Abstract
Background
The invasive species Xanthium spinosum has been used as a traditional Chinese medicine for many years. Unfortunately, no extensive molecular studies of this plant have been conducted.
Results
Here, the complete chloroplast (cp) genome sequence of X. spinosum was assembled and analyzed. The cp genome of X. spinosum was 152,422 base pairs (bp) in length, with a quadripartite circular structure. The cp genome contained 115 unique genes, including 80 PCGs, 31 tRNA genes, and 4 rRNA genes. Comparative analyses revealed that X. spinosum contains a large number of repeats (999 repeats) and 701 SSRs in its cp genome. Fourteen divergences (Π > 0.03) were found in the intergenic spacer regions. Phylogenetic analyses revealed that Parthenium is a sister clade to both Xanthium and Ambrosia and an early-diverging lineage of subtribe Ambrosiinae, although this finding was supported with a very weak bootstrap value.
Conclusion
The identified hotspot regions could be used as molecular markers for resolving phylogenetic relationships and species identification in the genus Xanthium.
Keywords: Nucleotide diversity; Divergence; Ambrosiinae: genetic markers, phylogenomics
Background
The structure of the majority of the flowering plant chloroplast (cp) genome consists of a pair of inverted repeats (IRs), along with large single-copy (LSC) and small single-copy (SSC) regions, and cp genome size ranges from 107 to 280 kb [1, 2]. With the emergence of next-generation sequencing technology [3], complete cp genome sequences are being extensively used to improve phylogenetic resolution at the interspecific level [4]. In addition, cp genomes have been found to contain polymorphic regions generated through genomic expansion, contraction, inversion, or gene rearrangement, and such sequences have been widely used as an effective tool for plant phylogenomic analyses [5].
The invasive species Xanthium spinosum belongs to the family Asteraceae and is within the subtribe Ambrosiinae (Heliantheae), which includes annual and perennial herbaceous plants [6]. It is native to South America and has been introduced to Canada, the United States, Central and South America, parts of Africa, the Middle East, Russia, China, Australia, and the Korean Peninsula [7–10]. The genus Xanthium has been widely used for various traditional medicinal treatments in multiple countries [11]. Parts of the X. spinosum plant are used for the treatment of cancer and diarrhea [12, 13], intermittent fever related to hydrophobia and rabies [14], and rheumatoid arthritis [15], and have antibacterial [14] and antiviral properties [14, 16–18]. Although several antimicrobial substances and their functions have been studied in X. spinosum over the past five decades, no exclusive genetic or genomic studies have been conducted to date.
Universal molecular markers such as the plastid genes rbcL and psbA and nuclear internal transcribed spacer (ITS) have been widely used for the rapid and precise identification of plant species but have proved unsuccessful for distinguishing very closely related species [19–21]. The genus Xanthium is commonly known as cocklebur, and is a close relative of the genus Ambrosia. The number of species in the genus Xanthium remains under debate, and this genus may include 5 to more than 20 species [22–25]. Phylogenetic analyses of several plastid and nuclear DNA markers have shown conflicting results for Xanthium and its relatives [11]. By contrast, Somaratne et al. (2019) used 46 cp protein-coding genes (PCGs) to resolve the phylogenetic positions of Xanthium and Parthenium and revealed that Parthenium is not an early-diverging lineage of the subtribe Ambrosiinae. However, most plant cp genomes contain highly conserved structures that are useful molecular markers for the identification of plant species in genome-wide evolutionary studies; such structures provide significant quantities of genetic information and can resolve taxonomic and phylogenetic relationships [26, 27].
In the present study, we examined both plastome evolution and the phylogenetic relationships within Heliantheae. For this purpose, we first sequenced and characterized the X. spinosum cp genome and compared it with the X. sibiricum cp genome as well as those of closely related species of Heliantheae. In addition, we identified hotspot regions of sequence variation and clarified the evolutionary dynamics among Xanthium species.
Results
General features of the cp genome and its organization
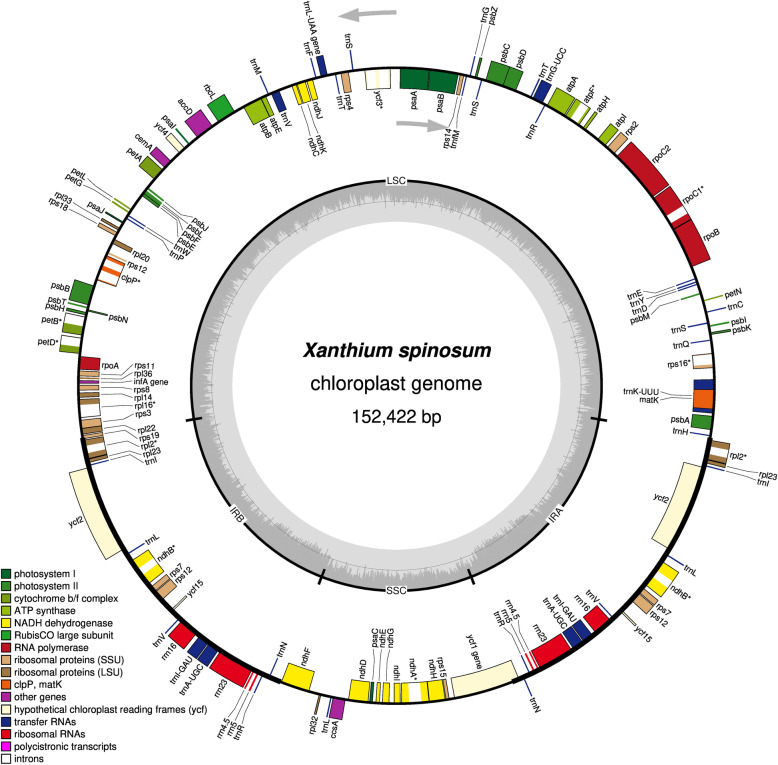
The complete cp genome of X. spinosum was 152,422 bp in length. The cp genome showed a typical quadripartite structure containing two short inverted repeats (IRa and IRb) (25,075 bp) separated by a small single-copy (SSC) region (18,083 bp) and a large single-copy (LSC) region (84,189 bp) (Fig. 1). The cp genome encodes 115 unique genes, including 80 PCGs, 31 transfer RNA (tRNA) genes, and 4 ribosomal RNA (rRNA) genes. Six protein-coding, six tRNA, and four rRNA genes were duplicated in the IR regions. The overall GC content of the cp genome was 37.4%, while those of LSC, SSC, and IR regions were 35.4, 31.2, and 43%, respectively (Table 1).
Fig. 1.
Gene map of Xanthium spinosum. Genes lying outside the outer circle are transcribed in a counter-clockwise direction, and genes inside this circle are transcribed in a clockwise direction. The coloured bars indicate known protein-coding genes, transfer RNA genes, and ribosomal RNA genes. The dashed, dark grey area in the inner circle denotes GC content, and the light grey area indicates genome AT content. LSC, large single-copy; SSC, small single-copy; IR, inverted repeat
Table 1.
Comparative analysis of chloroplast genome of X.spinosum and X. sibiricum
| Characteristics | X. spinosum | X. sibiricum | |
|---|---|---|---|
| Size (bp) | 152,422 | 151,897 | |
| LSC length (bp) | 84,189 | 83,847 | |
| SSC length (bp) | 18,083 | 17,900 | |
| IR length (bp) | 25,075 | 25,070 | |
| Total number of genes | 132 | 132 | |
| Protein-coding genes | 87 | 87 | |
| tRNA genes | 37 | 37 | |
| rRNA genes | 8 | 8 | |
| Duplicate genes | 17 | 17 | |
| GC content | Total (%) | 37.4 | 37.5 |
| LSC (%) | 35.4 | 35.5 | |
| SSC (%) | 31.2 | 31.4 | |
| IR (%) | 43 | 43 | |
| CDS (%) | 37.9 | 37.9 | |
| rRNA (%) | 55.2 | 55.2 | |
| tRNA (%) | 53 | 52.9 | |
| All genes (%) | 39.5 | 39.5 | |
| Protein-coding genes (%bp) | 51.65 | 51.74 | |
| All genes (%bp) | 72.5 | 72.89 | |
| Non-coding regions (%) | 27.5 | 27.11 |
Comparative analyses of Xanthium species
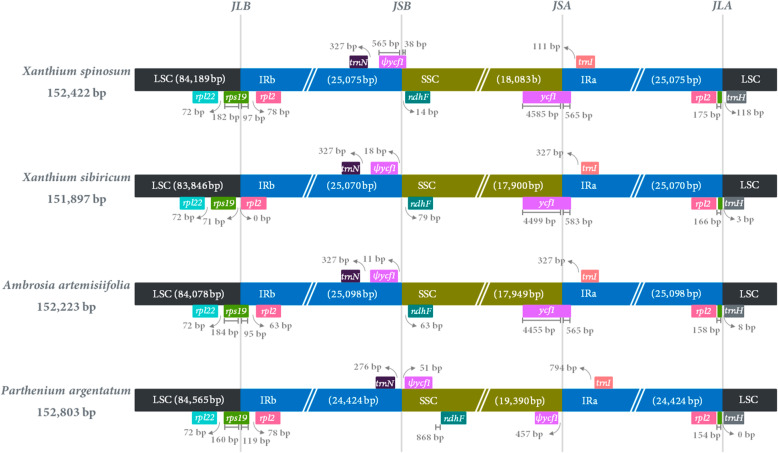
The borders of LSC-IRb and SSC-IRa in the cp genome of X. spinosum were compared to three other closely related species of Heliantheae, namely, X. sibiricum, Ambrosia artemisiifolia, and Parthenium argentatum [28, 29] (Fig. 2). An intact copy of the rps19 gene was present in the LSC/IRb borders of X. spinosum, A. artemisiifolia, and P. argentatum, as well as a shared 95 bp to 119 bp sequence in the IRb region adjacent to the rpl2 gene. By contrast, the X. sibiricum rps19 gene was completely shifted to the LSC region, 71 bp away from the IRb region, despite the rpl2 gene being present at the LSC/IRb border. In addition, 154–175 bp of the fragmented rps19 gene in all four species was present at the IRa/LSC, LSC/IRa regions or its border. On the other hand, ѱycf1 was present in the IRa/SSC border of X. spinosum, whereas it was located in the IRb or silenced in the SSC region of X. sibiricum and A. artemisiifolia, and was situated in the SSC region of the P. argentatum cp genome. The entire ndhF gene was present in the SSC region of all four cp genomes. Similarly, an intact ycf1 gene was present in the SSC/IRa region of all of the cp genomes analyzed, except P. argentatum, which has a 565 to 583 bp fragment of ycf1 in the IRa region. However, P. argentatum encodes two copies of ѱycf1 in its genome. The trnH gene sequences are located in the LSC region 0 to 118 bp from the IRa/LSC border in all cp genomes.
Fig. 2.
Comparison of the large single-copy (LSC), small single-copy (SSC) and inverted repeat (IR) border regions of four Heliantheae (Xanthium spinosum, X. sibiricum, Ambrosia artemisiifolia and Parthenium argentatum) chloroplast genomes. Ѱ indicates a pseudogene. The figure is not drawn to scale
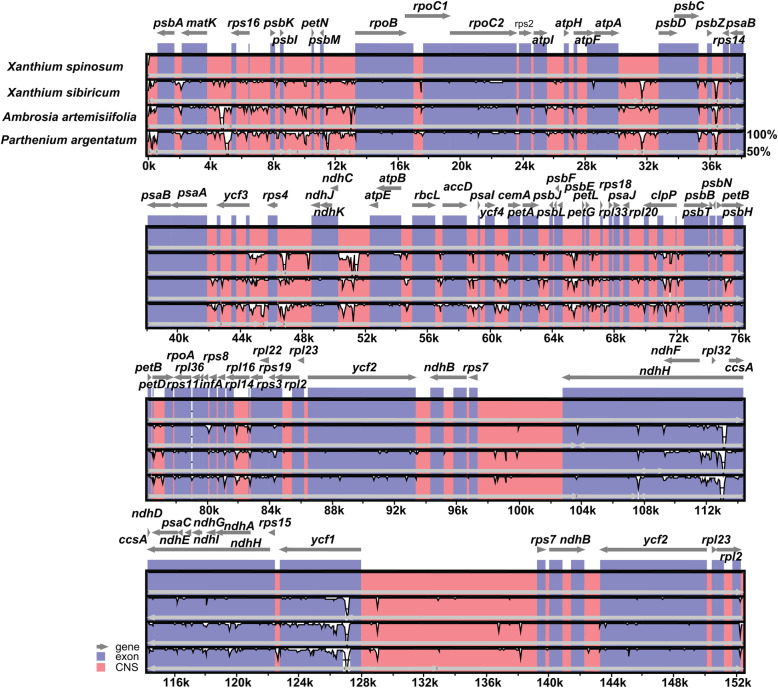
The cp genomic sequences of four Heliantheae species were analyzed using mVISTA software to detect variation among the sequences (Fig. 3). The sequence divergence differed markedly among regions. The data revealed that the non-coding region was more divergent than its coding counterparts. Relative to the LSC and SSC regions, IR regions of all cp genomes were less divergent.
Fig. 3.
Sequence alignment of four Heliantheae chloroplast genomes performed using the mVISTA program with Xanthium spinosum as a reference. The top grey arrow shows genes in order (transcriptional direction) and the position of each gene. A 70% cut-off was used for the plots. The y-axis indicates a percent identity of between 50 and 100%, and the red and blue areas indicate intergenic and genic regions, respectively
Repeat structure and SSR analyses
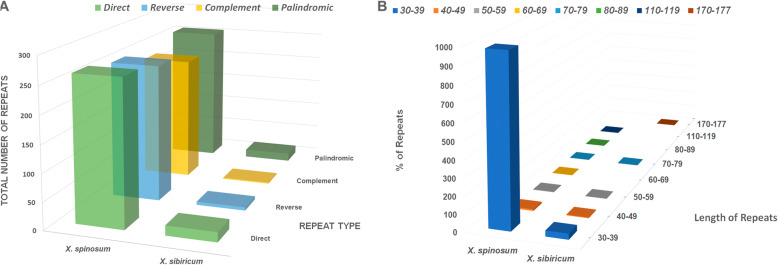
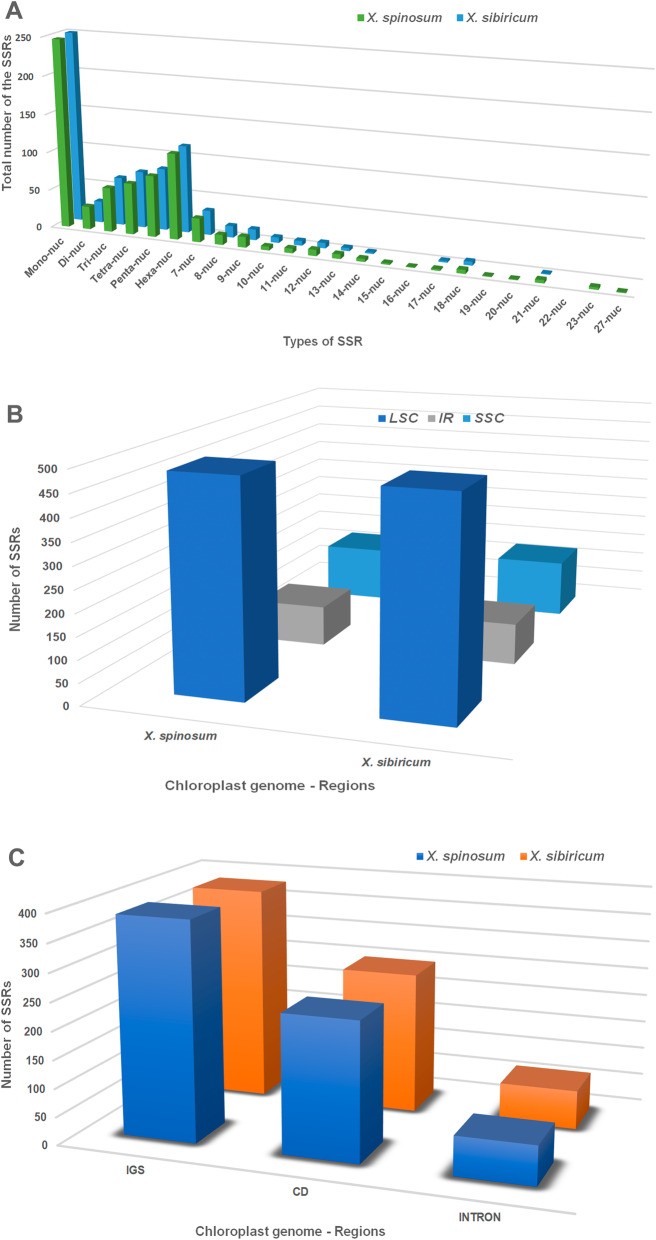
The presence of repeat sequences in the X. spinosum and X. sibiricum cp genomes was analyzed and the species were compared. Repeats in the X. spinosum cp genome consist of 264 forward, 256 palindromic, 251 reverse, and 228 complement. By contrast, X. sibiricum contained 18 forward, 15 palindromic, 6 reverse, and 2 complement repeats (Fig. 4a). In total, X. spinosum and X. sibiricum contain 999 repeats and 41 repeats, respectively. Among the 999 repeats identified in X. spinosum, repeats of 30–39 bp in length (983) were predominant in the cp genome; the longest repeat was 115 bp and was a palindrome sequence. Similarly, in X. sibiricum, 34 repeats were 30–39 bp in length, and the longest was a palindromic sequence of 177 bp (Fig. 4b).
Fig. 4.
Comparison he distribution of different repeat types in the Xanthium spinosum vs. X. sibiricum cp genomes. a The number of different types of repeats. F – forward repeats; R – Reverse repeats; P – palindromic repeats; C – complement repeats. b The length and the total number of repeat sequences present in their respective cp genomes
In total, 701 and 705 simple sequence repeats (SSRs) were identified in the X. spinosum and X. sibiricum cp genomes, respectively. The 701 SSRs in the X. spinosum cp genome included 247 (35.24%) mono-nucleotide repeats, 30 (4.3%) di-nucleotide repeats, 58 (8.3%) tri-nucleotide repeats, 67 (9.6%) tetra-nucleotide repeats, 80 (11.4%) penta-nucleotide repeats, 112 (15.98%) hexa-nucleotide repeats, 31 (4.42%) 7-nucleotide repeats, and 76 other repeats ranging from 8 nucleotides to 27 nucleotides (10.84%) (Fig. 5a). Similarly, the cp genome of X. sibiricum contained 250 (35.46%) mono-nucleotide repeats, 28 (3.97%) di-nucleotide repeats, 63 (8.94%) tri-nucleotide repeats, 74 (10.5%) tetra-nucleotide repeats, 81 (11.49%) penta-nucleotide repeats, 114 (16.18%) hexa-nucleotide repeats, 32 (4.54%) 7-nucleotide repeats, and 63 repeats with lengths from 8 nucleotides to 21 nucleotides (8.94%). Furthermore, the distributions of SSRs in the LSC, IR and SSC regions of X. spinosum and X. sibiricum indicated that the corresponding cp genomes contain 483 and 481 SSRs in the LSC, 91 and 93 in the IR, and 127 and 131 in the SSC regions (Fig. 5b). Likewise, SSRs were analyzed in the protein-coding (exon, protein-coding exon), intron and intergenic spacer (IGS) sequences of X. spinosum and X. sibiricum, which indicated that their cp genomes contain 244 and 252 SSRs in CDs, 69 and 69 in introns and 388 and 384 in IGS regions, respectively (Fig. 5c).
Fig. 5.
Comparison the presence of simple sequence repeats (SSRs) in the Xanthium spinosum vs. X. sibiricum cp genomes. a Distribution of different types of SSRs. b Presence of SSRs in the LSC, SSC, and IR regions. c Presence of SSRs in intergenic spacers, protein-coding regions, and intron regions
Nucleotide diversity analyses
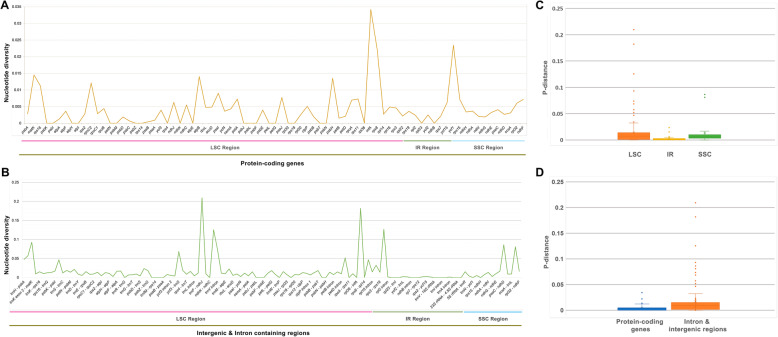
The nucleotide diversity of 208 regions was analyzed using DnaSP software, including 79 PCGs and 129 intergenic and intron regions in the cp genomes of X. spinosum and X. sibiricum. The most variable region was infA (0.03) among PCGs (Fig. 6a), and high variability was observed for the trnH-psbA (0.05), psbA-trnK (0.06), trnK exon2-matK (0.09), psbI-trnS (0.05), ycf3-trnS (0.07), trnF-ndhJ (0.21), ndhC-trnV (0.13), trnV intron (0.07), petD-rpoA (0.05), infA-rps8 (0.18), rpl14-rpl16 (0.05), rpl16-rps3 (0.03), psaC-ndhD (0.09) and trnL-rpl32 (0.08) genes in introns and intergenic regions (Fig. 6b; Table 2).
Fig. 6.
The genetic diversity based on Kimura’s two-parameter model. a The P-distance value of protein-coding genes (b) the P-distance value of intron and intergenic regions (c) Boxplots of P-distance value difference among LSC, IR and SSC regions (d) Boxplots of P-distance value differences between protein-coding genes and intron and intergenic regions
Table 2.
Mutational hotspots between X. spinosum and X. sibiricum
| S. No. | Region | Nucleotide diversity (Pi) | Total number of mutations | Region length (bp) |
|---|---|---|---|---|
| 1 | trnH - psbA | 0.04774 | 19 | 398 |
| 2 | psbA - trnK | 0.05714 | 12 | 210 |
| 3 | trnK exon 2 - matK | 0.09286 | 26 | 280 |
| 4 | psbI - trnS | 0.04667 | 7 | 150 |
| 5 | ycf3 - trnS | 0.06838 | 56 | 819 |
| 6 | trnF - ndhJ | 0.20940 | 49 | 234 |
| 7 | ndhC - trnV | 0.12551 | 123 | 980 |
| 8 | trnV intron | 0.07360 | 29 | 394 |
| 9 | petD - rpoA | 0.05181 | 10 | 193 |
| 10 | infA - rps8 | 0.18189 | 22 | 121 |
| 11 | rpl14 - rpl16 | 0.04673 | 5 | 107 |
| 12 | rpl16 - rps3 | 0.03226 | 5 | 155 |
| 13 | psaC - ndhD | 0.08621 | 10 | 116 |
| 14 | trnL - rpl32 | 0.08088 | 44 | 544 |
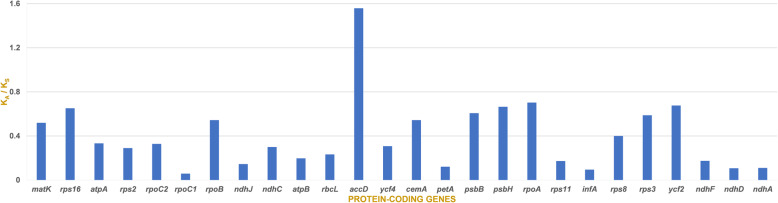
Synonymous (KS) and nonsynonymous (KA) substitution rate analyses
The synonymous and nonsynonymous substitution rates were evaluated for 79 PCGs in the X. spinosum and X. sibiricum cp genomes. The KA/KS ratios of nearly all genes were less than 1, except for the PCG accD (1.56) (Fig. 7).
Fig. 7.
Comparison the ratio of non-synonymous (KA) to synonymous (KS) substitutions of 79 protein-coding genes of Xanthium spinosum vs. X. sibiricum cp genomes
Positive selection analyses of the accD gene
Positive selection of the accD PCG in Heliantheae cp genome species was investigated using site-specific models with four comparisons (M0 vs. M3, M1 vs. M2a, M7 vs. M8, M8a vs. M8), using a likelihood ratio test (LRT) threshold of p ≤ 0.05 in EasyCodeML software. Among these models, M2a was the positive selective model and p (p0, p1 and p2) are the proportions of negative or purifying, neutral, and positive selection, respectively. The ω2 value of the accD gene was 3.70 in the M2a model. In addition, Bayes empirical Bayes (BEB) analyses were used to analyze the locations of consistent selective sites in the accD PCG using the M7 vs. M8 model comparison, and one site was found to potentially be under positive selection, with posterior probabilities greater than 0.95, while another site had probabilities greater than 0.99 (Table 3); the 2ΔLnL value was 25.91 and the p-value of LRT was 0 (Table 4).
Table 3.
Comparison of site models, positive selective amino acid loci and estimation of parameters for accD protein-coding genes in the Heliantheae species
| Protein-coding gene | Model | np | Ln L | Estimates of parameters | Model compared | LRT P-value |
Positive sites | |||
|---|---|---|---|---|---|---|---|---|---|---|
| accD | M3 | 45 | − 1567.812 | p: | 0.05791 | 0.85997 | 0.08211 | M0 vs. M3 | 0 | [] |
| ω: | 0.22724 | 0.22724 | 3.70209 | |||||||
| M0 | 41 | − 1597.602 | ω0: | 0.36846 | Not Allowed | |||||
| M2a | 44 | −1567.812 | p: | 0.91789 | 0 | 0.08211 | M1a vs. M2a | 0.000196255 | [] | |
| ω: | 0.22724 | 1 | 3.70208 | |||||||
| M1a | 42 | − 1576.349 | p: | 0.82957 | 0.17043 | Not Allowed | ||||
| ω: | 0.11911 | 1 | ||||||||
| M8 | 44 | −1567.829 | p0 = 0.91839 | p = 29.41052 | q = 99.00000 | M7 vs. M8 | 0.000002362 | 35 S 0.936, 42 R 0.963*, 98 H 0.701, 129 Q 0.685, 177 S 0.616, 181 S 0.765, 184 N 0.519, 185 A 1.000**, 187 A 0.905 | ||
| (p1 = 0.08161) | ω = 3.72034 | |||||||||
| M7 | 42 | − 1580.784 | p= | 0.07226 | q= | 0.08717 | Not Allowed | |||
| M8a | 43 | −1576.468 | p0 = 0.82999 | p = 13.67839 | q = 99.00000 | M8a vs. M8 | 0.000032257 | Not Allowed | ||
| (p1 = 0.17001) | ω = 1.00000 | |||||||||
Note:
[] – No data available
np represents the degree of freedom
Positively selected sites (* p > 95%; ** p > 99%)
Table 4.
Comparison of likelihood ratio test (LRT) statistics of positive selection models against their null models (2ΔLnL) for the accD gene
| Protein-coding genes | Comparison between models | 2ΔLnL | d.f. | p-value |
|---|---|---|---|---|
| accD | M0 vs M3 | 59.579136 | 4 | 0 |
| M1 vs M2A | 17.07219 | 2 | 0.0001963 | |
| M7 vs M8 | 25.91159 | 2 | 0.0000024 | |
| M8a vs M8 | 17.27995 | 1 | 0.0000323 |
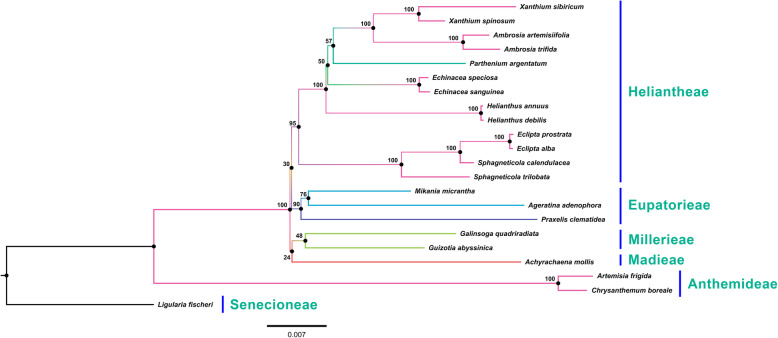
Phylogenetic analyses
In all, 71 PCGs from 21 cp genome sequences were selected for inferring phylogenetic relationships among closely related species of Heliantheae, and Ligularia fischeri (MG729822) was selected as an outgroup. A maximum likelihood tree was constructed using 71 concatenated PCGs in the cp genomes. The genus Xanthium was closely related to the genus Ambrosia (Fig. 8). Our analyses showed that Parthenium was a sister clade to both Xanthium and Ambrosia, and also an early-diverging lineage of the subtribe Ambrosiinae with a weak bootstrap value (57%).
Fig. 8.
Molecular phylogenetic tree based on 79 protein-coding genes of 21 Asteraceae chloroplast genomes. Achyrachaena mollis set as the outgroup. The tree was constructed by maximum likelihood analysis of the conserved regions using the RAxML program and the GTRI nucleotide model. The stability of each tree node was tested by bootstrap analysis with 1000 replicates. Bootstrap values are indicated on the branches, and the branch length reflects the estimated number of substitutions per 1000 sites
Discussion
The single circular cp genome structure of X. spinosum was similar to that of X. sibiricum with a typical quadripartite structure and equal GC content (37.45%) unevenly distributed across the cp genome. Relative to the LSC and SSC regions, the GC content is greater in IR regions across both cp genomes, possibly due to the presence of four extremely conserved rRNA genes with high GC content in these regions. The expansion and contraction of IR regions was the main cause of variation in cp genome size, and assessing these differences could shed light on the evolution of related taxa [30, 31]. The cp IR boundary regions of X. spinosum were compared to those of closely related species, and little difference was found, except for position changes in ѱycf1. The sizes of the four cp genomes (X. spinosum, X. sibiricum, A. artemisiifolia, and P. argentatum) were not affected. Moreover, the length of each region and the total genome size were similar to those of most plant cp genomes reported previously [32].
Repeat units, which are dispersed in cp genomes at high frequency, play a significant role in genome evolution [33–36]. Our comparative analyses of X. spinosum and S. sibiricum cp genomes showed a 24.4-fold higher level of repeats in X. spinosum. An earlier study reported that variation in the number and type of repeats may play a major role in plastome organization; however, we found no correlation between these large repeat regions and rearrangement endpoints [37]. SSRs, also known as microsatellite repeats [38, 39], are common in the cp genome, and these sequences display a high level of polymorphism, supporting their use as a genetic marker in previous investigations [40, 41]. The contents of different types of SSRs and their distributions among cp regions were similar in X. spinosum and X. sibiricum. Multiple definitions of repeat motifs and repeat number within motifs have been used in the literature; our SSR definition aligns with those of Bilgen et al. [42] and Karaca et al. [43].
The cp genomes of Xanthium showed less variation in non-coding regions than in their coding counterparts. The LSC region exhibited higher divergence levels than the IR and SSC regions (Fig. 6c). Specifically, the two IR regions were least divergent, perhaps due to the presence of four highly conserved rRNA sequences in those regions. The average nucleotide diversity (π) of intergenic regions was 0.0170, almost four times as high as that of PCGs (π = 0.004195), revealing that intergenic regions show greater divergence (Fig. 6d).
Not all PCGs are phylogenetically useful for determining taxonomic discrepancies [44]. In previous studies, several plastid and nuclear DNA markers from non-coding regions have been used to resolve the phylogenetic position of Xanthium species, leading to inconsistent results [11]. Hence, the use of the additional markers and broader taxonomic sampling are required to achieve greater phylogenetic resolution at low taxonomic levels [11, 45]. Therefore, in the present study, we proposed a set of 14 divergent regions between X. spinosum and X. sibiricum to resolve taxonomic discrepancies and provide a genetic barcode for the genus Xanthium. All of these regions are intergenic spacer regions, which might be useful for the development of molecular markers to use in phylogenetic and phylogeographic studies. The 14 sequences identified in the present study are extremely polymorphic compared to the sequences used in previous studies [6, 11, 45]. Based on our data, molecular markers can be developed for these intergenic regions that may be used for phylogenetic, phylogeographic, and barcoding studies of Xanthium. Moreover, this is the first report of the development of genetic markers based on these regions and their use to distinguish among Xanthium species. In addition, the nucleotide substitution rate and BEB analyses revealed that the accD gene may be under positive selection, and other positively selected sites detected in the present study may drive the accD PCG, supporting the occupation of various habitats [46, 47]. The earlier studies indicated that the gene accD encoded plastid beta carboxyl transferase subunit of acetyl-CoA carboxylase (ACCase) which is important for the proper chloroplast and as all stages of leaf growth [48], leaf longevity [49], fatty acid biosynthesis [50, 51] and embryo development [52]. Hence, the accD gene may have been involved in adaptation to specific ecological niches during the radiation of dicotyledonous plants [53].
Over the past few years, numerous plastid genome databases have been reported, offering an important foundation for resolving evolutionary, taxonomic, and phylogenetic questions in plants [54–60]. Our phylogenetic analyses showed that the genus Xanthium is most closely related to the genus Ambrosia. Several previous studies have used various methods including cladistic analyses [61, 62], cp restriction site variation assessments [63], and sequence analyses [11, 64] to understand the position of Xanthium, and these have shown that it is most closely related to Ambrosia species. Previous phylogenetic studies have shown that the genus Parthenium is an early-diverging lineage of the subtribe Ambrosiinae based on three plastid and two nuclear markers. We obtained consistent results, but with weak bootstrap support (57%). Somaratne et al. [6] suggested that Parthenium is not an early-diverging lineage of the subtribe Ambrosiinae, however, their phylogenetic analysis included only 46 cp PCGs. By contrast, we analyzed 71 PCGs in the present study, and the results suggest that Parthenium is an early-diverging lineage of subtribe Ambrosiinae.
Conclusion
We aimed to expand the molecular genetic resources available for the species X. spinosum through high-throughput sequencing and cp genome assembly. The structural characteristics of the X. spinosum cp genome is similar to other angiosperms. However, fourteen highly variable regions were detected and suggested as potential markers for future barcoding and phylogenetic studies of Xanthium species. Hence, the sequence data for the complete X. spinosum cp genome could be used as to distinguish among Xanthium species and resolve the phylogenetic relationships within the Ambrosiinae lineage.
Methods
DNA extraction and sequencing of Xanthium spinosum
Leaf material of Xanthium spinosum was obtained from Dr. George A Yatskievych, Curator, Plant Resources Center, University of Texas Herbarium (19–056), Austin, Texas, USA. Total genomic DNA was extracted using a modified cetyltrimethylammonium bromide method [65]. Illumina sequencing was carried out by LabGenomics, Seongnam, South Korea, using the Illumina HiSeq 2500 sequencing system. A paired-end library (150 × 2) was constructed with an insert size of 350 base pairs (bp). Read quality was analyzed with FastQC v0.11.9 [66] and low-quality reads were removed with Trimmomatic 0.39 [67]. The resultant clean reads were filtered using the GetOrganelle v1.6.0 pipeline (https://github.com/Kinggerm/GetOrganelle) to obtain plastid-like reads, and then the filtered reads were assembled de novo using SPAdes v3.12.0 [68]. The complete cp genome sequence of X. spinosum and its gene annotation were submitted to GenBank (MT668935).
Annotation of X. spinosum cp genome
The online program Dual Organellar GenoMe Annotator (DOGMA) was used to annotate the cp genome sequence of X. spinosum [69]. The initial annotation, putative starts, stops, and intron positions were fine-tuned through comparison with homologous genes in the closely related species X. sibiricum [6]. Transfer RNA genes were validated using tRNAscan-SE v1.21 with the default settings [70]. The program OGDRAW v1.3.1 was employed to draw a circular map of the X. spinosum cp genome [71].
Comparative cp genome analyses
The mVISTA program, which uses the Shuffle-LAGAN model, was employed to compare the cp genome of X. spinosum with three closely related cp genomes from X. sibiricum, Ambrosia artemisiifolia, and Parthenium argentatum using the X. spinosum annotation as a reference [72]. The boundaries between IR and SC regions of these species were also compared and investigated.
Repeat sequence and simple sequence repeats (SSRs) analyses
The program REPuter was used to predict the presence of repeat sequences in the X. spinosum and X. sibiricum cp genomes, including forward, reverse, palindromic, and complementary repeats [73]. The following parameters were used to identify repeats with REPuter: Hamming distance 3, minimum sequence identity of 90%, and repeat size > 30 bp. Phobos software v1.0.6 was employed to identify SSRs in the X. spinosum and X. sibiricum cp genomes; the match, mismatch, gap, and N positions parameters were set to 1, − 5, − 5, and 0, respectively [74]. For repeat and SSR marker analyses, only one IR region was used.
Anaglyses of genetic divergence
To analyze genetic divergence, the PCGs, intergenic, and intron-containing regions of the X. spinosum and X. sibiricum cp genomes were extracted and aligned independently using Geneious Prime v2020.1.2 (Biomatters, New Zealand). Genetic divergence between these Xanthium species was calculated based on nucleotide diversity (π) and the total number of polymorphic sites using DnaSP v5.10.01 [75]. For this analysis, gaps and missing data were excluded.
Characterization of substitution rates
To calculate the synonymous (KS) and nonsynonymous (KA) substitution rates, the cp genome of X. spinosum was compared to that of X. sibiricum. Corresponding single-functional PCG exons were extracted from both genomes and aligned independently using Geneious Prime v2020.1.2 (Biomatters, New Zealand). The aligned sequences were translated into protein sequences and analyzed using DnaSP v5.10.01 to obtain KA and KS substitution rates without stop codons.
Positive selection analyses
Positive selection (M2a and M8) and control (M1a, M7, and M8a) models provided in EasyCodeML software v1.21 [76] were used to identify the occurrence of positive selection (ω > 1) on the accD locus in Heliantheae cp genomes. The sequence of the accD gene was aligned using the program MAFFT v1.4.0 [77], and the maximum likelihood phylogenetic tree was constructed using RAxML v7.2.6 [78]. The site-specific model was used to calculate nonsynonymous (KA) and synonymous substitution (KS) rates using EasyCodeML. The codon substitution models M0, M1a, M2a, M3, M7, M8, and M8a were analyzed. The likelihood ratio test was used to identify positively selected sites in comparisons of M0 (one-ratio) vs. M3 (discrete), M1a (neutral) vs. M2a (positive selection), M7 (β) vs. M8 (β and ω > 1) and M8a ((β and ω = 1) vs. M8 using a site-specific model [76]. The likelihood ratio test (LRT) for these comparisons was used to evaluate the selection strength and p-values of less than 0.05 from the chi-square (χ2) test were considered significant. If the LRT p-values were significant (< 0.05), the Bayes Empirical Bayes (BEB) method was implemented to identify codons under positive selection. BEB values higher than 0.95 and 0.99 indicate sites that are potentially under positive selection and highly positive selection, respectively.
Phylogenetic tree analyses
A phylogenetic tree was constructed using 71 PCGs from 21 Asteroideae cp genomes, with L. fischeri as the outgroup. A total of 20 complete cp genome sequences were downloaded from the NCBI Organelle Genome Resource database. The aligned PCG sequences were saved in PHYLIP format using Clustal X v2.1 [79], and phylogenetic analyses were conducted based on the maximum likelihood (ML) method and the GTRI model using RAxML v7.2.6 with 1000 bootstrap replications [78].
Acknowledgments
Not applicable.
Abbreviations
- cp
Chloroplast
- LSC
Large single-copy
- SSC
Small single-copy
- IR
Inverted-repeat
- tRNA
Transfer RNA
- rRNA
Ribosomal RNA
- KS
Synonymous substitution
- KA
Nonsynonymous substitution
- ω
Nonsynonymous vs. synonymous ratio
- SSR
Simple sequence repeats
- LRT
Likelihood ratio test
- π
Nucleotide diversity
Authors’ contributions
SJP, JHK, and GR conceived and designed the experiments. GR and KP performed the experiments. GR analyzed the data and prepared a draft of the manuscript and figures. SJP, JHK, and GR modified the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by a project (PQ20180B009) entitled “Development of molecular markers for putative invasive alien” funded by the Research of Animal and Plant Quarantine Agency, South Korea.
Availability of data and materials
The dataset generated and or analysed during the current study is deposited in the genebank with accession number: MT668935. The phylogenetic genome datasets used and analysed in this study were retrieved from the National Center for Biotechnology Information Organelle Genome Resource Database.
Ethics approval and consent to participate
The plant was obtained from Dr. George A Yatskievych, Curator, Plant Resources Center, University of Texas Herbarium (19–056), Austin, Texas, USA with Institutional guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. [DOI] [PMC free article] [PubMed]
- 2.Palmer JD. Comparative organization of chloroplast genomes. Annu Rev Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- 3.Moore MJ, Dhingra A, Soltis PS, Shaw R, Farmerie WG, Folta KM, Soltis DE. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 2006;6:17. [DOI] [PMC free article] [PubMed]
- 4.Williams AV, Miller JT, Small I, Nevill PG, Boykin LM. Integration of complete chloroplast genome sequences with small amplicon datasets improves phylogenetic resolution in Acacia. Mol Phylogenet Evol. 2016;96:1–8. doi: 10.1016/j.ympev.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 5.Liu Q, Li X, Li M, Xu W, Schwarzacher T, Heslop-Harrison JS. Comparative chloroplast genome analyses of Avena: insights into evolutionary dynamics and phylogeny. BMC Plant Biol. 2020;20:406. doi: 10.1186/s12870-020-02621-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Somaratne Y, Guan D-L, Wang W-Q, Zhao L, Xu S-Q. Complete chloroplast genome sequence of Xanthium sibiricum provides useful DNA barcodes for future species identification and phylogeny. Plant Syst Evol. 2019;305(10):949–960. doi: 10.1007/s00606-019-01614-1. [DOI] [Google Scholar]
- 7.Amin S, Khan H. Revival of natural products: utilization of modern technologies. Curr Bioactive Compounds. 2016;12(2):103–106. doi: 10.2174/1573407212666160314195845. [DOI] [Google Scholar]
- 8.Robbins W. Alien plants growing without cultivation in California. Bulletin Calif Agric Exp Station. 1940;637:1–128. [Google Scholar]
- 9.Holm L, Pluncknett D, Pancho J, Herberger J. The world's worst weeds. Honolulu, USA: University Press of Hawaii; 1977. [Google Scholar]
- 10.Munz P, Keck D. A California flora and supplement. Berkeley, California, USA: University of California Press; 1973. [Google Scholar]
- 11.Tomasello S, Heubl G. Phylogenetic analysis and molecular characterization of Xanthium sibiricum using DNA barcoding, PCR-RFLP, and specific primers. Planta Med. 2017;83(11):946–953. doi: 10.1055/s-0043-106585. [DOI] [PubMed] [Google Scholar]
- 12.Romero M, Zanuy M, Rosell E, Cascante M, Piulats J, Font-Bardia M, Balzarini J, De Clerq E, Pujol MD. Optimization of xanthatin extraction from Xanthium spinosum L. and its cytotoxic, anti-angiogenesis and antiviral properties. Eur J Med Chem. 2015;90:491–496. doi: 10.1016/j.ejmech.2014.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salinas A. Ruiz RELd, Ruiz SO: sterols, flavonoids, and sesquiterpenic lactones from Xanthium spinosum (Asteraceae) Acta Farm Bonaer. 1998;17(4):297–300. [Google Scholar]
- 14.Ginesta-Peris E, Garcia-Breijo FJ, Primo-Yufera E. Antimicrobial activity of xanthatin from Xanthium spinosum L. Lett Appl Microbiol. 1993;18:206–208. doi: 10.1111/j.1472-765X.1994.tb00848.x. [DOI] [Google Scholar]
- 15.Yoon JH, Lim HJ, Lee HJ, Kim HD, Jeon R, Ryu JH. Inhibition of lipopolysaccharide-induced inducible nitric oxide synthase and cyclooxygenase-2 expression by xanthanolides isolated from Xanthium strumarium. Bioorg Med Chem Lett. 2008;18(6):2179–2182. doi: 10.1016/j.bmcl.2007.12.076. [DOI] [PubMed] [Google Scholar]
- 16.Willians RH, Martin FB, Henley ED, Swanson HE. Inhibitors of insulin degradation, metabolism. Clin Exp Rheumatol. 1959;8:99–113. [PubMed] [Google Scholar]
- 17.Naidenova E, Kolarova-Pallova I, Popov D, Dimitrova-Konaklieva S, Dryanovska-Noninska L. Isolation and obtaining of sesquiterpene lactones with antitumor properties – xanthinin, stizolicin, and solstitialin. Natl Oncol Cent Med Acad. 1988;41:105–106. [Google Scholar]
- 18.Cunat P, Primo E, Sanz I, Garcera MD, March MC, Bowers WS, Martinez-Pardo R. Biocidal activity of some Spanish Mediterranean plants. J Agric Food Chem. 1990;38(2):497–500. doi: 10.1021/jf00092a036. [DOI] [Google Scholar]
- 19.Turnne CY, Sanche SE, Hoban DJ, Karylowsky JA, Kabani AM. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J Clin Microbiol. 1999;37(6):1846–1851. doi: 10.1128/JCM.37.6.1846-1851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trobajo R, Mann DG, Clavero E, Evans KM, Vanormelingen P, McGregor RC. The use of partial cox1, rbcL and LSU rDNA sequences for phylogenetics and species identification within the Nitzschia palea species complex (Bacillariophyceae) Eur J Phycol. 2010;45(4):413–425. doi: 10.1080/09670262.2010.498586. [DOI] [Google Scholar]
- 21.Liu C, Liang D, Gao T, Pang X, Song J, Yao H, Han J, Liu Z, Guan X, Jiang K, et al. PTIGS-IdIt, a system for species identification by DNA sequences of the psbA-trnH intergenic spacer region. BMC Bioinformatics. 2011;12:S4. doi: 10.1186/1471-2105-12-S13-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Millspaugh C, Sherff E. Revision of the north American species of Xanthium. Field Mus Nat Hist Zool Ser. 1919;4:9–49. [Google Scholar]
- 23.Widder F: Die Arten der Gattung Xanthium. Beiträge zu einer Monographie. Repert Spec Nov Regn Veget 1923, 20:1–223.
- 24.Löve D, Dansereau L. Biosystematic studies on Xanthium: taxonomic appraisal and ecological status. Can J Bot. 1959;37:173–208. doi: 10.1139/b59-016. [DOI] [Google Scholar]
- 25.Strother J. Flora of North America. New York: Oxford University; 2006. Xanthium. [Google Scholar]
- 26.Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS One. 2011;6(5):e19254. [DOI] [PMC free article] [PubMed]
- 27.Sanita Lima M, Woods LC, Cartwright MW, Smith DR. The (in) complete organelle genome: exploring the use and nonuse of available technologies for characterizing mitochondrial and plastid chromosomes. Mol Ecol Resour. 2016;16(6):1279–1286. doi: 10.1111/1755-0998.12585. [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, Hahn FM, McMahan CM, Cornish K, Whalen MC. Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biol. 2009;9. [DOI] [PMC free article] [PubMed]
- 29.Amiryousefi A, Hyvonen J, Poczai P. The plastid genome sequence of the invasive plant common ragweed (Ambrosia artemisiifolia, Asteraceae) Mitochondrial DNA B. 2017;2(2):753–754. doi: 10.1080/23802359.2017.1390423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boudreau E, Turmel M. Gene rearrangements in Chlamydomonas chloroplast DNAs are accounted for by inversions and by the expansion/contraction of the inverted repeat. Plant Mol Biol. 1995;27(2):351–364. doi: 10.1007/BF00020189. [DOI] [PubMed] [Google Scholar]
- 31.Nazareno AG, Carlsen M, Lohmann LG. Complete chloroplast genome of Tanaecium tetragonolobum: the first bignoniaceae plastome. PLoS One. 2015;10(6):e0129930. [DOI] [PMC free article] [PubMed]
- 32.Zhang X, Zhou T, Kanwal N, Zhao YM, Bai GQ, Zhao GF. Completion of eight Gynostemma BL. (Cucurbitaceae) chloroplast genomes: Characterization, comparative analysis, and phylogenetic relationships. Front Plant Sci. 2017;8:1583. [DOI] [PMC free article] [PubMed]
- 33.Dong WP, Xu C, Cheng T, Lin K, Zhou SL. Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol Evol. 2013;5(5):989–997. doi: 10.1093/gbe/evt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu WZ, Kong HH, Zhou J, Fritsch PW, Hao G, Gong W. Complete chloroplast genome of Cercis chuniana (Fabaceae) with structural and genetic comparison to six species in Caesalpinioideae. Int J Mol Sci. 2018;19(5):1286. [DOI] [PMC free article] [PubMed]
- 35.Xie DF, Yu Y, Deng YQ, Li J, Liu HY, Zhou SD, He XJ. Comparative analysis of the chloroplast genomes of the Chinese endemic genus Urophysa and their contribution to chloroplast phylogeny and adaptive evolution. Int J Mol Sci. 2018;19(7):1847. [DOI] [PMC free article] [PubMed]
- 36.Shi HW, Yang M, Mo CM, Xie WJ, Liu C, Wu B, Ma XJ. Complete chloroplast genomes of two Siraitia merrill species: Comparative analysis, positive selection and novel molecular marker development. PLoS One. 2019;14(12):e0226865. [DOI] [PMC free article] [PubMed]
- 37.Liu Y, Huo NX, Dong LL, Wang Y, Zhang SX, Young HA, Feng XX, Gu YQ. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS One. 2013;8(2):e57533. [DOI] [PMC free article] [PubMed]
- 38.Ince AG, Karaca M, Elmasulu SY. New microsatellite and CAPS-microsatellite markers for clarifying taxonomic and phylogenetic relationships within Origanum L. Mol Breed. 2014;34(2):643–654. doi: 10.1007/s11032-014-0064-9. [DOI] [Google Scholar]
- 39.Ince AG, Karaca M, Onus AN. Polymorphic microsatellite markers transferable across Capsicum species. Plant Mol Biol Report. 2009;28(2):285–291. doi: 10.1007/s11105-009-0151-y. [DOI] [Google Scholar]
- 40.Smith JSC, Chin ECL, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Ziegle J. An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPS and pedigree. Theoritical applied Genetics. 1997;95:163–173. doi: 10.1007/s001220050544. [DOI] [Google Scholar]
- 41.Kawabe A, Nukii H, Furihata HY. Exploring the history of chloroplast capture in Arabis using whole chloroplast genome sequencing. Int J Mol Sci. 2018;19(2):602. [DOI] [PMC free article] [PubMed]
- 42.Karaca M, Bilgen M, Onus AN, Ince AG, Elmasulu SY. Exact tandem repeats analyzer (E-TRA): a new program for DNA sequence mining. J Genet. 2005;84(1):49–54. doi: 10.1007/BF02715889. [DOI] [PubMed] [Google Scholar]
- 43.Bilgen M, Karaca M, Onus AN, Ince AG. A software program combining sequence motif searches with keywords for finding repeats containing DNA sequences. Bioinformatics. 2004;20(18):3379–3386. doi: 10.1093/bioinformatics/bth410. [DOI] [PubMed] [Google Scholar]
- 44.Pfeil BE, Brubaker CL, Craven LA, Crisp MD. Phylogeny of Hibiscus and the tribe Hibisceae (Malvaceae) using chloroplast DNA sequences of ndhF and the rpl16 intron. Syst Bot. 2002;27(2):333–350. [Google Scholar]
- 45.Wallace LJ, Boilard SMAL, Eagle SHC, Spall JL, Shokralla S, Hajibabaei M. DNA barcodes for everyday life: routine authentication of natural health products. Food Res Int. 2012;49(1):446–452. doi: 10.1016/j.foodres.2012.07.048. [DOI] [Google Scholar]
- 46.Hu QM, Zhu Y, Liu Y, Wang N, Chen SL. Cloning and characterization of wnt4a gene and evidence for positive selection in half-smooth tongue sole (Cynoglossus semilaevis). Sci Rep-Uk. 2014;4:7167. [DOI] [PMC free article] [PubMed]
- 47.Dhar D, Dey D, Basu S, Fortunato H. Understanding the adaptive evolution of mitochondrial genomes in intertidal chitons. bioRxiv. 2020; 10.1101/2020.03.06.980664.
- 48.Kode V, Mudd EA, Iamtham S, Day A. The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 2005;44:237–244. doi: 10.1111/j.1365-313X.2005.02533.x. [DOI] [PubMed] [Google Scholar]
- 49.Madoka Y, Tomizawa K, Mizoi J, Nishida I, Nagona Y, Sasaki Y. Chloroplast transformation with modified accD operon increases acetyl-CoA carboxylase and causes extension of leaf longevity and increase in seed yield in tobacco. Plant Cell Physiol. 2002;43:1518–1525. doi: 10.1093/pcp/pcf172. [DOI] [PubMed] [Google Scholar]
- 50.Ohlrogge J, Browse J. Lipid biosynthesis. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sasaki Y, Nagano Y. Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci Biotechnol Biochem. 2004;68:1175–1184. doi: 10.1271/bbb.68.1175. [DOI] [PubMed] [Google Scholar]
- 52.Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D. Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol. 2011;155:1678–1689. doi: 10.1104/pp.110.168120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu S, Sablok G, Wang B, Qu D, Barbaro E, Viola R, et al. Plastome organization and evolution of chloroplast genes in Cardamine species adapted to contrasting habitats. BMC Genomics. 2015;16:306. doi: 10.1186/s12864-015-1498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boudreau E, Takahashi Y, Lemieux C, Turmel M, Rochaix JD. The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 1997;16(20):6095–6104. doi: 10.1093/emboj/16.20.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang JB, Tang M, Li HT, Zhang ZR, Li DZ. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol Biol. 2013;13. [DOI] [PMC free article] [PubMed]
- 56.Raman G, Park S. The complete chloroplast genome sequence of Ampelopsis: gene organization, comparative analysis, and phylogenetic relationships to other angiosperms. Front Plant Sci. 2016;7. [DOI] [PMC free article] [PubMed]
- 57.Zhang YJ, Du LW, Liu A, Chen JJ, Wu L, Hu WM, Zhang W, Kim K, Lee SC, Yang TJ, et al. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front Plant Sci. 2016;7. [DOI] [PMC free article] [PubMed]
- 58.Kahraman K, Lucas SJ. Comparison of different annotation tools for characterization of the complete chloroplast genome of Corylus avellana cv Tombul. BMC Genomics. 2019;20(1):874. [DOI] [PMC free article] [PubMed]
- 59.Li XQ, Zuo YJ, Zhu XX, Liao S, Ma JS. Complete chloroplast genomes and comparative analysis of sequences evolution among seven Aristolochia (Aristolochiaceae) medicinal species. Int J Mol Sci. 2019;20(5):1045. [DOI] [PMC free article] [PubMed]
- 60.Li CJ, Wang RN, Li DZ. Comparative analysis of plastid genomes within the Campanulaceae and phylogenetic implications. PLoS One. 2020;15(5):e0233167. [DOI] [PMC free article] [PubMed]
- 61.Karis P. Heliantheae sensu lato (Asteraceae), clades and classification. Plant Syst Evol. 1993;188:139–195. doi: 10.1007/BF00937727. [DOI] [Google Scholar]
- 62.Karis P. Cladistics of the subtribe Ambrosiinae (Asteraceae: Heliantheae) Syst Bot. 1995;20:40–54. doi: 10.2307/2419631. [DOI] [Google Scholar]
- 63.Miao B, Turner B, Mabry T. Systematic implications of chloroplast DNA variation in the subtribe Ambrosiinae (Asteraceae: Heliantheae) Am J Bot. 1995;82:924–932. doi: 10.1002/j.1537-2197.1995.tb15709.x. [DOI] [Google Scholar]
- 64.Martin MD, Quiroz-Claros E, Brush GS, Zimmer EA. Herbarium collection-based phylogenetics of the ragweeds (Ambrosia, Asteraceae) Mol Phylogenet Evol. 2018;120:335–341. doi: 10.1016/j.ympev.2017.12.023. [DOI] [PubMed] [Google Scholar]
- 65.Doyle J. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- 66.Andrews S: FASTQC. A quality control tool for high throughput sequence data 2010.
- 67.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20(10):714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20(17):3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 70.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33(Web Server issue):W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52(5–6):267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 72.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32(Web Server issue):W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29(22):4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mayer C, Leese F, Tollrian R. Genome-wide analysis of tandem repeats in Daphnia pulex--a comparative approach. BMC Genomics. 2010;11:277. doi: 10.1186/1471-2164-11-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 76.Gao F, Chen C, Arab DA, Du Z, He Y, Ho SYW. EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol Evol. 2019;9:3891–3898. doi: 10.1002/ece3.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 79.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset generated and or analysed during the current study is deposited in the genebank with accession number: MT668935. The phylogenetic genome datasets used and analysed in this study were retrieved from the National Center for Biotechnology Information Organelle Genome Resource Database.