Abstract
Background
Diarrhoea, affecting children in developing countries, is mainly caused by diarrheagenic Escherichia coli (DEC). This study principally aimed to determine the prevalence of DEC pathotypes and Extended-spectrum β-lactamase (ESBL) genes isolated from children under 5 years old with diarrhea.
Methods
A total of 320 diarrhoea stool samples were investigated. E. coli isolates were investigated for genes specific for enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC) and enterohemorrhagic E. coli (EHEC) using polymerase chain reaction (PCR). Furthermore, antimicrobial susceptibility testing, detection of antibiotic resistance-genes and phylogenetic typing were performed.
Results
Over all, DEC were isolated from 66/320 (20.6%) of the children with diarrhoea. EAEC was the predominant (47%), followed by typical EPEC (28.8%) and atypical EPEC (16.6%). Co-infection by EPEC and EAEC was detected in (7.6%) of isolates. However, ETEC, EIEC and EHEC were not detected. Phylogroup A (47%) and B2 (43.9%) were the predominant types. Multidrug-resistance (MDR) was found in 55% of DEC isolates. Extended-spectrum β-lactamase (ESBL) genes were detected in 24 isolates (24 blaTEM and 15 blaCTX-M-15). Only one isolate harbored AmpC β-lactamase gene (DHA gene).
Conclusion
The study concluded that, EAEC and EPEC are important causative agents of diarrhoea in children under 5 years. MDR among DEC has the potential to be a big concern.
Keywords: Diarrheagenic Escherichia coli (DEC), Pathotypes- ESBL, AmpC β-lactamase
Background
Diarrhoea is one of the top ten leading causes of death worldwide and the second in low-income countries in children under 5 years old [1]. Diarrheagenic Escherichia coli (DEC) is a very important cause of pediatric diarrhoea [2], particularly in developing countries, where these organisms are the main cause of diarrhoea affecting children under 5 years old [3]. The DEC is classified according to specific virulence characters into distinct pathotypes [2]. The epidemiology of the different DEC pathotypes varies according to the geographical variations even within the same area [3–5]. The most common pathotypes in developing countries are enteroaggregative E. coli (EAEC), enteropathogenic E. coli (EPEC) and enterotoxigenic E. coli (ETEC) [5]. The EPEC type is subdivided into typical EPEC (tEPEC), that carries both of the intimin gene (eae) and the bundle forming pili (bfp) genes, and atypical EPEC (aEPEC) which carries eae gene but lacks the bfp gene. While the eae gene is responsible for attachment and effacement of intestinal epithelial cells, bfp gene is encoded for adherence factors [6]. The EAEC, which is an emerging cause of diarrhoea in adults and children worldwide [7], can be determined by the plasmid-encoded gene probe pCVD432 which explain the aggregative phenotype [8]. Other pathotypes of E. coli, that display low levels of incidences such as enterohemorrhagic E. coli (EHEC) “a subtype of Shiga toxin-producing E. coli (stx1 and stx2)” and enteroinvasive E. coli (EIEC), can cause life-threatening diarrhoea [9]. DEC pathotypes can be determined by some techniques, such serotyping, phenotyping and molecular methods [9]. PCR is a highly sensitive and specific assay that gives fast and reliable results [9]. PCR assay can also categorize DEC into different phylogenetic groups [10]. Treatment with antimicrobials is recommended only for severely ill patients. Unfortunately, the treatment of E. coli is difficult due to the emergence of antibiotics resistant strains in the last decades [11]. The predominant mechanism of resistance is the hydrolysis of the antibiotic by beta-lactamases. The ability to produce B-lactamases, including Extended Spectrum Beta Lactamases (ESBL) and Amp-C B-lactamases is frequently acquired through large plasmids holding many different genes resistance (MDR) [12]. Intestinal E. coli is one of the important organisms harboring these genes [13]. Information about DEC pathotypes in Egypt is scarce, particularly in south Egypt because molecular techniques are not usually used to determine DEC in medical laboratories. The present study aimed to determine the prevalence, phylogenetic types and resistance patterns of DEC pathotypes among children under 5 years old with acute diarrhoea in Minia, South Egypt.
Methods
Study population
The cohort of this study included children less than 5 years of age with acute community-acquired diarrhoea, who visited the Pediatric departments of two major hospitals in Minia, Egypt; Minia university hospital and Minia governorate hospital as outpatients from December 2018 to May 2019. Acute diarrhoea was defined as passage of three or more loose or watery stools per day, plus one or more gastroenteritis symptoms as; nausea, vomiting, abdominal pain, fecal urgency, fever or cramps. Positive rotavirus samples were excluded from the study. Children with a history of antibiotic use in the last 2 weeks were excluded from the study.
Specimen collection, isolation, and identification of E. coli
A total of 320 stool samples were included in the study. One stool sample was collected from each patient in sterile container and transported to the microbiology laboratory in cool box for immediate examination. The samples were examined for consistency, presence of blood, mucous or occult blood. The samples were inoculated into MacConkey broth for enrichment at 37 °C for 24 h. The enrichments were streaked onto MacConkey agar (Oxoid, Basingstoke, United Kingdom). and incubated for 24 h at 37 °C. After that, lactose fermenters isolates were subcultured onto eosin methylene blue agar (EMB) agar (Oxoid, Basingstoke, United Kingdom). Colonies producing greenish metallic sheen on EMB agar were examined by biochemical tests including; IMViC (indole, methyl red, Voges-Proskauer, citrate utilization), urease agar, sugar fermentation and motility tests. In addition the identified E. coli isolates were confirmed by chromogenic media (CHROMagar™ Orientation, Paris, France). Samples containing E. coli only were included. Hemolytic activity was tested by culturing of isolates on blood agar. E. coli isolates were kept in trypticase soy broth (Oxoid, UK) with sterilized 20% glycerol at − 20 °C for further investigations.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the disk diffusion method and identified according to CLSI guidelines [14]. The used antimicrobial discs were; ceftriaxone (CRO) 30 μg, amoxicillin/clavulanic acid (AMC) 30 μg, ceftazidime (CAZ) 30 μg, meropenem 10 μg (MEM) 10 μg, amikacin (AK) 30 μg, sulphamethoxazole/ trimethoprim (SXT) 300 μg, cefoxitin 30 μg (FOX) and tetracyclin 30 mg (TE) (Thermo Scientific™ Oxoid, UK). Isolates with inhibition zone size ≤22 mm with (CAZ) and ≤ 25 mm with (CRO) were suggested to be ESBL-producers [14]. Double-Disc Synergy Test (DDST) was used for confirmation of ESBL production [15]. In addition, isolates showing an inhibitory zone diameter ≤ 18 mm with (FOX) were suspected to be AmpC β-lactamase producers [16]. Carbapenem inactivation method was used for confirmation of carbapenemase production in meropenem- non susceptible isolates [17]. Multiple drug resistance (MDR) was identified as the resistance to at least three different antimicrobial groups [18].
Molecular identification of diarrheagenic E. coli pathotypes
The stored E.coli isolates were streaked onto chromogenic media at 37 °C for 24 h. About 3–5 of isolated colonies from each isolate were inoculated onto trypticase soy broth tubes. at 37 °C for 18 h. About 200 μL broth of each isolate was centrifuged (8000 rpm) at 4 °C for 6 mins. The pellet was separated and processed for DNA extraction using GeneJET genomic DNA purification kit according to the manufacturer’s instructions (Thermo scientific, USA). For identification of DEC pathotypes, the following virulence genes were investigated using single PCR reactions; pCVD432 gene for enteroaggregative E. coli (EAEC) [19], eae gene for both types of enteropathogenic E.coli (typical enteropathogenic E.coli “tEPEC” and atypical enteropathogenic E.coli “aEPEC”) [20], bfpA gene for typical enteropathogenic E.coli (tEPEC) [21], LT gene, ST gene for enterotoxigenic E. coli (ETEC) [22] and ipaH gene for enteroinvasive E.coli (EIEC) [23]. While (stx1and stx2) genes for enterohemorrhagic E. coli (EHEC) were identified using multiplex PCR [24]. Each single PCR reaction was conducted in a final reaction of 25 μL containing 2 μL of template DNA (approximately 100 ng/μL) and 12.5 μL of master mix (Maxima Hot Start Green PCR Master Mix, Thermo scientific, USA), 10 pmol of each primer and 8.5 μL of nuclease-free water. However the multiplex PCR reaction was performed in a 25 μL reaction mixture containing 3 μL of purified DNA (approximately 100 ng/μL), 12.5 μL of Maxima Hot Start Green PCR Master Mix (Thermo scientific, USA), 1 μL (10 pmol) of each primer (Thermo scientific, USA) and 5.5 μL of nuclease free water. EPEC E2348/69, ETEC H10407, and EAEC positive strain from our laboratory served as the positive control [25].
Phylogenic analysis
DEC isolates were classified into different phylogenic types by triplex PCR using two genes (chuA and yjaA) and a DNA fragment TSPE4.C2 as described previously [11].
Detection of resistance genes
PCR assay was used for identification of resistant genes, blaTEM, blaSHV [26] and blaCTX-M-15 [27]. In addition, a multiplex PCR was done for AmpC β-lactamase genes (MOX, FOX, CIT and DHA) genes [28]. PCR assays were performed using Biometra, UNO II thermal cycler (Gottingen, Germany). Primers sequences and amplification parameters used in the study are described in (Table 1). Agarose gel electrophoresis (2%) was used to identify PCR products (Biometra Gottingen, Germany). Positive control was obtained from a previous research [29].
Table 1.
PCR primers used in the study
| Primer | Primers (5_ to 3_) | Amplicon size (bp) |
Annealing temperature °C | Reference |
|---|---|---|---|---|
| pCVD432 |
F-CTGGCGAAAGACTGTATCAT R-CAATGTATAGAAATCCGCTGTT |
630 | 58 | [19] |
| eae |
F-CTGAACGGCGATTACGCGAA R-CCAGACGATACGATCCAG |
917 | 58 | [20] |
| bfpA |
F-AATGGTGCTTGCGCTTGCTGC R-GCCGCTTTATCCAACCTGGTA |
326 | 58 | [21] |
| LT gene |
F-GGCGACAGATTATACCGTGC R-CGGTCTCTATATTCCCTGTT |
450 | 50 | [22] |
| ST gene |
F-ATTTTTMTTTCTGTATTRTCTT R-CACCCGGTACARGCAGGATT |
190 | 50 | [22] |
| ipaH |
F-GTTCCTTGACCGCCTTTCCGATACCGTC R-GCCGGTCAGCCACCCTCTGAGAGTAC |
600 | 52 | [23] |
| stx1 |
F-ATAAATCGCCATTCGTTGACTAC R-AGAACGCCCACTGAGATCATC |
180 | 50 | [24] |
| Stx2 |
F-GGCACTGTCTGAAACTGCTCC R-TCGCCAGTTATCTGACATTCTG |
255 | 50 | [22] |
| ChuA |
F- ACGAACCAACGGTCAGGAT R-TGCCGCCAGTACCAAAGACA |
279 | 59 | [10] |
| yjaA |
F-TGAAGTGTCAGGAGACGCTG R-ATGGAGAATGCGTTCCTCAAC |
211 | 59 | [10] |
| TspE4C2 |
F-GAGTAATGTCGGGGCATTCA R-CGCGCCAACAAAGTATTACG |
154 | 59 | [10] |
| blaTEM |
F-AAACGCTGGTGAAAGTA R-AGCGATCTGTCTAT |
822 | 58 | [25] |
| blaSHV |
F-ATGCGTTATATTCGCCTGTG R-TGCTTTGTTATTCGGGCCAA |
753 | 60 | [25] |
| CTX-M-15 |
F-CACACGTGGAATTTAGGGACT R-GCCGTCTAAGGCCATAAACA |
996 | 55 | [26] |
| MOX |
F-GCTGCTCAAGGAGCACAGGAT R-CAC ATT GAC ATA GGT GTG GTG C |
520 | ||
| FOX |
F-AAC ATG GGG TAT CAG GGA GAT G R-CAA AGC GCG TAA CCG GAT TGG |
190 | 64 | [27] |
| DHA |
F-AAC TTT CAC AGG TGT GCT GGG T R-CCG TAC GCA TAC TGG CTT TGC |
405 | ||
| CIT |
F-TGG CCA GAA CTG ACA GGC AAA R-TTT CTC CTG AAC GTG GCT GGC |
462 |
Statistical analysis
Demographic, clinical and laboratory data of the patients were analyzed using SPSS program for windows version 20.0 (IBM, USA). Categorical variables were analyzed using the chi-square test. P-values of < 0.05 were considered statistically significant.
Results
Demographic and clinical features of the study participants
Overall, 66 (20.6%) of isolates were identified as DEC among children with acute diarrhoea. Of these, 16/66 (24.2%) was aged< 1 year 22/66 (33.4%) of them were between 1 and 2 years old and 28/66 (42.4%) of them aged between 2 and 5 years. Clinical manifestations showed that, 18 (27.3%) of the cases pass more than 3 loose stools daily, while 48(72.7%) pass more than 5 loose stools daily. Presence of mucus and blood were reported in 58 (87.8%) and 14 (21.2%) of samples respectively. Vomiting was detected in 29 (43.9%) of cases and hemolytic activity was detected in 10 (15.2%) of cases.
The detected pathotypes in DEC strains
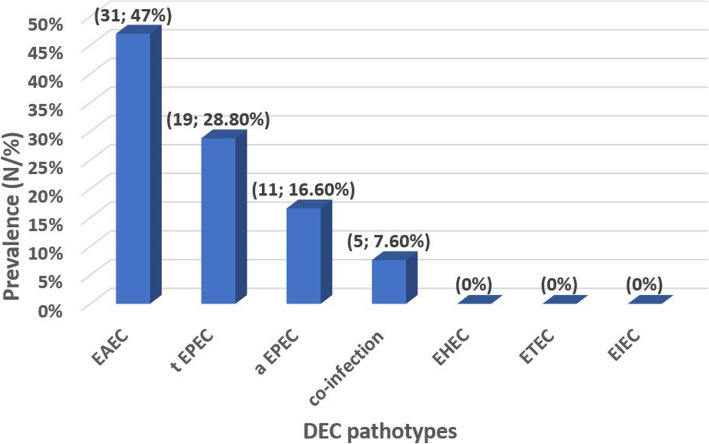
In the current study, 31/66 (47%) of isolates were identified as EAEC (positive pCVD432 gene), 19/66 (28.8%) of isolates were identified as tEPEC (positive eae gene+ bfpA gene) and 11/66 (16.6%) of isolates were identified as aEPEC (positive eae gene only). However, 5/66 (7.6%) of isolates had co-infection (4 isolates contain tEPEC + EAEC, while 1 isolate contains aEPEC + EAEC). EHEC, ETEC and EIEC were not detected in the study isolates (Fig. 1). Distribution of DEC types among age groups are presented in (Fig. 2) and the distribution of the clinical manifestations among the DEC pathotypes are summarized in (Table 2).
Fig. 1.
Prevalence of DEC detected in stool samples. DEC: diarrheagenic E. coli; EAEC: enteroaggregative E. coli tEPEC; typical enteropathogenic E. coli, aEPEC; A typical enteropathogenic E. coli; EIEC: enteroinvasive E. coli; EHEC: enterohemorrhagic E. coli; ETEC: enterotoxigenic E. coli
Fig. 2.
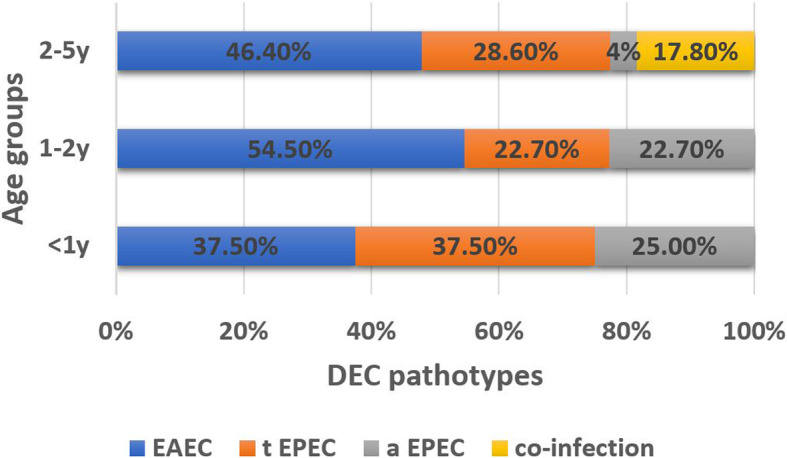
The distribution of DEC types among age groups. EAEC; enteroaggregative E. coli, tEPEC; typical enteropathogenic E. coli, aEPEC; A typical enteropathogenic E. coli. The proportion of EAEC and aEPEC were significantly higher in children under 2 years (p = 0.046 and 0.05, respectively)
Table 2.
Distribution of the clinical manifestations among the DEC pathotypes
| Clinical data | EAEC (N = 31) | tEPEC (N = 19) | aEPEC (N = 11) | Co-infection (N = 5) |
p-value |
|---|---|---|---|---|---|
| Diarrhoea> 3/day | 1 (3.2%) | 4 (21%) | 11 (100%) | 2 (40%) | 0.05 |
| Diarrhoea> 5/day | 30 (96.7%) | 15 (78.9%) | 0 (0%) | 3 (60%) | 0.08 |
| Vomiting | 8 (25.8%) | 9 (47.3%) | 7 (63.6%) | 5 (100%) | 0.27 |
| Blood | 10 (32.2%) | 0 (0%) | 1 (9.1%) | 3 (60%) | 0.21 |
| Mucus | 23 (74.2%) | 19 (100%) | 11 (100%) | 5 (100%) | 0.88 |
| Hemolytic activity | 4 (12.9%) | 1 (5.2%) | 3 (27.2%) | 2 (40%) | 0.27 |
EAEC enteroaggregative E. coli, tEPEC typical enteropathogenic E. coli, aEPEC A typical enteropathogenic E. coli
Distribution of phylogenetic groups among DEC isolates
The distribution of phylogenetic groups among the DEC isolates reveled the predominance of group A (31/66, 47%) followed by group B2 (29/66, 43.9%) then group D (6/66, 9.1%). However, group B1 was not detected. The distribution of phylogenetic groups among DEC types are presented in (Fig. 3).
Fig. 3.
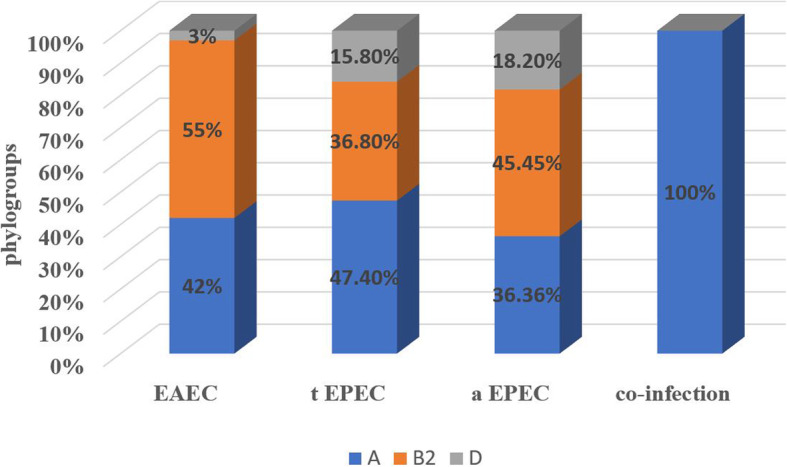
The distribution of phylogenetic groups among DEC types. EAEC; enteroaggregative E. coli, tEPEC; typical enteropathogenic E. coli, aEPEC; A typical enteropathogenic E. coli
Antimicrobial resistance of DEC isolates
Antibiotic resistance rates of the study isolates were as follows; 22.7, 24.2, 60.6, 62.1, 66.6%, 72.7, and 77.3%, of isolates were resistant to cefoxitin, amikacin, amoxicillin-clavulanate, trimethoprim-sulphamethoxazole, ceftriaxone, ceftazidime, and tetracycline respectively. On the other hand, resistance to meropenem was only (9.1%). Multiple drug resistance (MDR) was detected in 55% of isolates. ESBL production was identified using DDST in (60.6%) of isolates. Carbapenemase production was not detected. Antimicrobial susceptibility among different DEC types are presented in (Table 3).
Table 3.
Antimicrobial sensitivity patterns among different E. coli types
| Antibiotics | E coli types | |||||
|---|---|---|---|---|---|---|
| EAEC N = 31 |
t EPEC N = 19 |
a EPEC N = 11 |
Co-infection N = 5 |
Total N = 66 |
||
| MEM | Sensitive | 27 (87.10%) | 16 (84.21%) | 10 (90.9%) | 5 (100%) | 58 (87.9%) |
| Intermediate | 2 (6.45%) | 0 (0.0%) | 0(0.0%) | 0 (0.0%) | 2 (3%) | |
| Resistant | 2 (6.45%) | 3 (15.79%) | 1 (9.09%) | 0 (0.0%) | 6 (9.1%) | |
| CAZ | Sensitive | 4 (12.90%) | 3 (15.79%) | 0 (0.0%) | 0 (0.0%) | 7 (10.6%) |
| Intermediate | 6 (19.36%) | 4 (21.05%) | 1 (9.09%) | 0 (0.0%) | 11 (16.6%) | |
| Resistant | 21 (67.74%) | 12 (63.16%) | 10 (90.9%) | 5 (100.0%) | 48 (72.7%) | |
| CRO | Sensitive | 7 (22.58%) | 4 (21.05%) | 0 (0.0%) | 1 (20.0%) | 12 (18.2%) |
| Intermediate | 8 (25.81%) | 2 (10.53%) | 0 (0.0%) | 0 (0.0%) | 10 (15.2%) | |
| Resistant | 16 (51.61%) | 13 (68.42%) | 11 (100.0%) | 4 (80.0%) | 44 (66.6%) | |
| FOX | Sensitive | 21 (67.74%) | 10 (52.63%) | 4 (36.36%) | 4 (80.0%) | 39 (59.1%) |
| Intermediate | 4 (12.91%) | 5 (26.32%) | 3 (27.28%) | 0 (00.0%) | 12 (18.2%) | |
| Resistant | 6 (19.35%) | 4 (21.05%) | 4 (36.36%) | 1 (20.0%) | 15 (22.7%) | |
| AK | Sensitive | 13 (41.93%) | 10 (52.63%) | 8 (72.73%) | 4 (80.0%) | 35 (53%) |
| Intermediate | 8 (25.81%) | 5 (26.32%) | 1 (9.09%) | 1 (20.0%) | 15 (22.7%) | |
| Resistant | 10 (32.26%) | 4 (21.05%) | 2 (18.18%) | 0 (00.0%) | 16 (24.2%) | |
| SXT | Sensitive | 7 (22.58%) | 6 (31.58%) | 0 (00.0%) | 0 (0.0%) | 13 (19.7%) |
| Intermediate | 3 (9.68%) | 4 (21.05%) | 4 (36.36%) | 1 (20.0%) | 12 (18.2%) | |
| Resistant | 21 (67.74%) | 9 (47.37%) | 7 (63.64%) | 4 (80.0%) | 41 (62.1%) | |
| TE | Sensitive | 3 (9.68%) | 5 (26.32%) | 0 (00.0%) | 4 (80.0%) | 12 (19.2%) |
| Intermediate | 0 (00.0%) | 3 (15.79%) | 0 (00.0%) | 0 (00.0%) | 3 (4.5%) | |
| Resistant | 28 (90.32%) | 11(57.89%) | 11 (100.0%) | 1 (20.0%) | 51 (77.3%) | |
| AMC | Sensitive | 5 (16.13%) | 2 (10.53%) | 1 (9.09%) | 0 (00.0%) | 8 (12.1%) |
| Intermediate | 1 (3.22%) | 5 (26.32%) | 7 (63.64%) | 5 (100.0%) | 18 (27.3%) | |
| Resistant | 25 (80.65%) | 12 (63.15%) | 3 (27.27%) | 0 (00.0%) | 40 (60.6%) | |
sulfamethoxazole -Trimethoprim, (SXT); Amikacin (AK); Cefoxitin (FOX); Ceftriaxone (CRO); Meropenem (MEM); Tetracycline (TE); Ceftazidime (CAZ); Amoxicillin-clavulanate (AMC)
Β-lactamases identification
ESBL production was identified phenotypically in 40/66 (60.6%) of isolates, however molecular examination of ESBL genes revealed that only 24 isolates harbored genes. blaTEM gene was detected in 24/40 (60%), and blaCTX-M-15 was detected in 15/40 (37.5%) of ESBL-producers. Co-carriage of TEM and CTX-M-15 genes were occurred in 15/40 (37.5%), however none of the isolates was carrying SHV gene. Multiplex PCR was performed to detect AmpC β-lactamase genes in cefoxitin resistant isolates (15). Only one of them harbored one gene of AmpC β-lactamase genes (DHA gene) 1/15 (6.6%). The distribution of resistance genes among DEC types are presented in (Table 4).
Table 4.
The distribution of resistance genes among DEC types
| E. coli types | N |
TEM gene |
SHV gene |
CTX-M-15 gene |
AmpC genes |
|---|---|---|---|---|---|
| DEC | 66 | 24 (36.4%) | – | 15 (22.7%) | – |
| EAEC | 31 | 13 (41.9%) | – | 7 (22.6%) | – |
| tEPEC | 19 | 2 (10.5%) | – | 2 (10.5%) | – |
| aEPEC | 11 | 4 (36.4%) | – | 3 (27.2%) | – |
| Co-infection | 5 | 5 (100%) | – | 3 (60%) | 1 (20%) DHA gene |
EAEC enteroaggregative E. coli, tEPEC typical enteropathogenic E. coli, aEPEC A typical enteropathogenic E. coli
Discussion
Diarrheagenic E. coli (DEC (pathotypes are responsible for most of acute diarrhoeal episodes in children < 5 years old in developing countries [3]. The travel of persons from one country to another or even within the same region increases the risk of DEC transmission. So, knowledge about the epidemiology of these infections in each region or country is very important for health policy makers [30]. Data about DEC incidences from Egypt and North Africa using molecular methods is scarce. Therefore, the findings of the present study may facilitate further epidemiological and therapeutic prospects. DEC prevalence in this study (20.6%) agrees with other studies elsewhere [3, 5]. However lower prevalence was also reported in Nigeria (12.8%) [31] and Libya (8.6%) [4]. The prevalence of DEC among children aged< 1 year, aged 1–2 years and aged 2–5 years old were 24.2, 33.4 and 42.4% respectively. These findings agree with Zhou et al., [32], who reported that (52.0%) of cases were less than 2 years old and 48% aged from 2 to 5 years old [32]. EAEC was the most frequent isolated pathotype in the present study (47%) followed by EPEC, that agrees with several previous studies in other developing countries [3, 4, 30, 33, 34]. For many decades, tEPEC was more common than aEPEC, particularly in poor areas [35], however, the epidemiology of EPEC infections has shifted. In the last decade, aEPEC has become more frequent in high-income and also in developing countries [36, 37]. Less studies still report tEPEC as more prevalent than aEPEC and as an important cause of diarrhoea [32, 38], that agrees with our results where (28.8%) of isolates were identified as tEPEC and (16.6%) of isolates were identified as aEPEC. Occurrence of co-infection was also found in our study, where (5%) of isolates had co-infection (4 isolates “tEPEC + EAEC” and 1 isolate “aEPEC + EAEC”). Isolation of multiple enteric pathogens from the same patient in developing countries is not rare, thus, co-infection was also detected previously in several countries [32, 38].
Although ETEC was detected in several previous studies [3, 34], our study could not detect it. However, our finding is comparable with some previous studies in Egypt and other African countries, which reported that ETEC was poorly detected (3.2%) [30, 34]. EHEC and EIEC were also not detected in the current study that agrees with several previous studies in Egypt and other countries [34, 3, 4]. Several factors may lead to such differences in the prevalence rates of DEC pathotypes found in the present and previous studies including geographical locations, study subjects, and standard of sanitation. The incidence of different DEC pathotypes was not uniform in all age groups of the current study. EPEC was more common in children under 2 years old, while EAEC was more in children aged from 2 to 5 years old. This is consistent with previous studies reported that, EPEC is among the most important pathogens infecting children under 2 years old in the developing countries [3, 39]. The distribution of phylogenetic groups among the studied DEC pathotypes revealed the predominance of group A (47%) followed by B2 (43. 9%) then D (9.1%), however, group B1was not present among the study isolates. Phylogenetic group B2 was the most frequent among EAEC strains, that disagrees with a previous Egyptian study recorded group D as the predominant group in DEC and also in EAEC strains [37]. In EPEC strains of the present study, phylogroup A was the commonest followed by B2 and lastly group D, that disagrees with Bozcal et al., [40], who reported group D as the most prevalent group among EPEC strains [40]. Okeke et al., [41], reported that EAEC strains were mostly belonging to phylogenetic groups A, B1, D and only 4.7% of them belonged to group B2 [41]. This difference in phylogenetic groups among our EAEC isolates and other studies could be due to different ancestral origins of EAEC in each area. DEC strains can be reservoirs for antibiotic resistance genes [42]. Multiple drug resistance was detected in 51% of the current study isolates. The observed resistance might be a result of antimicrobials abuse, which is common in low-income countries, including Egypt [43]. Supporting this hypothesis, most of strains in the current study were resistant to the commonly used antibiotics in the study area, such as 3rd generation cephalosporins, that agrees with several previous studies [44]. Resistance to 3rd generation cephalosporins is caused mainly by ESBL production. In the current study ESBL production was identified phenotypically in 40/66 (60.6%) of DEC isolates, however molecular examination of ESBL genes revealed that blaTEM gene was detected in 24/66(36.3%), and blaCTX-M-15 was detected in 15/66 (22.7%) of DEC isolates. Co-carriage of blaTEM and blaCTX-M-15 genes was detected in 15/66 (22.7%). The frequencies of both genes in different pathotypes were similar. However, none of the isolates was carrying SHV gene. These findings disagree with Khoshvaght et al., [45], who reported high frequencies of blaTEM and blaCTX-M genes in EAEC isolates (78.9 and 63.1% respectively) [45]. Our study disagrees also with Ali et al., who reported that, more than 78% of EAEC and none of the EPEC isolates harbored ESBL genes [34]. However, Zhou et al., have reported a very high percentage (93.3%) of positive ESBL genes [32]. Out of cefoxitin resistant isolates, only one AmpC β-lactamase gene (DHA gene) was detected 1/15 (6.6%), that was higher than a previous report in Egypt [46].
Conclusion
This study provides updated data about the prevalence of DEC Pathotypes, which can help future epidemiological studies on DEC in Egypt and North Africa. The current findings can also help in setting appropriate diagnostic and therapeutic strategies against DEC pathotypes. EAEC and EPEC are important causative agents of diarrhoea in young Egyptian children, with the majority of isolates belong to phylogenetic groups A and B2. The emergence of Multiple-drug resistance among DEC strains has the potential to be a big public health concern in Egypt, particularly ESBL producers. BlaTEM and blaCTX-M-15 were the major genetic determinants among ESBL producing DEC strains. A very few reports have evaluated the importance of antibiotics for the treatment of DEC; however, our findings highlight the importance of continuous assessment of the resistance profile for the suitable selection of antibiotics.
Acknowledgements
We thank the health care workers in Minia university hospital and Minia general hospital for their cooperation.
Abbreviations
- aEPEC
A typical enteropathogenic E. coli
- bfpA
Bundle-forming pilus major subunit
- DEC
Diarrheagenic Escherichia coli
- EAEC
Enteroaggregative E. coli
- EHEC
Enterohemorrhagic E. coli
- EIEC
Enteroinvasive E. coli
- EPEC
Enteropathogenic E. coli
- ESBL
Extended-spectrum beta-lactamase
- ETEC
Enterotoxigenic E. coli
- LT
Heat labile
- ST
Heat stable
- Stx
Shiga toxin
- tEPEC
Typical enteropathogenic E. coli
Authors’ contributions
RMK designed the study and drafted the manuscript. ZAF, RMK, ESM, and SSA performed the experimental work and analyzed the data. DMM analyzed the clinical data. All authors have approved the final manuscript.
Funding
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Availability of data and materials
All data generated or analyzed during this study are included in this article and supplementary file.
Ethics approval and consent to participate
The study protocol was approved by the Medical Ethics Committee of Minia university Hospital, Faculty of Medicine, Minia, Egypt. Written informed consents were taken from patient’s parent or legal guardians.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO . The Top 10 Causes of Death. Geneva: World Health Organization; 2018. [Google Scholar]
- 2.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26(4):822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canizalez-Roman A, Flores-Villasenor HM, Gonzalez-Nunez E, Velazquez-Roman J, Vidal JE, et al. Surveillance of Diarrheagenic Escherichia coli strains isolated from diarrhea cases from children, adults and elderly at northwest of Mexico. Front Microbiol. 2016;7:1924. doi: 10.3389/fmicb.2016.01924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali MM, Mohamed ZK, Klena JD, Ahmed SF, Moussa TAA, Ghenghesh KS. Molecular characterization of diarrheagenic Escherichia coli from Libya. Am J Trop Med Hyg. 2012;86:866–871. doi: 10.4269/ajtmh.2012.11-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borujerdi SM, Ardakani MR, Elham Rezatofighi SE. Characterization of diarrheagenic Escherichia coli strains associated with diarrhea in children, Khouzestan, Iran. J Infect Dev Ctries. 2018;12(8):649–656. doi: 10.3855/jidc.9538. [DOI] [PubMed] [Google Scholar]
- 6.Kaper JB. Defining EPEC. Rev Microbiol. 1996;27:130–133. [Google Scholar]
- 7.Jensen BH, Olsen KE, Struve C, et al. Epidemiology and clinical manifestations of enteroaggregative Escherichia coli. Clin Microbiol Rev. 2014;27:614–630. doi: 10.1128/CMR.00112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panchalingam S, Antonio M, Hossain A, et al. Diagnostic microbiologic methods in the GEMS-1 case/control study. CID. 2012;55:S294–S302. doi: 10.1093/cid/cis754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/CMR.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wellington EM, Boxall AB, Cross P, Feil EJ, Gaze WH, Hawkey PM, et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect Dis. 2013;13(2):155–165. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 12.Falagas ME, Karageorgopoulos DE. Extended-spectrum beta-lactamase-producing organisms. J Hosp Infect. 2009;73(4):345–354. doi: 10.1016/j.jhin.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 13.Otter JA, Natale A, Batra R, Tosas Auguet O, Dyakova E, Goldenberg SD, Edgeworth JD. Individual- and community-level risk factors for ESBL Enterobacteriaceae colonization identified by universal admission screening in London. Clin Microbiol Infect. 2019;25(10):1259–1265. doi: 10.1016/j.cmi.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 14.CLSI . Performance Standards for Antimicrobial Disk Susceptibility Tests. M012 -A12. Wayne: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 15.Jacoby GA, Medeiros AA. More extended Spectrum beta lactamases. Antimicrob Agents Chemother. 1991;35(9):1697–1704. doi: 10.1128/AAC.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polsfuss S, Bloemberg GV, Giger J, Meyer V, Bottger EC, Hombach M. Practical approach for reliable detection of AmpC beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol. 2011;49(8):2798–2803. doi: 10.1128/JCM.00404-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS One. 2015;10:e0123690. doi: 10.1371/journal.pone.0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt H, Knop C, Franke S, Aleksic S, Heesemann J, Karch H. Development of PCR for screening of enteroaggregative Escherichia coli. J Clin Microbiol. 1995;33:701–705. doi: 10.1128/JCM.33.3.701-705.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid SD, Betting DJ, Whittam TS. Molecular detection and identification of intimin alleles in pathogenic Escherichia coli by multiplex PCR. J Clin Microbiol. 1999;37:2719–2722. doi: 10.1128/JCM.37.8.2719-2722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunzburg ST, Tornieporth NG, Riley LW. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J Clin Microbiol. 1995;33(5):1375–1377. doi: 10.1128/JCM.33.5.1375-1377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stacy-Phipps S, Mecca JJ, Weiss JB. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J Clin Microbiol. 1995;33:1054–1059. doi: 10.1128/JCM.33.5.1054-1059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sethabutr O, Venkatesan M, Murphy GS, Eampokalap B, Hoge CW, Echeverria P. Detection of Shigella and enteroinvasive Escherichia coli by amplification of the invasion plasmid antigen H DNA sequence in patients with dysentery. J Infect Dis. 1993;167:458–461. doi: 10.1093/infdis/167.2.458. [DOI] [PubMed] [Google Scholar]
- 24.Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2,eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36:598–602. doi: 10.1128/JCM.36.2.598-602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khairy RM, Mohamed ES, Abdel Ghany HM, Abdelrahim SS. Phylogenic classification and virulence genes profiles of uropathogenic E. coli and diarrhegenic E. coli strains isolated from community acquired infections. PLoS One. 2019;14(9):e0222441. doi: 10.1371/journal.pone.0222441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paterson DL, Hujer KM, Hujer AM, Yeiser B, Bonomo MD, Rice LB, et al. Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type β-lactamases. Antimicrob Agents Chemother. 2003;47:3554–3560. doi: 10.1128/AAC.47.11.3554-3560.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muzaheed DY, Adams-Haduch JM, Endimiani A, Sidjabat HE, Gaddad SM, Paterson DL. High prevalence of CTX-M-15–producing Klebsiella pneumoniae among inpatients and outpatients with urinary tract infection in southern India. J Antimicrob Chemother. 2008;61(6):1393–1394. doi: 10.1093/jac/dkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Youssef MM, Rizk HA, Hassuna NA. Phenotypic and genotypic characterization of extended-Spectrum β-lactamase-producing Enterobacteriaceae in asymptomatic Bacteriuria in pregnancy. Microb Drug Resist. 2019;25(5):731–738. doi: 10.1089/mdr.2018.0088. [DOI] [PubMed] [Google Scholar]
- 30.Konaté A, Dembélé R, Kagambèga A, et al. Molecular characterization of Diarrheagenic Escherichia Coli in children less than 5 years of age with diarrhea in Ouagadougou, Burkina Faso. Eur J Microbiol Immunol (Bp) 2017;7(3):220–228. doi: 10.1556/1886.2017.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ifeanyi CI, Ikeneche NF, Bassey BE, Al-Gallas N, Ben Aissa R, Boudabous A. Diarrheagenic Escherichia coli pathotypes isolated from children with diarrhea in the federal capital territory Abuja, Nigeria. J Infect Dev Ctries. 2015;9(2):165–174. doi: 10.3855/jidc.5528. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y, Zhu X, Hou H, et al. Characteristics of diarrheagenic Escherichia coli among children under 5 years of age with acute diarrhea: a hospital-based study. BMC Infect Dis. 2018;18:63. doi: 10.1186/s12879-017-2936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saka HK, Dabo NT, Muhammad B, García-Soto S. Ugarte-Ruiz M and Alvarez J Diarrheagenic Escherichia coli Pathotypes from children younger than 5 years in Kano state, Nigeria. Front Public Health. 2019;7:348. doi: 10.3389/fpubh.2019.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ali MMM, Ahmed SF, Klena JD, Mohamed ZK, Moussa TAA, Ghenghesh KS. Enteroaggregative Escherichia coli in diarrheic children in Egypt: molecular characterization and antimicrobial susceptibility. J Infect Dev Ctries. 2014;8(5):589–596. doi: 10.3855/jidc.4077. [DOI] [PubMed] [Google Scholar]
- 35.Sarantuya J, Nishi J, Wakimoto N, Erdene S, Nataro JP, Sheikh J, et al. Typical enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in Mongolian children. J Clin Microbiol. 2004;42:133–139. doi: 10.1128/JCM.42.1.133-139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buss SN, Leber A, Chapin K, et al. Multicenter evaluation of the bio fire film Array gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol. 2015;53:915–925. doi: 10.1128/JCM.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foster MA, Iqbal J, Zhang C, McHenry R, Cleveland BE, Romero-Herazo Y, Fonnesbeck C, Payne DC, Chappell JD, Halasa N, et al. Enteropathogenic and enteroaggregative E. coli in stools of children with acute gastroenteritis in Davidson County, Tennessee. Diagn Microbiol Infect Dis. 2015;83(3):319–324. doi: 10.1016/j.diagmicrobio.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh T, Das S, Ramachandran VG, Dar SA, Snehaa K, Saha R, Shah D. Spectrum of diarrhoeagenic Escherichia coli in paediatric population suffering from diarrhoea and as commensals in healthy children. Indian J Med Microbiol. 2017;35:204–210. doi: 10.4103/ijmm.IJMM_16_21. [DOI] [PubMed] [Google Scholar]
- 39.Nakhjavani FA, Emaneini M, Hosseini H, et al. Molecular analysis of typical and atypical enteropathogenic Escherichia coli (EPEC) isolated from children with diarrhoea. J Med Microbiol. 2013;62:191–195. doi: 10.1099/jmm.0.046516-0. [DOI] [PubMed] [Google Scholar]
- 40.Bozcal E, Eldem V, Aydemir S, Skurnik M. The relationship between phylogenetic classification, virulence and antibiotic resistance of extraintestinal pathogenic Escherichia coli in Izmir province, Turkey. PeerJ. 2018;6:e5470. doi: 10.7717/peerj.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okeke IN, Wallace-Gadsden F, Simons HR, Matthews N, Labar AS, Hwang J, et al. Multi-locus sequence typing of enteroaggregative Escherichia coli isolates from Nigerian children uncovers multiple lineages. PLoS One. 2010;5:e14093. doi: 10.1371/journal.pone.0014093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sommer MO, Church GM, Dantas G. The human microbiome harbors a diverse reservoir of antibiotic resistance genes. Virulence. 2010;1:299–303. doi: 10.4161/viru.1.4.12010. [DOI] [PubMed] [Google Scholar]
- 43.Ibrahim OM, Saber-Ayad M. Antibiotic misuse in different hospital wards (a pilot study in an Egyptian hospital) Asian J Pharm Clin Res. 2012;5:95–97. [Google Scholar]
- 44.Shahraki Zahedani SH, Sayadzai N. Frequency and antibiotic resistance pattern of Diarrheagenic Escherichia coli (DEC) strains isolated from children aged less than 10 years. Med Lab J. 2018;12(2):7–12. doi: 10.29252/mlj.12.2.7. [DOI] [Google Scholar]
- 45.Khoshvaght H, Haghi F, Zeighami H. Extended spectrum betalactamase producing Enteroaggregative Escherichia coli from young children in Iran. Gastroenterol Hepatol Bed Bench. 2014;7(2):131136. [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Agamy MH, El Mahdy TS, Shibl AM. Fecal Colonization with Extended-Spectrum Beta-Lactamase and AmpC-Producing Escherichia coli. Biomed Res Int. 2016;3704150:7. doi: 10.1155/2016/3704150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article and supplementary file.