Abstract
The novel coronavirus disease (COVID-19) that emerged in December 2019 had caused substantial morbidity and mortality at the global level within few months. It affected economies, stopped travel, and isolated individuals and populations around the world. Wildlife, especially bats, serve as reservoirs of coronaviruses from which the variant Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) emerged that causes COVID-19. In this review, we describe the current knowledge on COVID-19 and the significance of wildlife hosts in its emergence. Mammalian and avian coronaviruses have diverse host ranges with distinct lineages of coronaviruses. Recombination and reassortments occur more frequently in mixed-animal markets where diverse viral genotypes intermingle. Human coronaviruses have evolved through gene gains and losses primarily in interfaces where wildlife and humans come in frequent contact. There is a gap in our understanding of bats as reservoirs of coronaviruses and there is a misconception that bats periodically transmit coronaviruses to humans. Future research should investigate bat viral diversity and loads at interfaces between humans and bats. Furthermore, there is an urgent need to evaluate viral strains circulating in mixed animal markets, where the coronaviruses circulated before becoming adapted to humans. We propose and discuss a management intervention plan for COVID-19 and raise questions on the suitability of current containment plans. We anticipate that more virulent coronaviruses could emerge unless proper measures are taken to limit interactions between diverse wildlife and humans in wild animal markets.
Keywords: Animal reservoir, COVID-19, Interspecies transmission, MERS-CoV, SARS-CoV, SARS-CoV-2
1. Introduction
The recent outbreak of pneumonia in China in December 2019, was caused by a novel Betacoronavirus (2019-nCoV). It was confirmed by deep sequencing analysis of samples taken from lower respiratory tracts of patients (Chen et al., 2020, Huang et al., 2020, Wu et al., 2020a). So far, 2 954 222 (28 April) confirmed cases, 202 597 deaths have been reported in the world (WHO, 2020). The World Health Organization (WHO) declared a public health emergency due to this worldwide pneumonia outbreak (on 30th January 2020) and named this novel viral disease as Coronavirus Disease (COVID-19) on 11th February 2020. However, the International Committee on Taxonomy of Viruses (ICTV) recommended the virus name as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) due to its phylogenetic and taxonomic analysis (Gorbalenya et al., 2020, Peng et al., 2020). Full-genome sequencing and phylogenic analysis indicated that SARS-CoV-2 is different from the other Betacoronaviruses [Severe Acute Respiratory Syndrome (SARS) and Middle East respiratory syndrome (MERS)] associated with humans (Zhu et al., 2020). Therefore, the novelty of COVID-19 and its swift and sudden onset resulted in the loss of many lives before scientists were able to identify it and establish a preliminary containment plan. In addition, the vast spread of the virus into many countries revealed a serious gap in the biosecurity policies in all countries of the world and consequently shattered the illusion of a stable health security that many developed countries were experiencing.
In recent years, two novel coronaviruses SARS-CoV and MERS-CoV emerged and caused epidemics in 2002 and 2012 worldwide (WHO, 2019, WHO, 2003, Wu et al., 2020a). These CoVs were both of zoonotic origin and were transmitted between domestic or captive wild animals and people. Although it was widely suggested that civets were reservoirs of the SARS-CoV, detailed studies revealed that civets likely received the infections from other caged animals (Cui et al., 2019). Less is known about the MERS-CoV that was probably transmitted from bats to dromedaries at least 30 years earlier and continued to circulate within domestic camels, before adapting to humans and emerging as a novel virus in 2012 (Cui et al., 2019). Since Jan 25, 2020, the SARS-CoV-2 global outbreak was unprecedented in recent history, far exceeding the number of cases and deaths (18,398 deaths) in the H1N1 influenza pandemic from 2009 to 2010 (Morens et al., 2010, Viviani et al., 2011).
Wild animal wet markets played a significant role in the COVID-19 outbreak (Cui et al., 2019). It is highlighted broadly in the media and in the scientific literature, that humans became infected with SARS-CoV-2 through contact with animals in Huanan seafood wholesale market (Chen et al., 2020). More importantly, there is mounting evidence that diverse viruses have high frequency of reassortments at the interfaces between human, domestic and wildlife animals (e.g. in live animal markets) (Cui et al., 2019). This has been documented for other pathogenic viruses such as the Highly Pathogenic Avian Influenza H5N1 virus that emerged in 1996 (Takekawa et al., 2010) and coronaviruses such as SARS-CoV that emerged in 2002 (Cui et al., 2019).
To help in evaluating the role of live wild animal markets in the epidemiology of this disease, it is crucial to study this hypothesis critically and to determine the link between viral evolution and mixed animal markets, one of the major human-wild animal interfaces. Our goal here is to shed light on the ecological and evolutionary dynamics relating to the emergence of the SARS-CoV-2. Our breif review on the ecology and evolution of this novel coronavirus and its animal to human transmission, identify gaps in our understanding and highlight management strategies that could be useful in preventing outbreaks in humans, domestic animals and wildlife.
2. Novel human coronaviruses strains and nomenclature
2.1. Coronavirus classification
Coronaviruses (CoVs) are one of the largest positive sense RNA viruses that are about 30 kb in length and that belong to the order Nidovirales, family Coronaviridae, subfamily Coronavirinae. They generally circulate in animals and humans (Fehr and Perlman, 2015, Gorbalenya et al., 2006, Richman et al., 2016). The ICTV currently recognizes four genera of CoVs: Alphacoronavirus (α-CoV), Betacoronavirus (β-CoV), Gammacoronavirus (γ-CoV) and Deltacoronavirus (δ-CoV) (Fan et al., 2019, Paim et al., 2019). Coronaviruses may cause viral diseases in birds and mammals. The Alphacoronavirus and Betacoronavirus mainly infect humans and other mammals and cause diseases especially of respiratory and nervous systems (Table 1, Table 2), whereas Gammacoronavirus and Deltacoronavirus infect birds. Some of the latter two coronaviruses may cause diseases in mammals as well (Table 2) (Cui et al., 2019, Fehr and Perlman, 2015, Perlman and Netland, 2009, Weiss and Leibowitz, 2011, Woo et al., 2012, Yin and Wunderink, 2017).
Table 1.
Timeline of recorded humans’ coronaviruses.
| Virus strain | Country | Taxonomic group | Year | Reference |
|---|---|---|---|---|
| HCoV-229E | USA | I | 1962 | (Hamre and Procknow, 1966) |
| HCoV-OC43 | USA | II | 1965 | (Mcintosh et al., 1967) |
| HCoV-SARS | China | II | 2003 | (Drosten et al., 2003, Ksiazek et al., 2003) |
| HCoV-NL63* | Netherlands | I | 2004 | (van der Hoek et al., 2004) |
| HCoV-NL* | Netherlands | I | 2004 | (Fouchier et al., 2004) |
| HCoV-NH* | USA | I | 2005 | (Esper et al., 2005) |
| HCoV-HKU1 | Hong Kong | II | 2005 | (Woo et al., 2005) |
| HCoV-MERS | Saudi Arabia | II | 2012 | (de Groot et al., 2013, Zaki, 2012) |
| HCoV-SARS-CoV-2 | China | II | 2019 | (Chen et al., 2020, Huang et al., 2020, Wu et al., 2020a) |
Closely related.
Table 2.
Animals’ coronaviruses in the world.
| Virus strain | Species | Country | Taxonomic group | Year | Reference |
|---|---|---|---|---|---|
| Bat viruses | |||||
| Bt-MiCoV-1 | Miniopterus spp. | China | I | 2005 | (Poon et al., 2005) |
| Bt-CoV-CDPHE15 | USA | I | 2006 | (Fan et al., 2019) | |
| Bt-MiCoV-HKU8 | Miniopterus spp. | China | I | 2006 | (Chu et al., 2006, Ge et al., 2016) |
| Bt-PiCoV-HKU5 | Pipistrellus | Hong Kong | II | 2006 | (Woo et al., 2006) |
| Bt-TyCoV-HKU4 | Tylonycteris | Hong Kong | II | 2006 | (Woo et al., 2006) |
| Bt-RoCoV-HKU9 | Rousettus | China | II | 2006 | (Lau et al., 2010, Woo et al., 2007) |
| Bt-RhCoV-HKU2 | Rhinolophus | China | I | 2007 | (Lau et al., 2007) |
| Bt-CoV-HKU10 | China | I | 2012 | (Lau et al., 2012) | |
| Bt-Rf-CoV-HuB13 | Rhinolophus ferrumequinum | China | I | 2015 | (Wu et al., 2016) |
| BtMy-Sax11 | Myotis ricketti | China | I | 2016 | (Lin et al., 2017, Wu et al., 2016) |
| Bt-Ny-Sc13 | Nyctalus velutinus | China | I | 2016 | (Wu et al., 2016) |
| Bt-HpCoV-ZJ13 | China | II | 2016 | (Wu et al., 2016) | |
| Bt-ScCoV-512 | Scotophilus spp. | China | I | 2017 | (Lin et al., 2017) |
| Bt-KYNL63 | Kenya | I | 2017 | (Tao et al., 2017) | |
| Bt-EoCoV-GCCDC1 | Rousettus | China | II | 2017 | (Obameso et al., 2017) |
| Bat-CoV-RaTG13 | Rhinolophus affinis | China | II | 2020 | (Zhou et al., 2020) |
| Pangolins viruses | |||||
| Pangolin-CoV | Manis javanica | China | II | 2020 | (Lam et al., 2020) |
| Rat viruses | |||||
| MHV | Murine | Czechoslovakia | II | 1980 | (Bardos et al., 1980) |
| Rt-CoV-LRNV | Rat | China | I | 2015 | (Wang et al., 2015) |
| Rt-CoV-HKU24 | Rattus | China | II | 2015 | (Lau et al., 2015) |
| Hedgehog virus | |||||
| EriCoV-1 | Erinaceus europaeus | Germany | II | 2014 | (Corman et al., 2014) |
| Ferret viruses | |||||
| FRCoV | Ferret | Netherlands | I | 2011 | (Provacia et al., 2011) |
| Mink viruses | |||||
| MCoV | Mink | USA | I | 2011 | (Vlasova et al., 2011) |
| Pig Viruses | |||||
| TGEV | China | I | 2013 | (Zhang et al., 2013) | |
| PEDV | Porcine | China | I | 2016 | (Xu et al., 2016) |
| PorCoV-HKU15 | Pig | Hong Kong | IV | 2017 | (Woo et al., 2017) |
| Bovine viruses | |||||
| Bovine coronavirus (BCoV) | Bos taurus | USA | II | 1975 | (Kaye et al., 1975) |
| BCoV-like coronavirus | Ovis aries | Victoria | II | 1978 | (Tzipori et al., 1978) |
| BCoV-like coronavirus | Capra hircus | Spain | II | 1996 | (Munoz et al., 1996) |
| BCoV | Cows | II | 2010 | (Woo et al., 2010) | |
| Feline virus | |||||
| Feline coronavirus | Cats | I | 2010 | (Woo et al., 2010) | |
| Equine virus | |||||
| Equine coronavirus | Horses | II | 2010 | (Woo et al., 2010) | |
| Avian Viruses | |||||
| Infectious bronchitis virus (IBV) | Gallus gallus | III | 2007 | (Cavanagh, 2007) | |
| Pigeon coronavirus-IBV | Columbia livia | III | 2007 | (Cavanagh, 2007) | |
| Duck coronavirus | Anas platyrhynchos | III | 2007 | (Cavanagh, 2007) | |
| Goose coronavirus | Anser anser | III | 2007 | (Cavanagh, 2007) | |
| Turkey coronavirus | Meleagris gallopavo | III | 2007 | (Cavanagh, 2007) | |
| Pheasant coronavirus | Phasianus colchicus | III | 2007 | (Cavanagh, 2007) | |
| BuCoV-HKU11 | Bulbul | Hong Kong | IV | 2009 | (Woo et al., 2012, Woo et al., 2009) |
| ThCoV-HKU12 | Thrush | Hong Kong | IV | 2009 | (Woo et al., 2012, Woo et al., 2009) |
| MunCoV-HKU13 | Munia | Hong Kong | IV | 2009 | (Woo et al., 2012, Woo et al., 2009) |
| Wigeon coronavirus | Anas americana | Hong Kong | IV | 2011 | (Chu et al., 2011) |
| Heron coronavirus | Ardeola bacchus/speciosa | Cambodia | IV | 2011 | (Chu et al., 2011) |
| Duck coronavirus | Dendrocygna javanica | Cambodia | III | 2011 | (Chu et al., 2011) |
| Duck coronavirus | Aythya fuligula | Hong Kong | III | 2011 | (Chu et al., 2011) |
| Platalea minor | Hong Kong | IV | 2011 | (Chu et al., 2011) | |
| Phalacrocorax carbo | Hong Kong | IV | 2011 | (Chu et al., 2011) | |
| Heron coronavirus | Ardea cinerea | Hong Kong | IV | 2011 | (Chu et al., 2011) |
| Coronavirus | Anas acuta, Anas penelope, Anas clypeata, Anas crecca | Hong Kong | III & IV | 2011 | (Chu et al., 2011) |
| WiCoV-HKU20 | Wigeon | China | IV | 2012 | (Woo et al., 2012) |
| WECoV-HKU16 | China | IV | 2012 | (Woo et al., 2012) | |
| SpCoV-HKU17 | Sparrow | China | IV | 2012 | (Woo et al., 2012) |
| NHCoV-HKU19 | Night heron | China | IV | 2012 | (Woo et al., 2012) |
| CMCoV-HKU21 | Common moorhen | China | IV | 2012 | (Woo et al., 2012) |
| IBV | Poultry birds | China | IV | 2016 | (Zhao et al., 2016) |
| AvCoV-pigeon-67T | Pegion | Brazil | III | 2018 | (Martini et al., 2018) |
| AvCoV | Anseriformes Columbiformes | Norway | 2018 | (Miłek and Domańska, 2018) | |
| Charadriiformes | USA | III | 2018 | (Miłek and Domańska, 2018) | |
| IBV-like | Anseriformes Charadriiformes | Northern England, Sweden, Poland | 2018 | (Miłek and Domańska, 2018) | |
| Anseriformes | Korea, Madagascar, Beringia, Sweden | III | 2018 | (Miłek and Domańska, 2018) | |
| Passeriformes | Hong Kong | IV | 2018 | (Miłek and Domańska, 2018) | |
| IBV-like | Columbiformes | Brazil | |||
| Anseriformes Ciconiiformes Pelecaniformes | Hong Kong, Cambodia | III & IV | 2018 | (Miłek and Domańska, 2018) | |
| Aquatic animals | |||||
| BWCoV-SW1 | Beluga whale | USA | III | 2008 | (Mihindukulasuriya et al., 2008) |
2.2. Human coronaviruses (HCoVs) strains
Most of the HCoV infections are mild although epidemics of SARS-CoV and MERS-CoV (Table 3) in the last two decades collectively resulted in 10,000 cases with mortality rates of 10% and 37% respectively (de Groot et al., 2013, Ksiazek et al., 2003, WHO, 2019, WHO, 2003). Prior to the SARS-CoV outbreak, one alphacoronavirus HCoV-229E (which infects humans and bats) and one betacoronavirus HCoV-OC43 (which infects humans and cattle) were known. Both of these coronaviruses were identified in the 1960s and cause mild symptoms, such as colds, with the exception of infants, the elderly and the immune-compromised (Hamre et al., 1967, Reed, 1984, van der Hoek et al., 2004). However, in 2004–2005 two additional HCoVs (NL63 and HKU1) were detected from clinical specimens (Table 1) (Abdul-rasool and Fielding, 2010, Cui et al., 2019, Graham et al., 2013, Pyrc et al., 2007, Zhao et al., 2008).
Table 3.
Key events of coronaviruses in humans.
| Year | Country | HCoV strain | Event description | Fatality rate (%) | Reference |
|---|---|---|---|---|---|
| 2003 | China | (SARS-CoV) | very severe pneumonia cases | 10 | (Cheng et al., 2007, Drosten et al., 2003, Ksiazek et al., 2003, Tsang et al., 2003) |
| 2012 | Saudi Arabia | (MERS-CoV) | 1st case of MERS | 37 | (de Groot et al., 2013) |
| 2019 | China | SARS-CoV-2 | 1st case of COVID-19 | 6.9 | (Chen et al., 2020, Huang et al., 2020, WHO, 2020, Wu et al., 2020b) |
3. SARS-CoV-2 genome sequence identity and wildlife
The SARS-CoV-2 caused the deadly infections and spread all over the world at a rapid rate (Chen et al., 2020, Li et al., 2020, Peng et al., 2020, Zhou et al., 2020), so finding the source of SARS-CoV-2 and its main intermediate host is crucial. It is assumed that SARS-CoV-2 has originated from bats due to its close phylogenetic relationship with beta-genus lineage B bat SARS-CoV (Wan et al., 2020). Angiotensin-converting enzyme 2 (ACE2), the host cell receptor for SARS, has recently been demonstrated in mediating SARS-CoV-2 infection (Chai et al., 2020). Moreover, SARS-CoV-2 probably recognizes ACE2 receptors from a variety of animals such as palm civets (Wan et al., 2020).
The genome nucleotide sequence identity (96.2%) between SARS-CoV-2 and bat CoV (BatCoV RaTG13) in Rhinolophus affinis, indicating that R. affinis bats could be the natural source of the precursors of SARS-CoV-2 (Zhou et al., 2020). However, we may assume that there may be other intermediate hosts for transmission of virus between bats and humans due to differences in sequence identity. Examination of more than 1000 metagenomic samples from pangolins indicated that about 70% contained β CoV (Liu et al., 2019). One coronavirus from the same study had a genomic similarity of 99% with the SARS-CoV-2 genome. This suggested that the pangolin could be a possible intermediate host of SARS-CoV-2 (Wahba et al., 2020). It must be noted here that the pangolins in the study were from smuggling operations, suggesting that the animals were likely held in unhygienic conditions characteristic of illegal wildlife trade. SARS-CoV-2 displayed the structure with the “spike protein” in the membrane envelope (Li, 2017) and this spike glycoprotein (S) from CoV may bind to the host receptors to ease viral entry into target cells and is a primary determinant of cell tropism and pathogenesis (Belouzard et al., 2012, Hantak et al., 2019). There are four amino acid variations of S protein between SARS-CoV-2 and SARS-CoV. SARS-CoV-2 can bind to the ACE2 receptor from the cells from human and other animals (for human-human transmission and cross-species transmission), however, it cannot bind to the cells without ACE2 (Chai et al., 2020, Wan et al., 2020, Zhou et al., 2020). The high affinity between ACE2 and SARS-CoV-2 spike glycoprotein (S) proposed that the people with higher expression of ACE2 might be more vulnerable to SARS-CoV-2 (Guy et al., 2005, Zhao et al., 2020). The trans-membrane protease serine 2 (TMPRSS2) also contributed to the spike glycoprotein (S) priming of SARS-CoV-2, indicating that management of SARS-CoV-2 may be possible through TMPRSS2 inhibitor (Hoffmann et al., 2020, Peng et al., 2020).
4. Evolutionary process: Emergence of novel viruses
Coronaviruses are diverse and are capable of evolving rapidly. Viral emergence may be recognized as a two-step process: (i) introduction of the virus into one of more novel hosts and (ii) adaptation of virus into the novel host species. Rapid evolution of viruses may occur depending on the evolutionary potential of the virus and environmental conditions (Morse, 1991). Knowledge of the origin of virus is crucial because viruses mutate quickly and unpredictably making viral evolution difficult or impossible to predict (Morse, 1997). This is also the case with SARS-CoV-2. New viruses generally arise from closely related pre-existing viruses. Among emerging infections 75% are zoonotic, originating principally from wildlife (Cunningham, 2005, Daszak et al., 2000). Of the 1415 known human pathogens (including 217 viruses and prions), 61% are zoonotic (Taylor et al., 2001). Most of the major viral disease outbreaks were caused by RNA viruses with higher mutation rates compared to other kinds of microorganisms (Chan et al., 2013, Cheng et al., 2007). HCoVs have long genomes encoding diverse accessory proteins which may stimulate virus adaptation to specific hosts and suppression of immune responses by hosts (Forni et al., 2017). Understanding of evolutionary processes is very important in determining how an existing virus that usually infects animals would be able to cross over into humans (Morse, 1991).
4.1. Natural reservoirs of coronaviruses and their cross-species transmission
Comprehensive investigations are required about the natural zoonotic reservoirs of coronaviruses. It is likely that newly emerging coronaviruses originate from strains circulating in these reservoirs from years. They have the potential of cross-species transmission within domestic or captive, wild animals. Before spillover to humans these coronaviruses most likely adapted to intermediate hosts where there is no direct contact of humans to these natural zoonotic reservoirs. Furthermore, the wild animal wet markets provide enabling conditions for cross- species spillover of these coronaviruses and finally to infect the humans (Bolles et al., 2011).
4.1.1. Coronaviruses transmission from wild animals to humans
Animal coronaviruses have been identified since 1930s and different CoVs have been isolated from various infected animals before the first isolation of HCoV-229E from a patient (Ye et al., 2020). So far, various coronaviruses have been reported in mammalian and avian hosts (Table 2). Most widely studied and of common occurrence are coronaviruses reported in chickens, turkeys, cats, dogs, swine, cattle, mice, rats, rabbits, and humans (Guan et al., 2003, Swayne et al., 2004). The SARS-CoV was detected for the first time in animals during a study conducted in a live animal market (Guan et al., 2003). Viruses closely related to SARS-CoV were found in three different animal species, masked palm civet, raccoon dog and Chinese ferret badger in Shenzhen market, China (Wang et al., 2006). Subsequently, the detection of SARS-CoV infection in humans working at wild animal wet markets where civets were sold proposed that civets perhaps the source of human infections (Wang et al., 2018).
Coronaviruses are found in an exceptionally wide distribution in wild mammals including bats (Table 2) (Lau et al., 2010, Vijgen et al., 2005). Compared to human coronaviruses, the genetic diversity of bat coronaviruses is so huge that it is speculated that bats are the hosts of all mammalian coronaviruses (Drexler et al., 2010, Vijaykrishna et al., 2007) and this huge genetic diversity offers a chance for the emergence of novel animal and human coronaviruses. The occurrence of highly pathogenic coronaviruses such as SARS-CoV, MERS-CoV and severe acute diarrhea syndrome coronavirus (SADS-CoV) which shattered the livestock industry by infecting pigs (Zhou et al., 2018) were caused by coronaviruses of bat origin. Thus, the next viral disease events may be due to bats coronaviruses, changing ecological and epidemiological situations. Three HCoVs outbreaks (Table 3) including COVID-19 leading us that there is urgent need to study the reasons of coronaviruses emergence to avoid future outbreaks. Out of thirty eight, twenty two bat-borne viruses are found in China (Fan et al., 2019). Consequently, due to Chinese food culture (Fan et al., 2019) it is assumed that bat-borne CoVs can re-emerge and hotspot of next disease outbreak may be China.
The natural recombination between distantly related African bats (Triaenops afer and Hipposideros sp.) CoVs created NL63-like virus perhaps responsible for HCoV-NL63. Consequently, the interspecies recombination may perhaps contribute in CoVs evolution and emergence of novel coronaviruses with zoonotic potential with different genotypes for instance three genotypes in HCoV-HKU1 (Tao et al., 2017). In one longitudinal study of bat coronaviruses in a cave system from Yunan China, researchers found all known SARS-like coronaviruses (SARSr-CoVs) over a five year period (Cui et al., 2019). However, the direct progenitors of SARS-CoV have never been isolated from wild bats, suggesting that recombination can be occurring elsewhere. As of now, a lot of the evidence points towards wild animal wet markets where many species of animals ranging from mammals to fish are sold. These markets have been important in re-assortments of other types of viruses (e.g. well documented in the emergence of highly pathogenic avian influenza H5N1 subtype in mixed poultry markets) (Takekawa et al., 2010). Since, wild animal markets are not well studied; there must be a concerted effort to study existing CoV diversity and their variants in wild animals markets.
Furthermore, a wide variety of CoVs have been observed in birds (Table 2). However, birds are infected by Gamma and Deltacoronaviruses that are not adapted to mammalian hosts (Cui et al., 2019). Thus, birds can be precluded as possible hosts of progenitors of coronaviruses of human importance. Additionally molecular epidemiological research studies in bats, civets, pangolins or other forest mammals and birds, and complete genome sequencing will contribute to know the CoVs diversity and their evolutionary processes and histories to protect global health (Woo et al., 2009).
It is crucial to recognize animals that are susceptible to these coronaviruses as there are vast numbers of live animals kept and sold in wild animal wet markets (Wang et al., 2006). In the past, more than 10 mammalian species were proven susceptible to the SARS-CoV or related viral infections (Wang et al., 2006) including rats. Rats were involved in the spread of SARS-CoV during SARS outbreaks in Hong Kong (Ng, 2003), however, further studies are required to elucidate their potential role in CoVs transmission. Domestic poultry were excluded as reservoirs or linked with the spreading of SARS-CoV in the animal markets because avian species were not susceptible to SARS-CoV infection (Swayne et al., 2004, Weingartl et al., 2004).
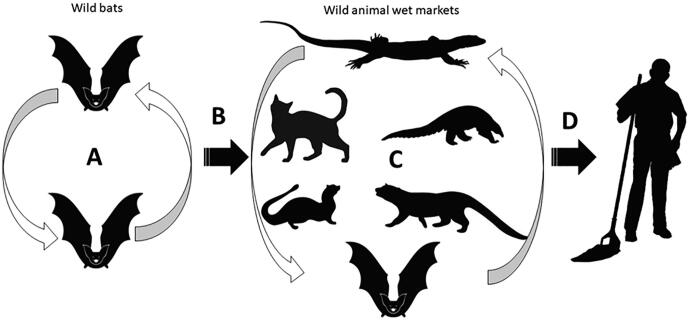
Diverse groups of wild animals are collected in wild animal wet markets across Asia, particularly southeastern and eastern Asia often transported over large distances and crammed together into cages. These animals are stressed and immunosuppressed and shed infective stages of pathogens from existing infections, while also being increasingly vulnerable to new infections from within the wet market system (Cunningham, 2020). Co-infection of more than one variant of a given virus increases the chances of reassortment of viruses that leads to the emergence of novel genotypes (Cunningham, 2020, Takekawa et al., 2010)(Fig. 1). The extent to which this has happened in coronaviruses is not clear, but this must have happened before the CoVs could adapt to other mammals in the wild animal wet market systems. Subsequent to this, the likely exposure of humans in the wild animal wet markets allowed the novel genotypes of CoVs to make the ‘jump’ and adapt to humans (Cui et al., 2019) like SARS-CoV-2.
Fig. 1.
Emergence of novel coronaviruses. A) Circulation of coronaviruses in wild bats. All coronavirus genotypes have been isolated from bats in Southeast Asia; B) Spillover from bats to wild animal wet markets through bats captured for sale. C) Reassortment and adaptation of diverse coronaviruses to wider range of wild animal species, including civets, mongooses or pangolins. D) Spillover and adaptation of novel coronavirus to humans associated with wild animal wet markets.
4.1.2. History of interspecies transmission of HCoVs
Human coronaviruses (Table 1) have a strong history of interspecies transmission and are of zoonotic animal origin (Rest and Mindell, 2003). HCoVs including SARS, MERS, OC43, 229E and new SARS-CoV-2 all evolved through likely host shifts from animals to humans and created coronavirus disease epidemics and pandemics. It is evident that OC43 had a high sequence similarity with bovine Betacoronavirus (BCoV) whereas 229E probably emerged from a bat Alphacoronavirus (Bolles et al., 2011, Kocherhans et al., 2001). However, investigations are still needed to confirm the origins of HCoV-NL63 and HCoV-HKU1. SARS, MERS and novel SARS-CoV-2 most likely have bat origin (Bolles et al., 2011, Song et al., 2005).
5. Anthropogenic factors that affect emergence
Ecological changes due to agricultural or economic development are among the most common known factors governing the emergence of novel viruses and their outbreak. Climate change deforestation and industrialization are main factors that are changing the habitats of wildlife and consequently permitting the displacement and mixing of various hosts that harbor coronaviruses and other pathogens (Chan et al., 2013). Urbanization poses significant threats to the ecology and population dynamics of animals in different natural ecosystems. Through urbanization, humans are encroaching further into the natural habitats of animals and creating overlapping areas where there are more chances of exchange of diseases between humans and wildlife. Humans are responsible for spreading pathogens into new areas or new populations through the travel and trade (Wu et al., 2017). Through the advances in travel and transportation technology, we are now globally connected and trading in distant regions to boost economies and consequently spreading viruses, vectors and hosts in distant regions (Wu et al., 2017). Through these different factors we are enhancing the intermingling of diverse viral genotypes in various host populations which affect the genetic make-up and immune responses of host populations. Ultimately, greater exposure to novel pathogens could lead to emergence and re-emergence of diseases sometimes in disparate geographic regions (Xu et al., 2012).
Human coronaviruses SARS-CoV, MERS-CoV and now the novel SARS-CoV-2 which is the cause of COVID-19 pandemic is believed to have originated in wild animals and bats are most likely the natural reservoir of these viruses (Forni et al., 2017). Due to current menace and future threats of viral diseases scientist are now more interested in the ecology and evolution of coronaviruses (Graham et al., 2013) to see the sink and sources in life cycle of coronaviruses for their management options and to stave off coronaviruses spillover in human populations.
Coronaviruses spread from bats to humans involves some conditions for instance dispersion of reservoir hosts, viral contamination of the hosts, exposure of recipient host to reservoir and susceptibility of recipient hosts. The overlapping areas of reservoir and recipient hosts describes the probable zones of high risk of transmission and infections of viruses in recipient hosts (Plowright et al., 2014), and this was the case in outbreak of novel coronavirus SARS-CoV-2 in live animal market of Wuhan, China, where live wild animals of all kinds including bats, pangolin and palm civets were on sale (Plowright et al., 2014).
6. Management of coronaviruses outbreaks
Little is known of the epidemiology of human infections with coronaviruses. Here, we propose management intervention in five major areas: i) mitigating or baning wildlife trade to avoid the exposure of coronaviruses to new hosts and new places ii) overlapping areas between domestic animals and wildlife due to deforestation and urbanization iii) managing or closing the wild animal wet markets where vast variety of wild and domestic animals are on sale which provide ideal condition for viruses reassortments and evolution of novel coronaviruses iv) managing livestock which may reduce the probability of exposure to viral diseases v) control strategies to reduce the chances of viruses spillover from animals to humans.
Moreover, investigations and continuous surveillance for new coronavirus pathogens in natural ecosystems, overlapping areas of wildlife and livestock, and especially in wild animal wet markets are necessary to stave off the concurrence of new viruses’ evolution and emergence and their spill over to humans.
7. Conclusion
Since the history of two HCoVs pandemics (SARS-CoV and MERS-CoV), many evidences showed that both have a bat origin and are spread to humans via intermediate hosts. Coronaviruses originating in bats most likely spilled over into wild animal wet markets where variant circulated and reasserted to produce the SARS-CoV-2 virus. Consequently, the role of intermediate hosts is very important to understand mechanisms of spillover and subsequent adaptation of viruses to humans. At present, COVID-19 has been spread worldwide. Conditions that increase the interactions at the interface between humans and wildlife such as wet markets exist throughout Southeast and East Asia, including China, constituting a risk for the emergence of novel coronaviruses. Determining transmission routes between wildlife and humans is a challenge and new molecular techniques are instrumental in improving our understanding emerging infectious diseases of significance to human’s health, domestic animal and wildlife. We suggest that the most effective way to prevent emergence of novel coronaviruses (or other viruses) is through a total ban of wild animal wet markets and associated wildlife trade, since contact between diverse viruses occur at these interfaces.
COVID-19 needs to be studied intensely as this is a global health threat. Careful surveillance is crucial due to the pandemic potential of SARS-CoV-2, to monitor future host adaptions, especially in wild animal wet markets, evolutionary process, infectivity, transmissibility, and pathogenicity. The knowledge of the origin, epidemiology, and duration of animal to humans and human to human transmission, and clinical spectrum of COVID-19 is critical for control strategies.
8. Learned lessons and recommendations
There is a gap in the research conducted on bats and a host of forest mammals, as reservoirs or intermediate hosts of coronaviruses. Future research should investigate viral loads, genetic variation and potential for reassortments, especially in countries where there is direct contact between humans and bats.
It is clear that big funds and large number of studies were dedicated for combating non-communicable diseases such as cancer, which claim a predictable number of lives via a predictable trend. But, after COVID-19, more money and research should be devoted to study unpredictable emerging pandemic diseases. As a result, inter-country research grants should be made available to research teams worldwide.
COVID-19 destroyed the fake sense of biosecurity, which was established in many countries in the past century. Consequently, this should trigger a comprehensive review process of the current health systems and emergency plans.
Ethical approval
Ethical approval is not required.
Funding
UAE University by UPAR grant # G00002604.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
The authors thank the UAE University for its support.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Nighat Perveen, Email: 201790740@uaeu.ac.ae.
Sabir Bin Muzaffar, Email: s_muzaffar@uaeu.ac.ae.
Mohammad Ali Al-Deeb, Email: m_aldeeb@uaeu.ac.ae.
References
- Abdul-rasool S., Fielding B.C. Understanding Human Coronavirus HCoV-NL63. Open Virol. J. 2010;4:76–84. doi: 10.2174/1874357901004010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardos V., Schwänzer V., Pesko J. Identification of Tettnang Virus (‘ Possible Arbovirus ’) as Mouse Hepatitis Virus ’. Intervirology. 1980;13:275–283. doi: 10.1159/000149135. [DOI] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolles M., Donaldson E., Baric R. SARS-CoV and emergent coronaviruses: Viral determinants of interspecies transmission. Curr. Opin. Virol. 2011;1:624–634. doi: 10.1016/j.coviro.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Chai, X., Hu, L., Zhang, Y., Han, W., Lu, Z., Ke, A., 2020. Specific ACE2 Expression in Cholangiocytes May Cause Liver Damage After 2019-nCoV Infection. bioRxiv. https://doi.org/10.1101/2020.02.03.931766.
- Chan J.F.W., To K.K.W., Tse H., Jin D.Y., Yuen K.Y. Interspecies transmission and emergence of novel viruses: Lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng V.C.C., Lau S.K.P., Woo P.C.Y., Kwok Y.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Leung C.Y.H., Gilbert M., Joyner P.H., Ng E.M., Tse T.M., Guan Y., Peiris J.S.M., Poon L.L.M. Avian Coronavirus in Wild Aquatic Birds. J. Virol. 2011;85:12815–12820. doi: 10.1128/JVI.05838-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Poon L.L.M., Chan K.H., Chen H., Guan Y., Yuen K.Y., Peiris J.S.M. Communication Coronaviruses in bent-winged bats (Miniopterus spp.) J. Gen. Virol. 2006;87:2461–2466. doi: 10.1099/vir.0.82203-0. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Kallies R., Philipps H., Göpner G., Müller A., Eckerle I., Brünink S., Drosten C., Drexler F. Characterization of a Novel Betacoronavirus Related to Middle East Respiratory Syndrome Coronavirus in European Hedgehogs. J. Virol. 2014;88:717–724. doi: 10.1128/JVI.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, A., 2020. COVID-19: the wildlife facts [WWW Document]. ZSL Inst. Zool. Available online: https//www.zsl.org/blogs/science/covid-19-the-wildlife-facts (accessed 16 April 2020).
- Cunningham A.A. A walk on the wild side—emerging wildlife diseases. BMJ. 2005;331:1214–1215. doi: 10.1136/bmj.331.7527.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P., Cunningham A.A., Hyatt A.D. Emerging infectious diseases of wildlife - Threats to biodiversity and human health. Science (80-.) 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A.M., Galiano M., Gorbalenya A.E., Memish Z.A., Perlman S., Poon L.L.M., Snijder E.J., Stephens G.M., Woo P.C.Y., Zaki A.M., Zambon M., Ziebuhr J. Middle East Respiratory Syndrome Coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J. Virol. 2013;87:7790–7792. doi: 10.1128/jvi.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Gloza-rausch F., Corman V.M., Muth D., Goettsche M., Seebens A., Niedrig M., Pfefferle S., Yordanov S., Zhelyazkov L., Hermanns U., Vallo P., Lukashev A., Mu M.A., Herrler G., Drosten C. Genomic Characterization of Severe Acute Respiratory Syndrome-Related Coronavirus in European Bats and Classification of Coronaviruses Based on Partial RNA-Dependent RNA Polymerase Gene Sequences. J. Virol. 2010;84:11336–11349. doi: 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Günther S., Preiser W., Van Der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A. Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Esper F., Weibel C., Ferguson D., Landry M.L., Jeffrey S., Esper F., Weibel C., Ferguson D., Landry M.L., Kahn J.S. Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J. Infect. Dis. 2005;191:492–498. doi: 10.1086/428138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Zhao K., Shi Z., Zhou P. Bat Coronaviruses in China. Viruses. 2019;1–14 doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, A.R., Perlman, S., 2015. Coronaviruses : An Overview of Their Replication and Pathogenesis, in: In Coronaviruses. Humana Press, New York, NY, pp. 1–23. https://doi.org/10.1007/978-1-4939-2438-7. [DOI] [PMC free article] [PubMed]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A.M., Hartwig N.G., Bestebroer T.M., Niemeyer B., Jong J.C.D., Simon J.H., Osterhaus A.D.M.E. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. USA. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Wang N., Zhang W., Hu B., Li B., Zhang Y., Zhou J., Luo C., Yang X., Wu L., Wang B., Zhang Y., Li Z., Shi Z. Coexistence of multiple coronaviruses in several bat colonies in an abandoned mineshaft. Virol. Sin. 2016;31:31–40. doi: 10.1007/s12250-016-3713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya, A.E., Baker, S.C., Baric, R.S., Groot, R.J. De, Gulyaeva, A.A., Haagmans, B.L., Lauber, C., Leontovich, A.M., 2020. Severe acute respiratory syndrome-related coronavirus: The species and its viruses – a statement of the Coronavirus Study Group. bioRxiv. https://doi.org/10.1101/2020.02.07.937862.
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: Evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: Strategies to control emerging coronaviruses. Nat Rev Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., Butt K.M., Wong K.L., Chan K.W., Lim W., Shortridge K.F., Yuen K.Y., Peiris J.S.M., Poon L.L.M. Isolation and Characterization of Viruses Related to the SARS Coronavirus from Animals in southern China. Science. 2003;302(80):276–279. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Guy J.L., Lambert D.W., Warner F.J., Hooper N.M., Turner A.J. Membrane-associated zinc peptidase families: comparing ACE and ACE2. Biochim. Biophys. Acta. 2005;1751:2–8. doi: 10.1016/j.bbapap.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D., Kindig D.A., Mann J. Growth and Intracellular Development of a New Respiratory Virus. J. Virol. 1967;1:810–816. doi: 10.1128/jvi.1.4.810-816.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D., Procknow J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- Hantak M.P., Qing E., Earnest J.T. Tetraspanins: Architects of Viral Entry and Exit Platforms. J. Virol. 2019;93:1–8. doi: 10.1128/JVI.01429-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M., Kleine-Weber, H., Krüger, N., Müller, M., Drosten, C., Pöhlmann, S., 2020. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv. https://doi.org/10.1101/2020.01.31.929042.
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye H.S., Yarbrough W.B., Reed C.J. Calf Diarrhea Coronavirus. Lancet. 1975;306:509. doi: 10.1016/S0140-6736(75)90591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocherhans R., Bridgen A., Ackermann M., Tobler K. Completion of the Porcine Epidemic Diarrhoea Coronavirus (PEDV) Genome Sequence. Virus Genes. 2001;23:137–144. doi: 10.1023/a:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret E., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.-E., Humphrey C.D., Shieh W.-J., Guarn J. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lam, T.T., Shum, M.H., Zhu, H., Tong, Y., 2020. Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China. bioRxiv. https://doi.org/10.1101/2020.02.13.945485.
- Lau S.K.P., Li K.S.M., Tsang A.K.L., Shek C., Wang M., Choi G.K.Y., Guo R. Recent Transmission of a Novel Alphacoronavirus, Bat Coronavirus HKU10, from Leschenault ’ s Rousettes to Pomona Leaf-Nosed Bats: First Evidence of Interspecies Transmission of Coronavirus between Bats of Different Suborders. J. Virol. 2012;86:11906–11918. doi: 10.1128/JVI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Poon R.W.S., Wong B.H.L., Wang M., Huang Y., Xu H., Guo R., Li K.S.M., Gao K., Chan K., Zheng B., Woo P.C.Y., Yuen K., Irol J.V. Coexistence of Different Genotypes in the Same Bat and Serological Characterization of Rousettus Bat Coronavirus HKU9 Belonging to a Novel Betacoronavirus Subgroup. J. Virol. 2010;84:11385–11394. doi: 10.1128/JVI.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Wang M., Lam C.S.F., Xu H., Guo R., Chan K., Zheng B., Yuen K. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Tsang A.K.L., Fan R.Y.Y., Luk H.K.H. Discovery of a Novel Coronavirus, China Rattus Coronavirus HKU24, from Norway Rats Supports the Murine Origin of Betacoronavirus 1 and Has Implications for the Ancestor of Betacoronavirus Lineage A. J. Virol. 2015;89:3076–3092. doi: 10.1128/JVI.02420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol. 2017;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., You Z., Wang Q., Zhou Z., Qiu Y., Luo R., Ge X. The epidemic of 2019-novel-coronavirus (2019-nCoV) pneumonia and insights for emerging infectious diseases in the future. Microbes Infect. 2020;22:80–85. doi: 10.1016/j.micinf.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Wang W., Hao Z., Wang Z., Guo W., Guan Q., Wang M., Wang H., Zhou R., Li M., Tang P., Wu J., Holmes E.C., Zhang Y. Extensive diversity of coronaviruses in bats from China. Virology. 2017;507:1–10. doi: 10.1016/j.virol.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Chen W., Chen J.-P. Viral Metagenomics Revealed Sendai Virus and Coronavirus Infection of Malayan Pangolins. Viruses. 2019;979:1–15. doi: 10.3390/v11110979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini M.C., Caserta L.C., Santos M.M.A.B., Ana C., Barnabé S., Durães-carvalho R., Padilla M.A., Simão R.M., Rizotto L.S., Simas P.V.M., Bastos J.C.S., Cardoso T.C., Felippe P.A.N., Ferreira H.L., Arns C.W., Martini M.C., Caserta L.C., Santos M.M.A.B., Ana C., Barnabé S., Durães-carvalho R., Padilla M.A., Simão R.M., Rizotto L.S., Simas V.M., Bastos J.C.S., Cardoso T.C., Felippe P.A.N., Ferreira H.L. Avian coronavirus isolated from a pigeon sample induced clinical disease, tracheal ciliostasis, and a high humoral response in day-old chicks. Avian Pathol. 2018;47:286–293. doi: 10.1080/03079457.2018.1442557. [DOI] [PubMed] [Google Scholar]
- Mcintosh B.Y.K., Dees J.H., Becker W.B., Kapikian A.Z., Chanock R.M. Recovery in tracheal organ culture of novel viruscs from patients with respiratory disease. Proc. Proc. Natl. Acad. Sci. 1967;57:933–940. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihindukulasuriya K.A., Wu G., Leger J.S., Nordhausen R.W., Wang D. Identification of a Novel Coronavirus from a Beluga Whale by Using a Panviral Microarray. J. Virol. 2008;82:5084–5088. doi: 10.1128/JVI.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miłek J., Domańska K.B. Coronaviruses in avian species – review with focus on epidemiology and diagnosis in wild birds. J Vet Res. 2018;62:249–255. doi: 10.2478/jvetres-2018-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M., Taubenberger J.K., Fauci A.S. The 2009 H1N1 Pandemic Influenza Virus: What Next? MBio. 2010;1:1–5. doi: 10.1128/mBio.00211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse S.S. The Public Health Threat of Emerging Viral Disease. J. Nutr. 1997;127:951–957. doi: 10.1093/jn/127.5.951S. [DOI] [PubMed] [Google Scholar]
- Morse S.S. Emerging Viruses: Defining the Rules for Viral Traffic. Perspect. Biol. Med. 1991;34:387–409. doi: 10.1353/pbm.1991.0038. [DOI] [PubMed] [Google Scholar]
- Munoz M., Alvarez M., Lanza I., Carmenes P. Role of enteric pathogens in the aetiology of neonatal diarrhoea in lambs and goat kids in Spain. Epidemiol. Infect. 1996;117:203–211. doi: 10.1017/s0950268800001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.K.C. Possible role of an animal vector in the SARS outbreak at Amoy Gardens. Lancet. 2003;362:570–572. doi: 10.1016/S0140-6736(03)14121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obameso J.O., Li H., Jia H., Han M., Zhu S., Huang C., Zhao Y., Zhao M., Bai Y., Yuan F., Zhao H., Peng X., Xu W., Tan W., Zhao Y., Yuen K., Liu W.J., Lu L., Gao G.F. The persistent prevalence and evolution of cross-family recombinant coronavirus GCCDC1 among a bat population: a two-year follow-up. Sci. China Life Sci. 2017;60:1357–1363. doi: 10.1007/s11427-017-9263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paim F.C., Bowman A.S., Miller L., Feehan B.J., Marthaler D., Saif L.J., Vlasova A.N. Epidemiology of Deltacoronaviruses (δ -CoV) and Gammacoronaviruses (γ -CoV) in Wild Birds in the United States. Viruses. 2019;11:1–11. doi: 10.3390/v11100897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Xu X., Li Y., Cheng L., Zhou X., Ren B. Transmission routes of 2019-nCoV and controls in dental practice. Int. J. Oral Sci. 2020;12:1–6. doi: 10.1038/s41368-020-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright R.K., Eby P., Hudson P.J., Smith I.L., Westcott D., Bryden W.L., Middleton D., Reid P.A., McFarlane R.A., Martin G., Tabor G.M., Skerratt L.F., Anderson D.L., Crameri G., Quammen D., Jordan D., Freeman P., Wang L.F., Epstein J.H., Marsh G.A., Kung N.Y., McCallum H. Ecological dynamics of emerging bat virus spillover. Proc. R. Soc. Biol. Sci. 2014;282:1–9. doi: 10.1098/rspb.2014.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L.M., Chu D.K.W., Chan K.H., Wong O.K., Ellis T.M., Leung Y.H.C., Lau S.K.P., Woo P.C.Y., Suen K.Y., Yuen K.Y., Guan Y., Peiris J.S.M. Identification of a Novel Coronavirus in Bats. J. Virol. 2005;79:2001–2009. doi: 10.1128/JVI.79.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provacia, L.B.V., Smits, S.L., Martina, B.E., Raj, V.S., Doel, P. v.d., Amerongen, G. v., Hanneke Moorman-Roest, A.D.M.E.O., Haagmans, B.L., 2011. Enteric Coronavirus in Ferrets, the Netherlands. Emerg. Infect. Dis. 17, 2–3. https://doi.org/10.1056/NEJMoa. [DOI] [PMC free article] [PubMed]
- Pyrc, K., Berkhout, B., Hoek, L. Van Der, 2007. The Novel Human Coronaviruses NL63 and HKU1. J. Virol. 81, 3051–3057. https://doi.org/10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed]
- Reed S.E. The behaviour of recent isolates of human respiratory coronavirus in vitro and in volunteers: Evidence of heterogeneity among 229E-related strains. J. Med. Virol. 1984;13:179–192. doi: 10.1002/jmv.1890130208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rest J.S., Mindell D.P. SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect. Genet. Evol. 2003;3:219–225. doi: 10.1016/j.meegid.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman D., Whitley R., Hayden F. 4th ed. ASM Press; Washington: 2016. Clinical Virology. [Google Scholar]
- Song H., Tu C., Zhang G., Wang S., Zheng K., Lei L. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne D.E., Suarez D.L., Spackman E., Tumpey T.M., Beck J.R., Erdman D., Rollin P.E., Ksiazek T.G. Domestic Poultry and SARS Coronavirus, Southern China. Emerg. Infect. Dis. 2004;10:5–7. doi: 10.3201/eid1005.030827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takekawa J.Y., Prosser D.J., Newman S.H., Muzaffar S.B., Hill N.J., Yan B., Xiao X., Lei F., Li T., Schwarzbach S.E., Howell J.A. Victims and vectors: highly pathogenic avian influenza H5N1 and the ecology of wild birds. Avi. Biol. Res. 2010;3:1–23. doi: 10.3184/175815510X12737339356701. [DOI] [Google Scholar]
- Tao Y., Shi M., Chommanard C., Queen K., Zhang J. Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History. J. Virol. 2017;91:1–16. doi: 10.1128/JVI.01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L.H., Latham S.M., Woolhouse M.E.J. Risk factors for human disease emergence. Phil. Trans. R. Soc. Lond. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang K.W., Ho P.L., Ooi G.C., Yee W.K., Wang T., Chan-Yeung M., Lam W.K., Seto W.H., Yam L.Y., Cheung T.M., Wong P.C. A Cluster of Cases of Severe Acute Respiratory Syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1977–1985. doi: 10.1056/NEJMoă66. [DOI] [PubMed] [Google Scholar]
- Tzipori S., Smith M., Makin T., McCaughan C. Enteric coronavirus-like particles in sheep. Aust. Vet. 1978;54:320–321. doi: 10.1111/j.1751-0813.1978.tb02478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J.M., Wolthers K.C., Wertheim-Van Dillen P.M.E., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D., Smith G.J.D., Zhang J.X., Peiris J.S.M., Chen H., Guan Y. Evolutionary Insights into the Ecology of Coronaviruses. J. Virol. 2007;81:4012–4020. doi: 10.1128/JVI.02605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moe E., Thoelen I., Wollants E., Lemey P., Vandamme A., Van Ranst M. Complete Genomic Sequence of Human Coronavirus OC43: Molecular Clock Analysis Suggests a Relatively Recent Zoonotic Coronavirus Transmission Event. J. Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viviani L., Assael B.M., Kerem E., Ecfs T., Hn A. Impact of the A (H1N1) pandemic influenza (season 2009–2010) on patients with cystic fibrosis. J. Cyst. Fibros. 2011;10:370–376. doi: 10.1016/j.jcf.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Vlasova A.N., Halpin R., Wang S., Ghedin E., Spiro D.J., Saif L.J. Molecular characterization of a new species in the genus Alphacoronavirus associated with mink epizootic catarrhal gastroenteritis. J. Gen. Virol. 2011;92:1369–1379. doi: 10.1099/vir.0.025353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba, L., Jain, N., Fire, A.Z., Shoura, M.J., Artiles, K.L., McCoy, M.J., Jeong, D.-E., 2020. An Extensive Meta-Metagenomic Search Identifies SARS-CoV-2-Homologous Sequences in Pangolin Lung Viromes. mSphere 5, e00160-20. https://doi.org/10.1128/mSphere.00160-20. [DOI] [PMC free article] [PubMed]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. J. Virol. 2020;94:e00127–e220. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shi Z., Zhang S., Field H., Daszak P., Eaton B.T. Review of Bats and SARS. Emerg. Infect. Dis. 2006;12:1834–1840. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Yang S.L.X., Zhang H.H.Y., Guo H., Luo C., Miller M., Zhu G., Chmura A.A., Hagan E., Zhang J.Z.Y., Peter L.W., Shi D.Z. Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol. Sin. 2018;33:104–107. doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin X., Guo W., Zhou R., Wang M., Wang C., Ge S., Mei S., Li M., Shi M., Holmes E.C., Zhang Y. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 2015;474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl H.M., Copps J., Drebot M.A., Marszal P., Smith G., Gren J., Andonova M., Pasick J., Kitching P., Czub M. Susceptibility of Pigs and Chickens to SARS Coronavirus. Emerg. Infect. Dis. 2004;10:179–184. doi: 10.3201/eid1002.030677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, S.R., Leibowitz, J.L., 2011. Coronavirus Pathogenesis, 1st ed, Advances in Virus Research. Elsevier Inc. 81, 85-164. https://doi.org/10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed]
- WHO, 2020. Coronavirus disease (COVID-2019) situation reports, Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/ (accessed on 29 April 2020).
- WHO, 2019. Middle East respiratory syndrome coronavirus (MERS-CoV). MERS monthly summary, November, 2019. Available online: https//www.who.int/emergencies/mers-cov/en/ (accessed 02 April 2020).
- WHO, 2003. Severe Acute Respiratory Syndrome (SARS). Available online: https//www.who.int/csr/sars/en/ (accessed 02 April 2020).
- Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K. Coronavirus Genomics and Bioinformatics Analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, P.C.Y., Lau, S.K.P., Chu, C. -m., Chan, K. -h., Tsoi, H. -w., Huang, Y., Wong, B.H.L., Poon, R.W.S., Cai, J.J., Luk, W. -k., Poon, L.L.M., Wong, S.S.Y., Guan, Y., Peiris, J.S.M., Yuen, K. -y., 2005. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 79, 884–895. https://doi.org/10.1128/jvi.79.2.884-895.2005. [DOI] [PMC free article] [PubMed]
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lai K.K.Y., Huang Y., Lee P., Luk G.S.M., Dyrting K.C., Chan K., Yuen K. Comparative Analysis of Complete Genome Sequences of Three Avian Coronaviruses Reveals a Novel Group 3c Coronavirus. J. Virol. 2009;83:908–917. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N., Bai R., Teng J.L.L., Tsang C.C.C., Wang M., Zheng B., Chan K., Yuen K. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Li K.S.M., Poon R.W.S., Wong B.H.L., Tsoi H., Yip B.C.K., Huang Y., Chan K., Yuen K. Molecular diversity of coronaviruses in bats. Virology. 2006;351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Tsang C., Lau C.C.Y., Wong P., Chow F.W.N., Fong J.Y.H., Yuen K. Coronavirus HKU15 in respiratory tract of pigs and fi rst discovery of coronavirus quasispecies in 5 ′ -untranslated region Animal surveillance. Emerg. Microbes Infect. 2017;6:e53. doi: 10.1038/emi.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Wang M., Lau S.K.P., Xu H., Poon R.W.S., Guo R., Wong B.H.L., Gao K., Tsoi H., Huang Y., Li K.S.M., Lam C.S.F., Chan K., Zheng B., Yuen K. Comparative Analysis of Twelve Genomes of Three Novel Group 2c and Group 2d Coronaviruses Reveals Unique Group and Subgroup Features. J. Virol. 2007;81:1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Tao Z., Tian J., Pei Y., Yuan M., Zhang Y., Dai F. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Perrings C., Kinzig A., Collins J.P., Minteer B.A., Daszak P. Economic growth, urbanization, globalization, and the risks of emerging infectious diseases in China: A review. Ambio. 2017;46:18–29. doi: 10.1007/s13280-016-0809-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Yang L., Ren X., He G., Zhang J., Yang J., Qian Z., Dong J., Sun L., Zhu Y., Du J., Yang F., Zhang S. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 2016;10:609–620. doi: 10.1038/ismej.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Davis C.T., Christman M.C., Rivailler P., Zhong H., Donis R.O., Lu G. Evolutionary history and phylodynamics of influenza A and B neuraminidase (NA) genes inferred from large-scale sequence analyses. PLoS One. 2012;7:e38665. doi: 10.1371/journal.pone.0038665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Zhang F., Yang W., Jiang T., Lu G., He B., Li X., Hu T., Chen G., Feng Y., Zhang Y., Fan Q., Feng J., Zhang H. Detection and characterization of diverse alpha- and betacoronaviruses from bats in China. Virol. Sin. 2016;31:69–77. doi: 10.1007/s12250-016-3727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., Yuan S., Yuen K., Fung S., Chan C., Jin D. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020;16:6–8. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wunderink R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2017;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A. Novel coronavirus—Saudi Arabia: human isolate. Int Soc Infect Dis. 2012:2012–2109. [Google Scholar]
- Zhang Q., Hu R., Tang X., Chen H., Wu B. Occurrence and investigation of enteric viral infections in pigs with diarrhea in China. Arch Virol. 2013;158:1631–1636. doi: 10.1007/s00705-013-1659-x. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Li S., Xue F., Zou Y., Chen C., Bartlam M., Rao Z. Structure of the Main Protease from a Global Infectious Human. J. Virol. 2008;82:8647–8655. doi: 10.1128/JVI.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhang H., Zhao J., Zhong Q., Jin J., Zhang G. Evolution of infectious bronchitis virus in China over the past two decades. J. Virol. 2016;96:1566–1574. doi: 10.1099/jgv.0.000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. 2020;202:756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X., Shi W., Zhang W., Zhu Y., Zhang Y., Mani S., Zheng X., Li B., Li J., Guo H., Pei G., An X., Chen J., Zhou L., Mai K., Wu Z., Li D., Anderson D.E., Zhang L., Li S., Mi Z., He T., Cong F., Guo P., Huang R., Luo Y., Liu X., Chen J., Huang Y., Sun Q. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X., Wang X., Hu B., Zhang L., Zhang W., Guo H., Jiang R., Liu M., Chen Y., Shen X., Wang X., Zhan F., Wang Y., Xiao G., Shi Z. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P. A Novel Coronavirus from Patients with Pneumonia in China, 2019. New Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]