Abstract
Carotenoids are lipid-soluble yellow to orange pigments produced by plants, bacteria, and fungi. They are consumed by animals and metabolized to produce molecules essential for gene regulation, vision, and pigmentation. Cave animals represent an interesting opportunity to understand how carotenoid utilization evolves. Caves are devoid of light, eliminating primary production of energy through photosynthesis and therefore limiting carotenoid availability. Moreover, the selective pressures that favor carotenoid-based traits, like pigmentation and vision, are relaxed. Astyanax mexicanus is a species of fish with river-adapted (surface) and multiple cave-adapted populations (i.e. Tinaja, Pachón, Molino). Cavefish exhibit regressive features, such as loss of eyes and melanin pigment, and constructive traits, like increased sensory neuromasts and starvation resistance. Here we show that unlike surface fish, Tinaja and Pachón cavefish accumulate carotenoids in the visceral adipose tissue (VAT). Carotenoid accumulation is not observed in Molino cavefish, indicating that it is not an obligatory consequence of eye loss. We used quantitative trait loci (QTL) mapping and RNA sequencing to investigate genetic changes associated with carotenoid accumulation. Our findings suggest that multiple stages of carotenoid processing may be altered in cavefish, including absorption and transport of lipids, cleavage of carotenoids into un-pigmented molecules, and differential development of intestinal cell types involved in carotenoid assimilation. Our study establishes A. mexicanus as a model to study the genetic basis of natural variation in carotenoid accumulation and how it impacts physiology.
Keywords: Cavefish, carotenoids, Astyanax mexicanus, evolution, adipose tissue, QTL mapping
Graphical Abstract
1. Introduction
Animals obtain carotenoids exclusively from their diet. Carotenoid metabolism produces the chromophore for all known visual systems (11-cis-retinal, Vitamin A) and an essential regulator of development and gene expression (retinoic acid). Differential processing and distribution of carotenoids is responsible for the spectacular range of yellow to red colors observed in bird plumage, fish pigment, and reptile skin (reviewed in Toews et al., 2017). In humans, carotenoid levels have been associated with the propensity for developing certain cancers, cardiovascular disease, and macular degeneration (reviewed in Eggersdorfer and Wyss, 2018). Despite interest in understanding natural variation in carotenoid processing and utilization, there are a limited number of genetic models.
Carotenoid-based traits are influenced by many variables. Animal diets differ in carotenoid abundance over space and time, making it difficult to make functional comparisons using wild populations. Moreover, how carotenoids are digested, absorbed, processed, and translocated likely involves many genes. Once carotenoids are released from the food matrix, they are mixed with bile salts and fats to form micelles that are absorbed into enterocytes by the action of lipid transporters. Carotenoids can be cleaved within the enterocyte or packaged into chylomicrons that are transferred into the lymphatic system. Chylomicrons are differentially routed to tissues based on tissue-specific expression of lipoprotein receptors.
Genetic variation at each of the steps of carotenoid assimilation has been linked to changes in carotenoid-based traits (reviewed in Moran et al., 2018; Toews et al., 2017). For example, mutations in CD36-family lipid transporters produce pale-colored silkworms due to reduced transport of β-carotene and lutein (Sakudoh et al., 2010, 2013). In salmon, the CD36-family lipid transporter SCARB1 is highly expressed in the midgut and genetic variation at this locus is associated with orange muscle color (Sundvold, Helgeland, Baranski, Omholt, & Våge, 2011). Mutations in genes encoding enzymes that convert carotenoids into unpigmented metabolites, such as β -carotene oxygenase (BCO1) and β-carotene-9′,10′-dioxygenase (BCO2), are linked with yellow phenotypes in chickens, sheep, and cows (Berry et al., 2009; Eriksson et al., 2008; Våge & Boman, 2010). In humans, single nucleotide polymorphisms in SCARB1 and BCO1 are associated with carotenoid levels in the serum (Borel et al., 2013; Hendrickson et al., 2012). Considering the potential health benefits of carotenoids, there is commercial interest in breeding animals that produce high carotenoid levels as well as understanding what causes variation in human carotenoid absorption.
Carotenoid utilization has played a role in adaptive radiation of birds, reptiles and fish. In addition to the role in vision, carotenoids are utilized for pigmentation. Red feather color in multiple bird species has been associated with CYP2J19, a gene that encodes a cytochrome P450 enzyme that likely coverts yellow carotenoids to red keto-carotenoids (Lopes et al., 2016; Mundy et al., 2016). The CYP2J19 gene is also present in some turtle lineages and is highly expressed in the retina and red portions of the shell (Twyman, Valenzuela, Literman, Andersson, & Mundy, 2016). Extensive research on the elaboration of carotenoid-based pigment in cichlid fish has revealed associations between carotenoid processing and mate-preference, social dominance, and oxidative stress (reviewed in Sefc et al., 2014). Genetic mapping and gene expression studies in cichlids indicate that genes that influence carotenoid metabolism (bco2), or the development of pigment-containing xanthaphore cells (csf1ra, edn3b, pax7) are associated with variation in carotenoid-based pigment (Diepeveen & Salzburger, 2011; O’Quin, Drilea, Conte, & Kocher, 2013; Salzburger, Braasch, & Meyer, 2007). However, the evolutionary trade-off of carotenoid coloration with other phenotypes dependent on carotenoids like vision and antioxidant function is difficult to dissect.
Studying carotenoid utilization in cave species offers a novel opportunity to disentangle the variables that contribute to the evolution of carotenoid-based traits. Cave environments are devoid of light, eliminating the favorability of pigmentation and vision, but also limiting carotenoid availability. Wolfe and Cornwell (1964) compared carotenoid levels in cave-adapted and non-cave-adapted species of crayfish found in the same cave in Kentucky. They found two pigments in the cave form (β-carotene and lutein) and eight in the surface form. The level of β-carotene was over six times higher in the cave-adapted form and the level of lutein was over twelve times higher. The authors concluded that the cave forms lost the ability to metabolize dietary carotenoids, but the genetic basis of this difference and its impact on other crayfish phenotypes was not investigated.
The Mexican tetra, Astyanax mexicanus, is a powerful model to study the genetic basis of evolution. It is a single species of fish that exists as a river-adapted form (surface fish) found in rivers from Central Mexico to Southern Texas and a cave-adapted form (cavefish) found in perpetually dark limestone caves of the Sierra de El Abra region in Northeastern Mexico (Elliott, 2015). Surface fish and cavefish have adapted to dramatically different diets; river fish consume plants and insects throughout the year (Lee & Lee, 2017), while most cavefish populations are dependent on external sources of food brought in by seasonal flooding or bat droppings (Turner, 2017). There are 29 known cave populations that are of polyphyletic origin. Of the three cave populations used in this study, those from the Tinaja and Pachón caves form a clade separate from Molino cavefish (Herman et al., 2018). The populations are interfertile and amenable to genetic manipulation (Stahl et al., 2019) allowing researchers to investigate the genetic changes associated with behavioral (Chin et al., 2018; Hyacinthe, Attia, & Rétaux, 2019; Lloyd et al., 2018; Yoshizawa et al., 2015), metabolic (Aspiras, Rohner, Martineau, Borowsky, & Tabin, 2015; Riddle, Aspiras, et al., 2018; Xiong, Krishnan, Peuß, & Rohner, 2018), and morphological (Gross & Powers, 2018; Lyon, Powers, Gross, & O’Quin, 2017) evolution utilizing the different cave populations as natural replicates.
While understanding eye and pigment regression has been a major focus in A. mexicanus research (Gore et al., 2018; Jeffery, Ma, Parkhurst, & Bilandžija, 2016; Klaassen, Wang, Adamski, Rohner, & Kowalko, 2018; Krishnan & Rohner, 2017), there is a dearth of knowledge on carotenoid processing. It has been noted that cavefish populations are more yellow to orange in appearance (Hinaux et al., 2011), but the molecular and genetic basis of this difference is unclear. Based on whole transcriptome comparison at multiple developmental stages, Pachón cavefish have greater expression of an enzyme that converts ß-carotene into colorless compounds (Bco2), a surprising finding considering Pachón larvae appear more yellow (Stahl & Gross, 2017). How carotenoid-processing evolved in cave-adapted forms of A. mexicanus, and its potential influence on other traits, has remained unexplored.
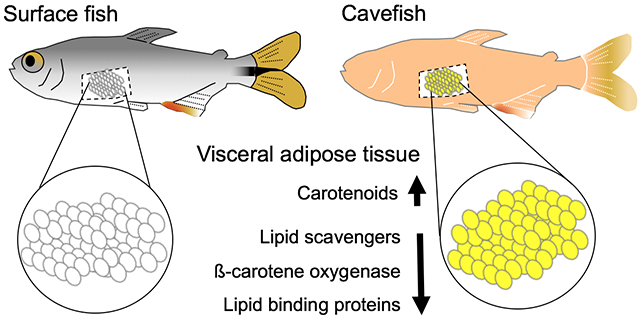
Here we present the first analysis of carotenoid processing in laboratory-raised A. mexicanus. We show that both surface fish and cavefish exhibit carotenoid-based pigment, but unlike surface fish, Pachón and Tinaja cavefish accumulate carotenoids in the visceral adipose tissue (VAT). Accumulation of carotenoids is not associated with eye loss or changes in the length of the digestive tract. Using genetic mapping, we discovered a quantitative trait locus associated with yellow VAT and identified coding mutations in a gene that could influence absorption of carotenoids by altering the number of absorptive cell types in the intestine. We found that Tinaja cavefish have reduced expression of genes important for carotenoid absorption, metabolism, and transport. Finally, we discuss how these differences may have evolved and the potential impact on cavefish physiology.
2. Materials and Methods
2.1. Fish husbandry and diet
Fish husbandry was performed according to (Elipot, Legendre, Père, Sohm, & Rétaux, 2014). Unless stated otherwise, fish were fed ad libitum with a combination of New Life Spectrum TheraA+ small fish formula and Artemia and housed at densities of less than, or equal to, two adult fish per liter of water. For fixed diet experiments fish were housed individually in 1.5L tanks and fed three pellets (approximately 6mg) of New Life Spectrum TheraA+ small fish formula once per day for greater than 4 months. The low-carotenoid diet consisted of Enchytraeus buchholzi (Fishgobble.com) fed ad libitum.
2.2. High Pressure Liquid Chromatography
We extracted pigments from fat samples of the same weight using the following protocol: we added equal parts hexane and methanol to the sample, vortexed the sample for two minute, added an equal volume of water, vortexed the sample for one minute, applied centrifugation at 4,000rpm for 10 minutes, and passed the upper phase through a 0.2-micron nylon filter. The samples were analyzed using an Agilent Technologies 1200 Analytical HPLC with C18 reversed-phase column (4.6mm × 150mm, 80A) with an isocratic mobile phase (acetone:water 50:50 v/v) at a flow rate of 1mL/minute for 45 minutes. UV absorbance was monitored with a Multi-Wavelength Detector (MWD) at 215nm. We included the following standards dissolved in hexane: HPLC grade retinol (Sigma-Aldrich R7632) and β-carotene (Sigma-Aldrich, 22040).
2. 3. Imaging
Images that were used to make comparisons between populations and conditions were taken with the same methods as described here. Figure 1B, C. The entire mass of organs was dissected out and imaged in PBS using the same illumination on a Leica M165SC stereomicroscope with DFC310 FX color camera. Figure 2. Samples of VAT were placed on a white background and imaged using a Cannon Powershot D12 digital camera mounted on a photography copy stand. An 18% photo grey card was included in each image for normalization. Figure 3. Samples of VAT from F2 hybrids were placed on a black background and images were taken using a Cannon Powershot D12 digital camera mounted on a photography copy stand. An 18% photo grey card was included in most images (see section 2.4). Figure 7. Fry were imaged with Leica DFC310 FX color camera. White balance was set using LAS 4.9 software and reference card (www.WhiBal.com). Images were taken in the same petri dish utilizing the white (A, D) and black (B,C,E,F) panes on the reference card as the background.
Figure 1. Astyanax mexicanus cavefish accumulate yellow pigment in the visceral adipose tissue (VAT).

Image of adult surface fish (A) and Tinaja cavefish (C) pointing out caudal fin (yellow arrow) and anal fin (white arrow). Image of the visceral organs of surface fish (B) and Tinaja cavefish (D). Enlarged image of VAT (B’, D’). E-F, HPLC chromatograms of pigments extracted from VAT of surface fish (E) and Tinaja cavefish (F) showing retention time and absorbance intensity (mAU) at 215nm. G, Bar graph comparing peak area in the HPLC chromatogram at the indicated retention time.
Figure 2. Ecotypic differences in A. mexicanus VAT color are independent of appetite and dependent on diet.

A, Fish were individually housed and consumed 6 mg of high-carotenoid food for greater than 4 months; figure shows fish and region of VAT sample (S, surface; T, Tinaja; P, Pachón; M, Molino). B, Fish were fed non-parasitic white grindal worms ad libitum for their entire lives; figure shows fish and region of VAT sample (S, surface; T, Tinaja; P, Pachón; M, Molino). C, Red intensity (iR) of VAT image from fish of the indicated populations and diet (fixed pellet: surface n= 2, Tinaja n = 7, Pachón n= 4, Molino n= 3; white worms: surface n= 8, Tinaja n = 10, Pachón n= 5, Molino n= 8). For box plots, median, 25th, 50th, and 75th percentiles are represented by horizontal bars and vertical bars represent 1.5 interquartile ranges (p-values from one-way ANOVA with Turkey’s post hoc test).
Figure 3. Eye regression is not linked with carotenoid accumulation in A. mexicanus.

A, Image of white fat sample (white dotted outline) from eyeless F2 surface/Tinaja hybrid. B, Image of yellow fat sample (white dotted outline) from eyed F2 hybrid fish. C, Plot showing left eye diameter and VAT red intensity of eyed F2 surface/Tinaja hybrid fish (n=99). Tau and p-value from Kendall rank correlation test. Results from right eye diameter: tau = −0.003, p-value = 0.97.
Figure 7. Maternal contribution of carotenoids in A. mexicanus cavefish.

A-C, Images of surface fish (S) and Pachón cavefish (P) larvae four days post fertilization. D-F, Images of hybrid larvae produced from pairing a Pachón cavefish female with surface fish male (P × S) and surface fish female with Pachón cavefish male (S × P).
2.4. Quantification of VAT color
We used MATLAB to obtain mean RGB values of a 16×16 pixel region of the VAT (Figure S2), and a 16×16 pixel region of an 18% photo grey card within the same image. We subtracted the grey card value from the sample value for each channel (mean sample R – mean grey card R = Rstand) to obtain standardized RGB values (Rstand, Bstand, Gstand). We then calculated normalized intensity of red: nIR = Rstand/ (Rstand + Gstand + Bstand). This measurement has been used as an estimate of carotenoid-based coloration in stickleback pelvic spines (Amundsen, Nordeide, Gjøen, Larsen, & Egeland, 2015). For F2 hybrid samples, we scored VAT color as either yellow or not yellow at the time of dissection. To obtain a more quantitative measure of VAT color in the F2 hybrids, we took images of the VAT and measured mean RGB level as described above in the images that had enough VAT to measure (n=97). For the majority of the images (n=62), a standard grey card was included, and we determined normalized red intensity (nIR). Images that did not include a grey card (n=35) were taken in the same light and we calculated red intensity (IR) without standardizing the values. The final set of measurements used for QTL and correlation analysis consisted of the combined normalized and non-normalized IR values (Figure S5). The same QTL peak was obtained if only normalized values were analyzed.
2.5. Quantitative trait loci (QTL) analysis
2.5.1. F2 hybrid population
We bred a surface fish female with a Pachón cavefish male to produce a clutch of F1 hybrids. We generated a population of surface/Pachón F2 hybrids by interbreeding individuals from this clutch. The F2 mapping population consisted of three clutches produced from breeding paired F1 surface/ Pachón hybrid siblings.
2.5.2. Genotype by sequencing
We extracted DNA from caudal tail fins using DNeasy Blood and Tissue DNA extraction kit (Qiagen). DNA was shipped to Novogene (Chula Vista, CA) for quality control analysis and sequencing. Samples that contained greater than 1.5 ug DNA, minimal degradation (as determined by gel electrophoresis), and OD260/OD280 ratio of 1.8 to 2.0 were used for library construction. Each genomic DNA sample (0.3~0.6 μg) was digested with Mse1, HaeIII, and EcoR1. Digested fragments were ligated with two barcoded adapters: a compatible sticky end with the primary digestion enzyme and the Illumina P5 or P7 universal sequence. All samples were pooled and size-selected after several rounds of PCR amplification to obtain the required fragments needed to generate DNA libraries. Concentration and insert size of each library was determined using Qubit® 2.0 fluorometer and Agilent® 2100 bioanalyzer respectively. Finally, quantitative real-time PCR (qPCR) was used to detect the effective concentration of each library. Qualified DNA libraries had an effective concentration of greater than 2 nM and were pooled by effective concentration and expected data production. Pair-end sequencing was then performed on Illumina® HiSeq platform, with the read length of 144 bp at each end. Raw Illumina genotype-by-sequencing reads were cleaned and processed through the process_shortreads command in the Stacks software package (Catchen, Amores, Hohenlohe, Cresko, & Postlethwait, 2011). The cleaned reads were aligned to the Astyanax mexicanus reference genome (AstMex102, INSDC Assembly GCA_000372685.1, Apr 2013) using the Bowtie2 software (Langmead & Salzberg, 2012). The aligned reads of 4 surface fish, 4 Tinaja cavefish and 4 F1 surface/Tinaja hybrids were manually stacked using the pstacks command. We then assigned the morphotypic origin of each allele and confirmed heterozygosity in the F1 samples using the cstacks command. Finally, we used this catalog to determine the genotypes at each locus in the F2 samples with the sstacks and genotypes command. This genotype database was formatted for use in R/qtl (Arends, Prins, Jansen, & Broman, 2010)
2.5.3. Linkage map construction
Using R/qtl, we selected for markers that were homozygous and had opposite genotypes in cavefish versus surface fish (based on three surface and three cave individuals). A linkage map was constructed from these loci using only the F2 population. All markers that were genotyped in less than 180 individuals were omitted, as well as all individuals that had poor marker genotyping (<1500 markers). Markers that did not conform to the expected allele segregation ratio of 1:2:1 were also omitted (p < 1e−10). These methods produced a map with 1839 markers, 219 individuals, and 29 linkage groups. Unlinked markers and small linkage groups (<10 markers) were omitted until our map consisted of the optimal 25 linkage groups (Astyanax mexicanus has 2n = 50 chromosomes (Kavalco & De Almeida-Toledo, 2007)). Each linkage group had 20–135 markers and markers were reordered by default within the linkage groups, producing a total map length of > 5000 cM. Large gaps in the map were eliminated by manually switching marker order to the best possible order within each linkage group (error.prob = 0.005). The final linkage map consisted of 1800 markers, 219 individuals and 25 linkage groups, spanning 2871 cM. Maximum spacing was 32.1 cM and average spacing was 1.6 cM.
2.5.4. QTL statistics
Using R/qtl we calculated the LOD of association using Haley-Knott regression and a binary model for VAT color scored as a binary trait (0=white, 1=yellow). and a normal model for VAT red intensity (IR). We assessed statistical significance of the LOD score by calculating the 95th percentile of genome-wide maximum penalized LOD score using 1000 random permutations (scanone). We estimated confidence intervals for the QTL using 1.5-LOD support interval (lodint) and 95% Bayesian credible interval (bayesint) expanded to the nearest genotyped markers.
2.6. Sequence analysis of candidate genes
Sequences were obtained from Ensembl gene builds for surface fish (Astyanax_mexicanus-2.0, INSDC Assembly GCA_000372685.2, Sep 2017) and Pachón cavefish (AstMex102, INSDC Assembly GCA_000372685.1, Apr 2013). Sequence comparisons between wild-caught fish were made using data from (Herman et al., 2018). Sequence alignments were produced using CLC Sequence Viewer 8. Predicted protein domains were obtained from the UniProt database (Bateman, 2019). Transmembrane prediction analysis was carried out using DTU bioinformatics TMHMM server v. 2.0
(http://www.cbs.dtu.dk/services/TMHMM/). (Sonnhammer, von Heijne, & Krogh, 1998).
2.7. RNA sequencing
2.7.1. Extraction and cDNA synthesis
Adult A. mexicanus were euthanized in 400ppm Tricane and the hindgut was immediately removed and homogenized in 0.3mL Trizol using a motorized pellet pestle and stored at −80°C. Total RNA was extracted using Zymo Research Direct-zol RNA MicroPrep with DNAse treatment according to the manufacturers protocol. LunaScript RT supermix kit with 1μg of RNA was used to synthesize cDNA. All samples were processed on the same day. Diluted cDNA samples (50ng/μL) were used for sequencing.
2.7.2. Sequencing and differential gene expression analysis
HiSeq Illumina sequencing was performed by Novogene (Chula Vista, CA). All samples were indexed and run as pools, providing an estimated 20–30 million single-end reads per sample.
Samples were processed using an RNA-seq pipeline implemented in the bcbio-nextgen project (https://bcbio-nextgen.readthedocs.org/en/latest/). Raw reads were examined for quality issues using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to ensure library generation and sequencing are suitable for further analysis.
Reads were aligned to Ensembl build 2 of the Astyanax mexicanus genome, augmented with transcript information from Ensembl release 2.0.97 using STAR (Dobin et al., 2013) with soft trimming enabled to remove adapter sequences, other contaminant sequences such as polyA tails and low quality sequences. Alignments were checked for evenness of coverage, rRNA content, genomic context of alignments and complexity using a combination of FastQC, Qualimap (García-Alcalde et al., 2012), MultiQC (https://github.com/ewels/MultiQC), and custom tools. Counts of reads aligning to known genes were generated by featureCounts (Liao, Smyth, & Shi, 2014) and used for further QC with the R package bcbioRNASeq [Steinbaugh MJ, Pantano L, Kirchner RD et al. bcbioRNASeq: R package for bcbio RNA-seq analysis [version 2; peer review: 1 approved, 1 approved with reservations]. F1000Research 2018, 6:1976 (https://doi.org/10.12688/f1000research.12093.2)]. In parallel, Transcripts Per Million (TPM) measurements per isoform were generated by quasialignment using Salmon (Bray, Pimentel, Melsted, & Pachter, 2015) for downstream differential expression analysis as quantitating at the isoform level has been shown to produce more accurate results at the gene level (Soneson, Love, & Robinson, 2016). Salmon output was imported into R using tximport (Soneson, Love, & Robinson, 2015) and differential gene expression analysis and data visualization was performed using R with the DEseq 2 package (Love, Huber, & Anders, 2014; R-core-team, 2019). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were done using clusterProfiler (Yu, Wang, Han, & He, 2012).
3. Results and Discussion
3.1. Carotenoid-based pigmentation in A. mexicanus river and cave morphotypes
A. mexicanus surface morphs exhibit orange coloration on the caudal and anal fins (Figure 1A). Tinaja, Pachón, and Molino cavefish also exhibit orange coloration on the caudal and anal fins, although it is less intense (Figure 1B, 2A). Based on laboratory observation, anal fin color may be associated with social interaction in surface fish. When housed together in the laboratory, the fish that exhibits the most aggressive behavior develops the brightest orange stripe on the anterior edge of the anal fin (Supplementary Video 1). Interestingly, if surface fish are housed individually, they each develop a bright orange stripe (Figure 2A). It is unclear if the amount of food consumed is the only factor in determining stripe formation (i.e. dominant fish obtain the most food), or if the cellular and biochemical pathways involved in behavior or social interaction directly influence carotenoid-processing. It is also unclear the extent to which the stripe itself influences surface fish social hierarchy, mate preference or other behaviors. However, comparing river- and cave-adapted populations reveals that while the genetic changes accompanying cave colonization have led to reduced or absent melanin pigment, some carotenoid-based pigment has persisted despite loss of the selective pressure for visual cues.
3. 2. Tinaja cavefish accumulate yellow pigment in the visceral adipose tissue (VAT)
To investigate carotenoid processing in A. mexicanus, we started by examining the color of the internal organs. We dissected lab-raised adult surface fish and Tinaja cavefish fed ad libitum with a combination of brine shrimp and commercially available pellet food that contains high amounts of β-carotene. We observed a striking difference in the color of the visceral adipose tissue (VAT) that surrounds the internal organs; the VAT of surface fish appears white (Figure 1B, B’) while in Tinaja cavefish it is characterized by an intense yellow color (Figure 1D, D’). The yellow color led us to hypothesize that it represents an accumulation of dietary carotenoids. To test this hypothesis, we examined the VAT in fish that were raised on a diet that is comparatively low in carotenoids: small white non-parasitic worms Enchytraeus buchholzi (grindal worms). We found that cavefish fed this diet do not have yellow VAT (Figure 2).
We considered that similar to cave-adapted crayfish, Tinaja cavefish may have lost the ability to produce unpigmented molecules like retinol from carotenoids. We extracted pigments from the VAT of an individual surface fish and Tinaja cavefish that were greater than one-year old and were fed a high-carotenoid diet their entire lives. We analyzed the samples using high-pressure liquid chromatography (HPLC, see Methods) and determined the mobility of retinol and β-carotene as standards within the same experiment. We observed compounds with the same retention times in surface fish and Tinaja cavefish samples (Figure 1E–F). Three peaks have retention times of less than 21 minutes (14.2, 15.6, 19.7), representing pigments with similar properties to retinol (elution time of 18.8 minutes) and five peaks have elution times of greater than 41 minutes (44.2, 44.7, 45.3, 46.1, 46.6), representing pigments with similar properties to β-carotene (elution time of 42.2). We found that the height and area of all but one peak (retention time 46.6) is greater in the Tinaja sample (Figure 1G). These results suggest that surface fish and cavefish have the same pigments in the VAT, but Tinaja may have a greater concentration. In line with the hypothesis that Tinaja cavefish accumulate carotenoids from their diet over time, we found that there is no difference in the amount of pigments extracted from the VAT in fish that are less than 6 months old (Figure S1).
3. 3. Carotenoid accumulation in multiple cave populations is independent of appetite
We next investigated the variables that influence carotenoid accumulation. We compared three A. mexicanus cave populations that evolved independently: Tinaja, Molino and Pachón. Tinaja and Molino cavefish experience prolonged nutrient deprivation in the wild, followed by a sudden influx of food during seasonal flooding. Differently, the Pachón cave is a perched pool that is less impacted by seasonal flooding and the cavefish have access to cave invertebrates (Espinasa et al., 2017). A previous study revealed that a mutation in melanocortin receptor 4 (mc4r) is fixed in the Tinaja and Molino cavefish and is associated with increased appetite; a potential adaptation to the fluctuating nutrients in these caves (Aspiras et al., 2015). Interestingly, mutations in mc4r are associated with increased appetite and carotenoid levels in humans (Borel et al., 2014).
To test if differences in appetite are sufficient to explain altered carotenoid content in cavefish, we analyzed the VAT of surface, Tinaja, Pachón and Molino populations after fish were individually housed as adults and consumed 6 mg of carotenoid-containing pellet food per day for more than four months (Figure 2A, C). While the color difference of the VAT is not as striking to fish fed ad libitum, Tinaja cavefish on the controlled diet have VAT that is visibly more yellow compared to surface fish (Figure 2A). To quantify the differences between populations, we compared the normalized red intensity (nIR) of the VAT as a proxy for carotenoid level (see Methods and Figure S2). We found that mean nIR of the VAT is greater in Tinaja compared to surface fish although the results are not statistically significant (Figure 2A, C, surface = 0.363, Tinaja = 0.377, p=0.85, ANOVA with Tukey’s HSD post hoc test). Pachón cavefish VAT appears more yellow and has nIR that is significantly greater than surface fish (Pachón = 0.400, p=0.04). Oppositely, Molino cavefish VAT appears white and the mean nIR is lower than in surface fish, although it is not statistically significant (mean nIR = 0.356, p = 0.99).
The yellow color of the VAT in Tinaja and Pachón is entirely dependent on diet; VAT nIR decreases in fish fed low-carotenoid white worms (Tinaja nIR = 0.378 (high), 0.366 (low), p = 0.60 ; Pachón nIR = 0.400 (high), 0.368 (low), p=0.01, ANOVA with Tukey’s HSD post hoc test). Similar to what we observed in Tinaja cavefish, in Pachón cavefish that are less than 6 months old, we did not find a difference compared to surface fish in the level of pigments extracted from the VAT and analyzed by HPLC (Figure S1). Differently, in Molino cavefish, the level of carotenoids with retention time similar to β-carotene is significantly less than in surface fish (p=0.03, one-way ANOVA with Tukey’s post-hoc test). Combined, our results suggest that carotenoids from the diet accumulate in the VAT of Tinaja and Pachón cavefish, but not Molino cavefish. While carotenoid accumulation is dependent on diet, differences in appetite are not sufficient to explain the differences between populations. Our findings raise the possibility that all three cave populations have altered carotenoid assimilation; resulting in accumulation in Tinaja and Pachón cavefish, and lower levels in Molino cavefish.
3. 4. Carotenoid accumulation is not associated with eye regression
Carotenoid metabolites are essential for vision. We considered that lack of eyes may lead to an accumulation of carotenoids that are unused in eyeless cavefish. However, the presence of white VAT in eyeless Molino cavefish refutes this hypothesis. Nevertheless, loss of the selective pressures that favor eye formation and function could, in principle, have led to accumulation of mutations or genetic drift in pathways important for production of carotenoid metabolites. Changes to these pathways could, in turn, play a role in eye degeneration. Eye loss in cavefish is likely the result of multiple genetic changes; there have been at least fourteen quantitative trait loci identified that account for variation in eye traits (Protas, Conrad, Gross, Tabin, & Borowsky, 2007). To further dissect the interaction between eye loss and carotenoid content, we took advantage of the ability to interbreed surface fish and cavefish to produce F1 hybrids that have small eyes, and F2 hybrids that vary in eye presence and eye size.
We found that one out of three surface/Pachón F1 hybrids we examined had markedly yellow VAT (Figure S3). This hybrid also had an unidentified growth in the abdomen, introducing an additional variable that may influence VAT color. Activation of the immune system in birds and fish can alter carotenoid metabolism (Brown, 2013; Toomey, Butler, & McGraw, 2010) and this may also occur in the Mexican tetra.
We interbred F1 hybrids and analyzed VAT color and eye diameter in surface/Pachón F2 hybrids that were individually housed and consumed the same amount per day for more than four months (Figure 3, Figure S3). In hybrids that had enough VAT to analyze (124/217), we scored color as yellow (n=12) or white (n=112, Figure 3A, B). We found that some surface/cave F2 hybrids that lacked eyes have white VAT and vice versa, some fish with eyes have yellow VAT (Figure 3A, B). In addition, we found that average eye diameter normalized to standard fish length does not differ between fish with yellow or white VAT (left eye: 0.062 vs 0.065, p-value = 0.23, right eye: 0.066 vs 0.062, p-value = 0.22, Wilcox-rank test). Furthermore, VAT IR does not correlate with relative diameter of the left or right eyes (Figure 3C, p-value = 0.67, 0.12, Kendall’s rank correlation). These results suggest that carotenoid accumulation is not a pleiotropic consequence of eye loss, substantiating the conclusion we reached based on the Molino cavefish.
3.5. Caeca number and standard body length correlate with VAT color
We considered that ecotypic differences in the gastrointestinal tract could influence carotenoid accumulation by altering digestion and absorption. For example, Pachón cavefish have altered gut motility at post-larval stages that results in slower transit of food; an adaptation that could increase nutrient assimilation (Riddle, Boesmans, Caballero, Kazwiny, & Tabin, 2018). We utilized adult surface/Pachón F2 hybrids to investigate whether the length of the gut or number of gastrointestinal appendages, called pyloric caeca, are linked with VAT color. There is not a significant difference in the relative length (gut length/standard fish length) of any region of the gut between hybrids with yellow VAT versus white VAT (stomach: 0.092 vs 0.093, p =.78, midgut: 0.32 vs 0.32, p =0.66, hindgut:0.15 vs 0.16, p = 0.26, total relative gut length: 0.68, 0.70, p=0.55, Wilcox-rank test). Furthermore, VAT IR does not correlate with relative length of the stomach, midgut, hindgut, or total gut (stomach: p= 0.31, midgut: p=0.17, hindgut: p=0.77, total gut: p=0.43, Kendall’s rank correlation). While we found no difference in pyloric caeca number in fish with white versus yellow VAT scored as a binary trait (white = 7.9, yellow = 7.6, p=0.50), we observed a negative correlation between pyloric caeca number and IR (p=0.03, Figure S4, Kendall’s rank correlation). The pyloric caeca are a major site of immune cell recruitment during infection and caeca number varies widely across fish species (Ballesteros et al., 2013; Townsend, 1944). The link between pyloric caeca development, the immune system, and carotenoid assimilation is currently unclear, but the differences we observed between cave and river morphotypes will allow future investigations aimed at understanding how these variables interact.
We found a positive correlation between standard fish length and VAT IR (p=0.0007, Figure S4). Carotenoid levels have been associated with size in other fish species. For example, in Midas Cichlids, gold morphotypes grow bigger than grey morphotypes (Barlow, 1973). The faster growth rate of gold morphs is due to dominance in food competition. Our results in A. mexicanus suggest that fish size and carotenoid-based traits may be associated independent of food competition, since the fish were individually housed and consumed the same amount. Metabolic or physiological traits that maximize nutrient assimilation and promote growth may be advantageous in cavefish and, in principle, could lead to increased accumulation of carotenoids.
3.6. Genetic mapping reveals quantitative trait loci associated with VAT color
To search for genes associated with carotenoid accumulation, we carried out a quantitative trait loci (QTL) analysis using F2 surface/Pachón hybrids (see Methods). For each fish, we scored VAT color as a binary trait (0 = white, 1 = yellow) and calculated VAT red intensity (IR, see Methods and Figure S2). For both methods of quantification, we identified a QTL on linkage group 16 with LOD scores of 8.0 and 4.62 at the peak marker, r164788, accounting for 25.53% and 17.73% of the phenotype variance respectively (Figure 4A, B, Figure S5). We plotted the phenotypes and genotypes at the peak marker position and determined the mean effect score for each genotype (Figure 4C, D). Most of the fish with yellow VAT are homozygous for the cave allele at this position (8 out of 12, Figure 4C). The cave allele appears to be recessive since the heterozygous genotype does not produce an intermediate effect score when scored as a binary trait (white/yellow: SS = 0.04, SC= 0.03, CC = 0.82, additive score = 0.39, dominance score = −0.40) or a quantitative trait (IR: SS = 0.357, SC=0.354, CC = 0.376, additive score = 0.0095, dominance score = −0.012).
Figure 4: Quantitative trait loci analysis of visceral adipose tissue color in A. mexicanus.

A, Results of genome wide LOD calculation of VAT color using Haley-Knott regression and binary model (0=white, 1= yellow). B, LOD score of markers on linkage group 16. Significance threshold of 5% (black dotted line) determined by calculating the 95th percentile of genome-wide maximum penalized LOD score using 1000 random permutations. Grey highlighted region indicates the 1.5-LOD support interval. The region between the markers that map to chromosome 3 is expanded to show the relative location of annotated genes. C, Phenotype score for each genotype at the peak marker r164788 (SS: homozygous surface, CS: heterozygous, CC: homozygous cave. Circles represent individual fish and bars indicated mean ± standard error. D. Effect plot showing phenotype values (mean ± standard error) for the indicated genotypes at the peak marker (r164788).
To identify candidate genes within the QTL associated with VAT color, we determined the location of the genotyped markers that define the 1.5-LOD support interval and the 95% Bayesian credible interval, on the surface fish genome (Table 1). Table 1 shows the locations of the peak marker (r164788), the markers that define the boundary of the 1.5-LOD support interval (r165238, r180290) and the markers that define the boundaries of the 95% Bayes credible interval (r179950, r9999). Two of the markers map to chromosome three and the others are on separate unplaced scaffolds. We identified the genes in the region between the markers on chromosome 3 (3: 2805128– 3751814, Figure 4B). There are eight genes in this region: clip2, gtf2ird1, dtx2, tmem132e, and four that are uncharacterized (Figure 4B, Table 2).
Table 1.
LOD score for VAT color QTL peak marker (r164788), markers defining the 1.5-LOD support interval (r165238, r180290), and 95% Bayes credible interval (r179950, r999). Location of the marker on the linkage map (LG, Linkage group) and surface fish genome (APW indicates unplaced scaffold).
| marker | LOD | LG: position | Chromosome: position |
|---|---|---|---|
| r165238 | 3.4 | 16:01.4 | APWO02000155: 987504 |
| r9999 | 7.3 | 16:11.0 | APWO02000113: 510973 |
| r164788 | 8 | 16:18.4 | APWO020000139:6870990 |
| r179950 | 7.8 | 16:20.1 | 3: 2805128 |
| r180290 | 5.8 | 16:23.5 | 3: 3751814 |
Table 2.
Gene IDs, name, and available description of genes between the VAT color QTL markers that map to surface fish chromosome 3.
| Gene stable ID | Gene name | Description |
|---|---|---|
| ENSAMXG00000007486 | dtx2 | E3 ubiquitin-protein ligase |
| ENSAMXG00000007478 | clip-2 | CAP-Gly domain containing linker protein 2 |
| ENSAMXG00000007488 | tmem132e | Transmembrane protein |
| ENSAMXG00000007482 | gtf2ird1 | GTF2I repeat domain containing 1 |
| ENSAMXG00000033138 | Retrotransposable element | |
| ENSAMXG00000037591 | Transposase derived nuclease | |
| ENSAMXG00000029351 | ||
| ENSAMXG00000038139 |
We considered which of the genes in the VAT color QTL could influence carotenoid processing, based on known activities. Clip2 and tmem132 encode proteins that likely function in neurons and have roles in microtubule stability (Vandeweyer, Van Der Aa, Reyniers, & Kooy, 2012) and hair cell function, respectively (Sanchez-Pulido & Ponting, 2018). Gtf2ird1 encodes a general transcription factor that is ubiquitously expressed. Dtx2 encodes an E3 ubiquitin ligase that is expressed in the mouse and human gastrointestinal (GI) tract (Smith et al., 2019)(GTEx Analysis Release V8 (dbGaP Accession phs000424.v8.p2)). DTX2 can promote Notch activation or degradation depending on the context and interaction with other proteins (Hori et al., 2004; Matsuno, Diederich, Go, Blaumueller, & Artavanis-Tsakonas, 1995; Puca, Chastagner, Meas-Yedid, Israël, & Brou, 2013). Notch signaling in the intestinal epithelium is important for maintenance of crypt-based columnar cells and controls the proportion of secretory and absorptive cell types (Tian et al., 2015). There is evidence that Notch stability and trafficking is controlled by E3 ligases in the intestine, but the role of DTX2 specifically has not been investigated (reviewed in Sancho, Cremona, & Behrens, 2015). Considering that intestine absorptive cell number could in principle influence carotenoid assimilation, we focused on A. mexicanus dtx2 as a candidate gene revealed by our QTL analysis of VAT color.
3.7. Coding changes in Dtx2 E3-ubiquitin ligase in A. mexicanus cavefish
We compared surface fish and Pachón cavefish Dtx2 using the available annotations for the surface fish and Pachón cavefish genomes (Figure 5A, Figure S6). Surface fish have two splice isoforms of Dtx2 and Pachón cavefish have one. We aligned the three predicted protein sequences and found a number of differences in Pachón Dtx2. At the N- and C-terminus of the protein is truncated by 39 and 12 amino acids respectively; there is a valine to isoleucine substitution (V166I) in the WWE protein domain (AA29–195), two substitutions (R441H and S443L) in the RING domain (AA435–497), a substitution (P398A) not in a specific domain, and additional amino acids that create a predicted transmembrane domain (Figure 5A,B). One of the insertions that creates the transmembrane domain represents a difference in the cDNA sequence and the addition of two exons, the other is an addition of a repeating 20 base pair (bp) genomic DNA sequence (CTGCAAACTTTTCGTCCT); surface fish have two repeats and Pachón cavefish have three.
Figure 5. Coding changes in Dtx2 E3-ubiquitin ligase in A. mexicanus cavefish.

A, Alignment of predicted protein sequences from surface fish and Pachón cavefish reference genomes. Protein domains are highlighted in purple (WWE, RING, TM: transmembrane). Asterisks indicate position of Pachón amino acid substitutions. B, Results from transmembrane prediction analysis using Pachón cavefish sequence (DTU bioinformatics TMHMM server v. 2.0 http://www.cbs.dtu.dk/services/TMHMM/).
The coding differences we identified are likely to impact Dtx2 function; the WWE domain binds to Notch ankyrin repeats (Zweifel, Leahy, & Barrick, 2005) and the RING domain specifies E3 ubiquitin ligase activity by interacting with E2 conjugating enzymes (Metzger, Pruneda, Klevit, & Weissman, 2014). To determine if the sequence differences we identified are present in other cave populations, we examined the dtx2 locus in wild-caught fish from two surface populations (Rio Choy and Rascón), three cavefish populations (Pachón, Tinaja, Molino), a closely related species (Astyanax aeneus), and a more distantly related species (Gymnocorymbus ternetz, white long fin) using data from Herman et al., 2018 (Table 3, Data S1).
Table 3.
Percentage of fish with the indicated coding changes in Dtx2. “d” indicates that one fish had a deletion at this position.
| repeating sequence | |||||||
|---|---|---|---|---|---|---|---|
| V116I | P398A | R441H | S443L | x2 | x3 | x4 | |
| White long fin (n=1) | 0 | 100 | 0 | 0 | 0 | 0 | 100 |
| A. Aeunus (n=2) | 0 | 100 | 0 | 0 | 0 | 0 | 100 |
| Río Choy surface fish (n=10) | 22 | 30d | 0 | 0 | 30 | 20 | 50 |
| Rascón surface fish (n=6) | 100 | 100 | 0 | 0 | 67 | 17 | 17 |
| Tinaja cavefish (n=9) | 100 | 100 | 0d | 89 | 0 | 50 | 50 |
| Pachón cavefish (n=10) | 100 | 100 | 100 | 100 | 0 | 0 | 100 |
| Molino cavefish (n=9) | 100 | 100 | 0 | 0 | 78 | 0 | 22 |
We found that the V116I substitution is present in 100% of the fish from each cavefish population and the Rascón surface fish population. The substitution is present in 22% of the Río Choy surface population and is not present in the two outgroups. We found that only some of the Río Choy surface fish have a proline at position 398, suggesting that an alanine at this position may be the ancestral state. The R441H substitution is specific to fish from the Pachón cave and the S443L substitution is only present in Pachón and Tinaja cavefish. We found wild-caught fish with two, three, and four repeats of the 20bp sequence we identified comparing the sequences from the annotated genomes of surface fish and Pachón cavefish. All of the wild-caught Pachón cavefish have four repeats (n=10). The predicted transmembrane domain in Pachón cavefish Dtx2 is due to a difference in splicing and consequent changes to the cDNA sequence; therefore, it is currently unclear if the transmembrane domain is present in other cavefish populations.
We found that the S443L substitution in the RING domain is the only change exclusively in cavefish that exhibit carotenoid accumulation (Tinaja and Pachón). This residue is conserved between surface fish and humans (Figure S7, Data S2) and therefore represents a strong candidate for future functional analysis. Numerous transmembrane ubiquitin ligases are present in the mammalian genome that regulate protein degradation and influence cellular processes from membrane trafficking to proliferation (reviewed in Nakamura, 2011). Future studies will be aimed at understanding how the changes we identified in Pachón Dtx2 influence protein structure, interaction with Notch, and development of intestinal cell types.
3.8. Carotenoid-associated genes near the VAT color QTL
To search for other genetic changes that could influence carotenoid accumulation in cavefish, we reviewed the literature and generated a list of fifty genes that have known roles in carotenoid processing or are linked with human variation in carotenoid levels (Table S1) (Bohn et al., 2017; Moran et al., 2018; Toews et al., 2017). None of these genes are on the unplaced scaffolds that were mapped in the VAT color QTL (Table 1). However, two of the genes, fatty acid elongase 2 (elovl2) and apolipoprotein A-I (apoa1a), are located on chromosome 3 approximately 6mbp and 9mbp from marker r180290, which defines one edge of the 1.5-LOD support interval. We compared the predicted protein sequence of Elovl2 and Apoa1a using the available Pachón and surface genomes. There are no coding changes in Apoa1a. We found that Elovl2 has two amino acid substitutions (A274V, V281I). However, when we examined the sequences in wild-caught fish, we found that all populations have a valine at position 274, including surface populations. Only two Río Choy surface fish and the outgroups have a valine at position 281. In summary, we found consistent sequence differences in the coding region of dtx2 between surface and cave populations but not in other genes near the QTL associated with carotenoid levels in humans like apoa1a or elovl2.
3.9. RNA sequencing of A. mexicanus hindgut reveals ecotypic differences in gene expression
To more closely examine the molecular basis of carotenoid accumulation, we sought to compare expression of carotenoid-associated genes between surface fish and cavefish. We noticed a sharp contrast between the color of the midgut and hindgut in both populations; the hindgut appears more orange suggesting it may be a primary site of carotenoid absorption (Figure S8A). We therefore performed RNA sequencing on hindguts from surface, Tinaja, and surface/Tinaja F1 hybrids (n=5 per population). Each sample had over 40 million reads and nearly 90% mapping rate to the A. mexicanus genome. We generated a list of normalized gene counts for each sample to control for differences in sequencing depth and then used the counts to make comparisons between populations. We detected over 20,000 genes (i.e. at least one read per gene) in each sample. Principal Component Analysis of normalized, variance stabilized data showed that the samples clustered by population (Figure S8B).
We performed differential gene expression analysis by first comparing surface fish and Tinaja cavefish samples using DESEq2’s pairwise Wald test. We found that 3373 genes are differentially expressed, 1612 upregulated, and 1761 downregulated in cavefish (adjusted p-value <0.05 and log2FoldChange ≥ 0.58). None of the genes within the VAT color QTL, including elovl2 and apoa1a, are differentially expressed. We examined the biological process gene ontologies (GO terms) of genes that have more than a 2-fold difference in expression between surface fish and Tinaja cavefish at an adjusted p-value of less than 0.05 (Figure S9). The top five significant GO terms in this set of 979 genes are: multicellular organism process, multicellular organism development, pancreas development, digestive system development, and specification of symmetry. These results suggest that the largest differences in expression may be linked to developmental processes rather than gut absorptive function.
To identify genetic pathways that may be altered but do not show large changes in gene expression, we performed gene set enrichment analysis (GSEA) using the Kyoto Encyclopedia of Genes and Genomes (KEGG) gene set. GSEA aggregates the per gene statistics across genes within a gene set, therefore making it possible to detect situations where all genes in a predefined set change in a small but coordinated way. Although none of the gene sets we identified have a significant adjusted p-value, we found that retinol metabolism has the fourth greatest enrichment score (Table S2). In summary, the largest differences in gene expression result from developmental pathways, yet there is some evidence for coordinated changes in metabolic pathways linked with carotenoid processing.
3.10. Altered expression of genes involved in carotenoid assimilation in A. mexicanus cavefish
We next compared expression of genes that are known to regulate carotenoid absorption or that have been associated with carotenoid levels in humans (Table S1). We found that five genes are differentially expressed: scarb1, bco1, isx, mttp, and apoea (Figure 6). Scarb1 is a transmembrane receptor expressed in enterocytes that detects and mediates absorption of micelles containing carotenoids. Bco1 cleaves ß -carotene into retinal within enterocytes. Intestine-specific homeobox gene (Isx) is a transcription factor that represses expression scarb1 and bco1, thereby forming a feedback loop that is responsive to changes in carotenoid levels (Lobo et al., 2010). We found that expression of isx is significantly higher in Tinaja (1.24-fold) and that scarb1 and bco1 are significantly lower (−2.63-fold and −2.84-fold, Figure 6A) consistent with the regulation of these genes by Isx. Decreased bco1 expression is associated with carotenoid accumulation in other species (reviewed in Bohn et al., 2017; Toews et al., 2017) and our findings suggest reduced bco1 expression could contribute to accumulation of carotenoids in cavefish. The regulatory points at which the feedback loop between Scarb1, Bco1, and Isx are altered are currently unclear but it is possible that an increased number of absorptive cells due to the changes in Dtx2 leads to greater carotenoid accumulation and isx expression on the whole tissue level.
Figure 6. Altered expression of carotenoid-processing genes in A. mexicanus cavefish.
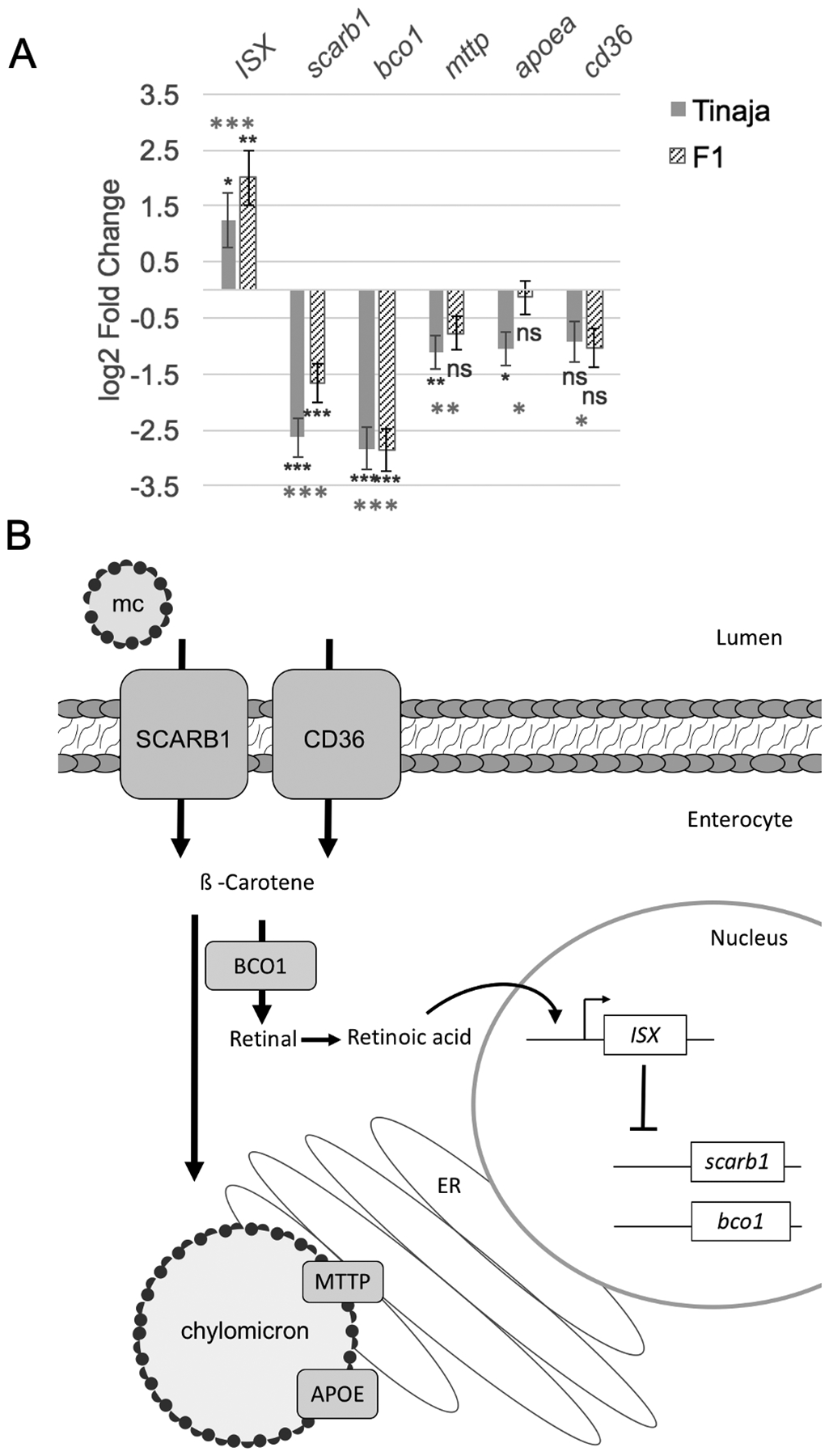
A, Bar graph showing log 2-Fold expression change compared to surface fish for the indicated gene in Tinaja cavefish hindguts and surface/Tinaja F1 hybrid hindguts (n=5 samples per population). Grey asterisks indicate significance from likelihood ratio test comparing all three sample types and black asterisks indicate significance from Wald test comparing to only surface fish (Significance code for adjusted p-value: *<.05 **<.005 ***<.0005, ns>.05). Error bars indicate standard error estimate for log2 fold change. B, Diagram of an enterocyte showing protein functions of the differentially expressed genes. Mixed micelles (mc) containing carotenoids are transported into enterocytes by lipid scavenger proteins SCARB1 and CD36. The carotenoid ß-carotene is cleaved by BCO1 into two molecules of retinal that can form retinoic acid. Retinoic acid binds directly to the ISX promoter to activate expression and ISX protein inhibits expression of scarb1 and bco1. Carotenoids can be packaged into chylomicrons that are formed in the endoplasmic reticulum (ER). MTTP and APOE are proteins associated with chylomicrons that are important for their formation and transport into the lymphatic system.
Tinaja cavefish also have reduced expression of microsomal triglyceride transfer protein (mttp) and apolipoprotein Eb (apoea); two genes that encode lipid-binding proteins. Mttp and Apoea function in formation and transport chylomicrons respectively and therefore influence lipid and carotenoid transport into the lymphatic system (Figure 6B). We considered that carotenoid-accumulation could be a pleiotropic consequence of selecting for genetic changes that alter lipid absorption or processing since Tinaja and Pachón cavefish accumulate fat at earlier stages of development and have more fat throughout their body as adults compared to surface fish (Aspiras et al., 2015; Hüppop, 2000; Xiong et al., 2018). Molino cavefish do not have yellow VAT, do not accumulate fat early, and have only moderately more fat than surface fish as adults (Xiong et al., 2018). However, reduced expression of lipid-binding proteins suggests that, if Tinaja have increased lipid absorption, they must achieve this through a novel mechanism. Examining the lipid absorption pathways at other stages of development and under various diets could help dissect how the differences between surface fish and cavefish arise.
To determine if the expression patterns we observed are influenced by dominant or recessive alleles, we used a likelihood ratio test to compare normalized counts between surface, Tinaja, and F1 hybrid samples (Figure 6A, Figure S10). We found that average isx expression level is highest in F1 hybrid samples (adjusted p-value = 0.004), but there is considerable variation; two of the samples have levels intermediate between surface and Tinaja and three of the samples are higher (Figure S10A). The average normalized count for the Isx target scarb1 is intermediate in the hybrid samples (adjusted p-value = 2×10−11, Figure S10B) and the other target, bco1, is similar to Tinaja (adjusted p-value = 1×10−15, Figure S10C). The average normalized counts for the lipid-binding proteins apoea and mttp are intermediate in the F1 hybrids (adjusted p-value = 0.014, 0.006 respectively), albeit the normalized mttp count is higher than the surface fish average in two of the F1 samples (Figure S10D,E). Using the likelihood ratio test we found that cd36, a scavenger receptor that similar to scarb1 is involved in carotenoid uptake has significantly lower expression in Tinaja and F1 hybrids (adjusted p-value =0.03, Figure S10F). In summary, we observed that cavefish have altered expression of multiple genes in the hindgut that could influence the absorption, metabolism, and circulation of carotenoids.
3. 11. Maternal contribution of carotenoids in A. mexicanus
We observed that Pachón cavefish larvae appear more yellow than surface fish larvae (Figure 7A–C). Since this difference is apparent prior to the onset of feeding, we hypothesized that carotenoids are more abundant in cavefish oocytes and carotenoid level in the yolk is therefore dependent on the female parent. Consistent with this hypothesis, we found that cave/surface F1 hybrids that result from surface fish females are not yellow, and those arising from cavefish females are yellow (Figure 7D–E). This suggests there is a difference in the carotenoid content in the eggs and provides an opportunity to determine if this impacts development by comparing hybrids produced from cave versus surface females.
4. Concluding remarks
When Astyanax mexicanus invaded the caves of the Sierra de El Abra, they found themselves trapped in a starkly different environment from their ancestral home in the river system. 200,000 years later, the populations have evolved strikingly different morphological and metabolic features. A major question is whether these cave traits result from genetic drift, selection, or pleiotropic interactions. Surface adapted A. mexicanus utilize carotenoids for vision and pigmentation; two traits that are not under selection in a perpetually dark cave. Despite this, we found that three independent cavefish populations (Tinaja, Pachón, and Molino) appear to have maintained some level of carotenoid-based pigment. Tinaja and Pachón cavefish populations also exhibit a pronounced difference in accumulation of carotenoids in the VAT; they may have evolved this trait in parallel, but it is currently unclear if there is a common molecular basis. In Tinaja cavefish, we observed reduced expression of genes linked with carotenoid levels in other species, suggesting that changes in the pathways controlling lipid absorption and carotenoid metabolism commonly alter carotenoid accumulation.
It is easy to imagine how altered lipid absorption or storage could be advantageous in the cave environment as a mechanism for starvation resistance, but could carotenoid accumulation specifically be advantageous? Cavefish have metabolic phenotypes that would be pathological in a human context including fatty liver (Aspiras et al., 2015), high blood sugar (Riddle, Aspiras, et al., 2018), and hypertrophic adipocytes (Xiong et al., 2018), yet do not appear to suffer damaging effects. In humans, the health benefits of a diet high in carotenoids are evident but the mechanisms of action are not entirely clear; carotenoids are thought to scavenge free-radicals in tissues and reduce oxidative damage (reviewed in Kaulmann and Bohn, 2014). In the VAT specifically, carotenoids control the transcriptional state of adipocytes and modulate inflammatory pathways (Luisa Bonet, Canas, Ribot, & Palou, 2015). A recent study comparing the immune system of surface- and cave-adapted A. mexicanus revealed that Pachón cavefish have fewer pro-inflammatory immune cells in the VAT, despite having hypertrophic adipocytes (Peuß et al., 2019). It is intriguing to hypothesize that accumulating carotenoids could provide protective benefits for cavefish. Our findings lay the groundwork to investigate this possibility and more broadly understand how natural variation in carotenoid processing impacts physiology.
Supplementary Material
Acknowledgments
Brian Martineu and Megan Peavey for fish husbandry, Fransisca Leal and Robert Peuß for assisting in fish dissections, Suzanne McGaugh for sharing sequencing data, and Hasreet Gill for writing MATLAB script. Work by JN Hutchinson at the Harvard Chan Bioinformatics Core was funded by the Harvard Medical School Tools and Technology Committee. This work was supported by grants from the National Institutes of Health [HD089934, DK108495].
Footnotes
The authors declare no conflicts of interest.
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Amundsen CR, Nordeide JT, Gjøen HM, Larsen B, & Egeland ES (2015). Conspicuous carotenoid-based pelvic spine ornament in three-spined stickleback populations-occurrence and inheritance. PeerJ. 10.7717/peerj.872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends D, Prins P, Jansen RC, & Broman KW (2010). R/qtl: high-throughput multiple QTL mapping: Fig. 1. Bioinformatics, 26(23), 2990–2992. 10.1093/bioinformatics/btq565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspiras AC, Rohner N, Martineau B, Borowsky RL, & Tabin CJ (2015). Melanocortin 4 receptor mutations contribute to the adaptation of cavefish to nutrient-poor conditions. Proceedings of the National Academy of Sciences, 112(31), 9668–9673. 10.1073/pnas.1510802112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros NA, Castro R, Abos B, Rodríguez Saint-Jean SS, Pérez-Prieto SI, & Tafalla C (2013). The Pyloric Caeca Area Is a Major Site for IgM+ and IgT+ B Cell Recruitment in Response to Oral Vaccination in Rainbow Trout. PLoS ONE, 8(6), e66118 10.1371/journal.pone.0066118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow GW (1973). Competition between Color Morphs of the Polychromatic Midas Cichlid Cichlasoma citrinellum. Science, 179(4075), 806–807. 10.1126/science.179.4075.806 [DOI] [PubMed] [Google Scholar]
- Bateman A (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Research, 47(D1), D506–D515. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry SD, Davis SR, Beattie EM, Thomas NL, Burrett AK, Ward HE, … Snell RG (2009). Mutation in bovine β-carotene oxygenase 2 affects milk color. Genetics. 10.1534/genetics.109.101741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn T, Desmarchelier C, Dragsted LO, Nielsen CS, Stahl W, Rühl R, … Borel P (2017). Host-related factors explaining interindividual variability of carotenoid bioavailability and tissue concentrations in humans. Molecular Nutrition & Food Research, 61(6), 1600685 10.1002/mnfr.201600685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel P, Desmarchelier C, Nowicki M, Bott R, Morange S, & Lesavre N (2014). Interindividual variability of lutein bioavailability in healthy men: Characterization, genetic variants involved, and relation with fasting plasma lutein concentration. American Journal of Clinical Nutrition. 10.3945/ajcn.114.085720 [DOI] [PubMed] [Google Scholar]
- Borel P, Lietz G, Goncalves A, Szabo de Edelenyi F, Lecompte S, Curtis P, … Reboul E (2013). CD36 and SR-BI Are Involved in Cellular Uptake of Provitamin A Carotenoids by Caco-2 and HEK Cells, and Some of Their Genetic Variants Are Associated with Plasma Concentrations of These Micronutrients in Humans. The Journal of Nutrition, 143(4), 448–456. 10.3945/jn.112.172734 [DOI] [PubMed] [Google Scholar]
- Bray N, Pimentel H, Melsted P, & Pachter L (2015). Near-optimal RNA-Seq quantification. BioRxiv. 10.1101/021592 [DOI] [PubMed] [Google Scholar]
- Brown AC (2013). Honesty and carotenoids in a pigmented female fish. ProQuest Dissertations and Theses. [Google Scholar]
- Catchen JM, Amores A, Hohenlohe P, Cresko W, & Postlethwait JH (2011). Stacks: building and genotyping Loci de novo from short-read sequences. G3 (Bethesda, Md.), 1(3), 171–182. 10.1534/g3.111.000240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JSR, Gassant CE, Amaral PM, Lloyd E, Stahl BA, Jaggard JB, … Duboue ER (2018). Convergence on reduced stress behavior in the Mexican blind cavefish. Developmental Biology, 441(2), 319–327. 10.1016/j.ydbio.2018.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepeveen ET, & Salzburger W (2011). Molecular Characterization of Two Endothelin Pathways in East African Cichlid Fishes. Journal of Molecular Evolution, 73(5–6), 355–368. 10.1007/s00239-012-9483-6 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, … Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29(1), 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggersdorfer M, & Wyss A (2018). Carotenoids in human nutrition and health. Archives of Biochemistry and Biophysics. 10.1016/j.abb.2018.06.001 [DOI] [PubMed] [Google Scholar]
- Elipot Y, Legendre L, Père S, Sohm F, & Rétaux S (2014). Astyanax Transgenesis and Husbandry: How Cavefish Enters the Laboratory. Zebrafish, 11(4), 291–299. 10.1089/zeb.2014.1005 [DOI] [PubMed] [Google Scholar]
- Elliott WR (2015). Cave Biodiversity and Ecology of the Sierra de El Abra Region. In Biology and Evolution of the Mexican Cavefish (pp. 59–76). 10.1016/B978-0-12-802148-4.00003-7 [DOI] [Google Scholar]
- Eriksson J, Larson G, Gunnarsson U, Bed’hom B, Tixier-Boichard M, Strömstedt L, … Andersson L (2008). Identification of the Yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genetics. 10.1371/journal.pgen.1000010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinasa L, Bonaroti N, Wong J, Pottin K, Queinnec E, & Rétaux S (2017). Contrasting feeding habits of post-larval and adult Astyanax cavefish. Subterranean Biology, 21(1), 1–17. 10.3897/subtbiol.21.11046 [DOI] [Google Scholar]
- García-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Götz S, Tarazona S, … Conesa A (2012). Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics, 28(20), 2678–2679. 10.1093/bioinformatics/bts503 [DOI] [PubMed] [Google Scholar]
- Gore AV, Tomins KA, Iben J, Ma L, Castranova D, Davis AE, … Weinstein BM (2018). An epigenetic mechanism for cavefish eye degeneration. Nature Ecology & Evolution, 2(7), 1155–1160. 10.1038/s41559-018-0569-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JB, & Powers AK (2018). A Natural Animal Model System of Craniofacial Anomalies: The Blind Mexican Cavefish. Anatomical Record. 10.1002/ar.23998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Analysis Release V8 (dbGaP Accession phs000424.v8.p2). (n.d.).
- Hendrickson SJ, Hazra A, Chen C, Eliassen AH, Kraft P, Rosner BA, & Willett WC (2012). β-Carotene 15,15′-monooxygenase 1 single nucleotide polymorphisms in relation to plasma carotenoid and retinol concentrations in women of European descent. The American Journal of Clinical Nutrition, 96(6), 1379–1389. 10.3945/ajcn.112.034934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman A, Brandvain Y, Weagley J, Jeffery WR, Keene AC, Kono TJY, … McGaugh SE (2018). The role of gene flow in rapid and repeated evolution of cave-related traits in Mexican tetra, Astyanax mexicanus. Molecular Ecology, 27(22), 4397–4416. 10.1111/mec.14877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinaux H, Pottin K, Chalhoub H, Père S, Elipot Y, Legendre L, & Rétaux S (2011). A Developmental Staging Table for Astyanax mexicanus Surface Fish and Pachón Cavefish. Zebrafish, 8(4), 155–165. 10.1089/zeb.2011.0713 [DOI] [PubMed] [Google Scholar]
- Hori K, Fostier M, Ito M, Fuwa TJ, Go MJ, Okano H, … Matsuno K (2004). Drosophila deltex mediates suppressor of hairless-independent and late-endosomal activation of notch signaling. Development. 10.1242/dev.01448 [DOI] [PubMed] [Google Scholar]
- Hüppop K (2000). How do cave animals cope with the food scarcity in caves? In Ecosystems of the World: Subterranean ecosystems. [Google Scholar]
- Hyacinthe C, Attia J, & Rétaux S (2019). Evolution of acoustic communication in blind cavefish. Nature Communications. 10.1038/s41467-019-12078-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery WR, Ma L, Parkhurst A, & Bilandžija H (2016). Pigment Regression and Albinism in Astyanax Cavefish. In Biology and Evolution of the Mexican Cavefish (pp. 155–173). 10.1016/B978-0-12-802148-4.00008-6 [DOI] [Google Scholar]
- Kaulmann A, & Bohn T (2014). Carotenoids, inflammation, and oxidative stress—implications of cellular signaling pathways and relation to chronic disease prevention. Nutrition Research, 34(11), 907–929. 10.1016/j.nutres.2014.07.010 [DOI] [PubMed] [Google Scholar]
- Kavalco KF, & De Almeida-Toledo LF (2007). Molecular Cytogenetics of Blind Mexican Tetra and Comments on the Karyotypic Characteristics of Genus Astyanax (Teleostei, Characidae). Zebrafish, 4(2), 103–111. 10.1089/zeb.2007.0504 [DOI] [PubMed] [Google Scholar]
- Klaassen H, Wang Y, Adamski K, Rohner N, & Kowalko JE (2018). CRISPR mutagenesis confirms the role of oca2 in melanin pigmentation in Astyanax mexicanus. Developmental Biology. 10.1016/j.ydbio.2018.03.014 [DOI] [PubMed] [Google Scholar]
- Krishnan J, & Rohner N (2017). Cavefish and the basis for eye loss. Philosophical Transactions of the Royal Society B: Biological Sciences, Vol. 372 10.1098/rstb.2015.0487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, & Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature Methods, 9(4), 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DS, & Lee DS (2017). Atlas of North American freshwater fishes, 1980- et seq. /. In Atlas of North American freshwater fishes, 1980- et seq. /. 10.5962/bhl.title.141711 [DOI] [Google Scholar]
- Liao Y, Smyth GK, & Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30(7), 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lloyd E, Olive C, Stahl BA, Jaggard JB, Amaral P, Duboué ER, & Keene AC (2018). Evolutionary shift towards lateral line dependent prey capture behavior in the blind Mexican cavefish. Developmental Biology, 441(2), 328–337. 10.1016/j.ydbio.2018.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo GP, Hessel S, Eichinger A, Noy N, Moise AR, Wyss A, … Von Lintig J (2010). ISX is a retinoic acid-sensitive gatekeeper that controls intestinal β,β-carotene absorption and vitamin A production. FASEB Journal. 10.1096/fj.09-150995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes RJ, Johnson JD, Toomey MB, Ferreira MS, Araujo PM, Melo-Ferreira J, … Carneiro M (2016). Genetic Basis for Red Coloration in Birds. Current Biology, 26(11), 1427–1434. 10.1016/j.cub.2016.03.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12), 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisa Bonet M, Canas JA, Ribot J, & Palou A (2015). Carotenoids and their conversion products in the control of adipocyte function, adiposity and obesity. Archives of Biochemistry and Biophysics. 10.1016/j.abb.2015.02.022 [DOI] [PubMed] [Google Scholar]
- Lyon A, Powers AK, Gross JB, & O’Quin KE (2017). Two - Three loci control scleral ossicle formation via epistasis in the cavefish astyanax mexicanus. PLoS ONE, 12(2). 10.1371/journal.pone.0171061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, & Artavanis-Tsakonas S (1995). Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. [DOI] [PubMed] [Google Scholar]
- Metzger MB, Pruneda JN, Klevit RE, & Weissman AM (2014). RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochimica et Biophysica Acta - Molecular Cell Research. 10.1016/j.bbamcr.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NE, Mohn ES, Hason N, Erdman JW, & Johnson EJ (2018). Intrinsic and extrinsic factors impacting absorption, metabolism, and health effects of dietary carotenoids. Advances in Nutrition. 10.1093/ADVANCES/NMY025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy NI, Stapley J, Bennison C, Tucker R, Twyman H, Kim K-W, … Slate J (2016). Red Carotenoid Coloration in the Zebra Finch Is Controlled by a Cytochrome P450 Gene Cluster. Current Biology, 26(11), 1435–1440. 10.1016/j.cub.2016.04.047 [DOI] [PubMed] [Google Scholar]
- Nakamura N (2011). The role of the transmembrane RING finger proteins in cellular and organelle function. Membranes. 10.3390/membranes1040354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Quin CT, Drilea AC, Conte MA, & Kocher TD (2013). Mapping of pigmentation QTL on an anchored genome assembly of the cichlid fish, Metriaclima zebra. BMC Genomics, 14(1), 287 10.1186/1471-2164-14-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peuß R, Box AC, Wang Y, Chen S, Krishnan J, Tsuchiya D, … Rohner N (2019). Single cell analysis reveals modified hematopoietic cell composition affecting inflammatory and immunopathological responses in Astyanax mexicanus; BioRxiv, 647255 10.1101/647255 [DOI] [Google Scholar]
- Protas M, Conrad M, Gross JB, Tabin C, & Borowsky R (2007). Regressive Evolution in the Mexican Cave Tetra, Astyanax mexicanus. Current Biology, 17(5), 452–454. 10.1016/j.cub.2007.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puca L, Chastagner P, Meas-Yedid V, Israël A, & Brou C (2013). α-arrestin 1 (ARRDC1) and β-arrestins cooperate to mediate Notch degradation in mammals. Journal of Cell Science. 10.1242/jcs.130500 [DOI] [PubMed] [Google Scholar]
- R-core-team. (2019). R: A Language and Environment for Statistical Computing. Retrieved from https://www.r-project.org/
- Riddle MR, Aspiras AC, Gaudenz K, Peuß R, Sung JY, Martineau B, … Rohner N (2018). Insulin resistance in cavefish as an adaptation to a nutrient-limited environment. Nature, 555(7698), 647–651. 10.1038/nature26136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle MR, Boesmans W, Caballero O, Kazwiny Y, & Tabin CJ (2018). Morphogenesis and motility of the Astyanax mexicanus gastrointestinal tract. Developmental Biology, 441(2), 285–296. 10.1016/j.ydbio.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakudoh T, Iizuka T, Narukawa J, Sezutsu H, Kobayashi I, Kuwazaki S, … Tsuchida K (2010). A CD36-related Transmembrane Protein Is Coordinated with an Intracellular Lipid-binding Protein in Selective Carotenoid Transport for Cocoon Coloration. Journal of Biological Chemistry, 285(10), 7739–7751. 10.1074/jbc.M109.074435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakudoh T, Kuwazaki S, Iizuka T, Narukawa J, Yamamoto K, Uchino K, … Tsuchida K (2013). CD36 homolog divergence is responsible for the selectivity of carotenoid species migration to the silk gland of the silkworm Bombyx mori. Journal of Lipid Research. 10.1194/jlr.M032771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W, Braasch I, & Meyer A (2007). Adaptive sequence evolution in a color gene involved in the formation of the characteristic egg-dummies of male haplochromine cichlid fishes. BMC Biology, 5(1), 51 10.1186/1741-7007-5-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, & Ponting CP (2018). TMEM132: An ancient architecture of cohesin and immunoglobulin domains define a new family of neural adhesion molecules. Bioinformatics. 10.1093/bioinformatics/btx689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho R, Cremona CA, & Behrens A (2015). Stem cell and progenitor fate in the mammalian intestine: Notch and lateral inhibition in homeostasis and disease. EMBO Reports. 10.15252/embr.201540188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefc KM, Brown AC, & Clotfelter ED (2014). Carotenoid-based coloration in cichlid fishes. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 173, 42–51. 10.1016/j.cbpa.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Hayamizu TF, Finger JH, Bello SM, McCright IJ, Xu J, … Ringwald M (2019). The mouse Gene Expression Database (GXD): 2019 update. Nucleic Acids Research. 10.1093/nar/gky922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, & Robinson MD (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research, 4, 1521 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, & Robinson MD (2016). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research, 4, 1521 10.12688/f1000research.7563.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, & Krogh A (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings / … International Conference on Intelligent Systems for Molecular Biology ; ISMB International Conference on Intelligent Systems for Molecular Biology. [PubMed] [Google Scholar]
- Stahl BA, & Gross JB (2017). A Comparative Transcriptomic Analysis of Development in Two Astyanax Cavefish Populations. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 10.1002/jez.b.22749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl BA, Jaggard JB, Chin JSR, Kowalko JE, Keene AC, & Duboué ER (2019). Manipulation of Gene Function in Mexican Cavefish. Journal of Visualized Experiments, (146). 10.3791/59093 [DOI] [PubMed] [Google Scholar]
- Sundvold H, Helgeland H, Baranski M, Omholt SW, & Våge D (2011). Characterisation of a novel paralog of scavenger receptor class B member I (SCARB1) in Atlantic salmon (Salmo salar). BMC Genetics, 12(1), 52 10.1186/1471-2156-12-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Biehs B, Chiu C, Siebel CW, Wu Y, Costa M, … Klein OD (2015). Opposing activities of notch and wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Reports. 10.1016/j.celrep.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toews DPL, Hofmeister NR, & Taylor SA (2017). The Evolution and Genetics of Carotenoid Processing in Animals. Trends in Genetics, 33(3), 171–182. 10.1016/j.tig.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Toomey MB, Butler MW, & McGraw KJ (2010). Immune-system activation depletes retinal carotenoids in house finches (Carpodacus mexicanus). Journal of Experimental Biology, 213(10), 1709–1716. 10.1242/jeb.041004 [DOI] [PubMed] [Google Scholar]
- Townsend LD (1944). Variation in the Number of Pyloric Caeca and Other Numerical Characters in Chinook Salmon and in Trout. Copeia, 1944(1), 52 10.2307/1438249 [DOI] [Google Scholar]
- Turner BJ (2017). Biology and Evolution of the Mexican Cavefish . Edited by Keene Alex C., Yoshizawa Masato, and McGaugh Suzanne E.. Academic Press; Amsterdam (The Netherlands) and Boston (Massachusetts): Elsevier. $99.95. xiv + 403 p.; ill.; index. ISBN: 978–0-12–802148-. The Quarterly Review of Biology, 92(4), 485–486. 10.1086/694993 [DOI] [Google Scholar]
- Twyman H, Valenzuela N, Literman R, Andersson S, & Mundy NI (2016). Seeing red to being red: conserved genetic mechanism for red cone oil droplets and co-option for red coloration in birds and turtles. Proceedings of the Royal Society B: Biological Sciences, 283(1836), 20161208 10.1098/rspb.2016.1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Våge DI, & Boman IA (2010). A nonsense mutation in the beta-carotene oxygenase 2 (BCO2) gene is tightly associated with accumulation of carotenoids in adipose tissue in sheep (Ovis aries). BMC Genetics. 10.1186/1471-2156-11-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeweyer G, Van Der Aa N, Reyniers E, & Kooy RF (2012). The contribution of CLIP2 haploinsufficiency to the clinical manifestations of the Williams-Beuren syndrome. American Journal of Human Genetics. 10.1016/j.ajhg.2012.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe DA, & Cornwell DG (1964). Carotenoids of Cavernicolous Crayfish. Science, 144(3625), 1467–1469. 10.1126/science.144.3625.1467 [DOI] [PubMed] [Google Scholar]
- Xiong S, Krishnan J, Peuß R, & Rohner N (2018). Early adipogenesis contributes to excess fat accumulation in cave populations of Astyanax mexicanus. Developmental Biology, 441(2), 297–304. 10.1016/j.ydbio.2018.06.003 [DOI] [PubMed] [Google Scholar]
- Yoshizawa M, Robinson BG, Duboué ER, Masek P, Jaggard JB, O’Quin KE, … Keene AC (2015). Distinct genetic architecture underlies the emergence of sleep loss and prey-seeking behavior in the Mexican cavefish. BMC Biology, 13, 15 10.1186/s12915-015-0119-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, & He QY (2012). ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS A Journal of Integrative Biology. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifel ME, Leahy DJ, & Barrick D (2005). Structure and Notch Receptor Binding of the Tandem WWE Domain of Deltex. Structure, 13(11), 1599–1611. 10.1016/j.str.2005.07.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


