Abstract
Leucine-rich repeat-containing G protein-coupled receptor 5 (Lgr5) has been identified as a marker of stem cells across multiple tissues. Lgr5-expressing cells are also regulators of tissue homeostasis and wound repair, and drivers of carcinogenic progression. The majority of information about Lgr5-expressing cells derives from genetically engineered mouse models. Human studies have been limited by a lack of specific reagents and experimental procedures for the purification of these cells. We recently demonstrated that antibody-based purification can be used to obtain viable LGR5-expressing cells from human primary tissues and patient derived organoids. Here, we provide detailed methods for the purification of these cells from colonic epithelial organoids generated from patient-derived tissues, from induced pluripotent stem cell (iPSC) derived intestinal organoids, and from freshly isolated patient tissue intestinal crypts. These methods will facilitate experimental analysis of human LGR5-expressing cells in development, wound healing, and cancer.
Keywords: Lgr5, Organoid, Colonoid, Enteroid, Antibody, MACS, Colon, Intestine, Stem cell, Human
1. Introduction
Leucine-rich repeat-containing G protein-coupled receptor 5 (Lgr5) has been characterized as a stem cell marker across multiple organ sites [1–4]. The LGR family of proteins act as receptors for R-spondin proteins, which function to potentiate Wnt/β-catenin signaling [5–11]. Lgr5 was first identified as a stem cell marker in the mouse intestine, where it marked a select subpopulation of cells at the base of the crypt [1]. Lineage tracing experiments have confirmed that Lgr5+ cells are actively cycling stem cells at the crypt base and give rise to differentiated cell lineages along the gastrointestinal tract. Lgr5 expressing cells are essential for tissue homeostasis, injury repair and regeneration in a number of tissues, including the pancreas, small intestine, stomach, and hair follicles [12–15]. Lineage tracing studies have also shown that Lgr5 marks a population of tumor initiating cells in precancerous adenoma lesions, which precede invasive cancer development [16]. Lgr5-expressing cells are implicated as the drivers of metastasis in colon cancer [17]. Due to their established role in cancer initiation and metastasis, Lgr5-expressing cells are currently under intense investigation as a target for chemotherapeutics [18, 19].
Many fundamental discoveries about stem cell biology have been made using mouse models genetically engineered to express an Lgr5 reporter [1]. Characterizing the biology of Lgr5 expressing cells in normal and tumor tissues from humans has been more challenging, due to a lack of methods for the accurate identification and purification of Lgr5-expressing cells, specifically due to unsuccessful efforts to generate effective LGR5-targeting antibodies [20]. RNA in situ hybridization has been utilized to detect LGR5-expressing cells in human tissues [21, 22]; however, this approach does not allow for the isolation of live cell populations. More recently, LGR5-expression reporter human organoids lines have been developed using gene editing techniques [23]; however, these methods are not broadly applicable to unmodified primary human tissues. We describe here the procedures for the dissociation of human organoids or fresh tissue crypts into viable single cells, labeling of cells with a magnetic bead-bound anti-LGR5 antibody, magnetic separation, and flow cytometry analysis (Fig. 1). Each aspect of this procedure required rigorous optimization, and we detail the culmination of that optimization process here. We have applied this procedure to human colonoids (normal and adenoma derived epithelial organoids) [24], human pluripotent stem cell (hPSC)-derived intestinal organoids, and freshly prepared normal colon tissue crypts [24]. The initiation and maintenance of colonoid or hPSC-derived organoid cultures is well established and previously described [25–28]. Primary colon organoids can be grown in conditions that are highly enriched in stem cells [1] or that recapitulate the tissue differentiation hierarchy [29]. Thus, patient-derived organoids provide a robust experimental platform for the interrogation of human LGR5 expressing cells. This protocol enables the antibody-based purification of viable LGR5-expressing cells, which will facilitate the experimental analysis of these cells in development, tissue homeostasis, wound repair, and carcinogenesis.
Fig. 1.
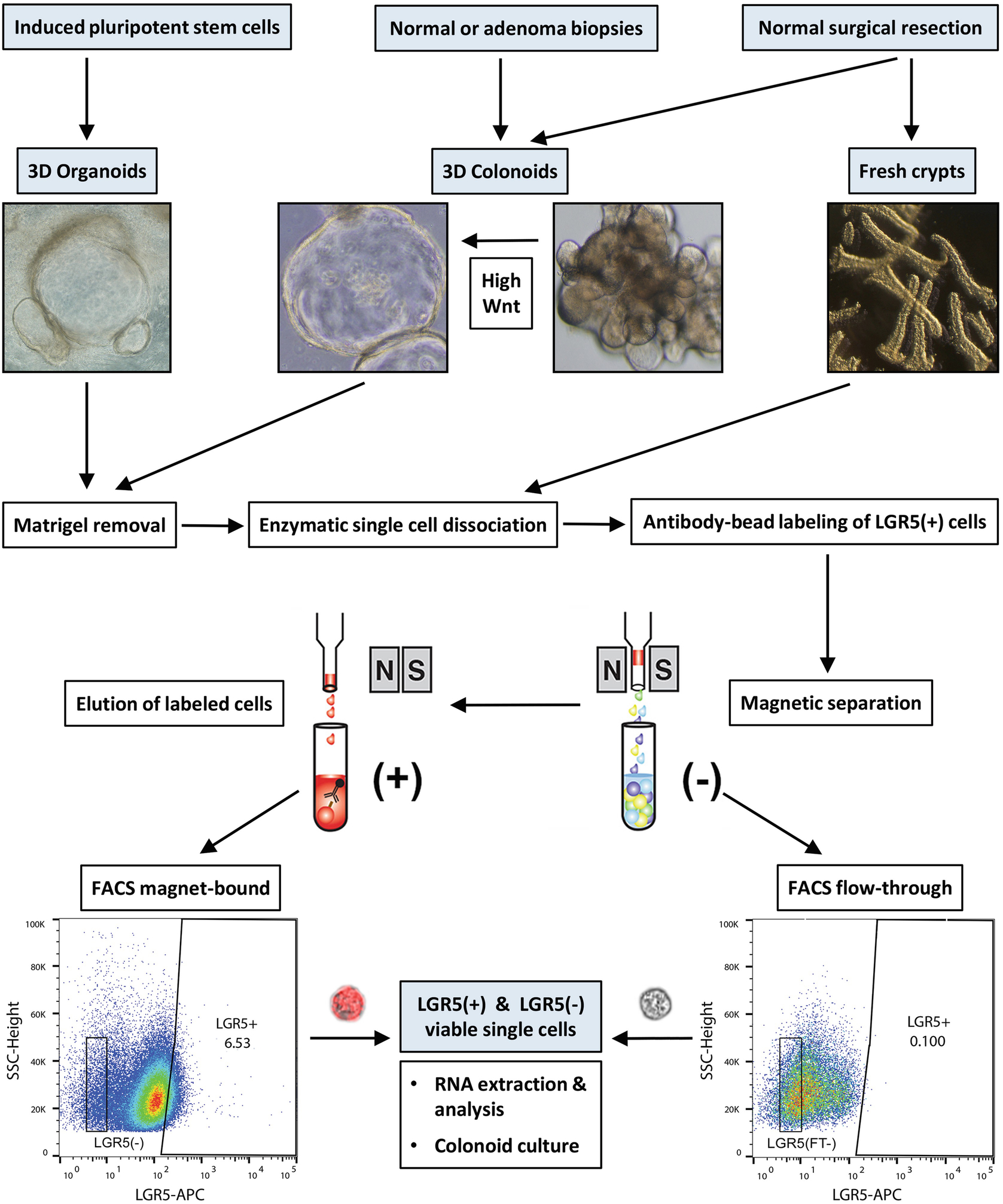
Graphical illustration of the strategy for the isolation of LGR5(+) cells. Outlined here are methods to isolate LGR5(+) cells from cultured human colonoids (normal and adenoma-derived), iPSC-derived organoids (composed of epithelium and mesenchyme), and from freshly isolated colonic tissue crypts. High-Wnt3a containing L-WRN medium is used to drive a thin-walled cystic morphology in the colonoid culture, enriching for the stem cell component. Matrigel is first removed, followed by single cell dissociation with a gentle enzyme preparation. Cells are labeled with anti-human LGR5 antibody-magnetic bead conjugate, followed by an anti-bead allophycocyanin (APC) stain. Where mesenchyme is present or contaminating, the epithelial marker EpCAM is used to discriminate epithelium. Cells are passed through magnetic columns to enrich for the LGR5-magnetic bead fraction, and FACS sorted for DAPI(−) and LGR5-APC(+) cells. Representative scatterplots are for cells isolated from an adenoma-derived colonoid. Final LGR5(+) and LGR5(−) single cells flow-imaged with the Amnis ImageStreamX (60× magnification; brightfield and APC fluorescence). (Content adapted from Dame et al., 2018 [24] with permission from Development)
2. Materials
All plasticware that comes into contact with tissue, organoids, or cells throughout the procedure is coated with 0.1% bovine serum albumen to minimize adherence of cells to plastic surfaces (see Note 1). Where noted, BSA-coated pipette tips are cut to minimize shearing of cells. Manipulations should be done on ice to maximize viability and epitope integrity. All solutions are kept cold in ice, with the exception of the 37 °C Tumor Dissociation Kit (TDK) enzyme preparation.
2.1. Removal of Matrigel™ from Cultured Colonoid Structures
Plasticware: 5 mL serological pipettes, 15 mL conical tubes.
Cell lifter.
Tube rotator.
Swinging bucket refrigerated centrifuge.
DPBS.
Y27632: 2.5 mM Y27632 stock solution in H2O (see Note 2).
2 mM EDTA-Y27632 solution: 2 mM ethylenediaminetetraacetic acid (EDTA) solution in DPBS, pH 7.3–7.4, containing 5 μM Y27632.
Enzyme Buffer (without enzymes): HBSS, 0.13 mM calcium, 0.9 mM magnesium, 5 μM Y27632 (see Note 3).
Human colonoids: cultures embedded in 10–50 μL-sized adherent droplets of 8 mg/mL Matrigel™ with 250 μL Matrigel™/well of a 6-well dish (see Notes 4 and 5).
2.2. Removal of Matrigel™ from Cultured iPSC-Derived Organoid Structures
4 mM EDTA-Y27632 solution (in place of 2 mM EDTA-Y27632 solution): 4 mM ethylenediaminetetraacetic acid (EDTA) solution in DPBS, pH 7.3–7.4, containing 5 μM Y27632.
Human iPSC organoids: cultures are embedded in 50–100 μL sized adherent droplets of 8 mg/mL Matrigel™ with 400 μL Matrigel™/well of a 6-well dish (see Note 5).
2.3. Single Cell Dissociation of Human Colonoids, Organoids or Fresh Colonic Crypts
Plasticware: 5 mL serological pipettes, 15 and 50 mL conical tubes.
Cell strainers: 100, 40, and 20 μm.
gentleMACS™ Dissociator.
37 °C incubator.
gentleMACS™ C Tubes.
Countess Automated Cell Counter.
Tumor Dissociation Kit (TDK) enzyme preparation: 400 μL enzyme H, 200 μL enzyme R, 50 μL enzyme A, add up to 10 mL total Enzyme Buffer including cell pellet volume (see Note 6).
Labeling Buffer: 2 mM EDTA-Y27632, 0.5% BSA.
Trypan Blue solution.
2.4. Antibody Labeling
2.4.1. LGR5 Antibody Labeling
Plasticware: 2 mL V-bottom Eppendorf tubes, cut-down P200 pipette tips, uncut P200 pipette tips.
MACS™ LS Column.
FcR Blocking Reagent.
Anti-human Lgr5-Microbead: microbead conjugated to monoclonal anti-human LGR5, rat IgG2b, clone 22H2.8.
Labeling Check Reagent-allophycocyanin (APC): protect from light.
Labeling Buffer: 2 mM EDTA-Y27632, 0.5% BSA.
Column Buffer: Enzyme Buffer, 0.5% BSA, 200 Kunitz units/mL DNase.
Flow Buffer: 2 mM EDTA, DPBS, 0.1% BSA, 10 μM Y27632.
2.4.2. EpCAM Labeling of iPSC-Derived Organoid or Freshly Isolated Crypt Cells
Plasticware: 2 mL V-bottom Eppendorf tubes, cut-down P200 pipette tips, uncut P200 pipette tips.
EpCAM antibody: phycoerythrin (PE)-conjugated anti-human EpCAM, mouse IgG2b κ, clone 9C4, protect from light.
EpCAM isotype control antibody: PE-mouse IgG2b κ, protect from light.
Labeling Buffer: 2 mM EDTA-Y27632, 0.5% BSA.
Flow Buffer: 2 mM EDTA, DPBS, 0.1% BSA, 10 μM Y27632.
Column Buffer: Enzyme Buffer, 0.5% BSA, 200 Kunitz units/mL DNase.
2.5. Magnetic Activated Cell Separation (MACS™) of LGR5(+) and (−) Fractions
Plasticware: 15 mL conical tubes, cut-down P200 pipette tips, uncut P200 and P1000 pipette tips.
MACS™ Separator permanent magnet.
20 μm cell strainer.
MACS™ LS Column: prepped from above Subheading 2.3.
Flow Buffer: 2 mM EDTA, DPBS, 0.1% BSA, 10 μM Y27632.
Column Buffer: Enzyme Buffer, 0.5% BSA, 200 Kunitz units/mL DNase.
2.6. Flow Analysis and FACS of LGR5(+) and (−) Fractions
Plasticware: uncut P200 and P1000 pipette tips, 2 mL V-bottom Eppendorf tubes.
Flow Buffer: 2 mM EDTA, DPBS, 0.1% BSA, 10 μM Y27632.
DAPI working solution: 100 μM 4′,6-diamidino-2-phenylindole dilactate in H2O (see Note 7).
RLT Lysis Buffer.
RNeasy Micro Kit with on-column DNase digestion.
Serum-free L-WRN conditioned medium (SF-LWRN): L-WRN 24-h-conditioned media in Advanced DMEM/F-12, 2 mM GlutaMax, 10 mM HEPES, 1× N-2 media supplement, 1× B-27 supplement minus vitamin A, 1 mM N-Acetyl-l-cysteine, 50 μg/mL Primocin, 5 ng/mL human EGF, and 5 ng/mL FGF-2.
Single Cell Culture Medium (SCC Medium) [23, 30–33]: Advanced DMEM/F-12, 50% SF-LWRN, 2 mM GlutaMax, 10 mM HEPES, 1× N-2 media supplement, 1× B-27 supplement minus vitamin A, 1 mM N-Acetyl-l-cysteine, 100 ng human EGF/mL, 10 mM Nicotinamide, 10 nM PGE2, 10 nM Gastrin, 10 μM Y27632, and 100 μg/mL Primocin. For normal colonoid cultures, supplement with 3 μM SB202190 and 500 nM A83–01. 5 μM CHIR99021, and 2 μM Jagged-1 are added for the first 2 days of single cell culture.
FACS Live Cell Collection Medium: SCC Medium with added 0.25 mg/mL Matrigel™.
2.7. Single Cell Colonoid-Forming Culture
3. Methods
Detailed in vitro culture methodologies are outlined for the tissue-derived epithelium-only colonoids/enteroids by Sugimoto and Sato [34], and for the iPSC-derived organoids [26, 35–37]. We further cultured adenoma-derived colonoids [28] under conditions which enriched for the LGR5(+) stem cell component [24] (see Note 4). Colonoids were established from tissue acquired by endoscopy, from surgical resection or from deceased donors, and according to protocols approved by the University of Michigan Institutional Review Board. Procedures for patient-derived epithelial colonoids begin at Subheading 3.1.1, for iPSC-derived organoids at Subheading 3.1.2, and for freshly isolated tissue colon crypts at Subheading 3.2. See schematic illustration for overview of entire procedure (Fig. 1).
3.1. Removal of Matrigel™
3.1.1. Removal of Matrigel™ from Cultured Colonoid Structures (See Note 9) (1+h)
Treat cultures with 10 μM Y27632 for at least 2.5 h prior to harvesting (see Note 2).
Remove culture medium from wells and wash with cold DPBS.
Transfer Matrigel™ droplets with cell lifter in cold 2 mM EDTA-Y27632 into 15 mL tube(s) (up to 1000 μL Matrigel™ per 15 mL). Triturate vigorously 10× in 10 mL with 5 mL serological pipette. Fill tube(s) to 15 mL with 2 mM EDTA-Y27632.
Incubate with slow rotation (approximately 15 rpm) for 15 min at 4 °C (see Note 10).
Triturate 20× with 5 mL serological pipette. Centrifuge at 250 × g for 3 min at 4 °C.
Aspirate supernatant (first wash) from the 15 mL tube(s) and combine pellet(s) by gently triturating 5× with a 5 mL serological pipette in 7 mL cold DPBS. Fill tube with DPBS and slow-spin at 100 × g to additionally remove dead single cells.
Aspirate supernatant (second wash). Add 7 mL cold DPBS, gently triturate 5× with a 5 mL serological pipette, and centrifuge at 250 × g at 4 °C.
Aspirate supernatant (third wash). Add 7 mL cold Enzyme Buffer (without enzymes), gently triturate 5× with a 5 mL serological pipette, and centrifuge at 250 × g at 4 °C.
Proceed to Subheading 3.2.
3.1.2. Removal of Matrigel™ from Cultured iPSC-Derived Organoid Structures (2+ h) (See Note 11)
Treat cultures with 10 μM Y27632 for at least 2.5 h prior to harvesting (see Note 2).
Remove culture medium from wells and wash with cold DPBS.
Transfer Matrigel™ droplets with cell lifter in cold 4 mM EDTA-Y27632 into 15 mL tube(s) (up to 1000 μL Matrigel™/15 mL). Triturate 10× in 10 mL with 5 mL serological pipette. Fill tube(s) to 15 mL with 4 mM EDTA-Y27632.
Incubate with slow rotation (approximately 15 rpm) for 30 min at 4 °C.
Triturate 30× with 5 mL serological pipette. Centrifuge at 300 × g for 5 min at 4 °C.
Aspirate supernatant (first wash). Wash pellet by triturating 20× with a 5 mL serological pipette in 7 mL cold DPBS. Fill tubes to 15 mL with DPBS. Centrifuge at 300 × g for 5 min at 4 °C.
Aspirate supernatant (second wash). Add 7 mL cold DPBS, gently triturate 10× with a 5 mL serological pipette, and centrifuge at 300 × g.
Aspirate supernatant (third wash). Add 7 mL cold Enzyme Buffer (without enzymes), gently triturate 5× with a 5 mL serological pipette. Slow-spin at 100 × g for 5 min at 4 °C to additionally remove dead single cells.
Proceed to Subheading 3.2.
3.2. Single Cell Dissociation of Human Colonoids, Organoids or Fresh Colonic Crypts (2+ h)
-
Aspirate supernatant and resuspend pellet in 37 °C warm TDK-enzyme Preparation to a final volume of 10 mL. Transfer to warm MACS™ C tube.
-
Colonoid/organoid cultures: up to a 0.5 mL pellet per 10 mL TDK-enzyme Preparation.
or
Freshly prepared tissue crypts (see Notes 5 and 12): Prior to enzyme digestion, wash newly isolated crypts once with Enzyme Buffer (without enzymes) and slow-spin at 100 × g for 5 min at 4 °C, aspirate buffer, and then add 10 mL TDK-enzyme Preparation per 1 mL of a dense crypt pellet.
-
Mechanically assist enzyme dissociation with a gentleMACS™ Dissociator using the program h_tumor_01, once at the beginning, and then every 15 min for 1 h. Finish with two h_tumor_01 program runs. Slowly rotate suspension at 37 °C between runs (see Note 13).
Place tube on ice. Triturate 30× with a 5 mL serological pipette and pipet over 100 μm strainer into a 50 mL tube containing 15 mL Labeling Buffer.
Triturate 5× with a 5 mL serological pipette and pipet over 40 μm strainer into a 50 mL tube containing 5 mL Labeling Buffer.
Centrifuge at 500 × g for 5 min at 4 °C.
Aspirate supernatant and resuspend pellet in 2 mL Labeling Buffer by triturating 30× with a P1000. Add 28 mL of Labeling Buffer and pipet over 20 μm strainer(s) into two 15 mL tubes (first wash).
Centrifuge at 500 × g for 5 min at 4 °C.
Aspirate supernatant and resuspend pellet in 2 mL Labeling Buffer by triturating 30× with a P1000. Add 13 mL of Labeling Buffer. Estimate cell count and viability by trypan blue exclusion (second wash) (see Note 14).
Centrifuge at 500 × g for 5 min at 4 °C.
- Aspirate supernatant and resuspend in 10 mL Labeling Buffer by triturating 5–10× with a 5 mL serological pipette. Aliquot the appropriate number of cells to control and sort 15 mL tubes and bring up to 10 mL per tube (third wash):
-
Control cell 15 mL tube: colonoid cells: 0.2–1.0 × 106 cells (cells for 1 FACS control) and iPSC organoid or fresh crypt cells: 0.6–3.0 × 106 cells (cells for 3 FACS controls).and
- Sort cell 15 mL tube: add remaining cells.
-
Pellet both tubes at 500 × g for 5 min at 4 °C.
3.3. LGR5 Antibody Labeling (1–1.5 h)
All manipulations are conducted on ice, while incubations are at 4 °C. During incubations, at 5 min intervals, gently tap tubes to disperse cells. Resuspend or mix cells with a cut-down or wide-bore P200 pipette tip to minimize sheering of cells. Volumes listed below are for ≤107 cells; adjust volumes 2× for 1–2 × 107; 3× for 2–3 × 107 and so on.
- Add 10 μL FcR Blocking reagent to empty 2 mL Eppendorf tubes (see Note 15).
-
Colonoid cells: two Eppendorf tubes (one “Sort” and one “No Stain” control).or
- iPSC organoid or crypt cells: four Eppendorf tubes (one “Sort” and three control tubes: “No Stain,” “EpCAM only,” and “EpCAM isotype”).
-
-
Aspirate supernatant from tubes in Subheading 3.2, step 11 and resuspend cell pellets in the following volumes of cold Labeling Buffer.
-
Colonoid cells: 70 μL (sort 15 mL tube) and 70 μL (control 15 mL tube).
or
iPSC organoid or crypt cells: 70 μL (sort 15 mL tube) and 210 μL (control 15 mL tube).
-
Add 70 μL of cells in Labeling Buffer to FcR-Eppendorf tubes (total volume now 80 μL per Eppendorf tube).
Incubate for 10 min at 4 °C. Set aside control Eppendorf tubes at 4 °C; periodically tap tubes to disperse cells.
During incubation prepare the LS column(s) (≤1 × 107 cells per column) for Subheading 3.4 (at least 45 min before use). Precoat the LS column by applying 2.5 mL of Column Buffer to column. After buffer begins to drip from column, stop end with syringe cap wrapped in Parafilm. Place at 4 °C.
Add 20 μL Lgr5-Microbeads to Eppendorf sort tube and gently mix (total volume now 100 μL).
Incubate for 15 min at 4 °C.
Add 1.7 mL Labeling Buffer to Eppendorf “Sort” tube (total volume now 1.8 mL).
Centrifuge Eppendorf “Sort” tube at 500 × g for 5 min at 4 °C.
Aspirate supernatant and resuspend pellet in 90 μL of Labeling Buffer.
Add 10 μL APC Labeling Check Reagent to Eppendorf “Sort” tube and gently mix (total volume now 100 μL). Protect cells from light henceforward.
Incubate for 10 min at 4 °C.
Centrifuge Eppendorf “Sort” tube at 500 × g for 5 min at 4 °C.
Aspirate supernatant and resuspend Eppendorf “Sort” tube and Eppendorf “No Stain” control tube with up to 1.8 mL Labeling Buffer (first wash).
Centrifuge at 500 × g for 5 min at 4 °C.
3.3.1. Colonoid Cells: No EpCAM Labeling Required
Aspirate supernatant and resuspend Eppendorf “No Stain” control tube in 1 mL Flow Buffer and place on ice.
Aspirate supernatant and resuspend Eppendorf “Sort” tube in 1.8 mL of Labeling Buffer (second wash).
Centrifuge at 500 × g for 5 min at 4 °C.
Aspirate supernatant and resuspend Eppendorf “Sort” tube in 1 mL Column Buffer by triturating 30× with an uncut P200 pipette tip and proceed with magnetic separation (MACS™).
Proceed to Subheading 3.4.
3.3.2. iPSC-Derived Organoid or Freshly Isolated Crypt Cells: EpCAM Labeling (See Note 16)
Aspirate supernatant and resuspend Eppendorf “No Stain” control tube in 1 mL Flow Buffer and place on ice.
Aspirate supernatant and resuspend the Eppendorf “EpCAM only” and “EpCAM isotype” tubes in 100 μL Labeling Buffer.
Add 1 μL EpCAM isotype antibody to “EpCAM isotype” Eppendorf tube, and mix well.
Add 4 μL EpCAM antibody to the “EpCAM only” Eppendorf tube and mix well (see Note 17).
Aspirate supernatant and resuspend the Eppendorf “Sort” tube in 400 μL Labeling Buffer per 1 × 107 cells.
Add 16 μL EpCAM antibody to the Eppendorf “Sort” tube and mix well (see Note 18).
Incubate tubes for 10 min at 4 °C.
Resuspend all Eppendorf tubes up to 1.8 mL of Labeling Buffer (first EpCAM wash).
Centrifuge at 500 × g for 5 min at 4 °C.
Repeat wash and centrifugation (second EpCAM wash).
Aspirate supernatant and resuspend the “EpCAM Only” and “EpCAM isotype” control Eppendorf tubes in 1 mL Flow Buffer and place on ice.
Resuspend Eppendorf “Sort” tube in 1 mL Column Buffer by triturating 30× with an uncut P200 pipette tip and proceed with magnetic separation (MACS™).
3.4. Magnetic Activated Cell Separation (MACS™) of LGR5(+) and (−) Fractions (1 h)
Snap column to the magnet. Remove column end cap to drain coating buffer. Position 15 mL flow-through collection tube on ice.
Place 20 μm strainer above column. If cells have settled since resuspension at Subheading 3.3.1, step 4 or 3.3.2, step 12, triturate cells 30× immediately with an uncut P200 pipette tip before applying to column to ensure that they are in single cell suspension. Apply 1 mL cell suspension with a P1000 uncut pipette tip (see Note 19).
After the entire cell volume has passed through, apply 3 mL cold Column Buffer (see Note 20).
Repeat 3 mL cold Column Buffer wash twice more, continuing to collect flow-through unbound fractions on ice. Perform a cell count and viability assessment of the flow-through fraction by trypan blue exclusion.
Remove column from magnet and place over 15 mL collection tube on ice. Apply 2.5 mL Column Buffer and vigorously flush out magnet-bound cells by firmly pushing plunger into column. Perform a cell count and viability assessment of the magnet-bound positive fraction by trypan blue exclusion.
Centrifuge both the flow-through and magnet-bound fractions at 500 × g for 10 min at 4 °C.
Resuspend both the flow-through and magnet-bound fractions in cold Flow Buffer with an P200 uncut pipette tip at concentrations approximately 3–5 × 106 cells/mL, depending on the specifications of your FACS instrument.
3.5. Flow Analysis and FACS of LGR5(+) and (−) Fractions (See Note 21)
Stain with 1 μM DAPI for 1 min prior to analysis for viability assessment: add 10 μL 100 μM DAPI working solution per mL cells.
Triturate cells 20× before analysis/sorting with P1000 uncut pipette tip to ensure cells are in a single cell suspension prior to analysis.
With unstained control cells, set forward and side scatter gating strategy to exclude debris and doublets.
Add DAPI to unstained control cells. Gate and exclude nonviable cells, including intermediate dying cells (see Note 22). Proceed to step 5 for colonoid-derived cells (no EpCAM).
-
For iPSC-derived organoid cells and fresh tissue crypt cells (EpCAM gating).
-
Add DAPI to EpCAM isotype-PE-stained cells to set gating threshold for EpCAM at 0.1% events based on EpCAM isotype-PE staining in DAPI(−) cells.
and
Add DAPI to EpCAM-PE-stained cells and. Analyze for DAPI(−) EpCAM(+) cells (see Note 23). Further LGR5 analyses should be gated for DAPI(−)EpCAM(+) cells.
-
Add DAPI to the MACS™ flow-through negative fraction. Set the threshold APC-positive gating at 0.01–0.1% APC (see Note 24).
FACS sort viable LGR5(−) cells from both flow-through negative and magnet-bound positive fractions, while collecting LGR5(+) cells from the magnet-bound positive fraction (see Note 25). See representative Flow scatter plots of LGR5(+) cells isolated from iPSC-derived organoids (Fig. 2).
- FACS cell collection options:
-
RNA analyses: FACS collect into 100 μL RLT Lysis Buffer for ≤5000 cells; 350 μL for 5000–50,000 cells. For ≥50,000 sorted cells, add 100 μL additional lysis buffer immediately after sort. Vortex for 1 min and place on ice for RNA processing (see Note 26). We extract RNA with RNeasy Micro Kit with on-column DNase digestion according to manufacturer’s instructions. We have used RNA extracted from these cells for RNA sequencing of LGR5(+) and LGR5(−) cells from patient-derived colonoids (Fig. 3).or
- Live-cell collection for analysis or culture: FACS collect approximately 2000 cells into 13 mL cold FACS Live Cell Collection Medium into a 15 mL tube. Proceed to Subheading 3.6 for culture of FACS single cells from colonoid cultures.
-
Fig. 2.
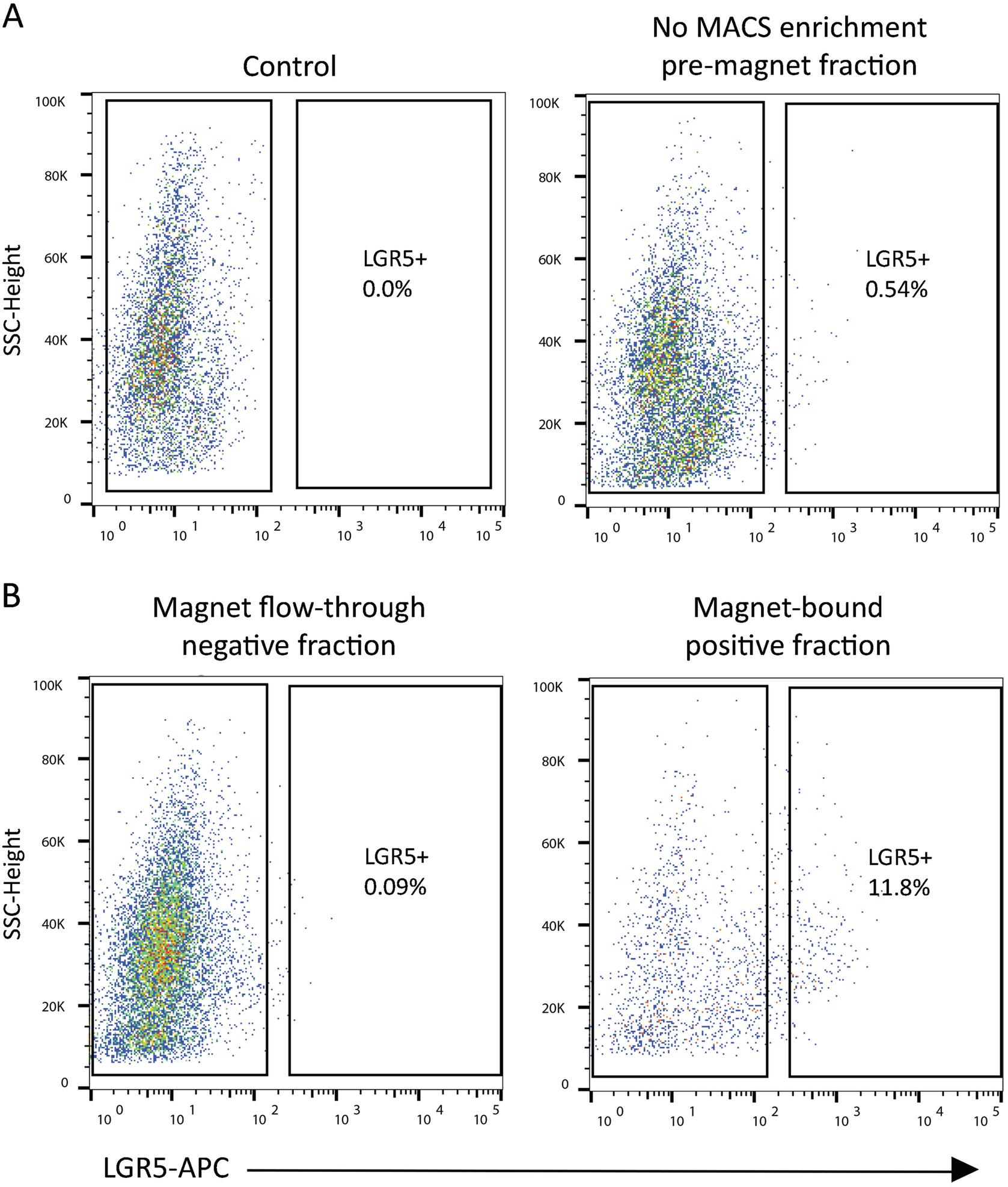
Isolation of LGR5(+) cells from human iPSC-derived organoids. Cells were first isolated on the live DAPI(−), EpCAM-PE(+) epithelial cell markers to discriminate epithelial cells from the associated mesenchymal iPSC cell lineage (scatter plots not shown). (a) Control FMO-stained cells are DAPI(−), and EpCAM-PE(+), minus stain for APC. Representative scatterplots of LGR5-APC events are shown before and (b) after magnetic bead separation (MACS). The flow-through effluent is depleted of LGR5-APC(+) cells, while the magnetic bead-bound fraction is enriched 20-fold over the pre-MACS fraction for LGR5-APC(+) cells
Fig. 3.
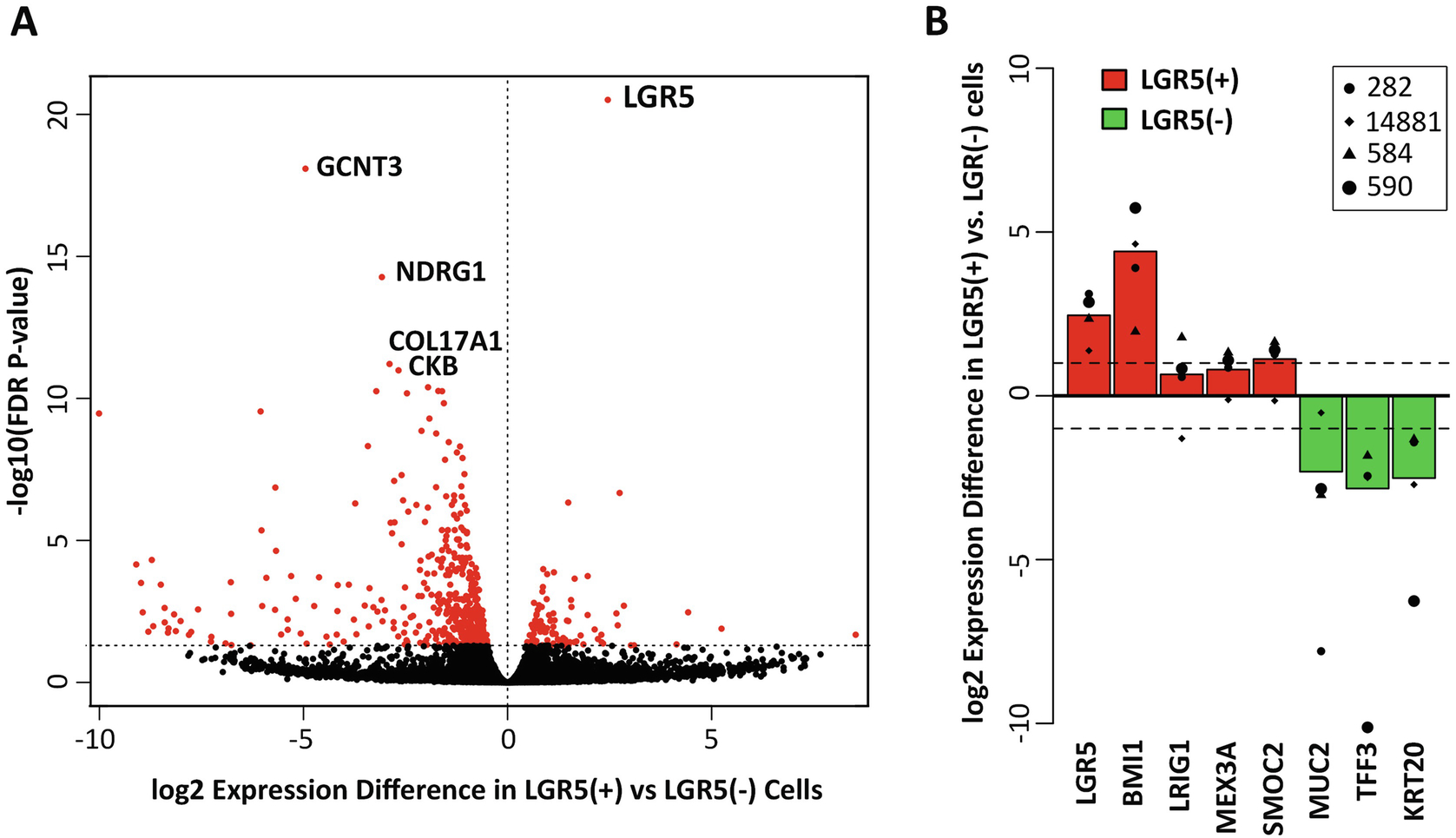
Transcriptomic analysis of isolated LGR5(+) vs LGR5(−) cells. Colonoid cultures were established from four patient-derived, genetically diverse, tubular adenomas (patient identifiers 14881, 282, 584, and 590). LGR5(+) cells were isolated and compared to LGR5(−) cells for differential gene expression across these four specimens. (a) FDR volcano plot of the log(2) ratio of gene expression between the LGR5(+) and LGR5(−) cells, based on the top 500 most variable genes. LGR5 had the highest level of statistical enrichment in the LGR5(+) cells (FDR, 3.8E–21) and was expressed an average of 5.5-fold higher compared with in LGR5(−) cells. (b) Log(2) fold change in gene expression between LGR5(+) and LGR5(−) cells for known markers of colon stem (red) and differentiated (green) cells. Stem cell markers associated with the colon, as well as other tissue-specific stem cell markers, including BMI1, MEX3A, and SMOC2, were upregulated in LGR5(+) cells, whereas known markers of colonic differentiation, including MUC2, TFF3, and KRT20, were downregulated. (Content adapted from Dame et al., 2018 [24] with permission from Development)
3.6. Single Cell Colonoid-Forming Culture
Pellet FACS sorted cells at 500 × g for 10 min at 4 °C.
Resuspend pellet in 22 μL SCC Medium (including cell pellet-residual supernatant volume) and mix with 88 μL ice-cold Matrigel™ to a final concentration of 8 mg/mL Matrigel™ (assuming Matrigel™ stock is 10 mg/mL) for a total of 110 μL Matrigel™-cell mixture.
Pipet 10–12 raised 10 μL Matrigel-cell drops (approximately 200 cells/10 μL Matrigel™) onto the well surface of a pre-warmed 12-well plate placed on a warm pack.
After 10 min, add 800 μL of warm SCC Medium.
Change medium every 2 days.
When small cyst-like structures are evident (6–8 days), change to standard LWRN medium, or continue in SCC Medium (see Note 8).
4. Notes
All plastic surfaces are coated in 0.1% bovine serum albumen in DPBS (tissue culture grade BSA). Prepare a 10% BSA stock solution in DPBS and sterilize with a low-protein binding syringe-filter. Dilute in sterile DPBS to prepare 0.1% BSA working solution. Store solutions at 4 °C for up to 6 months; can be reused multiple times. Coat by filling plastic components at room temperature for below time periods or overnight at 4 °C. Polypropylene surfaces (conical tubes, Eppendorf tubes, MACS™ C-tubes, P200 and P1000 pipette tips, cell strainers, and columns) require at least 20 min of coating time. Serological pipettes require 3 min of coating time. Coated plasticware can be stored at 4 °C for at least 2 weeks. Do not let surfaces dry. After coating, rinse once in DBPS.
Cultures are pretreated with the Rho-associated protein kinase (ROCK) inhibitor, Y27632, to reduce anoikis of dissociated single cells. Cultures are also routinely treated with Y27632 at passaging, while some colonoid cultures are maintained with 10 μM Y27632 in their regular growth medium. 2.5 mM stock solutions are prepared in sterile H2O and stored as per manufacturer’s instructions.
HBSS-containing standard calcium/magnesium is reduced 10:1 with HBSS that is calcium/magnesium-free. These cations are reduced to minimize stem cell differentiation but at sufficient levels to facilitate enzymatic activity.
To enrich cultures for LGR5(+) stem cells, we have transitioned adenoma colonoid cultures [28] from KGM-Gold to L-WRN conditioned medium, which is high in Wnt3a, R-spondin-3, and Noggin [38]. L-WRN can promote a shift from a differentiated budding phenotype to a stem-cell enriched cystic phenotype [24, 39–43]. These cystic-driven cultures can also serve as robust positive controls for the LGR5(+) cell isolation procedure. Cultures are gradually transitioned from KGM-Gold to L-WRN: 3 days post-passage switch for 5 days to 50/50 L-WRN, then 5 days 75/25, to finally 100% L-WRN. The complete L-WRN medium contains Advanced DMEM/F-12, 2 mM GlutaMax, 10 mM HEPES, 1× N-2 media supplement, 1× B-27 supplement minus vitamin A, 1 mM N-Acetyl-l-cysteine, 100 ng/mL EGF, 10 mM nicotinamide, and 100 μg/mL Primocin. For normal colonoid cultures, supplement with 10 μM SB202190, 500 nM A83–01, plus/minus 10 μM Y27632.
To isolate sufficient human LGR5(+) cells for experimental analysis, we recommend starting with at least 2 wells of a 6-well plate of dense cystic colonoids (500 μL Matrigel; 10E6 total starting cells), 15 wells of a 6-well plate of iPSC-derived organoids (3750 μL Matrigel; 15E6 total starting cells including mesenchymal cells), or a 9 cm2-sized colon resection of freshly isolated crypts (20E6 total starting cells). We have successfully scaled this procedure up to 30 wells of colonoids or to 25 cm2 of freshly resected colon tissue.
We found that both trypsin and the Neural Tissue Dissociation Kit were too aggressive for this application and removed some relevant cell surface epitopes. We employed a gentle enzyme preparation, Tumor Dissociation Kit. TDK enzymes H and R were reconstituted with warm Enzyme Solution described here, and enzyme A was reconstituted with Buffer A supplied with the kit. Enzymes were aliquoted in working volumes and stored at −80 °C according to manufacturer’s instructions.
10 mM DAPI dilactate stock solution prepared in H2O. Store at −20 °C for up to 9 months. Day of use prepare 100 μM working solution in H2O. Protect from light.
The serum-free Single Cell Culture Medium (SCC Medium) is employed for early initiation of cystic colonoids from FACS single cells. Once small cysts are evident (≥50 μm in size, after approximately 5–8 days), we have employed a serum-containing commercially available colonoid medium (IntestiCult™ Organoid Growth Medium), plus 10 μM Y27632, to enhance the establishment of colonoids from the starter cysts.
We have found that EDTA is not required for routine passaging of cultures. Instead, to passage colonoids: (1) 30× triturate the Matrigel™ pads with a P1000 pipette in cold DPBS (2 mL per 250 μL Matrigel™), (2) fill tube to 5 mL, (3) pellet at 300 × g 4 °C for 3 min, (4) 30× triturate pellet with a P200 pipette in the small volume of medium required to dilute the stock Matrigel™ to 8 mg/mL final, and (5) 15× triturate pellet with a P200 pipette in Matrigel™ before plating as 10–50 μL drops.
Incubate in 2 mM EDTA to remove Matrigel™ to prepare for enzymatic dissociation. Extend the EDTA incubation time period when colonoids/organoids have been in culture for periods more than 7 days (before next subculture); add 3 min for every subsequent day of culture.
iPSC-derived organoids require more vigorous methods to remove associated Matrigel™, owing to the mesenchymal contribution (30 min 4 mM EDTA). During the initial wash steps many Matrigel™ fragments with organoids will remain suspended in the supernatant after pelleting. Recover these fragments along with the pellet by permitting them to settle on ice (4 min) after centrifugations, before discarding the supernatant.
Fresh human intestinal crypts are prepared from surgical tissue resections as described by Jung et al. [27]. After crypts have been released from the tissue, keep on ice to preserve viability.
A customized program has been designed for this specific protocol which integrates program run steps (h_tumor_01) and incubations steps for use with the heated gentleMACS Dissociator.
We counted and estimated viability via trypan blue exclusion with an Invitrogen Countess Automated Cell Counter. Mix cell sample 1:1 with trypan blue solution and inject into automated hemocytometer slide. If necessary, adjust cell dilution to recommended instrument cell number range of accuracy. Use this process to also visually note single cell efficiency of digestion.
Eppendorf tubes containing FcR blocking reagent are prepared earlier during breaks in steps and stored at 4 °C.
LGR5 has been identified in both epithelial and stromal tissue by our laboratory [24] and others [44, 45]. iPSC-derived organoids have both an epithelial and mesenchymal component, and freshly isolated tissue crypts will have some percentage of contaminating stroma. EpCAM is used as a marker to separate epithelial cells from the stroma.
The EpCAM IgG2b κ isotype control antibody concentration was according to manufacturer’s instruction. The EpCAM antibody concentration, although ½ the concentration of the isotype control, was determined based on a concentration which would efficiently delineate a positive population without shifting the entire negative population of cells.
For cell numbers greater than 1 × 107 cells, adjust volume of buffer and EpCAM antibody 2× for 1–2 × 107 cells (i.e., 800 μL Labeling buffer/32 μL antibody), 3× for 2–3 × 107 cells, and so on.
Avoid bubbles in Column Solution which may cause blockage. If at any point the column appears to stop, place thumb over the top of the column and push down to pressurize restart.
Drain column completely before applying next volume.
Maintain cells on ice before analysis/FACS. A refrigerated sorting instrument is highly recommended.
When the viable cell population shifts with sort time, we readjust gating to exclude dying cells.
EpCAM gating also resulted in enhanced selection of viable cells. We have included EpCAM staining even with epithelium-only colonoids when added assurances of long-term viability are of high priority (i.e., FACS for subsequent single cell colonoid-forming efficiency).
The APC Check Reagent is highly specific to the microbead. The unbound column flow-through cells provide a robust negative control for the LGR5-microbead-APC conjugate. Rigorous FMO (fluorescence-minus-one) controls are suitable for samples that are not MACS-processed [DAPI(−), EpCAM(+)].
We observe an approximately tenfold plus enrichment of LGR5(+) cells by MACS, which results in a significantly abbreviated FACS time. We believe this reduced processing time likely translates into higher stem cell viability and possible preservation of the LGR5-bound moiety.
We extract RNA from lysed sorted cells with the RNeasy Micro Kit (with on-column DNase digestion) according to manufacturer instructions. We observe a significant enhancement in quantity and quality of RNA when extracted soon after FACS, as opposed to freezing for later extraction.
Acknowledgments
This work was supported by National Institute of Environmental Health Sciences of the National Institutes of Health (R01ES028802 and P30 ES017885 to J.A.C.); the University of Michigan Rogel Comprehensive Cancer Center Research Grant Fund (P30CA046592); the University of Michigan Center for Gastrointestinal Research (UMCGR) (5P30DK034933); the University of Michigan MCubed Program (to J.A.C.); the Ravitz Family Foundation (to J.A.C.); the Intestinal Stem Cell Consortium (U01DK103141 to J.R.S.), a collaborative research project funded by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK); and the National Institute of Allergy and Infectious Diseases (NIAID); and the NIAID Novel Alternative Model Systems for Enteric Diseases (NAMSED) consortium (U19AI116482 to J.R.S.). We thank Maliha Berner, Erica Katz, Caroline McCarthy, Gina Newsome, and Angeline Wu of the Translational Tissue Modeling Laboratory (TTML) of the Department of Internal Medicine, University of Michigan; Michael Dellheim of the University of Michigan BRCF Flow Cytometry Core; and Robin Kunkel of the Department of Pathology, University of Michigan, for graphic design.
References
- 1.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449(7165):1003–1007. 10.1038/nature06196.nature06196 [pii] [DOI] [PubMed] [Google Scholar]
- 2.de Visser KE, Ciampricotti M, Michalak EM, Tan DW, Speksnijder EN, Hau CS, Clevers H, Barker N, Jonkers J (2012) Developmental stage-specific contribution of LGR5(+) cells to basal and luminal epithelial lineages in the postnatal mammary gland. J Pathol 228(3):300–309. 10.1002/path.4096 [DOI] [PubMed] [Google Scholar]
- 3.Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgard R (2008) Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet 40(11):1291–1299. 10.1038/ng.239 [DOI] [PubMed] [Google Scholar]
- 4.Barker N, Rookmaaker MB, Kujala P, Ng A, Leushacke M, Snippert H, van de Wetering M, Tan S, Van Es JH, Huch M, Poulsom R, Verhaar MC, Peters PJ, Clevers H (2012) Lgr5(+ve) stem/progenitor cells contribute to nephron formation during kidney development. Cell Rep 2(3):540–552. 10.1016/j.celrep.2012.08.018 [DOI] [PubMed] [Google Scholar]
- 5.Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM, Clevers H (2012) Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488(7413):665–669. 10.1038/nature11308 [DOI] [PubMed] [Google Scholar]
- 6.de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, Stange DE, van Es JE, Guardavaccaro D, Schasfoort RB, Mohri Y, Nishimori K, Mohammed S, Heck AJ, Clevers H (2011) Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476(7360):293–297. 10.1038/nature10337 [DOI] [PubMed] [Google Scholar]
- 7.Fafilek B, Krausova M, Vojtechova M, Pospichalova V, Tumova L, Sloncova E, Huranova M, Stancikova J, Hlavata A, Svec J, Sedlacek R, Luksan O, Oliverius M, Voska L, Jirsa M, Paces J, Kolar M, Krivjanska M, Klimesova K, Tlaskalova-Hogenova H, Korinek V (2013) Troy, a tumor necrosis factor receptor family member, interacts with lgr5 to inhibit wnt signaling in intestinal stem cells. Gastroenterology 144(2):381–391. 10.1053/j.gastro.2012.10.048 [DOI] [PubMed] [Google Scholar]
- 8.Ruffner H, Sprunger J, Charlat O, Leighton-Davies J, Grosshans B, Salathe A, Zietzling S, Beck V, Therier M, Isken A, Xie Y, Zhang Y, Hao H, Shi X, Liu D, Song Q, Clay I, Hintzen G, Tchorz J, Bouchez LC, Michaud G, Finan P, Myer VE, Bouwmeester T, Porter J, Hild M, Bassilana F, Parker CN, Cong F (2012) R-Spondin potentiates Wnt/beta-catenin signaling through orphan receptors LGR4 and LGR5. PLoS One 7(7):e40976 10.1371/journal.pone.0040976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmon KS, Lin Q, Gong X, Thomas A, Liu Q (2012) LGR5 interacts and cointernalizes with Wnt receptors to modulate Wnt/beta-catenin signaling. Mol Cell Biol 32(11):2054–2064. 10.1128/mcb.00272-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, Boutros M, Cruciat CM, Niehrs C (2011) LGR4 and LGR5 are R-spondin receptors mediating Wnt/beta-catenin and Wnt/PCP signalling. EMBO Rep 12(10):1055–1061. 10.1038/embor.2011.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q (2011) R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A 108(28):11452–11457. 10.1073/pnas.1106083108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJ, van de Wetering M, Sojoodi M, Li VS, Schuijers J, Gracanin A, Ringnalda F, Begthel H, Hamer K, Mulder J, van Es JH, de Koning E, Vries RG, Heimberg H, Clevers H (2013) Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J 32(20):2708–2721. 10.1038/emboj.2013.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Es JH, Sato T, van de Wetering M, Lyubimova A, Yee Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, Martens ACM, Barker N, van Oudenaarden A, Clevers H (2012) Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol 14(10):1099–1104. 10.1038/ncb2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sigal M, Logan CY, Kapalczynska M, Mollenkopf HJ, Berger H, Wiedenmann B, Nusse R, Amieva MR, Meyer TF (2017) Stromal R-spondin orchestrates gastric epithelial stem cells and gland homeostasis. Nature 548(7668):451–455. 10.1038/nature23642 [DOI] [PubMed] [Google Scholar]
- 15.Hoeck JD, Biehs B, Kurtova AV, Kljavin NM, de Sousa EMF, Alicke B, Koeppen H, Modrusan Z, Piskol R, de Sauvage FJ (2017) Stem cell plasticity enables hair regeneration following Lgr5(+) cell loss. Nat Cell Biol 19(6):666–676. 10.1038/ncb3535 [DOI] [PubMed] [Google Scholar]
- 16.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H (2012) Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337(6095):730–735. 10.1126/science.1224676 [DOI] [PubMed] [Google Scholar]
- 17.de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, Anderson JE, Storm EE, Modrusan Z, Koeppen H, Dijkgraaf GJ, Piskol R, de Sauvage FJ (2017) A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature 543(7647):676–680. 10.1038/nature21713 [DOI] [PubMed] [Google Scholar]
- 18.Junttila MR, Mao W, Wang X, Wang BE, Pham T, Flygare J, Yu SF, Yee S, Goldenberg D, Fields C, Eastham-Anderson J, Singh M, Vij R, Hongo JA, Firestein R, Schutten M, Flagella K, Polakis P, Polson AG (2015) Targeting LGR5+ cells with an antibody-drug conjugate for the treatment of colon cancer. Sci Transl Med 7(314):314ra186 10.1126/scitranslmed.aac7433 [DOI] [PubMed] [Google Scholar]
- 19.Gong X, Azhdarinia A, Ghosh SC, Xiong W, An Z, Liu Q, Carmon KS (2016) LGR5-targeted antibody-drug conjugate eradicates gastrointestinal tumors and prevents recurrence. Mol Cancer Ther 15(7):1580–1590. 10.1158/1535-7163.MCT-16-0114 [DOI] [PubMed] [Google Scholar]
- 20.Barker N (2014) Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol 15(1):19–33. 10.1038/nrm3721 [DOI] [PubMed] [Google Scholar]
- 21.Baker AM, Graham TA, Elia G, Wright NA, Rodriguez-Justo M (2015) Characterization of LGR5 stem cells in colorectal adenomas and carcinomas. Sci Rep 5:8654 10.1038/srep08654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang BG, Lee BL, Kim WH (2013) Distribution of LGR5+ cells and associated implications during the early stage of gastric tumorigenesis. PLoS One 8(12):e82390 10.1371/journal.pone.0082390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimokawa M, Ohta Y, Nishikori S, Matano M, Takano A, Fujii M, Date S, Sugimoto S, Kanai T, Sato T (2017) Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature 545(7653):187–192. 10.1038/nature22081 [DOI] [PubMed] [Google Scholar]
- 24.Dame MK, Attili D, McClintock SD, Dedhia PH, Ouillette P, Hardt O, Chin AM, Xue X, Laliberte J, Katz EL, Newsome GM, Hill DR, Miller AJ, Tsai YH, Agorku D, Altheim CH, Bosio A, Simon B, Samuelson LC, Stoerker JA, Appelman HD, Varani J, Wicha MS, Brenner DE, Shah YM, Spence JR, Colacino JA (2018) Identification, isolation and characterization of human LGR5-positive colon adenoma cells. Development 145(6). 10.1242/dev.153049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H (2011) Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141(5):1762–1772. 10.1053/j.gastro.2011.07.050.S0016-5085(11)01108-5 [pii] [DOI] [PubMed] [Google Scholar]
- 26.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, Shroyer NF, Wells JM (2011) Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470(7332):105–109. 10.1038/nature09691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung P, Sato T, Merlos-Suarez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, Clevers H, Batlle E (2011) Isolation and in vitro expansion of human colonic stem cells. Nat Med 17(10):1225–1227. 10.1038/nm.2470.nm.2470 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Dame MK, Jiang Y, Appelman HD, Copley KD, McClintock SD, Aslam MN, Attili D, Elmunzer BJ, Brenner DE, Varani J, Turgeon DK (2014) Human colonic crypts in culture: segregation of immunochemical markers in normal versus adenoma-derived. Lab Investig 94(2):222–234. 10.1038/labinvest.2013.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujii M, Matano M, Toshimitsu K, Takano A, Mikami Y, Nishikori S, Sugimoto S, Sato T (2018) Human intestinal organoids maintain self-renewal capacity and cellular diversity in Niche-inspired culture condition. Cell Stem Cell 23(6):787–793.e786. 10.1016/j.stem.2018.11.016 [DOI] [PubMed] [Google Scholar]
- 30.Yin X, Farin HF, van Es JH, Clevers H, Langer R, Karp JM (2014) Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods 11(1):106–112. 10.1038/nmeth.2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Furstenberg RJ, Buczacki SJ, Smith BJ, Seiler KM, Winton DJ, Henning SJ (2014) Side population sorting separates subfractions of cycling and non-cycling intestinal stem cells. Stem Cell Res 12(2):364–375. 10.1016/j.scr.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, Scoville D, He XC, Mahe MM, Box A, Perry JM, Smith NR, Lei NY, Davies PS, Fuller MK, Haug JS, McClain M, Gracz AD, Ding S, Stelzner M, Dunn JCY, Magness ST, Wong MH, Martin MG, Helmrath M, Li L (2013) Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology 145(2):383–395.e321. 10.1053/j.gastro.2013.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung P, Sommer C, Barriga FM, Buczacki SJ, Hernando-Momblona X, Sevillano M, Duran-Frigola M, Aloy P, Selbach M, Winton DJ, Batlle E (2015) Isolation of human colon stem cells using surface expression of PTK7. Stem Cell Rep 5(6):979–987. 10.1016/j.stemcr.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugimoto S, Sato T (2017) Establishment of 3D intestinal organoid cultures from intestinal stem cells In: Koledova Z (ed) 3D Cell culture: methods and protocols. Springer, New York, pp 97–105. 10.1007/978-1-4939-7021-6_7 [DOI] [PubMed] [Google Scholar]
- 35.McCracken KW, Howell JC, Wells JM, Spence JR (2011) Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc 6(12):1920–1928. 10.1038/nprot.2011.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capeling MM, Czerwinski M, Huang S, Tsai YH, Wu A, Nagy MS, Juliar B, Sundaram N, Song Y, Han WM, Takayama S, Alsberg E, Garcia AJ, Helmrath M, Putnam AJ, Spence JR (2019) Nonadhesive alginate hydrogels support growth of pluripotent stem cell-derived intestinal organoids. Stem Cell Rep 12(2):381–394. 10.1016/j.stemcr.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finkbeiner SR, Hill DR, Altheim CH, Dedhia PH, Taylor MJ, Tsai YH, Chin AM, Mahe MM, Watson CL, Freeman JJ, Nattiv R, Thomson M, Klein OD, Shroyer NF, Helmrath MA, Teitelbaum DH, Dempsey PJ, Spence JR (2015) Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Rep. 10.1016/j.stemcr.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyoshi H, Stappenbeck TS (2013) In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc 8(12):2471–2482. 10.1038/nprot.2013.153. http://www.nature.com/nprot/journal/v8/n12/abs/nprot.2013.153.html#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onuma K, Ochiai M, Orihashi K, Takahashi M, Imai T, Nakagama H, Hippo Y (2013) Genetic reconstitution of tumorigenesis in primary intestinal cells. Proc Natl Acad Sci U S A 110(27):11127–11132. 10.1073/pnas.1221926110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, Korving J, van de Wetering M, Schwank G, Logtenberg M, Cuppen E, Snippert HJ, Medema JP, Kops GJ, Clevers H (2015) Sequential cancer mutations in cultured human intestinal stem cells. Nature 521(7550):43–47. 10.1038/nature14415 [DOI] [PubMed] [Google Scholar]
- 41.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H (2011) Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469(7330):415–418. 10.1038/nature09637.nature09637 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farin HF, Van Es JH, Clevers H (2012) Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143(6):1518–1529. e1517. 10.1053/j.gastro.2012.08.031 [DOI] [PubMed] [Google Scholar]
- 43.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T (2015) Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med 21(3):256–262. 10.1038/nm.3802 [DOI] [PubMed] [Google Scholar]
- 44.Boddupally K, Wang G, Chen Y, Kobielak A (2016) Lgr5 marks neural crest derived multi-potent oral stromal stem cells. Stem Cells 34(3):720–731. 10.1002/stem.2314 [DOI] [PubMed] [Google Scholar]
- 45.Lee J-H, Tammela T, Hofree M, Choi J, Marjanovic ND, Han S, Canner D, Wu K, Paschini M, Bhang DH, Jacks T, Regev A, Kim CF (2017) Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell 170(6):1149–1163. e1112. 10.1016/j.cell.2017.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]


