epidemiological data of the prevalence of N86Y-Y184F-D1246Y SNPs in Plasmodium falciparum multidrug resistance 1 (pfmdr1) in the malaria hotspot of Northern Nigeria.
Methods
Plasmodium falciparum-positive blood samples on Whatman-3MM filter papers were collected from 750 symptomatic patients from four states (Kano, Kaduna, Yobe and Adamawa) in Northern Nigeria, and genotyped via BigDye (v3.1) terminator cycle sequencing for the presence of three SNPs in pfmdr1. SNPs in pfmdr1 were used to construct NYD, NYY, NFY, NFD, YYY, YYD, YFD and YFY haplotypes, and all data were analysed using Pearson Chi square and Fisher’s exact (FE) tests.
Results
The prevalence of the pfmdr1 86Y allele was highest in Kaduna (12.50%, 2 = 10.50, P = 0.02), whilst the 184F allele was highest in Kano (73.10%, 2 = 13.20, P = 0.00), and the pfmdr1 1246Y allele was highest in Yobe (5.26%, 2 = 9.20, P = 0.03). The NFD haplotype had the highest prevalence of 69.81% in Kano (2 = 36.10, P = 0.00), followed by NYD with a prevalence of 49.00% in Adamawa, then YFD with prevalence of 11.46% in Kaduna. The YYY haplotype was not observed in any of the studied states.
Conclusion
The present study suggests that strains of P. falciparum with reduced sensitivity to the lumefantrine component of AL exist in Northern Nigeria and predominate in the North-West region.
Keywords: Anti-malarial drug resistance, P. falciparum, Single nucleotide polymorphisms, pfmdr1, Haplotypes
Background
Anti-malarial drug resistance is a major impediment to malaria chemotherapy in sub-Saharan Africa [1] largely because Plasmodium falciparum rapidly develops resistance to drugs [2]. Resistance to anti-malarial drugs occurs through drug-selection of spontaneous mutations in P. falciparum that confer tolerance to the drug [3]. The selection and spread of drug resistant P. falciparum is facilitated by the rapid genome replication rate and by a relatively high mutation rate per generation of the parasite [4, 5]. The speed of selection of mutants within parasite populations depends upon the pharmacokinetics of the drug itself and its degree of usage within a given host population [1]. For many anti-malarial drugs, molecular markers of parasite resistance are known. Surveillance of these markers in parasite populations can act as a proxy measure of the efficacy of drugs within that population, and can act as early warning signals of the emergence of resistance into new regions. Frequent and thorough molecular surveys of the prevalence of mutations associated with drug resistance can, therefore, inform regional drug policies.
Single nucleotide polymorphisms (SNPs) in the P. falciparum multidrug resistance gene (pfmdr1) have been shown to modulate the susceptibility of the parasite to the long acting partner drug in Artemisinin-based combination therapy (ACT) [6], but are not associated with resistance to artemisinin. Artemether-lumefantrine (AL) and artesunate-amodiaquine (AS-AQ) are two combinations commonly used in ACT in sub-Saharan Africa to treat uncomplicated malaria, but until now, there is no clear evidence of treatment failure from their use in the region. However, some genetic studies involving pfmdr1 have suggested opposing selective pressures following separate use of the drugs, in which parasites harbouring N86, 184F, D1246 pfmdr1 genotypes predominate in African countries that recommend AL as first line anti-malarial drug, whilst those carrying 86Y, Y184 and 1246Y pfmdr1 genotypes predominate in African countries that use AS-AQ as frontline anti-malarial therapy [7].
African P. falciparum isolates may carry the resistant allele of pfcrt encoding the amino acids CVIET at codons 72–76 as well as a variety of polymorphic pfmdr1 alleles which have originated and spread within the African continent [8–10]. The pfmdr1 gene is a structural homologue of the mammalian multidrug resistance gene encoding a P-glycoprotein homologue-1 (Pgh1) multi-drug resistant transporter [11] and is expressed into a PfMDR1 transporter located in the P. falciparum food vacuole.
Mutations in pfmdr1 are associated with reduced influx of diverse anti-malarial drugs reducing their intracellular accumulation [12, 13]. Single nucleotide polymorphisms (SNPs) in pfmdr1 are associated with resistance to aminoquinolines [14, 15]. Several codons in pfmdr1 have been putatively linked with changes in the parasite’s susceptibility to anti-malarial drugs, but codons N86Y, Y184F and D1246Y are uniquely associated with changes in sensitivity to lumefantrine (LUM) and amodiaquine (AQ) in sub-Saharan Africa [16]. While the pfmdr1 86Y allele was strongly associated with chloroquine (CQ) and amodiaquine (AQ) resistance [17, 18], 1246Y alleles were shown to confer resistance to quinine (QN) and possess the capacity to increase the parasite susceptibility to mefloquine (MQ), halofantrine (HF) and artemisinin (ART) [19, 20]. In the study of Reed et al. [21], the sensitivity of CQS D10 parasites to CQ was not affected by transfection of the parasites with pfmdr1 D1246Y mutation, but reduced by half, due to replacement of the mutation with a wild-type D10 pfmdr1 sequence on a different genetic background of the parasite (CQR 7G8).
The mutant pfmdr1 86Y and 1246Y alleles have also been linked to reduced sensitivity to AQ, whereas the wild-type pfmdr1 N86 and D1246 alleles are linked to reduced susceptibility against LUM [22, 23]. In Africa, the common use of AL and AS-AQ in the treatment of uncomplicated malaria has been linked with the emergence of pfmdr1 N86Y, Y184F and D1246Y SNPs [24], and the prevalence of these mutations are frequently used for evaluating changes in sensitivity to LUM and AQ partners in artemisinin-based combinations [7]. Several studies have shown that parasites carrying a combination of pfmdr1 N86, 184F, and D1246 (the “NFD” haplotype) display decreased susceptibility to AL and that treatment with AL can select for such a haplotype [25, 26].
Nigeria accounts for 25% of global cases of malaria and an estimated 50% of the country’s population suffer at least one episode of malaria every year, while under-five children experience an average of 2 to 4 attacks in a year [27]. Plasmodium falciparum is stably and perennially transmitted in all parts of the country [28], with transmission increased during the wet season compared to the dry [29, 30]. North-West and North–East Nigeria have so far been identified as hotspots of malaria in relation to the southern parts of the country due primarily to climatic and environmental conditions [31]. The North-West region of the country suffers a much higher P. falciparum transmission rate than the other regions including North-East Nigeria [32].
The frontline drug for malaria chemotherapy in the country was chloroquine until 2005 when it was withdrawn as a result of resistance [33]. Subsequently, the artemether-lumefantrine was recommended as the only first-line drug for the treatment of uncomplicated malaria in the regions. Unfortunately, several reports investigating molecular markers of anti-malarial resistance have suggested a massive reduction of parasite susceptibility to LUM component of AL [19, 24, 34, 35]. In Uganda, Dokomajilar et al. [34] showed a high prevalence of pfmdr1 N86, 184F, and D1246 alleles after treatment with AL, where the pattern persists even in patients that presented with clinical failure. A few years after, Mbogo et al. [24] genotyped 982 archived samples collected during 2003–2012 for pfmdr1 polymorphisms and reported a dramatic reduction in pfmdr1 86Y and 1246Y alleles over time. Similarly, the relationship between presence of mutations involving pfmdr1 86, 184 and 1246 codons and success of ultralow-dose mefloquine treatment was investigated in Gabon, where Mawili-Mboumba et al. [35] observed a low prevalence of pfmdr1 N86 allele but the prevalence of 184F and D1246 alleles was above 80% each. In Tanzania, Humphreys et al. [19] observed a high prevalence of pfmdr1 86Y, Y184 and 1246Y in patients who failed treatment with AQ, but observed the opposite in those who failed AL treatment. The prevalence of pfmdr1 polymorphisms in Nigeria was majorly reported from the Southern region, where a positive association between pfmdr1 N86, F184 and D1246 alleles and clinical failure was observed [36]. In contrast, a prevalence of 62.2% and 69.0% for pfmdr1 86Y and F184 allele, respectively, was also reported from the region [37] which was recently followed by another survey where the pfmdr1 86Y and 1246Y alleles had prevalence of 24% and 18.6%, respectively [38]. Yet, there is no valid baseline data involving pfmdr1 SNPs in both North-West and North–East Nigeria since the withdrawal of CQ and adoption of AL in Northern Nigeria. In this study, the distributions of the pfmdr1 N86Y, Y184F and D1246Y SNPs across the North-West and-East Nigeria were investigated.
Methods
Description of study sites
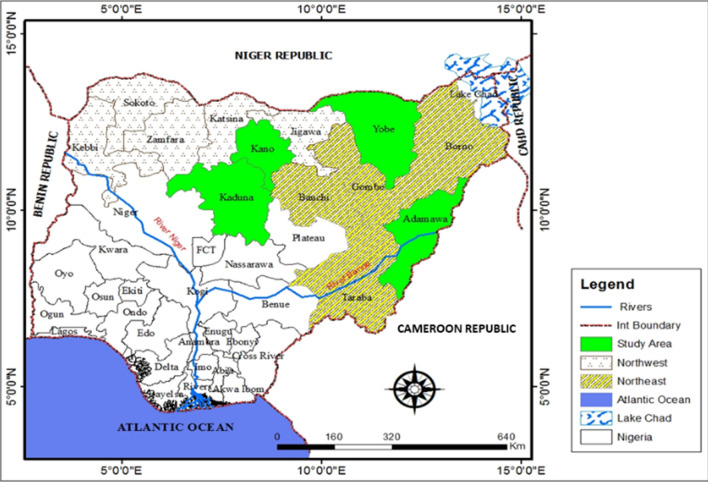
Nigeria’s North-West and North–East are two out of the six geo-political zones of Nigeria. The North-West is made up of seven states and is home to a population of over 35 million people whilst the North-East comprises six states with a population of over 18 million [39]. Two states from the North-West; Kano (longitude 7° 10′ E, 10° 35′ E and latitude 10° 25′ N, 13° 53′ N) and Kaduna (longitudes 7° 23′ E and 7° 29′ E and latitudes 10° 25′ N and 10° 36′ N) with a combined population of 15,450,244 were randomly selected for inclusion in this study while Yobe (longitude 13.5° E and latitude 11° N) and Adamawa (longitude 11° and 14° E and latitude 7° and 11° N) states with a combined population of 5,489,692 were similarly selected from the North-Eastern region [39]. Other relevant details about the study sites are indicated in Fig. 1.
Fig. 1.
A map of Nigeria showing the study sites for the surveillance of pfmdr1 N86Y-Y184F-D1246Y polymorphisms
Selection criteria
In this study, patients who presented with symptoms and confirmed of uncomplicated malaria across all ages, but did not take any anti-malarial drug 2 weeks previously before arrival to the facilities, were included, this is because previous treatment with any of the commonly used AL, QN and sulfadoxine-pyrimethamine (SP) anti-malarials may amplify establishment of drug resistant parasites, whilst those who presented with severe malaria were excluded.
Sample collection
Between June and November 2017, thick and thin film microscopy was used to confirm P. falciparum positivity of malaria symptomatic patients that attended selected health facilities within the study sites. The total number of samples collected from Kano, Kaduna, Adamawa and Yobe were 250, 150, 150 and 200 respectively. 10 µL of microscopically confirmed P. falciparum parasitized blood samples were spotted on four different positions onto Whatman-3MM filter papers and allowed to dry at room temperature. Each sample was placed in sachets containing desiccant, and was preserved in a refrigerator at 4 °C.
Genomic DNA isolation, amplification and genotyping of pfmdr1
Three discs (3 mm/disc) were punched from the P. falciparum-positive dried blood spots and the punch sterilized between each sample. The discs were used to extract genomic DNA using a QIAamp DNA Mini Kit (Qiagen Inc, Japan) according to the manufacturer’s instructions. Nested polymerase chain reaction (PCR) was carried out as described by Humphreys et al. [19] with slight modifications to the PCR cycle programs (Table 1). The nested PCR runs for the amplification of two separate pfmdr1 segments S1 and S2, spanning codons 86-184, and -1246 respectively were performed with a 10 μL final master mixture and 200 ηM primers, and 1U ExTaq polymerase (Takara-Bio, Japan). Two µL of isolated genomic DNA was added to each of the first PCR master mixtures and run in the thermocycler. At the completion of the first PCR runs, 1 μL of each of the resulting amplified fragments were further used as templates in 10 μL of secondary PCR reactions. The PCR reactions were run along with parasite free genomic DNA (negative) and 3D7 clone P. falciparum genomic DNA (positive) controls. Two μL of each PCR product was evaluated by electrophoresis on 1.5% agarose gels that were stained with Midori Green Advance (Bulldog Bio Inc, USA) and visualized under ultraviolet light. The remaining 8 μL nested PCR products were stored at − 30 °C. All nested amplicons were subsequently purified using the one step ExoSAP-IT (ThermoFisher Scientific, Japan) purification kit and the resulting products subjected to BigDye Terminator (v3.1) Cycle Sequencing (ThermoFisher Scientific, Japan). Sequences were analysed using BioEdit Sequence Alignment Editor (v7.0.5.3) while pfmdr1 SNPs were determined using MEGA5 software (Build#:5130611) in reference to the pfmdr1 sequence of P. falciparum deposited at the NCBI database [Accession Number X56851]. Consequently, the samples bearing mixed genotypes were excluded in the calculations of prevalence of pfmdr1 alleles and haplotypes. The prevalence was calculated as shown by the following formula:
Table 1.
Primers for Nested PCR of pfmdr1 Long and Short Fragments and Cycling Programs Adapted and Modified from the Work of Humphreys et al. [19]
| Primers | Amplicon sizes (bp) | PCR cycling programs |
|---|---|---|
|
Outer forward FN1/15_- AGGTTGAAAAAGAGTTGAAC Outer reverse REV/C15_-ATGACACCACAAACATAAAT |
578 |
34 cycles of 94 °C for 30 s; 55 °C for 30 s; and 65 °C for 1 min; then 65 °C for 5 min and 4 °C forever 34 cycles of 94 °C for 30 s; 55 °C for 1 min; and 65 °C for 1.5 min; then 65 °C for 5 min and 4 °C forever |
|
Outer forward MDRFR2F15_-GTGTATTTGCTGTAAGAGCT Outer reverse MDRFR2R15_-GACATATTAAATAACATGGGTTC |
958 | |
|
Nested forward MDR2/15_-ACAAAAAGAGTACCGCTGAAT Nested reverse NEWREV15_-AAACGCAAGTAATACATAAAGTC |
534 |
29 cycles of 94 °C 30 s; 60 °C for 30 s; and 65 °C for 1 min; then 65 °C for 5 min and 4 °C forever 28 cycles of 94 °C for 30 s; 60 °C for 30 s; and 65 °C for 1 min; then 65 °C for 5 min and 4 °C forever |
|
Nested forward MDRFR2F25_ CAGATGATGAAATGTTTAAAGATC Nested reverse MDRFR2R25_-TAAATAACATGGGTTCTTGACT |
864 |
Each PCR run was preceded by an initial denaturation at 94 °C for 3 min
Pfmdr1 N86Y-Y184F-D1246Y haplotypes
The pfmdr1 haplotypes used in this study were based on eight previously reported haplotypes associated with artemether lumefantrine-tolerance; pfmdr1 N86Y, Y184F and D1246Y single nucleotide polymorphisms in different P. falciparum populations from Africa [40, 41].
Statistical analysis
All data were statistically analysed using Pearson Chi square and Fisher’s exact (FE) tests of Graph-Pad Prism (8.1.0) and P values ˂ 0.05 were considered to be statistically significant.
Results
Prevalence of SNPs in pfmdr1 codons 86, 186 and 1246
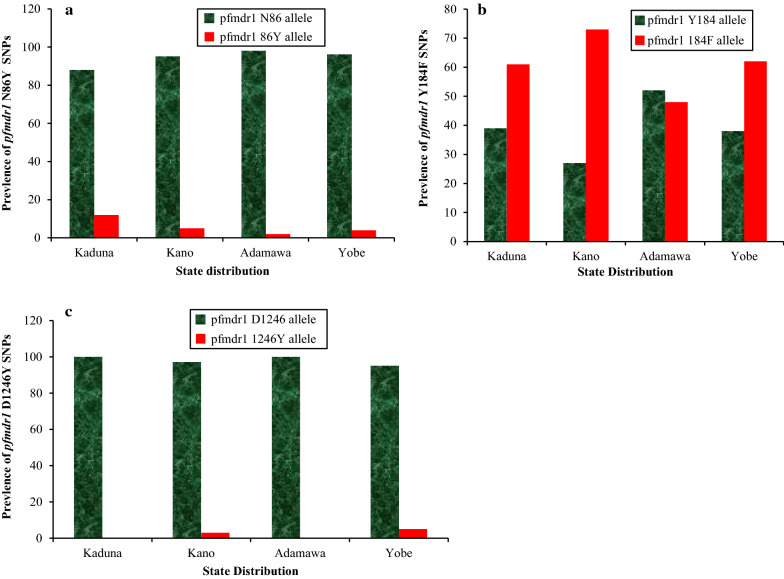
Of the 750 P. falciparum positive samples collected from the four states in Northern Nigeria, 500 were successfully genotyped for the pfmdr1 N86Y, Y184F and D1246Y alleles, possibly due to a very low parasite density in the remaining samples. Six of the genotyped pfmdr1 sequences were deposited in the GenBank with accession numbers MT472640, MT472641, MT472642, MT495456, MT495458, and MT495459 on the basis of presence or absence of mutations in the three codons of the gene. The prevalence of the mutant pfmdr1 86Y allele was observed to be significantly (P = 0.02) different across the states, with highest prevalence of 12.50% obtained in Kaduna state, followed by Kano with 4.68% and 2.00% in Adamawa state (Fig. 2a). The prevalence of the mutant pfmdr1 184F allele was 73.10% in Kano state, 61.46% in Kaduna state and 48.00% in Adamawa state (P = 0.00), Fig. 2b. Yobe state had the highest prevalence of the mutant pfmdr1 1246Y allele (5.26%) while Kaduna and Adamawa states had zero prevalence of this mutation (P = 0.03) (Fig. 2c).
Fig. 2.
Prevalence of pfmdr1 Single Nucleotide Polymorphisms (SNPs) across Four States of Northern Nigeria. The State-wise Distribution of pfmdr1 SNPs at codons 86, 184 and 1246 are shown in A, B and C respectively. Green and red bars in A, B and C (wild and mutant alleles, respectively) are significantly different at codons 86 (2 = 10.50, P = 0.02), 184 (2 = 13.20, P = 0.00) and 1246 (2 = 9.20, P = 0.03) of pfmdr1
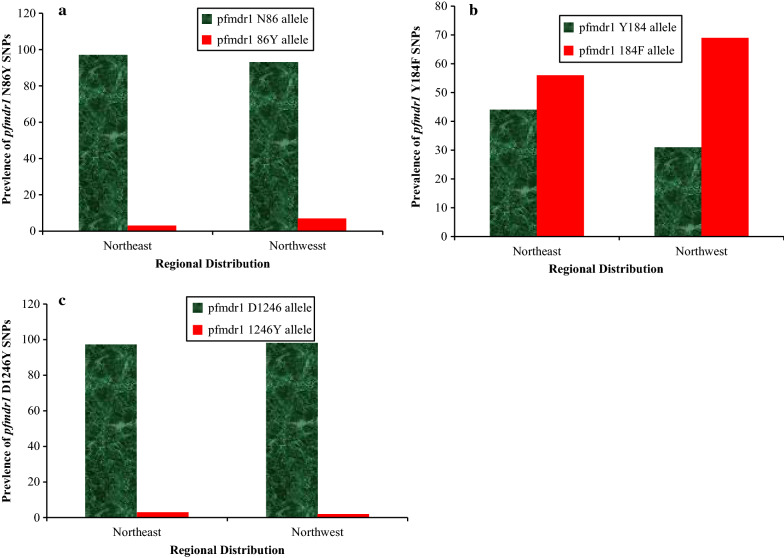
The regional distributions of the mutant pfmdr1 86Y, 184F and 1246Y alleles are shown in Fig. 3. Based on the results, an overall regional prevalence of 7.49% and 3.00% for the pfmdr1 86Y allele was recorded in North-West and North–East Nigeria, respectively. However, the observed difference was not significant (P = 0.19), Fig. 3a. Similarly, the prevalence of the pfmdr1184F and 1246Y mutants were not significantly different between the North-West and North–East Nigeria, whereas the prevalence of the 184F allele differed significantly between these two regions; 68.91% and 56.22%, in the North-West and North-East, respectively (P = 0.05) (Fig. 3b). The prevalence of the pfmdr1 1246Y allele in the North-West and North–East Nigeria was 1.930% and 3.000% respectively (P = 0.65) (Fig. 3c).
Fig. 3.
Prevalence of pfmdr1 Single Nucleotide Polymorphisms (SNPs) across North-East and –West Nigeria. The Regional Distribution of pfmdr1 SNPs at codons 86, 184 and 1246 are shown in A, B and C respectively. Green and red bars in A, B and C (wild and mutant alleles, respectively) are not significantly different at codons 86 (2 = 1.70, P = 0.19) and 1246 (2 = 3.60, P = 0.05) and significantly different at 184 (2 = 0.20, P = 0.65) of pfmdr1
Analysis of pfmdr1 haplotypes
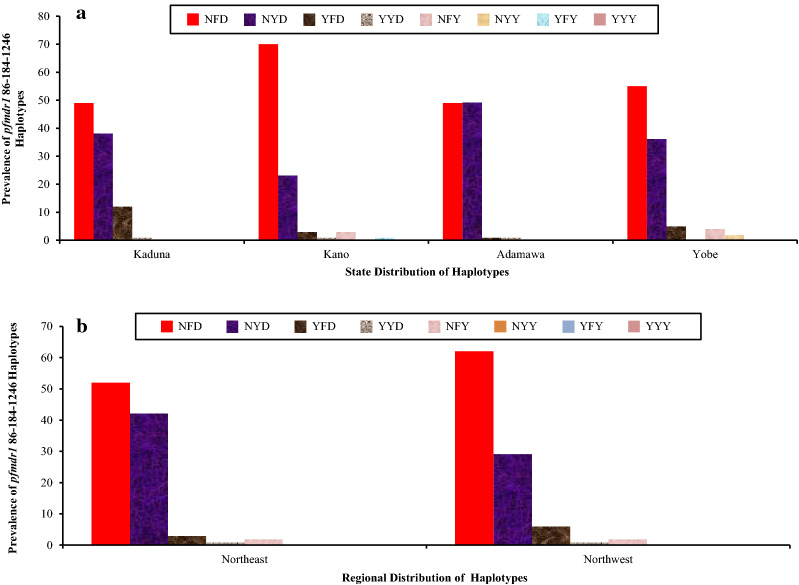
The distribution of pfmdr1 haplotypes in the four Northern Nigerian states is shown in Fig. 4a. The pfmdr1 N86Y, Y184F and D1246Y mutations were constructed into NYD, NYY, NFY, NFD, YYY, YYD, YFD and YFY haplotypes. Out of the 500 pfmdr1 samples genotyped, a total of 486 haplotypes were constructed (sub-divided into seven different types). As shown in the Fig. 3, there was a significant difference in the prevalences of all pfmdr1 haplotypes across the locations (P = 0.00); the NFD pfmdr1 haplotype was highest in Kano state with a prevalence of 69.81%, the NYD haplotype was highest in Adamawa with a prevalence of 49.00%, whilst the pfmdr1 YFD haplotype predominated in Kaduna with a prevalence of 11.46%. The pfmdr1 YYY haplotype was not detected (Fig. 4a). Figure 4b showed the distribution of the pfmdr1 haplotypes across North-West and North–East Nigeria, but in contrast to the states distribution, no significant difference was observed (P = 0.37). The pfmdr1 NFD haplotype was highest in the North-West with a prevalence of 61.96%, the NYD haplotype was highest in the North-East with a prevalence of 41.63%, and the YFD haplotype was highest in the North-West with a prevalence of 5.88%.
Fig. 4.
Prevalence of pfmdr1 86-184-1246 Haplotypes across Four States and Two Geopolitical Regions of Nigeria. The Distribution of pfmdr1 Haplotypes across the States and Regions are shown in A and B respectively. Red, Purple, Brown, Ash, Lilac and Empty bars in A and B (pfmdr1 NFD, NYD, YFD, YYD, NFY, NYY, YFY and YYY haplotypes, respectively) are significantly different across the states (2 = 36.10, P = 0.00) and not significantly different across the two regions (2 = 4.30, P = 0.37)
Discussion
Mutations in several genes, including pfmdr1, pfcrt and pfk13 are associated with variation in parasite sensitivity to a range of drugs [12, 42, 43]. The pfmdr1 mutations N86Y, Y184F and D1246Y SNPs are thought to modulate susceptibility to CQ, AL and AS-AQ [6]. It was observed that, pfmdr1 184F and 86Y alleles predominated in North-West Nigeria while 1246Y was higher in the North-East.
Alleles of pfmdr1 carrying the wild type N86 residue are associated with higher IC50 and IC90 values for LMF, MFQ and DHA, while the alternative 86Y residue seems to confer increased resistance against CQ and AQ [6]. Similarly, there are varying epidemiological reports on the prevalence and consequences of pfmdr1 N86Y polymorphisms from different parts of the world. For example, Ibraheem et al. [44] suggested that pfmdr1 mutations are geographically confined and have inconsistent distributions from one geographic region to another.
Following adoption of ACT in many African countries, some studies from West Africa have linked the prevalence of the pfmdr1 N86 allele to selection by AL [7]. Therefore, the high prevalence of pfmdr1 N86 allele observed in the present study, even with the absence of clinical data of patients, might be suggestive of possible AL pressure in all the states. In addition, the finding might also suggest that the efficacy of the LMF component of ACT is susceptible to the emergence of tolerance in the local P. falciparum populations, as the presence of pfmdr1 N86 is critical in the initiation of resistance to LMF in vivo and that its selection primarily follows reinfection and recrudescence events associated with the elimination stage of LMF, 4–5 days after artemether clearance [45].
Some reports have associated a rise in the prevalence of pfmdr1 86Y alleles with increasing CQ resistance [14, 46, 47]. The low prevalence of pfmdr1 86Y in Adamawa and Yobe raises the possibility that CQ may be effective against P. falciparum malaria in North-Eastern Nigeria once again, although this would be presumably tempered by CQ-resistance associated mutations in pfcrt, which was not assayed here. It is possible that the selection of pfmdr1 86Y allele in this region was aided by the cessation of CQ usage due to the emergence of resistance. The high prevalence of this mutation across Northern Nigeria may indicate that the efficacy of AL is at risk in this region, but raises the possibility that CQ may be effective in the chemotherapy of uncomplicated malaria here.
The pfmdr1 Y184F polymorphisms have been reported by various in vitro studies to be weakly associated with changes in anti-malarial drug response [6, 12, 48]. The IC50 of some anti-malarial drugs was shown to vary on the sole acquisition of either pfmdr1 N86 or 86Y alleles by parasite lines expressing wild type pfmdr1-Y184 or mutant 184F alleles [6]. Several epidemiological studies on the prevalence of pfmdr1 Y184F polymorphisms have shown that the Y184 allele is predominantly confined to East and Central Africa while the mutant 184F allele predominates in West Africa [7, 49–51]. Okell et al. [7] observed higher occurrence of pfmdr1 184F in West Africa than East and Central Africa after extracting data from 397 surveys measuring the prevalence or frequency of at least one pfmdr1 polymorphism in 30 countries. In Cameroon, the prevalence of pfmdr1 184F allele was shown to reduce drastically from 97.3 to 56% in 2003–2013 [50]. Achieng et al. [49] showed that between 1995 and 2003 prior to the introduction of ACT in Kenya, the prevalence of pfmdr1 Y184 was 100%, but declined to 99.3% between 2008 and 2014 post ACT introduction. In a study conducted in Ghana by Duah et al. [52] using archived filter paper blood spots from under-five children with uncomplicated malaria in 2003–2010, the prevalence of pfmdr1 184F was reported to increase steadily as 26–78%, 35–82%, 48–70%, and 40–80% for 2003–2004, 2005–2006, 2007–2008, and 2010, respectively. Indeed, reports of the high occurrence of the mutant pfmdr1 184F in West Africa were corroborated by the present findings in which it was observed that the prevalence of pfmdr1 184F was high in all the states, and especially in Kano, and that its prevalence is higher in North-West compared to North-East Nigeria. This mutation has been previously linked to a reduction in susceptibility to LUM and/or ART [53]. It is perhaps unsurprising that a relatively high prevalence of this mutation was found in regions, such as Tanzania in East Africa, where AL is first-line intervention against uncomplicated malaria. Thus, the high prevalence of mutant pfmdr1 184F obtained in the present study may simply reflect the relatively common use of AL treatment in this region.
The pfmdr1 D1246Y mutation affects P. falciparum susceptibility to various anti-malarials including QN, MFQ, (HF), CQ and ART, with the latter two drugs affected in a strain specific manner [16, 21]. The observed low prevalence of mutant pfmdr1 1246Y alleles compared to the wild type in this study is consistent with reports from Southern Nigeria [37] as well as other West and East African countries that adopted AL as a front-line anti-malarial therapy for uncomplicated malaria [7, 49]. Countries in Central Africa have observed an unsteady increase in the prevalence of the pfmdr1 D1246 allele, possibly due to the selective pressure of AS-AQ [42, 54].
Several reports from Africa have suggested that linkage between pfmdr1 N86Y/Y184F/D1246Y results in haplotypes with particular phenotypic characteristics that may be selected depending on the particular drugs that the population is exposed to [55, 56]. The occurrence of pfmdr1 NFD and NYD haplotypes, for example, may result from AL selection while the pfmdr1 YYY haplotype may be favoured in regions where parasites are exposed to AS-AQ, DHAP and CQ [12, 50]. The treatment of uncomplicated malaria with AL often selects pfmdr1 haplotypes bearing the N86 allele [51, 57]. A predominance of the pfmdr1 NFD haplotype was found in Northern Nigeria with Kano, and a complete absence of the pfmdr1 YYY haplotype. These findings are in line with the selective effect of AL on the NFD haplotype. Hence, the abundance of pfmdr1 NFD haplotype may suggest a loss of susceptibility to AL treatment by the parasites in the region. Resistance associated SNPs in pfk13 should be analysed in a subsequent follow-up study, so as to confirm the influence of pfmdr1 NFD haplotype on reduced AL sensitivity in the regions.
Conclusions
In conclusion, Kaduna and Kano States had higher prevalences of the pfmdr1 86Y allele than Yobe and Adamawa indicating differential selection in North-East and North-West Nigeria, possibly due to differing population density and malaria transmission intensity in these regions. Furthermore, there was a very high prevalence of pfmdr1 NFD and NYD haplotypes, which may suggest a reduction in susceptibility to AL in both regions. Overall, the scarcity of pfmdr1 YYY combined with a high prevalence of NFD haplotypes suggests there is a need for continuous pharmacovigilance surrounding the use of AL in these regions.
Acknowledgements
We are thankful to the administrators of Ahmadu Bello University, Zaria, Nigeria and deanery of School of Tropical Medicine and Global Health, Nagasaki University, Japan who were highly instrumental to the success and completion of this study.
Abbreviations
- SNPs
Single nucleotide polymorphisms
- pfmdr1
P. falciparum multidrug resistance gene-1
- pfcrt
P. falciparum chloroquine resistance transporter gene
- Pgh-1
P-glycoprotein homologue-1
- PfMDR1
P. falciparum multidrug resistance protein-1
- DNA
Deoxyribonucleic acid
- PCR
Polymerase chain reaction
- CIOMS
Council for International Organizations of Medical Sciences
- ACT
Artemisinin-based combination therapy
- CQ
Chloroquine
- AQ
Amodiaquine
- QN
Quinine
- HF
Halofantrine
- ART
Artemisinin
- MQ
Mefloquine
- AL
Artemether-lumefantrine
- AS-AQ
Artesunate-amodiaquine
- DHAP
Dihydroartemisinin-piperaquine
Authors’ contributions
MNS, AA, MSJ, BOY and HMSH conceived the idea, designed and performed the experiments, analysed the data and wrote the manuscript. MAI and EOB participated in the design of the experiment, supervised samples collection/preparation in the field and revised the manuscript. KK, KH, RC, TS and DKI supervised the laboratory experiments, participated in the overall interpretation of data and revision of the manuscript. The authors read and affirmed the final version of the manuscript.
Funding
The study was funded by the board of research of Ahmadu Bello University, Zaria, Nigeria.
Availability of data and materials
Authors assure that data will be available upon request following acceptance and publication of the article.
Ethics and consent to participate
The consent of patients or their guardians attending public health facilities was sought following explanation of the voluntary nature of participation. Prior to sample collection, ethical clearance was granted from the Ministries of Health of Kano, Adamawa, Yobe and Kaduna with references MOH/Off/797/T.1/285, S/MOH/1131/V.1, MOH/GEN/704/VOL.1 and MOH/ADM/744/VOL.1/537 respectively. The study was conducted in accordance with the 2016 international ethical guidelines for health-related research involving human subjects developed by the Council for International Organizations of Medical Sciences (CIOMS) in collaboration with the World Health Organization.
Consent for publication
Authors received the consent of patients or their guardians to use data for publication.
Competing interests
The authors of this study have no competing interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cravo P, Napolitano H, Culleton R. How genomics is contributing to the fight against artemisinin-resistant malaria parasites. Acta Trop. 2015;148:1–7. doi: 10.1016/j.actatropica.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Blasco B, Leroy D, Fidock DA, BenjaminBlasco DL. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat Med. 2017;23:917–928. doi: 10.1038/nm.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White N. Antimalarial drug resistance and combination chemotherapy. Biol Sci. 1999;354:739–749. doi: 10.1098/rstb.1999.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bopp SER, Manary MJ, Bright AT, Johnston GL, Dharia NV, Luna FL, et al. Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet. 2013;9:e1003293. doi: 10.1371/journal.pgen.1003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Culleton R, Abkallo HM. Malaria parasite genetics: doing something useful. Parasitol Int. 2015;64:244–253. doi: 10.1016/j.parint.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Veiga MI, Dhingra SK, Henrich PP, Straimer J, Gnädig N, Uhlemann AC, et al. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun. 2016;77:11553. doi: 10.1038/ncomms11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okell LC, Reiter LM, Ebbe LS, Baraka V, Bisanzio D, Watson OJ, et al. Emerging implications of policies on malaria treatment: Genetic changes in the Pfmdr-1 gene affecting susceptibility to artemether–lumefantrine and artesunate–amodiaquine in Africa. BMJ Glob Health. 2018;3:e000999. doi: 10.1136/bmjgh-2018-000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ariey F, Fandeur T, Durand R, Randrianarivelojosia M, Jambou R, Legrand E, et al. Invasion of Africa by a single pfcrt allele of South East Asian type. Malar J. 2006;5:34. doi: 10.1186/1475-2875-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muwanguzi J, Henriques G, Sawa P, Bousema T, Sutherland CJ. Lack of K13 mutations in Plasmodium falciparum persisting after artemisinin combination therapy treatment of Kenyan children. Malar J. 2016;36:1–6. doi: 10.1186/s12936-016-1095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearce RJ, Pota H, Evehe MSB, Bâ EH, Mombo-Ngoma G, Malisa AL, et al. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med. 2009;6:e1000055. doi: 10.1371/journal.pmed.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foote SJ, Kyle DE, Martin RK, Oduola AMJ, Forsyth K, Kemp DJ, et al. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature. 1990;345:255–258. doi: 10.1038/345255a0. [DOI] [PubMed] [Google Scholar]
- 12.Ferreira PE, Holmgren G, Veiga MI, Uhlén P, Kaneko A, Gil JP. PfMDR1: mechanisms of transport modulation by functional polymorphisms. PLoS One. 2007;6:e23875. doi: 10.1371/journal.pone.0023875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiling SJ, Rohrbach P. Monitoring PfMDR1 transport in Plasmodium falciparum. Malar J. 2015;14:270. doi: 10.1186/s12936-015-0791-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das S, Tripathy S, Chattopadhayay S, Das B, Kar Mahapatra S, Hati AK, et al. Progressive increase in point mutations associates chloroquine resistance: even after withdrawal of chloroquine use in India. Int J Parasitol Drugs Drug Resist. 2017;7:251–261. doi: 10.1016/j.ijpddr.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekihara M, Tachibana SI, Yamauchi M, Yatsushiro S, Tiwara S, Fukuda N, et al. Lack of significant recovery of chloroquine sensitivity in Plasmodium falciparum parasites following discontinuance of chloroquine use in Papua New Guinea. Malar J. 2018;17:434. doi: 10.1186/s12936-018-2585-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duraisingh MT, Cowman AF. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 2005;94:181–190. doi: 10.1016/j.actatropica.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Folarin OA, Bustamante C, Gbotosho GO, Sowunmi A, Zalis MG, Oduola AMJ, et al. In vitro amodiaquine resistance and its association with mutations in pfcrt and pfmdr1 genes of Plasmodium falciparum isolates from Nigeria. Acta Trop. 2011;120:224–230. doi: 10.1016/j.actatropica.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tinto H, Guekoun L, Zongo I, Guiguemdé RT, D’Alessandro U, Ouédraogo JB. Chloroquine-resistance molecular markers (Pfcrt T76 and Pfmdr-1 Y86) and amodiaquine resistance in Burkina Faso. Trop Med Int Health. 2008;13:238–240. doi: 10.1111/j.1365-3156.2007.01995.x. [DOI] [PubMed] [Google Scholar]
- 19.Humphreys GS, Merinopoulos I, Ahmed J, Whitty CJM, Mutabingwa TK, Sutherland CJ, et al. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother. 2007;51:991–997. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lekostaj JK, Natarajan JK, Paguio MF, Wolf C, Roepe PD. Photoaffinity labeling of the Plasmodium falciparum chloroquine resistance transporter with a novel perfluorophenylazido chloroquine. Biochemistry. 2008;47:10394–10406. doi: 10.1021/bi8010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 22.Mwai L, Ochong E, Abdirahman A, Kiara SM, Ward S, Kokwaro G, et al. Chloroquine resistance before and after its withdrawal in Kenya. Malar J. 2009;8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pickard AL, Wongsrichanalai C, Purfield A, Kamwendo D, Emery K, Zalewski C, et al. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47:2418–2423. doi: 10.1128/AAC.47.8.2418-2423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mbogo GW, Nankoberanyi S, Tukwasibwe S, Baliraine FN, Nsobya SL, Conrad MD, et al. Temporal changes in prevalence of molecular markers mediating antimalarial drug resistance in a high malaria transmission setting in Uganda. Am J Trop Med Hyg. 2014;91:54–61. doi: 10.4269/ajtmh.13-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baliraine FN, Rosenthal PJ. Prolonged selection of pfmdr1 polymorphisms after treatment of falciparum malaria with artemether-lumefantrine in Uganda. J Infect Dis. 2011;204:1120–1124. doi: 10.1093/infdis/jir486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sondo P, Derra K, Diallo Nakanabo S, Tarnagda Z, Kazienga A, Zampa O, et al. Artesunate-amodiaquine and artemether-lumefantrine therapies and selection of Pfcrt and Pfmdr1 alleles in Nanoro, Burkina Faso. PLoS One. 2016;11:e0151565. doi: 10.1371/journal.pone.0151565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.WHO . World Malaria Report 2018. Geneva: Word Health Organization; 2018. [Google Scholar]
- 28.Onwuemele A. An assesment of the spatial pattern of malaria infection in Nigeria. Int J Med Med Sci. 2014;6:80–86. doi: 10.5897/IJMMS2013.1006. [DOI] [Google Scholar]
- 29.Houben CH, Fleischmann H, Gückel M. Malaria prevalence in north-eastern Nigeria: a cross-sectional study. Asian Pac J Trop Med. 2013;6:865–868. doi: 10.1016/S1995-7645(13)60154-6. [DOI] [PubMed] [Google Scholar]
- 30.Samdi LM, Ajayi JA, Oguche S, Ayanlade A. Seasonal variation of malaria parasite density in paediatric population of Northeastern Nigeria. Glob J Health Sci. 2012;4:103–109. doi: 10.5539/gjhs.v4n2p103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okunlola OA, Oyeyemi OT. Spatio-temporal analysis of association between incidence of malaria and environmental predictors of malaria transmission in Nigeria. Sci Rep. 2019;9:17500. doi: 10.1038/s41598-019-53814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MIS. Nigeria Malaria Indicator Survey 2015. Final Report. 2017.
- 33.Umar RA, Hassan SW, Ladan MJ, Jiya MN, Abubakar MK, Nataala U. The associaion of K76T mutation in Pfcrt gene and chloroquine treatment failure in uncomplicated Plasmodium falciparum malaria in a cohort of Nigerian children. J Appl Sci. 2007;7:3696–3704. doi: 10.3923/jas.2007.3696.3704. [DOI] [Google Scholar]
- 34.Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother. 2006;50:1893–1895. doi: 10.1128/AAC.50.5.1893-1895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mawili-Mboumba DP, Kun JFJ, Lell B, Kremsner PG, Ntoumi F. Pfmdr1 alleles and response to ultralow-dose mefloquine treatment in gabonese patients. Antimicrob Agents Chemother. 2002;46:166–170. doi: 10.1128/AAC.46.1.166-170.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Happi CT, Gbotosho GO, Folarin OA, Sowunmi A, Hudson T, O’Neil M, et al. Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in nigerian children with uncomplicated falciparum malaria. Antimicrob Agents Chemother. 2009;53:888–895. doi: 10.1128/AAC.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oladipo OO, Wellington OA, Sutherland CJ. Persistence of chloroquine-resistant haplotypes of Plasmodium falciparum in children with uncomplicated Malaria in Lagos, Nigeria, four years after change of chloroquine as first-line antimalarial medicine. Diagn Pathol. 2015;10:41. doi: 10.1186/s13000-015-0276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dokunmu TM, Adjekukor CU, Yakubu OF, Bello AO, Adekoya JO, Akinola O, et al. Asymptomatic malaria infections and Pfmdr1 mutations in an endemic area of Nigeria. Malar J. 2019;18:218. doi: 10.1186/s12936-019-2833-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.UNDP. National Human Development Report 2018: Achieving Human Development in North East Nigeria. National Human Development Report; 2018. Timor-Lest, pp 1–116.
- 40.Kavishe RA, Paulo P, Kaaya RD, Kalinga A, Van Zwetselaar M, Chilongola J, et al. Surveillance of artemether-lumefantrine associated Plasmodium falciparum multidrug resistance protein-1 gene polymorphisms in Tanzania. Malar J. 2014;13:264. doi: 10.1186/1475-2875-13-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lobo E, De Sousa B, Rosa S, Figueiredo P, Lobo L, Pateira S, et al. Prevalence of pfmdr1 alleles associated with artemether-lumefantrine tolerance/resistance in Maputo before and after the implementation of artemisinin-based combination therapy. Malar J. 2014;13:300. doi: 10.1186/1475-2875-13-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berzosa P, Esteban-Cantos A, García L, González V, Navarro M, Fernández T, et al. Profile of molecular mutations in pfdhfr, pfdhps, pfmdr1, and pfcrt genes of Plasmodium falciparum related to resistance to different anti-malarial drugs in the Bata District (Equatorial Guinea) Malar J. 2017;16:28. doi: 10.1186/s12936-016-1672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ljolje D, Dimbu PR, Kelley J, Goldman I, Nace D, Macaia A, et al. Prevalence of molecular markers of artemisinin and lumefantrine resistance among patients with uncomplicated Plasmodium falciparum malaria in three provinces in Angola, 2015. Malar J. 2018;17:84. doi: 10.1186/s12936-018-2233-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ibraheem ZO, Abd Majid R, Noor SM, Sedik HM, Basir R. Role of different Pfcrt and Pfmdr-1 mutations in conferring resistance to antimalaria drugs in Plasmodium falciparum. Malar Res Treat. 2014;2014:950424. doi: 10.1155/2014/950424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sisowath C, Strömberg J, Mårtensson A, Msellem M, Obondo C, Björkman A, Gil JP, et al. In vivo selection of Plasmodium falciparum pfmdr1 86 N Coding Alleles by Artemether-Lumefantrine (Coartem) J Infect Dis. 2005;191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 46.Das S, Mahapatra SK, Tripathy S, Chattopadhyay S, Dash SK, Mandal D, et al. Double mutation in the pfmdr1 gene is associated with emergence of chloroquine-resistant Plasmodium falciparum malaria in Eastern India. Antimicrob Agents Chemother. 2014;58:5909–5915. doi: 10.1128/AAC.02762-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Chen J, Xie D, Monte-Nguba S, Eyi JUM, Matesa RA, et al. High prevalence of pfmdr1 N86Y and Y184F mutations in Plasmodium falciparum isolates from Bioko island, Equatorial Guinea. Pathog Glob Health. 2014;108:339–343. doi: 10.1179/2047773214Y.0000000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srimuang K, Miotto O, Lim P, Fairhurst RM, Kwiatkowski DP, Woodrow CJ, et al. Analysis of anti-malarial resistance markers in pfmdr1 and pfcrt across Southeast Asia in the Tracking Resistance to Artemisinin Collaboration. Malar J. 2016;15:541. doi: 10.1186/s12936-016-1598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Achieng AO, Muiruri P, Ingasia LA, Opot BH, Juma DW, Yeda R, et al. Temporal trends in prevalence of Plasmodium falciparum molecular markers selected for by artemether-lumefantrine treatment in pre-ACT and post-ACT parasites in western Kenya. Int J Parasitol Drugs Drug Resist. 2015;5:92–99. doi: 10.1016/j.ijpddr.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moyeh MN, Njimoh DL, Evehe MS, Ali IM, Nji AM, Nkafu DN, et al. Effects of drug policy changes on evolution of molecular markers of Plasmodium falciparum resistance to chloroquine, amodiaquine, and sulphadoxine-pyrimethamine in the South West Region of Cameroon. Malar Res Treat. 2018;2018:7071383. doi: 10.1155/2018/7071383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor AR, Flegg JA, Holmes CC, Guérin PJ, Sibley CH, Conrad MD, et al. Artemether-lumefantrine and dihydroartemisinin-piperaquine exert inverse selective pressure on Plasmodium falciparum drug sensitivity-associated haplotypes in Uganda. Open Forum Infect Dis. 2017;4:ofw229. doi: 10.1093/ofid/ofw229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duah NO, Matrevi SA, De Souza DK, Binnah DD, Tamakloe MM, Opoku VS, et al. Increased pfmdr1 gene copy number and the decline in pfcrt and pfmdr1 resistance alleles in Ghanaian Plasmodium falciparum isolates after the change of anti-malarial drug treatment policy. Malar J. 2013;12:377. doi: 10.1186/1475-2875-12-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang B, Wang Q, Deng C, Wang J, Yang T, Huang S, et al. Prevalence of CRT and mdr-1 mutations in Plasmodium falciparum isolates from Grande Comore island after withdrawal of chloroquine. Malar J. 2016;15:414. doi: 10.1186/s12936-016-1474-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Apinjoh TO, Mugri RN, Miotto O, Chi HF, Tata RB, Anchang-Kimbi JK, et al. Molecular markers for artemisinin and partner drug resistance in natural Plasmodium falciparum populations following increased insecticide treated net coverage along the slope of mount Cameroon: cross-sectional study. Infect Dis Poverty. 2017;6:136. doi: 10.1186/s40249-017-0350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tumwebaze P, Conrad MD, Walakira A, LeClair N, Byaruhanga O, Nakazibwe C, et al. Impact of antimalarial treatment and chemoprevention on the drug sensitivity of malaria parasites isolated from Ugandan children. Antimicrob Agents Chemother. 2015;59:3018–3030. doi: 10.1128/AAC.05141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tumwebaze P, Tukwasibwe S, Taylor A, Conrad M, Ruhamyankaka E, Asua V, et al. Changing antimalarial drug resistance patterns identified by surveillance at three sites in Uganda. J Infect Dis. 2017;215:631–635. doi: 10.1093/infdis/jiw614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malmberg M, Ngasala B, Ferreira PE, Larsson E, Jovel I, Hjalmarsson A, et al. Temporal trends of molecular markers associated with artemether- lumefantrine tolerance/resistance in Bagamoyo district. Tanzania. Malar J. 2013;12:103. doi: 10.1186/1475-2875-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Authors assure that data will be available upon request following acceptance and publication of the article.