Abstract
Background
An increasing number of studies have focused on the association between leptin, adiponectin levels and the risk as well as the prognosis of hepatocellular carcinoma. However, the reported results are conflicting.
Methods
A meta-analysis was performed to assess the correlation between leptin, adiponectin levels and risk and prognosis of hepatocellular carcinoma (CRD42020195882). Through June 14, 2020, PubMed, Cochrane Library and EMBASE databases were searched, including references of qualifying articles. Titles, abstracts, and main texts were reviewed by at least 2 independent readers. Stata 16.0 was used to calculate statistical data.
Results
Thirty studies were included in this meta-analysis and results showed that hepatocellular carcinoma group had significantly higher leptin levels than the cancer-free control group (SMD = 1.83, 95% CI (1.09, 2.58), P = 0.000), the healthy control group (SMD = 4.32, 95% CI (2.41, 6.24), P = 0.000) and the cirrhosis group (SMD = 1.85, 95% CI (0.70, 3.01), P = 0.002). Hepatocellular carcinoma group had significantly higher adiponectin levels than the healthy control group (SMD = 1.57, 95% CI (0.37, 2.76), P = 0.010), but no statistical difference compared with the cancer-free control group (SMD = 0.24, 95% CI (− 0.35, 0.82), P = 0.430) and the cirrhosis group (SMD = − 0.51, 95% CI (− 1.30, 0.29), P = 0.213). The leptin rs7799039 polymorphism was associated with increased risk of hepatocellular carcinoma (G vs A: OR = 1.28, 95% CI (1.10, 1.48), P = 0.002). There were linear relationships between adiponectin levels and the risk of hepatocellular carcinoma (OR = 1.066, 95% CI (1.03, 1.11), P = 0.001). In addition, the results showed that high/positive expression of adiponectin was significantly related to lower overall survival in hepatocellular carcinoma patients (HR = 1.70, 95% CI (1.22, 2.37), P = 0.002); however, there was no significantly association between the leptin levels and overall survival (HR = 0.92, 95% CI (0.53, 1.59), P = 0.766).
Conclusion
The study shows that high leptin levels were associated with a higher risk of hepatocellular carcinoma. Adiponectin levels were proportional to hepatocellular carcinoma risk, and were related to the poor prognosis.
Keywords: Leptin, Adiponectin, Hepatocellular carcinoma, Meta-analysis
Introduction
Hepatocellular carcinoma (HCC) accounts for the sixth most common cancer in the world and the fourth leading cause of cancer death globally [1], and has highly invasive and aggressive behavior [2]. Generally, HCC is secondary to liver cirrhosis or viral hepatitis. About 80% of patients newly infected with the virus will develop chronic infections, and 10–20% of them will develop liver cirrhosis within 20–30 years, and 1–5% of them will meet advanced liver cancer [3]. Studies have further identified obesity, particularly abdominal obesity, as a potential risk factor for HCC. Therefore, leptin and adiponectin, as the most plenteous and the best-studied obesity-related adipokines, may play an important role in the development of HCC [4, 5].
Leptin is well known as a regulator, which acts in energy expenditure and food intake. Research on leptin has shown that it not only plays a key role in metabolism, but also mediates tumor development by enhancing tumor angiogenesis, promoting cell proliferation, migration, invasion, and inhibiting tumor cell apoptosis [6]. Leptin acts on receptors expressed in many tissues, including the liver. Adiponectin, also known as AdipoQ, was first described by Arita et al. It has liver-protecting and anti-inflammatory properties [7, 8]. The beneficial effects of AdipoQ have been observed in alcoholic and non-alcoholic liver disease models [9]. Animal studies also proven that AdipoQ has a protective effect in liver cancer and cirrhosis [10, 11], and high AdipoQ levels in patients with cirrhosis and HCC are related to disease progression [12, 13].
In recent years, reports on the correlation between leptin, AdipoQ levels and HCC have gradually increased, and some studies have tried to clarify the role of the two in HCC. Unfortunately, the reported results are conflicting. Therefore, we conducted a meta-analysis to systematically determine the relationship between leptin, AdipoQ levels and HCC risk and prognosis.
Methods
The protocol for this meta-analysis is available in PROSPERO (CRD42020195882).
Literature search strategy
The PubMed, EMBASE, and Cochrane Library Databases were explored on June 14, 2020, limited to the English language. The following terms were searched in [Title/Abstract]: Adiponectin [MeSH], AdipoQ, Adipocyte Complement Related Protein 30 kDa, Adipose Most Abundant Gene Transcript 1, apM 1 Protein, ACRP30 Protein, Adipocyte, C1q and Collagen Domain Containing Protein, Leptin [MeSH], Obese Protein, Obese Gene Product, Ob Gene Product, Ob Protein, Liver Neoplasms [MeSH], Liver Neoplasm*, Hepatocellular Cancer*, Hepatic Cancer*, Liver Cancer*, Cancer of Liver. Besides, the reference lists of included articles were manually searched.
Study selection criteria
Inclusion criteria: (1) studies were focused on the correlation between leptin or AdipoQ levels and HCC risk, and provided full text and complete data in HCC patients and cancer-free controls (CFC), or (2) investigated the correlation between AdipoQ or leptin levels and the prognosis of HCC, and provided sufficient information to get the hazard ratio (HR) and 95% confidence intervals (CIs). Exclusion criteria: conference abstracts, case reports, comments, review, and experimental animal studies were excluded. When studies reported on the same or overlapping patient populations, only the study with the most complete data set and the most rigorous methodology was used.
Data extraction
Basic information from each study were extracted: the first author, year of publication, country, study design, study period, number of subjects, and sample source. Also, the following information was extracted in Table 1: the source of the case group, the source of the CFC group, age, gender, body mass index, measurement indicators, and detection methods. Table 2 summarized the following information: source of cases/controls, matching variables, single nucleotide polymorphisms, genotyping methods, frequency of cases, and control genotypes. In Table 3, the following information was selected: follow-up, measured indicators, detected methods, cut-off values, survival analysis, HR sources, and analysis methods.
Table 1.
Main characteristics of the studies examining the relationship between circulating leptin, adiponectin levels and HCC
| Author, Year, Country | Study design | Study period | the source of case group | the source of Cancer-free control group | Number | Mean age | Males | BMI | Sample source | Measured indicators | Detected method | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Abouzied, 2017, Egypt | C | – | HCC | Healthy controls | 25/25 | 57.7/29.2 | 18/23 | 21.7/22.2 | Serum | Leptin | ELISA | 5 |
| Aleksandrova, 2014, Europe | N | 2000–2006 | HCC | Healthy controls | 125/250 | 60.1/60.1 | 85/171 | 28.1/26.9 | Serum | Leptin and AdipoQ | ELISA | 8 |
| Ataseven, 2006, Yurkey | C | – | HBV-related HCC | HBV-related cirrhosis/ Healthy controls | 22/23/25 | 59.8/45.5/37.1 | 15/11/11 | – | Serum | Leptin | ELISA | 8 |
| Bakir, 2017, Egypt | S | 03/2016–11/2016 | HCV-related cirrhotic HCC | HCV-related cirrhosis | 50/40 | 53.2/50.7 | 29/25 | 24.5/26.1 | Serum | Leptin | ELISA | 5 |
| Bastard, 2018, France | N | 03/2006–07/2016 | Viral cirrhotic HCC | Viral cirrhosis | 56/96 | 59.8/58.9 | 34/61 | 25.6/25.8 | Serum | Leptin and AdipoQ | ELISA | 7 |
| Chen, CL, 2014, Taiwan | N | 01/1999–12/2004 | HBV-related HCC | Chronic hepatitis B | 187/374 | 52.4/52.2 | 154/311 | – | Plasma | Leptin and AdipoQ | ELISA | 8 |
| Chen, MJ, 2012, Taiwan | C | 01/2009–12/2009 | Viral HCC | Healthy controls | 65/165 | 58.8/47.7 | 47/112 | 24.7/24.4 | Serum | AdipoQ | RIA | 6 |
| Costantini,2013, Italy | C | – | HCV-related HCC | HCV-related cirrhosis / Chronic hepatitis C / Healthy controls | 26/30/30/20 | 70.0/68/63.4/60.9 | 18/14/15/9 | – | Serum | Leptin | ELISA | 8 |
| Feder, 2019, Germany | P | 05/2012–05/2015 | HCC | Healthy controls | 32/49 | – | – | – | Serum | AdipoQ | ELISA | 7 |
| Fukushima, 2010, Japan | P | 1993–2003 | HCV-related HCC | Chronic hepatitis C | 9/27 | 53.0/51.3 | 5/11 | – | Serum | Leptin (RIA) and HMW AdipoQ (ELISA) | 8 | |
| Hamdy, 2015, Egypt | S | 01/2014–12/2014 | HCV-related cirrhotic HCC | HCV-related cirrhosis | 61/29 | 52.3/52.3 | 51/21 | 33.7/36.7 | Serum | AdipoQ | ELISA | 8 |
| Khattab, 2012, Egypt | C | 02/2009–01/2010 | HCC | Chronic hepatitis C / Healthy controls | 147/147/320 | 43.9/41.6/42.9 | 114/115/201 | 24.9/25.1/25.3 | Serum | AdipoQ | ECLI | 6 |
| Kotani, 2009, Japan | N | 1990–1999 | HCC | Healthy controls | 59/334 | 63.5/62.7 | – | – | Serum | AdipoQ | ELISA | 5 |
| Liu, CJ, 2009, Taiwan | S | 01/2002–10/2005 | HBV-related HCC | HBV-related cirrhosis / Chronic hepatitis B/ Healthy controls | 120/40/120/116 | 50.7/50.3/30/53.8 | 100/29/63/67 | – | Serum | AdipoQ | ELISA | 8 |
| Liu, ZW, 2005, China | C | – | HCV-related cirrhotic HCC | HCV-related cirrhosis/ Chronic hepatitis C/ Healthy controls | 2/10/30/30 | 59.5/53.7/41/39.4 | 2/6/17/18 | 23.0/22.7/23.0/23.1 | Serum | Leptin | ELISA | 8 |
| Michikawa, 2013, Japan | N | 1993–2006 | Viral HCC | Chronic viral hepatitis | 90/117 | – | 62/80 | – | plasma | AdipoQ | ELISA | 8 |
| Radwan, 2019, Egypt | S | – | HCC | Chronic hepatitis C | 48/52 | 53.2/52.5 | 26/32 | 25.2/27.7 | Serum | AdipoQ | ELISA | 5 |
| Sadik, 2012, Egypt | C | 01/2008–02/2009 | HCV-related HCC | HCV-related cirrhosis/ Healthy controls | 69/36/21 | 59.1/53.0/55.8 | 43/23/13 | 28.0//27.1/29.1 | Serum | Leptin and AdipoQ | ELISA | 6 |
| Sumie, 2011, Japan | C | 01/1997–10/2007 | HCV-related HCC | Chronic hepatitis C | 97/97 | 67.4/61.2 | 67/67 | 22.5/23.1 | Serum | AdipoQ | ELISA | 7 |
| Voumvouraki, 2011, Greece | C | – | Viral cirrhotic HCC | Viral cirrhosis/ Chronic hepatitis C / Healthy controls | 38/34/44/60 | 62.0/60.0/53.0/−− | 25/12/8/−− | – | Serum | Leptin | ELISA | 8 |
| Wang, 2003, Taiwan | C | 01/2000–12/2000 | Viral cirrhotic HCC | Viral cirrhosis/ Healthy controls | 31/26/25 | 65.0/59.0/65.0 | 31/26/25 | 23.2/23.7/24.4 | Serum | Leptin | RIA | 6 |
P Cohort, S Cross-sectional, C Case-control, N Nested Case-control, HCC Hepatocellular carcinoma, HCV hepatitis C virus, HBV hepatitis B virus, ELISA Enzyme-linked immunosorbent assay, ECLI Electro-chemiluminescence immunoassay, RIA Radioimmunoassay, HMW AdipoQ High Molecular Weight Adiponectin, BMI body mass index, NOS Newcastle-Ottawa Scale
Table 2.
Main characteristics of the studies examining the relationship between leptin, adiponectin gene polymorphism and HCC
| Author, Year, Country | Study design | Case/control | Number | Matching variables | SNP | Samples source | Genotyping methods | Frequency of case genotype | Frequency of control genotype | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|
| Amer, 2017, Egypt | C | HCC/Healthy controls | 150/100 | age, sex and smoking rate | rs7799039 (leptin gene) | blood | PCR-RFLP technique | AA = 60; AG = 69; GG = 21; G allele =111; A allele =189 | AA = 49; AG = 47; GG = 4; G allele =54; A allele =146 | 7 |
| Zhang, 2018, China | C | HCC/Healthy controls | 575/921 | age and sex | rs7799039 (leptin gene) | blood | SNPscan™ | AA = 295; AG = 221; GG = 59; G allele = 339; A allele =811 | AA = 505; AG = 360; GG = 56; G allele = 472; A allele =1370 | 7 |
| Cai, 2013 China | C | HCC/Healthy controls | 200/200 | age and sex | rs1501299 (adiponectin gene) | blood | DNA sequencing | TT = 12; TG = 60; GG = 128; G allele = 316; T allele =84 | TT = 39; TG = 69; GG = 92; G allele = 253; T allele =147 | 8 |
C Case-control, SNP single-nucleotide polymorphisms, PCR-RFLP polymerase chain reaction-restriction fragment length polymorphism, NOS Newcastle-Ottawa Scale
Table 3.
Main characteristics of the studies examining the relationship between adiponectin, leptin levels and the prognosis of HCC
| Author, Year, Country | Study design | Study period | Number | Follow-up (months) | Sample | Measured indicators | Detected method | Cut-off value | Survival analysis | Source of HR | Analytic method | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Siegel, 2014, USA | P | 2008–2012 | 140 | 8 | Serum | AdipoQ; Leptin | RIA |
≥13.05 μg/ml vs<13.05 μg/ml; ≥7.9 ng/ml vs<7.9 ng/ml |
OS | Report | M;U | 7 |
| Shen, 2016, USA | P | 2008–2014 | 135 | 84 | Plasma | AdipoQ | ELISA | ≥13.10 μg/ml vs<13.10 μg/ml | OS | Report | M | 7 |
| Shin, 2014, Korea | P | 1996–2001 | 75 | 82.5 | Cytoplasmic | AdipoQ | IHC | Positive: > 0% of cells stained vs Negative: no cells stained | OS | Report | M | 6 |
| Watanabe, 2011, Japan | P | 2006–2008 | 33 | 39 | Plasma | Leptin | NA | ≥5.0 ng/ml vs<5.0 ng/ml | DFS | Report | M | 7 |
| Wang, 2014, Taiwan | P | 1999–2003 | 85 | NA | Cytoplasmic | AdipoQ | IHC | Low: - ~ + vs High: ++ ~ +++a | OS | SC | M | 7 |
| Wang, 2006, Taiwan | P | 1994–2003 | 68 | 31.7 | Cytoplasmic | Leptin | IHC | Low: - ~ + vs High: ++ ~ +++a | OS | SC | M | 6 |
ELISA Enzyme-linked immunosorbent assay, RIA Radioimmunoassay, IHC Immunohistochemistry, NA Not available, DFS Disease-free survival, OS Overall survival, SC Survival curves, U Univariate, M Multivariate. aSemiquantitative scoring system, NOS Newcastle-Ottawa Scale
Quality assessment
The Newcastle-Ottawa Scale was used to assess the quality of non-randomized controlled studies. The following three factors were considered: patient selection, comparability of study groups, and outcome evaluation. The highest score was 9 points, and studies with a score ≥ 7 were considered high-quality [14]. The above steps were independently cross-checked by two researchers (Zhang Lilong, Yuan Qixing), and all differences were handled by the senior author (Wang Weixing).
Statistical analysis
Stata 16.0 was used for statistical analysis. Dichotomous variables and continuous variables were compared by the odds ratio (OR) and standardized mean difference (SMD). The hazard ratio (HR) was calculated to assess the correlation between AdipoQ or leptin expression and the prognosis of HCC. In the case that the study only provided the Kaplan-Meier survival curve, used Engage Digitizer version 2.11 software to extract relevant values from the survival curve and calculate HR (95% CI) [15, 16]. For all analyses, the 95% confidence interval (CI) was used. P<0.05 was defined as statistically significant. The chi-squared test was calculated to evaluate the statistical heterogeneity between different studies. P > 0.1 and I2 < 50% indicated low heterogeneity where a fixed-effect model was used; otherwise, the random-effect model was adopted.
Furthermore, a 2-stage dose-response meta-analysis was performed to explore the association between different categories of leptin, AdipoQ levels and HCC risk [17, 18]. 1) The fixed-effect nonlinear model was constructed based on the restrictive cubic spline function (Knot =3). 2) According to the results of the heterogeneity test and nonlinear correlation test, the corresponding model was adopted.
Finally, for indicators with high heterogeneity, we conducted a sensitivity analysis by leaving one out method to check the robustness of the results and determine the source of heterogeneity. Besides, meta-regression analysis was performed to explore the potential sources of heterogeneity. For indicators containing more than 10 articles, we generated a funnel plot to visually check publication bias. Begg’s and Egger’s tests were also performed to quantitatively analyze publication bias, and P < 0.05 was considered statistically significant. If necessary, we verify the results of the publication bias by establishing a trim and fill funnel plot.
Results
Studies retrieved and characteristics
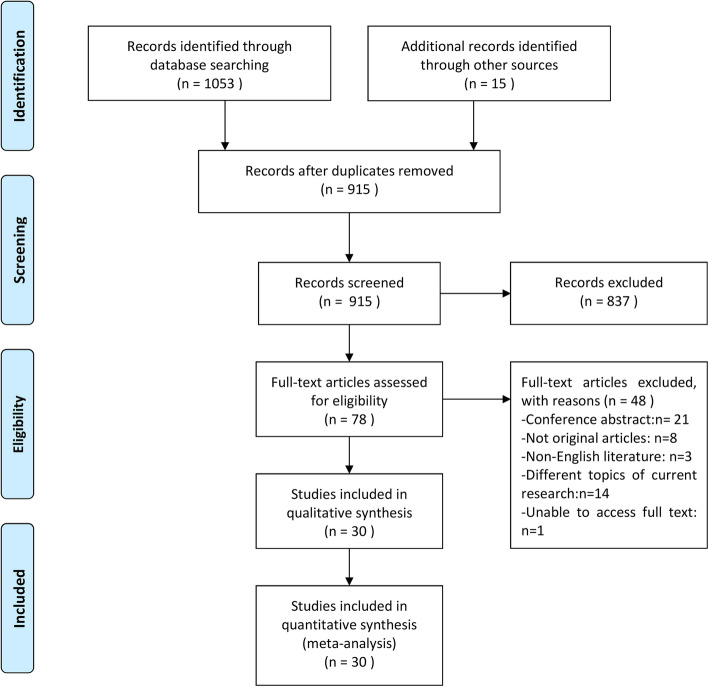
In this meta-analysis, we identified 1068 potentially eligible records and screened their titles and abstracts for inclusion. After reading the full text of 78 records in detail, 30 studies met our inclusion criteria (Fig. 1). Although Ebrahim’s article fits the research topic, it is excluded because the full text is not available [19]. Twenty-one articles [20–40] (10 case-control, 5 nested case-control, 4 cross-sectional, and 2 cohort studies) evaluated the relationship between Leptin or AdipoQ levels and HCC risk, and main characteristics were summarized in Table 1. Three case-control studies [41–43] assessed the relationship between Leptin or AdipoQ gene polymorphism and HCC risk, and main characteristics were reported in Table 2. Six articles [44–49] analyzed the relationship between AdipoQ or leptin expression and the prognosis of HCC, and the main characteristics were summarized in Table 3. Quality assessment of the included studies using the Newcastle–Ottawa scale was shown in Tables 1, 2, 3, and the scores ranged from 5 to 8. Twenty articles were awarded 7 or 8 points, and considered as high-quality; Six studies were awarded 6 points and four studies were awarded 5 points, which were considered as moderate quality.
Fig. 1.
Flow diagram of the meta-analysis search process
Association between circulating leptin levels and HCC risk
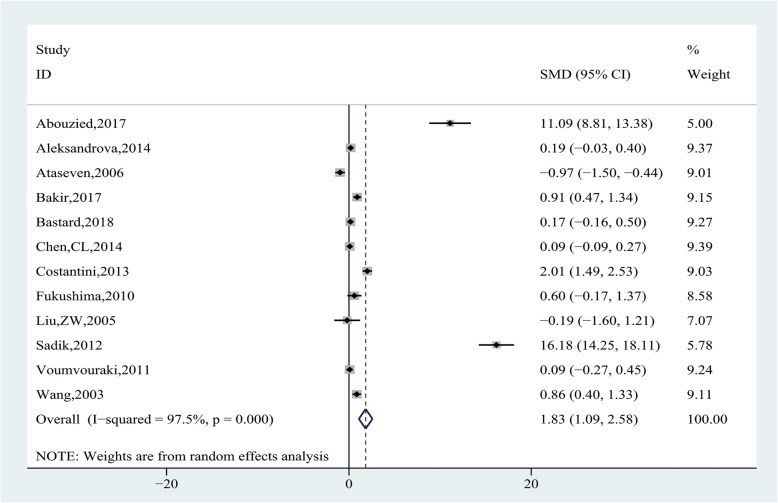
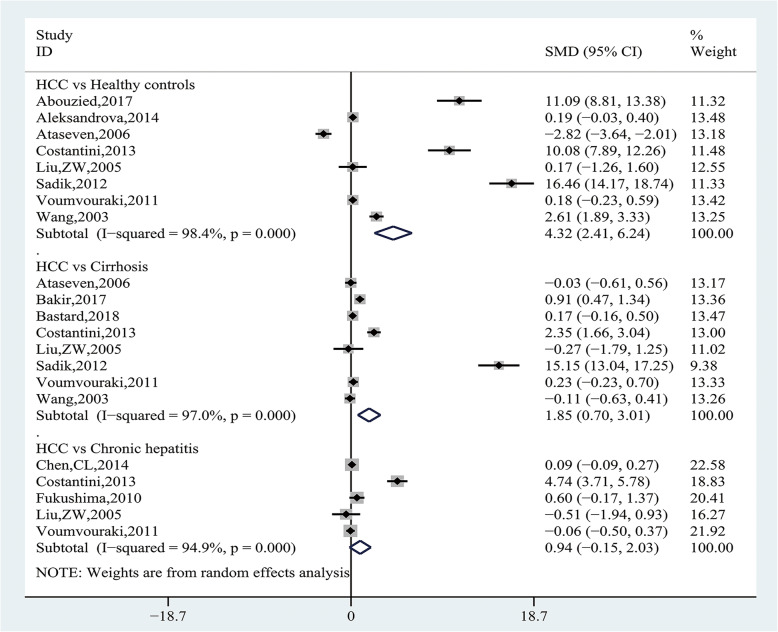
Pooling data of 12 studies [20–25, 27, 29, 34, 37–39] with 1896 participants assessed the association between leptin levels and HCC risk. Heterogeneity analysis showed that significant heterogeneity was observed among the studies (I2 = 97.5%, P = 0.000), and the random-effect model was applied. The results showed that leptin levels were significantly higher in the HCC group than CFC group (SMD = 1.83, 95% CI (1.09, 2.58), P = 0.000) (Fig. 2). Subgroup analysis, according to the source of CFC group, showed HCC group had significantly higher leptin levels than the healthy control group (SMD = 4.32, 95% CI (2.41, 6.24), P = 0.000) and the cirrhosis group (SMD = 1.85, 95% CI (0.70, 3.01), P = 0.002), but there was no statistical difference when compared with the chronic hepatitis group (SMD = 0.94, 95% CI (− 0.1, 2.03), P = 0.090) (Fig. 3 and Table 4). We further conducted subgroup analysis by the source of case group, and the results showed HCV-related cirrhotic HCC had significantly higher leptin levels than HCV-related cirrhosis (SMD = 0.82, 95% CI (0.40, 1.24), P = 0. 0.000), whereas there was no difference in other subgroups (Table 4).
Fig. 2.
Forest plot of comparing circulating leptin levels between the HCC and cancer-free control group
Fig. 3.
Forest plot of the subgroup analyses concerning circulating leptin levels
Table 4.
Subgroup analysis of the association between circulating leptin levels and HCC risk
| Variable | Included studies | Test of association | Test of heterogeneity | ||||
|---|---|---|---|---|---|---|---|
| SMD | 95%CI | P-value* | Modal | P-value | I2 | ||
| HCC vs Healthy controls | |||||||
| Overall | [23–25, 30, 37, 40–42] | 4.32 | 2.41, 6.24 | 0.000 | RE | 0.000 | 98.4% |
| HCC(unreported reason) vs Healthy controls | [23, 24] | 5.58 | −5.11, 16.26 | 0.306 | RE | 0.000 | 98.8% |
| Viral cirrhotic HCC vs Healthy controls | [37, 41, 42] | 1.02 | − 0.78, 2.79 | 0.263 | RE | 0.000 | 94.1% |
| HCV-related HCC vs Healthy controls | [30, 37, 40] | 8.87 | − 1.08, 1.82 | 0.081 | RE | 0.000 | 98.7% |
| HCC vs Cirrhosis | |||||||
| Overall | [25–27, 30, 37, 40–42] | 1.85 | 0.70, 3.01 | 0.002 | RE | 0.000 | 97.0% |
| HCV-related cirrhotic HCC vs HCV-related cirrhosis | [26, 37] | 0.82 | 0.40, 1.24 | 0.000 | FE | 0.145 | 53.0% |
| HCV-related HCC vs HCV-related cirrhosis | [30, 40] | 8.71 | −3.84, 21.25 | 0.174 | RE | 0.000 | 99.2% |
| Viral cirrhotic HCC vs Viral cirrhosis | [27, 41, 42] | 0.13 | −0.11,0.37 | 0.302 | FE | 0.591 | 0 |
| HCC vs Chronic hepatitis | |||||||
| Overall | [28, 30, 32, 37, 41] | 0.94 | −0.15, 2.03 | 0.090 | RE | 0.000 | 94.9% |
| HCV-related HCC vs Chronic hepatitis C | [30, 32, 37] | 1.63 | −1.39, 4.65 | 0.290 | RE | 0.000 | 96.0% |
| Viral cirrhotic HCC vs Chronic hepatitis C | [37, 41] | −0.10 | − 0.51,0.32 | 0.643 | FE | 0.561 | 0 |
| Ethnicity | |||||||
| Asian | [25, 28, 32, 37, 42] | 0.10 | −0.50, 0.70 | 0.751 | RE | 0.000 | 85.6% |
| Caucasian | [24, 27, 30, 41] | 0.58 | −0.06, 1.22 | 0.077 | RE | 0.000 | 93.3% |
| African | [23, 26, 40] | 9.36 | −1.27, 19.99 | 0.084 | RE | 0.000 | 99.3% |
| Sample size | |||||||
| < 100 | [23, 25, 26, 32, 37, 42] | 1.57 | 0.22, 2.91 | 0.022 | RE | 0.000 | 95.8% |
| ≥ 100 | [24, 27, 28, 30, 40, 41] | 2.23 | 1.21, 3.26 | 0.000 | RE | 0.000 | 98.4% |
| Mean age | |||||||
| < 60 | [23, 25–28, 32, 37, 40] | 2.87 | 1.57, 4.17 | 0.000 | RE | 0.000 | 98.2% |
| ≥ 60 | [24, 30, 41, 42] | 0.76 | 0.03, 1.49 | 0.040 | RE | 0.000 | 93.7% |
| Study design | |||||||
| Case-control | [23, 25, 30, 37, 40, 41] | 3.81 | 1.83, 5.79 | 0.000 | RE | 0.000 | 97.5% |
| Nested Case-control | [24, 27, 28] | 0.14 | 0.01, 0.26 | 0.035 | FE | 0.777 | 0.0% |
| Assay method | |||||||
| ELISA | [23–28, 30, 37, 40, 41] | 2.13 | 1.27, 2.99 | 0.000 | RE | 0.000 | 97.9% |
| RIA | [32, 42] | 0.79 | 0.39, 1.19 | 0.000 | FE | 0.570 | 0.0% |
| Alanine aminotransferase | |||||||
| < 70 U/L | [23, 26, 28, 32, 37, 40] | 4.42 | 2.26, 6.50 | 0.000 | RE | 0.000 | 98.6% |
| ≥ 70 U/L | [25, 27, 30, 41, 42] | 0.43 | −0.38, 1.23 | 0.296 | RE | 0.000 | 94.4% |
| Albumin | |||||||
| < 3.5 g/dl | [25, 26, 30, 40, 42] | 3.47 | 1.28, 5.66 | 0.002 | RE | 0.000 | 98.7% |
| ≥ 3.5 g/dl | [27, 28, 32, 41] | 0.12 | −0.02, 0.26 | 0.091 | FE | 0.633 | 0.0% |
RE Random-effects model, FE Fixed-effects model, HCC Hepatocellular carcinoma, HCV hepatitis C virus
*Statistically significant results were shown in bold
In addition, we also performed other subgroup analyses and the results were shown in Table 4. Stratification by ethnicity showed no significant difference in the HCC group and CFC group in Asian (SMD = 0.10, 95% CI (− 0.50, 0.70), P = 0.751), Caucasian (SMD = 0.58, 95% CI (− 0.06, 1.22), P = 0.077) and African population (SMD = 9.36, 95% CI (− 1.27, 19.99), P = 0.084). Stratification by sample size showed HCC group had significantly higher leptin levels than CFC group in both small (n < 100) sample numbers (SMD = 1.57, 95% CI (0.22, 2.91), P = 0.022) and large (n ≥ 100) sample numbers (SMD = 2.23, 95% CI (1.21, 3.26), P = 0.000). Stratification by mean age showed HCC group had significantly higher leptin levels than CFC group in both “< 60” (SMD = 2.87, 95% CI (1.57, 4.17), P = 0.000) and “≥ 60” (SMD = 0.76, 95% CI (0.03, 1.49), P = 0.040). Stratification by study design showed HCC group had significantly higher leptin levels than CFC group in both case-control studies (SMD = 3.81, 95% CI (1.83, 5.79), P = 0.000) and nested case-control studies (SMD = 0.14, 95% CI (0.01, 0.26), P = 0.035). Stratification by assay method revealed the HCC group had significantly higher leptin levels than the CFC group by both “ELISA” (SMD = 2.13, 95% CI (1.27, 2.99), P = 0.000) and “RIA” (SMD = 0.79, 95% CI (0.39, 1.19), P = 0.000). Stratification by Alanine aminotransferase (ALT) levels of HCC patients showed HCC group had significantly higher leptin levels than CFC group in “< 70 U/L” (SMD = 4.42, 95% CI (2.26, 6.50), P = 0.000), but not in the “≥ 70 U/L” (SMD = 0.43, 95% CI (− 0.38, 1.23), P = 0.296). Stratification by albumin levels of HCC patients showed HCC group had significantly higher leptin levels than CFC group in “< 3.5 g/dl” (SMD = 3.47, 95% CI (1.28, 5.66), P = 0.002), but not in the “≥ 3.5 g/dl” (SMD = 0.12, 95% CI (− 0.02, 0.26), P = 0.091).
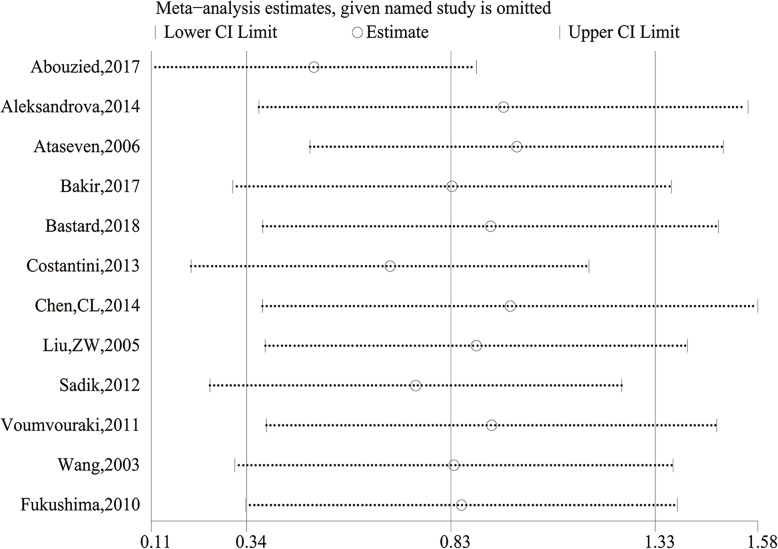
Meta-regression analysis showed that only the ethnicity (P = 0.004), not the source of control (P = 0.242) and case (P = 0.185), sample size (P = 0.735), mean age (P = 0.420), study design (P = 0.344), assay method (P = 0.606), ALT (P = 0.172) and albumin (P = 0.853), had significant impacts on the heterogeneity in the meta-analysis. To assess the impacts of each study on the overall meta-analysis, we carried out sensitivity analysis using the leave-one-out method. No substantial change of data on leptin levels was observed. Therefore, the results of our meta-analysis were relatively stable and credible (Fig. 4).
Fig. 4.
Sensitivity analysis of comparing circulating leptin levels between the HCC and cancer-free control group
A funnel plot representing SMDs of the leptin levels in the HCC group compared to the CFC group was used to evaluate publication bias. Through the visual inspection of the funnel plot, there was obvious asymmetry that indicated a possibility of publication bias (Fig. 5), which were supported by Begg’s tests (P = 0.034) and Egger’s tests (P = 0.025). Therefore, further verification by trim and fill funnel plot was employed to adjust for the potential publication bias. However, the pooled data regarding leptin that had been significant before the adjustment with the “trim and fill” method remained significant after the adjustment (SMD = 3.486, 95% CI (0.937–6.035), P < 0.05), indicating that this publication bias did not affect the pooled estimates.
Fig. 5.
Funnel plot of comparing circulating leptin levels between the HCC and cancer-free control group
Association between circulating AdipoQ levels and HCC risk
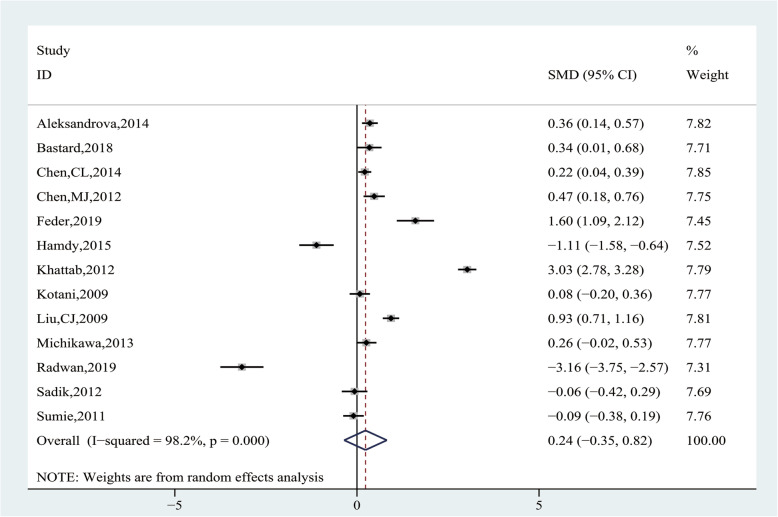
Pooling data of 13 studies [21, 24–26, 28, 30–33, 35–37, 40] with 2092 participants were evaluated on the association between AdipoQ levels and HCC risk. Heterogeneity analysis showed significant heterogeneity among the studies (I2 = 98.2%, P = 0.000), and the random-effect model was applied. There was no statistical difference in HCC and CFC group on AdipoQ levels(SMD = 0.24, 95% CI (− 0.35, 0.82), P = 0.430) (Fig. 6).
Fig. 6.
Forest plot of comparing circulating adiponectin levels between the HCC and cancer-free control group
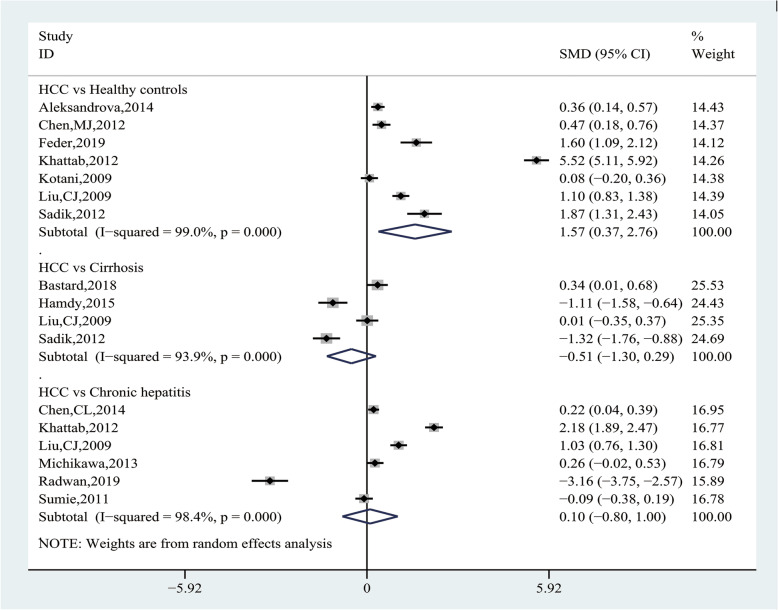
Subgroup analysis, according to the source of CFC group, showed HCC group had significantly higher AdipoQ levels than the healthy control group (SMD = 1.57, 95% CI (0.37, 2.76), P = 0.010), but there was no statistical difference compared with the chronic hepatitis group (SMD = 0.10, 95% CI (− 0.80, 1.00), P = 0.826) and the cirrhosis group (SMD = − 0.51, 95% CI (− 1.30, 0.29), P = 0.213) (Fig. 7 and Table 5). We further conducted subgroup analysis by the source of case group, and the results showed viral HCC had significantly higher AdipoQ levels than healthy controls (SMD = 1.11, 95% CI (0.44, 1.78), P = 0. 0.001), and HCV-related HCC had significantly lower AdipoQ levels than HCV-related cirrhosis (SMD = − 1.22, 95% CI (− 1.54, − 0.90), P = 0. 0.000), whereas there was no difference in other subgroups (Table 5).
Fig. 7.
Forest plot of the subgroup analyses concerning circulating adiponectin levels
Table 5.
Subgroup analysis of the association between circulating adiponectin levels and HCC risk
| Variable | Included studies | Test of association | Test of heterogeneity | ||||
|---|---|---|---|---|---|---|---|
| SMD | 95%CI | P-value* | Modal | P-value | I2 | ||
| HCC vs Healthy controls | |||||||
| Overall | [24, 29, 31, 34–36, 40] | 1.57 | 0.37, 2.76 | 0.010 | RE | 0.000 | 99.0% |
| HCC(unreported reason) vs Healthy controls | [24, 31, 34, 35] | 1.88 | −0.31, 4.08 | 0.092 | RE | 0.000 | 99.5% |
| Viral HCC vs Healthy controls | [29, 36, 40] | 1.11 | 0.44, 1.78 | 0.001 | RE | 0.000 | 90.8% |
| HCC vs Cirrhosis | |||||||
| Overall | [27, 33, 36, 40] | −0.51 | −1.30, 0.29 | 0.213 | RE | 0.000 | 93.9% |
| Viral cirrhotic HCC vs Viral cirrhosis | [27, 33] | −0.37 | −1.80, 1.05 | 0.607 | RE | 0.000 | 95.9% |
| HCV-related HCC vs HCV-related cirrhosis | [33, 40] | −1.22 | −1.54, −0.90 | 0.000 | FE | 0.531 | 0.0% |
| HCC vs Chronic hepatitis | |||||||
| Overall | [28, 34, 36, 38, 39, 43] | 0.10 | −0.80, 1.00 | 0.826 | RE | 0.000 | 98.4% |
| HCC(unreported causes) vs Chronic hepatitis C | [34, 39] | − 0.48 | −5.71, 4.75 | 0.857 | RE | 0.000 | 99.6% |
| HCV-related HCC vs Chronic hepatitis C | [28, 43] | 0.08 | −0.22,0.38 | 0.599 | RE | 0.068 | 70.0% |
| Viral HCC vs Chronic viral hepatitis | [28, 36, 38, 43] | 0.35 | −0.08, 0.78 | 0.108 | RE | 0.000 | 91.8% |
| Ethnicity | |||||||
| Asian | [28, 29, 35, 36, 38, 43] | 0.31 | 0.02, 0.61 | 0.036 | RE | 0.000 | 88.3% |
| Caucasian | [24, 27, 31] | 0.73 | 0.11, 1.35 | 0.022 | RE | 0.000 | 90.3% |
| African | [33, 34, 39, 40] | −0.32 | −2.93, 2.29 | 0.811 | RE | 0.000 | 99.5% |
| Sample size | |||||||
| < 200 | [27, 31, 33, 39, 40, 43] | 0.76 | 0.03, 1.50 | 0.042 | RE | 0.000 | 98.5% |
| ≥ 200 | [24, 28, 29, 34–36, 38] | −0.40 | −1.34, 0.54 | 0.403 | RE | 0.000 | 97.1% |
| Mean age | |||||||
| < 60 | [27–29, 33, 34, 36, 39, 40] | 0.10 | −0.85, 1.05 | 0.833 | RE | 0.000 | 98.8% |
| ≥ 60 | [24, 35, 43] | 0.13 | −0.14, 0.39 | 0.362 | RE | 0.037 | 69.7% |
| Study design | |||||||
| Nested Case-control | [24, 27, 28, 35, 38] | 0.25 | 0.14, 0.36 | 0.000 | EE | 0.585 | 0.0% |
| Case-control | [29, 34, 40, 43] | 0.84 | −0.74, 2.12 | 0.298 | RE | 0.000 | 99.2% |
| Cross-sectional | [33, 36, 39] | −1.10 | −3.46, 1.26 | 0.361 | RE | 0.000 | 99.0% |
| Sample source | |||||||
| Serum | [24, 27, 29, 31, 33–36, 39, 40, 43] | 0.23 | −0.51, 0.97 | 0.540 | RE | 0.000 | 98.5% |
| Plasma | [28, 38] | 0.23 | 0.08, 0.38 | 0.003 | FE | 0.795 | 0.0% |
| Assay method | |||||||
| ELISA | [24, 27, 28, 31, 33, 35, 36, 38–40, 43] | −0.03 | −0.45, 0.40 | 0.901 | RE | 0.000 | 95.7% |
| Non-ELISA | [29, 34] | 1.75 | −0.75, 4.26 | 0.170 | RE | 0.000 | 99.4% |
| Alanine aminotransferase | |||||||
| < 70 U/L | [28, 33, 36, 40, 43] | 0.00 | −0.53, 0.53 | 0.992 | RE | 0.000 | 94.8% |
| ≥ 70 U/L | [27, 34, 39] | 0.08 | −2.96, 3.12 | 0.958 | RE | 0.000 | 99.5% |
| Albumin | |||||||
| < 3.5 g/dl | [33, 34, 40] | 0.62 | −1.98, 3.22 | 0.639 | RE | 0.000 | 98.5% |
| ≥ 3.5 g/dl | [27, 28] | 0.24 | 0.09,0.40 | 0.002 | RE | 0.000 | 99.4% |
RE Random-effects model, FE Fixed-effects model, HCC Hepatocellular carcinoma, HCV hepatitis C virus
*Statistically significant results were shown in bold
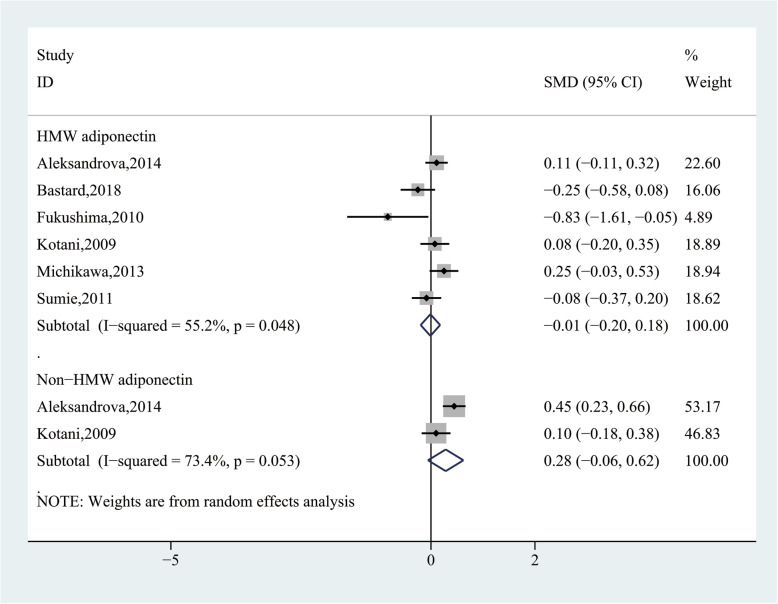
Subgroup analysis, according to the molecular-weight of AdipoQ, showed no significant difference about high-molecular-weight AdipoQ (SMD = − 0.01, 95% CI (− 0.20, 0.18), P = 0.911) and non-high-molecular-weight AdipoQ (SMD = 0.28, 95% CI (− 0.06, 0.62), P = 0.103) levels in the HCC group and CFC group (Fig. 8). In addition, we also performed other subgroup analysis and the results were shown in Table 5. Stratification by ethnicity showed HCC group had significantly higher AdipoQ levels than CFC group in Asian (SMD = 0.31, 95% CI (0.02, 0.61), P = 0.036) and Caucasian population (SMD = 0.73, 95% CI (0.11, 1.35), P = 0.022), but not in African population (SMD = − 0.32, 95% CI (− 2.93, 2.29), P = 0.811). Stratification by sample size showed HCC group had significantly higher AdipoQ levels than CFC group in small (n < 200) sample numbers (SMD = 0.76, 95% CI (0.03, 1.50), P = 0.042), but not in large (n ≥ 200) sample numbers (SMD = − 0.40, 95% CI (− 1.34, 0.54), P = 0.403). Stratification by mean age showed no significant difference in the HCC group and CFC group in both “< 60” (SMD = 0.10, 95% CI (− 0.85, 1.05), P = 0.833) and “≥ 60” (SMD = 0.13, 95% CI (− 0.14, 0.39), P = 0.362). Stratification by study design showed HCC group had significantly higher AdipoQ levels than CFC group in nested case-control studies (SMD = 0.25, 95% CI (0.14, 0.36), P = 0.000), but not in case-control studies (SMD = 0.84, 95% CI (− 0.74, 2.12), P = 0.298), and cross-sectional studies (SMD = − 1.10, 95% CI (− 3.46, 1.26), P = 0.361). Stratification by the sample source revealed HCC group had significantly higher AdipoQ levels than CFC group in the source of plasma (SMD = 0.23, 95% CI (0.08, 0.38), P = 0.003), but not in the source of serum (SMD = 0.23, 95% CI (− 0.51, 0.97), P = 0.540). Stratification by assay method revealed no significant difference in the HCC group and CFC group by both “ELISA” (SMD = − 0.03, 95% CI (− 0.45, 0.40), P = 0.901) and “Non-ELISA” (SMD = 1.75, 95% CI (− 0.75, 4.26), P = 0.170). Stratification by ALT levels of HCC patients showed no significant difference in the HCC group and CFC group in both “< 70 U/L” (SMD = 0.00, 95% CI (− 0.53, 0.53), P = 0.992) and “≥ 70 U/L” (SMD = 0.08, 95% CI (− 2.96, 3.12), P = 0.958). Stratification by albumin levels of HCC patients showed HCC group had significantly higher AdipoQ levels than CFC group in “≥ 3.5 g/dl” (SMD = 0.24, 95% CI (0.09,0.40), P = 0.002), but not in the “< 3.5 g/dl” (SMD = 0.62, 95% CI (− 1.98, 3.22), P = 0.639). In addition, we also found that AdipoQ levels in HCC patients were not related to gender(man vs woman: SMD = − 0.29, 95% CI (− 0.69, 0.11), P = 0.153) and vascular invasion (present vs absent: SMD = 0.19, 95% CI (− 0.11, 0.49), P = 0.208).
Fig. 8.
Forest plot of the subgroup analyses based on the molecular-weight of adiponectin between the HCC and cancer-free control group
Khattab et al. [30] found that AdipoQ levels in HCC with the size of nodules≥5 cm were significantly higher than<5 cm (24.2 ± 2.1 vs 20.8 ± 3.8, P = 0.009), whereas, AdipoQ levels were not related to TNM stages, number of nodules and lymph node metastasis. Feder et al. [27] discovered that AdipoQ levels were no statistical difference in HCC and colorectal liver metastases patients, and negatively related to steatosis grade, but not correlate with inflammation or fibrosis score. Sadik et al.35 reported that AdipoQ levels of cirrhotic HCC were significantly higher than the noncirrhotic HCC group, whereas leptin was not.
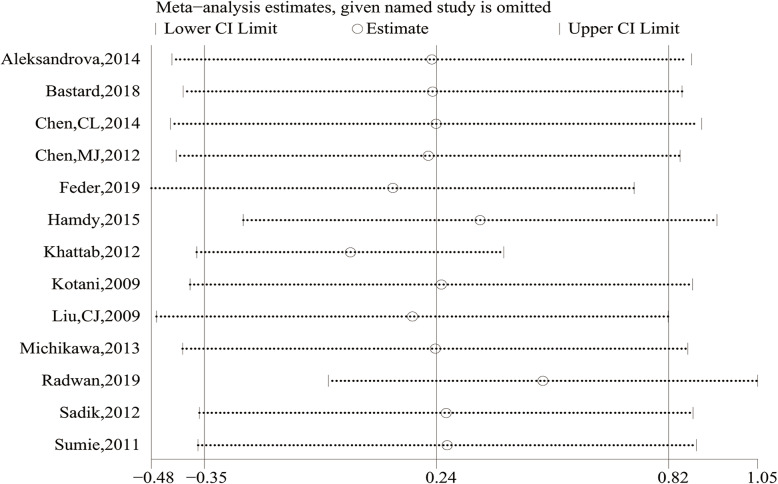
Meta-regression analysis showed that the source of control (P = 0.150) and case (P = 0.579), ethnicity (P = 0.338), sample size (P = 0.140), mean age (P = 0.540), study design (P = 0.283), assay method (P = 0.092), source of sample (P = 0.993), ALT(P = 0.544) and albumin (P = 0.575) had no significant effects on the heterogeneity in the meta-analysis. We also carried out sensitivity analysis using the leave-one-out method, and no substantial change of data on AdipoQ levels were observed, therefore, the results of our meta-analysis were relatively stable and credible (Fig. 9).
Fig. 9.
Sensitivity analysis of comparing circulating adiponectin levels between the HCC and cancer-free control group
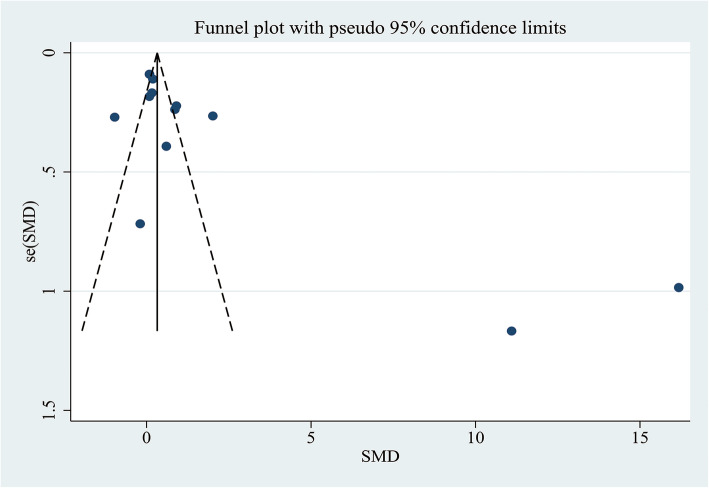
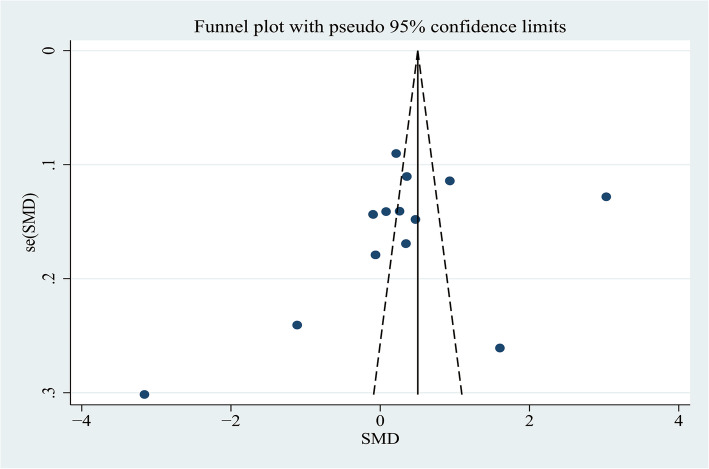
A funnel plot representing SMDs of the AdipoQ levels in the HCC group compared to the CFC group was used to assess publication bias. Through the visual inspection of the funnel plot, there was obvious asymmetry that indicated a possibility of publication bias (Fig. 10), which were not supported by the Begg’s tests (P = 0.300) and Egger’s tests (P = 0.142); therefore, further verification by trim and fill funnel plot was employed to adjust for the potential publication bias. The result of the “trim and fill” method revealed that no trimming was performed and the data was unchanged, suggesting that there was no significant publication bias.
Fig. 10.
Funnel plot of comparing circulating adiponectin levels between the HCC and cancer-free control group
Association between leptin, AdipoQ gene polymorphism and HCC risk
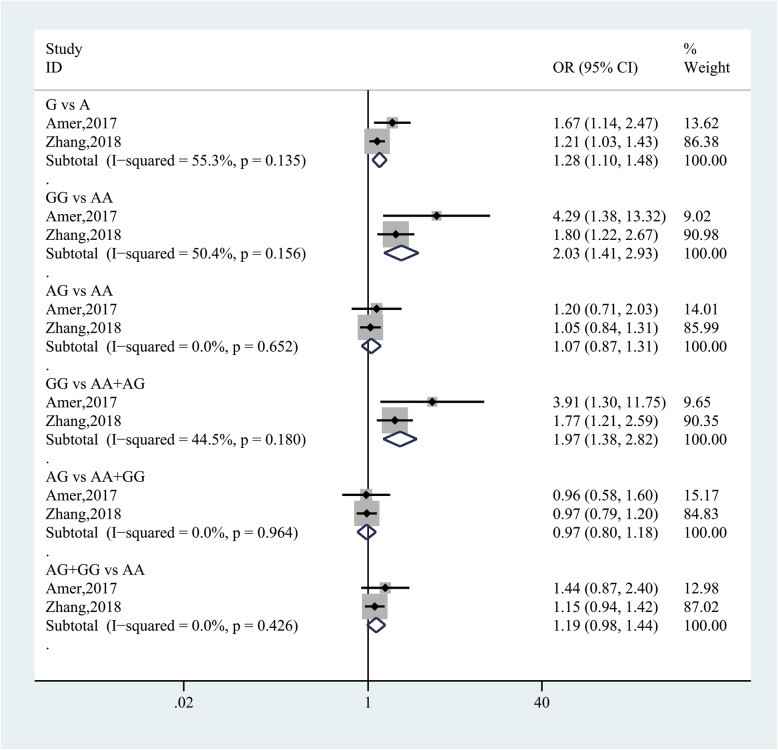
Pooling data from 2 studies [41, 43] with 1746 participants evaluated the association between leptin rs7799039 and HCC risk. In the allele model analysis, the G allele was significantly associated with an increased risk in HCC (G vs A: OR = 1.28, 95% CI (1.10, 1.48), P = 0.002). In the codominant model analysis, the GG genotype was associated with a 2.03-fold elevated risk in HCC (GG vs AA: OR = 2.03, 95% CI (1.41, 2.93), P = 0.000), whereas the AG genotype was not (AG vs AA: OR = 1.07, 95% CI (0.87, 1.31), P = 0.505). In the recessive model analysis, the GG genotype was associated with a 1.97-fold increased risk in HCC (GG vs AA+AG: OR = 1.97, 95% CI (1.38, 2.82), P = 0.000). However, in the overdominant and dominant model analysis, the AG and AG + GG genotypes were no significantly associated with the risk in HCC (AG vs AA+GG: OR = 0.97, 95% CI (0.80, 1.18), P = 0.770; AG + GG vs AA: OR = 1.19, 95% CI (0.98, 1.44), P = 0.078). There was no significant heterogeneity in the above results (Fig. 11).
Fig. 11.
Forest plot for the association between leptin rs7799039 and HCC risk
As for AdipoQ, Cai et al. [42] found that the AdipoQ rs1501299 was associated with the increased susceptibility to HCC, and the additive model showed that the GT and GG genotypes were significantly associated with an increased risk in HCC (GT vs TT: OR = 2.83, 95% CI (1.36, 5.89), P = 0.006; GG vs TT: OR = 4.52, 95% CI (2.25, 9.11), P = 0.001). In the dominant model analysis, the GG + GT genotypes were associated with a 3.8-fold elevated risk in HCC(GG + GT vs TT: OR = 3.795, 95% CI (1.92, 7.49), P = 0.001). However, the rs266729, rs822395, rs822396, and rs2241766 were not significantly associated with HCC. Unfortunately, we just retrieved one study that evaluated the association of AdipoQ gene polymorphism with HCC, so we failed to perform a related meta-analysis.
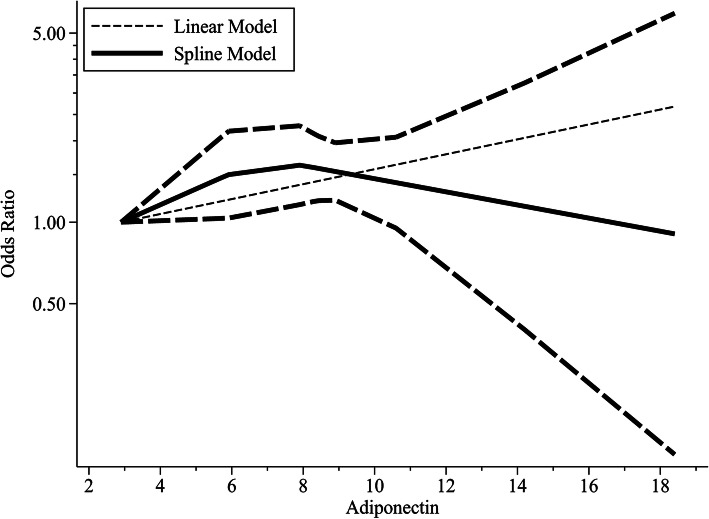
Dose-response of circulating AdipoQ, leptin levels and HCC risk
Pooling data from 4 studies [21, 25, 32, 35] with 1507 participants showed that there was a linear dose-response relationship between circulating AdipoQ levels and HCC risk (Pnon-linearity = 0.233). We defined the increment in 1 μg/ml AdipoQ levels as a unit to show the trend more clearly. The trends were significant for increasing HCC risk per one unit increase of AdipoQ (OR = 1.066, 95% CI (1.03, 1.11), P = 0.001; Fig. 12), without significant heterogeneity (Pheterogeneity = 0.338).
Fig. 12.
Dose-response associations of circulating adiponectin levels and HCC risk
As for leptin, Aleksandrova et al. [21] and Chen et al. [25] both confirmed that circulating leptin levels were no significant dose-response trend in the development of HCC. Unfortunately, we only found two studies, and unable to perform a meta-analysis.
Association between AdipoQ, leptin and survival in HCC
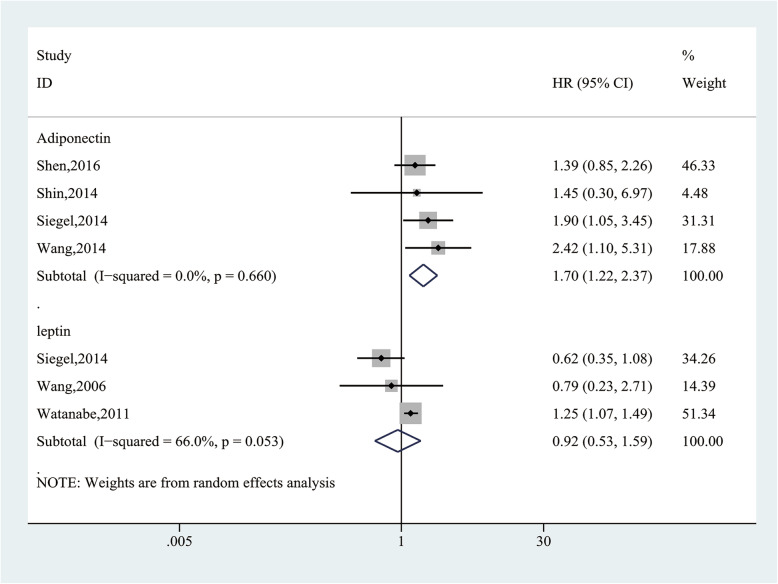
Pooling data of 4 studies [44–46, 48] with 435 participants analyzed the association between AdipoQ expression and survival in HCC. Heterogeneity analysis showed that no heterogeneity was observed among the studies (I2 = 0%, P = 0.660), the fixed-effect model was applied. The results showed that high/positive expression of AdipoQ was significantly related to lower overall survival (OS) in HCC patients (HR = 1.70, 95% CI (1.22, 2.37), P = 0.002; Fig. 13).
Fig. 13.
Forest plot of the relationship between adiponectin, leptin expression and survival in HCC
Pooling data of 3 studies [44, 47, 49] with 241 participants measured the association between leptin expression and prognosis for HCC. Heterogeneity analysis showed that significant heterogeneity was observed among the studies (I2 = 66.0%, P = 0.053), the random-effect model was applied. The results showed that high/positive expression of leptin was not significantly associated with prognosis in HCC patients (HR = 0.92, 95% CI (0.53, 1.59), P = 0.766; Fig. 13).
Discussion
In the past two decades, many studies have explored the relationship between the abnormal expression of leptin and AdipoQ and various obesity-related cancers. In 2016, a meta-analysis of 107 articles [50] was conducted to investigate AdipoQ levels in various malignancies and found that AdipoQ levels in certain cancers (including acute leukemia, multiple myeloma, breast cancer, colorectal cancer, and uterine cancer, endometrial cancer, prostate cancer, thyroid cancer, tongue cancer, and gastroesophageal cancer) were significantly reduced, while HCC was significantly higher than the CFC group. However, the meta-analysis involved only 7 articles on HCC. Song et al. [51] analyzed 9 Chinese and English studies and got the same result. Our results showed that the AdipoQ levels of HCC patients were significantly higher than that of the healthy control group, but there is no significant difference in the AdipoQ levels compared with the CFC group. So far, no meta-analysis on leptin and liver cancer risk has been performed. Our results indicated that the leptin levels of liver cancer patients were significantly higher than that of the CFC group, healthy control group, and liver cirrhosis group. In addition, comparing the different sources of the HCC group and the CFC group, the results were different. Therefore, we can conclude that AdipoQ and leptin levels in patients with chronic hepatitis and cirrhosis have changed compared with healthy controls, which is consistent with Buechler’s conclusion [13].
Because the patients included in this study were recruited from different backgrounds of gender, race, other demographic parameters, and the overall health of the individual, a high degree of heterogeneity was observed in this meta-analysis, and the study results should be interpreted with caution. We performed meta-regression, subgroup analysis, and sensitivity analysis to determine the source of heterogeneity. The meta-regression results of leptin indicated that the heterogeneity came from race, and the subgroup analysis showed that the heterogeneity was related to the source of the control group, study design, assay method, and baseline albumin levels. In the pooled analysis of AdipoQ, the source of heterogeneity was not found by the meta-regression, and the heterogeneity was linked to the control group, study design, average age, and sample source by the subgroup analyses.
Many single nucleotide polymorphisms have been found in the leptin gene. The earliest is the LEP rs7799039 polymorphism, a SNP identified in the 50 untranslated regions of the leptin gene [52]. Research has been conducted on the tumor and recommendations have been made. It may affect the transcription levels and leptin expression [53]. Some previous meta-analysis showed that LEP rs7799039 polymorphism confers cancer risk [54–56]. However, no meta-analysis was performed to explore the association between LEP rs7799039 gene polymorphism and HCC risk. In this study, we found that the LEP rs7799039 polymorphism was related to the susceptibility to HCC. Unfortunately, too few studies were included, and conclusions should be confirmed by higher-quality studies.
Through dose-response, we can more clearly explore the relationship between AdipoQ and leptin and liver cancer risk. In 2019, Yoon et al. [57] found that AdipoQ levels were significantly associated with a decreased risk of breast cancer, colorectal cancer, and endometrial cancer, while leptin was associated with an increased risk of cancers such as endometrial cancer and kidney cancer. However, our results indicated that the increase in AdipoQ levels was of great significance for increasing the risk of HCC. There were only 2 dose-response studies between leptin and HCC risk, so the meta-analysis was abandoned.
Leptin and AdipoQ may not only be closely related to the occurrence of cancer, but closely associated with the prognosis of cancer. Our findings indicated that the high/positive expression of AdipoQ in liver cancer patients was significantly associated with decreased OS, which is similar to a meta-analysis of 10 studies, which revealed that increased AdipoQ expression is associated with poor prognosis in cancer patients (including HCC patients) [58]. It is worth noting that in this meta-analysis, the high/positive expression of leptin was not significantly associated with the prognosis of HCC patients.
High levels of leptin may play an important role in promoting cancer cell migration, proliferation, survival, and angiogenesis [59, 60]. This is achieved by activating the Janus kinase/signal transducer and activator of transcription, phosphatidylinositol 3-kinase, mitogen-activated protein kinase, and extracellular signal-regulated kinase signaling pathway [61, 62], which is considered related to oncogenes [63, 64].. Leptin can also promote the development of liver fibrosis, steatosis, and pro-inflammatory [4, 65]. In addition, Mittenbuhler et al. the leptin signal is found in obesity as the promoter of HCC [66].
Many studies have found that AdipoQ has significant anti-proliferative, anti-cancer activity, and anti-inflammatory effects [67]. Nazmy et al. have found that the tumor suppressor activity of AdipoQ can provide the body with more anti-HCC effects by preventing the decrease of p53 expression, the reactivation of TNF-related apoptosis-inducing ligand signals, and the induction of apoptosis pathways [68]. Al-Gayyar et al. revealed that AdipoQ can completely prevent the increase of HCC-induced sulfatase-2, improve HCC-induced tumor invasion markers, matrix metalloproteinase 9, syndecan-1 and fibroblast growth Factor-2 induced by HCC, decrease the expression of NF-κB and tumor necrosis factor α (TNF-α) induced by HCC, to achieve liver protection [69]. Manieri’s research shows that AdipoQ can activate two proteins in hepatocytes, p38α and AMP-activated protein kinase, which can prevent cell proliferation and damage tumor growth [70]. Our finding that elevated AdipoQ is associated with a high risk of liver cancer and poor prognosis seems to be contradictory to the above view. Some mechanisms have been proposed to resolve this contradiction. 1) AdipoQ resistance: Even if AdipoQ is expressed in large amounts, it may not be able to prevent poor prognosis caused by the down-regulation of AdipoQ receptors or dysfunction of AdipoQ signaling pathway. Many HCC patients suffer from cirrhosis or fibrosis, both of which are related to the down-regulation of AdipoQ receptors in the liver and the decrease of AdipoQ clearance rate, which leads to AdipoQ resistance state [71]. Similarly, the expression of AdipoQ could have been enhanced to compensate for the progression of HCC, but due to the overall deterioration of the patient’s physical condition, higher AdipoQ levels were ineffective. 2) AdipoQ stimulates AKT-mediated activation of cancer cells, which is an important predictor of low survival rate [48, 72].
Some limitations should be considered in this meta-analysis. First, due to the high degree of heterogeneity, some results should be interpreted with caution. Secondly, few studies have been conducted to explore the correlation between leptin, AdipoQ gene polymorphism and HCC risk. The results of LEP rs7799039 gene polymorphism and HCC risk need more confirmation. Third, we partially extracted HR from the survival curve of the original article, which may introduce some small errors. Finally, almost all studies in the meta-analysis measured leptin and AdipoQ levels only once, and did not show long-term changes in the course of HCC.
Conclusions
This study showed that high leptin levels were associated with a higher risk of HCC, which may be a useful biomarker for early detection of HCC. The AdipoQ levels were directly proportional to the risk of HCC with a linear dose-response relationship and were associated with poor prognosis, which may be a potential biomarker for evaluating the prognosis of HCC.
Acknowledgments
The authors thank all the medical staff who contributed to the maintenance of the medical record database.
Abbreviations
- HCC
Hepatocellular carcinoma
- HCV
Hepatitis C virus
- HBV
Hepatitis B virus
- CFC
Cancer-free control
- AdipoQ
Adiponectin
- SMD
Standardized mean difference
- OR
Odds ratio
- HR
Hazard ratio
- CI
Confidence interval
- NOS
Newcastle-Ottawa Scale
- ALT
Alanine aminotransferase
- ELISA
Enzyme-linked immunosorbent assay
- RIA
Radioimmunoassay
- OS
Overall survival
Authors’ contributions
ZL and WW conceived and designed the study. ZL, YQ, and CD were responsible for the collection and assembly of data, data analysis, and interpretation. ZL was involved in writing the manuscript. LM and DW revised the manuscript. All the work was performed under DW and WW instruction. All authors read and approved the final manuscript.
Funding
The authors received no specific funding for this work.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wenhong Deng and Weixing Wang contributed equally to this work.
Contributor Information
Wenhong Deng, Email: wenhongdeng@whu.edu.cn.
Weixing Wang, Email: sate.llite@163.com.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Liang H, Xiong Z, Li R, Hu K, Cao M, Yang J, et al. BDH2 is disintegrated in hepatocellular carcinoma and acts as a tumor suppressor regulating cell apoptosis and autophagy. J Cancer. 2019;10(16):3735–3745. doi: 10.7150/jca.32022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueno Y, Sollano JD, Farrell GC. Prevention of hepatocellular carcinoma complicating chronic hepatitis C. J Gastroenterol Hepatol. 2009;24(4):531–536. doi: 10.1111/j.1440-1746.2009.05814.x. [DOI] [PubMed] [Google Scholar]
- 4.Wang SN, Lee KT, Ker CG. Leptin in hepatocellular carcinoma. World J Gastroenterol. 2010;16(46):5801–5809. doi: 10.3748/wjg.v16.i46.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buechler C, Wanninger J, Neumeier M. Adiponectin, a key adipokine in obesity related liver diseases. World J Gastroenterol. 2011;17(23):2801–2811. doi: 10.3748/wjg.v17.i23.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stolzenberg-Solomon RZ, Newton CC, Silverman DT, Pollak M, Nogueira LM, Weinstein SJ, et al. Circulating leptin and risk of pancreatic cancer: a pooled analysis from 3 cohorts. Am J Epidemiol. 2015;182(3):187–197. doi: 10.1093/aje/kwv041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arita Y, Kihara S, Ouchi N, Takahashi M, Maeda K, Miyagawa J, et al. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem Biophys Res Commun. 1999;257(1):79–83. doi: 10.1006/bbrc.1999.0255. [DOI] [PubMed] [Google Scholar]
- 8.Wang ZV, Scherer PE. Adiponectin, the past two decades. J Mol Cell Biol. 2016;8(2):93–100. doi: 10.1093/jmcb/mjw011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adolph TE, Grander C, Grabherr F, Tilg H. Adipokines and non-alcoholic fatty liver disease: multiple interactions. Int J Mol Sci. 2017;18(8). [DOI] [PMC free article] [PubMed]
- 10.Kamada Y, Matsumoto H, Tamura S, Fukushima J, Kiso S, Fukui K, et al. Hypoadiponectinemia accelerates hepatic tumor formation in a nonalcoholic steatohepatitis mouse model. J Hepatol. 2007;47(4):556–564. doi: 10.1016/j.jhep.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 11.Saxena NK, Fu PP, Nagalingam A, Wang J, Handy J, Cohen C, et al. Adiponectin modulates C-jun N-terminal kinase and mammalian target of rapamycin and inhibits hepatocellular carcinoma. Gastroenterology. 2010;139(5):1762–1773, 1773.e1761–1765. doi: 10.1053/j.gastro.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wieser V, Moschen AR, Tilg H. Adipocytokines and hepatocellular carcinoma. Dig Dis. 2012;30(5):508–513. doi: 10.1159/000341702. [DOI] [PubMed] [Google Scholar]
- 13.Buechler C, Haberl EM, Rein-Fischboeck L, Aslanidis C. Adipokines in liver cirrhosis. Int J Mol Sci. 2017;18(7). [DOI] [PMC free article] [PubMed]
- 14.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283(15):2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 15.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834. doi: 10.1002/(SICI)1097-0258(19981230)17:24<2815::AID-SIM110>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 16.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo J, Astrup A, Lovegrove JA, Gijsbers L, Givens DI, Soedamah-Muthu SS. Milk and dairy consumption and risk of cardiovascular diseases and all-cause mortality: dose-response meta-analysis of prospective cohort studies. Eur J Epidemiol. 2017;32(4):269–287. doi: 10.1007/s10654-017-0243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenland S, Longnecker MP. Methods for trend estimation from summarized dose-response data, with applications to meta-analysis. Am J Epidemiol. 1992;135(11):1301–1309. doi: 10.1093/oxfordjournals.aje.a116237. [DOI] [PubMed] [Google Scholar]
- 19.Ebrahim MA, Shams MEE, Al-Gayyar MMH, El-Shishtawy MM. Circulating adiponectin in hepatocellular carcinoma: a possible risk factor for development and progression. Eur J Clin Med Oncol. 2012;4(4).
- 20.Abouzied MM, Nazmy MH, Mohamed RM, Fawzy MA, Eltahir HM. Diagnostic utility of leptin and insulin-like growth factor binding protein-2 in hepatocellular carcinoma of diabetic and non-diabetic Egyptian patients. Trop J Pharm Res. 2017;16(1):211–218. doi: 10.4314/tjpr.v16i1.28. [DOI] [Google Scholar]
- 21.Aleksandrova K, Boeing H, Nothlings U, Jenab M, Fedirko V, Kaaks R, et al. Inflammatory and metabolic biomarkers and risk of liver and biliary tract cancer. Hepatology. 2014;60(3):858–871. doi: 10.1002/hep.27016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakir AS, Al Swaff R, Rasmy HS, Mandoor A. Circulating leptin in patients with liver cirrhosis and hepatocellular carcinoma. Res J Pharm, Biol Chem Sci. 2017;8(3):341–350. [Google Scholar]
- 23.Bastard JP, Fellahi S, Audureau E, Layese R, Roudot-Thoraval F, Cagnot C, et al. Elevated adiponectin and sTNFRII serum levels can predict progression to hepatocellular carcinoma in patients with compensated HCV1 cirrhosis. Eur Cytokine Netw. 2018;29(3):112–120. doi: 10.1684/ecn.2018.0413. [DOI] [PubMed] [Google Scholar]
- 24.Chen CL, Yang WS, Yang HI, Chen CF, You SL, Wang LY, et al. Plasma adipokines and risk of hepatocellular carcinoma in chronic hepatitis B virus-infected carriers: a prospective study in Taiwan. Cancer Epidemiol Biomark Prev. 2014;23(8):1659–1671. doi: 10.1158/1055-9965.EPI-14-0161. [DOI] [PubMed] [Google Scholar]
- 25.Chen MJ, Yeh YT, Lee KT, Tsai CJ, Lee HH, Wang SN. The promoting effect of adiponectin in hepatocellular carcinoma. J Surg Oncol. 2012;106(2):181–187. doi: 10.1002/jso.23059. [DOI] [PubMed] [Google Scholar]
- 26.Costantini S, Capone F, Maio P, Guerriero E, Colonna G, Izzo F, et al. Cancer biomarker profiling in patients with chronic hepatitis C virus, liver cirrhosis and hepatocellular carcinoma. Oncol Rep. 2013;29(6):2163–2168. doi: 10.3892/or.2013.2378. [DOI] [PubMed] [Google Scholar]
- 27.Feder S, Kandulski A, Schacherer D, Weiss TS, Buechler C. Serum adiponectin levels do not distinguish primary from metastatic liver tumors. Anticancer Res. 2020;40(1):143–151. doi: 10.21873/anticanres.13935. [DOI] [PubMed] [Google Scholar]
- 28.Fukushima N, Kuromatsu R, Arinaga-Hino T, Ando E, Takata A, Sumie S, et al. Adipocytokine involvement in hepatocellular carcinoma after sustained response to interferon for chronic hepatitis C. Hepatol Res. 2010;40(9):911–922. doi: 10.1111/j.1872-034X.2010.00699.x. [DOI] [PubMed] [Google Scholar]
- 29.Hamdy K, Al Swaff R, Hussein HA, Gamal M. Assessment of serum adiponectin in Egyptian patients with HCV-related cirrhosis and hepatocellular carcinoma. J Endocrinol Investig. 2015;38(11):1225–1231. doi: 10.1007/s40618-015-0379-3. [DOI] [PubMed] [Google Scholar]
- 30.Khattab MA, Eslam M, Mousa YI, Ela-Adawy N, Fathy S, Shatat M, et al. Association between metabolic abnormalities and hepatitis C-related hepatocellular carcinoma. Ann Hepatol. 2012;11(4):487–494. doi: 10.1016/S1665-2681(19)31462-0. [DOI] [PubMed] [Google Scholar]
- 31.Liu CJ, Chen PJ, Lai MY, Liu CH, Chen CL, Kao JH, et al. High serum adiponectin correlates with advanced liver disease in patients with chronic hepatitis B virus infection. Hepatol Int. 2009;3(2):364–370. doi: 10.1007/s12072-008-9111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu ZW, Zhang N, Han QY, Zeng JT, Chu YL, Qiu JM, et al. Correlation of serum leptin levels with anthropometric and metabolic parameters and biochemical liver function in Chinese patients with chronic hepatitis C virus infection. World J Gastroenterol. 2005;11(22):3357–3362. doi: 10.3748/wjg.v11.i22.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michikawa T, Inoue M, Sawada N, Sasazuki S, Tanaka Y, Iwasaki M, et al. Plasma levels of adiponectin and primary liver cancer risk in middle-aged Japanese adults with hepatitis virus infection: a nested case-control study. Cancer Epidemiol Biomark Prev. 2013;22(12):2250–2257. doi: 10.1158/1055-9965.EPI-13-0363. [DOI] [PubMed] [Google Scholar]
- 34.Radwan HA, Hamed EH, Saleh OM. Significance of serum adiponectin and insulin resistance levels in diagnosis of Egyptian patients with chronic liver disease and HCC. Asian Pac J Cancer Prev. 2019;20(6):1833–1839. doi: 10.31557/APJCP.2019.20.6.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadik NA, Ahmed A, Ahmed S. The significance of serum levels of adiponectin, leptin, and hyaluronic acid in hepatocellular carcinoma of cirrhotic and noncirrhotic patients. Hum Exp Toxicol. 2012;31(4):311–321. doi: 10.1177/0960327111431091. [DOI] [PubMed] [Google Scholar]
- 36.Voumvouraki A, Koulentaki M, Notas G, Sfakianaki O, Kouroumalis E. Serum surrogate markers of liver fibrosis in primary biliary cirrhosis. Eur J Intern Med. 2011;22(1):77–83. doi: 10.1016/j.ejim.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Sumie S, Kawaguchi T, Kuromatsu R, Takata A, Nakano M, Satani M, et al. Total and high molecular weight adiponectin and hepatocellular carcinoma with HCV infection. PLoS One. 2011;6(11):e26840. doi: 10.1371/journal.pone.0026840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ataseven H, Bahcecioglu IH, Kuzu N, Yalniz M, Celebi S, Erensoy A, et al. The levels of ghrelin, leptin, TNF-alpha, and IL-6 in liver cirrhosis and hepatocellular carcinoma due to HBV and HDV infection. Mediat Inflamm. 2006;2006(4):78380. doi: 10.1155/MI/2006/78380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotani K, Wakai K, Shibata A, Fujita Y, Ogimoto I, Naito M, et al. Serum adiponectin multimer complexes and liver cancer risk in a large cohort study in Japan. Asian Pac J Cancer Prev. 2009;10(Suppl):87–90. [PubMed] [Google Scholar]
- 40.Wang YY, Lin SY. Leptin in relation to hepatocellular carcinoma in patients with liver cirrhosis. Horm Res. 2003;60(4):185–190. doi: 10.1159/000073231. [DOI] [PubMed] [Google Scholar]
- 41.Amer T, El-Baz R, Mokhtar AR, El-Shaer S, Elshazli R, Settin A. Genetic polymorphisms of IL-23R (rs7517847) and LEP (rs7799039) among Egyptian patients with hepatocellular carcinoma. Arch Physiol Biochem. 2017;123(5):279–285. doi: 10.1080/13813455.2017.1320680. [DOI] [PubMed] [Google Scholar]
- 42.Cai X, Gan Y, Fan Y, Hu J, Jin Y, Chen F, et al. The adiponectin gene single-nucleotide polymorphism rs1501299 is associated with hepatocellular carcinoma risk. Clin Transl Oncol. 2014;16(2):166–172. doi: 10.1007/s12094-013-1056-7. [DOI] [PubMed] [Google Scholar]
- 43.Zhang S, Jiang J, Chen Z, Wang Y, Tang W, Liu C, et al. Investigation of LEP and LEPR polymorphisms with the risk of hepatocellular carcinoma: a case-control study in eastern Chinese Han population. Onco Targets Ther. 2018;11:2083–2089. doi: 10.2147/OTT.S153931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siegel AB, Goyal A, Salomao M, Wang S, Lee V, Hsu C, et al. Serum adiponectin is associated with worsened overall survival in a prospective cohort of hepatocellular carcinoma patients. Oncology. 2015;88(1):57–68. doi: 10.1159/000367971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen J, Yeh CC, Wang Q, Gurvich I, Siegel AB, Santella RM. Plasma adiponectin and hepatocellular carcinoma survival among patients without liver transplantation. Anticancer Res. 2016;36(10):5307–5314. doi: 10.21873/anticanres.11103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin E, Yu YD, Kim DS, Won NH. Adiponectin receptor expression predicts favorable prognosis in cases of hepatocellular carcinoma. Pathol Oncol Res. 2014;20(3):667–675. doi: 10.1007/s12253-014-9747-0. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe N, Takai K, Imai K, Shimizu M, Naiki T, Nagaki M, et al. Increased levels of serum leptin are a risk factor for the recurrence of stage I/II hepatocellular carcinoma after curative treatment. J Clin Biochem Nutr. 2011;49(3):153–158. doi: 10.3164/jcbn.10-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang SN, Yang SF, Tsai HH, Lee KT, Yeh YT. Increased adiponectin associated with poor survival in hepatocellular carcinoma. J Gastroenterol. 2014;49(9):1342–1351. doi: 10.1007/s00535-013-0898-7. [DOI] [PubMed] [Google Scholar]
- 49.Wang SN, Yeh YT, Yang SF, Chai CY, Lee KT. Potential role of leptin expression in hepatocellular carcinoma. J Clin Pathol. 2006;59(9):930–934. doi: 10.1136/jcp.2005.035477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei T, Ye P, Peng X, Wu LL, Yu GY. Circulating adiponectin levels in various malignancies: an updated meta-analysis of 107 studies. Oncotarget. 2016;7(30):48671–48691. doi: 10.18632/oncotarget.8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song RR, Gu XL. Serum adiponectin levels may be associated with the pathogenesis of hepatocellular carcinoma. Tumour Biol. 2015;36(4):2983–2992. doi: 10.1007/s13277-014-2931-8. [DOI] [PubMed] [Google Scholar]
- 52.Skibola CF, Holly EA, Forrest MS, Hubbard A, Bracci PM, Skibola DR, et al. Body mass index, leptin and leptin receptor polymorphisms, and non-hodgkin lymphoma. Cancer Epidemiol Biomark Prev. 2004;13(5):779–786. [PubMed] [Google Scholar]
- 53.Hoffstedt J, Eriksson P, Mottagui-Tabar S, Arner P. A polymorphism in the leptin promoter region (−2548 G/A) influences gene expression and adipose tissue secretion of leptin. Horm Metab Res. 2002;34(7):355–359. doi: 10.1055/s-2002-33466. [DOI] [PubMed] [Google Scholar]
- 54.Liu P, Shi H, Huang C, Shu H, Liu R, Yang Y, et al. Association of LEP A19G polymorphism with cancer risk: a systematic review and pooled analysis. Tumour Biol. 2014;35(8):8133–8141. doi: 10.1007/s13277-014-2088-5. [DOI] [PubMed] [Google Scholar]
- 55.Yang Y, Liu P, Guo F, Liu R, Yang Y, Huang C, et al. Genetic G2548A polymorphism of leptin gene and risk of cancer: a meta-analysis of 6860 cases and 7956 controls. J BUON. 2014;19(4):1096–1104. [PubMed] [Google Scholar]
- 56.Liu Y, Wu H, Zhu Y, Gao Y. Genetic association between leptin-2548G/A polymorphism and risk of cancer: a meta analysis. Int J Clin Exp Med. 2015;8(1):448–455. [PMC free article] [PubMed] [Google Scholar]
- 57.Yoon YS, Kwon AR, Lee YK, Oh SW. Circulating adipokines and risk of obesity related cancers: a systematic review and meta-analysis. Obes Res Clin Pract. 2019;13(4):329–339. doi: 10.1016/j.orcp.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 58.Ye J, Liang Z, Liang Q, Zhang J, Mao S, Liang R. Adiponectin is associated with poor prognosis in carcinoma patients: evidence from a meta-analysis. Lipids Health Dis. 2015;14(1). [DOI] [PMC free article] [PubMed]
- 59.Saxena NK, Sharma D, Ding X, Lin S, Marra F, Merlin D, et al. Concomitant activation of the JAK/STAT, PI3K/AKT, and ERK signaling is involved in leptin-mediated promotion of invasion and migration of hepatocellular carcinoma cells. Cancer Res. 2007;67(6):2497–2507. doi: 10.1158/0008-5472.CAN-06-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Q, Lui VW, Yeo W. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Future Oncol. 2011;7(10):1149–1167. doi: 10.2217/fon.11.95. [DOI] [PubMed] [Google Scholar]
- 61.Dupont J, Reverchon M, Cloix L, Froment P, Ramé C. Involvement of adipokines, AMPK, PI3K and the PPAR signaling pathways in ovarian follicle development and cancer. Int J Dev Biol. 2012;56(10–12):959–967. doi: 10.1387/ijdb.120134jd. [DOI] [PubMed] [Google Scholar]
- 62.Wauman J, Tavernier J. Leptin receptor signaling: pathways to leptin resistance. Front Biosci (Landmark Ed) 2011;16:2771–2793. doi: 10.2741/3885. [DOI] [PubMed] [Google Scholar]
- 63.El-Habr EA, Levidou G, Trigka EA, Sakalidou J, Piperi C, Chatziandreou I, et al. Complex interactions between the components of the PI3K/AKT/mTOR pathway, and with components of MAPK, JAK/STAT and Notch-1 pathways, indicate their involvement in meningioma development. Virchows Arch. 2014;465(4):473–485. doi: 10.1007/s00428-014-1641-3. [DOI] [PubMed] [Google Scholar]
- 64.Pietrzyk L, Torres A, Maciejewski R, Torres K. Obesity and obese-related chronic low-grade inflammation in promotion of colorectal cancer development. Asian Pac J Cancer Prev. 2015;16(10):4161–4168. doi: 10.7314/APJCP.2015.16.10.4161. [DOI] [PubMed] [Google Scholar]
- 65.Polyzos SA, Kountouras J, Zavos C, Deretzi G. The potential adverse role of leptin resistance in nonalcoholic fatty liver disease: a hypothesis based on critical review of the literature. J Clin Gastroenterol. 2011;45(1):50–54. doi: 10.1097/MCG.0b013e3181ec5c66. [DOI] [PubMed] [Google Scholar]
- 66.Mittenbuhler MJ, Sprenger HG, Gruber S, Wunderlich CM, Kern L, Bruning JC, et al. Hepatic leptin receptor expression can partially compensate for IL-6Ralpha deficiency in DEN-induced hepatocellular carcinoma. Mol Metab. 2018;17:122–133. doi: 10.1016/j.molmet.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perrier S, Jarde T. Adiponectin, an anti-carcinogenic hormone? A systematic review on breast, colorectal, liver and prostate cancer. Curr Med Chem. 2012;19(32):5501–5512. doi: 10.2174/092986712803833137. [DOI] [PubMed] [Google Scholar]
- 68.Nazmy EA, El-Khouly OA, Zaki MMA, Elsherbiny NM, Said E, Al-Gayyar MMH, et al. Targeting p53/TRAIL/caspase-8 signaling by adiponectin reverses thioacetamide-induced hepatocellular carcinoma in rats. Environ Toxicol Pharmacol. 2019;72:103240. doi: 10.1016/j.etap.2019.103240. [DOI] [PubMed] [Google Scholar]
- 69.Al-Gayyar MM, Abbas A, Hamdan AM. Chemopreventive and hepatoprotective roles of adiponectin (SULF2 inhibitor) in hepatocelluar carcinoma. Biol Chem. 2016;397(3):257–267. doi: 10.1515/hsz-2015-0265. [DOI] [PubMed] [Google Scholar]
- 70.Manieri E, Herrera-Melle L, Mora A, Tomás-Loba A, Leiva-Vega L, Fernández DI, et al. Adiponectin accounts for gender differences in hepatocellular carcinoma incidence. J Exp Med. 2019;216(5):1108–1119. doi: 10.1084/jem.20181288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corbetta S, Redaelli A, Pozzi M, Bovo G, Ratti L, Redaelli E, et al. Fibrosis is associated with adiponectin resistance in chronic hepatitis C virus infection. Eur J Clin Investig. 2011;41(8):898–905. doi: 10.1111/j.1365-2362.2011.02498.x. [DOI] [PubMed] [Google Scholar]
- 72.Barb D, Neuwirth A, Mantzoros CS, Balk SP. Adiponectin signals in prostate cancer cells through Akt to activate the mammalian target of rapamycin pathway. Endocr Relat Cancer. 2007;14(4):995–1005. doi: 10.1677/ERC-06-0091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.