Abstract
Human-induced pluripotent stem cells (hiPSCs) derived cardiomyocytes (hiPSC-CMs) have been explored for cardiac regeneration and repair as well as for the development of in vitro 3D cardiac tissue models. Existing protocols for cardiac differentiation of hiPSCs utilize a 2D culture system. However, the efficiency of hiPSC differentiation to cardiomyocytes in 3D culture systems has not been extensively explored. In the present study, we investigated the efficiency of cardiac differentiation of hiPSCs to functional cardiomyocytes on 3D nanofibrous scaffolds. Coaxial polycaprolactone (PCL)-gelatin fibrous scaffolds were fabricated by electrospinning and characterized using scanning electron microscopy (SEM) and fourier transform infrared (FTIR) spectroscopy. hiPSCs were cultured and differentiated into functional cardiomyocytes on the nanofibrous scaffold and compared with 2D cultures. To assess the relative efficiencies of both the systems, SEM, immunofluorescence staining and gene expression analyses were performed. Contractions of differentiated cardiomyocytes were observed in 2D cultures after 2 weeks and in 3D cultures after 4 weeks. SEM analysis showed no significant differences in the morphology of cells differentiated on 2D versus 3D cultures. However, gene expression data showed significantly increased expression of cardiac progenitor genes (ISL-1, SIRPA) in 3D cultures and cardiomyocytes markers (TNNT, MHC6) in 2D cultures. In contrast, immunofluorescence staining showed no substantial differences in the expression of NKX-2.5 and α-sarcomeric actinin. Furthermore, uniform migration and distribution of the in situ differentiated cardiomyocytes was observed in the 3D fibrous scaffold. Overall, our study demonstrates that coaxial PCL-gelatin nanofibrous scaffolds can be used as a 3D culture platform for efficient differentiation of hiPSCs to functional cardiomyocytes.
Keywords: Induced pluripotent stem cells, cardiomyocytes, polycaprolactone, gelatin, nanofibrous scaffolds, stem cell differentiation
1. Introduction
Cardiovascular diseases (CVDs) are the leading cause of mortality worldwide. In the United States alone, an estimated 48% of adults suffer from some form of CVD [1]. Cell-based approaches using adult stem/progenitor cells have been used for the treatment of CVDs in many pre-clinical and clinical studies [2]. Unfortunately, the use of these stem cells has been challenging on account of their limited availability and low differentiation to functional cardiomyocytes post-transplantation [3]. In contrast, since their discovery, human-induced pluripotent stem cells (hiPSCs), have overcome this limitation and hence, opened up new avenues in the field of cardiovascular regenerative medicine [4]. Moreover, the use of cardiomyocytes differentiated from hiPSCs (hiPSC-CMs) have become increasingly popular as in vitro model systems for cardiac developmental biology studies [5], disease modeling [6–8], and toxicology studies [9, 10], as well as for the development of a new generation cell-based therapies for CVDs [11].
Indeed, to conduct these studies, many protocols have been standardized for the efficient differentiation of hiPSCs to functional cardiomyocytes [12]. However, the majority of these studies have been conducted using 2-dimensional (2D) culture systems. Moreover, recent reports, have clearly shown that hiPSC-CMs differentiated in 2D cultures do not completely resemble adult cardiomyocytes [13–15]. Similarly, it has been proposed that 3D cultures may represent a better model system to recapitulate in vivo differentiation of hiPSCs [16]. Among different 3D culture systems, previous reports have shown enhanced differentiation of hiPSCs to chondrogenic cells [17], definitive endoderm cells [18], pancreatic β-cells [19], and neuronal cells [20], when cultured on 3-dimensional (3D) scaffolds. Adipose-derived stem cells [21] and mouse pluripotent stem cells [22] have also been shown to have improved differentiation to functional cardiomyocytes on 3D scaffolds. A detailed study investigating the efficiency of cardiac differentiation of hiPSCs on 3D scaffolds is currently lacking.
Synthetic polymers like polycaprolactone (PCL), poly (glycerol sebacate) (PGS), poly (lactic-co-glycolic acid) (PLGA) and poly (lactic acid) (PLA) have been commonly used for the synthesis of nanofibrous scaffolds for culture and differentiation of hiPSCs [23]. Among these, PCL, an FDA approved polymer, has been well-established for the development of long-term controlled release drug delivery systems [24, 25]. Also, compared to other synthetic polymers, PCL degrades more slowly, resulting in a more gradual formation of acidic by-products, which are also rapidly cleared upon formation in culture [26]. However, although studies have reported cardiac differentiation of hiPSCs on these scaffolds, some studies have reported a negative effect of these polymers on cell viability and function due to the inherent hydrophobicity and poor biocompatibility of the polymer, as well as lack of cell adhesion sites [20, 27]. Biopolymers like fibrin, collagen, and gelatin have been commonly used as a substrate for culturing hiPSCs in 2D, mainly due to their biomimetic properties [23]. However, their use in the fabrication of 3D scaffolds has been limited due to their poor mechanical strength and rapid degradation in prolonged culture [28, 29]. Hence, the fabrication of a stable, nanofibrous scaffold with superior biomimetic and mechanical properties will be essential to understanding the differentiation of hiPSCs to functional cardiomyocytes in 3D cultures.
In this study, we have fabricated and characterized a coaxial PCL-gelatin nanofibrous scaffold with a gelatin shell and a PCL core [27, 30]. Using this nanofibrous scaffold, we have established a protocol for culturing and differentiating hiPSCs into functional cardiomyocytes in a 3D microenvironment. Additionally, we compared the cardiac differentiation of hiPSCs in 3D and 2D cultures, to understand the relative efficiencies of differentiation and maturation of hiPSC-CMs in vitro in the two culture systems.
2. Materials and methods
2.1. Preparation of polycaprolactone (PCL)-gelatin coaxial nanofibrous scaffold
PCL-gelatin coaxial randomly aligned fibrous scaffolds were fabricated using electrospinning [27, 30, 31]. Gelatin (12% w/v) and PCL (8%) were dissolved in 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP, Sigma-Aldrich, MA). The PCL solution was fed to the inner tube of the coaxial spinneret with the flow-rate of 1 ml/hr, while the gelatin solution was fed to the outer needle at a flow-rate of 4 ml/hr. The distance between the spinneret tip and the grounded rotating collector was kept at 20 cm and the voltage applied at the tip was 20 kV. Scaffolds were thoroughly dried and cut into circular patches of 8- or 12-mm diameter before crosslinking in a 7 mM 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC, Sigma-Aldrich, MO) solution in 100% ethanol overnight to stabilize the gelatin shell. The patches were then disinfected by soaking in 70% ethanol for 24 hours, followed by three rinses in sterile PBS and one rinse in Essential 8™ culture medium.
2.2. Measurement of scaffold porosity
Dry scaffold porosity was calculated by cutting the as-spun scaffolds into discs using a 10 mm biopsy punch. The thickness of each scaffold was measured using digital calipers. Scaffolds were weighed (Mettler Toledo Precision Balance) and the density of the scaffold (ρscaffold) calculated. Porosity of the scaffold quantified by comparing the scaffold’s density to a theoretical solid of the same PCL-Gelatin composition (% Porosity = (ρsolid − ρscaffold) / ρsolid) (ρsolid = 1.233g/cm3). Six scaffolds were measured with porosity presented as mean ± standard deviation.
2.3. Culturing of hiPSCs on a 3D scaffold
Maintaining hiPSCs in 2D cultures
The hiPSC cell line (SCVI840), re-programmed from peripheral blood mononuclear cells (PBMCs), used in the study was obtained from the Stanford Cardiovascular Institute (SCVI) Biobank and the Stem Cell Core Facility of Genetics, Stanford University. The hiPSCs were cultured and maintained as previously described [32]. Briefly, the hiPSCs were thawed and cultured in Essential 8™ Medium (E8, Thermo Fisher Scientific, MA) on Vitronectin XF (Stem Cell Technologies, Canada)-coated 6-well plates (Greiner, NC). The medium was supplemented with 10 μM of Rho-associated protein kinase (ROCK) inhibitor (Y-27632, TOCRIS, MN) for the first 24 hours of culture. Once the cultures attained a confluence of >80% in the dish, the cells were passaged onto Matrigel® (354277, Corning, NY) coated 12-well plates for 2D culture or onto the PCL-gelatin scaffolds for 3D cultures.
Preparing the 3D scaffold for the plating of hiPSCs
PCL-gelatin scaffolds were coated with Matrigel® (Cat. No. 354277, Corning, NY) for 1–2 hours. The Matrigel®-coated patches were then placed on top of a sterile N-terface® (Winfield Labs, TX), which was further placed on a sterile surgical sponge (Hydrosorb: Carwil Corp, New London, CT) pre-soaked in E8 medium supplemented with 10 μM ROCK inhibitor in 94-mm dishes. hiPSCs cultured in 2D culture plates were dissociated into single cells by incubating with Gentle Cell Dissociation Reagent (Stem Cell Technologies, Canada) in E8 medium supplemented with 10 μM ROCK inhibitor to get a concentrated cell suspension containing of 0.2 million cells/100 μl, which was then added dropwise onto individual 3D patches (0.2 million cells per 12 mm patch). The seeded patches were incubated for 7–8 hrs to ensure cell attachment then transferred to a 12-well plate containing E8 medium, with one patch per well.
2.4. Cardiac differentiation of hiPSCs
Cardiac differentiation of hiPSCs was performed using a Cardiomyocyte Differentiation Kit (Thermo Fisher Scientific, MA), as per the manufacturer’s instructions [33]. Briefly, on Day 0 (D0) of differentiation, the hiPSC colonies, with 50–60% confluence, were incubated with Cardiomyocyte Differentiation Medium A for 48 hrs. On Day 2 (D2), the medium was replaced with Cardiomyocyte Differentiation Medium B for another 48 hrs. From Day 4 (D4) onwards, the cells were cultured in Cardiomyocyte Maintenance Medium. The medium for the cultures was changed every other day. The morphological changes during cardiac differentiation of hiPSCs were assessed by phase-contrast imaging using a Leica DM IL LED microscope (Leica Microsystems, Germany). Videos were recorded to monitor the contractility of functional cardiomyocytes and quantitative analysis was performed post-acquisition. For quantitative assessment of the beating frequencies of differentiated cardiomyocytes, the beats per minute were manually counted at different days of differentiation in four-patches or wells from three independent experiments.
2.5. Scanning electron microscopy (SEM) analysis
To assess the morphology, hiPSCs were cultured and differentiated on glass coverslips or electrospun scaffolds and processed as previously described [34]. Briefly, the cells were fixed in 4% paraformaldehyde (PFA) for 24 hours at 4°C and dehydrated sequentially in 50%, 70%, 85%, 90% and 100% ethanol gradients. The samples were finally dried using a graded series of hexamethyldisilazane (HMDS, Sigma-Aldrich, MO) in ethanol. After processing, the patches were affixed to SEM stubs using carbon tape and sputter-coated with gold-palladium (Pelco Model 3). For characterization of the acellular scaffolds, scaffolds were cut, affixed to SEM stubs and sputter-coated with gold-palladium coating. All samples were imaged on the Nova NanoSEM 400 microscope (FEI, OR) at 5 kEV. The images acquired were analyzed using ImageJ software to determine the fiber diameter, pore size, and pore area. The pore size is the interfiber distance and the pore area is the area measured between two fibers. Ten images were used to analyze the above parameters and the mean values are calculated.
2.6. Fourier-transform infrared spectroscopy (FTIR) studies
Surface chemical analysis of the patches was assessed using attenuated total reflection Fourier transform infrared spectroscopy (ATR-FTIR) in the range of 400–4000 cm−1 (Thermo Nicolet Nexus 670 FTIR spectrometer, MN). Approximately 25–40 scans were performed on gelatin, PCL and PCL-gelatin coaxial scaffolds.
2.7. Immunofluorescence studies
Immunofluorescence imaging was performed on undifferentiated and differentiated cells cultured on 3D patches and on coverslips (2D). The cultured cells were washed twice with DPBS and processed for immunostaining as described previously [35]. Briefly, the cells were fixed in 4% paraformaldehyde for 15 minutes at RT, permeabilized using 0.2% Triton X-100 and incubated in a blocking buffer containing 1% bovine serum albumin (BSA, Sigma-Aldrich, MO). The cells were incubated with the primary antibody overnight at 4°C, followed by the secondary antibody for 1 hour at RT in the dark. The cells were counterstained with DAPI (Thermo Fisher Scientific, MA) and the coverslips were mounted over glass slides using ProLong™ Gold Antifade Mountant (Life Technologies, MA). The cells were then imaged on the Olympus FV3000 (Olympus Life Sciences, PA) confocal microscope. For cells differentiated in 3D, the patches were fixed in 4% paraformaldehyde, embedded in Optimal Cutting Temperature (OCT) compound, frozen, and sectioned at 7 μm using the Leica CM 1950 cryostat (Leica Biosystems, Germany). The sections were mounted onto glass slides and immunostaining was performed as described above. The antibodies used for immunostaining are as follows: rabbit anti-Oct4 (1:200, Thermo Fisher Scientific, MA, Cat # PA5–27438), mouse anti-SSEA4 (1:200, Thermo Fisher Scientific, MA, Cat #MA1–021), rabbit anti-Troponin T (1:200, Sigma-Aldrich, MO, Cat # HPA017888), mouse anti-Sarcomeric Alpha-Actin (1:200, Sigma-Aldrich, MO, Cat #A7811), rabbit anti-NKX-2.5 (1:200, Thermo Fisher Scientific, MA, Cat # PA5–49431), anti-mouse Alexa Fluor® 488 (1:1000, Cell Signaling Technology, MA, Cat # 4408S), anti-rabbit Alexa Fluor® 594 (1:1000, Cell Signaling Technology, MA, Cat # 8889S).
2.8. Cell viability assay by immunofluorescence
The viability of cells was determined using the LIVE/DEAD™ cell imaging kit (Molecular Probes, Life Technologies Corp., CA) per the manufacturer’s instructions. Briefly, hiPSCs cultured on 3D scaffolds were incubated with an equal volume of 2X staining solution for 20 min at 37°C. The cells were washed in PBS and fresh medium was added to the cells. The samples (n = 3) were imaged at 488 nm and 570 nm for green and red fluorescence, respectively.
2.9. Lactate Dehydrogenase (LDH) Release Assay
The cytotoxic effect of the coaxial patches on hiPSCs was determined by measuring the LDH released by the cells cultured in 2D or 3D for 72 hours. The spent medium from 2D and 3D cultures were collected and the LDH release was estimated using the In vitro toxicology assay kit, lactic dehydrogenase based (Cat# TOX7–1KT, Sigma-Aldrich Inc., MO, USA) per the manufacturer’s protocol. Medium incubated at 37°C for 48 hrs without cells was used as a control. Background absorbance and primary absorbance was measured at 690 nm and 490 nm, respectively, using the 2030 Multilabel Reader, Victor™ x3, spectrophotometer (PerkinElmer Inc., MA, USA). The assay was performed in 96-well plates in quadruplicate (n=4) and data obtained was analyzed on WorkOut 2.5 (build 0428, PerkinElmer Inc., MA, USA), by subtracting background absorbance from primary absorbance.
2.10. Gene expression analysis
Cellular gene expression profiles were analyzed on days 0, 7 and 28 of differentiation in culture. The cells were lysed in TRIzol (Invitrogen. MA) and total RNA was isolated using the Direct-zol RNA Miniprep kit (Zymo Research, CA) as per the manufacturer’s instructions. The quantity and purity of the RNA was assessed using a NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, MA). First-strand cDNA was synthesized using the RT2 First Strand Kit (330404, Qiagen, MD) as per the manufacturer’s instructions with the RT reaction being performed for 1 hour at 37°C. qRT-PCR was performed using Qiagen RT2 SYBR Green ROX qPCR Mastermix (330523, Qiagen, MD) according to the manufacturer’s protocol. The reaction was performed on a QuantStudio 3 (Applied Biosystems, MA) using QuantStudio design and analysis software V.1.4.1. Gene expression was normalized to the geometric mean of three housekeeping genes- β-Actin, beta-2 microglobulin, and RPL13a and the relative expression was calculated relative to undifferentiated cells, D0, using the 2−ΔΔCt method. Data are expressed as mean ± SD, n = 3.
2.11. Statistical analysis
Statistical significance was assessed by one-way ANOVA followed by Tukey’s HSD post-hoc test or by two-tailed Student’s t-test to obtain p-values. All values were represented as mean ± SD, and p<0.05 was considered statistically significant.
3. RESULTS
3.1. Characterization of coaxial PCL-gelatin nanofibrous scaffold
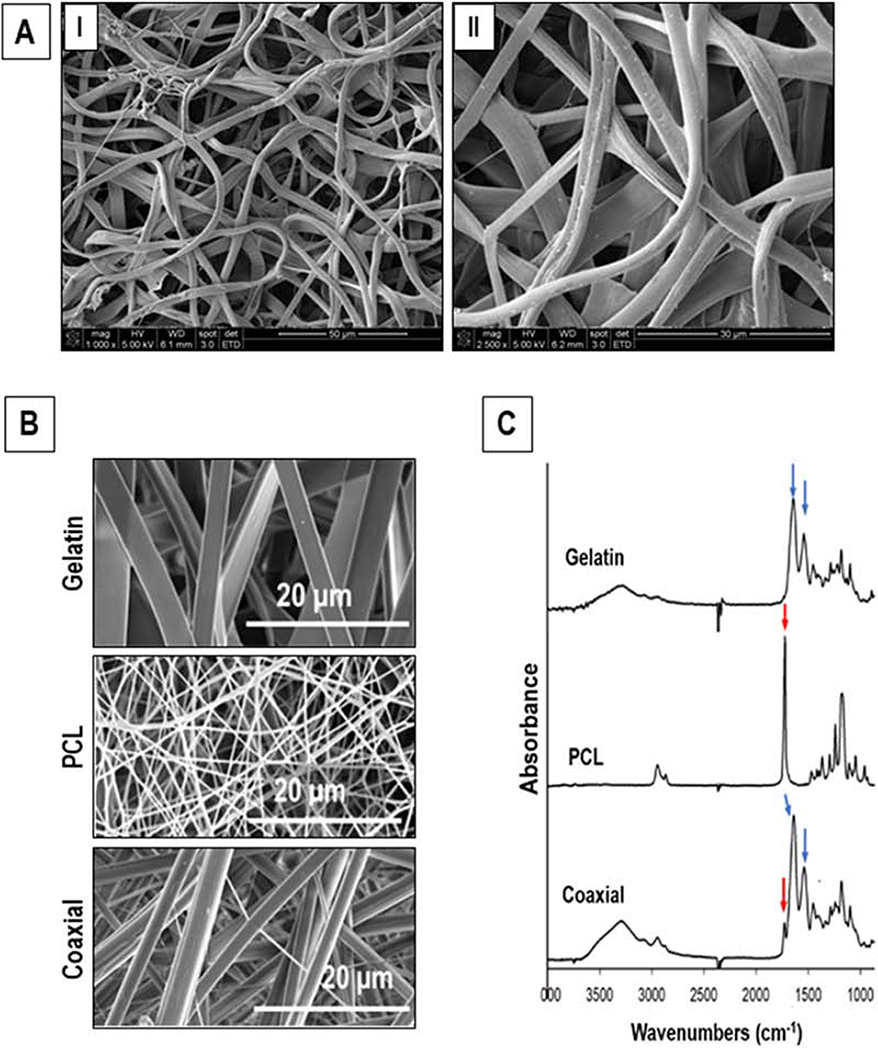
SEM images of the coaxial PCL-gelatin fibrous scaffold showed a random alignment of the fibers within the patch (Fig. 1A). The average fiber diameter within the scaffold was found to be 2.39 ± 0.66 μm. Furthermore, these fibrous scaffolds had a mean porosity of 82.5 + 6.2 %, while the pore area and interfiber diameter of these scaffolds was 392.36 ± 194.65 μm2 and 21.6 ± 6.74 μm, respectively. Additionally, the coaxial fibers appeared to be more cylindrical (Fig. 1B) compared to nanofibers fabricated with only gelatin or PCL. FTIR spectrum for the gelatin scaffold showed strong characteristic peaks at 1650 and 1540 cm−1 (Fig. 1C, blue arrows) corresponding to the amide bonds (I and II, respectively) of gelatin. PCL scaffolds displayed a characteristic peak at 1724 cm−1 corresponding to its carbonyl group (Fig. 1C, red arrow). The FTIR spectrum of the coaxial PCL-gelatin scaffold was dominated by the amide I and II peaks, with a smaller peak at 1724 cm−1 (Fig. 1C, blue and red arrows, respectively). These results indicated the presence of both PCL and gelatin spectrums in the coaxial scaffolds.
Figure 1.
Characterization of the electrospun scaffold. (A) SEM images showing random alignment of PCL-gelatin coaxial scaffolds. (B) SEM images of gelatin, PCL and PCL-gelatin coaxial scaffolds. (C) ATR-FTIR analyses of gelatin, PCL, and coaxial nanofibrous scaffolds.
3.2. Morphological assessment of hiPSCs cultured in 2D and 3D microenvironment
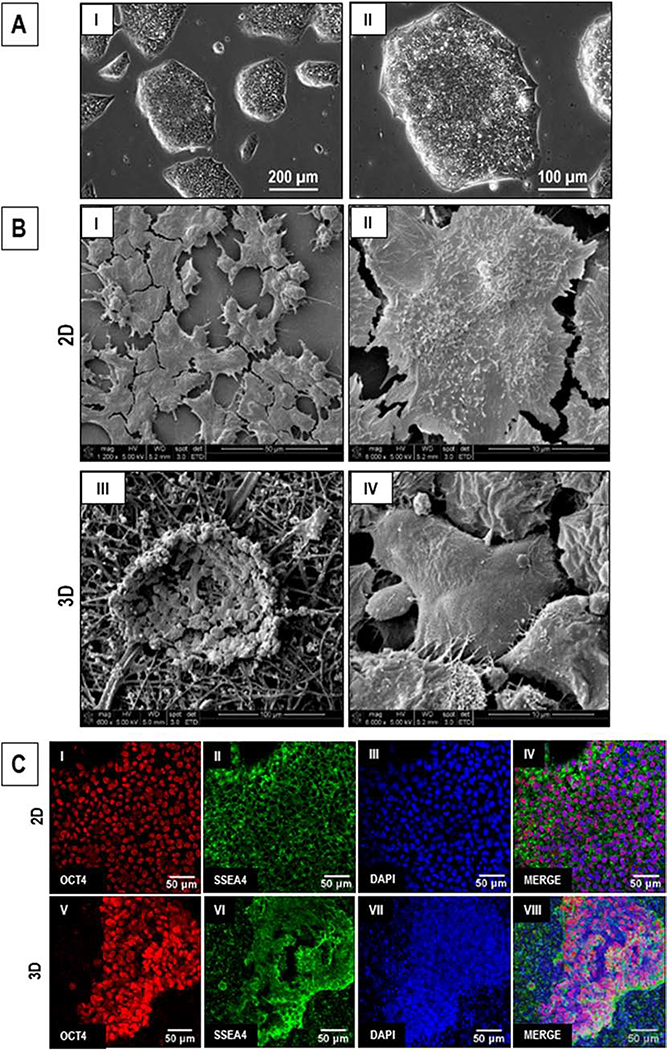
hiPSCs cultured on 2D tissue culture plates showed 60–70% confluence in 48–72 hours (Fig. 2A). While the hiPSCs cultured in 2D showed a flattened morphology and formation of monolayer colonies (Fig. 2B, I-II), the hiPSCs cultured on 3D PCL-gelatin scaffolds showed the formation of individual dispersed colonies (Fig. 2B, III-IV). No significant difference was observed in the expression of pluripotency markers, OCT4 (localized to the nucleus) and SSEA4 (localized in the cell membrane), in hiPSCs cultured in 2D versus 3D cultures (Fig. 2C), as assessed by immunofluorescence staining. These data indicate that hiPSCs cultured on the 3D scaffolds retained their pluripotency.
Figure 2.
Comparative assessment of hiPSCs cultured in 2D and on 3D PCL-gelatin coaxial scaffolds. (A) Phase-contrast images of hiPSC colonies after 72 hr in culture. Scale: I: 200 μm; II: 100 μm. (B) SEM images of hiPSCs cultured in 2D (I-II) and 3D (III-IV) cultures. Scale: I: 50 μm; II, IV: 10 μm; III: 100 μm. (C) Fluorescent images showing the expression of PSC markers, OCT4 and SSEA4, in the hiPSCs cultures in 2D (I-IV) and 3D (V-VIII). Scale: 50 μm.
3.3. Viability of hiPSCs cultured on 3D scaffolds
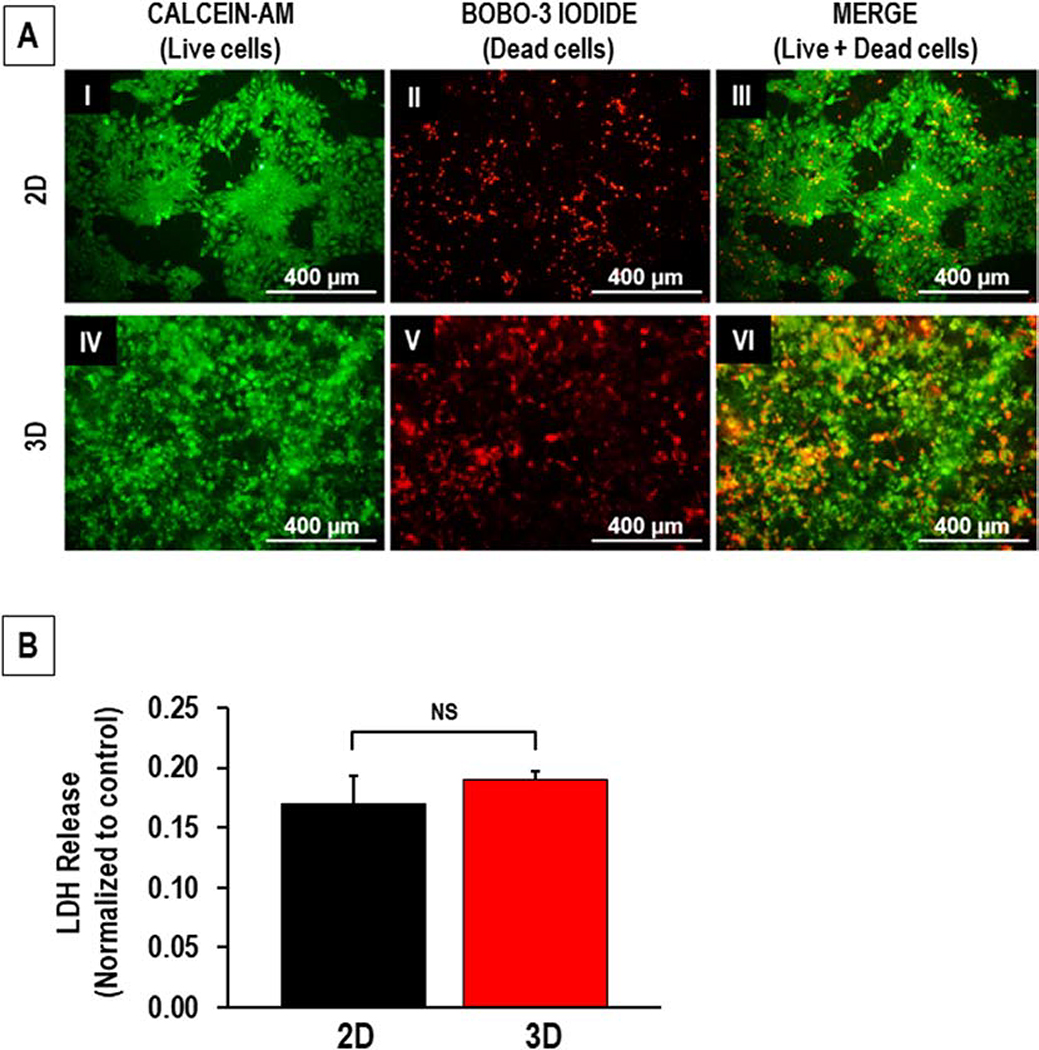
Cell viability was assessed following the culture of hiPSCs on 3D scaffolds for 72 hours. A majority of the cells in 2D and 3D cultures stained positive for Calcein-AM (green), indicating the presence of live cells; however, positive staining for BOBO-3 iodide (red) was also detected in both cultures, indicative of dead cells (Fig. 3A). Furthermore, measurement of the LDH released by cells cultured in 2D and 3D cultures did not vary significantly (Fig. 3B) indicating that the cells remained viable in the scaffold and that the coaxial nature of the scaffold is non-toxic to the cells.
Figure 3:
(A) Staining for viability of hiPSCs cultured in 2D (I-III) and 3D (IV-VI) cultures. Representative fluorescence images showing the live (Calcein-AM, green) and dead (BOBO-3 iodide, red) cells in 2D and 3D cultures. Scale: Scale: 400 μm. (B) Lactate dehydrogenase (LDH) release measured in 2D and 3D cultures. n = 4, NS: *p > 0.05.
3.4. Morphological assessment of cardiac differentiation in 2D and 3D cultures
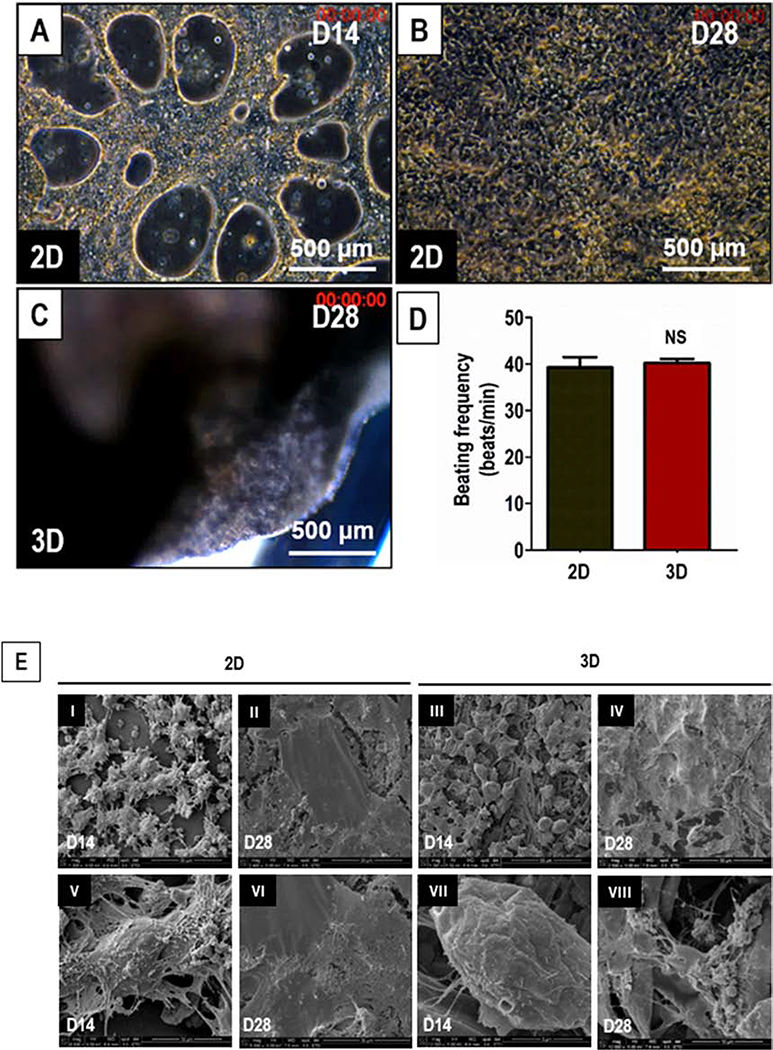
Following induction of cardiac differentiation of hiPSCs in 2D cultures, small clusters of functional contracting cardiomyocytes were observed by D8. By D14, contracting ‘sheets’ of cardiomyocytes was observed in 100% of the wells (Fig. 4A, Supplementary Movie 1). By D28, the cardiomyocytes formed a continuous syncytium and showed synchronous contractions (Fig. 4B, Supplementary Movie 2). In the case of hiPSCs differentiated on 3D PCL-gelatin scaffolds, spontaneous macroscopic contractions were observed from D21 and by D28, 89% of the scaffolds showed spontaneous contractions (Fig. 4C, Supplementary Movie 3). No significant differences in the beating frequencies were observed in hiPSCs differentiated on 2D versus 3D cultures (Fig. 4D).
Figure 4.
Morphological and functional assessment of cardiac differentiation of hiPSCs in 2D and 3D cultures. (A) Phase-contrast image of hiPSCs differentiated to cardiomyocytes in 2D cultures on D14 (A) and D28 (B), and 3D cultures on D28 (C). Scale: 500 μm. (D) Quantitative assessment of beating frequency of the hiPSCs differentiated to functional cardiomyocytes in 2D culture (black) and 3D cultures (red). (E) SEM images showing the surface morphology of the cells differentiated from hiPSCs cultured and differentiated to functional cardiomyocytes in 2D (I-II, V-VI) and 3D ( III-IV, VII-VIII) cultures on D14 and D28. Scale: I-IV: 50 μm; V & VI: 30 μm; VII: 5 μm; and VIII: 10 μm.
Changes in cell morphology during cardiac differentiation in 2D versus 3D cultures were assessed by SEM analysis. On D14, SEM images of the 2D and 3D cultures did not show any significant differences in their morphology (Fig. 4E, I, III, V and VII). However, in the case of 3D cultures, the cells appeared to have penetrated the scaffold and wrapped themselves around the individual fibers (Fig. 4E, VII-VIII). On D28, the differentiated cardiomyocytes formed a monolayer with good cell-cell contact in both 2D and 3D cultures (Fig. 4E, II, IV, VI and VIII). Increased levels of ECM secretions were observed by D28 as compared to D14 in both the 2D and 3D cultures. Taken together, these results showed differentiation of hiPSCs to functional cardiomyocytes on the scaffolds comparable to that achieved in 2D culture.
3.5. Immunofluorescence imaging for cardiac differentiation in 2D and 3D cultures
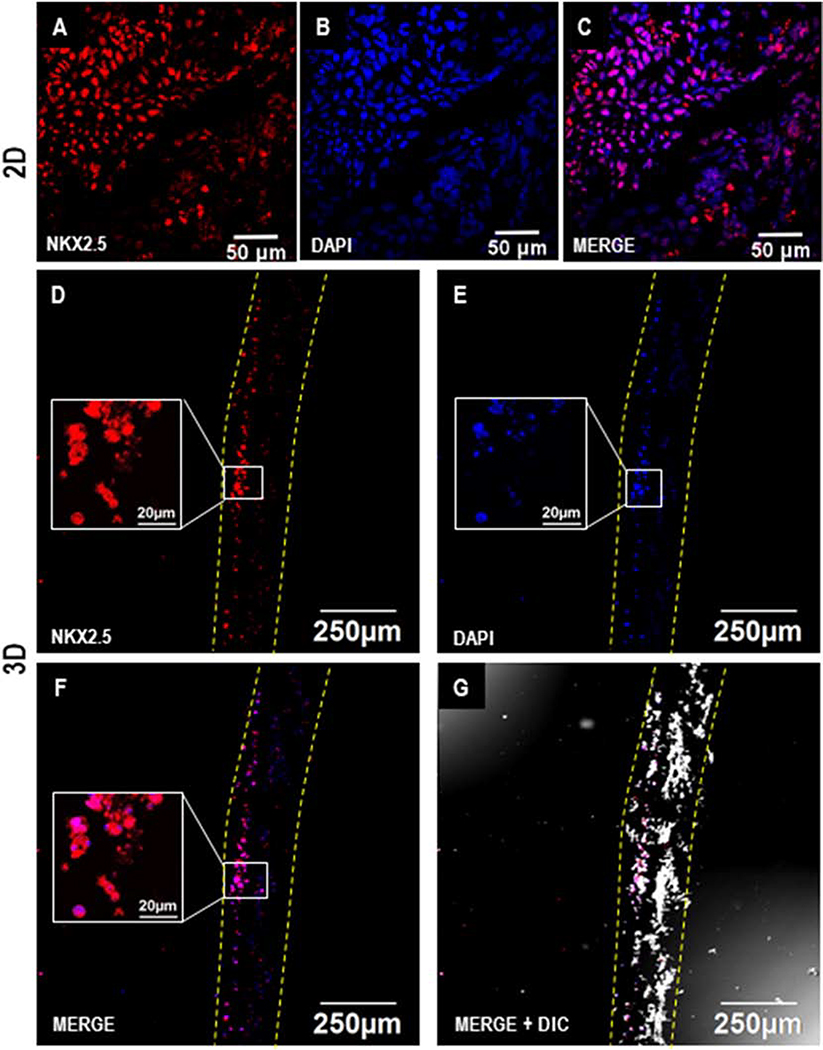
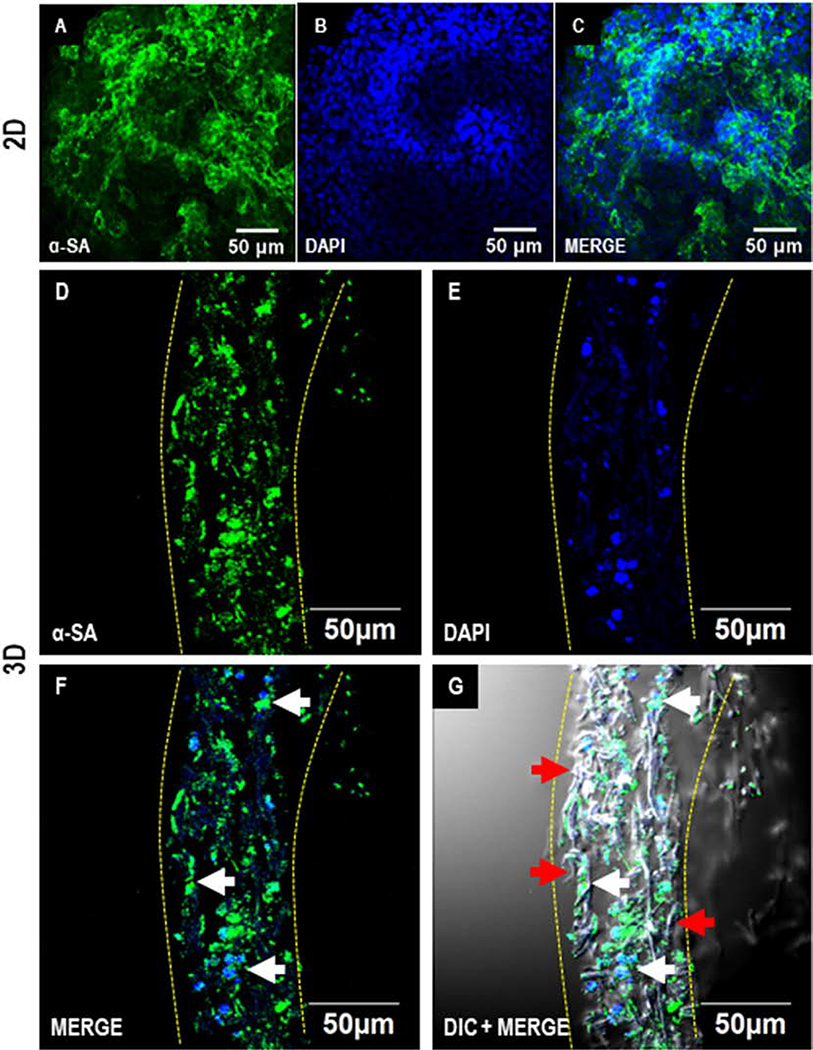
Following the cardiac differentiation of hiPSCs, the expression of cardiac lineage marker NKX-2.5 was observed in the differentiated cells in both 2D and 3D cultures by D7 (Fig. 5). NKX2.5 is a hallmark transcription marker in cardiac progenitor cells [33]. Furthermore, on D28, the expression of cardiomyocyte marker sarcomeric alpha-actinin was observed in a majority of the differentiated cells in 2D as well as 3D cultures (Fig. 6). Initially, D7 cross sections of the 3D differentiated cultures demonstrated migrations of the cells to only a few microns (Fig. 5 D-G). Remarkably, by D28 the cells had migrated uniformly throughout the depth of the scaffold (Fig. 6 D-G). These observations indicated a gradual migration of differentiating hiPSCs into the scaffold. Taken together, these results demonstrate that cardiomyocytes differentiated from hiPSCs in situ on 3D scaffolds expressed stage-specific markers associated with differentiation of cardiomyocytes and also were distributed uniformly in the patch.
Figure 5.
Immunofluorescence analysis of cardiac differentiation of hiPSCs in 2D and 3D cultures on D7. Confocal images showing the expression of cardiac progenitor marker NKX-2.5 on D7 during cardiac differentiation of hPSCs in 2D (A-C) and 3D (D-G). Scale: A-C: 50 μm; D-G: 250 μm; inset figures (D-F): 20 μm. Dotted yellow lines represent the coaxial scaffold.
Figure 6.
Immunofluorescence analysis of cardiac differentiation of hiPSCs in 2D and 3D cultures on D28. Confocal images showing the expression of cardiomyocyte marker sarcomeric alpha-actinin (α-SA) in hiPSCs differentiated in 2D (A-C) and 3D (D-G). Scale: 50 μm. Dotted yellow lines represent the coaxial scaffold. Red and white arrows indicate fibers and cells, respectively.
3.6. Assessment of gene expression during cardiac differentiation in 2D and 3D cultures
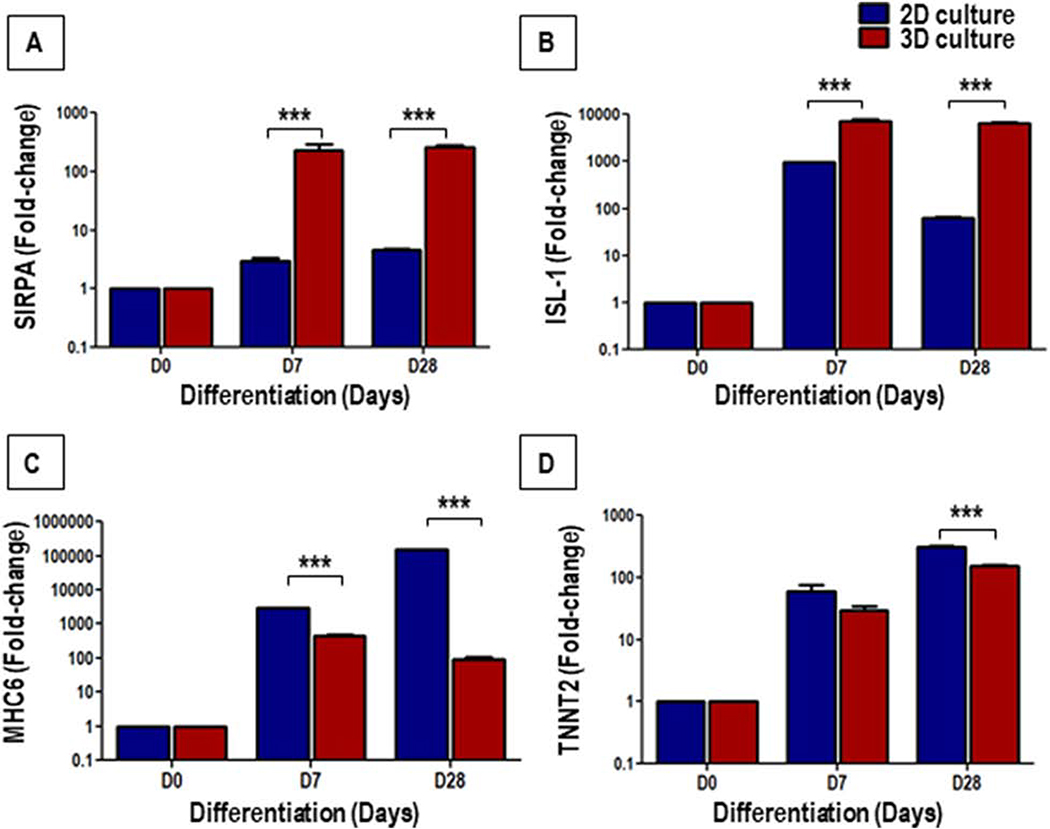
Gene expression analysis by qRT-PCR showed a significant up-regulation in the expression of Cardiac Progenitor (CP)-associated genes, SIRPA and ISL-1, and cardiomyocyte-associated genes, MHC6 and TNNT2, on D7 and D28, in both 2D and 3D cultures (Fig. 7). However, compared to 2D cultures, the CP-associated genes, SIRPA (Fig. 7A) and ISL-1 (Fig. 7B) were significantly up-regulated in the 3D cultures on both D7 and D28, as compared to their 2D counterparts. On the other hand, the expression of the cardiomyocyte-associated genes, MHC6 (Fig. 7C) and TNNT2 (Fig. 7D) was significantly up-regulated in the 2D cultures when compared to the 3D cultures on D28. These data demonstrate an enhanced differentiation of hiPSCs to cardiac progenitors in 3D cultures, when compared to their 2D counterparts. On the other hand, the data clearly showed a significantly higher expression of cardiomyocyte-specific genes in 2D cultures compared to 3D cultures.
Figure 7.
Gene expression analysis of cardiac progenitors (CP) and cardiomyocytes markers during cardiac differentiation of hiPSCs in 2D and 3D cultures. The bar graph shows the fold-change in expression of the CP-associated genes, SIRPA (A) and ISL1 (B), and cardiomyocyte-associated genes, MHC6 (C) and TNNT2 (D), in 2D and 3D cultures on D0, D7 and D28 of cardiac differentiation relative to D0 (undifferentiated hiPSCs) expression. Values are represented as mean ± SEM, n = 3; ***p < 0.001; D represent days.
4. Discussion
This study demonstrated the effect of coaxial PCL-gelatin nanofibrous scaffolds on the growth and differentiation of hiPSCs to functional cardiomyocytes in vitro. To the best of our knowledge, this study is the first to demonstrate the differentiation of hiPSCs to functional cardiomyocytes in 3D cultures using coaxial nanofibrous scaffolds. While no significant differences were observed in culturing undifferentiated hiPSCs in 2D and 3D cultures, a delay in the appearance of functional cardiomyocytes was observed in the latter. On the other hand, while 3D cultures showed increased expression of cardiac progenitor-associated genes, 2D cultures showed increased expression of cardiomyocyte-associated genes. Furthermore, a gradual migration into the scaffold and homogenous distribution of the differentiated hiPSC-CMs was observed in 3D cultures.
Differentiation of hiPSCs to functional cardiomyocytes in 2D cultures has been well-studied [36]. While 2D differentiation of hiPSCs is efficient, numerous studies have reported that these culture systems do not mimic in vivo differentiation [37–39]. Several parameters may contribute to this, including the 2D culture surface stiffness (which ranges in GPa, while the tissue tensile strength ranges around kPa) and lack of 3D interaction between neighboring cells[40, 41]. Recent studies showed a comparative assessment for differentiation of stem cells into cardiomyocytes [42, 43] and other cell types [37, 44] in 2D and 3D cultures, demonstrated that 3D culture systems more closely resembled the in vivo mechanism. Additionally, culturing cardiomyocytes in a 3D culture, showed improved contractile function, increased mitochondrial numbers, and improved maturation and functionality [39, 42, 43]. Although, different 3D culture systems have been developed for culturing hiPSC-CMs [41, 43], an efficient 3D culture system to obtain reproducible differentiation of hiPSCs to functional cardiomyocytes is still lacking.
In this study, we have successfully developed a random coaxial scaffold comprised of nanofibers with a PCL core and gelatin shell for the culture and cardiac differentiation of hiPSCs. The use of electrospun PCL-based scaffolds for cardiac applications has previously been well-established, mainly due to the superior mechanical properties of PCL, including tensile strength and stiffness [22, 45]. Additionally, fibrous scaffolds made from PCL have been used for the differentiation of hiPSCs to neural cells as well as pancreatic β-cells [19, 20]. However, reports have suggested a negative effect of PCL fibers on cell viability and function on account of its hydrophobicity and gradual degradation resulting in the formation of toxic acidic by-products, like caproic acid [20, 46]. On the other hand, when PCL was combined with natural polymers like gelatin or collagen, the scaffolds have been shown to have improved biocompatibility [47]. A core-shell structure, wherein PCL is enclosed within a gelatin shell has been shown to exhibit biocompatibility similar to gelatin scaffolds, without significant changes to the mechanical properties of the PCL scaffold [27]. In agreement with these previous reports, we observed no significant effect on cell viability following the culture of hiPSCs in the PCL-gelatin coaxial scaffolds indicated by similar LDH profiles in our 2D and 3D cultures.
In terms of morphology, we did not observe any striking differences in the morphology of the hiPSCs differentiated in 2D or 3D cultures. Consistent with our observations, a previous study demonstrated that mouse embryonic stem cells cultured on PCL nanofibrous scaffolds also showed no significant differences in morphology of cells cultured in 2D or 3D cultures [22]. While we observed cardiac differentiation of hiPSCs in 2D cultures as previously reported [33], we also observed spontaneously contracting ‘cardiac patches’ in culture following differentiation of hiPSCs cultured on 3D scaffolds. Interestingly, we have observed visible contractions at a significantly earlier time (D8), when iPSCs were differentiated to functional cardiomyocytes in 2D cultures as compared to 3D cultures. However, no significant differences were observed in the beating frequency of cardiomyocytes four weeks (D28) after induction of differentiation between the two culture systems.
We have observed a delay in the appearance of macroscopic contractions in hiPSCs differentiated on 3D scaffolds (D21), when compared to 2D cultures (D10). Contrary to our findings, a recent study reported the appearance of macroscopic contractions by D14, following differentiation of hiPSCs to cardiomyocytes on polyvinyl alcohol (PVA) scaffolds fabricated by gas foaming [48]. The delayed detection of contractions on our 3D scaffolds could be due to a few reasons including the ability to visualize contractions. The cells cultured on the scaffold in this study could not be visualized under phase-contrast light microscopy, thus the precise time of the initiation of the beating of the cardiomyocytes could not be determined. In contrast, small clusters of cardiomyocytes showing weak contractions could easily be identified in 2D cultures as early as D8 (data not shown). It is also plausible that the force of contraction generated by the cardiomyocytes at D14 was not strong enough to translate into macroscopic contractions on the 3D scaffolds. It has been well-established that the force of contraction generated by cardiomyocytes increases with their maturation [49, 50] and duration in culture [35]. Hence, the delay in appearance of contractions on our 3D scaffold may be indicative of an immature state of the cardiomyocytes differentiated from hiPSCs on 3D scaffolds or relative stiffness of the coaxial scaffold compared to the normal extracellular matrix.
When compared to corresponding 2D cultures, the 3D cultures had a significantly higher expression of CP-associated genes, while the cardiomyocyte-associated genes were expressed (on both D7 and D28) at significantly lower levels indicating efficient differentiation of hiPSCs to CPs on 3D scaffolds, but not to cardiomyocytes. Our observation is consistent with another study comparing the differentiation of hiPSC to cardiomyocytes in 2D and 3D cultures [42]. In this study, however, the decreased expression of cardiomyocyte-associated genes during later days was attributed to the differentiation of hiPSCs to other cardiac cell types like endothelial cells, in addition to cardiomyocytes. Furthermore, appearance of other differentiated cell types like fibroblasts, in addition to cardiomyocytes during cardiac differentiation of hiPSCs has been observed by other groups [33] and also by our group (data not shown). Since we did not employ any cardiomyocyte enrichment strategies in our differentiation protocol, it is possible that the lower expression of cardiomyocyte-associated genes is due to the presence of other differentiated cell types in 3D cultures and not necessarily due to the lack of cardiomyocyte maturation.
On the other hand, contradictory to our observations, a few reports have shown an up-regulation of cardiac genes following the differentiation of mouse iPSCs into cardiomyocytes [22] or human embryonic stem cell-derived CPs [51], cultured on PCL/fibrin-based scaffolds. Importantly, recent studies have shown improved differentiation, maturation, and contractile gene expression following the culture of hiPSC-derived cardiomyocytes in 3D cultures [34, 52, 53], as compared to 2D cultures. These studies have implicated several factors in the enhanced maturation of the cardiomyocytes in 3D cultures, such as (a) matrix composition, (b) fiber diameter and alignment, (c) matrix stiffness, (d) density of cells seeded onto the scaffold, (e) cells used for co-culture, and (f) electrical or mechanical stimulation [34, 50, 54–56]. However, a comprehensive understanding of how these factors affect the differentiation of hiPSCs to functional cardiomyocytes is currently lacking. We, therefore, believe that, while differentiation of hiPSCs to functional cardiomyocytes on 3D coaxial scaffolds has been demonstrated in the present study, in the future, it is imperative to decipher the effect of these extracellular factors on the differentiation of hiPSCs to functional cardiomyocytes on scaffolds to improve the efficiency of 3D culture systems.
Another intriguing observation found in our study was the migration and distribution of hiPS cells throughout the depth of the scaffold following cardiomyocyte differentiation on the 3D scaffolds. The hiPSC-CMs were able to migrate without the use of inducing factors; this has not been reported previously [57]. Also, per our expectations, a gradual migration and uniform distribution of the differentiated cells in the scaffold was observed during in situ cardiac differentiation since the pore size of the coaxial scaffold used in our study was greater than hiPSCs (~10–20 μm). Also, previous studies have demonstrated that during differentiation, hiPSCs undergo epithelial-to-mesenchymal transition followed by mesenchymal to epithelial transition in a stage-specific manner [58]. Using in vitro and in vivo models, a higher migration potential of mesenchymal cells as compared to the epithelial cells has been well-established [59]. Hence, it is possible that in situ differentiation of hiPSCs to functional cardiomyocytes enabled the cells to migrate into the scaffold during the process of epithelial-to-mesenchymal transition (EMT), which occurs during the process of differentiation. However, deciphering the molecular mechanism underlying this observation is beyond the scope of the present study.
Conclusions
Overall, our study demonstrated that coaxial PCL-gelatin nanofibrous scaffolds can be used as a 3D platform for the culture and differentiation of hiPSCs to functional cardiomyocytes. Furthermore, efficient migration and uniform distribution of differentiated cells was observed on 3D scaffolds. In summary, the results from our study could be applied towards developing future strategies to differentiate stem cells in a 3D microenvironment and for tissue-engineering approaches for mending broken hearts.
Supplementary Material
Highlights.
hiPSCs can be cultured on an electrospun coaxial PCL-gelatin nanofibrous scaffolds
hiPSCs can be differentiated into functional cardiomyocytes on 3D scaffolds
In situ differentiation enables migration and uniform distribution of cells into the scaffold
Acknowledgments
This work was supported by the National Institute of Health HL136232 (MK) and OSU start-up funds to MK. We thank the Stanford Cardiovascular Institute (SCVI) Biobank for providing hiPSCs. We would also like to thank the Ohio State University Campus Microscopy Imaging Facility for helping with SEM and confocal microscopy studies. These facilities are supported in part by grant P30 CA016058, National Cancer Institute, Bethesda, MD, USA.
Footnotes
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Das SR, Delling FN, Djousse L, Elkind MSV, Ferguson JF, Fornage M, Jordan LC, Khan SS, Kissela BM, Knutson KL, Kwan TW, Lackland DT, Lewis TT, Lichtman JH, Longenecker CT, Loop MS, Lutsey PL, Martin SS, Matsushita K, Moran AE, Mussolino ME, O’Flaherty M, Pandey A, Perak AM, Rosamond WD, Roth GA, Sampson UKA, Satou GM, Schroeder EB, Shah SH, Spartano NL, Stokes A, Tirschwell DL, Tsao CW, Turakhia MP, VanWagner LB, Wilkins JT, Wong SS, Virani SS, E. American Heart Association Council on, C. Prevention Statistics, S. Stroke Statistics, Heart Disease and Stroke Statistics-2019 Update: A Report From the American Heart Association, Circulation 139(10) (2019) e56–e528. [DOI] [PubMed] [Google Scholar]
- [2].Duelen R, Sampaolesi M, Stem Cell Technology in Cardiac Regeneration: A Pluripotent Stem Cell Promise, EBioMedicine 16 (2017) 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Guo X, Bai Y, Zhang L, Zhang B, Zagidullin N, Carvalho K, Du Z, Cai B, Cardiomyocyte differentiation of mesenchymal stem cells from bone marrow: new regulators and its implications, Stem Cell Res Ther 9(1) (2018) 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Musunuru K, Sheikh F, Gupta RM, Houser SR, Maher KO, Milan DJ, Terzic A, Wu JC, G. American Heart Association Council on Functional, B. Translational, Y. Council on Cardiovascular Disease in the, C. Council on, N. Stroke, Induced Pluripotent Stem Cells for Cardiovascular Disease Modeling and Precision Medicine: A Scientific Statement From the American Heart Association, Circ Genom Precis Med 11(1) (2018) e000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhu Z, Huangfu D, Human pluripotent stem cells: an emerging model in developmental biology, Development 140(4) (2013) 705–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kussauer S, David R, Lemcke H, hiPSCs Derived Cardiac Cells for Drug and Toxicity Screening and Disease Modeling: What Micro- Electrode-Array Analyses Can Tell Us, Cells 8(11) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ebert AD, Liang P, Wu JC, Induced pluripotent stem cells as a disease modeling and drug screening platform, J Cardiovasc Pharmacol 60(4) (2012) 408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sayed N, Liu C, Wu JC, Translation of Human-Induced Pluripotent Stem Cells: From Clinical Trial in a Dish to Precision Medicine, J Am Coll Cardiol 67(18) (2016) 2161–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kopljar I, Lu HR, Van Ammel K, Otava M, Tekle F, Teisman A, Gallacher DJ, Development of a Human iPSC Cardiomyocyte-Based Scoring System for Cardiac Hazard Identification in Early Drug Safety De-risking, Stem Cell Reports 11(6) (2018) 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Huang CY, Liu CL, Ting CY, Chiu YT, Cheng YC, Nicholson MW, Hsieh PCH, Human iPSC banking: barriers and opportunities, J Biomed Sci 26(1) (2019) 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Park M, Yoon YS, Cardiac Regeneration with Human Pluripotent Stem Cell-Derived Cardiomyocytes, Korean Circ J 48(11) (2018) 974–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mummery CL, Zhang J, Ng ES, Elliott DA, Elefanty AG, Kamp TJ, Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview, Circ Res 111(3) (2012) 344–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Karakikes I, Ameen M, Termglinchan V, Wu JC, Human induced pluripotent stem cell-derived cardiomyocytes: insights into molecular, cellular, and functional phenotypes, Circ Res 117(1) (2015) 80–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Goversen B, van der Heyden MAG, van Veen TAB, de Boer TP, The immature electrophysiological phenotype of iPSC-CMs still hampers in vitro drug screening: Special focus on IK1, Pharmacol Ther 183 (2018) 127–136. [DOI] [PubMed] [Google Scholar]
- [15].Tu C, Chao BS, Wu JC, Strategies for Improving the Maturity of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes, Circ Res 123(5) (2018) 512–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Machiraju P, Greenway SC, Current methods for the maturation of induced pluripotent stem cell-derived cardiomyocytes, World J Stem Cells 11(1) (2019) 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu J, Nie H, Xu Z, Niu X, Guo S, Yin J, Guo F, Li G, Wang Y, Zhang C, The effect of 3D nanofibrous scaffolds on the chondrogenesis of induced pluripotent stem cells and their application in restoration of cartilage defects, PLoS One 9(11) (2014) e111566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hoveizi E, Khodadadi S, Tavakol S, Karima O, Nasiri-Khalili MA, Small molecules differentiate definitive endoderm from human induced pluripotent stem cells on PCL scaffold, Appl Biochem Biotechnol 173(7) (2014) 1727–36. [DOI] [PubMed] [Google Scholar]
- [19].Abazari MF, Soleimanifar F, Nouri Aleagha M, Torabinejad S, Nasiri N, Khamisipour G, Amini Mahabadi J, Mahboudi H, Enderami SE, Saburi E, Hashemi J, Kehtari M, PCL/PVA nanofibrous scaffold improve insulin-producing cells generation from human induced pluripotent stem cells, Gene 671 (2018) 50–57. [DOI] [PubMed] [Google Scholar]
- [20].KarbalaeiMahdi A, Shahrousvand M, Javadi HR, Ghollasi M, Norouz F, Kamali M, Salimi A, Neural differentiation of human induced pluripotent stem cells on polycaprolactone/gelatin bi-electrospun nanofibers, Mater Sci Eng C Mater Biol Appl 78 (2017) 1195–1202. [DOI] [PubMed] [Google Scholar]
- [21].Safaeijavan R, Soleimani M, Divsalar A, Eidi A, Ardeshirylajimi A, Comparison of random and aligned PCL nanofibrous electrospun scaffolds on cardiomyocyte differentiation of human adipose-derived stem cells, Iran J Basic Med Sci 17(11) (2014) 903–11. [PMC free article] [PubMed] [Google Scholar]
- [22].Chen Y, Zeng D, Ding L, Li XL, Liu XT, Li WJ, Wei T, Yan S, Xie JH, Wei L, Zheng QS, Three-dimensional poly-(epsilon-caprolactone) nanofibrous scaffolds directly promote the cardiomyocyte differentiation of murine-induced pluripotent stem cells through Wnt/beta-catenin signaling, BMC Cell Biol 16 (2015) 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Qasim M, Arunkumar P, Powell HM, Khan M, Current research trends and challenges in tissue engineering for mending broken hearts, Life Sci 229 (2019) 233–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mondal D, Griffith M, Venkatraman SS, Polycaprolactone-based biomaterials for tissue engineering and drug delivery: Current scenario and challenges, International Journal of Polymeric Materials and Polymeric Biomaterials 65(5) (2016) 255–265. [Google Scholar]
- [25].Woodruff MA, Hutmacher DW, The return of a forgotten polymer—Polycaprolactone in the 21st century, Progress in Polymer Science 35(10) (2010) 1217–1256. [Google Scholar]
- [26].Wong DY, Hollister SJ, Krebsbach PH, Nosrat C, Poly(epsilon-caprolactone) and poly (L-lactic-co-glycolic acid) degradable polymer sponges attenuate astrocyte response and lesion growth in acute traumatic brain injury, Tissue Eng 13(10) (2007) 2515–23. [DOI] [PubMed] [Google Scholar]
- [27].Blackstone BN, Drexler JW, Powell HM, Tunable engineered skin mechanics via coaxial electrospun fiber core diameter, Tissue Eng Part A 20(19–20) (2014) 2746–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang L, Wu S, Cao G, Fan Y, Dunne N, Li X, Biomechanical studies on biomaterial degradation and co-cultured cells: mechanisms, potential applications, challenges and prospects, J Mater Chem B 7(47) (2019) 7439–7459. [DOI] [PubMed] [Google Scholar]
- [29].Goyal R, Guvendiren M, Freeman O, Mao Y, Kohn J, Optimization of Polymer-ECM Composite Scaffolds for Tissue Engineering: Effect of Cells and Culture Conditions on Polymeric Nanofiber Mats, J Funct Biomater 8(1) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Blackstone BN, Hahn JM, McFarland KL, DeBruler DM, Supp DM, Powell HM, Inflammatory response and biomechanical properties of coaxial scaffolds for engineered skin in vitro and post-grafting, Acta biomaterialia 80 (2018) 247–257. [DOI] [PubMed] [Google Scholar]
- [31].Drexler JW, Powell HM, Regulation of electrospun scaffold stiffness via coaxial core diameter, Acta biomaterialia 7(3) (2011) 1133–9. [DOI] [PubMed] [Google Scholar]
- [32].Lin Y, Linask KL, Mallon B, Johnson K, Klein M, Beers J, Xie W, Du Y, Liu C, Lai Y, Zou J, Haigney M, Yang H, Rao M, Chen G, Heparin Promotes Cardiac Differentiation of Human Pluripotent Stem Cells in Chemically Defined Albumin-Free Medium, Enabling Consistent Manufacture of Cardiomyocytes, Stem Cells Transl Med 6(2) (2017) 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu Q, Van Bortle K, Zhang Y, Zhao M-T, Zhang JZ, Geller BS, Gruber JJ, Jiang C, Wu JC, Snyder MP, Disruption of mesoderm formation during cardiac differentiation due to developmental exposure to 13-cis-retinoic acid, Scientific Reports 8(1) (2018) 12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Khan M, Xu Y, Hua S, Johnson J, Belevych A, Janssen PM, Gyorke S, Guan J, Angelos MG, Evaluation of Changes in Morphology and Function of Human Induced Pluripotent Stem Cell Derived Cardiomyocytes (HiPSC-CMs) Cultured on an Aligned-Nanofiber Cardiac Patch, PLoS One 10(5) (2015) e0126338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kumar N, Dougherty JA, Manring HR, Elmadbouh I, Mergaye M, Czirok A, Greta Isai D, Belevych AE, Yu L, Janssen PML, Fadda P, Gyorke S, Ackermann MA, Angelos MG, Khan M, Assessment of temporal functional changes and miRNA profiling of human iPSC-derived cardiomyocytes, Sci Rep 9(1) (2019) 13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mummery CL, Zhang J, Ng ES, Elliott DA, Elefanty AG, Kamp TJ, Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview, Circ Res 111(3) (2012) 344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Centeno EGZ, Cimarosti H, Bithell A, 2D versus 3D human induced pluripotent stem cell-derived cultures for neurodegenerative disease modelling, Mol Neurodegener 13(1) (2018) 27–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Duval K, Grover H, Han L-H, Mou Y, Pegoraro AF, Fredberg J, Chen Z, Modeling Physiological Events in 2D vs. 3D Cell Culture, Physiology (Bethesda) 32(4) (2017) 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [39].Pontes Soares C, Midlej V, de Oliveira MEW, Benchimol M, Costa ML, Mermelstein C, 2D and 3D-organized cardiac cells shows differences in cellular morphology, adhesion junctions, presence of myofibrils and protein expression, PLoS One 7(5) (2012) e38147–e38147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Moles MD, Scotchford CA, Ritchie AC, Development of an elastic cell culture substrate for a novel uniaxial tensile strain bioreactor, Journal of biomedical materials research. Part A 102(7) (2014) 2356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Edmondson R, Broglie JJ, Adcock AF, Yang L, Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors, Assay Drug Dev Technol 12(4) (2014) 207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Branco MA, Cotovio JP, Rodrigues CAV, Vaz SH, Fernandes TG, Moreira LM, Cabral JMS, Diogo MM, Transcriptomic analysis of 3D Cardiac Differentiation of Human Induced Pluripotent Stem Cells Reveals Faster Cardiomyocyte Maturation Compared to 2D Culture, Scientific Reports 9(1) (2019) 9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zuppinger C, 3D Cardiac Cell Culture: A Critical Review of Current Technologies and Applications, Frontiers in Cardiovascular Medicine 6(87) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Meier F, Freyer N, Brzeszczynska J, Knöspel F, Armstrong L, Lako M, Greuel S, Damm G, Ludwig-Schwellinger E, Deschl U, Ross JA, Beilmann M, Zeilinger K, Hepatic differentiation of human iPSCs in different 3D models: A comparative study, Int J Mol Med 40(6) (2017) 1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Guex AG, Frobert A, Valentin J, Fortunato G, Hegemann D, Cook S, Carrel TP, Tevaearai HT, Giraud MN, Plasma-functionalized electrospun matrix for biograft development and cardiac function stabilization, Acta biomaterialia 10(7) (2014) 2996–3006. [DOI] [PubMed] [Google Scholar]
- [46].S. V.S, M. P.V, Degradation of Poly(ε-caprolactone) and bio-interactions with mouse bone marrow mesenchymal stem cells, Colloids and Surfaces B: Biointerfaces 163 (2018) 107–118. [DOI] [PubMed] [Google Scholar]
- [47].Hu Y, Feng B, Zhang W, Yan C, Yao Q, Shao C, Yu F, Li F, Fu Y, Electrospun gelatin/PCL and collagen/PCL scaffolds for modulating responses of bone marrow endothelial progenitor cells, Exp Ther Med 17(5) (2019) 3717–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Dattola E, Parrotta EI, Scalise S, Perozziello G, Limongi T, Candeloro P, Coluccio ML, Maletta C, Bruno L, De Angelis MT, Santamaria G, Mollace V, Lamanna E, Di Fabrizio E, Cuda G, Development of 3D PVA scaffolds for cardiac tissue engineering and cell screening applications, RSC Advances 9(8) (2019) 4246–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bedada FB, Wheelwright M, Metzger JM, Maturation status of sarcomere structure and function in human iPSC-derived cardiac myocytes, Biochim Biophys Acta 1863(7 Pt B) (2016) 1829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ribeiro AJ, Ang YS, Fu JD, Rivas RN, Mohamed TM, Higgs GC, Srivastava D, Pruitt BL, Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness, Proc Natl Acad Sci U S A 112(41) (2015) 12705–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhang D, Shadrin IY, Lam J, Xian HQ, Snodgrass HR, Bursac N, Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes, Biomaterials 34(23) (2013) 5813–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jabbour R, Owen T, Reinsch M, Pandey P, Terracciano C, Weinberger F, Eschenhagen T, Harding S, BS27 Development and preclinical testing of a large heart muscle patch, Heart 105(Suppl 6) (2019) A157. [Google Scholar]
- [53].Li J, Minami I, Shiozaki M, Yu L, Yajima S, Miyagawa S, Shiba Y, Morone N, Fukushima S, Yoshioka M, Li S, Qiao J, Li X, Wang L, Kotera H, Nakatsuji N, Sawa Y, Chen Y, Liu L, Human Pluripotent Stem Cell-Derived Cardiac Tissue-like Constructs for Repairing the Infarcted Myocardium, Stem Cell Reports 9(5) (2017) 1546–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Besser RR, Ishahak M, Mayo V, Carbonero D, Claure I, Agarwal A, Engineered Microenvironments for Maturation of Stem Cell Derived Cardiac Myocytes, Theranostics 8(1) (2018) 124–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tallawi M, Rai R, Boccaccini AR, Aifantis KE, Effect of substrate mechanics on cardiomyocyte maturation and growth, Tissue Eng Part B Rev 21(1) (2015) 157–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, Pabon L, Reinecke H, Murry CE, Growth of engineered human myocardium with mechanical loading and vascular coculture, Circ Res 109(1) (2011) 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Moyes KW, Sip CG, Obenza W, Yang E, Horst C, Welikson RE, Hauschka SD, Folch A, Laflamme MA, Human embryonic stem cell-derived cardiomyocytes migrate in response to gradients of fibronectin and Wnt5a, Stem Cells Dev 22(16) (2013) 2315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li X, Pei D, Zheng H, Transitions between epithelial and mesenchymal states during cell fate conversions, Protein Cell 5(8) (2014) 580–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].von Gise A, Pu WT, Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease, Circ Res 110(12) (2012) 1628–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.