Abstract
Background: The current treatment of Alzheimer's disease (AD) is far from adequate. AD can be treated by inhibiting either β-amyloid protein deposition or acetylcholinesterase enzyme activity. Many treatments for AD are directed at these 2 targets. In the present study, the phytoconstituents of Carissa carandas chloroform leaf extract were identified by gas chromatography-MS/MS analysis, and in silico molecular docking studies were performed to evaluate their potential against AD.
Objectives: The present study aimed to identify the possible anti-Alzheimer's activity of novel phytoconstituents isolated from C carandas.
Methods: The powdered leafy material was subjected to successive Soxhlet extraction using 3 different solvents: n-hexane, chloroform, and methanol. The chloroform extract was subjected to gas chromatography-MS/MS analysis, and the observed chromatogram revealed the presence of 48 chemical constituents. Among them, 42 new phytoconstituents are reported in this plant for the first time. The gas chromatography-MS/MS–identified phytoconstituents were evaluated by iGEMDOCK software against AD targets of β-amyloid fibril (protein data bank ID: 2LMN) and recombinant human acetylcholinesterase (protein data bank ID: 3LII) ligands, and their anti-AD potential were compared with those of known inhibitors of galantamine and curcumin.
Results: On the basis of results from both docking assays, the 5 compounds with the highest docking energy were further analyzed using in silico admetSAR web portal modeling for the evaluation of parameters such as intestinal absorption, blood-brain barrier permeation, carcinogenicity, and acute oral toxicity.
Conclusions: The chloroform leaf extract of C carandas was found to contain constituents that have affinities for the 2 targets tested; that is, amyloid β and acetylcholinesterase. The best docking scores were found for 7 compounds: 1-heneicosanol; N-nonadecanol-1; cholesta-4,6-dien-3-ol, (3beta); di-n-octyl phthalate; 7,9-di-tert-butyl-1-oxaspiro(4,5)deca-6,9-diene-2,8-dione; 6-undecyl-5,6-dihydro-2H-pyran-2-one, and phenol, 2,4-di-t-butyl-6-nitro compounds, and these compounds were therefore suggested to be promising anti-AD lead compounds. Further, the target leads were subjected to ligplot analysis for their 2-dimensional representation of hydrogen bonding and hydrophobic interactions. Thus, the results obtained from the in silico study of C carandas leaf extract using these computational approaches indicate the presence of phytoconstituents that have affinities for the selected 2 targets of AD. (Curr Ther Res Clin Exp. 2020; 81:XXX–XXX)
Key words: Acetylcholinesterase, anti-Alzheimer's activity, amyloid β, Carissa carandas
Graphical abstract
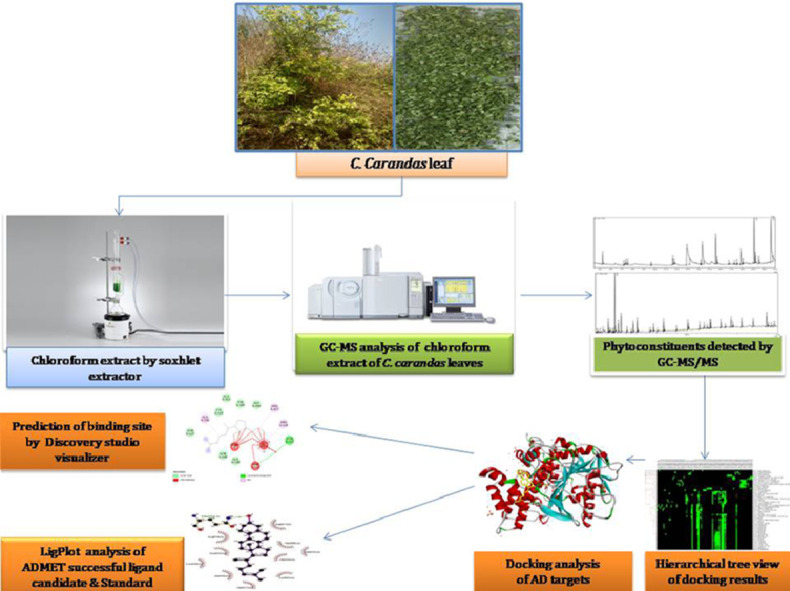
Introduction
Carissa carandas Linn belongs to the family of Apocynaceae, which represents the 89 species found in India. It is a large dichotomous, evergreen branched shrub with a short stem and a strong thorn in pairs. These species grow usually up to 3 to 5 m in height; they are stout, woody, and sometimes climb to the top of tall trees.1 Different plant parts such as roots, leaves, and berries have been used in traditional medicine for treating various diseases such as sore throat, diarrhea, stomachache, anorexia, burning sensation, syphilitic pain, mouth ulcer, fever, epilepsy, and scabies, and other well-known therapeutic uses of this plant include antibacterial, analgesic, anti-inflammatory, antipyretic, antioxidant, hepatoprotective, adaptogenic, antidiabetic, antihyperlipidemic, and anticonvulsant activities; in vitro DNA damage inhibition; and histamine-releasing activity.2, 3, 4, 5, 6, 7, 8, 9, 10 A literature survey revealed that Carissa species have ethnomedicinal uses in South Africa because of their effects on the central nervous system. In particular, the leaf part of C carandas Linn shows sedative, anticonvulsant, and neuropharmacological effects.11, 12, 13, 14 Hence, in silico approaches were used herewith to identify the chemical constituents present in C carandas that have amyloid-β (Aβ) and acetylcholinesterase (AChE) inhibition effects.
Alzheimer's disease (AD) is a progressive neurodegenerative disease and characterized by cognitive impairment, psychosis, hyperactivity, aggressive behavior, and depression in patients.15 Late-onset or sporadic AD is currently cured by preventing AChE or Aβ, both of which have serious side effects.16, 17, 18 Histopathologic examination of brain tissues exposed to enzymes that produce and utilize acetylcholine (Ach) show considerable reduction in the level of Ach in patients with AD.19,20 AChE inhibitors such as donepezil, rivastigmine, and galantamine block the effects of AChE in the synapse, which causes side effects such as nausea, vomiting, anorexia, diarrhea, and bradycardia.21 Memantine is an N-methyl D-aspartate receptor antagonist approved in Europe and the United States for the treatment of moderate-to-severe AD, but it has side effects of headache, insomnia, constipation, fatigue, dizziness, auditory hallucinations, and mild allergies.22,23
There is a high demand to develop new anti-AD drugs. In recent years, the use of medicinal plants in the treatment of several diseases has gained considerable importance. Plants are considered among the main sources of therapeutically active compounds. According to the World Health Organization, 85% to 90% of the world population relies on traditional medicine to meet their health care needs.24 It is a well-known fact that plants produce secondary metabolites that show therapeutic activity.25 The secondary metabolites from Opuntia ficus-indica cladode exhibited neuroprotective effects in rats with aluminum chloride-induced neurotoxicity.26 In a previous study, in silico investigation of anti-AD compounds present in methanolic extract of Neolamarckia cadamba bark using gas chromatography (GC)-MS/MS revealed the probable lead compounds against the AD targets of Aβ and AChE.27 The main objective of this investigation was to analyze and assess the secondary metabolites from the chloroform extract of C carandas leaves for their anti-AD potential by performing in silico molecular docking studies against Aβ and AChE targets and comparing the current GC-MS/MS data with the previously reported chemical constituents of the same plant. Thus, our research will enable to identify novel chemical constituents of more potent, possible anti-AD lead molecules than the existing compounds.
Materials and Methods
Chemicals and instrumentation
All solvents were purchased from Loba Chemie Pvt Ltd, Mumbai, India. These solvents were of analytical grade and used without any purification. Chromatographic analysis was performed on a GCMS-QP2010 Plus (Shimadzu, Toshvin Analytical Pvt. Ltd., Mumbai, India) GC-MS/MS system.
Collection and authentication of plant material
Fresh leaves of C carandas were collected in February 2018 from hills of Amarkantak, Anuppur district, Madhya Pradesh, India. The collected leaves were washed with running tap water to remove sticky sand particles and other impurities and finally washed with sterile distilled water. The plant material was pulverized to form powder and then stored it in an airtight container. The plant material was authenticated by a taxonomist, and a herbarium specimen sample voucher (DOB/Herb/Apoc-01) has been deposited at the Department of Botany, Indira Gandhi National Tribal University, Amarkantak, India.
Soxhlet extraction
The powdered leafy material (100 g/250 mL) was extracted on a Soxhlet extractor by using 3 different solvents successively, namely n-hexane, chloroform, and methanol, at 50°C to 75°C for 12 hours to extract nonpolar to polar compounds. The collected extracts were dried in a water bath at 60°C and stored in a refrigerator at 4°C up to 3 months.
GC-MS/MS instrument conditions
In GC-MS/MS analysis, the injection temperature was maintained at 280°C, the analytical injection volume of the solution was 2.0 µL, and the split ratio was 1:3. The oven temperature program was set initially at 50°C (maintained for 1 minute) and increased at the rate of 7.5°C/min up to 300°C for 10 minutes. Highly pure helium was used as the inert gas, and the temperature of the ion source was maintained at 250°C; the interface temperature was set at 300°C. The mass spectrometer was used for quantitative analysis in the acquisition mode. Mass spectral data were obtained for the entire range of m/z 50 to 1000 to obtain the spectrum of the target analyte fragments.
Identification of new compounds from C carandas
The phytoconstituents currently detected by GC-MS/MS were compared with the reported phytoconstituents of C carandas.5,28, 29, 30, 31, 32, 33
Hardware specification
Molecular docking studies were performed using an Intel Core operating system (Intel Corp, Santa Clara, California) running Windows 10 (Microsoft Corp, Redmond, Washington).
Software specifications
The receptor protein structure is available at https://www.rcsb.org. Receptors were screened by PyRx 0.8 software downloaded from https://pyrx.software.informer.com. The structures of the compounds detected by GC-MS/MS were drawn using a trial licensed version of Chemdraw 19.1 (PerkinElmer, Waltham, Massachusetts). iGEMDOCK V2.1 available at http://gemdock.life.nctu.edu.tw was used for molecular docking studies. Docking verifications were performed by Discovery studio visualizer V20 from https://www.3dsbiovia.com. The online translation of SMILES was performed by https://cactus.nci.nih.gov/. In silico studies were carried out by the admetSAR web portal at www.lmmd.ecust.edu.cn, and the LigPlot analysis was performed by LigPlot + V 2.1 free academic license version acquired from European Bioinformatics Institute (Hinxton, United Kingdom).
Preparation of the receptors
The structural receptors of Aβ (protein data bank ID: 2LMN) and AChE (protein data bank ID: 3LII) structures were obtained from the protein data bank. All nonprotein molecules and all other alternative atomic sites were separated from the protein receptor structures by using the PyRx 0.8 software.34
Preparation of ligands
The ligand preparation process consists of various steps such as conversions, corrections, and variations of the structures, elimination, and lead optimization. Two-dimensional structures of the ligands and GC-MS/MS tentatively identified compounds were drawn using Chemdraw and then converted to 3-dimensional mol format.35 Conversion of SMILES structure from 3-dimensional format was performed using the cactus smile converter tool. The addition of hydrogen atoms, removal of heteroatoms, neutralization of charged groups, generation of ionization states and tautomers, filtration, alternative chirality, geometry optimization, low-energy ring conformers, and final conversion and saving of the output allows to implicitly screen a compound database and predict the strongest binders based on their scoring functions.36,37
Molecular docking studies
Molecular docking is the process by which 2 molecules fit together in 3-dimensional space; it is a key tool in structural biology and computer-aided drug design.38,39 To obtain an accurate docking, a stable standard dock was used as a default standard dock setting on which the following parameters were set: population size (n = 200), generations (n = 70), and number of solutions (n = 2). The best pose ligands were selected on the basis of their best conformation that allows the lowest free binding energy.40
For protein-ligand docking studies, the selected iGEMDOCK provides an interactive interface for preparing the binding site and ligand docking status, post docking analysis, monitoring the progress, ranking, and visualization of the screened compounds by combining the pharmacologic interactions and the energy-based scoring function.41 The results obtained from this docking software were analyzed to study the binding energy and the interactions of the docked structure. For most docking study tools, users usually need to prepare the structure of the binding site and ligand. The selected software provides a straightforward method to derive the binding site from the bounded ligand, and it can also automatically consider the effects of hydrogen atoms on the binding sites.
In silico absorption, distribution, metabolism, excretion, and toxicity
To determine which lead compounds have drug-likeness characteristics and should thus become a focus of further analysis, we performed in silico absorption, distribution, metabolism, excretion, and toxicity (ADMET) assays for the compounds with the most promising structural backbones.42 ADMET studies have shown properties that are necessary and helpful in developing new natural compounds with enhanced pharmacokinetic and pharmacodynamic properties. ADMET prediction includes intestinal absorption, blood-brain barrier (BBB) penetration, carcinogenicity, and acute oral toxicity of the best-5 phytoconstituents that likely possessed drug-likeness using admetSAR server.43,44
LigPlot analysis
The LigPlot program automatically generates schematic 2-dimensional representations of protein-ligand complexes from standard protein data bank file input. The output is a color or black-and-white file that provides intermolecular interactions such as hydrogen bonds and hydrophobic interactions and their strengths. This program is completely general for any ligand and can also be used to show other types of interactions in proteins and nucleic acids.45 The LigPlot analysis was used to understand the in-depth interaction pattern between the docked ligands and the active site residues.46
Results
Compound identification by GC-MS/MS
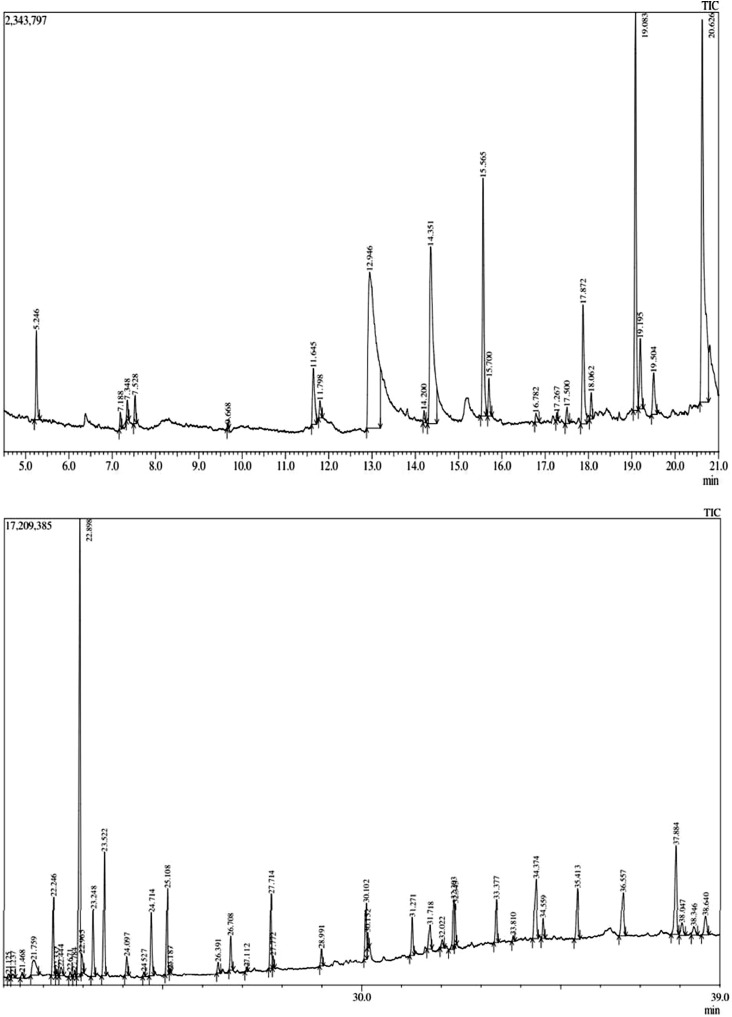
The characteristic mass fragments of GC-MS/MS chromatogram allow to quantitatively and qualitatively evaluate the compounds according to their retention times; the results revealed the presence of 48 phytoconstituents from the chloroform leaf extract of C carandas as shown in Supplemental Table 1 (in the online version at doi:XXXXXXXXXX) and Figure 1. MS data of all the phytoconstituents and their fragmentation pattern are shown in Supplemental Figure 1 (in the online version at doi:XXXXXXXXXX). The identification and structural elucidation of tentatively identified compounds were performed using NIST 14.lib and WILEY 8.lib.
Fig 1.
GC-MS/MS Chromatogram of chloroform extract of C. carandas leaves.
Identification of new compounds from C carandas
The GC-MS/MS-detected phytoconstituents in the present study were compared with the reported phytoconstituents of C carandas and are shown in Supplemental Table 2 (in the online version at doi:XXXXXXXXXX). Among the identified phytoconstituents, only 6 phytoconstituents were reported earlier in the literature, namely decane, dodecane, heptadecane, eicosane, heneicosane, and squalene. However, the remaining 42 phytoconstituents have not been previously described as the constituents of any part of C carandas.
Ligand preparation
Supplemental Figure 2 (in the online version at doi:XXXXXXXXXX) shows the prepared ligands present in the chloroform leaf extract of C carandas using GC-MS/MS.
Molecular docking study
From the identified phytoconstituents, the best pose compounds were predicted for their interaction against AD targets of Aβ protein (protein data bank ID: 2LMN) and AChE (protein data bank ID: 3LII). Supplemental Table 1 (in the online version at doi:XXXXXXXXXX) shows the binding energy between ligand-receptor interactions of all the tentatively identified phytoconstituents. The ligand molecules having the highest binding affinity with the receptors were chosen as the potential lead compounds and were confirmed through iGEMDOCK studies. These iGEMDOCK studies revealed that the phytocompounds from C carandas chloroform leaf extract possess anti-AD potential.
Top dock score phytoconstituents interaction with 2LMN
Twelve phytoconstituents that exhibited the highest docking score and lowest binding energy are compared with standard curcumin (–110.22 kcal/mol). These include octacosane (–152.58); hexacosane (–148.89); carbonic acid, eicosyl vinyl ester (–148.18); 1-heneicosanol (–135.38); N-nonadecanol-1 (–127.65); eicosane (–126.24); 3-eicosene, (E) (–125.79); 1-nonadecene (–120.26); tetratetracontane (–119.09); 9-octadecene, (E) (–114.18); squalene (–112.92); E-14-hexadecenal (–110.99); and hexadecanal (–110.67).
Top dock score phytoconstituents’ interaction with 3LII
Twenty-four phytoconstituents that exhibited the highest docking score and lowest binding energy were compared with standard galantamine (–74.5). These include cholesta-4,6-dien-3-ol, (3beta) (–92.38); di-n-octyl phthalate (–91.43); 7,9-Di-tert-butyl-1-oxaspiro(4,5)deca-6,9-diene-2,8-dione (–87.60); 6-Undecyl-5,6-dihydro-2H-pyran-2-one (–87.31); phenol, 2,4-di-t-butyl-6-nitro (–86.65); 1-heneicosanol (–85.99); octacosane (–85.85); heneicosane (–85.49); squalene (–83.72); 3-eicosene, (E) (–83); eicosane (–82.67); palmitaldehyde, diallyl acetal (–82.55); vitamin E (–82.11); methyl 3,4-O-ethylidenehexopyranoside (–81.57); hexacosane (–81.29); N-nonadecanol-1 (–81.06); 1-nonadecene (–80.41); 2-hexadecen-1-ol, 3,7,11,15-tetramethyl-, [R-[R*,R*-(E)]] (–79.98); 2(4H)-benzofuranone, 5,6,7,7a-tetrahydro-6-hydroxy-4,4,7 a-trimethyl-, (6s-cis) (–79.62); 9-octadecene, (E) (–77.29); carbonic acid, eicosyl vinyl ester (–76.78); 3-bromocholest-5-ene (–76.68); 1,2-benzenedicarboxylic acid, dibutyl ester (–76.53), and neophytadiene (–75.54).
Prediction of binding sites
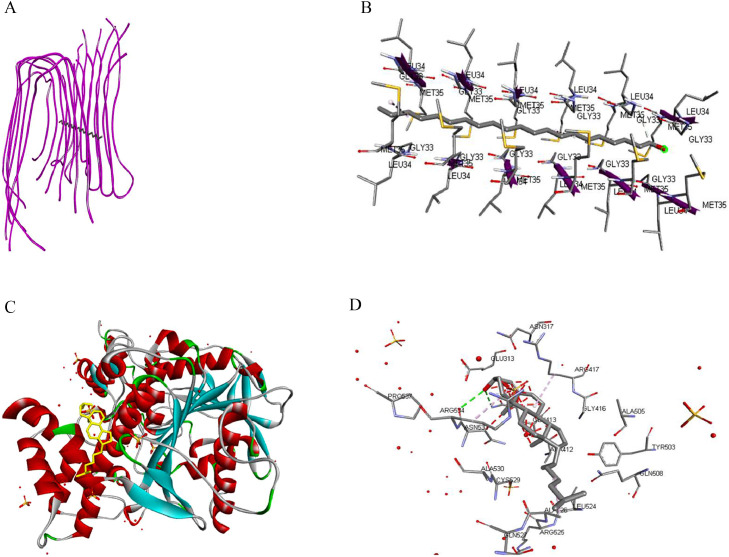
Hierarchical clustering of iGEMDOCK postscreening analysis of the interaction profile for AD targets with phytoconstituents of C carandas chloroform leaf extract and standard is shown in Supplemental Figure 3 (in the online version at doi:XXXXXXXXXX). Chain interaction and hydrophobic binding chain of lead compounds with 2LMN and 3LII protein are shown in Figure 2. The binding sites present in 2LMN and 3LII protein receptors were predicted with the least binding energy, indicating an active site where targeted ligand molecules can bind or interact, as shown in Figure 3. These binding interactions between ligand-receptor active sites were predicted by Discovery studio visualizer software.
Fig 2.
Molecular docking view: (A) Chain 2LMN interacting with 1-heneicosanol. (B) Hydrophobic binding chain of 2LMN protein with 1-heneicosanol. (C) Chain 3LII interacting with cholesta-4,6-dien-3-ol, (3beta). (D) Hydrophobic binding chain of 3LII protein with cholesta-4,6-dien-3-ol, (3beta).
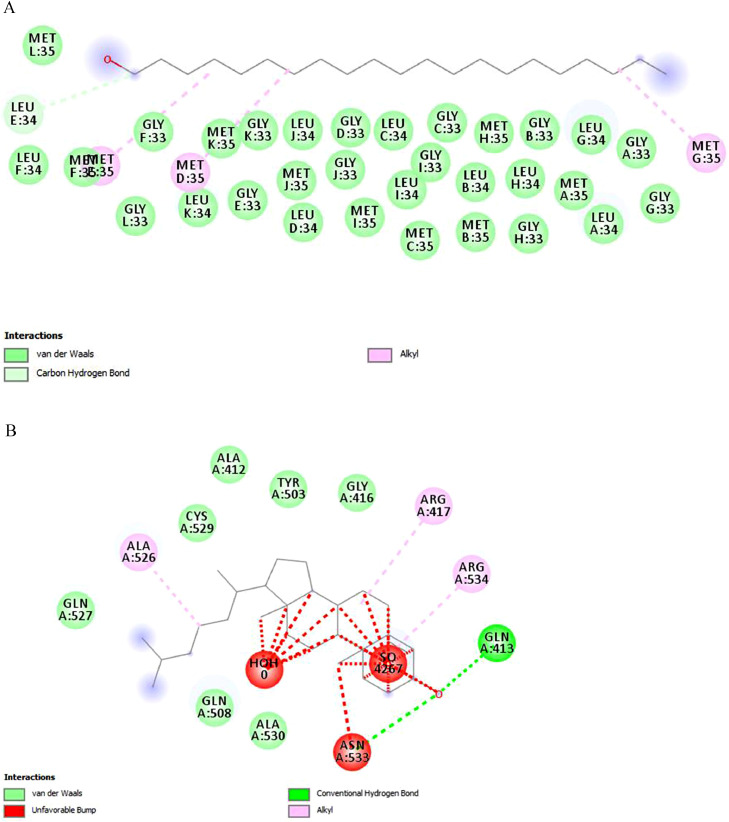
Fig 3.
2D view of binding interaction: (A) 2LMN with 1-heneicosanol. (B) 3LII with cholesta-4,6-dien-3-ol, (3beta).
In silico ADMET studies
Based on the docking study, the best-5 potential ligands of the selected extract were studied for their ADMET prediction, which includes intestinal absorption, BBB penetration, carcinogenicity, and acute oral toxicity as shown in Table 1.
Table 1.
In silico ADMET predictions of top 5 possible compounds.
| Top-5 compounds with | Dock score rank | Compound No. | Intestinal absorption | BBB permeation | Caricino genicity | Acute oral toxicity (kg/mol) |
| 2LMN - Structural model for amyloid fibrils | ||||||
| Octacosane | 1 | 41 | + | + | + | 2.118 |
| Hexacosane | 2 | 37 | + | + | + | 2.125 |
| Carbonic acid, eicosyl vinyl ester | 3 | 45 | + | + | + | 2.370 |
| 1-Heneicosanol | 4 | 36 | + | + | – | 2.213 |
| n-Nonadecanol-1 | 5 | 40 | + | + | – | 2.223 |
| 3LII - Recombinant human acetylcholinesterase | ||||||
| Cholesta-4,6-dien-3-ol, (3beta) | 1 | 46 | + | + | – | 3.150 |
| Di-n-octyl phthalate | 2 | 38 | + | + | – | 3.372 |
| 7,9-Di-tert-butyl-1-oxaspiro(4,5)deca-6,9-diene-2,8-dione | 3 | 29 | + | + | – | 2.255 |
| 6-Undecyl-5,6-dihydro-2H-pyran-2-one | 4 | 39 | – | + | – | 3.374 |
| Phenol, 2,4-di-t-butyl-6-nitro | 5 | 19 | + | + | – | 1.951 |
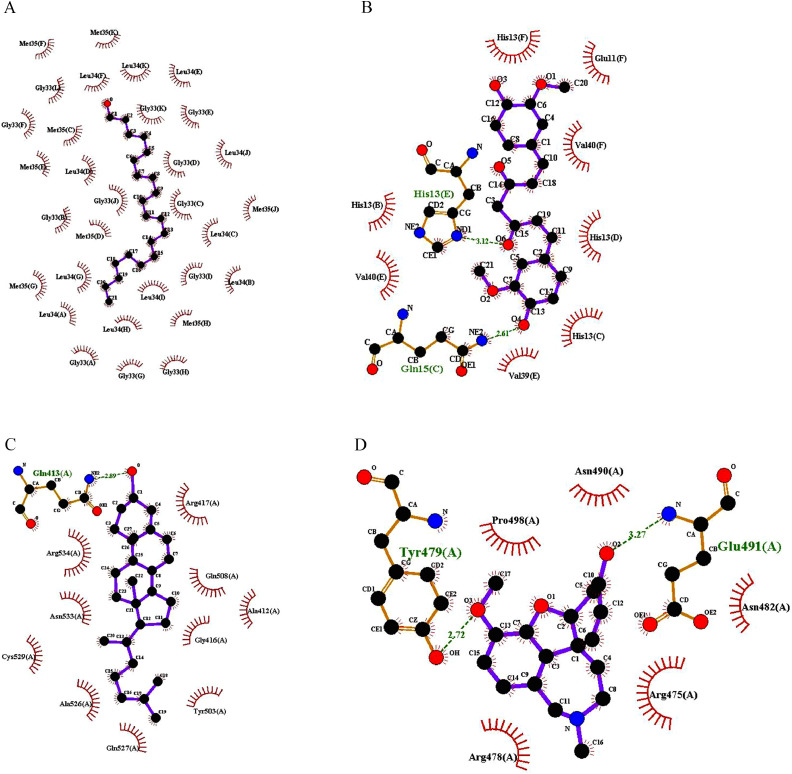
LigPlot analysis
The LigPlot analysis was performed to analyze hydrogen bond interaction and hydrophobic interactions of the compounds with positive results in in silico analysis and compared with their respective standards are shown in Table 2 and Figure 4. The ligand 1-heneicosanol with 2LMN interaction has 31 hydrophobic interactions and no H-bond interaction; similarly, the ligand cholesta-4,6-dien-3-ol, (3beta) with 3LII has 10 hydrophobic interactions and 1 H-bond interaction. Amino acid residues such as Gly33(A), Gly33(B), Gly33(C), Gly33(D), Gly33(E), Gly33(F), Gly33(G), Gly33(H), Gly33(I), Gly33(J), Gly33(K), Gly33(L), Leu34(A), Leu34(B), Leu34(C), Leu34(D), Leu34(E), Leu34(F), Leu34(G), Leu34(H), Leu34(I), Leu34(J), Leu34(K), Met35(C), Met35(D), Met35(E), Met35(F), Met35(G), Met35(H), Met35(J), and Met35(K) were found to be form 2LMN molecular interactions with 1-heneicosanol, whereas Ala412(A), Ala526(A), Arg417(A), Arg534(A), Asn533(A), Cys529(A), Gln508(A), Gln527(A), Gly416(A), Tyr503(A) and Glu413(A) were found to have 1 H-bond interaction with amino acid residues, such as cholesta-4,6-dien-3-ol, (3beta) with 3LII.
Table 2.
Ligplot analysis: protein-ligand interactions of lead molecule with standard.
| No. | Complex | Ligplot analysis |
||
| H-bond interactions |
Hydrophobic contacts | |||
| Amino acid | Distance (Å) | Amino acid | ||
| 1 | 2LMN complex with 1-heneicosanol | – | – | Gly33(A), Gly33(B), Gly33(C), Gly33(D), Gly33(E), Gly33(F), Gly33(G), Gly33(H), Gly33(I), Gly33(J), Gly33(K), Gly33(L), Leu34(A), Leu34(B), Leu34(C), Leu34(D), Leu34(E), Leu34(F), Leu34(G), Leu34(H), Leu34(I), Leu34(J), Leu34(K), Met35(C), Met35(D), Met35(E), Met35(F), Met35(G), Met35(H), Met35(J), Met35(K). |
| 2 | 2LMN complex with curcumin | Gln15(C) | 2.61 | Glu11(F), His13(B), His13(C), His13(D), His13(F), Val39(E), Val40(E), Val40(F). |
| His13(E) | 3.12 | |||
| 3 | 3LII complex with cholesta-4,6-dien-3-ol, (3beta) | Gln413(A) | 2.89 | Ala412(A), Ala526(A), Arg417(A), Arg534(A), Asn533(A), Cys529(A), Gln508(A), Gln527(A), Gly416(A), Tyr503(A). |
| 4 | 3LII complex with galantamine | Tyr479(A) | 2.72 | Arg478(A), Arg475(A), Asn482(A), Pro498(A), Asn490(A). |
| Glu491(A) | 3.27 | |||
Fig 4.
Ligplot analysis intermolecular interaction: (A) 2LMN with 1-heneicosanol. (B) 2LMN with curcumin. (C) 3LII with cholesta-4,6-dien-3-ol, (3beta). (D) 3LII with galantamine.
Discussion
Powdered leaves of C carandas were subjected to Soxhlet extraction with n-hexane, chloroform, and methanol solvents successively to separate the phytoconstituents of nonpolar and polar in nature. GC-MS/MS analyses of the chloroform extract revealed the presence of 48 phytoconstituents. Among them, 42 phytoconstituents are detected for the first time in this investigation as parts of the C carandas leaf phytoconstituents. All these phytoconstituents were identified according to their different retention times, and their mass spectral data were compared with the National Institute of Standards and Technology database and Wiley library. AD is a dynamic neurodegenerative disease with age-related symptoms, including cognitive impairment, psychosis, hyperactivity, aggressive behavior, and depression.15 Aβ proteins are present generally in the soluble form in the cerebrospinal fluid and blood; however, in patients with AD, Aβ is fibrillated as the main constituent of amyloid plaques in the brain.47 The fibrillation of Aβ is considered as the main event in AD pathology, which causes the death of nerve cells and leads to cognitive and behavioral decline, which is the main characteristic feature of AD.48 Aβ aggregates of nonpathogenic proteins cause toxicity to neuronal cells, and its clearance has not yet been fully understood.49,50 The identification of compounds that can defibrillate fibrillar Aβ or inhibits Aβ fibrillation is expected to be of therapeutic and preventive value in AD.51 AChE inhibitors retard the metabolic degradation of Ach, thus optimizing the availability of Ach for cell communication.52 AChE inhibitors are indicated for the treatment of mild-to-moderate AD.53 There are only 4 medications, namely donepezil, tacrine, galantamine, and rivastigmine, available in the market that are approved by Food and Drug Administration. Among them, donepezil alone has been approved for the treatment of severe symptoms of AD.54
All 48 phytoconstituents were screened against 2 AD receptors (2LMN and 3LII) on the basis of their high binding affinity for their respective targets. Twelve phytoconstituents showed the least binding energy against Aβ, and 24 phytoconstituents exhibited the least binding energy against AChE when compared with their standard: curcumin and galantamine, respectively. The lowest binding energy results were considered for further postdocking analysis.55 The top-possible 5 phytoconstituents with the best docking score of Aβ and AChE were screened further for their drug-likeness through the admetSAR prediction, which revealed that 2 out of the 5 compounds tested for Aβ target; that is, 1-heneicosanol and N-nonadecanol-1. These compounds do not have carcinogenicity and have better BBB permeation, intestinal absorption, and less oral toxicity. Top-possible 5 phytoconstituents that were tested for AChE target, namely cholesta-4,6-dien-3-ol, (3beta); di-n-octyl phthalate; 7,9-di-tert-butyl-1-oxaspiro(4,5)deca-6,9-diene-2,8-dione; 6-undecyl-5,6-dihydro-2H-pyran-2-one, and phenol, 2,4-di-t-butyl-6-nitro, do not have any carcinogenicity and show better BBB permeation, intestinal absorption, and less oral toxicity. Thus, these 7 phytoconstituents from C carandas chloroform leaf extract showed the highest binding affinity and better selectivity scores with both the therapeutic targets of AD and proved to be a potential lead molecule for the anti-AD effect.
Conclusions
The present study showed that important bioactive phytocompounds of C carandas resolved by GC-MS/MS analysis possess anti-AD potential against the 2 AD targets of Aβ and AChE. Thus, this type of computational analysis (in silico molecular docking study) helps to understand the presence of phytoconstituents that have binding affinities for the selected 2 targets of AD. Exploring the natural compounds for the search of lead molecules by virtual screening methods by using molecular docking analysis reduces side effects, cost, and time in drug discovery. Screening of the ligands based on pharmacokinetic properties is of utmost importance because it will reduce the failure of most drugs at the clinical stage. Therefore, this study reveals 7 potential natural compounds that have good pharmacokinetic properties and binding energy to 2 AD receptors (2LMN and 3LII), and hence are suggested as possible drugs for the treatment of AD after further research. These identified lead compounds can be further modified and optimized for better AD drugs. These screened compounds could be a great source for the development of new AD drugs (see the Appendix in the online version at doi:XXXXXXXXXX).
Acknowledgments
The authors thank taxonomist Dr Ravindra Shukla, assistant professor, Department of Botany, Indira Gandhi National Tribal University, Amarkantak, for plant authentication. The authors also thank the head of the Nanotechnology Research Centre, Department of Nanotechnology, SRM Institute of Technology, Chennai, India, for providing gas chromatography MS/MS facility as well as V. Chandrabose who assisted with English language editing and Cosmic Strands Language Editing services.
The present research work was carried out by P. Subash, who is currently pursuing his PhD under the guidance and supervision of Dr K. S. Rao, Department of Pharmacy, Indira Gandhi National Tribal University, Amarkantak, India.
Conflicts of Interest: The authors have indicated that they have no conflicts of interest regarding the content of this article.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.curtheres.2020.100615.
Appendix. Supplementary materials
References
- 1.Mishra CK, Shrivastava B, Sasmal D. Pharmacognostical Standarization and Phytochemical Identification of Fruit and Root of Carissa carandas Linn. Int J Pharm Pharm Sci. 2013;5(3):347–350. [Google Scholar]
- 2.Parvin MN. Phytochemical screening, antinociceptive, anthelmintic and cytotoxicity studies of the leaves of Carissa carandas Linn. (Family: Apocynaceae) Int. J. Sci. 2018;4(5):119. [Google Scholar]
- 3.Agarwal T, Singh R., Shukla AD, Waris I. In Vitro Study of Antibacterial activity of Carissa carandas leaf extracts. Pelagia Research Library. 2012;2(1):36–40. [Google Scholar]
- 4.Bhaskar VH, Balakrishnan N. Analgesic, anti-Inflammatory and antipyretic activities of Pergularia daemia and Carissa carandas. DARU. J. Pharm. Sci. 2009;17:168–174. [Google Scholar]
- 5.Hegde K, Joshi AB. Hepatoprotective effect of Carissa carandas Linn root extract against CCl 4 and paracetamol induced hepatic oxidative stress. Indian J Exp Biol. 2009;47(8):660–667. [PubMed] [Google Scholar]
- 6.Arifa M, Fareed S, Hussain T, Ali M. Adaptogenic Activity of Lanostane Triterpenoid Isolated from Carissa carandas Fruit against Physically and Chemically Challenged Experimental Mice. Phcog J. 2013;5:216–220. [Google Scholar]
- 7.Verma K, Shrivastava D, Kumar G. Antioxidant activity and DNA damage inhibition in vitro by a methanolic extract of Carissa carandas (Apocynaceae) leaves. J Taibah Univ. Sci. 2015;9(1):34–40. [Google Scholar]
- 8.Joglekar SN, Gaitonde BB. Histamine Releasing Activity of Carissa carandas Roots (Apocyaneceae) The Japanese journal of Pharmacology. 1970;20:367–372. doi: 10.1254/jjp.20.367. [DOI] [PubMed] [Google Scholar]
- 9.Itankar PR, Lokhande SJ, Verma PR, Arora SK, Sahu RA, Patil AT. Antidiabetic Potential of Unripe Carissa carandas Linn. Fruit Extract. J. Ethnopharmacol. 2011;135(2):430–433. doi: 10.1016/j.jep.2011.03.036. [DOI] [PubMed] [Google Scholar]
- 10.Sumbul S, Ahmed SI. Anti-Hyperlipidemic activity of Carissa carandas (Auct.) leaves extract in egg yolk induced hyperlipidemic Rats. J. Basic Appl Sci. 2012;8:124–134. [Google Scholar]
- 11.Hegde K, Thakker SP, Joshi AB, Shastry CS, Chandrashekhar KS. Anticonvulsant activity of Carissa carandas Linn. Root extract in experimental mice. Trop J Pharm Res. 2009;8(2):117–125. [Google Scholar]
- 12.Stafford GI, Pedersen ME, Staden JV, Jager AK. Review on Plants with CNS-Effects Used in Traditional South African Medicine against Mental Diseases. J. Ethnopharmacol. 2008;119:513–537. doi: 10.1016/j.jep.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Shinde M., Ritu G., Sanjay C. Anticonvulsant and Sedative activities of Extracts of Carissa carandas Leaves. J. drug deliv. ther. 2018;8(5):369–373. [Google Scholar]
- 14.Saha R, Hossain L, Bose U, Rahman AA. Neuropharmacological and Diuretic Activities of Carissa carandas Linn. Leaf. Pharmacologyonline. 2010;2:320–327. [Google Scholar]
- 15.Hope T, Keene J, Fairburn CG, Jacoby R, Mcshane R. Natural history of behavioural changes and psychiatric symptoms in Alzheimer's disease: A longitudinal study. Br J Psychiatry. 1999;174:39–44. doi: 10.1192/bjp.174.1.39. [DOI] [PubMed] [Google Scholar]
- 16.Mcgleenon BM., Dynan KB, Passmore AP. Acetyl cholinesterase inhibitors in Alzheimer's disease. Br J Clin Pharmacol. 1999;48:471–480. doi: 10.1046/j.1365-2125.1999.00026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma S., Nehru B., Saini A. Inhibition of Alzheimer's amyloid-beta aggregation in-vitro by carbenoxolone: Insight into mechanism of action. Neurochem. Int. 2017;108:481–493. doi: 10.1016/j.neuint.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Casey DA, Antimisiaris D, Brien JO. Drugs for Alzheimer's disease: Are they effective? P & T. 2010;354:208–211. [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbard BM., Fentonm GW, Anderson JM. A Quantitative Histological Study of Early Clinical and Preclinical Alzheimer's disease. Neuropathol. Appl. Neurobiol. 1990;16(2):111–121. doi: 10.1111/j.1365-2990.1990.tb00940.x. [DOI] [PubMed] [Google Scholar]
- 20.Hope T, Keene J, Fairburn C, Mcshane R, Jacoby R. Behaviour Changes in Dementia 2 : Are There Behavioural Syndromes ? Int J. Geriat. Psychiatry. 1997;12(24):1074–1078. doi: 10.1002/(sici)1099-1166(199711)12:11<1074::aid-gps696>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 21.Lanctot KL, Herrmann N, Yau KK, Khan LR, Liu BA, Loulou MM, Einarson TR. Efficacy and safety of cholinesterase inhibitors in Alzheimer's disease: A meta-analysis. Can. Med. Assoc. J. 2003;169(6):557–564. [PMC free article] [PubMed] [Google Scholar]
- 22.Veerman SRT, Schulte PFJ, Smith JD, Haan LD. Memantine augmentation in Clozapine-Refractory Schizophrenia: A Randomized, Double-Blind, Placebo-Controlled Crossover Study. Psychol. Med. 2016;46(9):1909–1921. doi: 10.1017/S0033291716000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman AJ, Papadakis K, Csernansky J, Litman R, Volavka J, Jia XD, Gage A. A Randomized, Placebo-Controlled Study of Memantine as Adjunctive Treatment in Patients with Schizophrenia. Neuropsychopharmacol. 2009;34:1322–1329. doi: 10.1038/npp.2008.200. [DOI] [PubMed] [Google Scholar]
- 24.Okigbo RN, Anuagasi CL, Amadi JE. Advances in selected medicinal and aromatic plants indigenous to Africa. J. Med. Plant Res. 2009;3(2):86–95. [Google Scholar]
- 25.Ji SP. Ethnobotanical approaches of traditional medicine studies: Some experiences from Asia. Pharm. Bio. 2001;139:74–79. doi: 10.1076/phbi.39.s1.74.0005. [DOI] [PubMed] [Google Scholar]
- 26.El-Hawary SS, Sobeh M, Badr WK, Abdelfattah MA, Ali ZY, El-Tantawy ME, Rabeh MA, Wink M. HPLC-PDA-MS/MS profiling of secondary metabolites from Opuntia ficus-indica cladode, peel and fruit pulp extracts and their antioxidant, neuroprotective effect in rats with aluminum chloride induced neurotoxicity. Saudi J. Biol. Sci. 2020;27(10):2829–2838. doi: 10.1016/j.sjbs.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao KS, Subash P. In silico exploration of anti-Alzheimer's compounds present in methanolic extract of Neolamarckia cadamba bark using GC-MS/MS. Arab. J. Chem. 2020;13(7):6246–6255. [Google Scholar]
- 28.Rao MRK, Anisha G, Prabhu K, Shil S, Vijayalakshmi N. Preliminary Phytochemical and Gas Chromatography-Mass Spectrometry study of one medicinal plant Carissa carandas. Drug Invitation Today. 2019;12(7):1629–1630. [Google Scholar]
- 29.Anupama N, Madhumitha G, Rajesh KS. Role of Dried Fruits of Carissa carandas as anti-inflammatory agents and the analysis of Phytochemical Constituents by GC-MS. BioMed Research International. 2014:1–6. doi: 10.1155/2014/512369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aslam F, Rasool N, Riaz M, Zubair M, Rizwan K, Abbas M, Bukhari TH, Bukhari IH. Antioxidant, Haemolytic activities and GC-MS Profiling of Carissa carandas Roots. Int J of Phytomedicine. 2011;3:567–578. [Google Scholar]
- 31.Pino JA, Marbot R, Vazuez C. Volatile Flavor Constituents of Karanda (Carissa carandas L.) Fruit. J Essent. Oil Res. 2004;16:432–434. [Google Scholar]
- 32.Galipallia S, Patela NK, Prasannaa K, Bhutania KK. Activity-guided investigation of Carissa carandas (L.) roots for anti-inflammatory constituents. Nat Prod Res. 2015;29(17):1–5. doi: 10.1080/14786419.2014.989846. [DOI] [PubMed] [Google Scholar]
- 33.Bhandane BS, Patil RH. Isolation, Purification and Characterization of antioxidant steroid derivative methanolic extract of Carissa carandas (L.) Leaves. Biocatalysis and Agricultural Biotechnology. 2017;10:216–223. [Google Scholar]
- 34.Dallakyan OS, Olson AJ. Small-Molecule Library Screening by Docking with PyRx. Chemical Biology. 2015;19:1263. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 35.Mendelsohn LD. ChemDraw 8 ultra, windows and Macintosh versions. J. Chem. Inf. Model. 2004;44:2225–2226. [Google Scholar]
- 36.Suri C, Naik PK. Elucidating the precise interaction of reduced and oxidized states of neuroglobin with Ubc12 and Cop9 using molecular mechanics studies. Int J Fundam Appl Sci. 2012;1:74–77. [Google Scholar]
- 37.Usha T, Tripathi P, Pande V, Middha SK. Molecular docking and quantum mechanical studies on pelargonidin-3-glucoside as renoprotective ACE inhibitor. ISRN Computational Biology. 2013:1–4. [Google Scholar]
- 38.Palleti JD, Jyothsna P, Muppalaneni NB, Chitti S. Virtual screening and molecular docking analysis of Zap-70 Kinase inhibitors. International Journal of Chemical and Analytical Science. 2011;29:1208–1211. [Google Scholar]
- 39.Ladokun OA, Abiola A, Okikiola D, Ayodeji F. GC-MS and molecular docking studies of Hunteria umbellata methanolic extract as a potent anti-diabetic. Inform. Med. Unlocked. 2018;13:1–8. [Google Scholar]
- 40.Kandeel M., Kitade Y. Computational analysis of siRNA recognition by the Ago2 PAZ domain and identification of the determinants of RNA-induced gene silencing. PLoS ONE. 2013;8:2. doi: 10.1371/journal.pone.0057140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu KC, Chen YF, Lin SR, Yang JM. iGEMDOCK : a graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinformatics. 2011;12(1):1–11. doi: 10.1186/1471-2105-12-S1-S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Obianyo O, Dall E, Du Y, Haian F, Liu X, Kang SS, Song M, Shan PY, Cabrele C, Schuber M, Xiaoguang L, Wang JZ, Brandstetter H, Ye K. Inhibition of delta-secretase improves cognitive functions in mouse models of Alzheimer's disease. Nature Communications. 2017:1–17. doi: 10.1038/ncomms14740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alhazmi MI. Molecular docking of selected phytocompounds with H1N1 proteins. Bioinformation. 2015;114:196–202. doi: 10.6026/97320630011196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Lee PW, Tang Y. AdmetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012;5211:3099–3105. doi: 10.1021/ci300367a. [DOI] [PubMed] [Google Scholar]
- 45.Wallace AC, Laskowski RA, Thornton JM. LigPlot: A program to generate schematic diagrams of protein-ligand interactions. Protein Engineering, Design and Selection. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 46.Bharatham KBN, Park KH, Lee K. Binding mode analyses and pharmacophore model development for sulfonamide chalcone derivatives, a new class of α-glucosidase inhibitors. J. Mol. Graph. Model. 2008;26:1202–1212. doi: 10.1016/j.jmgm.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Seubert P, Pelfrey CV, Esch F, Lee M, Dovey H, Davis D, Sinha S, Schlossmacher M, Whaley J, Swindlehurst C. Isolation and quantification of soluble Alzheimer's β-peptide from biological Fluids. Letters of Nature. 1992;359:325–327. doi: 10.1038/359325a0. [DOI] [PubMed] [Google Scholar]
- 48.Lorenzo A, Yankner BA. β-Amyloid Neurotoxicity Requires Fibril Formation and Is Inhibited by Congo Red. Proc Natl Acad Sci U S A. 1994;91:12243–12247. doi: 10.1073/pnas.91.25.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent cytotoxicity of aggregates implies a common origin for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 50.Citron M. Emerging Alzheimer's disease therapies: Inhibition of β-Secretase. Neurobiology of Aging. 2002;23:1017–1022. doi: 10.1016/s0197-4580(02)00122-7. [DOI] [PubMed] [Google Scholar]
- 51.Chauhan NKC, Wegiel WJ, Malik MN. Walnut extract inhibits the fibrillization of amyloid beta-protein and also defibrillizes its preformed fibrils. Current Alzheimer Research. 2004;1:183–188. doi: 10.2174/1567205043332144. [DOI] [PubMed] [Google Scholar]
- 52.Moss DE, Perez RG, Kobayashi H. Cholinesterase inhibitor therapy in Alzheimer's disease: the limits and tolerability of irreversible CNS-selective acetyl cholinesterase inhibition in primates. Journal of Alzheimer's disease. 2017;55:1285–1294. doi: 10.3233/JAD-160733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trinh NH, Hoblyn J, Mohanty S, Yaffe K. Efficacy of cholinesterase inhibitors in the treatment of neuropsychiatric symptoms and functional impairment in Alzheimer disease: a meta-analysis. Journal of the American Medical Association. 2003;289(2):210–216. doi: 10.1001/jama.289.2.210. [DOI] [PubMed] [Google Scholar]
- 54.Mehta M, Adem A, Sabbagh M. New Acetylcholinesterase inhibitors for Alzheimer's Disease. International Journal of Alzheimer's disease. 2012:1–7. doi: 10.1155/2012/728983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kandagalla S, Sharath BS, Bharath BR, Hani U, Manjunatha H. Molecular docking analysis of curcumin analogues against kinase domain of ALK5. In Silico Pharmacology. 2017;5(15):1–9. doi: 10.1007/s40203-017-0034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.