Highlights
-
•
Approximately 650,000 people will be diagnosed this year with cancers of the oral cavity and pharynx worldwide.
-
•
The absence of biomarkers for the disease early detection contributes to the late diagnosis.
-
•
Despite some advances with regards to treatment, overall survival has not significantly improved in decades.
-
•
We have shown that increased relative mRNA expression of KLK5 to LEKTI is associated with disease’s poor outcome.
-
•
This work supports the relative expression of KLK5 to LEKTI as a valuable prognostic marker.
Keywords: OSCC, Kallikrein 5, LEKTIb, SPINK5, Prognostic marker
Abbreviations: BrdU, 5-Bromo-2-Deoxyuridine; DMEM, Dulbecco's Modified Eagle's Medium; DAPI, (4′,6-diamidino-2-phenylindole); FBS, Fetal Bovine Serum; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; GDC, Genomic Data Commons; HNSCC, Head and Neck Squamous Cell Carcinoma; HNSCCs, Head and Neck Squamous Cell Carcinomas; HRP, Horseradish peroxidase; H&E, Hematoxylin and Eosin; hrKLK5, Human Recombinant Kallikrein 5; IgG, Immunoglobulin G; KLKs, Kallikreins; KLK5, Kallikrein 5; KLK7, Kallikrein 7; LEKTI, Lympho-Epithelial Kazal-Type related Inhibitor; Mrna, messenger RNA; O.M., human Oral Mucosa; OSSC, Oral Squamous Cell Carcinoma; PAR2, Protease-activated receptor 2 (PAR2); P.D.C., Poorly Differentiated Carcinoma; P.L., Premalignant Lesions; RT., Room Temperature; SPINK5, Serine Peptidase Inhibitor Kazal type 5; W.D.C., Well Differentiated Carcinoma
Abstract
Background
Oral squamous cell carcinoma (OSCC) remains a challenging cancer to treat despite all the advances of the last 50 years. Kallikrein 5 (KLK5) is among the serine proteases implicated in OSCC development. However, whether the activity of KLK5 promotes carcinogenesis is still controversial. Moreover, knowledge regarding the role of the KLK5 cognate inhibitor, Lympho-Epithelial Kazal-Type related Inhibitor (LEKTI), in OSCC is scarce. We have, thus, sought to investigate the importance of KLK5 and LEKTI expression in premalignant and malignant lesions of the oral cavity.
Methods
KLK5 and LEKTI protein expression was evaluated in 301 human samples, which were comprised of non-malignant and malignant lesions of the oral cavity. Moreover, a bioinformatic analysis of the overall survival rate from 517 head and neck squamous cell carcinoma (HNSCC) samples was performed. Additionally, to mimic the uncovered KLK5 to serine peptidase inhibitor (SPINK5) imbalance, the KLK5 gene was abrogated in an OSCC cell line using CRISPR-Cas9 technology. The generated cell line was then used for in vivo and in vitro carcinogenesis related experiments.
Results
LEKTI was found to be statistically downregulated in OSCCs, with increased KLK5/SPINK5 mRNA ratio being associated with a shorter overall survival (p = 0.091). Indeed, disruption of KLK5 to SPINK5 balance through the generation of KLK5 null OSCC cells led to smaller xenografted tumors and statistically decreased proliferation rates following multiple time points of BrdU treatment in vitro.
Conclusion
The association of increased enzyme/inhibitor ratio with poor prognosis indicates KLK5 to SPINK5 relative expression as an important prognostic marker in OSCC.
Introduction
Oral Squamous cell carcinoma (OSCC) accounts for more than 95% of all oral cancers and is characterized by a delayed clinical detection, poor prognosis, and a modest 5-year survival rate (1). Therefore, it is of upmost importance to investigate novel molecular mechanisms and markers for its diagnosis and prognosis.
Kallikrein-related peptidases (KLKs) constitute a large family of secreted serine proteases that are expressed in multiple tissues (2). KLKs are involved in many tightly regulated physiological functions and their altered expression or regulation has been consistently implicated in wide range of cancers ([3], [4]–5). Interaction with endogenous protease inhibitors is one of the key mechanisms that control KLKs’ proteolytic activity (6,7).
KLK5 plays a crucial role in skin desquamation through the proteolytic processing of corneodesmosomal proteins (8,9). This process is tightly regulated via direct inhibition of KLK5 by Lympho-Epithelial Kazal Type-related Inhibitor (LEKTI) (8). LEKTI is a large protein encoded by the Serine Peptidase Inhibitor Kazal type 5 (SPINK5) gene and is comprised of 15 Kazal-type serine protease inhibitory domains (10). Altered KLK5 expression, which includes inconsistencies regarding its increase or decrease, has been previously reported in head and neck squamous cell carcinomas (HNSCCs) ([11], [12], [13], [14]–15). Moreover, KLK5 and SPINK5 are part of a gene signature found to predict lymph node metastasis in HNSCCs ([16], [17]–18).
In this study, we have investigated whether the relative expression of KLK5 and LEKTI, in malignant lesions of the oral cavity, might be useful for developing near future prognostic markers for this disease. Indeed, using in vivo and in vitro approaches, we have shown that an increase in KLK5 expression relative to SPINK5 expression elicited signaling mechanisms leading to disease aggressiveness.
Material and methods
Human tissue samples
The retrospective cohort study included formalin-fixed and paraffin-embedded samples of human oral mucosa (O.M., non-neoplastic oral samples diagnosed with fibrous hyperplasia, n = 50), premalignant lesions (P.L., oral leukoplakia, n = 50) and OSCCs (well differentiated OSCC, W.D.C., n = 122; and poorly differentiated OSCC, P.D.C., n = 79), retrieved from the archives of the Division of Oral and Maxillofacial Pathology, Department of Stomatology, School of Dentistry, University of Sao Paulo. The samples were collected between 2010 and 2018. Specimen inclusion criteria: 29–96 years old patients, regardless of gender or ethnicity. Exclusion criteria: lip and/or oropharyngeal lesions. Dr. Braz-Silva and Dr. Sales performed a blinded histopathological validation of the samples included in the study.
Cells
The HNSCC cell lines Cal 27, UD-SCC-2, HN6, and HN30 were a kind gift from Dr. J. Silvio Gutkind (University California San Diego, San Diego, CA, USA). Cal 27 and HN6 are derived from human tongue squamous cell carcinomas (19,20). HN30 is derived from a pharyngeal squamous cell carcinoma (20). UD-SCC-2 is derived from a hypopharyngeal squamous cell carcinoma (21). Cells were cultured in Dulbecco's modified Eagle's Medium (DMEM) (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS) (Merck, St. Louis, MO, USA), 0.434 mg/mL glutamine, and an antibiotic-antimycotic reagent (Thermo Fisher Scientific).
Antibodies and recombinant protein
The following primary antibodies were used in this study: mouse anti-SPINK5 (3 µg/ml – Clone HPA009067, Merck); rabbit anti-Kallikrein 5 (3 µg/ml – Cat#: ab28565 Abcam Plc, Cambridge, UK) and anti-BrdU antibody (Clone #5292 – 0.48 µg/mL, Cell Signaling Technology, Danvers, MA, USA). Recombinant Domain 6 of LEKTI inhibitor was generated as previously described (8).
LEKTI and KLK5 immunohistochemistry
Six μm sections were cut from each specimen and mounted on glass slides. Tissue sections were deparaffinized in xylene, rehydrated and stained either with hematoxylin and eosin (H&E) or immunostained for LEKTI or KLK5. Briefly, the samples were subjected to antigen retrieval by boiling in 0.01 M sodium citrate buffer, pH6.0, for 12 min, and the endogenous peroxidase activity was quenched by incubating the tissues in 3% hydrogen peroxide in methanol for 20 min at room temperature (RT). The sections were blocked for 1 h at RT with 2% bovine serum albumin in PBS and incubated overnight at 4 °C with the primary antibodies. Bound antibodies were visualized using appropriate biotin-conjugated secondary antibodies and the Vectastain ABC Elite kit (Vector Laboratories, Burlingame, CA, USA), before incubation in 3,3′-diaminobenzidine (25 mg/mL, Merck) and H2O2. Subsequently, the sections were counterstained with hematoxylin, dehydrated, cleared in xylene and coverslips mounted with Permount (Thermo Fisher Scientific). Negative controls were carried out using non-immune anti-rabbit IgG (3 µg/ml, Cat#: 011–000–003, Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA) for LEKTI and for KLK5. Images were acquired using an Olympus VS120 slide scanner (Olympus Corporation, Tokyo, Japan). Image J software (22) was used to quantify the total epithelial area and the stained epithelial area of each sample. Briefly, for each slide stained for LEKTI or KLK5, ratios between the total epithelial area (μm²) and immunolabeled epithelial area (μm²) were calculated and plotted as a percentage (%).
Bioinformatic analysis of TCGA samples
The Head-Neck Squamous Cell Carcinoma (TCGA-HNSCC) dataset for Cancer Genomics (23) was analyzed for mRNA expression levels of SPINK5 and KLK5 genes and clinical features. Data were downloaded from Genomic Data Commons (GDC) on June 2020. Overall survival analysis of 517 HNSCC samples was done using Kaplan-Meier, logrank and Cox regression. Samples were classified as low/high SPINK5 and/or KLK5 if gene expression was in the bottom 25% or the top 25% of HNSCC samples, respectively. In addition, a dataset of 249 OSCCs was analyzed for mRNA expression levels of SPINK5 and KLK5 genes (W.D.C. samples comprise low and intermediate histological grades, respectively, G1 and G2, n = 201; and P.D.C. comprises high histological grades, G3 and G4, n = 48).
KLK5 and SPINK5 mRNA expression analysis in HNSCC cell lines
HNSCC cell lines were plated in 6 well plates and cultivated overnight in DMEM containing 10% FBS. Total RNA was purified from 1 × 106 cells using Trizol reagent (Thermo Fisher Scientific), as recommended by the manufacturer. Reverse transcription and PCR amplification (RT-PCR) were performed using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific), as recommended by the manufacturer (Merck) using gene-specific primers for human KLK5 and SPINK5 genes. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control gene. Amplicons were analyzed by agarose gel electrophoresis. Primer sequences were as follows:
KLK5 Forward: 5′- GTGGGTGCTCTGTGCTCTGA - 3′;
KLK5 Reverse: 5′- TCCTGGTTGCTCCCAGAGG - 3′;
SPINK5 Forward: 5′- TGTGTGCCAGTGTGTTCAAA - 3′;
SPINK5 Reverse: 5′- GGAGTCGTCCATTCCTCACA - 3′;
GAPDH Forward: 5′- AGATCCCTCCAAAATCAAGTGG - 3′;
GAPDH Reverse: 5′- GGCAGAGATGATGACCCTTTT - 3′;
Generation of KLK5 knockout cells
To disrupt the KLK5 genes in Cal 27 cells, two 20-nt target DNA sequences preceding a 5′-NGG PAM sequence at signal peptide region within exon 1 in the genomic KLK5 locus (NC_000019.10) were selected for generating single-guide RNA (sgRNA) for SpCas9 targets using the CRISPR design website http://crispr.mit.edu/ (24). The two target sequences are as follows:
Guide 1: 5′-AGCAAGACCCCCCTGGATGT-3′
Guide 2: 5′-TGGCTACAGCAAGACCCCCC-3′
To express SpGuides in the targeted cells, sense and antisense oligos of the two target sequences were annealed and cloned into the LentiCRISPR v2 vector by BsmB1 (New England Biolabs, Boston, MA, USA), respectively. The lentiCRISPR v2 vector (25) was purchased from Addgene (Cat. 52961; Cambridge, MA, USA). All clones were confirmed by DNA sequencing using the following primer: 5′-GGACTATCATATGCTTACCG-3′, from the sequence of U6 promoter that drives expression of sgRNAs. Both synthesis of primers and oligos and sequencing of PCR products and clones were performed at the Food and Drug Administration - Center for Biologics Evaluation and Research (CBER) Facility for Biotechnology Resources at National Institutes of Health (Bethesda, MD, USA).
The generated LentiCRISPR constructs were transfected together with the packaging plasmid psPAX2 (Addgene: 12260), and the envelope plasmid pMD2.G (Addgene: 12259) into HEK293T cells using lipofectamine 3000 as directed by the manufacturer (Thermo Fisher Scientific). After 24 h, the supernatant containing viral particles was used to transduce Cal 27 cells. After transduction, puromycin-selected clones were screened by PCR amplification of the targeted genomic region followed by Sanger DNA sequencing for analysis of mutations. Mutants were selected based on occurrence of mutations that generated a frameshift or major disruption of the signal peptide region. In the selected clones, expression of KLK5 was determined by Western blot analyses.
Cell proliferation analysis – BrdU incorporation assay
Wild-type or KLK5 KO Cal 27 cells were plated at 5 × 104 cells per well in 24 well plates and cultivated overnight in DMEM containing 10% FBS. Cells were then incubated with 30 µg/mL of BrdU (5-Bromo-2-Deoxyuridine; Merck) at 37 °C for 1, 2, and 3 h. Subsequently, the cells were fixed with ice cold 70% ethanol for 5 min, washed with PBS for 5 min before the addition of 1.5 M HCl and incubated for 30 min at RT. Cells were then washed with PBS and blocked (5 µg/mL Normal Donkey IgG – Jackson Immunoresearch Laboratories Inc. -in PBS plus 1% BSA) for 1 hour at RT before incubation with anti-BrdU antibody (Clone #5292 – 0.48 µg/mL, Cell Signaling Technology, Danvers, MA, USA) overnight at 4 °C. After incubation, cells were rinsed thoroughly in PBS and incubated for 45 min at RT with donkey anti-mouse IgG F(ab)′2-Alexa 488 antibody diluted in PBS (2 µg/mL, Thermo Fisher Scientific). Cells were then rinsed in PBS and mounted with DAPI (4′,6-diamidino-2-phenylindole) Fluoromount-G (Electron Microscopy Sciences, Hatfield, PA). Cells incubated without primary antibody served as controls, and they were all negative. All samples were analyzed using a DMI 6000B fluorescence microscope (Leica Microsystems; Heidelberg, Germany). Five representative images were acquired from each sample. Image J software was used to obtain the percentage (%) of cells stained with BrdU out of the total number of cells stained with DAPI (22). The percentage of BrdU positive cells was used as a measure of proliferation.
Scratch assay
Wild-type or KLK5 KO Cal 27 cells were plated at 1 × 106 cells per well in 6 well plates and cultivated for 16 h in DMEM containing 10% FBS. During this period, Cal 27 WT cells were treated or not with recombinant LEKTI Domain 6 (1.2 µM), a KLK5 cognate inhibitor, in order to mimic KLK5 KO Cal 27 cells. A 10 µL plastic tip was used to scratch the cells in the middle of each well. Images were acquired at 0, 24, 48 and 72 h after scratching (time 0) using an inverted microscope coupled with a camera (Leica CTR 6000). Images were analyzed using the FIJI image processing package (26), where the area between the edges of the risk were measured every time the images were acquired. The wound closure area was calculated using the following the formula: Wound Closure Area = Area [T0] – Area [Tn]
Tumor xenograft
Males and females’ Swiss Nude mice were obtained from the Transgenic Mice Animal Facility of the Ribeirao Preto Medical School, University of São Paulo. All the experiments were approved by the Ethics Committee for Animals Usage (Process #: 127/2019). Four-week-old Swiss Nude mice were injected in both flanks with 2 × 106 cells, in serum free DMEM, per site. The left flank was injected with WT Cal 27 cells, while the right flank was injected with KLK5 KO Cal 27 cells. Mice were periodically inspected and 40 days after injection the size of the resulting tumor was measured every 3 days. Tumor width and length were measured using a caliper and volume values were calculated using the following the formula (27):
Mice were euthanized, using overdose of chemical anesthetics, about 11 weeks after transplant and tumors were removed for histological analysis. Tumor lesion ≥ 20 mm was considered as the endpoint for the experiment.
Statistical analysis
Results were analyzed using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA) or R (R Project 4.0). In the analysis of LEKTI and KLK5 protein expression, statistical differences were assessed by Kruskal Wallis test followed by Dunn's Multiple Comparison test. In the analysis of LEKTI and KLK5 mRNA expression statistical differences were assessed by Mann–Whitney U test. Kaplan–Meier, logrank test, and the multivariate Cox regression model was used in overall survival analysis comparing enzyme/inhibitor, each individual gene, tumor stage, gender and race. SPINK5 and KLK5 expression, TNM staging, gender, and race were used in the cox regression model. The analysis of KLK5 protein expression in low LEKTI and high LEKTI samples was assessed by two-way ANOVA. Chi-Square test was used to assess differences in patients harboring low LEKTI and high LEKTI protein expression. In the analysis of wound closure two-way ANOVA with Tukey's Multiple Comparisons tests were used. Unpaired t-test was used to assess differences in BrdU incorporation between Cal 27 WT and Cal 27 KO cells. Two tail paired t-test was used to assess differences in tumor weight between Cal 27 WT and Cal 27 KO cells.
Results
LEKTI expression is decreased in OSCCs
To investigate the expression of LEKTI and KLK5, we have performed immunohistochemical analysis of 301 human oral samples of which 201 were OSCCs. The results showed that LEKTI and KLK5 (Fig. 1, respectively black and red arrows) are expressed in the outermost layer of the epithelia in O.M. (oral mucosa), in which there is absence of dysplastic cells in the basal layer of the epithelia (Fig. 1B and 1 C; O.M.) and in P.L. (premalignant epithelial lesions of the oral cavity) (Fig. 1E and 1 F; oral dysplasia, P.L.). These premalignant lesions have dysplastic cells (atypical keratinocytes) in the bottom third of the epithelia that comprises the basal and spinous cell layers (P.L., black arrowheads show atypical keratinocytes). In OSCC samples, LEKTI and KLK5 are expressed by differentiated cells in W.D.C. (well differentiated carcinomas), which are polygonal and cytoplasm-abundant cells surrounding the keratin pearls (Fig. 1H and 1 I; W.D.C, asterisk shows keratin pearls), while in P.D.C. (poorly differentiated carcinomas), which show increased numbers of spindle-like cells, atypical mitosis and nuclear pleomorphism, expression is confined to islands or isolated polygonal-like cells (Fig. 1K and 1 L; P.D.C, yellow arrowhead indicates nuclear pleomorphism while blue arrowhead indicates atypical mitosis). Human oral mucosa biopsies (Supplementary Fig. 1 A–D), as well as murine skin biopsies from LEKTI-null and WT mice (Supplementary Fig. 1 E–H), were incubated with anti-LEKTI antibody (Supplementary Fig. 1 A, F and H), anti-KLK5 antibody (Supplementary Fig. 1 C) and non-immune IgG (negative controls, Supplementary Fig. 1 B and D).
Fig. 1.
KLK5 expression decreases in OSCCs bearing higher levels of LEKTI
A, D, G and J) Representative H&E staining of oral mucosa (A, O.M., n = 50), premalignant hyperkeratotic lesion (D, P.L., n = 50), well-differentiated OSCC (G, W.D.C., n = 122), and poorly differentiated OSCC (J, P.D.C., n = 79), bar: 25 µm. B, E, H and K) Representative IHC staining of LEKTI in O.M. (B), P.L. (E), W.D.C. (H), and P.D.C. (K), bar: 25 µm. C, F, I and L) Representative IHC staining of KLK5 in O.M. (C), P.L. (F), W.D.C. (I), and P.D.C. (L), bar: 25 µm. Black arrows indicate LEKTI-immunostained areas; Red arrows indicate KLK5-immunostained areas; Black arrowheads indicate dysplastic cells; Yellow arrowheads indicate nuclear polymorphism; Blue arrowheads indicate atypical mitosis; Asterisks indicate keratin pearls. M) Quantification of LEKTI expression in each experimental group,% of immunostained area. O.M., circles; P.L., squares; W.D.C., triangles; P.D.C., inverted triangles. Kruskal Wallis test followed by Dunn's Multiple Comparison test; p<0.0005[***]. N) Quantification of KLK5 expression in each experimental group,% of immunostained area. O.M., circles; P.L., squares; W.D.C., triangles; P.D.C., inverted triangles. Kruskal Wallis test followed by Dunn's Multiple Comparison test; p<0.05[*]; p<0.0005[***]. O) KLK5 protein expression in O.M (n = 50, circles), P.L. (n = 50, triangles) and O.S.C.C.s. samples (n = 201, diamonds) harboring low LEKTI (up to 1% of O.M. immunostaining, black circles, black triangles and black diamonds) and high LEKTI expression (>1% of O.M. immunostaining, gray circles, gray triangles and gray diamonds). Statistical analysis: ANOVA; p<0.05 [*]; p<0.005 [**].
Quantitative analysis of the immunohistochemistry data (Fig. 1M) showed that LEKTI is abundantly expressed in oral mucosa (O.M.; circles) and in premalignant lesions (P.L.; squares). Interestingly, its expression is severely downregulated in well differentiated carcinomas (W.D.C.; triangles) and it is barely detectable in poorly differentiated carcinomas (P.D.C.; inverted triangles). However, there is no statistical difference between LEKTI expression in W.D.C and P.D.C. Likewise, KLK5 expression is reduced in OSCCs, but no significant difference was found between W.D.C.s and P.D.C.s. (compare triangles with inverted triangles).
To further investigate modulation of KLK5 and LEKTI protein levels, we evaluated KLK5 expression in samples (O.M., P.L. and OSCCs) with low and high LEKTI expression. The results showed that overall, in non-neoplastic (O.M.) and in pre-neoplastic (P.M.L.) tissues, the amount of KLK5 protein increase with the increase in LEKTI expression (Fig. 1O, compare black circles with gray circles, and black triangles with gray triangles). However, in malignancy, samples that express higher levels of LEKTI do not show the same increase in KLK5 expression (Fig. 1O, compare black lozenges with gray lozenges).
Table 1 displays clinical characteristics of the included OSCC patients according to either high or low LEKTI expression.
Table 1.
Clinical characteristics of included OSCC patients.
| Clinical Information | High Lekti | Low Lekti | ||||||
|---|---|---|---|---|---|---|---|---|
| Total Cases | 41 | 142 | ||||||
| % | % | p.value | q.value | |||||
| Gender | 0,88 | 1,00 | ||||||
| Female | 31,71 | 34,51 | ||||||
| Male | 68,29 | 65,49 | ||||||
| Age | 0,01 | 0,05 | ||||||
| 0–59 | 19,51 | 44,37 | ||||||
| ≥60 | 70,73 | 45,07 | ||||||
| No Information | 9,76 | 10,56 | ||||||
| Ethnicity | 0,06 | 0,28 | ||||||
| Caucasian origin | 65,85 | 71,13 | ||||||
| African american origin | 12,20 | 19,72 | ||||||
| Asian origin | 0,00 | 1,41 | ||||||
| No information | 21,95 | 7,75 | ||||||
| Smoking status | 0,98 | 1,00 | ||||||
| Smokers | 41,46 | 40,85 | ||||||
| Non-smokers | 29,27 | 32,39 | ||||||
| Ex-smokers | 2,44 | 2,11 | ||||||
| No information | 26,83 | 24,65 | ||||||
| Tumor site | 0,04 | 0,21 | ||||||
| Tongue | 19,51 | 33,10 | ||||||
| Palate | 9,76 | 5,63 | ||||||
| Maxilla | 2,44 | 1,41 | ||||||
| Mandibule (including retromolar trigone) | 29,27 | 9,15 | ||||||
| Floor of mouth | 2,44 | 16,20 | ||||||
| Lip | 2,44 | 4,23 | ||||||
| Gum | 0,00 | 2,11 | ||||||
| Alveolar border | 17,07 | 13,38 | ||||||
| Mucosa Jugal | 7,32 | 6,34 | ||||||
| Multiplos sites | 2,44 | 0,70 | ||||||
| Other | 4,88 | 2,82 | ||||||
| No information | 2,44 | 4,93 | ||||||
n = 182 due to a loss to follow-up.
Increased relative mRNA expression of KLK5 to LEKTI is associated with poor outcome
To investigate whether alterations in protein expression were accompanied by changes in mRNA levels, a bioinformatic analysis comparing mRNA levels of SPINK5 (Fig. 2A), KLK5 (Fig. 2B) and the relative expression of KLK5 to LEKTI (Fig. 2C) on TCGA data were carried out. Samples included a dataset of 517 HNSCCs, in which the OSCC type is included, from the HNSCC-TCGA comprised of: (i) low and intermediate histological grades, respectively, G1 and G2 (W.D.C., n = 388, red bars); and (ii) high histological grades, respectively, G3 and G4 (P.D.C., n = 129, blue bars). Accordingly, SPINK5 and KLK5 mRNA levels were decreased in P.D.C. samples, when compared to W.D.C. specimens (Fig. 2A and B). Interestingly, increased relative KLK5 to LEKTI mRNA levels were found in P.D.C. samples (Fig. 2C). Moreover, in order to assure that the results obtained with the HNSCC dataset represent the OSCC patients as well, 249 OSCCs were analyzed for SPINK5 and KLK5 mRNA levels (Supplementary Fig. 2 A and B; W.D.C. samples comprises low and intermediate histological grades, respectively, G1 and G2, n = 201; and P.D.C. comprises high histological grades, G3 and G4, n = 48).
Fig. 2.
SPINK5, KLK5 and KLK5 to SPINK5 mRNA levels decrease in high-grade HNSCCs.
A–C) Bioinformatic analysis comparing mRNA levels of SPINK5 (A), KLK5 (B) and KLK5 to SPINK5 (C) in a dataset of 517 OSCCs from the HNSCC-TCGA (red bars, W.D.C. comprises low and intermediate histological grades, respectively, G1 and G2; blue bars, P.D.C., comprises high histological grades G3 and G4; Mann–Whitney U test, p = 6 × 10−5 for SPINK5 mRNA levels, p = 2 × 10−8 for KLK5 mRNA levels and p = 0.002 for KLK5/SPINK5 mRNA levels).
Since LEKTI and KLK5 protein and mRNA expression were altered in both OSCC, it was of interest to investigate whether the relationship between KLK5 and LEKTI expression has any impact on overall survival. Therefore, a bioinformatic analysis of 517 HNSCC samples was carried out using the HNSC-TCGA database. The analysis showed that high (Fig. 3A, SPINK.cat=High) SPINK5 mRNA levels, which corresponds to higher 25% of mRNA expression for SPINK5, was associated with increased overall survival. On the other hand, the absolute increase and/or decrease in the levels of KLK5 mRNA was not statistically correlated with overall survival (Supplementary Fig. 2). Importantly, the high relative expression of enzyme to inhibitor, which corresponds to the higher 25% (Fig. 3B; KLK5/SPINK5.cat=High) of KLK5 to LEKTI mRNA expression, was associated with decreased overall survival using Kaplan-Meier and Multivariate Cox analysis (Fig. 3B and Table 2).
Fig. 3.
Higher relative KLK5 to SPINK5 (KLK5/SPINK5) mRNA expression is correlated with decreased overall survival in HNSCCs.
A and B) Bioinformatic analysis comparing overall survival of 517 HNSCC samples. Low (SPINK5.cat=Low) and high (SPINK.cat=High) SPINK5 mRNA levels correspond to, respectively, the lower 25% and higher 25% of mRNA expression for SPINK5 (A). Similarly, low and high relative expression of enzyme to inhibitor correspond to, respectively, the lower 25% (KLK5/SPINK5.cat=Low) and higher 25% (KLK5/SPINK5.cat=High) KLK5 to SPINK5 mRNA levels (B). Statistical analysis: Kaplan-Meier logrank test.
Table 2.
Results of multivariate Cox regression model.
| Variable tested | coef | exp(coef) | s.e.(coef) | z-value | p-value |
|---|---|---|---|---|---|
| SPINK5 | −3.072e-06 | 1.000e+00 | 5.588e-06 | −0.550 | 0.58242 |
| KLK5 | −9.371e-07 | 1.000e+00 | 8.637e-06 | −0.108 | 0.91360 |
| KLK5.SPINK5 | 3.590e-03 | 1.004e+00 | 1.235e-03 | 2.907 | 0.00365 |
| ajcc_pathologic_stageStage II | 6.766e-01 | 1.967e+00 | 4.797e-01 | 1.410 | 0.15842 |
| ajcc_pathologic_stageStage III | 8.207e-01 | 2.272e+00 | 4.816e-01 | 1.704 | 0.08835 |
| ajcc_pathologic_stageStage IVA | 1.192e+00 | 3.292e+00 | 4.593e-01 | 2.594 | 0.00948 |
| ajcc_pathologic_stageStage IVB | 1.713e+00 | 5.544e+00 | 5.949e-01 | 2.879 | 0.00399 |
| ajcc_pathologic_stageStage IVC | 3.037e+00 | 2.084e+01 | 1.146e+00 | 2.650 | 0.00805 |
| sex male | −3.285e-01 | 7.200e-01 | 1.502e-01 | −2.187 | 0.02873 |
| race asian | 4.980e-01 | 1.645e+00 | 1.108e+00 | 0.450 | 0.65301 |
| race black or african american | 4.224e-01 | 1.526e+00 | 1.032e+00 | 0.410 | 0.68217 |
| race not reported | 4.093e-01 | 1.506e+00 | 1.080e+00 | 0.379 | 0.70469 |
| race white | 1.664e-01 | 1.181e+00 | 1.009e+00 | 0.165 | 0.86907 |
Supplementary Fig. 3 shows overall survival and ratio of KLK5/SPINK5 mRNA levels according to disease staging (Supplementary Fig. 3 A and B, respectively). In order to rule out tumor stage or other characteristics effect on KLK5/SPINK5 mRNA association with overall survival in TCGA data, we used a Cox regression model with KLK5/SPINK5 mRNA expression, each individual gene, tumor stage, gender and race. The higher KLK5/SPINK5 mRNA was associated with the worst survival, independently of other variables (Table 2), while SPINK5 association with survival is not significant after correcting for the ratio. In addition, Supplementary Table 1 shows HNSC-TCGA full clinical characteristics comparing clinical features of 25% top and 25% bottom expressors for SPINK5 (23). Altogether, our analysis suggests that KLK5 and SPINK5 interaction is more important than each gene alone on HNSCC outcome.
KLK5/LEKTI imbalance leads to increased tumor growth in vivo
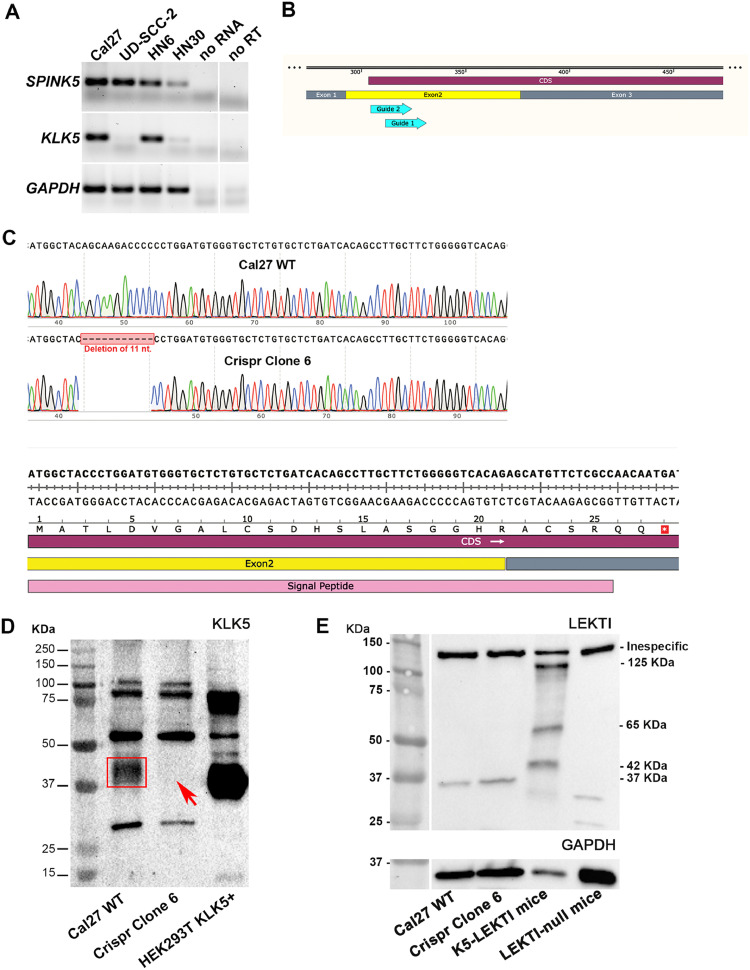
Because LEKTI is known to directly inhibit KLK5 (8,28), we used CRISPR-Cas9 technology to knockout KLK5 and thus disturb the KLK5/LEKTI cellular balance. To select a HNSCC cell line for these assays we tested a panel of four HNSCC cell lines (Cal 27, UD-SCC-2, HN6 and HN30) for the expression of LEKTI and KLK5. Both Cal 27 and HN6 cells lines were positive for the expression of both LEKTI and KLK5 (Fig. 4A). The Cal 27 cell line was shown to express both LEKTI and KLK5 mRNAs and was amenable to KLK5 CRISPR knockout and clone selection (Fig. 4B–D). In addition, LEKTI expression does not alter following KLK5 ablation in Cal27 cell line (Clone 6, 37 KDa, Fig. 4E). Wild-type (KLK5 WT) or knockout (KLK5 KO) Cal 27 cells were then used for in vitro and in vivo experiments.
Fig. 4.
Generation of CRISPR-driven KLK5 knockout OSCC cells
A) RT-PCR analysis of SPINK5 and KLK5 mRNA expression in HNSCC cell lines. Lanes 1–4 are Cal 27, UD-SCC-2, HN6 and HN30, respectively. No RNA was added in lane 5, and reverse transcriptase was omitted from the reactions in lane 6. B) Schematic diagram for the KLK5-specific guide RNA design. Blue arrows show guide RNAs 1 and 2 that target KLK5 Exon 2 (yellow box). Coding sequence is shown in purple and exons one and two are shown in gray and yellow boxes. C) Sequence analysis of viable KLK5 KO OSCC cell line (clone 6) exon 2 amplicon showing 11 nucleotide deletion (upper panel, red boxed sequence) and a premature stop codon (lower panel, red-boxed white asterisk). D) KLK5 western blot shows KLK5 protein band (red rectangle, lane 2) and the absence of KLK5 protein (red arrow, lane 3) in clone 6 lysate supernatant. Molecular weight standards and positive control using HEK293 cells overexpressing KLK5 are shown in lanes one and four, respectively. E) LEKTI western blot shows the 37-KDa LEKTI fragment in both KLK5 WT and KLK5 KO OSCC cell line (Clone 6) in vitro. Murine skin from mice overexpressing LEKTI in the epidermis (K5-LEKTI transgenic mouse) and murine skin from LEKTI-null mice were used, respectively, as positive and negative controls (8).
To access migration and proliferation rates in vitro, scratch wound healing assay (Fig. 5A–L) was used. KLK5 WT (Fig. 5A, D, G and J), KLK5 KO (Fig. 5B, E, H and K) and rLEKTI-treated KLK5 WT (Fig. 5C, F, I and L) confluent cell monolayers were scratched and wound closure was evaluated for 72 h. Wound closure of both KLK5 KO and rLEKTI-treated KLK5 WT OSCC cells was partially inhibited at 48 h and 72 h following scratch (Fig. 5M). KLK5 deficiency also impaired cell proliferation as shown by decreased BrdU incorporation in KLK5 KO cells following 1 h, 2 h and 3 h BrdU treatment (Fig. 5N).
Fig. 5.
LEKTI expressing KLK5 KO OSCC cells display decreased malignant characteristics.
A–L) Representative images showing that KLK5 KO OSCC cells partially inhibit wound closure (dashed white lines, wound edges). Inhibition of KLK5 with recombinant LEKTI treatment leads to a similar inhibition (dashed white lines, compare C, F, I and L with A, D, G and J) in wild-type cells. M) Quantification of the wound closure shows a delay, in KLK5 KO (red columns) and rLEKTI-treated KLK5 WT (gray columns) OSCC cells, when compared to the KLK5 WT OSCC cells (black columns), after 48 h and 72 h. Statistical analysis: two-way ANOVA and Tukey's Multiple Comparisons tests (p<0.02 = [*] and p<0.0001 = [***]). N) Quantification of BrdU positive cells using immunofluorescence shows that KLK5 KO OSCC cells (red column) have a decreased proliferation rate when comparing with the KLK5 WT OSCC cells (black column) following 1 h, 2 h and 3 h BrdU incubation. Statistical analysis: two-way ANOVA and Sidak's Multiple Comparisons tests, p = 0,0254 [*]; p = 0.0002 [***]; p<0,0001 [****]. O and P) KLK5 WT (black dashed circle) and KLK5 KO (red dashed circle) cells were xenografted, respectively, into the left and right flanks of nude mice (O, n = 8) and tumor growth was monitored every 3 days, starting at 40 days post injection, for 41 days. KLK5 WT (P, black) and KLK5 KO (P, red) tumors were followed for approximately 11 weeks when the endpoint was reached, and the mice were euthanized. Q and R) Images of dissected wild-type (Q, black asterisks) and KLK5 KO (Q, red asterisks) tumors. Necropsied tumors were weighed and plotted (R, KLK5 WT tumor, black dots; KLK5 KO tumor red dots). Statistical analysis: two-tailed paired t-test, p<0.005 [**]. S–W) Representative images of KLK5 WT and KLK5 KO tumors following anti-KLK5 and anti-LEKTI immunohistochemistry. As expected, KLK5 immunostaining is present only in the WT tumors (black arrows) LEKTI staining was more abundant in KLK5 WT tumors, however, it is present in both wild-type (T, black arrows) and KLK5 KO (W, black arrow) tumors. Negative controls were incubated in the absence of primary antibodies using KLK5 WT (U) and KLK5 KO (X) tumors.
Next, KLK5 WT and KLK5 KO OSCC cells were xenografted into athymic mice to investigate whether the impaired wound closure and decreased proliferation rate would have an impact on tumor progression in vivo (Fig. 5O; KLK5 WT cells were subcutaneously injected in the left flank and area of tumor is illustrated with black dotted circle; KLK5 KO cells were injected in the right flank and area of tumor is illustrated with red dotted circle). Tumors were followed for approximately 8 weeks and the absence of KLK5 inhibited xenograft tumor growth of human OSCC cells (Fig. 5P; compare black and red lines, showing KLK5 WT and KLK5 KO tumor volumes, respectively). Indeed, tumor dissection following euthanasia showed a significant difference in the size (Fig. 5Q; compare black and red asterisks, representing KLK5 WT and KLK5 KO tumors, respectively) and the weight (Fig. 5R; compare black and red circles, showing KLK5 WT and KLK5 KO tumor weight, respectively) from KLK5 WT and KLK5 KO derived tumors. Xenografted tumors were then fixed and processed for routine histology. KLK5 immunohistochemistry staining, as expected, was negative for KLK5 KO derived tumors (Fig. 5S and V, respectively, KLK5 WT and KLK5 KO tumors; black arrows show KLK5 staining). Interestingly, different from KLK5 KO cells cultured in petri dishes, LEKTI staining was severely decreased in KLK5 KO derived tumors, yet it was still detectable (Fig. 5T and W, respectively, KLK5 WT and KLK5 KO tumors; black arrows show LEKTI staining).
Discussion
In the western world, oral squamous cell carcinoma remains as one of the most important life-threatening diseases (29). Indeed, low 5-year survival rates is associated with currently available methods for treatment and diagnosis of the OSCCs (30). Therefore, the aim of this study was to evaluate the expression of KLK5 and LEKTI in malignant lesions of the oral cavity in order to test the hypothesis that the relative expression of the enzyme to its inhibitor (KLK5 to LEKTI) represents a promising tool to be used as a prognostic marker in OSCCs and thus contribute to a near future personalized precision medicine. To this aim, we have shown here that the relative expression of KLK5 to LEKTI in OSCC is associated with decreased overall survival. Moreover, disturbance of KLK5/LEKTI balance through the abrogation of KLK5 expression leads to decreased tumor growth in vivo.
We have found that the decrease in the absolute expression of LEKTI is associated with malignancy progression and decreased overall survival in OSCCs. Our results, therefore, are in agreement with previous studies where SPINK5 genetic expression levels were found to be downregulated in head and neck squamous cell carcinoma (HNSCCs) [13–15]. However, different reports diverge with regards to the increase, or decrease, in the KLK5 expression levels in OSCCs ([11], [12]–13). In the present study, we observed a decrease in KLK5 absolute expression between non-malignant and malignant oral tissues at protein levels alone. Of relevance, protease to protease inhibitor balance has been reported as marker for malignant disorders (31,32). Importantly, in this study, bioinformatic analysis of 475 OSCC samples showed that increased relative expression of the enzyme to its inhibitor (KLK5 to LEKTI) is, indeed, associated with a decrease in overall survival. Moreover, in carcinoma samples expressing high LEKTI levels (higher than 1% of the O.M. LEKTI expression), KLK5 expression is decreased when compared to non-malignant samples that also express higher levels of LEKTI. Alternatively, malignant and/or non-malignant samples that express lower LEKTI levels do not show a modulation in KLK5 expression. These results might explain contradictory findings regarding KLK5 expression in OSCC when it is evaluated independently of LEKTI (11–13). In addition, differences in KLK5 expression might also be explained by the cohort constitution in the different studies, where sub-site lesion origin and histopathological differentiation may vary. Divergences in the absolute quantitation of kallikreins (KLKs) emphasize the advantage of evaluating the expression of the protease relative to its inhibitor as a better indication of the “free protease” in the tissue/lesion. In fact, our in-silico analysis of 517 HNSCC samples, revealed that overall survival was associated with increased expression of KLK5 relative to LEKTI.
Hypothesizing that the relationship between KLK5 and LEKTI could be key to understanding the divergences in KLK5 expression levels in OSCCs, the KLK5 to LEKTI balance in an OSCC cell line expressing both KLK5 and LEKTI was disrupted by using CRISPR-Cas9 technology to knock-out KLK5 gene expression. Of importance, KLK5 ablation alone did not modulated LEKTI expression in OSCC cell line. We found that KLK5 KO OSCC cells exhibit impaired growth/migration in a wound healing assay and decreased proliferation. Disruption of KLK5 and LEKTI proteolytic balance, which have an established role in epithelial desquamation (8,9), could contribute to enhanced cell-cell junctional integrity explaining the observed reduced migration of KLK5 deficient and rLEKTI treated cells (33). Although increased KLK5 expression has been associated with several cancers, studies on its impact on cell proliferation are scarce. Nonetheless, knockdown of KLK5 in PC9 lung adenocarcinoma cells also cause inhibition of cellular proliferation (34). Moreover, our results showing impaired growth/migration and proliferation in KLK5 KO OSCC cells corroborate observations that LEKTI can inhibit proliferation and migration of esophageal cancer cells (35). Indeed, KLK5 KO derived tumor showed decreased expression of LEKTI and, therefore, corroborate the findings in which KLK5 deficiency and LEKTI overexpression are associated with decreased and increased tumor progression in other xenograft models (12,35).
Taken together our data supports the relative expression of KLK5 to LEKTI as a valuable prognostic marker since the increased enzyme (KLK5) to inhibitor (LEKTI) expression in human patients and the KLK5 to LEKTI unbalance in genetically modified OSCC cell line are associated with a poor prognosis and aggravated tumor progression. Therefore, this study contributes to the development of biomarkers for this disease.
Additional information
The study presented herein was approved by the Ethics Committee of the Hospital das Clinicas of Ribeirao Preto Medical School – University of Sao Paulo (HCFMRP) under the protocol number 50533515.6.0000.5440. Given the Brazilian legislations, as the work was done with paraffin-embedded non-identified samples, there was no need for consent forms. Additionally, the study was performed in accordance with the Declaration of Helsinki.
CRediT authorship contribution statement
Márcia Gaião Alves: Investigation, Formal analysis. Márcio Hideki Kodama: Investigation, Formal analysis. Elaine Zayas Marcelino da Silva: Investigation, Formal analysis, Writing - original draft. Bruno Belmonte Martinelli Gomes: Investigation, Formal analysis. Rodrigo Alberto Alves da Silva: Investigation. Gabriel Viliod Vieira: Investigation, Formal analysis. Vani Maria Alves: Investigation. Carol Kobori da Fonseca: Formal analysis. Ana Carolina Santana: Formal analysis. Nerry Tatiana Cecílio: Investigation. Mara Silvia Alexandre Costa: Investigation. Maria Célia Jamur: Writing - review & editing. Constance Oliver: Writing - review & editing. Thiago Mattar Cunha: Conceptualization, Resources. Thomas H. Bugge: Conceptualization, Supervision, Writing - review & editing. Paulo Henrique Braz-Silva: Conceptualization, Resources, Supervision. Leandro M. Colli: Conceptualization, Methodology, Formal analysis, Investigation. Katiuchia Uzzun Sales: Conceptualization, Methodology, Writing - review & editing, Supervision, Formal analysis, Investigation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Funding
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (São Paulo Research Foundation; FAPESP, 2014/06316-2) and Fundação de Apoio ao Ensino, Pesquisa e Assistência do Hospital das Clínicas da Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo (FAEPA) research grants to KUS. EZMS - post-doctoral fellowship FAPESP #: 2016/13228-8. MGA, MHK, GVV, RAS - master's fellowship from CAPES (Brazilian Ministry of Education). BBG - master's fellowship FAPESP#: 2016/16715-7 and ACS - PhD's fellowship CNPq#: 166166/2014. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
The authors are grateful to Elisa Santos, from the Division of Oral and Maxillofacial Pathology, Department of Stomatology, School of Dentistry University of Sao Paulo, Sao Paulo, SP; and to Jose Augusto Maulin, from the Department of Cell and Molecular Biology and Pathogenic Bioagents, FMRP-USP, Ribeirao Preto, SP; for excellent technical assistance. This work was supported by the Intramural Research Program at the National Institute of Dental and Craniofacial Research, NIH.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100970.
Appendix. Supplementary materials
References
- 1.Chi A.C., Day T.A., Neville B.W. Oral cavity and oropharyngeal squamous cell carcinoma–an update. CA Cancer J. Clin. 2015;65(5):401–421. doi: 10.3322/caac.21293. [DOI] [PubMed] [Google Scholar]
- 2.Shaw J.L., Diamandis E.P. Distribution of 15 human kallikreins in tissues and biological fluids. Clin. Chem. 2007;53(8):1423–1432. doi: 10.1373/clinchem.2007.088104. [DOI] [PubMed] [Google Scholar]
- 3.Kryza T., Silva M.L., Loessner D., Heuzé-Vourc'h N., Clements J.A. The kallikrein-related peptidase family: dysregulation and functions during cancer progression. Biochimie. 2016;122:283–299. doi: 10.1016/j.biochi.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Filippou P.S., Karagiannis G.S., Musrap N., Diamandis E.P. Kallikrein-related peptidases (KLKs) and the hallmarks of cancer. Crit. Rev. Clin. Lab. Sci. 2016;53(4):277–291. doi: 10.3109/10408363.2016.1154643. [DOI] [PubMed] [Google Scholar]
- 5.Sotiropoulou G., Pampalakis G., Diamandis E.P. Functional roles of human kallikrein-related peptidases. J. Biol. Chem. 2009;284(48):32989–32994. doi: 10.1074/jbc.R109.027946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goettig P., Magdolen V., Brandstetter H. Natural and synthetic inhibitors of kallikrein-related peptidases (KLKs) Biochimie. 2010;92(11):1546–1567. doi: 10.1016/j.biochi.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalinska M., Meyer-Hoffert U., Kantyka T., Potempa J. Kallikreins – the melting pot of activity and function. Biochimie. 2016;122:270–282. doi: 10.1016/j.biochi.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deraison C., Bonnart C., Lopez F., Besson C., Robinson R., Jayakumar A. LEKTI fragments specifically inhibit KLK5, KLK7, and KLK14 and control desquamation through a pH-dependent interaction. Mol. Biol. Cell. 2007;18(9):3607–3619. doi: 10.1091/mbc.e07-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGovern J.A., Meinert C., de Veer S.J., Hollier B.G., Parker T.J., Upton Z. Attenuated kallikrein-related peptidase activity disrupts desquamation and leads to stratum corneum thickening in human skin equivalent models. Br. J. Dermatol. 2017;176(1):145–158. doi: 10.1111/bjd.14879. [DOI] [PubMed] [Google Scholar]
- 10.Mägert H.J., Ständker L., Kreutzmann P., Zucht H.D., Reinecke M., Sommerhoff C.P. LEKTI, a novel 15-domain type of human serine proteinase inhibitor. J. Biol. Chem. 1999;274(31):21499–21502. doi: 10.1074/jbc.274.31.21499. [DOI] [PubMed] [Google Scholar]
- 11.Pettus J.R., Johnson J.J., Shi Z., Davis J.W., Koblinski J., Ghosh S. Multiple kallikrein (KLK 5, 7, 8, and 10) expression in squamous cell carcinoma of the oral cavity. Histol. Histopathol. 2009;24(2):197–207. doi: 10.14670/hh-24.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson J.J., Miller D.L., Jiang R., Liu Y., Shi Z., Tarwater L. Protease-activated receptor-2 (PAR-2)-mediated Nf-κB activation suppresses inflammation-associated tumor suppressor microRNAS in oral squamous cell carcinoma. J. Biol. Chem. 2016;291(13):6936–6945. doi: 10.1074/jbc.M115.692640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leusink F.K., van Diest P.J., Frank M.H., Broekhuizen R., Braunius W., van Hooff S.R. The co-expression of kallikrein 5 and kallikrein 7 associates with poor survival in non-HPV oral squamous-cell carcinoma. Pathobiology. 2015;82(2):58–67. doi: 10.1159/000381904. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez H.E., Gujrati M., Frederick M., Henderson Y., Arumugam J., Spring P.W. Identification of 9 genes differentially expressed in head and neck squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 2003;129(7):754–759. doi: 10.1001/archotol.129.7.754. [DOI] [PubMed] [Google Scholar]
- 15.Ye H., Yu T., Temam S., Ziober B.L., Wang J., Schwartz J.L. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9(69) doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roepman P., Wessels L.F., Kettelarij N., Kemmeren P., Miles A.J., Lijnzaad P. An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat. Genet. 2005;37(2):182–186. doi: 10.1038/ng1502. [DOI] [PubMed] [Google Scholar]
- 17.Roepman P., Kemmeren P., Wessels L.F., Slootweg P.J., Holstege F.C. Multiple robust signatures for detecting lymph node metastasis in head and neck cancer. Cancer Res. 2006;66(4):2361–2366. doi: 10.1158/0008-5472.CAN-05-3960. [DOI] [PubMed] [Google Scholar]
- 18.van Hooff S.R., Leusink F.K., Roepman P., Baatenburg de Jong R.J., Speel E.J., van den Brekel M.W. Validation of a gene expression signature for assessment of lymph node metastasis in oral squamous cell carcinoma. J. Clin. Oncol. 2012;30(33):4104–4110. doi: 10.1200/JCO.2011.40.4509. [DOI] [PubMed] [Google Scholar]
- 19.Gioanni J., Fischel J.L., Lambert J.C., Demard F., Mazeau C., Zanghellini E. Two new human tumor cell lines derived from squamous cell carcinomas of the tongue: establishment, characterization and response to cytotoxic treatment. Eur. J. Cancer Clin. Oncol. 1988;24(9):1445–1455. doi: 10.1016/0277-5379(88)90335-5. [DOI] [PubMed] [Google Scholar]
- 20.Yeudall W.A., Crawford R.Y., Ensley J.F., Robbins K.C. MTS1/CDK4I is altered in cell lines derived from primary and metastatic oral squamous cell carcinoma. Carcinogenesis. 1994;15(12):2683–2686. doi: 10.1093/carcin/15.12.2683. [DOI] [PubMed] [Google Scholar]
- 21.Balló H., Koldovsky P., Hoffmann T., Balz V., Hildebrandt B., Gerharz C.D. Establishment and characterization of four cell lines derived from human head and neck squamous cell carcinomas for an autologous tumor-fibroblast in vitro model. Anticancer Res. 1999;19(5B):3827–3836. [PubMed] [Google Scholar]
- 22.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Network C.G.A. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen M.M., Jørgensen J.T., Binderup T., Kjaer A. Tumor volume in subcutaneous mouse xenografts measured by microCT is more accurate and reproducible than determined by 18F-FDG-microPET or external caliper. BMC Med. Imaging. 2008;8:16. doi: 10.1186/1471-2342-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sales K.U., Masedunskas A., Bey A.L., Rasmussen A.L., Weigert R., List K. Matriptase initiates activation of epidermal pro-kallikrein and disease onset in a mouse model of Netherton syndrome. Nat. Genet. 2010;42(8):676–683. doi: 10.1038/ng.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zini A., Czerninski R., Sgan-Cohen H.D. Oral cancer over four decades: epidemiology, trends, histology, and survival by anatomical sites. J. Oral Pathol. Med. 2010;39(4):299–305. doi: 10.1111/j.1600-0714.2009.00845.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhong L., Liu Y., Wang K., He Z., Gong Z., Zhao Z. Biomarkers: paving stones on the road towards the personalized precision medicine for oral squamous cell carcinoma. BMC Cancer. 2018;18(1):911. doi: 10.1186/s12885-018-4806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogel L.K., Saebø M., Skjelbred C.F., Abell K., Pedersen E.D., Vogel U. The ratio of matriptase/HAI-1 mRNA is higher in colorectal cancer adenomas and carcinomas than corresponding tissue from control individuals. BMC Cancer. 2006;6:176. doi: 10.1186/1471-2407-6-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kit O.I., Frantsiyants E., Kozlova L.S., Bandovkina V.A., Evgeniy N., Kolesnikov E.N., Kozhushko M.A., Kazieva T.B., Averkin M. Protease/protease inhibitor balance in blood plasma to predict postoperative complications in operated patients with pancreatic head cancer. J. Clin. Oncol. 2018;36:258. [Google Scholar]
- 33.Jiang R., Shi Z., Johnson J.J., Liu Y., Stack M.S. Kallikrein-5 promotes cleavage of desmoglein-1 and loss of cell-cell cohesion in oral squamous cell carcinoma. J. Biol. Chem. 2011;286(11):9127–9135. doi: 10.1074/jbc.M110.191361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma H., Hockla A., Mehner C., Coban M., Papo N., Radisky D.C. PRSS3/Mesotrypsin and kallikrein-related peptidase 5 are associated with poor prognosis and contribute to tumor cell invasion and growth in lung adenocarcinoma. Sci. Rep. 2019;9(1):1844. doi: 10.1038/s41598-018-38362-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q., Lv Q., Bian H., Yang L., Guo K.L., Ye S.S. A novel tumor suppressor SPINK5 targets Wnt/β-catenin signaling pathway in esophageal cancer. Cancer Med. 2019;8(5):2360–2371. doi: 10.1002/cam4.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.