Abstract
Gap Junction Protein Alpha 1 (GJA1) belongs to the gap junction family and has been widely studied in cancers. We evaluated the role of GJA1 in cervical cancer (CC) using public data from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) database. The difference of GJA1 expression level between CC and normal tissues was analyzed by the Gene Expression Profiling Interactive Analysis (GEPIA), six GEO datasets, and the Human Protein Atlas (HPA). The relationship between clinicopathological features and GJA1 expression was analyzed by the chi-squared test and the logistic regression. Kaplan–Meier survival analysis and Cox proportional hazard regression analysis were used to assessing the effect of GJA1 expression on survival. Gene set enrichment analysis (GSEA) was used to screen the signaling pathways regulated by GJA1. Immune Cell Abundance Identifier (ImmuCellAI) was chosen to analyze the immune cells affected by GJA1. The expression of GJA1 in CC was significantly lower than that in normal tissues based on the GEPIA, GEO datasets, and HPA. Both the chi-squared test and the logistic regression showed that high-GJA1 expression was significantly correlated with keratinization, hormone use, tumor size, and FIGO stage. The Kaplan–Meier curves suggested that high-GJA1 expression could indicate poor prognosis (p = 0.0058). Multivariate analysis showed that high-GJA1 expression was an independent predictor of poor overall survival (HR, 4.084; 95% CI, 1.354-12.320; p = 0.013). GSEA showed many cancer-related pathways, such as the p53 signaling pathway and the Wnt signaling pathway, were enriched in the high-GJA1-expression group. Immune cell abundance analysis revealed that the abundance of CD8 naive, DC, and neutrophil was significantly increased in the high-GJA1-expression group. In conclusion, GJA1 can be regarded as a potential prognostic marker of poor survival and therapeutic target in CC. Moreover, many cancer-related pathways may be the critical pathways regulated by GJA1. Furthermore, GJA1 can affect the abundance of immune cells.
1. Introduction
Cervical cancer (CC) is the fourth most common cancer in females, with 570,000 new cases and 311,000 deaths estimated for 2018 worldwide [1]. The vast majority of CC is in sub-Saharan Africa and South-Eastern Asia, and there were 98,900 new cases and 30,500 deaths for 2015 in China [1, 2]. Although the incidence and mortality rate of cervical declined due to vaccination, screening, and the control of precancerous lesions, it is even higher in some rural areas in China with the characteristics of young age (peak 45-49) and late stage [3–5]. In clinical, patients with early-stage CC (stages I to IIA) are mainly treated with surgery, whereas those with late-stage CC (stages IIB to IV) are treated with chemoradiotherapy [6]. However, the recurrence rate of CC is approximately 20%–25%, and the 5-year relative survival rate of CC remains poor in China (overall 45.4%), and the data from a retrospective population-based study in Hong Kong also showed that 5-year relative survival rates were 90.9%, 71.0%, 41.7%, and only 7.8% for the stages I, II, III, and IV, respectively [7–9]. Therefore, it is crucial to identify sensitive and specific biomarkers that could predict CC prognosis and serve as a target for CC treatment.
GJA1 maps to 6q22.31. GJA1 is also known as Connexin43 and belongs to the gap junction family [10]. Gap junction family plays an essential role in determining cellular phenotypes [10]. The role of gap junction family in cancers has been widely studied, among which GJA1 is one of the most studied genes. Firstly, GJA1 has been reported to play a suppressive role in tumorigenesis [11–13]. GJA1 could facilitate the transmission of cAMP, leading to increased p27 levels and reduced tumor growth by gap junctional intercellular communication [11]. Moreover, GJA1 could inhibit cell proliferation by inhibiting the Wnt signaling pathway [12, 13]. Secondly, although GJA1 has suppressive effects on cancer, many articles have reported the overexpression of GJA1 in metastatic tumors [14, 15]. Thirdly, GJA1 could also accelerate cancer progression by promoting cell migration via activating p38 [16, 17]. Also, high-GJA1 mRNA expression levels could indicate a more unsatisfactory outcome in cancer [18].
Based on the TCGA database and the GEO database, we firstly explored the difference of GJA1 expression level between CC and normal tissues. Also, we analyzed the relationship between GJA1 expression and the clinicopathological features of patients with CC and its prognostic value in CC. To gain further insight into the biological pathways and immune cell changes involved in CC pathogenesis related to GJA1, GSEA and immune cell abundance analysis were performed.
2. Materials and Methods
2.1. RNA-Sequencing Patient Data in the TCGA Database
The gene expression data (304 cases, workflow type: HTSeq-FPKM-UQ) and corresponding clinical information were downloaded from TCGA database (https://portal.gdc.cancer.gov) by TCGAbiolinks package [19]. The details of clinical information are shown in Table 1. Besides, it should be emphasized that survival information, including vital status and overall survival (OS) time, was directly taken from the articles of Liu et al. published in Cell in 2018 [20].
Table 1.
Clinical characteristics of patients with CC in TCGA database.
| Clinical characteristics | Subgroup | Frequency | Percentage (%) |
|---|---|---|---|
| Total | 304 | ||
| Age | Range:20-88 (average: 48.2, median: 46) | ||
| BMI | Range: 13.4-70.5 (average: 27.9, median: 26.6) | ||
| Histology | Squamous cell carcinoma (SCC) | 252 | 89.0 |
| Adenomas and adenocarcinoma (ACC) | 31 | 11.0 | |
| Keratinization | No | 119 | 68.4 |
| Yes | 55 | 31.6 | |
| Hormone use | No | 89 | 56.7 |
| Yes | 68 | 43.3 | |
| Hysterectomy | Other | 8 | 4.8 |
| Hysterectomy | 159 | 95.2 | |
| Tumor size | T1+T2 | 211 | 87.6 |
| T3+T4 | 30 | 12.4 | |
| Lymph node | N0 | 133 | 68.9 |
| N1 | 60 | 31.1 | |
| ∗FIGO stage | ≤IIA2 (early stage) | 188 | 63.3 |
| ≥IIB (late stage) | 109 | 36.7 | |
| Differentiation grade | ≤G2 | 153 | 56.3 |
| ≥G3 | 119 | 43.7 | |
| Lymphovascular invasion | Absent | 71 | 47.3 |
| Present | 79 | 52.7 | |
| Distant metastasis | No | 273 | 89.8 |
| Yes | 31 | 10.2 | |
| Vital status | Alive | 233 | 76.6 |
| Dead | 71 | 23.4 |
∗FIGO stage: there were five samples only described as FIGO II without specific stages. We classified these samples as ≤IIA2 in this study. BMI: body mass index; FIGO: the International Federation of Gynecology and Obstetrics.
2.2. Relative GJA1 Expression Level between CC and Normal Tissues
GEPIA (http://gepia.cancer-pku.cn/) was used to access the mRNA expression levels of GJA1 in CC (based on TCGA database) and normal tissues (based on GTEx project) [21]. limma package [22] was used to explore whether the expression of GJA1 is significantly different between CC and normal tissues in six GEO datasets: GSE39001 [23], GSE52903 [24], GSE63514 [25], GSE6791 [26], GSE7803 [27], and GSE9705 [28]. Meeting the criteria of |LogFC | >1 and adjusted p value < 0.05, GJA1 could be considered differentially expressed between CC and normal tissues. Additionally, we explored the expression of GJA1 at the protein level by the HPA database [29].
2.3. Survival Analysis
We matched the GJA1 expression data with survival information derived from Liu et al. [20]. The patients in cohorts were divided into two groups according to the median expression levels of GJA1 (the low-GJA1-expression group and the high-GJA1-expression group). The survival and survminer packages were used for survival analysis and visualization, and then the Kaplan-Meier survival curve was obtained. Also, we conducted a stratified analysis and explored the prognostic value of GJA1 in squamous cell carcinoma (SCC) patients, early-stage patients (FIGO stage ≤ IIA2), and late-stage patients (FIGO stage ≥ IIB).
2.4. Univariate and Multivariate Cox Regression Analyses
The Cox proportional hazard regression model was used to conduct univariate and multivariate analyses. The hazard ratio (HR) value and 95% confidence interval (CI) were calculated. In univariate analysis, the independent predictive value of the clinicopathological parameters and GJA1 expression on survival was evaluated. Multivariate Cox analysis was conducted to compare the influence of GJA1 expression on survival along with other characteristics as categorical variables. Variables with p value ≤ 0.05 or close to 0.05, including age, BMI, tumor size, lymph node, FIGO stage, lymphovascular invasion, and distant metastasis, were entered in the multivariate Cox regression analysis as categorical variables. The cut-off value of GJA1 expression was set based on the median expression value. The data were analyzed by using survival and survminer packages and visualized by forestplot package in R.
2.5. Gene Set Enrichment Analysis (GSEA)
GSEA (version 4.0.3) was used to explore the signaling pathways related to GJA1 in cervical cancer [30]. Gene expression enrichment analysis was carried out between different phenotypes determined by the GJA1 expression level. The annotated gene set was selected (c2.cp.kegg.v7.1.symbols.gmt) as the reference gene set. Gene set permutations were performed 1,000 times for analysis. The normalized enrichment score (NES), nominal p value, and false discovery rate (FDR) q-value were used to sort the pathways enriched in each group. Pathways with NES > 1, nominal p value < 0.05, and FDR q − value < 0.25 were selected out.
2.6. Immune Cell Abundance Analysis
ImmuCellAI is a tool to estimate the abundance of 24 immune cells from gene expression datasets [31]. We used this tool to survey the infiltration difference of immune cells between the low- and the high-GJA1 expression groups.
2.7. Statistical Analysis
All statistical analyses were performed with IBM SPSS statistical software (version 26.0) and R software (version 3.6.3), and p < 0.05 was used to be as the significance level. The differences in GJA1 expression between the two groups were compared by Wilcoxon test. Chi-squared (χ2) test and logistic regression analysis were used to evaluate the relationship between GJA1 expression and clinicopathological parameters. Kaplan–Meier analysis and log-rank test were used to explore the relationship between survival rates and the GJA1 expression level. A Cox proportional hazard regression model was used for univariate and multivariate survival analysis.
3. Results
3.1. The Difference of GJA1 Expression between CC and normal tissues
GEPIA was used to explore the mRNA expression levels of GJA1 in CC and normal tissues, showing that GJA1 expression is significantly decreased in CC tissues (Figure S1A). Similar results were observed in five of the six GEO datasets.(Table S1). On the protein level, the expression of GJA1 was also higher in normal tissues than in CC tissues by using the HPA database (Figure S1B). In addition, the expression level of GJA1 was different in groups classified according to histology (p < 0.001, Figure 1(a)) and FIGO stage (p = 0.018, Figure 1(b)). In a stratified analysis of SCC patients, we found the same results that the GJA1 expression level was related to the FIGO stage (p = 0.038, Figure 1(d)).
Figure 1.
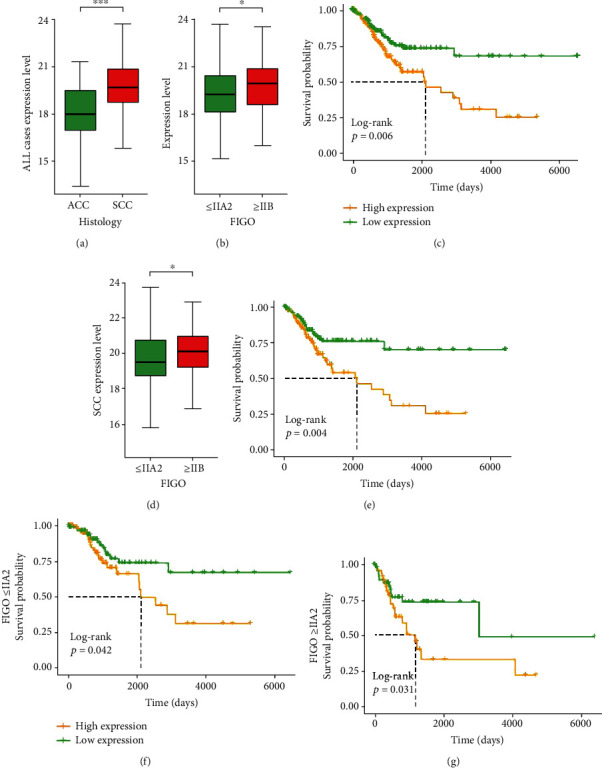
The expression of GJA1 and its association with clinicopathological parameters based on TCGA data: (a) histology type; (b) FIGO stage of all CC patients; (c) impact of GJA1 expression on OS in all CC patients; (d) FIGO stage of SCC patients; (e) impact of GJA1 expression on OS in all SCC patients; (f) impact of GJA1 expression on OS in patients with FIGO stage ≤ IIA2; (g) impact of GJA1 expression on OS in patients with FIGO stage ≥ IIB. TCGA: The Cancer Genome Atlas; FIGO: the International Federation of Gynecology and Obstetrics; CC: cervical cancer; OS: overall survival; SCC: squamous cell carcinoma; ACC: adenomas and adenocarcinomas; GJA1: gap junction protein alpha 1; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
3.2. High Expression of GJA1 in CC Is Related to Poor Overall Survival
We evaluated the relationship between GJA1 expression and prognosis of CC patients by Kaplan–Meier risk estimates. Compared to the low-GJA1 expression, the high-GJA1 expression was significantly associated with poor overall survival (p = 0.006, Figure 1(c)). The median OS of the high-GJA1-expression group was 627.5 days (range: 0-5271 days), while for the low-GJA1-expression group, the median OS was 734.5 days (range: 0-6408 days). The 5-year survival rate of patients in the low-GJA1-expression group (32.9%) was also higher than that in the high-GJA1-expression group (25.0%). In the stratified analysis of SCC patients, early-stage patients (FIGO ≤ IIA2), and late-stage patients (FIGO ≥ IIB), the high-GJA1 expression group all showed poor prognosis, and the p value was 0.004, 0.042, and 0.031, respectively (Figures 1(e)–1(g)).
3.3. The relationship between GJA1 Expression and Clinicopathological Variables
To further explore the relationship between GJA1 expression and clinicopathological parameters, the clinical data of 304 CC samples with GJA1 expression data were analyzed from the TCGA database. Using chi-squared (χ2) test, the high expression level of GJA1 was significantly correlated with keratinization (p = 0.045), hormone use (p = 0.019), tumor size (0.042), and FIGO stage (p = 0.003) (Table 2). Multiple logistic regression analysis showed that the increased expression of GJA1 in CC was significantly correlated with keratinization (OR, 1.987; 95% CI, 1.011-3.905; p = 0.046), hormone use (OR, 0.457; 95% CI, 0.236-0.885; p = 0.020), tumor size (OR, 2.234; 95% CI, 1.013-4.927; p = 0.046), and FIGO stage (OR, 2.062; 95% CI, 1.274-3.338; p = 0.003) (Table 3).
Table 2.
Relationships between GJA1 expression and clinicopathological parameters in CC.
| Clinicopathological parameters | GJA1 expression | Total | p value | |
|---|---|---|---|---|
| Low (n = 152) | High (n = 152) | |||
| Age∗ | ||||
| Median | 46.0 | 46.5 | 46.0 | 0.485 |
| Interquartile range | (38.0-55.0) | (38.0-59.0) | (38.0-57.0) | |
| BMI∗ | ||||
| Median | 26.9 | 25.8 | 26.6 | 0.246 |
| Interquartile range | (23.1-33.3) | (22.6-31.2) | (22.6-32.4) | |
| Keratinization | ||||
| No | 56 | 63 | 119 | 0.045 |
| Yes | 17 | 38 | 55 | |
| Hormone use | ||||
| No | 45 | 44 | 89 | 0.019 |
| Yes | 47 | 21 | 68 | |
| Hysterectomy# | ||||
| Other | 4 | 4 | 8 | 0.734 |
| Hysterectomy | 89 | 70 | 159 | |
| Tumor size | ||||
| ≤T2 | 119 | 92 | 211 | 0.042 |
| ≥T3 | 11 | 19 | 30 | |
| Lymph node | ||||
| N0 | 79 | 54 | 133 | 0.063 |
| N1 | 27 | 33 | 60 | |
| FIGO stage | ||||
| ≤IIA2 | 106 | 82 | 188 | 0.003 |
| ≥IIB | 42 | 67 | 109 | |
| Differentiation grade | ||||
| ≤G2 | 74 | 79 | 153 | 0.151 |
| ≥G3 | 68 | 51 | 119 | |
| Lymphovascular invasion | ||||
| No | 40 | 31 | 71 | 0.697 |
| Yes | 42 | 37 | 79 | |
| Distant metastasis | ||||
| No | 141 | 132 | 273 | 0.088 |
| Yes | 11 | 20 | 31 | |
∗Mann-Whitney U test. #Fisher's exact test.
Table 3.
GJA1 expression correlated with clinicopathological parameters (logistic regression).
| Clinicopathological parameters | Total (N) | OR | 95% confidence interval | p value | |
|---|---|---|---|---|---|
| Lower limit | Upper limit | ||||
| Age (continuous) | |||||
| 304 | 1.007 | 0.991 | 1.024 | 0.393 | |
| BMI (continuous) | |||||
| 259 | 0.982 | 0.950 | 1.015 | 0.280 | |
| Keratinization | |||||
| Yes vs. no | 174 | 1.987 | 1.011 | 3.905 | 0.046 |
| Hormone use | |||||
| Yes vs. no | 157 | 0.457 | 0.236 | 0.885 | 0.020 |
| Hysterectomy | |||||
| Yes vs. no | 167 | 0.787 | 0.190 | 3.257 | 0.740 |
| Tumor size | |||||
| ≥T3 vs. ≤T2 | 241 | 2.234 | 1.013 | 4.927 | 0.046 |
| Lymph node | |||||
| N1 vs. N0 | 193 | 1.788 | 0.967 | 3.308 | 0.064 |
| FIGO stage | |||||
| ≥IIB vs. ≤IIA2 | 297 | 2.062 | 1.274 | 3.338 | 0.003 |
| Differentiation grade | |||||
| ≥G3 vs. ≤G2 | 272 | 0.703 | 0.434 | 1.138 | 0.151 |
| Lymphovascular invasion | |||||
| Yes vs. no | 150 | 1.137 | 0.597 | 2.165 | 0.697 |
| Distant metastasis | |||||
| Yes vs. no | 304 | 1.942 | 0.896 | 4.207 | 0.092 |
3.4. The Effect of GJA1 Expression on Survival Based on Univariate and Multivariate Analyses
The univariate and multivariate Cox proportional hazard regression analyses were performed to investigate whether the high-GJA1 expression is an independent predictor of poor survival in CC patients. After excluding patients with incomplete data, we included 120 patients with CC for the multivariate Cox proportional hazard regression analysis. The univariate analysis showed that BMI (HR, 0.953; 95% CI, 0.911-0.996; p = 0.034), tumor size (HR, 3.643, 95% CI, 1.922-6.904, p < 0.001), lymph node (HR, 2.695; 95% CI, 1.358-5.349; p = 0.0046), FIGO stage (HR, 1.861; 95% CI, 1.166-2.970; p = 0.009), lymphovascular invasion (HR, 10.041; 95% CI, 2.361-42.700; p = 0.002), distant metastasis (HR, 3.141; 95% CI, 1.866-5.289; p < 0.001), and high-GJA1 expression (HR, 1.943; 95% CI, 1.202-3.140; p = 0.007) were important predictors of survival (Table 4). As the p value is close to 0.05, age was also included in multivariate analysis. The results showed that tumor size (HR, 6.181; 95% CI, 1.219-31.334; p = 0.028), lymphovascular invasion (HR, 5.910; 95% CI, 1.124-31.059; p = 0.036), and the high-GJA1 expression (HR, 4.084; 95% CI, 1.354-12.320; p = 0.013) were the important independent predictors of poor overall survival of CC (Figure 2 and Table 4).
Table 4.
Associations between overall survival and clinicopathological characteristics in CC using univariate analysis and multivariate analysis.
| Parameters | Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | |||
| Lower | Upper | Lower | Upper | |||||
| Age (continuous) | 1.017 | 0.999 | 1.035 | 0.057 | 0.959 | 0.910 | 1.011 | 0.123 |
| BMI (continuous) | 0.953 | 0.911 | 0.996 | 0.034 | 0.976 | 0.894 | 1.066 | 0.593 |
| Tumor size (≥T3 vs. ≤T2) | 3.643 | 1.922 | 6.904 | <0.001 | 6.181 | 1.219 | 31.334 | 0.028 |
| Lymph node (N1 vs. N0) | 2.695 | 1.358 | 5.349 | 0.005 | 2.256 | 0.636 | 8.008 | 0.208 |
| FIGO stage (≥IIB vs. ≤IIA2) | 1.861 | 1.166 | 2.970 | 0.009 | 0.317 | 0.042 | 2.403 | 0.266 |
| Lymphovascular invasion (yes vs. no) | 10.041 | 2.361 | 42.700 | 0.002 | 5.910 | 1.124 | 31.059 | 0.036 |
| Distant metastasis (yes vs. no) | 3.141 | 1.866 | 5.287 | <0.001 | 1.043 | 0.257 | 4.240 | 0.953 |
| GJA1 expression level (high vs. low) | 1.943 | 1.202 | 3.140 | 0.007 | 4.084 | 1.354 | 12.320 | 0.013 |
Likelihood ratio test = 28.37 on 8 df, p = 0.0004, N = 120, number of events = 18.
Figure 2.
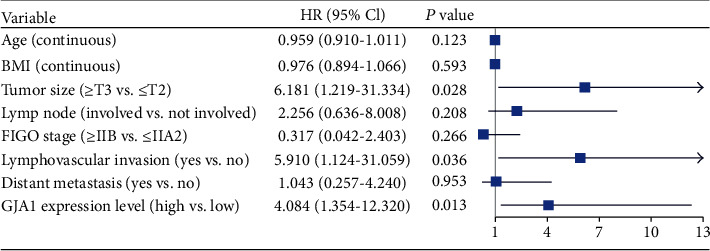
Forest plot for the multivariate Cox proportional hazard regression model. High-GJA1 expression was an independent predictor of poor survival rate (HR, 4.084; 95% CI, 1.354-12.320; p = 0.013). GJA1: Gap Junction Protein Alpha 1; HR: hazard ratio; CI: confidence interval; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
3.5. Identification of GJA1-Related Signaling Pathways by GSEA
To identify signaling pathways that are differentially activated in CC, we conducted the Gene Set Enrichment Analysis (GSEA) between low- and high-GJA1-expression datasets. We selected out 30 significantly enriched signaling pathways based on the standard that is NES > 1, nominal p value < 0.05, and FDR q − value < 0.25 (Table S2). Figure 3 shows 14 cancer-related pathways enriched in high-GJA1-expression groups, including many well-known pathways, such as the P53 signaling pathway, the Wnt signaling pathway, and the MAPK signaling pathway.
Figure 3.
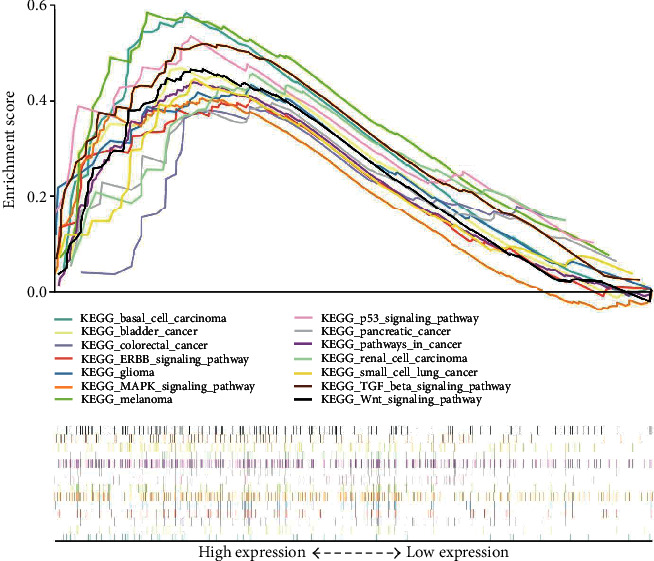
A merged enrichment plot from gene set enrichment analysis (GSEA) including enrichment score and gene sets. 14 cancer-related pathways are shown here.
3.6. Immune Cell Abundance Analysis
ImmuCellAI was used to analyze the infiltration difference of immune cells between the low- and the high-GJA1-expression group. We found that the abundance of CD8 naive, DC, and neutrophil is significantly increased in the high-GJA1-expression group, while the abundance of gamma delta and Th1 is significantly decreased in the low-GJA1-expression group (Figure 4).
Figure 4.
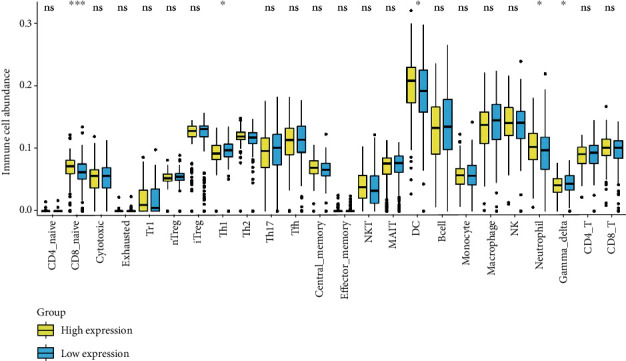
Immune cell abundance analysis between the low-GJA1-expression group and the high-GJA1-expression group. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
4. Discussion
The role of GJA1 in cancers has been widely studied in recent years. GJA1 has two opposite effects on cancers as it can act as an oncogene or a tumor suppressor gene. Many studies have unveiled the suppressive roles of GJA1 in tumorigenesis. In the colorectal cancer cell line (HT 29), Sirnes et al. found overexpression of GJA1 could inhibit Wnt signaling by interacting with β-catenin, thus inhibiting the growth of tumor cells [32]. Huang et al. identified that GJA1 could restrain glioblastoma development by reducing the anti-apoptotic Bcl-2 [13]. Similarly, overexpression of GJA1 was also found to inhibit the growth of lung cancer cells (LH7) [33]. Many studies have also reported the role of GJA1 in promoting cancer progression. Tang et al. reported that the overexpression of GJA1 could promote lymph node metastasis and peritoneal metastasis in gastric cancer [34, 35]. Zhao et al. found that high-GJA1 expression could indicate poor prognosis of gastric cancer patients based on five GEO datasets [18]. Ogawa et al. found that the silencing of GJA1 was associated with reduced invasion, migration, and metastasis [36]. From the Affymetrix analysis, Teleki et al. reported that high expression of GJA1 was associated with a reduced relapse-free survival (RFS) and overall survival (OS) in ER-negative breast cancer patients [37]. Moreover, the study by Stoletov et al. confirmed this observation using analysis from the Oncomine database [38]. Also, there are some researches on the impact of GJA1 on CC. Sun et al. found that HPV E6 may regulate GJA1 trafficking in cervical tumor cells, resulting in inhibition of the formation of gap junctions [39]. Using whole-genome microarray data, Cheng et al. showed that GJA1 was a critical gene for CC invasion and metastasis [40].
Using high-throughput RNA-sequencing data from the TCGA database, we aimed to explore the relationship between clinicopathological parameters and GJA1 expression and determine the role of GJA1 expression in CC progression, especially as a prognostic factor for CC. We also tried to screen signaling pathways related to GJA1 in CC to understand the underlying mechanism involved in the regulation of CC development by GJA1. In addition, we tried to explore GJA1 expression's impact on the abundance of the immune cells. First, we compared the expression of GJA1 in CC and normal tissues by GEPIA. The expression level of GJA1 in CC tissues was significantly lower than that in normal tissues. Six GEO datasets were used to verify this result, and the results from five datasets are the same as those from GEPIA. The HPA database also showed that the expression of GJA1 is higher in normal tissues than that in CC tissues at the protein level. These results suggest that GJA1 may be a tumor suppressor gene and play an important role in the progression of CC. In addition, GJA1 expression levels were different in groups classified by histology and FIGO stage. Further, chi-squared (χ2) test showed that the high expression level of GJA1 was significantly correlated with keratinization, hormone use, tumor size, and FIGO stage. Some literatures reported that GJA1 was associated with metastasis in a variety of tumors. Lin et al. found that the expression of GJA1 was higher in metastatic breast tumors than in primary breast tumors [41]. Wang et al. also showed that GJA1 could be used as a marker of metastasis in prostate cancer [42].
The Kaplan–Meier survival analysis showed that the high-GJA1-expression group indicated a worse prognosis than the low-GJA1-expression group. In the stratified analysis of SCC patients, early-stage patients (FIGO ≤ IIA2), and late-stage patients (FIGO ≥ IIB), we found the same result. Using five GEO datasets, Zhao et al. reported that high expression level of GJA1 was associated with poor prognosis in gastric cancer, which is similar to our result [18].
The univariate analysis showed that BMI, tumor size, lymph node, FIGO stage, lymphovascular invasion, distant metastasis, and high-GJA1expression were important predictors of survival. Importantly, multivariate analysis showed that tumor size, lymphovascular invasion, and the high-GJA1 expression were the important independent predictors of poor overall survival of CC. This result demonstrated the potential of GJA1 to become a prognosis biomarker of CC.
To further investigate the functions of GJA1 in CC, we performed GSEA using the TCGA data and 30 pathways were selected out based on established standards. The result showed that many cancer-related pathways, such as the p53 signaling pathway, the Wnt signaling pathways, and the MAPK signaling pathway, were enriched in the high-GJA1-expression group. Evidence has shown that the proliferation, migration, and invasion of CC cells can be modulated by regulating the p53 signaling pathway [43, 44]. Wnt signaling, which is needed for cell proliferation and differentiation, has been found to play a critical role in CC carcinogenesis [45, 46]. Activation of this pathway was shown to be related to the poor prognosis of CC patients [46]. MAPK signaling pathways regulate a variety of cellular activities, including proliferation, differentiation, survival, and death [47]. Many studies have reported that the interaction between HPV and the MAPK signaling pathway indirectly explains its role in CC [48, 49]. In addition, the TGF-β signaling pathway was enriched in the high-GJA1-expression group, which participates in the regulation of a variety of immune cells, such as NK cell, neutrophils, and macrophage [50]. Thus, we analyzed the differences of immune cells between the low-GJA1-expression group and the high-GJA1-expression group and found that the abundance of CD8 naive cell, DC, and neutrophil were significantly increased in the high-GJA1-expression group.
5. Conclusions
In conclusion, high GJA1 expression may be a potential prognostic molecular marker of poor survival in CC. Moreover, the p53 signaling pathway, Wnt signaling pathway, and MAPK signaling pathways may be the critical pathways regulated by GJA1. Moreover, GJA1 can affect the abundance of immune cells to a certain extent. Further experimental validation should be performed to prove the biologic impact of GJA1 in CC.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant number: 81974410).
Data Availability
Publicly available datasets were analyzed in this study. These can be found in The Cancer Genome Atlas (https://portal.gdc.cancer.gov/); the NCBI Gene Expression Omnibus (GSE39001, GSE52903, GSE63514, GSE6791, GSE7803, GSE9750).
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Authors' Contributions
Silu Meng, Xinran Fan, and Jianwei Zhang contributed equally to this work. Silu Meng, Xinran Fan, and Jianwei Zhang designed the overall idea of this study, analyzed the data, prepared the figures and tables, and wrote the drafts of the paper. Ran An checked the statistical results. Shuang Li supervised this study and revised the drafts of the paper.
Supplementary Materials
Supplementary Figure 1: comparison of GJA1 expression between cervical cancer and normal using GEPIA and The Human Protein Atlas.
Supplementary Table 1: information of selected six GEO datasets and GJA1 expression in these datasets. Supplementary Table 2: gene sets enriched in the high-GJA1-expression phenotype.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P. D., et al. Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Yu S., Yang C. S., Li J., et al. Cancer prevention research in China. Cancer Prevention Research (Philadelphia, Pa.) 2015;8(8):662–674. doi: 10.1158/1940-6207.CAPR-14-0469. [DOI] [PubMed] [Google Scholar]
- 4.Jiang X., Tang H., Chen T. Epidemiology of gynecologic cancers in China. Journal of Gynecologic Oncology. 2018;29(1) doi: 10.3802/jgo.2018.29.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W., Zheng R., Zhang S., et al. Cancer incidence and mortality in China, 2013. Cancer Letters. 2017;401:63–71. doi: 10.1016/j.canlet.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 6.Marth C., Landoni F., Mahner S., et al. Cervical cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28(supplement_4):iv72–iv83. doi: 10.1093/annonc/mdx220. [DOI] [PubMed] [Google Scholar]
- 7.Heintz A. P. M., Odicino F., Maisonneuve P., et al. Carcinoma of the fallopian tube. FIGO 26th Annual Report on the Results of Treatment in Gynecological Cancer. International Journal of Gynecology & Obstetrics. 2006;95:S145–S160. doi: 10.1016/S0020-7292(06)60032-5. [DOI] [PubMed] [Google Scholar]
- 8.Zeng H., Zheng R., Guo Y., et al. Cancer survival in China, 2003-2005: a population-based study. International Journal of Cancer. 2015;136(8):1921–1930. doi: 10.1002/ijc.29227. [DOI] [PubMed] [Google Scholar]
- 9.Cheung F. Y., Mang O. W., Law S. C. A population-based analysis of incidence, mortality, and stage-specific survival of cervical cancer patients in Hong Kong: 1997-2006. Hong Kong Medical Journal. 2011;17(2):89–95. [PubMed] [Google Scholar]
- 10.Wu J. I., Wang L. H. Emerging roles of gap junction proteins connexins in cancer metastasis, chemoresistance and clinical application. Journal of Biomedical Science. 2019;26(1):p. 8. doi: 10.1186/s12929-019-0497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y. W., Morita I., Ikeda M., Ma K. W., Murota S. Connexin43 suppresses proliferation of osteosarcoma U2OS cells through post-transcriptional regulation of p27. Oncogene. 2001;20(31):4138–4149. doi: 10.1038/sj.onc.1204563. [DOI] [PubMed] [Google Scholar]
- 12.Ai Z., Fischer A., Spray D. C., Brown A. M. C., Fishman G. I. Wnt-1 regulation of connexin43 in cardiac myocytes. The Journal of Clinical Investigation. 2000;105(2):161–171. doi: 10.1172/JCI7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang R. P., Hossain M. Z., Huang R., Gano J., Fan Y., Boynton A. L. Connexin 43 (cx43) enhances chemotherapy-induced apoptosis in human glioblastoma cells. International Journal of Cancer. 2001;92(1):130–138. doi: 10.1002/1097-0215(200102)9999:9999<::AID-IJC1165>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 14.Kanczuga-Koda L., Sulkowski S., Lenczewski A., et al. Increased expression of connexins 26 and 43 in lymph node metastases of breast cancer. Journal of Clinical Pathology. 2006;59(4):429–433. doi: 10.1136/jcp.2005.029272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ming J., Zhou Y., du J., et al. miR-381 suppresses C/EBPα-dependent Cx43 expression in breast cancer cells. Bioscience Reports. 2015;35(6) doi: 10.1042/BSR20150167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behrens J., Kameritsch P., Wallner S., Pohl U., Pogoda K. The carboxyl tail of Cx43 augments p38 mediated cell migration in a gap junction-independent manner. European Journal of Cell Biology. 2010;89(11):828–838. doi: 10.1016/j.ejcb.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh S., Kumar A., Tripathi R. P., Chandna S. Connexin-43 regulates p38-mediated cell migration and invasion induced selectively in tumour cells by low doses of γ-radiation in an ERK-1/2-independent manner. Carcinogenesis. 2014;35(2):383–395. doi: 10.1093/carcin/bgt303. [DOI] [PubMed] [Google Scholar]
- 18.Zhao X., Yu C., Zheng M., Sun J. Prognostic value of the mRNA expression of gap junction α members in patients with gastric cancer. Oncology Letters. 2019;18:1669–1678. doi: 10.3892/ol.2019.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colaprico A., Silva T. C., Olsen C., et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Research. 2016;44(8) doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J., Lichtenberg T., Hoadley K. A., et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie M. E., Phipson B., Wu D., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43(7):p. e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Espinosa A. M., Alfaro A., Roman-Basaure E., et al. Mitosis is a source of potential markers for screening and survival and therapeutic targets in cervical cancer. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0055975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina-Martinez I., Barrón V., Roman-Bassaure E., et al. Impact of gene dosage on gene expression, biological processes and survival in cervical cancer: a genome-wide follow-up study. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0097842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.den Boon J. A., Pyeon D., Wang S. S., et al. Molecular transitions from papillomavirus infection to cervical precancer and cancer: role of stromal estrogen receptor signaling. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(25):E3255–E3264. doi: 10.1073/pnas.1509322112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pyeon D., Newton M. A., Lambert P. F., et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Research. 2007;67(10):4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhai Y., Kuick R., Nan B., et al. Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Research. 2007;67(21):10163–10172. doi: 10.1158/0008-5472.CAN-07-2056. [DOI] [PubMed] [Google Scholar]
- 28.Scotto L., Narayan G., Nandula S. V., et al. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: potential role in progression. Genes, Chromosomes & Cancer. 2008;47(9):755–765. doi: 10.1002/gcc.20577. [DOI] [PubMed] [Google Scholar]
- 29.Ponten F., Jirstrom K., Uhlen M. The Human Protein Atlas--a tool for pathology. The Journal of Pathology. 2008;216(4):387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A., Tamayo P., Mootha V. K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao Y. R., Zhang Q., Lei Q., et al. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Advanced Science. 2020;7(7):p. 1902880. doi: 10.1002/advs.201902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sirnes S., Bruun J., Kolberg M., et al. Connexin43 acts as a colorectal cancer tumor suppressor and predicts disease outcome. International Journal of Cancer. 2012;131(3):570–581. doi: 10.1002/ijc.26392. [DOI] [PubMed] [Google Scholar]
- 33.Xu H. T., Li Q. C., Zhang Y. X., et al. Connexin 43 recruits E-cadherin expression and inhibits the malignant behaviour of lung cancer cells. Folia Histochemica et Cytobiologica. 2008;46(3):315–321. doi: 10.2478/v10042-008-0057-9. [DOI] [PubMed] [Google Scholar]
- 34.Tang B., Peng Z. H., Yu P. W., Yu G., Qian F. Expression and significance of Cx43 and E-cadherin in gastric cancer and metastatic lymph nodes. Medical Oncology. 2011;28(2):502–508. doi: 10.1007/s12032-010-9492-5. [DOI] [PubMed] [Google Scholar]
- 35.Tang B., Peng Z. H., Yu P. W., et al. Aberrant expression of Cx43 is associated with the peritoneal metastasis of gastric cancer and Cx43-mediated gap junction enhances gastric cancer cell diapedesis from peritoneal mesothelium. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0074527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogawa K., Pitchakarn P., Suzuki S., et al. Silencing of connexin 43 suppresses invasion, migration and lung metastasis of rat hepatocellular carcinoma cells. Cancer Science. 2012;103(5):860–867. doi: 10.1111/j.1349-7006.2012.02228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teleki I., Szasz A. M., Maros M. E., et al. Correlations of differentially expressed gap junction connexins Cx26, Cx30, Cx32, Cx43 and Cx46 with breast cancer progression and prognosis. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0112541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoletov K., Strnadel J., Zardouzian E., et al. Role of connexins in metastatic breast cancer and melanoma brain colonization. Journal of Cell Science. 2013;126(4):904–913. doi: 10.1242/jcs.112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun P., Dong L., MacDonald A., et al. HPV16 E6 controls the gap junction protein Cx43 in cervical tumour cells. Viruses. 2015;7(10):5243–5256. doi: 10.3390/v7102871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng Y., Ma D., Zhang Y., Li Z., Geng L. Cervical squamous cancer mRNA profiles reveal the key genes of metastasis and invasion. European Journal of Gynaecological Oncology. 2015;36(3):309–317. [PubMed] [Google Scholar]
- 41.Lin Z. J., Ming J., Yang L., du J. Z., Wang N., Luo H. J. Mechanism of regulatory effect of microRNA-206 on connexin 43 in distant metastasis of breast cancer. Chinese Medical Journal. 2016;129(4):424–434. doi: 10.4103/0366-6999.176071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.WANG Y., GUO W., XU H., et al. An extensive study of the mechanism of prostate cancer metastasis. Neoplasma. 2018;65(2):253–261. doi: 10.4149/neo_2018_161217N648. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y., Li L., Liu Y., et al. RECK inhibits cervical cancer cell migration and invasion by promoting p53 signaling pathway. Journal of Cellular Biochemistry. 2018;119(4):3058–3066. doi: 10.1002/jcb.26441. [DOI] [PubMed] [Google Scholar]
- 44.Kashyap V. K., Dan N., Chauhan N., et al. VERU-111 suppresses tumor growth and metastatic phenotypes of cervical cancer cells through the activation of p53 signaling pathway. Cancer Letters. 2020;470:64–74. doi: 10.1016/j.canlet.2019.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bahrami A., Amerizadeh F., ShahidSales S., et al. Therapeutic potential of targeting Wnt/β-catenin pathway in treatment of colorectal cancer: rational and progress. Journal of Cellular Biochemistry. 2017;118(8):1979–1983. doi: 10.1002/jcb.25903. [DOI] [PubMed] [Google Scholar]
- 46.Bahrami A., Hasanzadeh M., ShahidSales S., et al. Clinical significance and prognosis value of Wnt signaling pathway in cervical cancer. Journal of Cellular Biochemistry. 2017;118(10):3028–3033. doi: 10.1002/jcb.25992. [DOI] [PubMed] [Google Scholar]
- 47.Kim E. K., Choi E. J. Pathological roles of MAPK signaling pathways in human diseases. Biochimica et Biophysica Acta. 2010;1802(4):396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 48.Chakrabarti O., Veeraraghavalu K., Tergaonkar V., et al. Human papillomavirus type 16 E6 amino acid 83 variants enhance E6-mediated MAPK signaling and differentially regulate tumorigenesis by notch signaling and oncogenic Ras. Journal of Virology. 2004;78(11):5934–5945. doi: 10.1128/JVI.78.11.5934-5945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Contreras-Paredes A., De la Cruz-Hernandez E., Martinez-Ramirez I., Duenas-Gonzalez A., Lizano M. E6 variants of human papillomavirus 18 differentially modulate the protein kinase B/phosphatidylinositol 3-kinase (akt/PI3K) signaling pathway. Virology. 2009;383(1):78–85. doi: 10.1016/j.virol.2008.09.040. [DOI] [PubMed] [Google Scholar]
- 50.Batlle E., Massague J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019;50(4):924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: comparison of GJA1 expression between cervical cancer and normal using GEPIA and The Human Protein Atlas.
Supplementary Table 1: information of selected six GEO datasets and GJA1 expression in these datasets. Supplementary Table 2: gene sets enriched in the high-GJA1-expression phenotype.
Data Availability Statement
Publicly available datasets were analyzed in this study. These can be found in The Cancer Genome Atlas (https://portal.gdc.cancer.gov/); the NCBI Gene Expression Omnibus (GSE39001, GSE52903, GSE63514, GSE6791, GSE7803, GSE9750).


