To the Editor:
The upper airway microbiota develops over the first year of life with alterations in the natural development of the upper airway associated with increased risk for lower respiratory illnesses and wheeze during the first few years of life1. While studies have linked the increased presence of Streptococcus, Haemophilus and Moraxella in the infant airway with recurrent wheeze1, there remains a lack of objective data linking airway microbiota and airway measurements.
Lung function has been successfully measured during infancy with several studies demonstrating a link between decreased flows (airway obstruction) and later development of wheeze. In the Tucson Children’s Respiratory Study2, children with wheezing prior to the age of 3 years and a diagnosis of asthma at the age of 6 years, had significantly lower lung function during infancy (measured as maximal flow at functional residual capacity or Vmax FRC) compared to children who did not wheeze. These findings combined with the established link between reduced lung function in infancy and wheeze during the first years of life2, suggest that airway obstruction is established early in life. While an association between airway bacteria and infant lung function has not been explored, adults with asthma have increased airway hyper-responsiveness when decreased bacterial diversity is present in the airway3. To determine if infant pulmonary function testing could be associated with microbiota data, we performed a pilot study utilizing the first 22 samples obtained from our larger cohort described below. We hypothesized that a decrease in overall bacterial diversity in the upper airway is associated with lower lung function. To test this hypothesis, we prospectively obtained upper airway samples and lung function measurements in infants at 3 months of life.
Children included in this analysis were enrolled in a larger study evaluating the impact of the microbiota on airway inflammation and lung function during the first year of life. Full-term neonates (≥37 weeks gestation age) born to women with a history of physician diagnosed asthma, in addition to previous or current use of inhaled corticosteroids and/or albuterol, were recruited within 48 hours of birth and followed prospectively for the first year of life. Newborns with respiratory distress, requiring oxygen, and/or genetic diseases were excluded. At three months of age, nasopharyngeal (NP) swabs were obtained on the same day that sedated infant lung function testing was performed. Questionnaires regarding wheezing episodes and medication utilization were collected monthly for the first three months, then bi-monthly for the remaining 9 months of the study. Wheeze was defined as physician diagnosed wheezing and use of a bronchodilator for that episode of wheeze. This study included 16 participants without wheeze, and 6 participants with wheeze. 16S rRNA gene sequencing was performed on bacterial DNA isolated from each sample. Sequence processing was performed utilizing mothur (v.1.33.3) software and OTUs were generated using Swarm (v 2.1.9). OTUs with a relative abundance less than twice as high in samples as negative controls were removed as potential contaminants. An abundance filter was replied in which OTUs that did not compose at least 1% of at least one sample were removed. QIIME2 (2018.8) was utilized to analyze alpha and beta diversity. Students t-tests were performed to analyze differentially abundant bacterial genera.
Analyses were performed to determine associations between the variables of interest, using SAS v9.4 (SAS Institute, Cary, NC). Student’s t-tests were performed to analyze continuous data between groups, correlation analyses for comparing continuous variables, and logistic regression models for binary distributions.
Within the group of infants that did not wheeze during the first year of life (n=16), the alpha diversity (Shannon diversity) of the three month samples was higher compared to the infants with early wheeze (n=6) (p=0.03). Beta diversity was also measured to examine the variability between the microbiota of those with early wheeze versus those without wheeze, and found to be significant (weighted Unifrac PERMANOVA p=0.05). Participants with early wheeze had a microbiota dominated by Moraxella (relative abundance of early wheeze: 86% vs. no wheeze: 42%, adjusted p=0.01), with lower abundance of Corynebacterium (early wheeze: 8% vs. no wheeze: 22%, adjusted p=0.01), compared to those without early wheeze. There was no significant difference in the relative abundance of other bacteria present at ≥ 1% of abundance (Haemophilus, Streptococcus, Staphylococcus, Dolosigranulum, and Neisseria).
Infants underwent sedated infant lung function testing, prior to their first respiratory illness and their first diagnosis of wheeze, based on standardized protocols using the Carefusion MasterScreen™ BabyBody (Hoechberg, Germany).4 Median age at time of testing was 108 days. Measurements included: functional residual capacity (FRC), forced expiratory volume in 0.5 seconds (FEV0.5), forced vital capacity (FVC), forced expiratory flow at 50% and 75% of FVC (FEF 50 and FEF75), and forced expiratory flows between 25% and 75% of FVC (FEF25–75%). Because the demographics varied between study participants, values were adjusted for age, length, gender, and race. Our findings support previous findings, with the wheeze group having a lower FEV0.5 (mean and range z-score of wheeze group: −0.78 (−2.05, −0.06) vs. 0.20 (−1.31, 1.65), p=0.01). When examining associations between lung function measurements and airway diversity, independent of wheeze status, we found that infants with lower Shannon diversity had lower FEV0.5 (R2=0.26, p=0.01) (Figure 1); lower FEF25–75% (R2 0.11, p=0.05); and a trend for lower FEF50 (R2=0.07, p=0.08). We did not find an association between Shannon diversity and other lung function measurements. Associations between beta-diversity and lung function values were not significant but there was a trend for an association between unweighted Unifrac measurements and FEV0.5 (p=0.08). We next performed a logistic regression analysis to determine if differences in the abundance of the most abundant bacteria were associated with changes in lung function values. An association was found between lower FEV0.5 z-scores and Moraxella when Moraxella abundance was ≥ 75% of total bacterial abundance, (0.29 vs. −0.50; p=.03). No other associations were found between specific bacteria and lung function values.
Figure 1:
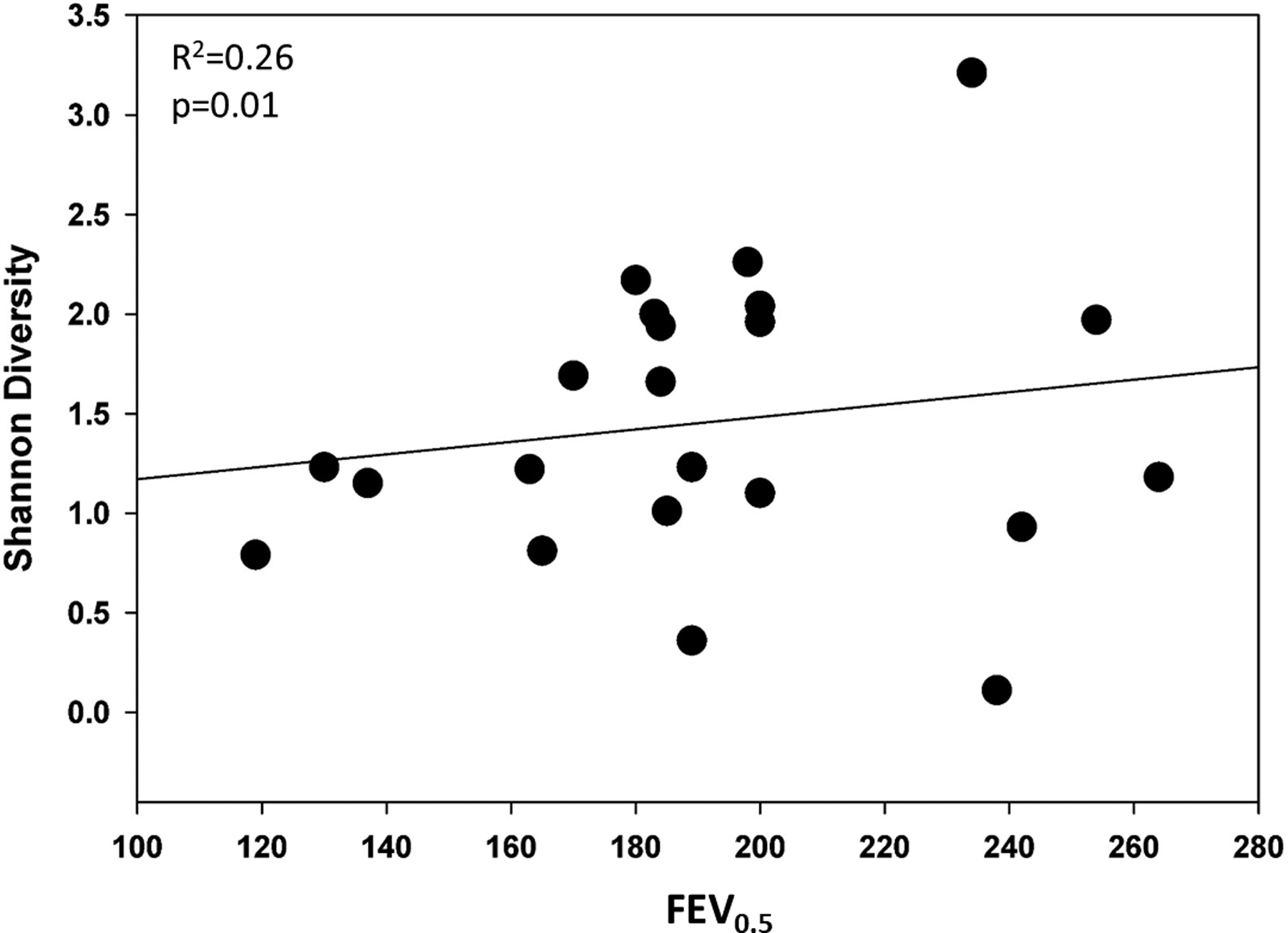
Association between Shannon diversity and FEV0.5. Adjusted for age, length, gender and race.
This study has a number of advantages and some limitations. The prospective design allowed us to obtain data about wheezing over the first year of life thereby decreasing recall bias. Samples were obtained on the same day as lung function testing eliminating a delay in the association between the presence of bacteria and lung function measurements. While most studies have evaluated the upper airway of Caucasian children, our study contains a racially diverse population (15 African-American, 4 Caucasian, 3 other) and may provide further insight into differing microbial profiles of a diverse population of children. Our study had a high rate of furry pet exposure in the no wheeze group (9/16) versus the wheeze group (0/6). It is possible that this exposure alters the airway microbiota, provides protection against early wheeze, as reported in previous studies.5 Finally, our results should be interpreted with caution due to the small sample size and the observational study design. Because this is a pilot study we could not confidently adjust for multiple comparisons such as mode of delivery, smoke exposure, and/or gender. However, we were able to look at each comparison as an independent variable. There was not a difference in the microbiota composition when each demographic value was analyzed independently. It is possible that a larger sample size will reveal microbiota differences.
We previously demonstrated that some differences in microbiota exist between nasal pharyngeal and bronchoalveolar lavage samples obtained in the same participant.6 A limitation of our study is the use of upper airway samples as a surrogate of the lower airway. Obtaining lower airway samples is not ethically possible in healthy infants due to the need for sedation when performing bronchoscopies. Given this, it cannot be concluded that these bacteria are directly causing lung function changes. However, it is possible that bacteria located in the upper airway are migrating into the lower airway, causing local inflammation and leading to the airway abnormalities. Future studies should focus on prospectively following neonates starting at birth to determine if the presence of a high abundance of Moraxella leads to early airway inflammation and ultimately obstructive lung disease.
In contrast to previous reports, our findings did not find a high abundance of Streptococcus in the upper airway of children with early wheeze. We found that higher bacterial diversity in the upper airway is linked with higher pulmonary function measurements. We also found that a high abundance of Moraxella is associated with early wheeze and lower FEV0.5 measurements. While we did not find a predictive value of Moraxella abundance to determine lower lung function values, we believe that the addition of more samples to our cohort will help us determine if a link exists, and could be utilized as a marker for future development of early wheeze.
Acknowledgments
The following grants supported this research:
National Institute for Allergy, Infections Disease (NIAID) K23 Career development Award 1 K23 AI135094-01
K-12 Indiana University School of Medicine (IUSM) Indiana Pediatric Scientist Award (IPSA) program through the Child Health Research Career Development Award (CHRCDA). 1K12HD068371-01A1
Showalter Trust Award
American Academy of Allergy, Asthma and Immunology (AAAAI) Foundation Faculty Development Award
Children’s Clinical Research Center, a member of the Indiana Clinical and Translational Sciences Institute supported by grant UL1TR001108 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award.
Lilly Physician Scientist Initiative Award
Author Disclaimers: Dr. Davis reports grants from Eli Lilly Physician Scientist Award during the conduct of the study. All other others have nothing to disclose.
References
- 1.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell host & microbe. 2015;17(5):704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taussig LM, Wright AL, Holberg CJ, Halonen M, Morgan WJ, Martinez FD. Tucson Children’s Respiratory Study: 1980 to present. The Journal of allergy and clinical immunology. 2003;111(4):661–675; quiz 676. [DOI] [PubMed] [Google Scholar]
- 3.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, Woyke T, Allgaier M, Bristow J, Wiener-Kronish JP, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. The Journal of allergy and clinical immunology. 2011;127(2):372–381 e371–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beydon N, Davis SD, Lombardi E, Allen JL, Arets HG, Aurora P, Bisgaard H, Davis GM, Ducharme FM, Eigen H, et al. An official American Thoracic Society/European Respiratory Society statement: pulmonary function testing in preschool children. American journal of respiratory and critical care medicine. 2007;175(12):1304–1345. [DOI] [PubMed] [Google Scholar]
- 5.Fall T, Lundholm C, Ortqvist AK, Fall K, Fang F, Hedhammar A, Kampe O, Ingelsson E, Almqvist C. Early Exposure to Dogs and Farm Animals and the Risk of Childhood Asthma. JAMA pediatrics. 2015;169(11):e153219. [DOI] [PubMed] [Google Scholar]
- 6.Kloepfer KM, Deschamp AR, Ross SE, Peterson-Carmichael SL, Hemmerich CM, Rusch DB, Davis SD. In children, the microbiota of the nasopharynx and bronchoalveolar lavage fluid are both similar and different. Pediatric pulmonology. 2018;53(4):475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]


