Abstract
Background
Undifferentiated pleomorphic sarcoma (UPS) is the most frequent, aggressive and less-characterized sarcoma subtype. This study aims to assess UPS molecular characteristics and identify specific therapeutic targets.
Methods
High-throughput technologies encompassing immunohistochemistry, RNA-sequencing, whole exome-sequencing, mass spectrometry, as well as radiomics were used to characterize three independent cohorts of 110, 25 and 41 UPS selected after histological review performed by an expert pathologist. Correlations were made with clinical outcome. Cell lines and xenografts were derived from human samples for functional experiments.
Findings
CD8 positive cell density was independently associated with metastatic behavior and prognosis. RNA-sequencing identified two main groups: the group A, enriched in genes involved in development and stemness, including FGFR2, and the group B, strongly enriched in genes involved in immunity. Immune infiltrate patterns on tumor samples were highly predictive of gene expression classification, leading to call the group B ‘immune-high’ and the group A ‘immune-low’. This molecular classification and its prognostic impact were confirmed on an independent cohort of UPS from TCGA. Copy numbers alterations were significantly more frequent in immune-low UPS. Proteomic analysis identified two main proteomic groups that highly correlated with the two main transcriptomic groups. A set of nine radiomic features from conventional MRI sequences provided the basis for a radiomics signature that could select immune-high UPS on their pre-therapeutic imaging. Finally, in vitro and in vivo anti-tumor activity of FGFR inhibitor JNJ-42756493 was selectively shown in cell lines and patient-derived xenograft models derived from immune-low UPS.
Interpretation
Two main disease entities of UPS, with distinct immune phenotypes, prognosis, molecular features and MRI textures, as well as differential sensitivity to specific anticancer agents were identified. Immune-high UPS may be the best candidates for immune checkpoint inhibitors, whereas this study provides rational for assessing FGFR inhibition in immune-low UPS.
Funding
This work was partly founded by a grant from La Ligue.
Keywords: Sarcomas, Genomics, Immunology, Proteomics, Preclinical, Therapeutic targets
Research in context.
Evidence before this study
Undifferentiated pleomorphic sarcoma (UPS) is one of the most aggressive and least characterized sarcoma subtypes. We searched PubMed with the terms “undifferentiated pleomorphic sarcomas (UPS)” OR “malignant fibrous histiocytoma” OR “pleomorphic sarcoma” AND “molecular profiling” OR “next generation sequencing” AND “proteomics” NOT “review” for translational studies in humans published in English up to March 31, 2020. Apart from the genomic characterization of sarcomas from the TCGA study, studies investigating molecular features of UPS were essentially based on array gene-expression analysis and comparative-genomic-hybridization. None of them aimed to integrate genomic, immunological and radiomic features to decipher the biological landscape of the specific group of UPS.
Added value of this study
By using a multi-omics platform, we identified and confirmed in three independent cohorts of UPS patients two main disease entities with distinct immune phenotypes, prognosis, molecular features, and MRI textures, as well as differential sensitivity to specific anticancer agents, in vitro and in vivo. The first group was enriched in stemness pathways and oncogenes such as FGFR2, the other one characterized by a specific highly-inflammatory micro-environment.
Implications of all the available evidence
We report a study integrating genomic, proteomic and radiomic data to characterize a rare and devastating disease with an unmet medical need. This comprehensive classification improves our understanding of UPS and helps building new selection strategies in patient care, with direct therapeutics potential.
Alt-text: Unlabelled box
1. Introduction
Since its first description in 1964 [1] malignant fibrous histiocytoma (MFH) has been considered the most common soft tissue sarcoma (STS) of adults. Main distinctive features of MFH are an apparent lack of specific differentiation and predominant pleomorphic patterns [2]. MFH has been considered as a heterogeneous group of tumors made up ‘by default’, including non-mesenchymatous and other mesenchymatous subtypes, and 'true' undifferentiated pleomorphic sarcoma after careful expert review [3]. In 2002, the WHO reclassified the group under undifferentiated pleomorphic sarcoma (UPS) [4]. Currently, UPS represents a diagnosis of exclusion based on the absence of a specific line of differentiation after careful histological examination and judicious use of ancillary techniques, including immuno-histo-chemistry (IHC) and molecular biology to exclude any differential diagnosis [5,6].
Despite adequate loco-regional treatment, up to 40% of patients will develop metastatic disease, with the worse progression-free survival in palliative stage among all histological sarcoma subtypes [7].
Existing omics studies have essentially assessed MFH/UPS among other sarcoma subtypes and were based on array gene-expression analysis and comparative-genomic-hybridization [8], [9], [10]. More recently, the TCGA study gave an integrated genomic landscape of sarcomas, including UPS and myxofibrosarcoma (MFS), and identified a subgroup associated with immune infiltrates [11]. Altogether, these studies have allowed preliminary characterization of UPS, including genomic instability and lack of commitment into mesenchymal differentiation pathways, but failed to identify pathways and targets with therapeutic potential. We report here the results of a multi-omics analysis dedicated to centrally-reviewed, high-grade, non-pretreated primary UPS, with the aims to deeply assess their molecular characteristics, better classify these entities, and identify specific therapeutic targets.
2. Methods
2.1. Patients and tumor samples
Patients inclusion criteria included 1) diagnosis of undifferentiated pleomorphic sarcoma after histological review performed by a soft tissue sarcoma referral pathologist of the French Sarcoma Group; 2) high grade defined as grade 2–3 according to the FNLCC grading system; 3) no treatment done before sampling; 4) informed consent obtained for the analysis. A Tissue Micro Array (TMA) was produced from Formalin-Fixed Paraffin-Embedded (FFPE) tumor blocks of 110 patients with non-pretreated surgically resected primary high-grade UPS that constitute the prognostic cohort. Three spots for each tumor sample were assessed. FFPE tumor blocks, frozen tumor and matched normal tissue samples of 25 patients with non-pretreated, surgically resected, primary, high grade UPS treated at Institut Bergonié were collected for the omic cohort.
2.2. Statistics
Quantitative variables were described using the median and range, and qualitative variables were described using frequency and percentage. Metastasis-free survival (MFS) and overall survival (OS) were defined from date of diagnosis to time of first metastasis or death (from any cause) and to death (from any cause), respectively.
Patients alive and progression-free were censored at the date of last follow-up. Survival endpoints were analyzed using the Kaplan-Meier method. Prognostic factors for MFS were identified by univariate and multivariate analyses using a Cox regression model. Variables tested in univariate analysis included: age, gender, tumor size, FLNCC grade, and CD8+ cell density. Variables associated with a p-value < 0.05 in the univariate analysis were planned to be included in the multivariate analysis. Statistical analyses were performed using SPSS software (SPSS Version 21.0, IMB Corp., Armonk, USA) for clinical data and R for biological data [12], P < 0.05 indicated statistical significance.
2.3. Immunohistochemistry
IHC was performed on FFPE tumor samples using automated protocols on a Ventana BenchMark Ultra platform (Roche, Bâle, Switzerland). Monoclonal primary antibodies for CD8 (Spring Bioscience Cat# M3164, RRID: AB_1660846), PD-1 (Abcam Cat# ab52587, RRID: AB_881954), IDO1 (Cell Signaling Technology Cat# 86630, RRID:AB_2636818), CD68 (Agilent Cat# M0876, RRID: AB_2074844), CD163 (Leica Biosystems Cat# NCL-L-CD163, RRID: AB_2756375) and FGFR2 (Cell Signaling Technology Cat# 23328, RRID: AB_2798862) were used. Amplification and detection steps were performed with an Ultraview kit and 3,3′-diaminobenzidine was used as a chromogen. CD68/CD163 double staining was performed on a Ventana Discovery Ultra platform (Roche, Bâle, Switzerland) and image analysis as described previously [13]. Image analysis was performed under the supervision of a pathologist (JA) to detect the density of CD8 and PD-1 positive cells in the tumor areas and the proportion of IDO1, CD68 and CD163 stained surface of tumor slides or spots, as previously described [14]. IDO1 staining on TMA slides was evaluated semi-quantitatively by a trained pathologist (JA) including percentage (0–100) and intensity (0 = null, 1 = low, 2 = moderate, 3 = strong) of staining on tumor spots, and a H-score (0–100) was defined by percentage x intensity of staining. FGFR2 staining on tumor slides was evaluated semi-quantitatively by a trained pathologist (RP), and included percentage (0–100) and intensity (0 = null, 1 = low, 2 = moderate, 3 = strong) of staining on tumor cells. M1 macrophages were defined as CD68+ CD163- cells and M2 macrophages as CD163+ cells.
2.4. Cell culture and reagents
The UPS cell lines used in this study were derived from human UPS surgical specimens after obtaining written, informed patient consent and Institut Bergonié Institutional Review Board approval. Briefly, following surgical resection, fresh tumor tissue was minced with scissors and then digested with 200 IU/ml type II collagenase (Roche) in serum-free RPMI medium overnight. After digestion, isolated cells and pieces were washed and seeded in a 25cm2 plastic flask. Four UPS cell lines were successfully established from tumor samples with different profiles: IB106 and JR588, KN473 and IB119. Each cell line was characterized by array comparative genomic hybridization every 10 passages until p50 to verify that its genomic profile was still representative of the originating tumor sample. No drift in the cell line maintenance or genetic imbalances were shown along passages. Cells were grown in RPMI medium 1640 GlutaMAX™ Supplement (Life Technologies, Carlsbad USA) in the presence of 10% (v/v) fetal bovine serum and Penicillin/Streptomycin 1% (Dutscher, Merignac, France), in flasks. Cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2. Cells were routinely passaged every 2 or 3 days and all the experiments were performed with cell lines between passages 25 and 60. JNJ-42756493, a pan-FGFR inhibitor, was provided by Johnson&Johnson (New Brunswick, USA), AZD4547 and BGJ398, two other pan-FGFR inhibitors, were purchased from Selleck Chemicals (Houston, USA), prepared as a 10 mmol/L stock solution in DMSO and stored at −20 °C for in vitro studies. Cultured cells were treated with medium changes without antibiotics and fresh drug additions as indicated in the figure legends. Two lentiviral vectors containing FGFR2 shRNA were purchased from Sigma-Aldrich (Saint-Louis, USA) (TRCN0000218493 shFGFR2–1 −5′- AGCCCTGTTTGATAGAGTATA-3′ and TRCN0000231051 shFGFR2–2 −5′- TTAGTTGAGGATACCACATTA-3′). Viral particles were produced by calcium phosphate transfection of 293T cells. Briefly, three plasmids encoding the viral envelope (pCMV-VSV-G, RRID: Addgene_8454), a packaging vector (psPAX2, RRID: Addgene_12,260)) and pLKO-shRNA FGFR2 were co-transfected in 293T cells (ATCC Cat# CRL-3216, RRID: CVCL_0063). After 6 h, medium was replaced by OptiMEM medium and supernatant containing lentiviral particles was collected after 40 h, filtered and stored at −150 °C. Functional titer was determined by transduction of cells with serial dilutions of virus and subsequent evaluation of viability after 3 days of puromycin selection (1 µg/ml). UPS cell lines were then infected overnight at a multiplicity of 1 or 5 infectious units per cell in presence of polybrene 8 µg/ml. Apoptotic cells were then analyzed by flow cytometry after Annexin V/Propidium Iodide incorporation 72 h after infection.
2.5. RNA isolation and RT-qPCR
Total RNA from UPS patients and cell lines were isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the supplier's instructions. The purity and concentration of isolated RNA were determined by spectrophotometer NanoDropTM and capillary electrophoresis using Bioanalyser 2100 (Agilent Technologies Inc.,Santa Clara, USA) was carried out to determine the RNA integrity number (RIN). For RTqPCR, cDNA was prepared from 500 ng total RNA using the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Carlsbad, USA). To analyze the FGFRs expression, the following human TaqMan Gene Expression Assays were used: RPLP0 Hs999999_m1, FGFR1 Hs00241111_m1, FGFR2 Hs01552918_m1, FGFR3 Hs00179829_m1, FGFR4 Hs00242558_ m1. Each gene was analyzed in duplicate with 2.5 ng cDNA in three independent experiments with an ABI PRISM 7500 Fast Detection System instrument (Applied Biosystems, Thermo Fisher Scientific, Waltham, USA). RPLP0 was used as a reference gene. Reaction conditions are the following: pre-incubation of 50 °C for 2 min, initial denaturation at 95 °C for 10 min and 40 cycles of: 95 °C/15 s, 60 °C/1 min. The relative comparative method was used to analyze RT-qPCR data and fold change are calculated using the Universal Human Reference RNA (Agilent Technologies Inc., Santa Clara, USA).
2.6. Western blot
Treated and control cells were harvested in 60 μl of radio-immuno-precipitation assay (RIPA) lysis buffer. The lysate was centrifuged (13 000 rpm, 15 min, 4 °C), and the supernatant was stored at −80 °C. Equal amounts of total protein (30 μg) were electrophoresed on 12% or 10% sodium dodecyl sulfate polyacrylamide gels and transferred onto polyvinylidene difluoride membranes. The blots were probed overnight at 4 °C with an anti- FGFR2 (Cell Signaling Technology Cat# 23,328, RRID: AB_2,798,862), anti-actin (Sigma-Aldrich Cat# A3853, RRID: AB_262,137), anti-phospho-FGFR2 (ser 782) (Thermo Fisher Scientific Cat# PA5–64,796, RRID: AB_2,662,677), anti-phospho-Erk (Thr202/Tyr204) (Cell Signaling Technology Cat# 4370, RRID: AB_2,315,112) and anti-Erk (Abcam Cat# ab17942, RRID: AB_2,297,336) primary antibody diluted in PBST (DPBS 10X (GibcoTM) after 1X dilution; 0.1% Tween-20) with 5% bovine serum albumin. The horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology Inc. Dallas, USA) was diluted 1:10,000. Bound antibodies were visualized on Fusion Fx7 imaging system (Fisher Bioblock Scientific, Waltham, USA) using the ImmobilonTM Western enhanced chemiluminescence detection kit (Millipore Corporation, Billerica, USA). The resulting bands were analyzed and quantified using ImageJ 1.48v software (RRID: SCR_003070, National Institutes of Health, Bethesda, USA).
2.7. Cell viability assay
Cells were seeded in triplicate at 4000 cells/well into 96-well plates, cultured with fresh growth medium for at least 24 h and treated with a range of increasing concentrations of drugs for 72 h. Cell viability was assessed by MTT (2-deoxyglucose (2-DG) and 3–4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide) assay (Sigma-Aldrich, St. Quentin Fallavier, France) at a final concentration of 0.5 mg/mL and 3 h of incubation. Then, supernatant was discarded, 100 μL of dimethyl sulfoxide (DMSO) was added and the absorbance at 570 nm was monitored using a Flexstation 3 Plate reader (Molecular Devices, Sunnyvale, USA), using 630 nm as a reference. The half maximal inhibitory concentration (IC50) was calculated with GraphPad Prism software version 5.0 for Windows (GraphPad Software, RRID: SCR_002798, La Jolla, USA). Each experiment was repeated at least 3 times.
2.8. Cell cycle analysis
The cell cycle distribution was studied by examining DNA content using fluorescence-activated cell sorting (FACS) and analyzed using Cell Quest Pro-software (BD Biosciences, San Jose, USA). About 3 × 105 cells were seeded in complete medium in 6-well plates. After 6 h, cells were washed once with PBS and medium without FBS were added to synchronize cells overnight. Then, the cells were treated for 24 h in medium with FBS and JNJ-42,756,493 at the IC50 concentration, centrifuged at 1500 g for 5 min, and washed with PBS. The cells were then fixed with 70% ethanol at −20 °C overnight. Following ethanol removal, the cells were washed with PBS. Next, 300 μl of a Propidium Iodide (PI) and ribonuclease-containing solution were added to the cells and then analyzed by FACS. The data were analyzed with FlowJo v.7.6.3 software (RRID: SCR_008520), and the results were expressed in terms of percentage of cells in a given phase of the cell cycle based on 3 independent experiments.
2.9. Animal studies
Induction of tumor patient derived xenograft (PDX) was performed by subcutaneous implantation of fragments of fresh surgically resected UPS tumor of approximately 3 mm3 into the right flanks of Ragγ2C-/- mice (MGI Cat# 3829712, RRID: MGI:3829712). PDX are maintained by successive animal to animal passages, the seventh and tenth generation for JR588 and KN473 respectively were used for this experiment. After successful engraftment of previously established PDX (with a mean of 18 and 30 days for engraftment of JR588 and KN473, respectively), mice were randomized according to tumor volume to either vehicle alone (10% hydroxypropyl-β-cyclodextrin) or vehicle containing JNJ-42756493, administered by oral gavage daily at 30 mg/kg for 3 weeks for PDX JR588 and 4 weeks for PDX KN473. Tumors were measured regularily until first tumors reached 2000 mm3. At the end of the experiments, mice were euthanized, and tumors were excised. Tumor progression was analyzed with GraphPad Prism software. Experiments have been performed in agreement with Bioethics Law No 2004–800 and the Ethics Charter from the National Institute of Cancer (INCa).
2.10. Sequencing protocols, bioinformatics analysis, proteomics and radiomics
See Supplementary Material.
2.11. Study ethic approval
Institutional ethic review board approval and patient informed consent have been obtained for this study. Animal care and procedures were approved by the Institutional Animal Care and Use Committee Office of Bordeaux University, France (APAFIS #8415).
2.12. Role of the funding source
The funder has no role in study design, data collection, analyses, interpretation or writing of the report.
3. Results
3.1. T cell infiltration has a prognostic value in UPS
We have previously shown that UPS are characterized by variable levels of CD8+ T Lymphocyte (TL) infiltrates [14]. We then aimed to assess the prognostic impact of CD8+ cell density by IHC in a series of 110 primary non-pretreated UPS (Fig. S1, Table S1). Median follow up was 115.2 months (95%CI: 77.8–152.5). There was a significant better MFS and a trend for better OS in patients with high CD8+ cell density in tumor (Fig. S2A and S2B). On multivariate analysis, high CD8+ cell density in tumor remained an independent prognostic factor associated with MFS (Cox proportional-hazards model, p = 0.02) (Table S2). These data suggest that analysis of tumor immune infiltrates could allow sub-classifying UPS patients into groups with distinct metastatic behavior and prognosis. This led us to hypothesize that these tumor immune infiltrates are associated with distinct underlying genomic events.
3.2. RNA-sequencing allows reproducible classification of UPS with distinct immune infiltrates and prognostic significance
We selected 25 additional UPS samples for full exome and RNA-sequencing based on the availability of frozen material (Fig. S1, Table S1). Unsupervised consensus and hierarchical clustering of RNA-sequencing data identified three groups of patients with associated gene-clusters (Fig. 1A, Fig. S3A and S3B). The analyses of differential gene expression between the two main groups A and B identified 1405 genes (Table S3). We observed that group A was mainly enriched in genes that play a crucial role in normal development, stemness and oncogenesis (Table S3). Such genes include LHX8, which encodes an important mediator of stem cell fate [15], LRRN1, implicated in pluripotency maintenance of stem cells [16], LGR5, which encodes a seven transmembrane spanning receptor that has recently been found to be a stem cell marker [17], and BMP5, a member of the TGF-beta family involved in development and differentiation [18]. Interestingly, FGFR2 was one of the top genes up-regulated in group A (fold-change: 23.5). Group B was strongly enriched in genes and gene-sets belonging to inflammatory response and IFNgamma response immune pathways, CD8+ TL and monocyte, but also Natural Killer, CD4+ helper TL, activated dendritic cell, memory BL, as well as regulatory TL immune cell signatures, indicating a complete immune representation in situ. Most up-regulated genes included MARCO, TIMD4, TIGIT, CD27, IFNG, CD8B, PDCD1, CD3D, IDO1 and numerous cytokines, together with DKK1, a key inhibitor of Wnt/beta-catenine pathway (Tables S3 and S4).
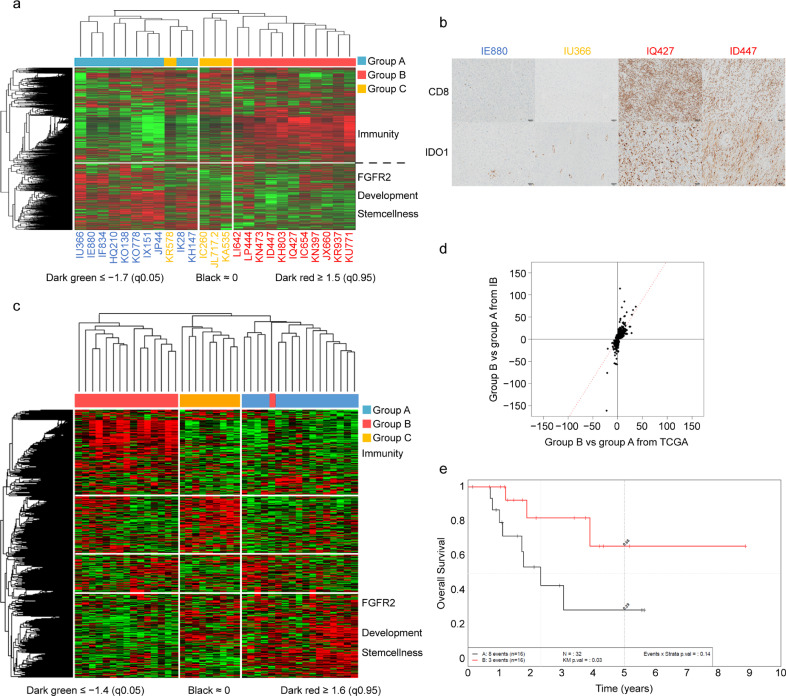
Fig. 1.
Gene expression profiling allows reproducible classification of UPS with distinct immune infiltrates and prognostic significance (A) Unsupervised Hierarchical Clustering of RNA-sequencing in UPS from Institut Bergonié (n = 25); 3 groups of patients are identified, with 3 associated gene-clusters. (B) Expression of CD8 (upper panel) and IDO1 (lower panel) by IHC staining shows concordance with RNA profiling group. Left: a sample belonging to group A negative for CD8 and IDO1 on IHC (IE880), middle: a sample belonging to group C negative for CD8 and IDO1 on IHC (IU366), right: 2 samples belonging to group B positive for CD8 and IDO1 on IHC (IQ427 and ID447) (magnification × 100). (C) Unsupervised Hierarchical Clustering of RNA-sequencing in UPS from TCGA consortium (n = 41); 3 groups of patients are identified, with 3 associated gene-clusters. (D) Analysis of Agreement between Differential EXpression of genes in group A vs B UPS from Institut Bergonié and group A vs B UPS from TCGA consortium reveals a very high correlation (1405 genes, Spearman = 0.83, Pearson = 0.73). (E) Overall survival (OS) of UPS patients from TCGA consortium according to gene expression is significantly different in group A (immune-low) versus group B (immune-high) (n = 32) (p = 0.03).
We then assessed immune infiltrates on related tumor samples by IHC and found a significantly higher proportion of tumor-infiltrating CD8+ TL in UPS from group B than group A and C. The most significant predictors of group B were CD8 and IDO1 (Fig. 1B, Fig. S4A, S4B, S4C, S4D, and S4E). This led us to call group B “immune-high”, group A “immune-low” and group C, less well characterized, “other”. Because group C was too small to have enough statistical robustness, we further focused on the two main groups of immune-high and immune-low UPS.
In order to test the robustness of our model, we then assed differential gene-expression of an independent cohort of 41 UPS from TCGA consortium (Fig. S1) [11]. We also identified three groups with associated gene-clusters (Fig. 1C, Fig. S5, Table S5). Analysis of AGreement between Differential Expression (AGDEX) of the 25 UPS from our cohort and the 41 UPS from TCGA cohort revealed a very high correlation between clusters (Spearman = 0.83, Pearson = 0.73) (Fig. 1D). Group A from TCGA cohort was enriched in genes involved in stemness, notably LHX8, LRRN1 and transcription factors found in group A from our cohort, and demonstrated a differential expression of FGFR2. Furthermore, group B from TCGA cohort was enriched in immune clusters and genes including IDO1, IFNG, TIMD4, PDCD1, CD3D, CD8B as well as TIGIT and CTLA4, together with DKK1.
We finally assessed the prognostic role of our gene expression classification on clinical follow-up data from TCGA cohort and showed that immune-low UPS had a significantly worst overall survival than immune-high UPS (log rank, p = 0.03) (Fig. 1E).
3.3. Opposing immune phenotypes are characterized by different gene copy number alteration patterns in UPS
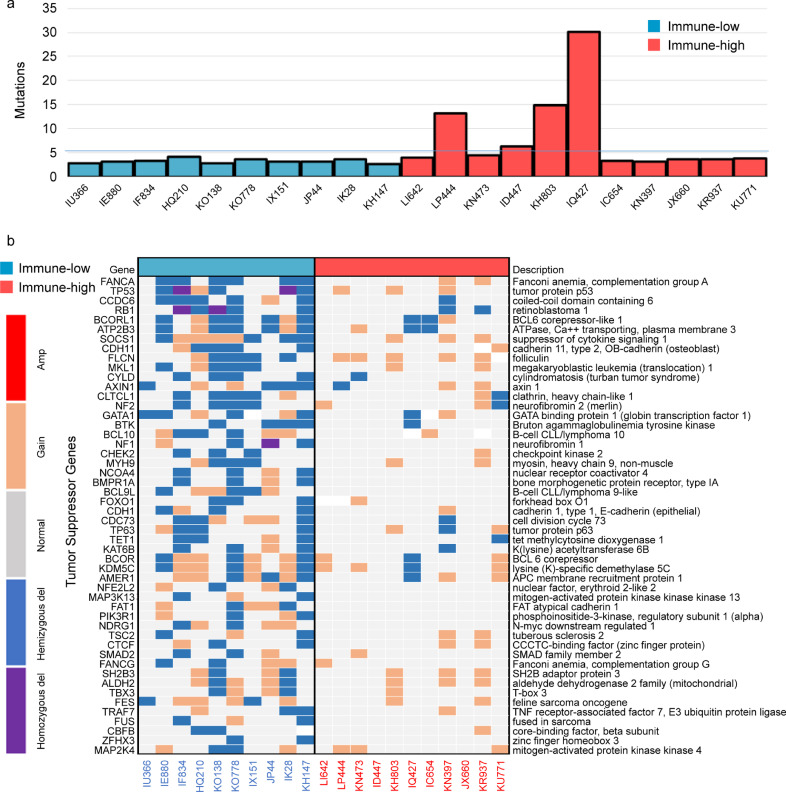
There is growing evidence that underlying genomic alterations may drive the composition of the tumor micro-environment in solid tumors [19,20]. We then assessed the relationship of somatic mutation and gene copy number alteration (CNA) rates with immune-high and immune-low gene expression profiles in our UPS cohort. Overall, mutation burden in the whole series was low (median: 4.3 mutations/Mb). However, all UPS samples with mutation burden > 5 mutations/Mb belonged to the immune-high group (Fig. 2A), with no relevant recurrent mutation in either of each group (Table S6). The UPS samples from the immune-low group were characterized by a significantly higher rate of gene CNA (OR = 10.8 for homozygous deletions, OR = 6.9 for amplifications) (Fig. S6A, S6B, and S6C, Table S7). There was notably a significant enrichment in deletions of tumor suppressor genes belonging to DNA repair, cell cycle, apoptosis, PI3K/mTOR, and Wnt/beta-catenin pathways (Fig. 2B). We also retrieved a significantly higher number of non-recurrent fusion genes in immune-low UPS (OR = 1.96, p < 0.01) (Table S8).
Fig. 2.
The UPS classification is consistent from tumor immune infiltration patterns to underlying genomic events (A) Frequency of mutations per Megabase (MB) in immune-low and immune-high UPS from Institut Bergonié, ordered according to gene expression clustering (n = 21) (B) Gene focal somatic copy-number alterations, focusing on deletions in tumor suppressor genes (TSG) in immune-low and immune-high UPS from Institut Bergonié, ordered according to gene expression clustering (n = 21). Enrichment in TSG deletions is found in immune-low UPS.
3.4. Integrative analyses reveal high transcriptome-proteome correlation in clustering of UPS
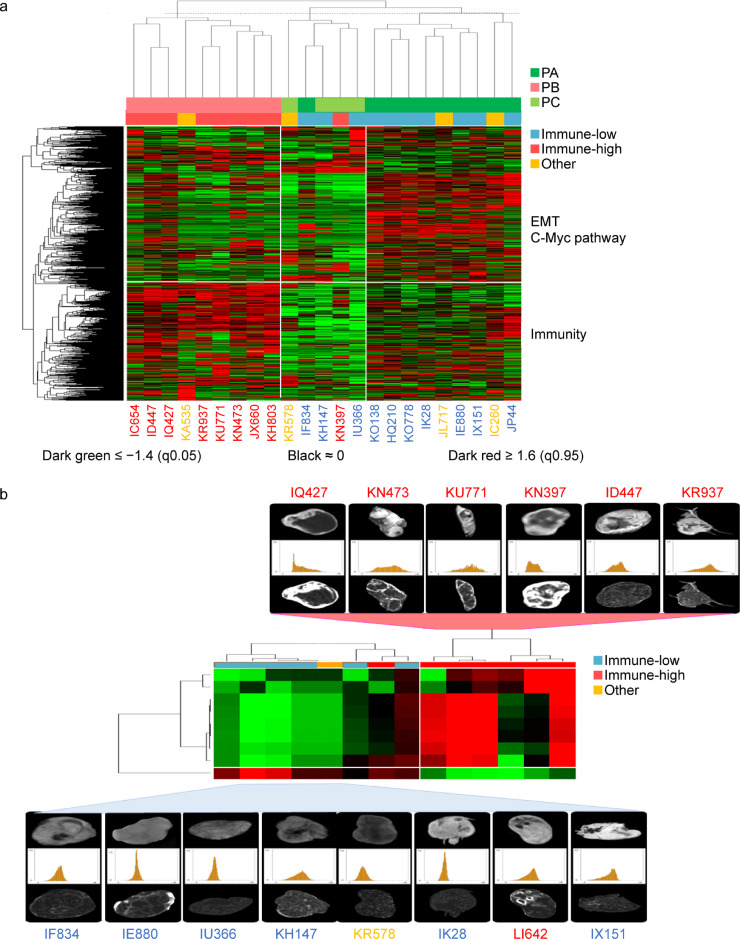
Proteins are the main effectors of most cellular reactions. We therefore explored whether proteomics analysis could recapitulate our UPS genetic classification. Unsupervised consensus clustering and hierarchical clustering of protein expression identified three proteomics groups, that we named PA (n = 9), PB (n = 10) and PC (n = 4). We found a high agreement between Proteomics and RNA-sequencing clusterings (precision: 82%) (Fig. 3A), and good correlation between protein/gene couples both differentially expressed in each analysis (Fig. S7A and Table S9). Notably, the PB group was significantly enriched in immune response pathways (Fig. S7B).
Fig. 3.
The UPS classification includes specific proteomic profiles and predictive radiomic features (A) Unsupervised Hierarchical Clustering of Protein Expression in UPS from Institut Bergonié (n = 23); 3 groups of patients are identified, PA, PB, PC, with 3 associated clusters of proteins and 565 proteins with differential expression (FC ≥ 2, p = 0.01). Corresponding color labels of samples according to RNA-sequencing profiling are shown. There is a high agreement on classification labels (precision: 82%) between proteomics and RNA-sequencing clusterings. (B) Hierarchical Clustering of UPS patients according to the 9-feature radiomic signature allows discriminating immune-high UPS from non-immune-high UPS (specificity: 100%, sensitivity: 86% and accuracy: 93%) (n = 14). Corresponding color labels of samples according to RNA-sequencing profiling are shown.
3.5. Nine IRM-based radiomic features can discriminate immune-high UPS
Radiomics represents a promising non-invasive way to characterize tumor molecular features [21]. We therefore explored if radiomic features could recapitulate our UPS molecular classification. We analyzed imaging from 14 patients from our UPS cohort with available pre-treatment MRI. After slice-by slice segmentation, signal intensities were discretized into a matrix of 64 radiomics features (Table S10). We identified a 9-feature radiomic signature that could discriminate the immune-high UPS from the others with a specificity of 100% (7/7), a sensitivity of 86% (6/7) and an accuracy of 93% (13/14) (Fig. 3B).
3.6. Targeting the fibroblast growth factor receptor 2 is associated with anti-tumor activity in immune-low UPS
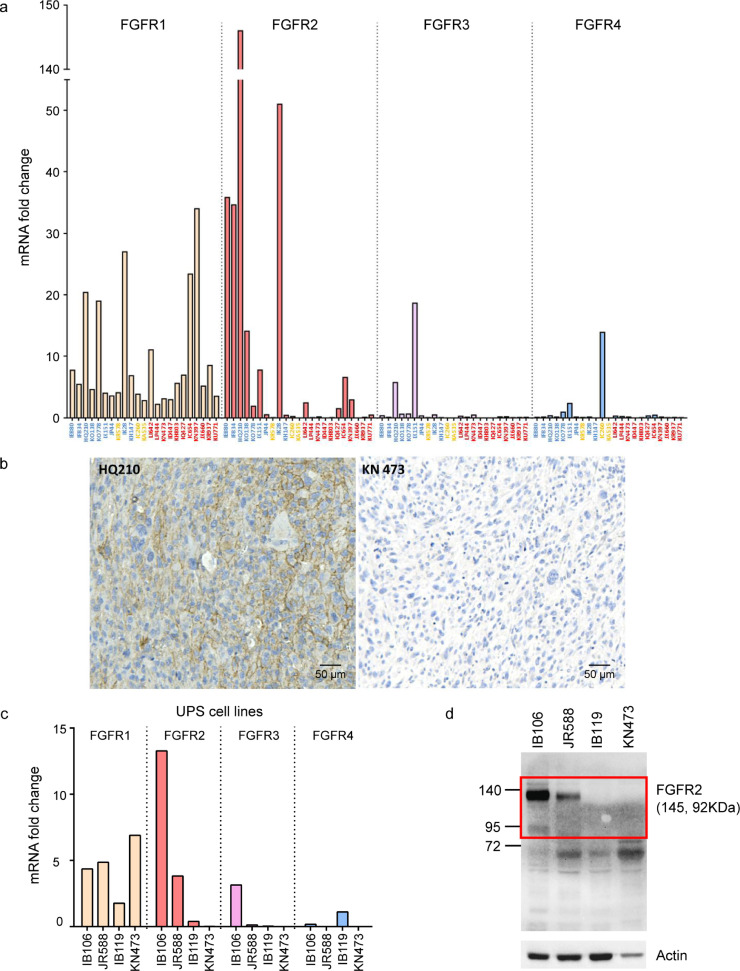
We finally focused on potential for therapeutic targets in immune-low UPS. Physiologically, precise regulation of FGFR expression is required in a wide range of processes in embryonic development, stem cell maintenance, and differentiation [22]. Given a crucial role in driving oncogenic growth in several cancers [23] and the current clinical development of several highly specific FGFR inhibitors, we considered FGFR2 as an interesting therapeutic target. By using quantitative RT-qPCR and IHC, we first confirmed FGFR2 overexpression in immune-low UPS compared to others. Tumor samples and respective associated cell lines had the same FGFR1, 2, 3, 4 expression profiles (Fig. 4A, 4B and 4C). We then investigated the importance of FGFR2 signaling in vitro and in vivo. ShRNA to FGFR2 resulted in a strong induction of apoptosis in cells derived from immune-low UPS but not in cells derived from immune-high UPS (Fig. S8A and S8B).
Fig. 4.
Expression of FGFR in UPS (A) mRNA expression of FGFR1,2,3,4 by RT-qPCR in UPS from Institut Bergonié; samples are ordered on axis by group A (blue), B (red) and C (yellow) according to gene expression clustering (n = 23); FGFR1 is randomly expressed in the 3 groups, FGFR2 is particularly overexpressed in group A whereas FGFR3 and 4 are weakly expressed in the whole cohort; (B) Expression of FGFR2 by Immunohistochemistry (IHC) staining shows concordance with mRNA expression by RT-qPCR. Left: sample belonging to group A positive for FGFR2 on IHC; right: sample belonging to group B negative for FGFR2 on IHC (magnification × 100); (C) Left: mRNA expression of FGFR1,2,3,4 by RT-qPCR in 4 UPS cell lines; IB106 is derived from patient HQ210 (group A/immune-low UPS); IB119 is derived from patient IC260 (group C/Other UPS);, KN473 from patient KN473 (group B/immune-high UPS) and JR588 from a subsequent patient not included in the NGS analysis with a FGFR2-overexpressing immune-low UPS. Right: Protein expression of FGFR2 by Western blot in the same 4 UPS cell lines shows concordance with mRNA expression by RT-qPCR.
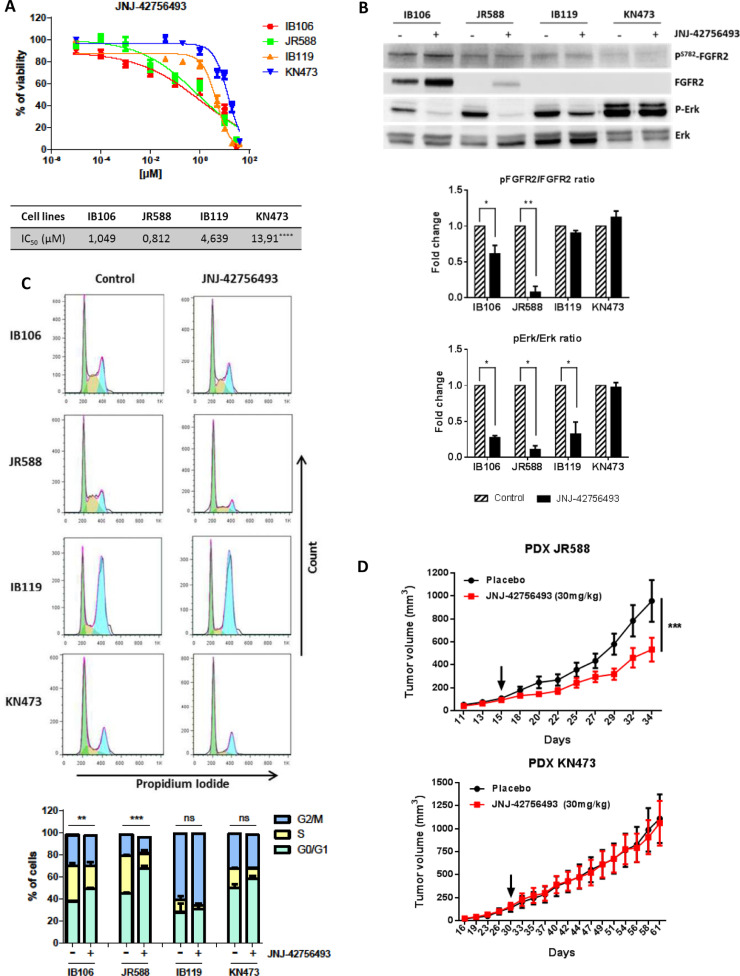
We then assessed several specific FGFR inhibitors and demonstrated that they all potently decreased in vitro viability, FGFR2 phosphorylation and downstream signaling, and induced G0-G1 arrest in cells derived from immune-low UPS, specifically (Fig. 5A, 5B, 5C, and Fig. S9). Finally, we showed that pharmacological FGFR inhibition significantly impaired the growth of patient derived xenografts (PDX) from immune-low UPS but had no effect on PDX from immune-high UPS (Fig. 5D).
Fig. 5.
Therapeutic potential of FGFR inhibition in a specific subgroup of UPS (A) Assessment of cell viability with pan-FGFR inhibitor JNJ-42756493 in 4 UPS cell lines; Growth curves indicate growth inhibition and IC50 of the 4 UPS cell lines after JNJ-42756493 treatment for 72 h (n = 6) (****p < 0.0001, one-way ANOVA). (B) FGFR-inhibitor induces MAPK pathway inhibition in FGFR2 overexpressing cell lines; Phospho-FGFR2/FGFR2 ratio and Phospho-Erk /Erk ratio decrease in FGFR2 overexpressing cell lines after 24 h of treatment with JNJ-42756493 at 1 µM (n = 3) (*p < 0.05 and **p < 0.01, two-way ANOVA) (Western-blot). (C) Activity of pan-FGFR inhibitor (JNJ-42756493) on cell cycle of 4 UPS cell lines; Top: cell-cycle profiles after 24 h of treatment with or without JNJ-42756493 at the IC50 analyzed by Propidium Iodide incorporation and flow cytometry; Bottom: cell cycle phase distributions were analyzed with FlowJo software and presented as mean ± SEM of 3 independent experiments (n = 3) (**p < 0.01 and ***p < 0.001, ns: not significant, two-way ANOVA). (D) Antitumoral effect of a pan-FGFR inhibitor (JNJ-42756493) in two Patient-Derived Xenograft (PDX) models of UPS; Mice were randomly assigned to receive 30 mg/kg of drug or vehicle; Tumor volume progression curves were drawn over 3 to 4 weeks of JNJ-42756493 treatment, arrows represent treatment beginning; The data points represent an average of 11 mice for the JR588 PDX (top) and 8 mice for the KN473 PDX (bottom) per condition (bars, SEM) (***p < 0.001, two-way ANOVA).
4. Discussion
This study is the first to use a multi-omics platform to characterize UPS and identify potential therapeutic targets in this heterogeneous and poorly understood disease.
One important result from this study is the reproducible identification of an immune-high subgroup of UPS with a prognostic significance. Early transcriptomic studies [8,10] have identified a subgroup of inflammatory MFH/UPS, corroborated by most recent studies, albeit non-specifically dedicated to UPS.[11,24] Together, these data bring light to modest results obtained with checkpoint inhibitors in unselected STS patients to date. The PEMBROSARC study assessing pembrolizumab and metronomic cyclophosphamide has reported negative results in unselected STS.[14] However, very low expression of PD-L1, low and variable expression of CD8 and high M2 macrophage infiltrates on tumor samples from the study could explain this primary resistance. In the other hand, four among 10 patients with UPS included in the SARC028 study assessing pembrolizumab in specific subtypes of sarcoma had a partial response.[25] Unfortunately, no assessment of tumor micro-environment was done at this time, limiting interpretation of this heterogeneity.[26] Our UPS classification echoes well a recent collaborative work that shed light on STS immune infiltrates heterogeneity.[27] In this study, gene-expression profiling of more than 600 sarcomas of different histologies, including UPS, allowed establishing an immune-based classification of five different phenotypes: immune-low (A and B), immune high (D and E) and highly-vascularized (C). Samples from patients included in the SARC028 trial were then analyzed and classified accordingly. Very interestingly, patients whose tumor sample belong to the class E - which had the highest immune infiltrates - had the highest response rate and improved survival.[27] In this regards, patients with immune-high UPS may certainly be the best candidates for immune checkpoint inhibitors.
We report a strong correlation between high level of CNA and low immune infiltrates in UPS. Our results echo recent work from Steele et al. who reported significant enrichment for immune-related pathways in a subset of undifferentiated sarcomas (US) characterized by higher mutational burden, as well as another subset of US characterized by a high rearrangement level.[24] Several studies have investigated how underlying genomic alterations may drive the composition of the tumor micro-environnement in solid tumors. Recent publications in other diseases have showed correlation between mutation burden and cytotoxic immune infiltrates, but also association of high level of CNA with low immune infiltrates and worse response to immune checkpoint blockade [19,20,28,29].
Interestingly, we found a reproducible representation of development and stemness pathways activation in immune-low UPS. Previous studies suggest that transformation of mesenchymal stem cells into UPS involves genomic instability together with loss of cell cycle regulators,[30,[31] but also alterations of PI3K/Akt, Notch and Wnt/beta-catenin pathways [32], [33], [34]. These pathways were up-regulated in immune-low UPS, as a result of gene overexpression and CNA. Importantly, PI3K/Akt and Wnt/beta-catenin pathways have been associated with immune exclusion in several models, including sarcomas. [34], [35], [36], [37] These data advocate for further assessment of PI3K/Akt and Wnt/beta-catenin pathway targeting in immune-low UPS.
Developing efficient therapeutic strategies to target stemness is challenging. Physiologically, FGFR signaling is required for sustaining self-renewal and pluripotency of human embryonic stem cells,[38] and FGFR signaling has been implicated in a variety of tissue stem cell activities, including bone marrow mesenchymal stem cells[39] and hematopoietic stem cells.[40] FGFR2 is a tyrosine kinase receptor that activates several oncogenic pathways such as MAPK or PI3K/Akt pathways. Our preclinical findings support the hypothesis that deregulated FGFR2 signaling is an important oncogenic pathway in the initiation and/or maintenance of immune-low UPS, and support recent clinical data showing significant activity of FGFR inhibitors in FGFR mRNA-overexpressing solid tumors without apparent FGFR gene fusion or mutation.[41]
Radiomics is a promising field for systematizing and predicting tumor biology.[21] Radiomics approach could be particularly relevant for soft tissue sarcomas, and especially UPS because of their marked inter-individual but also intra-tumoral heterogeneity. Here we show that nine radiomic features from basic conventional MRI sequences provide the basis for a radiomic signature that could accurately select patients with immune-high UPS on their pre-therapeutic imaging. Reproducibility of contouring and standardization of algorithms are key issues in dissemination of such promising technics. These exploratory data warrant further validation on a larger independent cohort of UPS patients.
This study has several limits. It is mainly descriptive, with the primary aim to give an integrated landscape of UPS. Only two of the three groups have been characterized due to small numbers. However, these two groups are robust and reproducible. Main genomic events described are CNA and functional studies rely on overexpression of target genes whereas proteomics analysis was limited by available library, and did not include FGFR2, but altered pathways were consistent among all different platforms used.
This UPS classification brings out several questions. The first is the availability of genomics in routine, and the need of reproducible surrogates to select immune-high from immune-low UPS patients for therapeutic decision. Our study also shows that numerous other brakes are expressed in immune-high UPS, such as LAG3, TIGIT, TIMD4, IDO1, beside PD-L1, whereas other pathways may be of importance in immune-low UPS, such as PI3K/Akt and Wnt/beta-catenin pathways. Another question of interest is on the best immuno-modulatory combinations to use in these different subgroups. Finally, this classification relies on differential immune infiltrates and pathway activations but underlying causal events of these differences remain to be determined. Preclinical and clinical collaborations will be key to address these questions.
This study provides the most comprehensive omics characterization of true primary UPS to date, revealing two main subsets with distinct genomic, immunologic, proteomic and radiomic features, and direct therapeutic potentials. This study improves the biological understanding of UPS and brings to light the reasons for mixed results obtained with checkpoint inhibitors in unselected STS to date.[25,26] These data directly help building new strategies to improve patient care. Since tumor infiltration by CD8+ TL correlates with better response to immune checkpoint inhibitors and patient outcome,[42] patients with immune-high UPS may certainly be the best candidates for immune checkpoint inhibitors. Moreover, radiomics could be a powerful tool to identify these patients from pre-therapeutic MRI. Finally, our results provide rationale for assessing FGFR inhibition as single agents or in combination in immune-low UPS. In the ongoing MULTISARC study, metastatic STS patients progressing after standard first line doxorubicin are offered whole-exome and RNA-sequencing (NCT03784014). Patients with immune-high UPS will be proposed immune checkpoint-based therapy whereas patients with FGFR2-overexpressing UPS will be proposed a specific FGFR inhibitor.
Contributors
Study design : MT, CL, SV, AL, AI
Conducting experiments: all authors
Acquiring data: all authors
Analyzing data: all authors
Writing the manuscript: MT, CL, AI
Reviewing the manuscript: all authors
Data sharing
Raw data are deposited in the Genome-phenome Archive database of the EMBL-EBI (European Molecular Biology Laboratory – European Bioinformatics Institute): https://www.ebi.ac.uk/ega/studies/EGAS00001004612; https://www.ebi.ac.uk/ega/datasets/EGAD00001006355. Request for data access must be send to Data Access Committee: https://www.ebi.ac.uk/ega/dacs/EGAC00001001707.
Declaration of Competing Interests
JA reports personal fees from AstraZeneca, Roche, BMS, Bayer and MSD outside the submitted work. SS reports employment by AstraZeneca outside the submitted work. TH reports employment by Nantomics and AstraZeneca outside the submitted work, and patents use from Nantomics relevant to this work. AI reports grants and personal fees from Bayer, Roche, grants from MSD, AstraZeneca, Pharmamar, personal fees from Springworks, Ipsen, and non-financial support from Merck outside the submitted work. The other authors declare no conflict of interest.
Acknowledgments
This work was supported by a grant from La Ligue. The authors thank Valérie Vélasco for technical support and Pr Jean-Michel Coindre for pathological review and cohort constitution supervision. The results published here are part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103131.
Appendix. Supplementary materials
References
- 1.O'Brien J.E., Stout .AP. Malignant fibrous xanthomas. Cancer. 1964;17:1445–1455. doi: 10.1002/1097-0142(196411)17:11<1445::aid-cncr2820171112>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 2.Weiss S.W., Enzinger F.M. Malignant fibrous histiocytoma: an analysis of 200 cases. Cancer. 1978;41(6):2250–2266. doi: 10.1002/1097-0142(197806)41:6<2250::aid-cncr2820410626>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 3.Fletcher C.D. Pleomorphic malignant fibrous histiocytoma: fact or fiction? A critical reappraisal based on 159 tumors diagnosed as pleomorphic sarcoma. Am J Surg Pathol. 1992;16(3):213–228. [PubMed] [Google Scholar]
- 4.Fletcher C.D. The evolving classification of soft tissue tumours: an update based on the new WHO classification. Histopathology. 2006;48(1):3–12. doi: 10.1111/j.1365-2559.2005.02284.x. [DOI] [PubMed] [Google Scholar]
- 5.Coindre J.M., Mariani O., Chibon F. Most malignant fibrous histiocytomas developed in the retroperitoneum are dedifferentiated liposarcomas: a review of 25 cases initially diagnosed as malignant fibrous histiocytoma. Mod Pathol. 2003;16(3):256–262. doi: 10.1097/01.MP.0000056983.78547.77. [DOI] [PubMed] [Google Scholar]
- 6.Guillou L., Aurias A. Soft tissue sarcomas with complex genomic profiles. Virchows Arch. 2010;456(2):201–217. doi: 10.1007/s00428-009-0853-4. [DOI] [PubMed] [Google Scholar]
- 7.Savina M., Le Cesne A., Blay J.Y. Patterns of care and outcomes of patients with METAstatic soft tissue SARComa in a real-life setting: the METASARC observational study. BMC Med. 2017;15(1):78. doi: 10.1186/s12916-017-0831-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baird K., Davis S., Antonescu C.R. Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res. 2005;65(20):9226–9235. doi: 10.1158/0008-5472.CAN-05-1699. [DOI] [PubMed] [Google Scholar]
- 9.Carneiro A., Francis P., Bendahl P.O. Indistinguishable genomic profiles and shared prognostic markers in undifferentiated pleomorphic sarcoma and leiomyosarcoma: different sides of a single coin. Lab Invest. 2009;89(6):668–675. doi: 10.1038/labinvest.2009.18. [DOI] [PubMed] [Google Scholar]
- 10.Gibault L., Perot G., Chibon F. New insights in sarcoma oncogenesis: a comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics. J Pathol. 2011;223(1):64–71. doi: 10.1002/path.2787. [DOI] [PubMed] [Google Scholar]
- 11.Network Cancer Genome Atlas Research. Electronic address edsc, Cancer Genome Atlas Research N. Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell. 2017;171(4):950–965. doi: 10.1016/j.cell.2017.10.014. e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Team RC. R . 2017. A language and environment for statistical computing. [Google Scholar]
- 13.Ou D., Adam J., Garberis I. Influence of tumor-associated macrophages and HLA class I expression according to HPV status in head and neck cancer patients receiving chemo/bioradiotherapy. Radiother Oncol. 2019;130:89–96. doi: 10.1016/j.radonc.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 14.Toulmonde M., Penel N., Adam J. Use of PD-1 Targeting, Macrophage Infiltration, and IDO Pathway Activation in Sarcomas: a Phase 2 Clinical Trial. JAMA Oncol. 2018;4(1):93–97. doi: 10.1001/jamaoncol.2017.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou C., Yang G., Chen M. Lhx6 and Lhx8: cell fate regulators and beyond. FASEB J. 2015;29(10):4083–4091. doi: 10.1096/fj.14-267500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao C.H., Wang Y.H., Chang W.W. Leucine-rich repeat neuronal protein 1 regulates differentiation of embryonic stem cells by post-translational modifications of pluripotency factors. Stem Cells. 2018;36(10):1514–1524. doi: 10.1002/stem.2862. [DOI] [PubMed] [Google Scholar]
- 17.Leung C., Tan S.H., Barker N. Recent advances in Lgr5(+) stem cell research. Trends Cell Biol. 2018;28(5):380–391. doi: 10.1016/j.tcb.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Shih H.Y., Hsu S.Y., Ouyang P. Bmp5 regulates neural crest cell survival and proliferation via two different signaling pathways. Stem Cells. 2017;35(4):1003–1014. doi: 10.1002/stem.2533. [DOI] [PubMed] [Google Scholar]
- 19.Davoli T., Uno H., Wooten E.C., Elledge S.J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355(6322) doi: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rizvi N.A., Hellmann M.D., Snyder A. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambin P., Leijenaar R.T.H., Deist T.M. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 22.Dvorak P., Dvorakova D., Koskova S. Expression and potential role of fibroblast growth factor 2 and its receptors in human embryonic stem cells. Stem Cells. 2005;23(8):1200–1211. doi: 10.1634/stemcells.2004-0303. [DOI] [PubMed] [Google Scholar]
- 23.Babina I.S., Turner N.C. Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer. 2017;17(5):318–332. doi: 10.1038/nrc.2017.8. [DOI] [PubMed] [Google Scholar]
- 24.Steele C.D., Tarabichi M., Oukrif D. Undifferentiated sarcomas develop through distinct evolutionary pathways. Cancer Cell. 2019;35(3):441–456. doi: 10.1016/j.ccell.2019.02.002. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tawbi H.A., Burgess M., Bolejack V. Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma (SARC028): a multicentre, two-cohort, single-arm, open-label, phase 2 trial. Lancet Oncol. 2017;18(11):1493–1501. doi: 10.1016/S1470-2045(17)30624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toulmonde M., Italiano A. PD-1 inhibition in sarcoma still needs investigation. Lancet Oncol. 2018;19(1):e6. doi: 10.1016/S1470-2045(17)30921-X. [DOI] [PubMed] [Google Scholar]
- 27.Petitprez F., de Reynies A., Keung E.Z. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577(7791):556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 28.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan T.A., Wolchok J.D., Snyder A. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2015;373(20):1984. doi: 10.1056/NEJMc1508163. [DOI] [PubMed] [Google Scholar]
- 30.Helman L.J., Meltzer P. Mechanisms of sarcoma development. Nat Rev Cancer. 2003;3(9):685–694. doi: 10.1038/nrc1168. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez R., Rubio R., Menendez P. Modeling sarcomagenesis using multipotent mesenchymal stem cells. Cell Res. 2012;22(1):62–77. doi: 10.1038/cr.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larue L., Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene. 2005;24(50):7443–7454. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 33.Wang C.Y., Wei Q., Han I. Hedgehog and Notch signaling regulate self-renewal of undifferentiated pleomorphic sarcomas. Cancer Res. 2012;72(4):1013–1022. doi: 10.1158/0008-5472.CAN-11-2531. [DOI] [PubMed] [Google Scholar]
- 34.Matushansky I., Hernando E., Socci N.D. Derivation of sarcomas from mesenchymal stem cells via inactivation of the Wnt pathway. J Clin Invest. 2007;117(11):3248–3257. doi: 10.1172/JCI31377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spranger S., Bao R., Gajewski T.F. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 36.Peng W., Chen J.Q., Liu C. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6(2):202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.George S., Miao D., Demetri G.D. Loss of PTEN Is associated with resistance to anti-PD-1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity. 2017;46(2):197–204. doi: 10.1016/j.immuni.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu C., Inokuma M.S., Denham J. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19(10):971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 39.Eom Y.W., Oh J.E., Lee J.I. The role of growth factors in maintenance of stemness in bone marrow-derived mesenchymal stem cells. Biochem Biophys Res Commun. 2014;445(1):16–22. doi: 10.1016/j.bbrc.2014.01.084. [DOI] [PubMed] [Google Scholar]
- 40.Itkin T., Ludin A., Gradus B. FGF-2 expands murine hematopoietic stem and progenitor cells via proliferation of stromal cells, c-Kit activation, and CXCL12 down-regulation. Blood. 2012;120(9):1843–1855. doi: 10.1182/blood-2011-11-394692. [DOI] [PubMed] [Google Scholar]
- 41.Schuler M., Cho B.C., Sayehli C.M. Rogaratinib in patients with advanced cancers selected by FGFR mRNA expression: a phase 1 dose-escalation and dose-expansion study. Lancet Oncol. 2019;20(10):1454–1466. doi: 10.1016/S1470-2045(19)30412-7. [DOI] [PubMed] [Google Scholar]
- 42.Daud A.I., Loo K., Pauli M.L. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest. 2016;126(9):3447–3452. doi: 10.1172/JCI87324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.