Abstract
Purpose:
High-dose-rate (HDR) brachytherapy is an established technique to be used as monotherapy option or focal boost in conjunction with external beam radiation therapy (EBRT) for treating prostate cancer. Radiation source path reconstruction is a critical procedure in HDR treatment planning. Manually identifying the source path is labor intensive and timely inefficient. Recent years, magnetic resonance imaging (MRI) becomes a valuable imaging modality for image-guided HDR prostate brachytherapy due to its superb soft-tissue contrast for target delineation and normal tissue contouring. The purpose of this study is to investigate a deep-learning-based method to automatically reconstruct multiple catheters in MRI for prostate cancer HDR brachytherapy treatment planning.
Methods:
Attention gated U-Net incorporated with total variation (TV) regularization model was developed for multi-catheter segmentation in MRI. The attention gates were used to improve the accuracy of identifying small catheter points, while TV regularization was adopted to encode the natural spatial continuity of catheters into the model. The model was trained using the binary catheter annotation images offered by experienced physicists as ground truth paired with original MRI images. After the network was trained, MR images of a new prostate cancer patient receiving HDR brachytherapy were fed into the model to predict the locations and shapes of all the catheters. Quantitative assessments of our proposed method were based on catheter shaft and tip errors compared to the ground truth.
Results:
Our method detected 299 catheters from 20 patients receiving HDR prostate brachytherapy with a catheter tip error of 0.37±1.68 mm and a catheter shaft error of 0.93±0.50 mm. For detection of catheter tips, our method resulted in 87% of the catheter tips within an error of less than ±2.0 mm, and more than 71% of the tips can be localized within an absolute error of no greater than 1.0 mm. For catheter shaft localization, 97% of catheters were detected with an error of less than 2.0 mm, while 63% were within 1.0 mm.
Conclusions:
In this study, we proposed a novel multi-catheter detection method to precisely localize the tips and shafts of catheters in 3D MRI images of HDR prostate brachytherapy. It paves the way for elevating the quality and outcome of MRI-guided HDR prostate brachytherapy.
Keywords: Magnetic resonance imaging, prostate brachytherapy, catheter detection, deep learning, total variation regularization
1. INTRODUCTION
Prostate cancer is the second leading cause of cancer deaths for men in the United States1. Approximately one in nine men will be diagnosed with prostate cancer in their lifetime. High-dose-rate (HDR) brachytherapy, in conjunction with external beam radiation therapy (EBRT), is currently adopted to treat patients with intermediate- to high-risk disease2. In HDR prostate brachytherapy, radioactive source which deliver high doses of radiation is dwelling inside the interstitial catheters in prostate gland to achieve excellent dose conformity to target while sparing adjacent organs such as rectum and bladder3. Treatment planning is a critical procedure in temporary interstitial HDR brachytherapy, requiring accurate target and organs-at-risk (OARs) delineations and optimal catheter reconstructions4. At present, this procedure is commonly assisted by transrectal ultrasound (TRUS) or X-ray computed tomography (CT)5,6. However, intrinsic limitations exist in TRUS- and CT-based treatment planning, which could potentially have a significant impact on the dose delivery. In TRUS, the visualization of the prostate gland and inserted catheters is suboptimal due to the limited image spatial resolution and ultrasonic shadows from the inserted catheters. While CT can improve the visualization of catheters by using metallic fiducials, CT suffers from poor soft-tissue contrast, highly deteriorating the visualization of the prostate.
Thanks to the advances in magnetic resonance imaging (MRI), pelvic MRI has emerged as a routine imaging modality to assist the staging of prostate cancer during the past decades7. MRI has demonstrated superiority to TRUS and CT in assessing the extent of the local disease and extracapsular extension attribute to its high spatial resolution, sensitivity, and specificity. Superb soft-tissue contrast in MRI provides better anatomic visualization of prostate gland and its surrounding tissues such as urinary sphincter, neurovascular bundles, the rectum, and the bladder, facilitating the delineation of target and OARs, which has potential to reduce the gastrointestinal and urinary toxicity of brachytherapy. Furthermore, the therapeutic ratio could be significantly improved by incorporating MRI into prostate brachytherapy due to MRI’s capability of differentiating benign and malignant prostate tumors. Currently, T2-weighted MRI images are acquired before the procedure and then used as guidance for target definition and/or fused to intraoperative TRUS images for guiding the catheter insertion. After the intraoperative TRUS-guided insertion of catheters, MRI images are obtained for target and OARs delineation. Commonly, CT images are also acquired for treatment planning, and MRI-CT registration is then done to transfer the contours from MRI to planning CT. Effects are needed to minimize the catheter movements during the logistical transfer of patients and reduce the MRI-CT fusion errors to optimize dosimetry. Conversely, one effective way is to acquire MRI images only, avoiding the fusion errors and reducing the logistical transfer uncertainties, since the malignant lesions and surrounding tissues appear distinctively in MRI. MRI has been proposed to be used as the image-guidance in focal or ultrafocal treatment because of the potential to visualize the dominant lesions by MRI8–10. Dedicated MRI-guided HDR brachytherapy suite has been developed for feasibility studies of interstitial catheters insertion under the guidance of MRI and subsequent MRI-based prostate HDR brachytherapy treatment planning11–13. The aim is to optimize the dose to the target volume with the visualization of dominant malignant nodule offered by MRI. Studies have demonstrated dosimetric parameters such as dose to the target can be achieved without increase in toxicity14,15. However, this approach has not been widely adopted in clinical practices since the overall procedural time is significantly prolonged, including the increase of the operating room (OR) time in intraoperative MRI guidance as well as its associated logistical time. One of the big challenges is that the dark and diffuse appearance of the catheters in MRI makes it difficult for catheter digitization either done by physicists manually or by segmentation algorithms automatically. For instance, blood vessels appear dark in T2-weighted MRI in a same way as catheters16,17, which can be easily confused with catheters. Therefore, robust catheter detection methods are highly desired to improve the efficiency of catheter reconstruction in MRI-guided HDR prostate brachytherapy.
Studies have been conducted extensively for catheter detection, a majority of which were focused on single catheter localization18–20. The approaches can be classified into two categories: 1) traditional model-based segmentation algorithms, and 2) machine learning based methods. In traditional approaches, like blood vessels and nerves, catheters are considered as the tubular tree-like structures21,22. These methods generally can be used in different imaging modalities including ultrasound (US)19, CT20,23, and MRI19. Zhao et al.20 presented an automatic catheter tracking method in 3D US images by firstly applying a line filter method to enhance the contrast of the catheter against the background for regions-of-interest (ROIs) selection using tubularness information, and then incorporating Kalman filter into a catheter fitting model with the random sample consensus algorithm which was originally developed by Uherčík et al.19 to detect the catheters within ROIs. Mastmeyer et al.19 investigated a mechanical model-based method for segmentation of gynecologic brachytherapy catheter detections in MRI images. In this method, an angular spring model was used to model a catheter as series of short rods attached to one another using torsion springs which become more angulated from the catheter tip toward the catheter end. Given recent advances in machine learning techniques, automatic segmentation methods have been developed based on machine learning algorithms, especially, deep convolutional neural networks24–34. Deep learning-based catheter segmentation algorithms are recently reported in CT35, MRI17,18, and US36–38 for biopsy and brachytherapy. While these methods are inspiring, reliable catheter detection approaches have to be further developed specifically for digitizing the catheters in MRI-guided HDR prostate brachytherapy.
In this paper, we propose a novel deep learning-based method, namely, deeply supervised attention U-Net incorporated with total variation (TV) regularization model, for multi-catheter segmentation in MRI. The attention gates were used to improve the accuracy of small catheter points, while TV regularization was adopted to encode the natural spatial continuity of catheters into the model. The approach is reliable and computationally efficient, and will facilitate treatment planning in MRI-guided HDR prostate brachytherapy.
2. MATERIALS AND METHODS
2.A. Method overview
U-Net39, a fully convolutional network (FCN), has been demonstrated as a robust and efficient algorithm for medical image segmentation. In U-Net, image features are extracted in multiple image scales, which are then merged to combine fine- and coarse-level dense predictions through skip connection. The entire model is trained by using Adam optimizer to minimize the loss function. In this way, global scale is used for modeling the relationship between tissues by the coarse-level feature maps that are obtained through downsampling from input images to capture sufficient semantic contextual information. It can achieve superb prediction accuracy for large objects vary little in shape. However, for small objects with large shape variability, it shows relatively high false-positive rate. Attention gates have been proposed to be integrated into the U-Net to improve the accuracy of predicting small objects in the image that vary largely in shapes40,41. Instead of adding additional models to separate subsequent segmentation steps for different ROIs, the integrating of attention gates in a U-Net ends up with a single model that can identify prune features that are relevant to specific regions and progressively suppress irrelevant background regions without cropping ROIs. Attention gates incorporated into the U-Net architecture is suitable for catheter segmentation where catheters are small objects in MRI images. Furthermore, in this work, total variation (TV) regularization is integrated into the attention gated U-Net to maintain catheter’s shape continuity in nature. By applying different weights, the gradients around x-, y-, z-axis of a catheter in 3D space could be considered differently. The architecture of the proposed method is schematically shown in Fig. 1. In the training stage, paired 3D volumetric MRI image and 3D volumetric binary catheter image containing the locations of catheters are fed into the model to obtain optimal parameters of the model. The volumetric sizes of both MRI and binary catheter images are 512×512×128. Manually contoured catheter images by experienced physicists were collected as ground truth for training the network. Once the model is well trained, in the detection stage, given a 3D MRI image, the catheters can be automatically delineated in the image by our proposed method.
FIG. 1.

The architecture of the proposed network.
2.B. Deep supervised attention U-Net
As shown in Fig.1, the deep supervised attention U-Net consists of encoding, decoding, and attention gates, and deep supervision paths. Encoding path is constructed by convolution layers and max-pool layers. In each convolution layer, features are extracted through convolution operations followed by rectified linear unit (ReLU) activation and batch normalization (BN). The convolutional kernels used in U-Net are 3D kernels with a size of 3×3×3. The detailed implementation of the attention gates can refer to recent studies40,41. In brief, as shown in Fig. 1, the attention gated connection is adopted to highlight salient features and preserve only relevant activations to a specific task. As shown in Fig.2, Input feature maps are firstly concatenated through linear transformations using channel-wise 1×1×1 convolutions. The concatenated intermediate tensor is then passing through ReLU, 1×1×1 convolution, and sigmoid operations. The attention gates are obtained by element-wise multiplication of input feature maps and the attention coefficients that are learned to focus on a subset of ROIs.
FIG. 2.
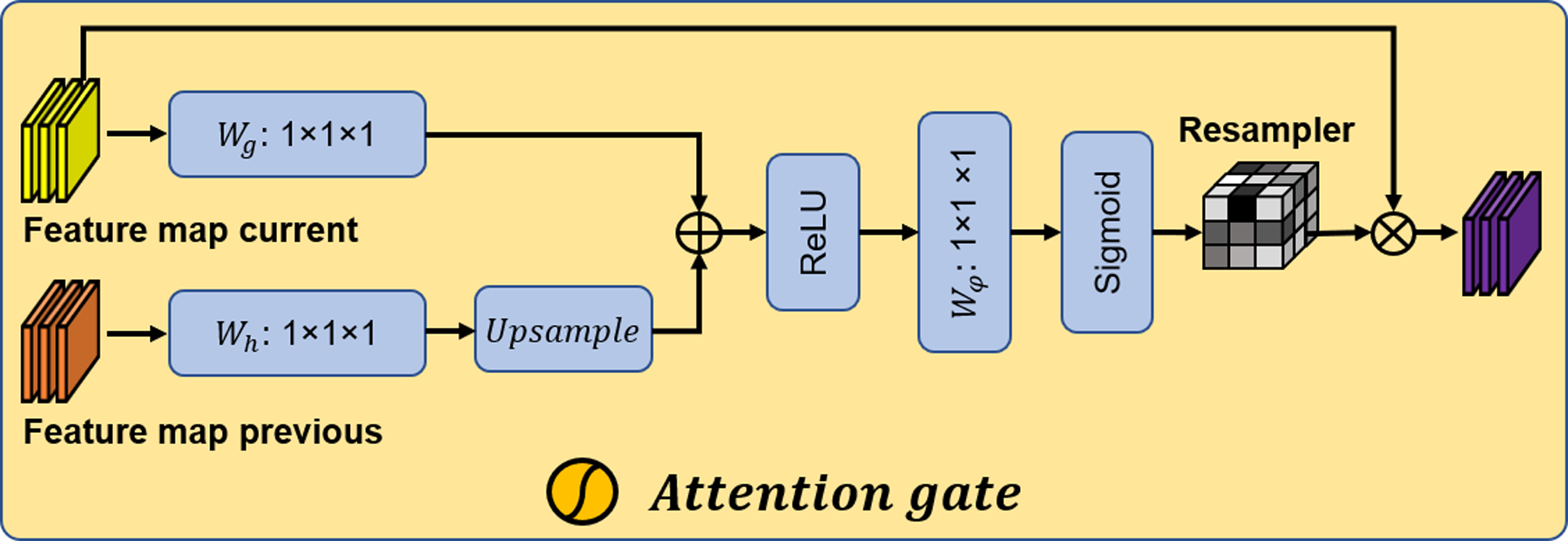
The schematic of the attention gate.
In decoding path, feature maps are extended using deconvolution, convolution, ReLU, and BN operations. A 2×2×2 stride is adopted in deconvolutions to output feature maps with consistent size. The decoding path and the encoding path are bridged using skip connections that integrated attention gates. These bridges in a fully convolutional network enhance the capability of each path for obtaining both high-frequency information like textural features and low-frequency information like structural features in original image.
For deep supervision path, feature maps extracted from the lower and middle stages pass through deconvolution, ReLU, and sigmoid functions to output images that have the same size as the catheter images. These output feature maps are then combined with the ground truth for constructing loss functions used for network training with Adam optimizer.
The compound loss function in this work consists of three parts. The first one is voxel-wise binary cross entropy (BCE) loss function, which can be expressed as the following format:
| (1) |
where, Gj and Pj, respectively, are the jth voxel in the ground truth image G and the prediction image P.
The second part is the Dice loss, which is defined as:
| (2) |
where, V is the volume of the region within the contours of G and P.
Dice losses encourage similarity while logistic losses penalize dissimilarity. Therefore, combining logistic loss and Dice loss functions, both concepts can be adopted to optimize network training.
The third part is the loss created by TV regularization which considers the natural continuity of catheter shaft along one direction. The details of TV regularization-based loss and the compound loss function are discussed in the following section.
2.C. TV regularization
TV regularization has been widely used in image processing for noise removal based on the principle that excessive and possibly spurious signals have high total variation. Compared to linear smoothing, TV based method removes noise but meanwhile can preserve edges through reduce the total variation. The TV norm is defined as the following format:
| (3) |
where, ∇C(x) is the gradient of an image C at a pixel x.
In general, TV regularization is isotropic in 3D space. In this study, as we can see in Fig.3, the catheters indicated by the yellow lines in MRI images show the strong continuities along their shaft directions. Thus, additive weights are incorporated into the TV regularization to highlight the gradient in z-axis (catheter shaft direction) more than that in x- and y-axis. The weighted TV can be defined as:
| (4) |
where, ω is the weight vector that has different coefficients along x-, y-, and z-axis.
FIG. 3.
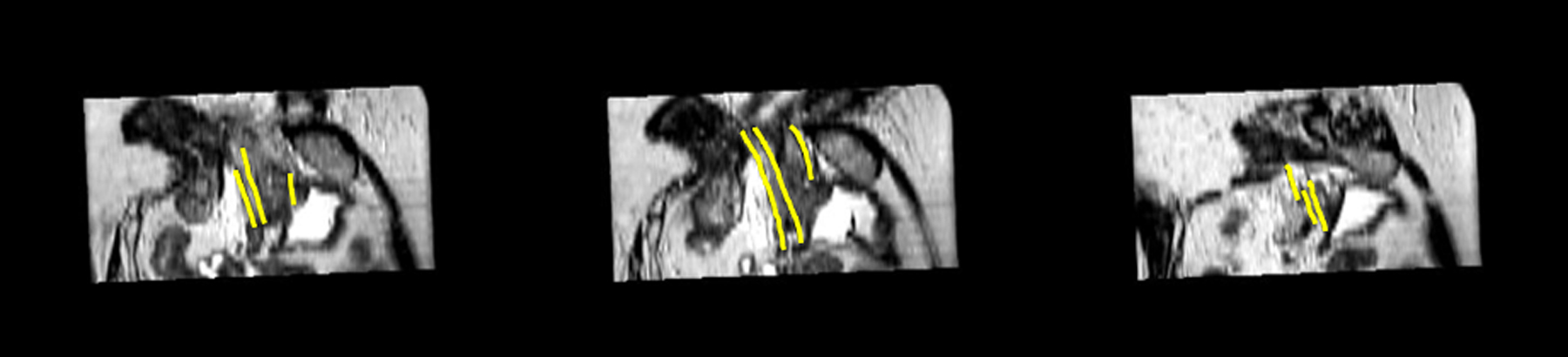
MRI 2D sagittal images with annotations of catheter shaft. The yellow lines indicate catheters.
Integrating BCE loss (1), Dice loss (2), and weighted TV (4), the compound loss function can be express as:
| (5) |
where l denotes the stage of our network, λl is the weight of the lth stage’s loss, μ and η are the parameters that balance the BCE lose, Dice loss, and TV regularization. The parameters including μ, η, and λl are set empirically to achieve optimal performance. Five-fold cross validation was adopted to evaluate the settings. The original data (20 patients) was randomly partitioned into five equal sized groups (four patients per group). A single group was retained as the validation data for testing the model, while the remaining four groups were used as the training data. This cross-validation process was repeated five times to make sure each group was used exactly once as the validation data.
2.D. Image data acquisition and preprocessing
A retrospective study was conducted on a cohort of 20 prostate cancer patients receiving HDR brachytherapy. T2-weighted MRI images were acquired on a Siemens Aera 1.5 T MRI scanner after intraoperative TRUS guided catheter insertion. The sequence parameters were: spin echo, TE 101 ms, TR ranging from 5360 to 7500 ms, patient position being FFS and flip angle being 160 degree. The voxel size is 0.9766 mm × 0.9766 mm × 1.0 mm. After the MRI images were acquired, experienced physicists conducted catheter digitization, which were then collected as a paired data set. As an example, Fig.4 shows the MRI 2D axial image with catheters (Fig. 4(A)) and its annotations (Fig.4(B)), and catheter-only image (Fig. 4(C)).
FIG. 4.

An example MRI 2D axial image with catheters (A), its annotations (B) and catheter-only image (C).
2.E. Implementation and evaluation metrics
The proposed method was implemented based on the widely used deep learning framework Tensorflow. The hyperparameter values for Eq. (5) are listed as follows: λl = 0.8l − 1, μ = 0.5, and η = 0.1. In order to enlarge the data variation, data augmentation was used to prepare for the training data. The methods which were used for data augmentation include rotation (0°, ±10° and ±20°) in axial plane, flipping along x-(axial), y-(sagittal), and z-(coronal) axes, scaling (1, 1.125, 1.25) and rigid warp (original +2 rigid warp), resulting in 360 times more training datasets. Therefore, a total of 5760 augmented MRI and corresponding contour pairs were used for training model. During training, for each iteration, a batch of randomly selected 20 pairs that fit the 32 GB memory of the NVIDIA Tesla V100 GPU was fed into the model to learn the learnable parameters via Adam gradient optimizer. The learning rate for Adam optimizer that used in our algorithm during the training was set to 2e-4, and the model is trained and tested on Ubuntu 18.04 environment with python 3.7. The network was trained for 200 epochs. Training required approximately 3.5 hours. Once the network was trained, the catheter delineation in MRI images of a new arrival patient can be performed within 1 seconds.
In this study, to evaluate the proposed method, a leave-one-out cross validation approach was adopted. For each experiment, one patient’s images were randomly selected as test data, while the rest of the patients’ images were used as training data. The experiment repeated 20 times to have each patient’s images used as test data exactly once. To quantitatively assess the catheter delineation results, the metrics including catheter tip error, catheter shaft error, and catheter detection accuracy were calculated. For catheter tip localization, we define the tip error as follow:
| (6) |
where, n is the total number of catheters in the MRI images, gi and pi are respectively the ground true and the predict tips of the i-th catheter.
In the catheter shaft detection, the catheter cross-sectional images were extracted from MRI 2D axial images, then all the catheter locations in each cross-sectional image were calculated for computing the catheter shaft error using the following equation:
| (7) |
where, m is the total number of all the cross-sectional contours of catheters in all the MRI 2D axial slices, Ck is the ground-truth center location of the k-th catheter contour, and is the predicted center.
Catheter detection accuracy is defined as:
| (8) |
where α is the percentage of the ground truth length of catheter inserted, si and ri are respectively the ground true and the predict length of the i-th catheter, and #{M} is the total element number in the set M.
Furthermore, commonly used metrics including precision, recall, and F1 are calculated to quantify the performance of our proposed method, which are defined as:
| (9) |
| (10) |
| (11) |
3. RESULTS
3.A. Overall performance
Fig. 5 shows visualization of catheter locations from 5 different slices of 2D MRI axial images in an example patient dataset. Each column in Fig.5 show a single slice. The top row is the MRI images, the second row is the ground-truth catheter delineations, the predicted catheter contours are shown in third row, and the bottom row shows the fused ground-truth and predicted catheter delineations. It is shown that our prediction of catheters reaches high agreement with the ground truth, except for a few incorrect positions. The blue arrow indicates the redundant catheter location that is shown in our prediction but not in the ground-truth image, while the yellow arrow shows the missing catheter location that was not predicted by our model. To qualitatively assess the performance of our model for contouring the catheters in 3D MRI images, the results of another example patient are shown in 3D view in Fig. 6. Figs. 6(A)–(C) respectively show the manual delineation (ground truth), reconstructed catheter image by the proposed method, and the fusion of the manual delineation and reconstructed catheter image. To closely investigate the precision of our prediction, the zoom-in view of the dashed line area in Fig. 6(C) is shown in Fig. 6(D). We can see, the catheters reconstructed by our model match well with the ground truth.
FIG. 5.

Visualization of catheter locations in 2D MRI axial slices. The rows from top to down shows the MRI images, catheter images of ground truth, generated by the proposed method, fusion of ground truth and generated, respectively. The columns from left to right respectively corresponds to the 1st, 10th, 20th, 30th, and 40th slice of the MRI images. The yellow arrow indicates the missed localization, while the blue arrow shows the incorrect localization.
FIG. 6.
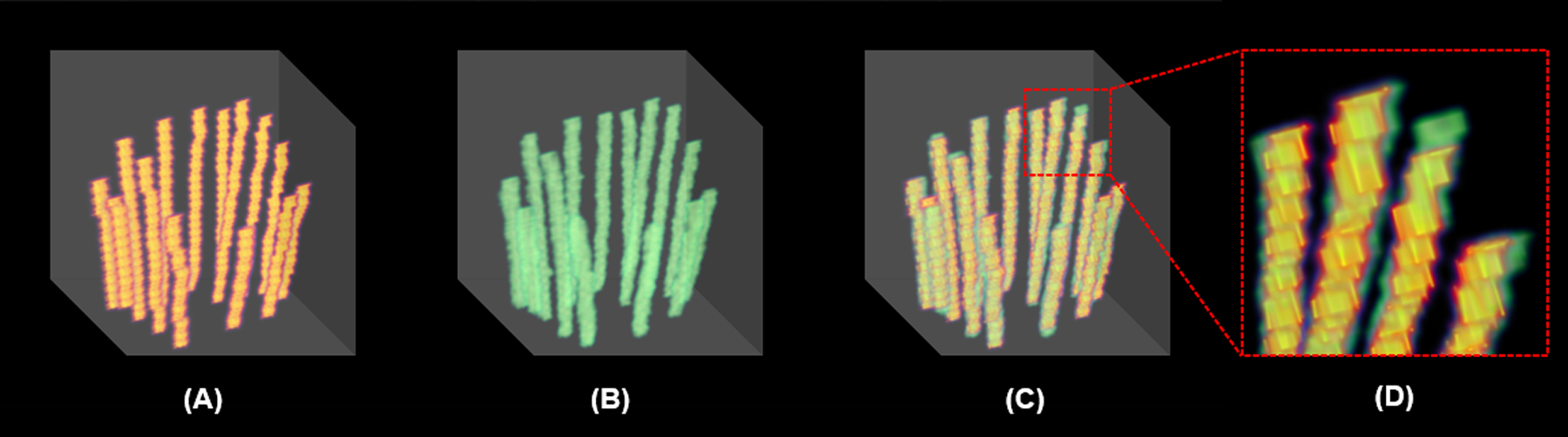
Multiple catheters in 3D view. (A) manual delineation (ground truth), (B) reconstructed catheter image by the proposed method, (C) fusion of the manual delineation and reconstructed catheter image, (D) zoom-in image of the red dashed-line square in (C).
For comparison, we also conducted all the experiments using conventional U-Net39. Table 1 summarizes the overall quantitative results of both U-Net and our proposed method from the entire testing data including catheter tip error, catheter shaft error, and accuracy. For accuracy, we computed two different α values in Eq. (8), α = 10%, and α = 15%, respectively. In total, 299 pieces of catheters from 20 patients receiving HDR prostate brachytherapy. Our proposed method detects the catheters with a shaft error of 0.93±0.50 mm and a tip error of 0.37±1.68 mm, which shows that high detection accuracy can be achievable with our proposed method compared to the ground truth. The catheters (Nucletron ProGuide Sharp 5F) that are generally used in our HDR prostate brachytherapy have a size of 1.6 mm in diameter and 240.0 mm in length. With a tolerance of 10% error in each predicted catheter length referred to its corresponding ground truth, a detection accuracy of 81.3% is achievable, while the accuracy can further reach as high as 93.3% with a tolerance of 15% error. Our proposed method gained significant improvements (p < 0.05) in all the quantities including tip error, shaft error, and accuracies with thresholds of 10% and 15%, respectively. Table 2 shows the metrics including precision, recall, and F1 for both U-Net based and our proposed methods. Our proposed method achieved better precision, recall and F1 than U-Net based method.
TABLE 1.
Overall quantitative results achieved by the U-Net based and our proposed methods.
| Method | errortip (mm) | errorshaft (mm) | accuracy (α=10%) | accuracy (α=15%) |
|---|---|---|---|---|
| U-Net | 0.55±2.26 | 1.05±0.71 | 72.2% | 88.6% |
| Proposed | 0.37±1.68 | 0.93±0.50 | 81.3% | 93.3% |
| p value (U-Net vs. proposed) |
<0.01 | <0.01 | <0.01 | <0.01 |
TABLE 2.
Precision, recall, and F1 of catheter shaft localization achieved by the U-Net based and our proposed methods.
| Method | precision | recall | F1 |
|---|---|---|---|
| U-Net | 0.93 | 0.80 | 0.86 |
| Proposed | 0.96 | 0.86 | 0.91 |
3.B. Catheter tip detection
The catheter tip detection error compared to ground truth is shown in Fig.7 for both the proposed and U-Net based methods, in which, Fig. 7(A) shows the standard difference between predicted tips and their corresponding ground truth tips. The differences range from −5 mm to 5 mm for our proposed method, while U-Net based method has differences range from −6 mm to 6 mm. The majority of the errors for the proposed method lies within ±1.0 mm. The percentage distribution of absolute errors is shown in Fig. 7(B). Around 87% of the catheter tips can be detected by the proposed method within an error of ±2.0 mm, while more than 71% of the tips can be localized within an absolute error of 1.0 mm.
FIG. 7.
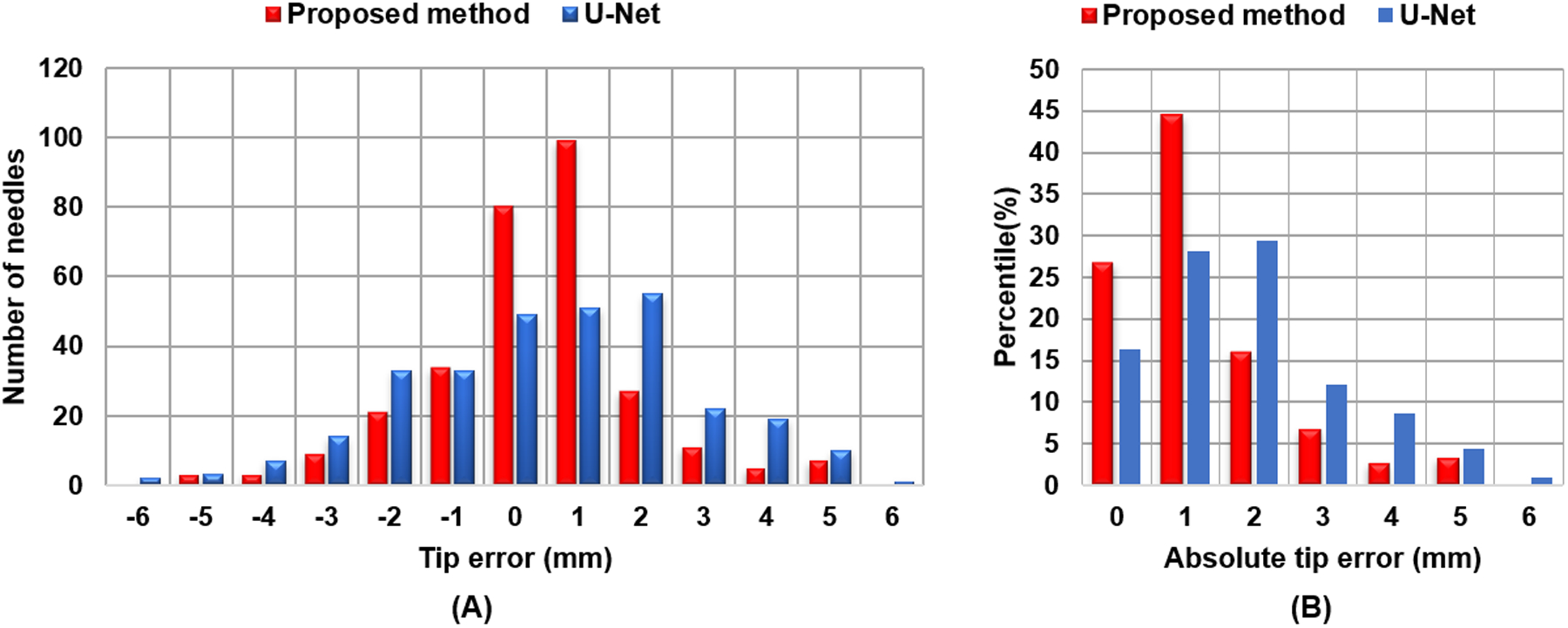
Catheter tip error distribution. (A) standard difference, (B) percentile of absolute difference.
3.C. Catheter shaft localization
Fig. 8 shows the catheter shaft localization error for all the 20 patients in our study achieved by the proposed and U-Net based methods. In the majority of patients, the catheter shaft detection mean error is less than 1.0 mm. We further computed the overall catheter shaft localization error. In total, using our proposed method, 97% of catheters were detected with an error of less than 2.0 mm, while 63% of the catheter shafts can be localized within 1.0 mm.
FIG. 8.

The catheter shaft localization error.
4. DISCUSSION AND CONCLUSIONS
Accurate dosimetry of HDR prostate brachytherapy relies on many factors, among which, the precision of catheter digitalization is a critical factor. Manual delineation is currently executed by physicists in clinic. It is tedious, labor intensive, and time consuming. Generally, it takes about 10–20 minutes for an experienced physicist to manually digitize all catheters in HDR prostate brachytherapy. Automatic catheter digitization is highly desired. However, in prostate brachytherapy, automatic segmentation is a challenge task due to the large number of inserted catheters.
Compared to CT images which are generally used for catheter digitization, identifying catheters in MRI images has its own intrinsic difficulty. Plastic catheters are the most widely used type in prostate brachytherapy. However, in MRI images, these plastic catheters appear as black regions because of no MRI signal is emitted by this type of material, which leads to the high possibility of confusion among these catheters with other tubular structures that also appear black, for example, blood vessels16,17. Moreover, if the catheter is not completely surrounded by tissues, it will be even more difficult to identify the catheter and estimate the shape solely relying on the dark appearance. To tackle these challenges, in our proposed method, we firstly used a 3D U-Net architecture aiming to maintain the 3D features of the catheters within the network. Secondly, deep supervised attention strategy was adopted to enhance the capability of 3D U-Net for small object detection. Thirdly, total variation regularization was considered to add the continuity feature of each catheter. The results show that our proposed method can achieve high accuracy of catheter digitization.
A limitation of the study is patient MRI images that have obvious artifacts such as intensity non-uniformity and motion artifacts were excluded for training and testing. Methods have been proposed to reduce these artifacts42–45, for instance, intensity non-uniformity can be corrected using N4ITK46. Nevertheless, in real clinical scenario, clinicians may have to use MRI images with certain artifacts for treatment planning due to certain constraints. Therefore, an ideal multi-catheter segmentation algorithm should still be able to deliver good performance when given those type of MRI images. To apply our proposed method for these images with strong artifacts, one way is to use modern pre-processing approaches to reduce the artifacts46. Another way, which is one of our future investigations, is to use a two-stage deep learning method that integrating artifacts-reduction network into our proposed segmentation network, in which, the first stage is designed to remove artifacts before the images passing through the segmentation stage. Another limitation of this work is that, in order to explore spatial and structural information of catheters, a 3D deep learning architecture was adopted. To some extent, it increases computational costs like GPU memory compared to that in a 2D manner. Our future investigation is to develop a computationally efficient network architecture.
In summary, in this study, a novel deep learning-based method, namely, deep supervised attention U-Net with TV regularization, was implemented and validated for multi-catheter detection in MRI images acquired post-insertion of catheters under the guidance of TRUS in MRI-guided HDR prostate brachytherapy. Quantitative evaluations include catheter tip localization error, catheter shaft detection error, and catheter detection accuracy were made using the manual contours done by experienced physicists as ground truth. Compared to very recent study on catheter segmentation in MRI-guided gynecologic brachytherapy using conventional deep convolutional neural network model34, our proposed method achieves higher accuracy and precision attribute to our advanced network architecture. Catheter reconstruction is a critical process during the treatment planning. The precision and accuracy of catheter delineation have a significant impact on the dose calculation, that will directly affect the accuracy of dose delivery compared to the prescription dose. It has potential to implement highly automated catheter digitalizing in MRI-guided HDR prostate brachytherapy treatment planning using the proposed approach. It can help removing the time-expensive manual delineation procedure which is generally done by physicists. To some extent, it can also speed up the entire process and may also avoid human error.
ACKNOWLEDGMENTS
This research is supported in part by the National Cancer Institute of the National Institutes of Health under Award Number R01CA215718 (XY), the Department of Defense (DoD) Prostate Cancer Research Program (PCRP) Award W81XWH-17-1-0438 (TL), W81XWH-17-1-0439 (AJ), and W81XWH-19-1-0567 (XD), and Dunwoody Golf Club Prostate Cancer Research Award (XY), a philanthropic award provided by the Winship Cancer Institute of Emory University.
Footnotes
DISCLOSURES
The authors declare no conflicts of interest.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians. 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 2.Morton GC. The emerging role of high-dose-rate brachytherapy for prostate cancer [published online ahead of print 2005/07/07]. Clin Oncol (R Coll Radiol). 2005;17(4):219–227. [DOI] [PubMed] [Google Scholar]
- 3.Yamada Y, Rogers L, Demanes DJ, et al. American Brachytherapy Society consensus guidelines for high-dose-rate prostate brachytherapy [published online ahead of print 2012/01/24]. Brachytherapy. 2012;11(1):20–32. [DOI] [PubMed] [Google Scholar]
- 4.Rylander S, Buus S, Pedersen EM, Bentzen L, Tanderup K. Dosimetric impact of contouring and needle reconstruction uncertainties in US-, CT- and MRI-based high-dose-rate prostate brachytherapy treatment planning [published online ahead of print 2017/03/13]. Radiother Oncol. 2017;123(1):125–132. [DOI] [PubMed] [Google Scholar]
- 5.Pinto F, Totaro A, Palermo G, et al. Imaging in prostate cancer staging: present role and future perspectives. Urol Int. 2012;88(2):125–136. [DOI] [PubMed] [Google Scholar]
- 6.Peikari M, Chen TK, Lasso A, Heffter T, Fichtinger G, Burdette EC. Characterization of ultrasound elevation beamwidth artifacts for prostate brachytherapy needle insertion. Med Phys. 2012;39(1):246–256. [DOI] [PubMed] [Google Scholar]
- 7.Villeirs GM, De Meerleer GO. Magnetic resonance imaging (MRI) anatomy of the prostate and application of MRI in radiotherapy planning. Eur J Radiol. 2007;63(3):361–368. [DOI] [PubMed] [Google Scholar]
- 8.Peters M, van Son MJ, Moerland MA, et al. MRI-Guided Ultrafocal HDR Brachytherapy for Localized Prostate Cancer: Median 4-Year Results of a feasibility study [published online ahead of print 2019/03/31]. Int J Radiat Oncol Biol Phys. 2019;104(5):1045–1053. [DOI] [PubMed] [Google Scholar]
- 9.Alayed Y, D’Alimonte L, Helou J, et al. MRI assisted focal boost integrated with HDR monotherapy study in low and intermediate risk prostate cancer (MARS): Results from a phase II clinical trial [published online ahead of print 2019/10/01]. Radiother Oncol. 2019;141:144–148. [DOI] [PubMed] [Google Scholar]
- 10.Hosni A, Carlone M, Rink A, Menard C, Chung P, Berlin A. Dosimetric feasibility of ablative dose escalated focal monotherapy with MRI-guided high-dose-rate (HDR) brachytherapy for prostate cancer [published online ahead of print 2016/12/06]. Radiother Oncol. 2017;122(1):103–108. [DOI] [PubMed] [Google Scholar]
- 11.Maenhout M, Peters M, van Vulpen M, et al. Focal MRI-guided salvage high-dose-rate brachytherapy in patients with radiorecurrent prostate cancer. Technol Cancer Res Treat. 2017;16(6):1194–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menard C, Susil RC, Choyke P, et al. MRI-guided HDR prostate brachytherapy in standard 1.5T scanner [published online ahead of print 2004/07/28]. Int J Radiat Oncol Biol Phys. 2004;59(5):1414–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maenhout M, Peters M, Moerland MA, et al. MRI guided focal HDR brachytherapy for localized prostate cancer: Toxicity, biochemical outcome and quality of life [published online ahead of print 2018/08/23]. Radiother Oncol. 2018;129(3):554–560. [DOI] [PubMed] [Google Scholar]
- 14.Murgic J, Chung P, Berlin A, et al. Lessons learned using an MRI-only workflow during high-dose-rate brachytherapy for prostate cancer. Brachytherapy. 2016;15(2):147–155. [DOI] [PubMed] [Google Scholar]
- 15.Buus S, Rylander S, Hokland S, et al. Learning curve of MRI-based planning for high-dose-rate brachytherapy for prostate cancer. Brachytherapy. 2016;15(4):426–434. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Fan Z, Carroll TJ, et al. Three-dimensional T2-weighted MRI of the human femoral arterial vessel wall at 3.0 Tesla. Invest Radiol. 2009;44(9):619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaffino P, Pernelle G, Mastmeyer A, et al. Fully automatic catheter segmentation in MRI with 3D convolutional neural networks: application to MRI-guided gynecologic brachytherapy [published online ahead of print 2019/07/05]. Phys Med Biol. 2019;64(16):165008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehrtash A, Ghafoorian M, Pernelle G, et al. Automatic needle segmentation and localization in MRI with 3-D convolutional neural networks: application to MRI-targeted prostate biopsy. IEEE Trans Med Imaging. 2018;38(4):1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uherčík M, Kybic J, Liebgott H, Cachard C. Model fitting using RANSAC for surgical tool localization in 3-D ultrasound images. Ieee T Bio-Med Eng. 2010;57(8):1907–1916. [DOI] [PubMed] [Google Scholar]
- 20.Görres J, Brehler M, Franke J, et al. Intraoperative detection and localization of cylindrical implants in cone-beam CT image data. Int J Comput Assist Radiol Surg. 2014;9(6):1045–1057. [DOI] [PubMed] [Google Scholar]
- 21.Mastmeyer A, Pernelle G, Ma R, Barber L, Kapur T. Accurate model-based segmentation of gynecologic brachytherapy catheter collections in MRI-images. Med Image Anal. 2017;42:173–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, Cachard C, Liebgott H. Automatic needle detection and tracking in 3D ultrasound using an ROI-based RANSAC and Kalman method. Ultrasonic Imaging. 2013;35(4):283–306. [DOI] [PubMed] [Google Scholar]
- 23.Dise J, Liang X, Scheuermann J, et al. Development and evaluation of an automatic interstitial catheter digitization tool for adaptive high-dose-rate brachytherapy. Brachytherapy. 2015;14(5):619–625. [DOI] [PubMed] [Google Scholar]
- 24.Dong X, Lei Y, Tian S, et al. Synthetic MRI-aided multi-organ segmentation on male pelvic CT using cycle consistent deep attention network [published online ahead of print 2019/10/22]. Radiother Oncol. 2019;141:192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong X, Lei Y, Wang T, et al. Automatic multiorgan segmentation in thorax CT images using U-net-GAN [published online ahead of print 2019/02/28]. Med Phys. 2019;46(5):2157–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu YB, Liu S, Li HH, Yang DS. Automatic and hierarchical segmentation of the human skeleton in CT images. Physics in Medicine and Biology. 2017;62(7):2812–2833. [DOI] [PubMed] [Google Scholar]
- 27.Fu YB, Mazur TR, Wu X, et al. A novel MRI segmentation method using CNN-based correction network for MRI-guided adaptive radiotherapy. Medical Physics. 2018;45(11):5129–5137. [DOI] [PubMed] [Google Scholar]
- 28.Lei Y, Dong X, Tian Z, et al. CT prostate segmentation based on synthetic MRI-aided deep attention fully convolution network [published online ahead of print 2019/11/21]. Med Phys. 2019;in press, doi: 10.1002/mp.13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei Y, Tian S, He X, et al. Ultrasound prostate segmentation based on multidirectional deeply supervised V-Net [published online ahead of print 2019/05/11]. Med Phys. 2019;46(7):3194–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei Y, Wang T, Tian S, et al. Male pelvic multi-organ segmentation aided by CBCT-based synthetic MRI [published online ahead of print 2019/12/19]. Phys Med Biol. 2019;in press, doi: 10.1088/1361-6560/ab63bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, Lei Y, Tian S, et al. Deeply supervised 3D fully convolutional networks with group dilated convolution for automatic MRI prostate segmentation [published online ahead of print 2019/02/01]. Med Phys. 2019;46(4):1707–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T, Lei Y, Tang H, et al. A learning-based automatic segmentation and quantification method on left ventricle in gated myocardial perfusion SPECT imaging: A feasibility study [published online ahead of print 2019/01/30]. J Nucl Cardiol. 2019;In press, doi: 10.1007/s12350-019-01594-2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang T, Lei Y, Tian S, et al. Learning-based automatic segmentation of arteriovenous malformations on contrast CT images in brain stereotactic radiosurgery [published online ahead of print 2019/05/06]. Med Phys. 2019;46(7):3133–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Xin J, Yang X, et al. Deep morphology aided diagnosis network for segmentation of carotid artery vessel wall and diagnosis of carotid atherosclerosis on black-blood vessel wall MRI [published online ahead of print 2019/07/30]. Med Phys. 2019;46(12):5544–5561. [DOI] [PubMed] [Google Scholar]
- 35.Lee H, Mansouri M, Tajmir S, Lev MH, Do S. A deep-learning system for fully-automated peripherally inserted central catheter (PICC) tip detection. J Digit Imaging. 2018;31(4):393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pourtaherian A, Zanjani FG, Zinger S, Mihajlovic N, Ng GC, Korsten HH. Robust and semantic needle detection in 3D ultrasound using orthogonal-plane convolutional neural networks. Int J Comput Assist Radiol Surg. 2018;13(9):1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, He X, Tian Z, et al. Multi-needle Detection in 3D Ultrasound Images Using Unsupervised Order-graph Regularized Sparse Dictionary Learning. IEEE Trans Med Imaging. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Lei Y, Qiu RL, et al. Multi-needle Localization with Attention U-Net in US-guided HDR Prostate Brachytherapy. Med Phys. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Paper presented at: International Conference on Medical image computing and computer-assisted intervention 2015. [Google Scholar]
- 40.Oktay O, Schlemper J, Folgoc LL, et al. Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:180403999. 2018. [Google Scholar]
- 41.Schlemper J, Oktay O, Schaap M, et al. Attention gated networks: Learning to leverage salient regions in medical images. Med Image Anal. 2019;53:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hedley M, Yan H, Rosenfeld D. Motion artifact correction in MRI using generalized projections. IEEE Trans Med Imaging. 1991;10(1):40–46. [DOI] [PubMed] [Google Scholar]
- 43.Atkinson D, Hill DL, Stoyle PN, et al. Automatic compensation of motion artifacts in MRI. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 1999;41(1):163–170. [DOI] [PubMed] [Google Scholar]
- 44.Duffy BA, Zhang W, Tang H, et al. Retrospective correction of motion artifact affected structural MRI images using deep learning of simulated motion. 2018. [Google Scholar]
- 45.Vovk U, Pernus F, Likar B. A review of methods for correction of intensity inhomogeneity in MRI. IEEE Trans Med Imaging. 2007;26(3):405–421. [DOI] [PubMed] [Google Scholar]
- 46.Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310. [DOI] [PMC free article] [PubMed] [Google Scholar]


