Abstract
Food and waterborne protozoan pathogens can cause serious disease in people. Three common species Cryptosporidium parvum, Giardia enterica and Toxoplasma gondii can contaminate diverse shellfish species, including commercial oysters. Current methods of protozoan detection in shellfish are not standardized, and few are able to simultaneously identify multiple species. Here, we present a novel metabarcoding assay targeting the 18S rRNA gene followed by next generation sequencing (NGS) for simultaneous detection of Cryptosporidium spp., Giardia spp. and T. gondii spiked into oyster samples. We further developed a bioinformatic pipeline to process and analyze 18S rRNA data for protozoa classification. The ability of the NGS assay to detect protozoa was later compared with conventional PCR. Results demonstrated that background amplification of oyster and other eukaryotic DNA competed with that of protozoa for obtained sequence reads. Sequences of target protozoans were obtained across all spiking levels; however, low numbers of target sequences in negative controls imply that a threshold for true positives must be defined for assay interpretation. While this study focused on three target parasites, the ability of this approach to detect numerous known and potentially unknown protozoan pathogens make it a promising screening tool for monitoring protozoan contamination in food and water.
Keywords: 18S rRNA, Metabarcoding, Cryptosporidium parvum, Giardia enterica, Toxoplasma gondii, Oysters
1. Introduction
Protozoan parasites can cause significant human disease worldwide. Three protozoa commonly associated with food- or waterborne illness in people are Cryptosporidium spp. Giardia spp. and Toxoplasma gondii (Bahia-Oliveira et al., 2017; Betancourt, 2019; Boarato-David et al., 2017). Cryptosporidiosis and giardiasis result in gastrointestinal disease that can be self-limiting in immunocompetent adults, but may lead to severe gastroenteritis, dehydration and death in young children or immunocompromised patients (DuPont, 2013; Khalil et al., 2018). Toxoplasma infection is usually asymptomatic or manifests with flu-like symptoms; however, mortality can occur in immunocompromised individuals, and infections in pregnant women can result in fetal loss, birth defects, learning deficits, blindness or possibly mental illness later in life (Dubey, 2013). Toxoplasma and certain species of Cryptosporidium and Giardia are zoonotic, with transmission occurring from animals to humans. C. parvum and G. enterica have many animal hosts, whereas the only known definitive hosts for T. gondii are felids. Transmission of protozoan parasite cysts and oocysts typically occurs through fecal contamination of food and water (Dorny et al., 2009).
C. parvum and T. gondii oocysts, and G. enterica cysts (here after called (oo)cysts collectively) have been detected in bivalve shellfish worldwide (Koutsoumanis et al., 2018; Ryan et al., 2019; Shapiro et al., 2019b). Bivalves can accumulate biological and chemical contaminants through filter-feeding. T. gondii and C. parvum oocysts are incredibly robust in the environment and can survive in seawater for up to six months and a year, respectively (Hohweyer et al., 2013). Further investigation into the longevity of G. enterica cysts is required; however, the presence of cysts in shellfish and other marine organisms suggests some resistance to salinity (Fayer et al., 2004). Contaminated commercial shellfish, such as oysters, are a particularly notable health concern due to their tendency to be eaten undercooked or raw. Incidence of shellfish-vectored protozoal illnesses is likely underestimated due to underreporting of mild cases, difficulty determining the source of foodborne illnesses, and lack of standardized pathogen detection (World Health Organization, 2010).
Presently, there is no standard approach for detection of protozoa in shellfish (Hohweyer et al., 2013; Robertson, 2007). Commonly used methods include direct fluorescence antibody (DFA) tests and polymerase chain reaction (PCR). DFA tests use monoclonal antibodies specific to protozoan (oo)cysts, but DFA testing can be limited in specificity when there is high degree of (oo)cyst structural similarity between protozoan species. For example, in the case of Cryptosporidium, DFA tests cannot distinguish between species (Hohweyer et al., 2013). Additionally, DFA cannot be used to identify T. gondii because monoclonal antibodies specific to its oocyst wall have yet to be optimized (Hohweyer et al., 2013). Because of these limitations, DFA is often supplemented with faster, more specific PCR methods. Simplex or multiplex PCR can amplify target protozoan gene(s) in order to identify the species, as well as for genotype and sub-genotype determination. However, the number of protozoan species that can be identified simultaneously with PCR methods is limited (Hohweyer et al., 2013). A single assay that can identify all pathogenic protozoa within a shellfish matrix would be a vastly more efficient diagnostic and surveillance tool for food safety and public health applications.
Metabarcoding is used for high throughput taxonomic identification of multiple species within a sample (Peters et al., 2018). It is a widely used method for characterizing bacterial populations, targeting the ubiquitous bacterial 16S ribosomal RNA (rRNA) gene (Allan, 2014). While recent studies have shown that eukaryotic organisms can be similarly characterized by targeting the 18S rRNA gene, it is still an emerging technique. This method has been used mainly to identify protozoa in fecal and water samples, but it has yet to be applied to shellfish (Marzano et al., 2017; Moreno et al., 2018). Moreover, due to the novelty of metabarcoding for detection of protozoan pathogens, there is a lack of available bioinformatics resources for processing and classifying protozoan 18S rRNA amplicons.
In this study, we developed a novel metabarcoding assay targeting the 18S rRNA gene for simultaneous targeted amplification of Cryptosporidium spp., Giardia spp. and T. gondii in oysters followed by next generation sequencing. We further developed a bioinformatic pipeline to process and analyze 18S rRNA metabarcode data. We applied and validated these techniques through systematic spiking of protozoan (oo)cysts in oyster homogenates and hemolymph and compared the ability of metabarcoding to detect targeted protozoa with conventional multiplexed PCR.
2. Materials and methods
2.1. Metabarcoding assay development
A primer pair was designed to target conserved nucleotide sequences flanking a variable (V4) region within the 18S rRNA gene of C. parvum, T. gondii, and G. enterica (Table 1). Due to substantive differences in the G. enterica 18S rRNA sequence, an additional reverse primer was designed to improve amplification of the G. enterica 18S rRNA V4 region. Primers were designed by aligning the 18S rRNA gene sequences from C. parvum (GenBank: L16996.1), G. lamblia (GenBank: AF473852.1), T. gondii (GenBank: L37415.1), Saccharomyces cerevisiae (GenBank: Z75578.1), and Crassostrea virginica (GenBank: X60315.1). The inclusion of S. cerevisiae was to identify homologous variable regions among different protozoa using pre-defined variable regions from a characterized eukaryote organism (Hadziavdic et al., 2014); inclusion of C. virginica was to assess the possibility of amplification of the host (oyster) 18S rRNA. The V4 region is the longest variable region of the 18S rRNA gene consistently identified in eukaryotes and was chosen as the target amplicon to maximize taxonomic resolution (Hadziavdic et al., 2014). Sequence alignment and digital primer validation was performed using Geneious 11.0.5 (Kearse et al., 2012). Complementation between the primers and aligned sequences were determined, and the expected amplicon sizes and % GC-content were determined (Table 1).
Table 1.
Primer sequences used for amplification of the 18S rRNA V4 region of target protozoa (Cryptosporidium parvum, Giardia enterica, and Toxoplasma gondii) or oyster (Crassostrea virginica), expected amplicon size and GC content for each product.
| Primer IDa | Sequence (5′-3′) |
|---|---|
| General 18S V4 Forward | GCC GCG GTA ATT CCA GCT C |
| General 18S V4 Reverse 1 | ATY YTT GGC AAA TGC TTT CGC |
| Giardia 18S V4 Reverse 2 | ATA CGG TGG TGT CTG ATC GC |
| Target | Gene-specific PCR Amplicon Size (bp) | Gene-specific PCR Amplicon Size with overhangs (bp)a | Dual-indexing PCR Amplicon Size (bp)a | Percent GC |
|---|---|---|---|---|
| T. gondii | 385 | 452 | 521 | 42.0% |
| C. parvum | 366 | 433 | 502 | 28.0% |
| G. enterica | 315 | 382 | 451 | 71.4% |
| C. virginica | 381 | 448 | 517 | 49.6% |
Incorporation of overhangs in the gene-specific primer adds 67 bp to the overall fragment length, and incorporation of sequences in dual-indexing PCR adds another 69 bp. However, only the non-overhang, gene-specific amplicon and relevant indices are sequenced.
Primer validation was completed by assessing DNA amplification using a high-fidelity polymerase, KAPA HiFi polymerase (KAPA Biosystems/Roche, Basel, Switzerland). Manufacturer recommended reaction mixtures and cycling conditions were used, as well as the provided PCR reagents. Purified C. parvum, T. gondii and G. enterica stock preparations were used as templates (see Section 2.6). Primers were tested in singleplex and multiplex with single-species and mixed-species templates, respectively. PCR amplification was considered successful when bands of expected amplicon sizes for mixed templates were visualized on 2% agarose gel, stained with RedSafe (FroggaBio; Toronto, ON, Canada). Additionally, mixed-template amplicons from one gel were isolated using the QIAquick gel extraction kit (Qiagen) and submitted for Sanger sequencing at the University of Guelph's Advanced Analysis Center (Guelph, ON, Canada) for sequence analysis and molecular confirmation.
2.2. Synthetic template (gBlock) design
To address possible PCR amplification bias, we implemented the addition of known copy numbers of synthetic DNA templates (gBlocks) for each of the protozoan parasites. gBlocks were designed with assistance from Integrated DNA Technologies (IDT) Inc. (Coralville, IA) by creating synthetic sequences that shared identical GC content and nucleotide length for the expected 18S rRNA amplicons of each parasite (Figs. S1.1–3). Sufficient random nucleotide variation was inserted into the gBlock products (internal to the conserved primer binding sites) that allowed their differentiation from target protozoa. Addition of the gBlocks was intended to provide insight into template amplification biases by comparing the starting copy number (500 copies each) to the number of gBlocks detected.
2.3. Library preparation and next generation sequencing
The library preparation protocol was adapted from the Illumina 16S Metagenomic Sequencing Library Preparation guide (Illumina, 2013). Library preparation consisted of 2 PCR steps: a multiplex PCR using the three newly designed primers targeting the 18S rRNA gene that were modified with prescribed overhang sequences (Table 1), followed by a dual-indexing PCR using Nextera XT v2 Index 1 and 2 primers. PCR was performed using the KAPA HiFi polymerase as described above. Products were purified after each PCR step with Mag-Bind RxnPure Plus (Omega Bio-Tek; Norcross, GA) using modified manufacturer protocols with a product:bead volume ratio of 1:1.2 to select for amplicons >200 bp in the initial PCR, and a volume ratio of 1:0.8 to select amplicons >250 bp from the second PCR.
Quantification, normalization, and library 2 × 250 bp sequencing using the Illumina MiSeq (v2 kit, PE500; Illumina) were performed by the University of Guelph's Agriculture and Food Laboratory (Guelph, ON, Canada). The DNA concentrations of the samples were measured using a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). To equalize sample read distribution the library was normalized by diluting samples to a standard concentration (20 nM) prior to pooling for sequencing. Normalization dilution factors were provided for each sample to correct for the absolute read counts for target protozoa and oyster sequences as described below.
2.4. Bioinformatic analysis
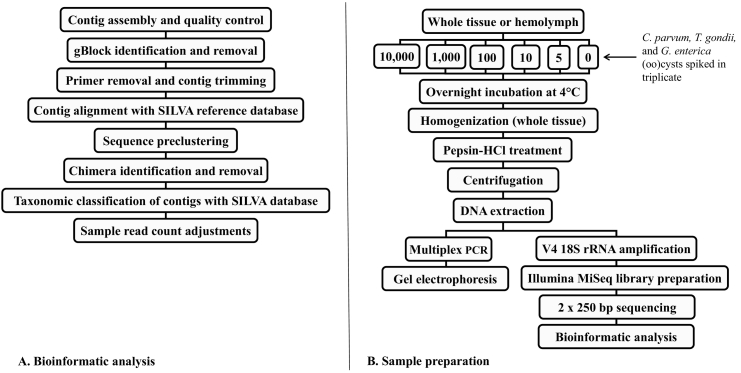
A novel bioinformatics pipeline was developed to preprocess the 18S rRNA 2 × 250 bp sequencing data produced by the Illumina MiSeq platform (Fig. 1A).
Fig. 1.
Flow diagrams depicting the bioinformatics pipeline (A) and oyster spiking experiment (B). Whole oyster tissue homogenates and hemolymph samples were spiked (in triplicate) with mixtures of protozoan (oo)cysts for metabarcoding assay validation.
2.4.1. Contig assembly and gBlock detection
The forward and reverse reads for each sample were assembled to form contigs using Paired End reAd mergeR (PEAR) (Zhang et al., 2014). The assembled contigs had a maximum length of 500 bp, a minimum length of 50 bp, and a minimum overlap of 10 bp. To avoid low quality pairs, the contig ends were trimmed following two consecutive bases with a Phred+33 score of below Q30 and their quality assessed using FastQC (Andrews, 2010).
The synthetic gBlock reads were identified via a localized BLASTn search and separated from the rest of the sequences before further processing and classification in mothur (Altschup et al., 1990).
2.4.2. Sequence preprocessing
The remaining sequences were further processed and primers were trimmed from the contigs using mothur. These steps were modifications of the Illumina MiSeq 16S rRNA standard operation procedure (SOP) (Kozich et al., 2013). To streamline contig alignment, a custom 18S V4 alignment database was adapted from the rRNA database, SILVA (Quast et al., 2013). Using the V4 region of the model organism S. cerevisiae (GenBank: Z75578) as an alignment reference, a mothur-compatible version of SILVA (release 132) was pared down to only the 18S V4 region. Contigs were preclustered and chimeric sequences were removed using VSEARCH (Rognes et al., 2016).
2.4.3. Taxonomic classification and read count adjustments
Taxonomic assignments were achieved using the RDP Naïve Bayesian Classifier method (Wang et al., 2007) using a bootstrap confidence score of 80%. The custom 18S V4 SILVA database was used as the reference database for taxonomic classification to the genus level (Quast et al., 2013). The raw contig counts for each target protozoan parasite and oyster reads were corrected for dilution during the normalization step prior to MiSeq sequencing. To account for possible amplification bias, the gBlocks counts for each sample were also used to correct parasite reads based on the known copy number of starting gBlocks (500 copies).
2.5. Pilot trials for reducing oyster (background) DNA amplification
Two reagents, bleach and pepsin-HCl, were assessed for their ability to reduce downstream amplification of oyster DNA that could limit the sequencing assay's ability to detect target protozoa accurately.
In the first bleach trial, 10,000 heat-inactivated parasites (C. parvum, G. enterica, and T. gondii (oo)cyst in PBS) were incubated in duplicate at room temperature with 10% household bleach (6% sodium hypochlorite) for 20 min. Due to preliminary results showing a negative impact on Giardia amplification, a second trial included 10,000 live G. enterica cysts incubated in duplicate at room temperature in 10%, 5%, 1%, 0.1%, or 0% bleach at a volume ratio of 1:9 for 5 min. This second trial was performed to confirm that bleach was responsible for impeding both live and heat-inactivated Giardia cyst DNA amplification.
Pepsin-HCl is a method that is often used to digest shellfish samples for pathogen detection. Preliminary findings suggested a potential reduction of oyster DNA amplification by this digest solution treatment. To further evaluate this effect, hemolymph (oyster circulatory fluid) was collected through aspiration with a needle and treated using a modified pepsin-HCl digestion protocol as described for whole oyster tissue homogenates (Willis et al., 2014). Both pepsin-HCl treated and untreated hemolymph and homogenate samples were spiked with 5 μl of parasite stock DNA solutions (50 uL of eluted DNA from 10,000 oocysts) and subjected to nucleic acid extraction, multiplex PCR and gel electrophoresis as described above.
Qualitative assessment for amplification of protozoa and oyster DNA following bleach or pepsin-HCl treatment was achieved via multiplex PCR followed by gel electrophoresis, as described above. Quantitative data on oyster and target protozoa sequence reads were subsequently obtained by applying the newly developed 18S metabarcoding assay and bioinformatics pipeline.
2.6. Metabarcoding assay validation: Oyster sample preparation and parasite spiking
Eastern oysters (Crassostrea virginica) (N = 36) were obtained from commercial grocery stores. Hemolymph (n = 18) was aspirated from oysters using a sterile needle and syringe inserted into the adductor muscle. Individual hemolymph samples were pooled, and 1 ml aliquots were placed into 2-ml microcentrifuge tubes. Whole oyster tissue from one individual (n = 18) were shucked and placed in separate 50-ml conical tubes followed by brief homogenization using the Omni-homogenizer (Omni International Inc., GA, USA) to slightly open the tissue before protozoa spiking. Hemolymph and whole tissue samples were spiked with different dilutions of C. parvum, T. gondii and G. enterica (oo)cyst mixtures (Fig. 1B). C. parvum (Iowa isolate) and G. enterica (H3 isolate) (oo)cysts were obtained from the Sterling Parasitology Laboratory at University of Arizona (Tucson, AZ, USA) and Waterborne™ Inc. (New Orleans, LA, USA), respectively. T. gondii oocysts were produced as previously described (Fritz et al., 2012) and generously provided by David Arranz-Solís at University of California, Davis, USA.
Following parasite spiking, samples were stored at 4 °C overnight, and whole tissue homogenates were prepared following mechanical homogenization and a pepsin-HCl digestion procedure as previously described (Willis et al., 2014). After pepsin-HCl digestion, pellets were washed with water, PBS eluting fluid and again with water. The final pellet was resuspended in 100 μl water for nucleic acid extraction. The spiked hemolymph samples (1 ml) were mixed with 0.5 ml of pepsin-HCl solution and treated in the same manner as whole tissues. In addition to the spiked oyster samples, reagent negative controls were also prepared for PCR and sequencing (in duplicate). Hemolymph and homogenate samples without protozoa spike were included to assess for presence of background protozoan pathogen contamination in the oysters.
DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, CA, USA) as previously described (Shapiro et al., 2019a). In brief, 180 μl of ATL buffer was added to 100 μl sample and subjected to a single freeze and thaw cycle (4 min each in liquid nitrogen and in boiling water). The sample was mixed with 40 μl proteinase K and incubated overnight at 56 °C. The nucleic acids were eluted with 50 μl 10% AE buffer and then used for protozoan parasite detection via either, metabarcoding and bioinformatic analysis or by conventional PCR as described below. For metabarcoding, a positive control was included that consisted of a parasite cocktail of 1000 T. gondii and Giardia (oo)cysts and 100,000C. parvum oocysts (higher concentrations of C. parvum were needed to produce a clearly visible band on gel electrophoresis). Five hundred copies of each gBlock were also added to all samples except for the negative reagent PCR controls.
2.7. Conventional PCR
To compare the results of the next generation sequencing assay with more conventional pathogen detection techniques, protozoa DNA was also targeted for amplification using a nested, multiplexed conventional PCR approach as previously described (Shapiro et al., 2019a). Detection of protozoan parasites were evaluated through visualization of PCR product at target bp size by gel electrophoresis.
3. Results
3.1. Reduction of oyster DNA amplification
Pepsin-HCl digested oyster homogenates did not yield any oyster DNA amplification following treatment with any of the tested bleach concentrations, including homogenates not treated with bleach (0% control) (Fig. S2). In contrast, an amplicon was visualized for hemolymph-derived DNA at all added concentrations of bleach, although the intensity of the visualized band did decrease with increasing concentrations of bleach (Fig. S2).
When testing the effect of bleach on heat-inactivated protozoan oocyst DNA amplification, treatment with 10% bleach for 20 min did not inhibit C. parvum or T. gondii amplification via PCR; however, DNA from G. enterica cysts was not amplified. In a subsequent experiment with different bleach concentrations (0.1%, 1%, 5%, 10%) and shorter contact time (5 min) with live G. enterica cysts, DNA amplification was still absent across all testing conditions (Table S1). Due to the inhibitory effect of bleach on G. enterica amplification, this reagent was not considered further for reducing oyster DNA amplification in metabarcoding experiments. Protozoan DNA was not negatively impacted by the pepsin-HCl treatment, with amplification products still visible on gel electrophoresis.
3.2. Metabarcoding assay validation: application in spiked oyster samples
3.2.1. Next generation sequencing – bioinformatics results
3.2.1.1. Sequence preprocessing
Quality control metrics for spiked hemolymph and homogenate NGS results were obtained from Illumina Basespace. The percent of clusters passing Illumina's chastity filter (%PF) was 89.30 ± 0.55%, and the percentage of base calls greater than or equal to Q30 was 83.3%. Approximately 9.5 million contigs from a total of 54 samples were successfully assembled and trimmed with PEAR, with a mean contig length of 369 bp. Of the obtained contigs, 7.8 million contigs were successfully classified into eukaryotic taxa. The most abundant taxon was the Ostreidae (edible oyster) family, consisting of 64% of the total classified reads. For target protozoa, 10.6% of reads were classified as Toxoplasma, 10.0% of reads were classified as Giardia and 7.2% of reads were classified as Cryptosporidium. The remaining reads were classified into 400 other eukaryotic genera.
A low level of each of the three protozoa (<10 reads) were detected in the negative PCR reagent control (Fig. S9). Approximately 95% of the positive control reads were classified as the expected spiked protozoa (Fig. S9).
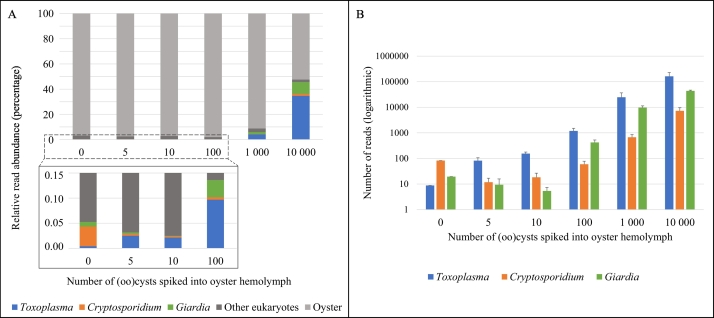
3.2.1.2. Spiked oyster hemolymph
The most abundant taxon for all the hemolymph samples was the oyster (Fig. 2A). Less than 1% of the classified reads for the hemolymph samples spiked with 0, 5, 10, and 100 (oo)cysts consisted of the spiked protozoan parasites. The three protozoa were detected in all hemolymph samples, including unspiked oysters (Fig. 2B). In general, the read counts for all three protozoa increased with the number of spiked (oo)cysts; however, the read counts for both Cryptosporidium and Giardia were higher in the unspiked hemolymph than the 5 and 10 (oo)cyst-spiked samples.
Fig. 2.
Relative abundance (A) and total number (B) of sequence reads obtained from oyster hemolymph samples spiked with increasing numbers of Cryptosporidium parvum, Toxoplasma gondii, and Giardia enterica (oo)cysts (performed in triplicate). Error bars represent one standard deviation from the mean.
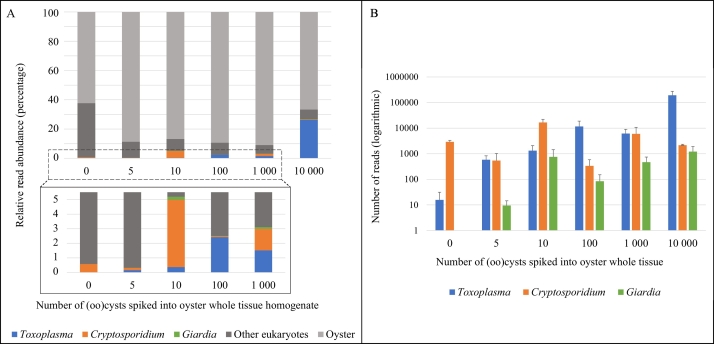
3.2.1.3. Spiked oyster homogenate
Similar to the results in hemolymph, oyster was the most abundant taxon for all homogenate samples (Fig. 3A). Spiked protozoan reads consisted of less than 1% of the total classified reads for the unspiked and 5 (oo)cyst homogenate samples. Toxoplasma and Cryptosporidium were detected in all homogenate samples, while Giardia was detected in all but the unspiked homogenate (Fig. 3B). Unlike the spiked hemolymph, the protozoa read counts in the homogenate samples did not show a general trend of increasing along with the number of spiked (oo)cysts.
Fig. 3.
Relative abundance (A) and total number (B) of sequence reads obtained from oyster whole tissue homogenate samples spiked with increasing numbers of Cryptosporidium parvum, Toxoplasma gondii, and Giardia enterica (oo)cysts (performed in triplicate). Error bars represent one standard deviation from the mean.
3.2.2. Conventional PCR results
When nested PCR was applied on pepsin-HCl treated hemolymph, C. parvum and T. gondii could be detected at a concentration of 5 (oo)cysts per hemolymph, whereas the lowest detection limit for G. enterica was 100 (oo)cysts per hemolymph (Table 2). In pepsin-HCl treated homogenate samples, the amplification of parasites via nested PCR was observed at spiking levels of 5, 100 and 1000 (oo)cysts per sample for T. gondii, C. parvum and G. enterica, respectively (Table 2).
Table 2.
Detection of parasites by conventional multiplex PCR (mPCR) assay in hemolymph and whole tissue pepsin-HCl digestion spiking experiments. The lowest level of detection is indicated in bold. Each hemolymph or homogenate replicate is comprised of matrix from one oyster.
| (Oo)cysts/sample | (Oo)cysts/reaction | Amplification/replicates tested |
|||
|---|---|---|---|---|---|
| Cryptosporidium | Giardia | Toxoplasma | |||
| Hemolymph | 10,000 | 1000 | 3/3 | 3/3 | 3/3 |
| 1000 | 100 | 3/3 | 3/3 | 3/3 | |
| 100 | 10 | 2/3 | 1/3 | 3/3 | |
| 10 | 1 | 1/3 | 0/3 | 3/3 | |
| 5 | 0.5 | 2/3 | 0/3 | 3/3 | |
| 0 | 0 | 0/3 | 0/3 | 1/3a | |
| Whole tissue homogenate | 10,000 | 1000 | 3/3 | 1/3 | 3/3 |
| 1000 | 100 | 1/3 | 1/3 | 3/3 | |
| 100 | 10 | 1/3 | 0/3 | 3/3 | |
| 10 | 1 | 0/3 | 0/3 | 2/3 | |
| 5 | 0.5 | 0/3 | 0/3 | 3/3 | |
| 0 | 0 | 0/3 | 0/3 | 0/3 | |
T. gondii amplification was found in one of three un-spiked hemolymph samples by mPCR targeting the 18S rRNA gene. A different PCR analysis targeting the B1 gene was attempted to evaluate the polymorphism of T. gondii target bands in spiked and un-spiked samples (Grigg and Boothroyd, 2001); however, T. gondii was not amplified in the B1 assay.
4. Discussion
All three protozoan parasites, C. parvum, G. enterica, and T. gondii were successfully detected in the spiked oyster hemolymph and homogenate matrices using the novel NGS assay and bioinformatics pipeline. While metabarcoding should not yet be used as a quantitative tool due to variables including primer bias, multitemplate PCR bias, and varying copy numbers, studies have shown that the read abundance of fungi and zooplankton can reflect relative estimated biomass (Peters et al., 2018; Porter and Hajibabaei, 2018). A similar result was seen with the hemolymph samples, with protozoan counts increasing with the number of spiked (oo)cysts (Fig. 2B). However, for the homogenate samples, there was not an easily discernable trend between the numbers of (oo)cysts spiked and the resulting parasite counts (Fig. 3B). Results for both hemolymph and homogenate samples showed target protozoan reads were dominated by host oyster and other eukaryotic reads.
When developing our metabarcoding assay, we anticipated that high levels of oyster DNA could impede amplification and obtaining sequence reads of target protozoan sequences. Oyster DNA that can be amplified by the 18S V4 targeted PCR could reduce the availability of primers and other PCR reagents required to amplify the spiked protozoa DNA; subsequently, oyster 18S V4 amplicons that undergo library preparation can compete with protozoan 18S V4 amplicons for sequencing reagents and sequence reads, possibly requiring greater sequencing depth to adequately detect lower ranges of (oo)cyst or protozoan spike-ins. A pepsin-HCl digestion step was incorporated into the sample preparation prior to DNA extraction in an effort to degrade oyster tissue/DNA and maximize target amplification (Fig. 1B). Digestion of shellfish tissue has been reported as an effective tool for improving detection of Cryptosporidium and Giardia species with DFA (Robertson and Gjerde, 2008). Unfortunately, this method yielded inconsistent results when used for reducing the amplification of oyster DNA in this study. Our small-scale oyster hemolymph and homogenate pepsin-HCl digestion experiments initially showed promise, with pepsin-HCl treated samples yielding no visible amplification oyster products on gel electrophoresis and reduced oyster sequence reads via NGS (Figs. S3 and S4); however, in the full-scale spiking studies, oyster reads were overwhelmingly abundant. These discrepancies in results between the pilot study and the validation experiment are not clear; it is possible that the oyster matrix is sufficiently different among the oysters we used (e.g. size, reproductive status, etc.) that the effect of pepsin-HCl on host DNA integrity is not consistent. Our attempts to use bleach for reduction of oyster DNA amplification were also unsuccessful, as even very low concentrations and a short contact time led to significant inhibition of G. enterica amplification.
Additional steps to minimize the currently overwhelming presence of host oyster DNA and maximize detection of target protozoa could improve the sensitivity of this assay. Depletion of Abundant Sequences by Hybridization (DASH) is a method of reducing unwanted background amplification from competing species. In this approach, clustered regularly interspaced short palindromic repeats (CRISPR) associated (Cas)9 nucleases complexed with single guide RNAs (sgRNAs) target the unwanted sequences for cleavage (Gu et al., 2016). Another possible technique could be the use of “blocking primers”, DNA oligos that target and bind to an unwanted sequence and prevent amplification (Vestheim and Jarman, 2008).
An important finding from our metabarcoding data was that target protozoa sequence reads were detected in the negative controls (PCR reagents with no added oyster or protozoa templates) as well as oyster samples that were not spiked. While the latter finding may be due to laboratory contamination, it is also possible that these oysters naturally accumulated protozoan (oo)cysts in the wild. The acute sensitivity of NGS methods illustrates a need for this approach to implement a minimum threshold of protozoan read numbers to reliably conclude presence of pathogen contamination. For example, the chosen cutoff can be based on a multiplier (for example 10× or 100×) of background protozoa read levels detected in the negative PCR reagent controls. Determining a positive protozoan detection cutoff depends on the desired diagnostic specificity and sensitivity for the assay. A higher cutoff will minimize false positives (clean oysters falsely deemed contaminated); however, some true positives (contaminated oysters) could be missed. A lower cutoff may increase the number of false positives but will minimize false negatives. In terms of protecting public health, an assay erring on the side of caution with a lower cutoff will maximize contamination detection.
To compare the detection of protozoa in spiked oyster samples between metabarcoding and conventional nested PCR, a cutoff of 100 reads was chosen as ‘positive’ detection using NGS. This cutoff was based on approximately 10 X the highest protozoan read count (9.5) detected in the negative PCR reagent control (Fig. S9). Using this approach, nested PCR appeared to be more sensitive than the NGS assay for detection of all three protozoan (oo)cysts spiked into oyster hemolymph (Table 2, Fig. 2B). The NGS assay results for spiked homogenate suggest it was more sensitive than PCR for detection of Cryptosporidium and Giardia, and equivalently sensitive for detection of Toxoplasma (Table 2, Fig. 3B). While our data suggest that the NGS-based assay is not definitively more sensitive than mPCR, its powerful multispecies detection may provide additional analytical advantages. In addition to conventional PCR which was used to compare parasite detection in this study, molecular tools such as digital droplet PCR (ddPCR) and real time PCR (qPCR) have been reported (Yang et al., 2014) and should be systematically evaluated in parallel with NGS for protozoa detection in shellfish.
Previous studies have revealed that because NGS-based methods have variations and biases in the PCR and sequencing steps, the final read counts do not quantitatively reflect the composition of the original sample (Amend et al., 2010; Porter and Hajibabaei, 2018). Adjustment steps were taken to minimize the effect of such biases on the number of total calculated reads for target protozoans. The Illumina MiSeq protocol includes a library normalization step to equalize read distribution during sequencing; as a result, the relative number of sequenced reads per sample may no longer reflect the relative protozoan (oo)cyst spike-in for the same samples. To correct for this, the normalization dilution coefficients were used to adjust raw read counts following taxonomic classification. Further, to address possible primer and amplification bias, a consistent number of three gBlocks (500 copies, corresponding with each targeted protozoan) was added to all samples except for the negative reagent controls. The ratio of the initial gBlock copy number to the final gBlock sequence read counts was calculated and used to adjust the final read counts for the equivalent protozoan amplicons.
It is likely that the differences in 18S rRNA gene copy numbers between the parasites also had an impact on the final read counts (Gong and Marchetti, 2019). The estimated 18S rRNA gene copy numbers for C. parvum (4 sporozoites per oocyst), G. enterica (2 trophozoites per cyst), and T. gondii (8 sporozoites per oocyst) are ~5, ~120, and ~ 880, respectively (Guay et al., 1992; Le Blancq et al., 1991; Taghi-Kilani et al., 1994). The high 18S rRNA gene copy number for Toxoplasma may have contributed to its high read counts in the spiked hemolymph samples (Fig. 2B). It should be noted that the G. enterica amplicon had a considerably higher GC content (71%), which is a factor known to influence PCR amplification rate (Kumar and Kaur, 2014). High %GC templates can form dimers and secondary structures that hinder amplification. A separate G. enterica 18S V4 region-specific reverse primer was included in the nested PCRs to improve amplification, and a GC stabilizing buffer was also included.
Metabarcoding-based identification of protozoan pathogens remains a relatively novel method for monitoring food or water-borne pathogens (Hino et al., 2016; Moreno et al., 2018). In 2018, Moreno et al. used 18S rRNA metabarcoding to identify several waterborne protozoan pathogens in irrigation water samples. G. enterica and T. gondii were both detected at very low levels (<5 reads). C. parvum was not identified in the water samples using metabarcoding, despite being detectable using DFA (Moreno et al., 2018). Due to the very low pathogen read counts detected by Moreno et al. (2018), additional control samples would have been useful for discerning between true presence of parasites in the water samples and laboratory contamination.
5. Conclusion
Of the few published examples of metabarcoding-based identification of protozoan pathogens, this study represents the first time this technique had been applied on a complex food matrix. The newly developed metabarcoding method offers a promising approach for protozoan pathogen surveillance in shellfish; however, developing additional steps to reduce oyster DNA amplification would be advantageous. Due to the highly sensitive nature of NGS and prevalent nature of DNA in laboratory environments that routinely work with the targeted pathogens, a threshold ‘positive’ level of parasite reads should also be considered for result interpretation if this method is adapted for food safety monitoring purposes.
Funding sources
This work was supported by the Natural Science and Engineering Research Council of Canada (Discovery award numbers 401134 (PI Shapiro) and 400095 (PI Feng), the Canada Excellence Research Chair in Aquatic Epidemiology, and the University of Guelph's Departments of Mathematics and Statistics and Integrative Pathobiology.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Joyce Rousseau and Lezlie Rueda for their assistance preparing the spiked oyster samples and the metabarcode libraries. We are also grateful for Cynthia Mitchell's assistance with the collection and processing of oysters from Prince Edward Island. Many thanks to Dr. Monica Antenos, Allison MacKay, Katrina Watson, and Ed Reyes for aid with laboratory techniques and supplies. We would also like to thank David Arranz-Solís for generously providing T. gondii oocysts.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fawpar.2020.e00096.
Appendix A. Supplementary materials
Supplementary materials
References
- Allan E. Unrestricted access to microbial communities. Virulence. 2014;5(3):397–398. doi: 10.4161/viru.28057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschup S.F., Gish W., Pennsylvania T., Park U. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amend A.S., Seifert K.A., Bruns T.D. Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Mol. Ecol. 2010;19(24):5555–5565. doi: 10.1111/j.1365-294X.2010.04898.x. [DOI] [PubMed] [Google Scholar]
- Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ Retrieved from Babraham Institute website.
- Bahia-Oliveira L., Gomez-Marin J., Shapiro K. Toxoplasma gondii. In J. B. Rose & B. Jiménez-Cisneros (Eds.), Global Water Pathogen Project. 2017. http://www.waterpathogens.org/book/toxoplasma-gondii Retrieved from.
- Betancourt W. (2019). Cryptosporidium spp. in J. B. Rose & B. Jiménez-Cisneros (Eds.), Global Water Pathogen Project. 2019. https://www.waterpathogens.org/book/cryptosporidium Retrieved from.
- Boarato-David E., Guimarães S., Cacciò S. Giardia duodenalis. In J. B. Rose & B. Jiménez-Cisneros (Eds.), Global Water Pathogen Project. 2017. https://www.waterpathogens.org/book/giardia-duodenalis Retrieved from.
- Dorny P., Praet N., Deckers N., Gabriel S. Emerging food-borne parasites. Vet. Parasitol. 2009;163(3):196–206. doi: 10.1016/j.vetpar.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Dubey J.P. The history and life cycle of Toxoplasma gondii. In Toxoplasma Gondii: The Model Apicomplexan - Perspectives and Methods: Second Edition (Second Edi). 2013 doi: 10.1016/B978-0-12-396481-6.00001-5. [DOI] [Google Scholar]
- DuPont H.L. Giardia: both a harmless commensal and a devastating pathogen. J. Clin. Investig. 2013;123(6):2352–2354. doi: 10.1172/JCI69932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayer R., Dubey J.P., Lindsay D.S. Zoonotic protozoa: from land to sea. Trends Parasitol. 2004;20(11):531–536. doi: 10.1016/j.pt.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Fritz H., Barr B., Packham A., Melli A., Conrad P.A. Methods to produce and safely work with large numbers of Toxoplasma gondii oocysts and bradyzoite cysts. J. Microbiol. Methods. 2012;88(1):47–52. doi: 10.1016/j.mimet.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W., Marchetti A. Estimation of 18S gene copy number in marine eukaryotic plankton using a next-generation sequencing approach. Front. Mar. Sci. 2019;6(APR):1–5. doi: 10.3389/fmars.2019.00219. [DOI] [Google Scholar]
- Grigg M.E., Boothroyd J.C. Rapid identification of virulent type I strains of the protozoan pathogen Toxoplasma gondii by PCR-restriction fragment length polymorphism analysis at the B1 gene. J. Clin. Microbiol. 2001;39(1):398–400. doi: 10.1128/JCM.39.1.398-400.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Crawford E.D., O'Donovan B.D., Wilson M.R., Chow E.D., Retallack H. Depletion of abundant sequences by hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016;17(1):1–13. doi: 10.1186/s13059-016-0904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guay J., Huot A., Gagnon S., Tremblay A., Levesque R.C. Physical and genetic mapping of cloned ribosomal DNA from Toxoplasma gondii: primary and secondary structure of the 5S gene. Gene. 1992;114:165–171. doi: 10.1016/0378-1119(92)90570-f. [DOI] [PubMed] [Google Scholar]
- Hadziavdic K., Lekang K., Lanzen A., Jonassen I., Thompson E.M., Troedsson C. Characterization of the 18s rRNA gene for designing universal eukaryote specific primers. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0087624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino A., Maruyama H., Kikuchi T. A novel method to assess the biodiversity of parasites using 18S rDNA Illumina sequencing; parasitome analysis method. Parasitol. Int. 2016;65(5):572–575. doi: 10.1016/j.parint.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Hohweyer J., Dumètre A., Aubert D., Azas N., Villena I. Tools and methods for detecting and characterizing Giardia, Cryptosporidium, and Toxoplasma parasites in marine mollusks. J. Food Prot. 2013;76(9):1649–1657. doi: 10.4315/0362-028X.JFP-13-002. [DOI] [PubMed] [Google Scholar]
- Illumina. (2013). Document 15044223 b: 16S Metagenomic Sequencing Library Preparation. Retrieved from https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil I.A., Troeger C., Rao P.C., Blacker B.F., Brown A., Brewer T.G. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob. Health. 2018;6(7):e758–e768. doi: 10.1016/S2214-109X(18)30283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsoumanis K., Allende A., Alvarez-Ordóñez A., Bolton D., Bover-Cid S., Chemaly M. Public health risks associated with food-borne parasites. EFSA J. 2018;16(12) doi: 10.2903/j.efsa.2018.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 2013;79(17):5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Kaur J. Primer based approach for PCR amplification of high GC content gene: Mycobacterium gene as a model. Mol. Biol. Int. 2014;2014:1–7. doi: 10.1155/2014/937308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Blancq S.M., Korman S.H., van Der Ploeg L.H.T. Frequent rearrangements of rRNA-encoding chromosomes in Giardia lamblia. Nucleic Acids Res. 1991;19(16):4405–4412. doi: 10.1093/nar/19.16.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzano V., Mancinelli L., Bracaglia G., Del Chierico F., Vernocchi P., Di Girolamo F. “Omic” investigations of protozoa and worms for a deeper understanding of the human gut “parasitome”. PLoS Negl. Trop. Dis. 2017;11(11):1–19. doi: 10.1371/journal.pntd.0005916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Y., Moreno-Mesonero L., Amorós I., Pérez R., Morillo J.A., Alonso J.L. Multiple identification of most important waterborne protozoa in surface water used for irrigation purposes by 18S rRNA amplicon-based metagenomics. Int. J. Hyg. Environ. Health. 2018;221(1):102–111. doi: 10.1016/j.ijheh.2017.10.008. [DOI] [PubMed] [Google Scholar]
- Peters L., Spatharis S., Dario M.A., Dwyer T., Roca I.J.T., Kintner A. Environmental DNA: a new low-cost monitoring tool for pathogens in salmonid aquaculture. Front. Microbiol. 2018;9(DEC):1–10. doi: 10.3389/fmicb.2018.03009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter T.M., Hajibabaei M. Scaling up: a guide to high-throughput genomic approaches for biodiversity analysis. Mol. Ecol. 2018;27(2):313–338. doi: 10.1111/mec.14478. [DOI] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(D1):590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson L.J. The potential for marine bivalve shellfish to act as transmission vehicles for outbreaks of protozoan infections in humans: a review. Int. J. Food Microbiol. 2007;120(3):201–216. doi: 10.1016/j.ijfoodmicro.2007.07.058. [DOI] [PubMed] [Google Scholar]
- Robertson L.J., Gjerde B. Development and use of a pepsin digestion method for analysis of shellfish for Cryptosporidium oocysts and Giardia cysts. J. Food Prot. 2008;71(5):959–966. doi: 10.4315/0362-028X-71.5.959. [DOI] [PubMed] [Google Scholar]
- Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;2016(10):1–22. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan U., Hijjawi N., Feng Y., Xiao L. Giardia: an under-reported foodborne parasite. Int. J. Parasitol. 2019;49(1):1–11. doi: 10.1016/j.ijpara.2018.07.003. [DOI] [PubMed] [Google Scholar]
- Shapiro Karen, Bahia-Oliveira L., Dixon B., Dumètre A., de Wit L.A., VanWormer E., Villena I. Environmental transmission of Toxoplasma gondii: Oocysts in water, soil and food. Food and Waterborne Parasitology. 2019;15 doi: 10.1016/j.fawpar.2019.e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro K., Kim M., Rajal V.B., Arrowood M.J., Packham A., Aguilar B., Wuertz S. Simultaneous detection of four protozoan parasites on leafy greens using a novel multiplex PCR assay. Food Microbiol. 2019;84(April):103252. doi: 10.1016/j.fm.2019.103252. [DOI] [PubMed] [Google Scholar]
- Taghi-Kilani R., Remacha-Moreno M., Wenman W.M. Three tandemly repeated 5S ribosomal RNA-encoding genes identified, cloned and characterized from Cryptosporidium parvum. Gene. 1994;142(2):253–258. doi: 10.1016/0378-1119(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Vestheim H., Jarman S.N. Blocking primers to enhance PCR amplification of rare sequences in mixed samples - a case study on prey DNA in Antarctic krill stomachs. Front. Zool. 2008;5:1–11. doi: 10.1186/1742-9994-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis J.E., McClure J.T., McClure C., Spears J., Davidson J., Greenwood S.J. Bioaccumulation and elimination of Cryptosporidium parvum oocysts in experimentally exposed eastern oysters (Crassostrea virginica) held in static tank aquaria. Int. J. Food Microbiol. 2014;173:72–80. doi: 10.1016/j.ijfoodmicro.2013.11.033. [DOI] [PubMed] [Google Scholar]
- World Health Organization . In: Safe Management of Shellfish and Harvest Waters: Minimizing Health Risks From Sewage-Contaminated Shellfish. Rees G., Pond K., Kay D., Bartram J., Domingo J. Santo, editors. World Health Organization; 2010. [Google Scholar]
- Yang R., Paparini A., Monis P., Ryan U. Comparison of next-generation droplet digital PCR (ddPCR) with quantitative PCR (qPCR) for enumeration of Cryptosporidium oocysts in faecal samples. Int. J. Parasitol. 2014;44(14):1105–1113. doi: 10.1016/j.ijpara.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Zhang J., Kobert K., Flouri T., Stamatakis A. PEAR: a fast and accurate Illumina paired-end reAd mergeR. Bioinformatics. 2014;30(5):614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials