Abstract
Purpose
Rare cancers are defined by an incidence of <6 out of 100 000 cases per year. They are under-represented in clinical research including tumour molecular analysis. The aim of Arcagen is to generate a multinational database integrating clinical and molecular information of patients with rare cancers.
Patients and methods
We present the retrospective feasibility cohort of patients with rare cancers, with previously collected tumour samples available from any stage. Molecular analysis was performed using FoundationOne CDx for all histologies except for sarcoma where FoundationOne Heme was used. Clinical data including demographic data, medical history, malignant history, treatment and survival data were collected.
Results
Eighty-seven patients from three centres were screened; molecular data were obtained for 77 patients (41 sarcomas, 9 yolk sac tumours, 14 rare head and neck cancers, 13 thymomas). The median age at the time of diagnosis was 48 (range 28–85). Most patients had reportable genomic alterations (89%). The most common alterations were linked to cell cycle regulation (TP53, RB1, CDKN2A/B deletions and MDM2 amplification). Multiple activating single-nucleotide variants (SNVs) could be detected in the RAS/RAF family. The tumour mutational burden status was globally low across all samples with a median of 3 Muts/MB (range 0–52). Only 4 cases (ie, 4.7% of tumours) had direct actionable mutations for a treatment approved in Europe within the patient’s tumour type.
Conclusion
The Arcagen project aims to bridge the gap and improve knowledge of the molecular landscape of rare cancers by prospectively recruiting up to 1000 patients.
Keywords: genomics, rare cancers, yolk sac tumour, sarcoma, thymoma, salivary gland tumours, precision medicine
Key questions.
What is already known about this subject?
Rare cancers are extremely heterogeneous (>300 histological subtypes).
Little is known regarding the molecular landscape of rare cancers.
Represent 20% of cancer diagnostic but 30% of death due to cancer.
What does this study add?
Confirming the importance of the TP53 signalling pathway in sarcoma.
Low actionability (below 10% for rare cancer).
Confirming need for more clinical research on rare cancers.
How might this impact on clinical practice?
Feasibility of recruiting patients with rare cancer for clinical research to understand better the molecular landscape and actionability of this patient population.
Introduction
Rare cancers are defined as histological diagnoses with an incidence of <6 out of 100 000 cases per year. They include >300 histological subtypes and may affect all organs.1 Altogether, they represent 20% of adult cancer incidence but 30% of cancer mortality.2 The mortality of rare cancers may be related to frequent misdiagnosis, delays in diagnosis and treatment, limited research efforts and lack of optimal guidelines.3–5
Rare cancers are under-represented in the research programmes exploring genomic alterations of cancer. Therefore, little is known regarding prevalence of clinically actionable genomic alterations in rare cancers in general, although sarcomas and brain tumours are starting to be better characterised.6–8 A better understanding of this population may bring new opportunities for diagnosis and treatment, and potential to improve patient outcome. Indeed, some rare cancers have become paradigmatic models for personalised medicine. Progress with the understanding of tumour biology and the subsequent development of targeted therapies enabled major therapeutic advances in gastrointestinal stromal tumours and thyroid cancers.9 10 Rare tumours now often represent strategic opportunities to develop new medications, with several examples of important treatments made available by these models, such as BRAF inhibitors or TRK inhibitors.11
Translational and clinical research on rare tumours has long been hindered by organisational difficulties linked to dispersal of patients to clinical centres of minimal experience, inaccurate diagnosis and limited efforts for tumour collection. Aiming to address these shortcomings, EURACAN is a European Reference Network dedicated to rare cancers, bringing together 66 centres from 17 countries as of January 2020. Dedicated research groups exist within EURACAN to overcome these bottlenecks.12
The translational research infrastructure associated with the research initiatives of EURACAN is SPECTA, a collaborative European platform led by EORTC.
To improve knowledge on rare cancers, EORTC, in collaboration with EURACAN developed the translational research project Arcagen, aiming to perform molecular profiling on clinically annotated tumour samples from 1000 patients with rare cancers, to improve the biological understanding of these tumours. We present here the results of the first step of this project, an analysis of retrospectively collected data and samples from sites within the EURACAN network. This first milestone of the project includes analysis of 100 samples, collected from 87 patients.
Materials and methods
Patient selection
To test the feasibility of this project, three centres from the EURACAN network were selected: Centre Leon Berard (Lyon, France), Bergonie Institute (Bordeaux, France) and Hospices Civils de Lyon (France).
We selected four rare cancer domains for the retrospective part of the study: sarcoma, thoracic malignancies, ovarian cancer and head and neck cancer. From those four domains, we chose to include patients with a diagnosis of undifferentiated pleomorphic sarcoma (UPS) and myxofibrosarcoma (MFS), ovarian yolk sac tumour (YST), thymomas and rare histologies of head and neck cancers including ethmoidal, paranasal sinus and nasal cavity adenocarcinomas, based on sample availabilities in the sites’ biobanks (see figure 1 for clinical characteristics).
Figure 1.

Clinical description of the population with number of patients (total/evaluable). NPS, nasopharynx and paranasal sinus; YST, ovarian yolk sac tumour.
All tumour samples were assessed by expert pathologists within their disease types, at the site level, to confirm the diagnosis.
All samples were analysed by Foundation Medicine Inc laboratories (FMI, Penzberg, Germany and Cambridge, Massachusetts, USA) (see online supplemental information).
esmoopen-2020-001075supp001.pdf (43.9KB, pdf)
Results
Project description and sample workflow—feasibility
One hundred samples from 87 patients were included in the retrospective pilot. Quality control (QC) was performed by the pathologist from the hospitals.
From the first 100 samples received, 22 failed (22% of failure). A second set of slides cut from the same block was requested to the site and was available for 17 out of 22 cases. The success rate for those 17 cases was 41% (n=7 out of 17) (online supplemental figure 1a). Thus, the global failure rate was 15% (15 failures out of the original 100 samples) due to two main reasons: sample quality issue (60%, n=9) or not enough material after DNA/RNA extraction (40%, n=6).
esmoopen-2020-001075supp002.pdf (289.4KB, pdf)
Out of the 100 samples, 43 came from patients with sarcoma, and the failure rate of the FoundationOne Heme test was 4.6% (n=2) (online supplemental figure 1a).
Results were reported via the FMI clinical reports with a median turnaround time of 16 days (range 9–157) for FoundationOne CDx and 15 days (range 10–106) for FoundationOne Heme. The 145 days turnaround time was due to a test failure on the first sample and the request of a second sample to the clinical site.
Clinical characteristics of the patient population
Seventy-seven patients (85 samples) had available molecular and clinical data and were included in the analysis. Forty-one patients (53.2%) were diagnosed with sarcoma, 9 (11.7%) with YST, 14 (18.2%) with rare head and neck cancers and 13 (16.9%) with thymoma. The baseline characteristics of the patients are described in figure 1. All clinical characteristics are limited to the evaluable population.
Molecular data were available for 41 patients with sarcoma. The median age at the time of diagnosis of our population was 69 (range 33–85). The reported histology was MFS for 19 patients and UPS for 22 patients. Samples were collected from the surgical resection of the primary tumour for 40 patients and from a local recurrence for 1 patient. Eleven patients (27%) had received treatment, radiotherapy (four, 9.7%) or chemotherapy (seven, 17%), before the sample was taken. A recurrence of the disease was reported for 14 patients (34%), 12 of which had a grade 3 tumour at the time of diagnosis and median time to recurrence from the primary diagnosis was 10 months (range 1–23 months).
Thymomas are among the rarest cancers, with an incidence of 0.15 cases per 100 000 person-years.13 In the pilot phase, molecular data were available for 13 patients. The median age at diagnosis was 53 (range 33–75). Histological subgroup as per 2015 WHO classification and Masaoka-Koga stage as assessed on surgical specimens can be found in online supplemental table 1. Most of the patients never smoked (9 out of 11), consistent with the absence of increased risk with smoking.13 Paired primary and metastatic samples were analysed for six patients, four patients had only primary tissue and three only metastatic lesion from the pleura. All patients underwent surgery, with a R0 status for 11 patients (85%) and R1 status for 2 patients (15%). Nine patients (69%) had received perioperative treatment either radiotherapy only (one case), chemotherapy only (two cases) or a combination of both (six cases). A recurrence of the disease was reported in 10 patients (77%), 4 of which had a grade 4 (A or B) at the time of diagnosis.
esmoopen-2020-001075supp003.pdf (243.9KB, pdf)
Nine patients with rare ovarian cancer were evaluable. Median age at diagnosis was 26 years (range 17–36) with a diagnosis of YST. All patients underwent surgery and chemotherapy and all samples were from the surgical resection of the primary tumour. Patients were diagnosed at different International Federation of Gynecology and Obstetrics stages; three with stage 1, one with stage 2, three with stage 3 and two unknown. A recurrence of the disease was reported for two patients (22%) with a median of 9 months (range 9–30), and only one patient died from disease within 23 months of diagnosis.
Eighteen patients with rare head and neck cancer were screened, and molecular data were available for 14. Median age at diagnosis was 60 (range 21–82) with a 6:1 male:female ratio. Histology at diagnosis was variable with 13 patients with a tumour located in the nasopharynx and paranasal sinuses (adenocarcinoma n=9, squamous cell carcinoma n=1, esthesioneuroblastoma n=2 and undifferentiated carcinoma n=1) and 1 patient with a salivary gland adenocarcinoma. Eleven patients underwent surgical resection, none with neo-adjuvant chemotherapy, and one patient received adjuvant radiotherapy. One patient with ethmoidal adenocarcinoma presented a local recurrence after 19 months. Out of the 14 patients for whom molecular data were available, 8 (57%) were non-smokers.
Molecular alterations, tumour mutational burden and microsatellite instability status, based on the clinical reports from FMI
From the FMI reports, most patients had reportable genomic alterations (89%, figure 2A). Recurrent pathways, such as in TP53 signaling or RTK pathways are highlighted in figure 2A and 2B.
Figure 2.
Overview of the significant molecular alterations found in the pilot study. (A) Waterfall plot highlighting the most common significant alterations per tumour types, sorted by molecular pathways. Only samples where molecular alterations in those specific genes were detected are represented. (B) TP53 signalling alterations in sarcoma (myxofibrosarcoma, n=19, in blue; undifferentiated pleomorphic sarcoma, n=22, in orange).
p53 signalling pathway
Molecular alterations on genes involved in the cell cycle were the most recurrent alterations in this pilot study. We identified TP53, RB1 and CDKN2A/B deletions as well as MDM2 amplification (figure 2A). This was especially noticeable in the sarcoma and rare head and neck population.
For sarcoma, we found significant molecular alterations in genes related to p53 signalling in 15 MFS samples (78.9%) and 15 UPS samples (68%) (figure 2B, blue bars for MFS, orange bars for UPS). More specifically regarding TP53, deletions were found in 3 MFS (15.8%) and 2 UPS (9.1%) and single-nucleotide variant (SNV) in 6 MFS (31.6%) and 12 UPS (54.5%). For RB1 in MFS, three samples had a deletion (15.8%) and one had an SNV (5.3%). In UPS, four samples had alterations in the RB1 gene as well (one was a deletion (4.5%) and three were SNV (13.6%)). CDKN2A/B was deleted in four MFS (21.1%) and three UPS (13.6%). Finally, some amplifications were also identified, specifically MDM2 was amplified in one MFS sample (5.3%), CCND1 amplified in one MFS sample (5.3%) and CCNE1 amplified in one UPS sample (4.5%) (figure 2B). The pattern of alterations was different between the two sarcoma subtypes; however, the majority of samples showed some form of pathway alteration, highlighting the central role of this pathway for sarcoma development.
Similarly, multiple alterations in this pathway were found within the head and neck samples, with 10 SNV in TP53 (55%), 2 in RB1 (11%) and 1 SNV, 1 DEL in CDKN2A/B (11%). Genes involved in the cell cycle pathway are the most frequently altered according to data previously reported in rare head and neck series but are interestingly also the most frequently altered pathway in head and neck squamous cell carcinoma (HNSCC).14–16
In contrast to more common epithelial ovarian cancer and as anticipated, no molecular alterations were identified in the p53 or homologous recombination deficiency (HRD) pathways for the YSTs. The only alteration found in the thymoma samples was one deletion of CDKN2A/B.
RTK-RAS/RAF-PI3K pathway
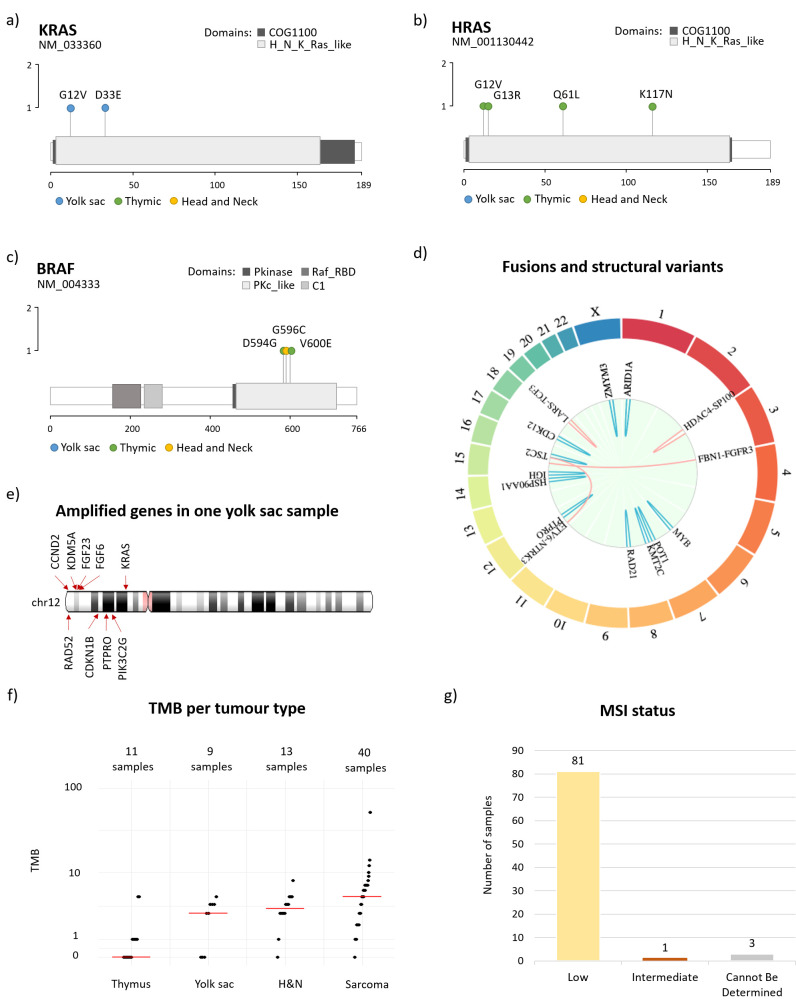
Multiple SNVs were detected in the RAS/RAF family (figure 3).
Figure 3.
Lollipop plot represented the mutations found in the RAS and RAF family. (A) Molecular alterations in KRAS, (B) molecular alterations in HRAS, (C) molecular alterations in BRAF. (D) Summary of fusions (red) and rearrangements (blue) found in all tumour types. Genes are localised on the outer ring. (E) Amplifications on chromosome 12p found in one yolk sac sample, suggesting a bigger chromosomal aberration. (F) Plot showing the tumour mutational burden (TMB) repartition within the four disease types in the pilot study. TMB could not be determined for two thymomas, one head and neck and one sarcoma sample and those are excluded from the plot. (G) Microsatellite instability status (MSI) across all samples.
In the YST samples,15–17 we found KRAS alterations (D33E and G12V, figure 3A) in two out of nine patients (22%). We also found a canonical KIT mutation in exon 17, codon 816, which was previously identified as a recurrent mutation for ovarian germ cell tumours and potentially actionable with drugs like avapritinib or ripretinib.17 18
Regarding the thymoma samples, we identified several mutations in this family: one patient with multiple HRAS canonical mutations19–22: G12V (hotspot, activating mutation), subclonal, G13R (hotspot, activating mutation) and subclonal, K117N (activating mutation); and one patient with a HRAS Q61L mutation conserved between primary and recurrent sample (thymus and pleura, figure 3B). In this population, we also observed several mutations in the RAF family: one mutation in BRAF V600E (activating mutation) and one in BRAF D594G (impaired kinase activity, figure 3C). Another BRAF mutation (G596C) was identified in a patient with a nasopharynx and paranasal sinus (NPS) adenocarcinoma (head and neck cohort). This specific mutation is located within the kinase domain of the BRAF protein, and results in decreased BRAF kinase activity, but was shown in vitro to activate downstream MEK and ERK in combination with CRAF.23
In sarcoma, a KRAS amplification and a HRAS amplification were each identified in one UPS sample (9% of UPS patients) (figure 3A, B).
Fusion and other rearrangements
Four fusions were identified in patients with sarcoma (two in UPS and two in MFS cases), two of which were actionable (ETV6-NTRK3 and FBN1-FGFR3) (figure 3D). It is important to note that sarcoma samples were analysed with FoundationOne Heme, which is expected to have higher sensitivity for certain fusion events as it includes analysis of RNA.
Rearrangements (or structural variants) were mostly found in sarcoma (four in MFS: IGH-15q25, KTM2C exon 45, ZMYM3 exon 12, POT1 intron 6; and 5 in UPS: RAD21 exon 8, KTM2C exon 45, TSC2 intron 5, HSP90AA1, PTPRO exon 22), rare histologies of head and neck (three cases: CDK12 exon 12, ARID1A exon 5 and MYB intron 14) as well as one in YST (ARID1A exon 19) (figure 3D). Similarly, multiple amplifications were found in regions of chromosome 12p for a yolk sac tumour sample (figure 3E), with specific amplifications detected in clinically relevant genes CCND2, KDM5A, FGF23, FGF6 and KRAS as well as amplifications of unknown significance in CDKN1B, PIK3C2G, PTPRO, RAD52 genes (all located in 12p). This might suggest the presence of bigger chromosomal aberration that would require further confirmation by an orthogonal molecular technique.
Tumour mutational burden and microsatellite status
The tumour mutational burden (TMB) status was low across all samples with a median of 3 Muts/MB, range 0–52 (figure 3F).
All samples but one (from an esthesioneuroblastoma) were microsatellite instability status low/stable (eg, mismatch repair proficient) (figure 3G).
Globally, most samples presented with a limited number of molecular alterations. This is particularly noticeable in YST, with five out of nine patients with no reportable alterations and three out of nine patients with only one alteration.
A complementary analysis of the molecular data can be found in online supplemental information.24–30
Discussion
Feasibility
Rare cancers represent 22% of all human cancers, and 30% of the cancer deaths. Projects describing the molecular landscape of common cancers and less frequently rare cancers have been recently reported by consortia such as The Cancer Genome Atlas (TCGA) or International Cancer Genome Consortium.31 These programmes provide description of molecular landscapes and may suggest actionable targets. However, rare cancers are under-represented in these studies: even if such analysis include some rare cancer population, their number are always limited. Describing the molecular landscape of rare cancers is therefore a considerable collective task which is of major importance to fill the knowledge gap and to improve the poor outcome generally associated with rare cancers.
Relevant molecular information was generated during this pilot phase and was associated with the clinical data already collected by the clinical sites. The archival FFPE (Formalin-Fixed Paraffin-Embedded) material was collected between 2001 and 2018. The failure rate of 15% is consistent with other studies and clinical trials, especially for use of retrospective material. This suggests that for some prospective cases, if an FFPE sample is not usable for next-generation sequencing an alternative option, such as liquid biopsy, should be used as a rescue pathway. Another advantage of using liquid biopsy, especially for advanced patients for whom an updated biopsy is not always clinically possible, is to have an up-to-date overview of the cancer.
This pilot study confirmed the feasibility of collecting material from patients with rare cancer from EURACAN sites for molecular analysis in a time frame compatible with recurrences after initial surgery. The prospective phase is currently recruiting patients all over Europe, and the failure rate on patient level, after adding liquid biopsy, dropped to 1.6%. Of the 190 patients analysed, 132 were successful using FFPE material (69.4%), and 55 were rescued with liquid biopsy (29%) (cut-off date 31 July 2020).
Clinical implications of this pilot study
The analysis performed by FMI include possible actionability of clinically relevant alterations. Those alterations can be either directly actionable (approved in Europe for the given molecular alteration) within the disease type, or in a different disease entity. It also includes potential clinical trials recruiting patients with the specific genomic alteration. Within this study, only 4.7% of rare cancers had directly actionable mutations, with already tested targeted therapy available within the same disease entity (3 sarcoma cases out of 41 (7.3%) and 1 rare head and neck case out of 14 (7.1%)): 1 patient had a NTRK fusion, 1 patient had an EGFR exon 20 insertion and 2 patients had FGFR fusion/amplification (figure 4A, green bars).
Figure 4.
(A) Number of samples with actionable mutations and therapy approved in the European Union. (B) Actionable mutations in Arcagen rare cancer cohort and FMI common cancer cohort. (C) Number of samples with actionable mutations per mutation type. FMI, Foundation Medicine Inc; HNSCC, head and neck squamous cell carcinoma; SNV, single-nucleotide variant; TMB, tumour mutational burden.
However, if we look into global actionability (independently of disease type), a European Medicines Agency-approved treatment for that specific molecular alteration could be recommended for almost 44.7% of the patients, including 2 YSTs (22.2%), 18 sarcomas (43.9%), 8 rare head and neck cancers (57.1%) and 7 thymomas (53.8%) (figure 4A, yellow bars). The recommendations encompass several pathways from CDK4/6 inhibitors, RTKI (Receptor Tyrosine Kinase Inhibitor), PARPi (Poly(ADP-ribose) polymerase inhibitor), mammalian target of rapamycin inhibitors or immune checkpoint inhibitors. Finally, clinical trials enrollment for drugs not yet approved for similar molecular alterations could be recommended for 3.5% of the patients, that is, three patients with rare histology of head and neck cancer (figure 4A, orange bars).
A closer look at the type of molecular alterations that were used for treatment recommendation highlighted that for rare cancers, half of the recommendations within the disease type (two out of four cases) are linked to fusions (figure 4B, dark blue bar), one recommendation based on indel and the last one based on amplification. However, recommendations based on other disease types are based on SNVs (17 cases, 45.7%), amplification (12 cases, 34.2%), indels (5 cases, 14.2%), rearrangements (1 case, 2.8%) or high TMB (1 case, 2.8%) (figure 4B).
In order to compare our results with data obtained from more common cancers, we selected 49 406 patients from the FMI database (run with the FoundationOne CDx platform), including: 11 389 breast cancer, 1626 HNSCC, 2838 melanoma, 1210 kidney cancer, 20 442 lung carcinoma, 5653 ovarian carcinoma and 6248 pancreatic carcinoma. In this population, we found that therapy recommendations within the same tumour type was possible for 58% of the patients (figure 4C, green area) and clinical trial enrollment was suggested for 18% of the patients (figure 4C, orange area). Recommendations for therapy available within a different tumour entity was possible for 15% of the patients (figure 4C, yellow area), leaving only 9% of the patients for which no therapy or clinical trial was recommended, versus almost 52% in the rare cancer population (figure 4C, grey area).
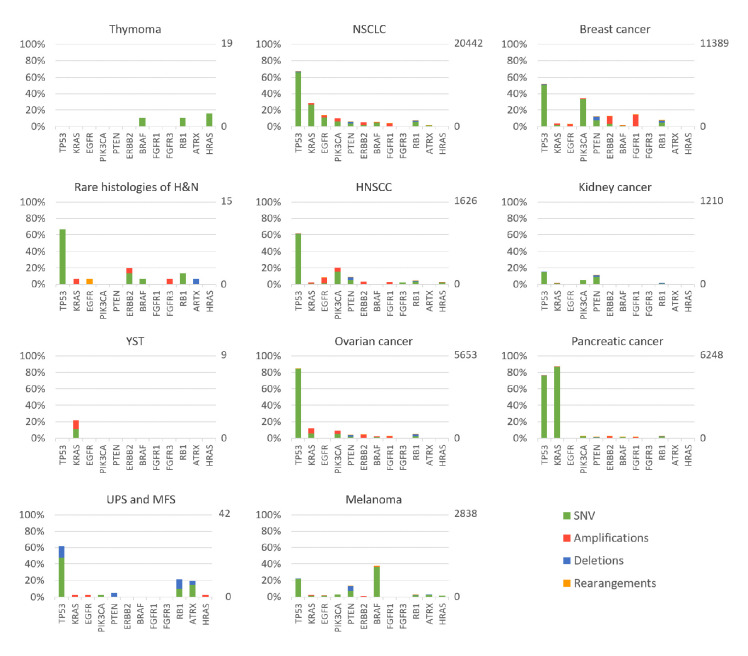
This raises the question of the relevance of using a panel defined for common solid tumours to determine the molecular landscape of rare cancers. In this project, two different panels were used; as the F1H is for sarcomas and does more specifically focus on fusions found in sarcomas. As described in figure 5, the landscape and types of molecular alterations varies tremendously between cancer types (figure 5, rare cancers in the first column, common cancers in columns 2 and 3). In common cancer, common driver alterations have been heavily studied and clinically targeted. These include multiple SNVs (eg, EGFR or KRAS G12C mutation in non-small-cell lung carcinoma), as well as amplification (ERBB2 in breast cancer).
Figure 5.
Summary of gene alteration frequencies for rare tumours from Arcagen cohort (first column) and common tumours provided by FMI (second and third column). Percentages and number of samples for each tumour type is presented on y-axis, left and right from the graph, respectively. MFS, myxofibrosarcoma; NSCLC, non-small-cell lung carcinoma; SNV, single-nucleotide variants; UPS, undifferentiated pleomorphic sarcoma; YST, yolk sac tumor.
Given the presence of mutations overlapping in haematological and solid tumours, and across solid tumour types, more broad panels encompassing all types of alterations could be proposed for all cancer types.
Sarcoma
Historically, MFS was considered a subset of UPS, however they have been recently separated based on their different clinical and pathological characteristics.32–35 A recent paper from TCGA36 revisited this separation based on molecular profile. Indeed, after analysis of 44 UPS and 17 MFS, they concluded that those are not 2 distinct populations but a spectrum of tumour entity, with different amounts of myxoid stroma. From our smaller dataset, we can conclude that both MFS and UPS have a strong dependency on cell cycle dysregulation. However, alterations in the RAS/RAF-PI3K pathways were mostly limited to the MFS population. This potentially points to different genetic alterations in the two subgroups, which will be further explored in the prospective phase.
Yolk sac tumours
YSTs account for 20% of ovarian germ cell tumours, which represent around 3% of all ovarian cancer. They usually affect adolescent and young women, which is consistent with the young median age of the pilot population.
In YST, we confirmed the low mutational rate and the lack of mutations in the HRD pathways, in contrast to 48% of the high-grade serous carcinoma.37 All samples were MSS (microsatellite stable) with no alterations in POLE (DNA Polymerase Epsilon) and their TMB scores were low. The low mutational rate (possibly linked to reduced tumour heterogeneity) is consistent with the encouraging response to chemotherapy, especially platinum-based chemotherapy, seen in this patient population, even in relapse cases.38
In contrast to what was already published, we did not identify any significant amplification in CDK nor PI3K/AKT in our limited cohort. We did, however, confirm in one out of nine samples (11%) a mutation in KIT (previously described as the most significantly mutated gene in dysgerminomas), and in one out of nine samples (11%), an amplification of genes located in chromosome 12p.17
Chromosome 12p gain is the most frequent copy-number variant in ovarian-germ cell tumours17 and amplification in chromosome 12p has been described in testicular germ cell tumours, where gain of the short arm of chromosome 12 is crucial for invasive growth and renders the tumour independent from the supportive Sertoli cells.39 This amplification in 12p was detected only in one YST sample, therefore increasing the sample size may be needed to potentially detect more cases.
Finally, we identified alterations in ARID1A in two samples: one rearrangement within exon 19 and one mutation Q538* (on exon 3). The SWI/SNF (SWItch/Sucrose Non-Fermentable) complexes are frequently mutated across histological subtypes of cancer (up to 20%), with ARID1A the most frequently mutated gene within this family. to date, no direct treatment is associated with alterations in ARID1A, but ARID1A mutations have been shown to be more frequent in clear cell carcinoma.40 This epigenetic vulnerability could potentially be targeted with HDAC or EZH2 inhibitors.41 Similarly, ARID1A mutations have been shown to sensitise ovarian clear cell carcinomas cell lines to BET inhibitors, in vitro and in vivo.42
Thymomas
Mutations in the Ras/Raf pathways were identified in 4 out of 13 patients (30%), identifying a potential therapeutic target, especially in the light of available RAF and more recently KRAS inhibitors. We did not find any TP53 mutations, and only one RB1 deletion, contrary to already published data.43 Enrollement of patient with thymoma in the prospective cohort will help generating more robust data.
Rare head and neck tumours
The most frequently altered genes are participating in cell cycle regulation (TP53, RB1, CDKN2A/B), which is consistent with previous publications.16 From our small dataset, we conclude that the most frequent alteration in rare head and neck tumours overlap with the alterations found in the common HNSCC.14 This could allow access of rare cancer entities to clinical trials testing molecular driven treatments in HNSCC.
Conclusions
The pilot study shows that comprehensive molecular characterisation of rare tumours is important, since the molecular landscape of these tumours is much less known than more common cancers. Fewer actionable targets and therapeutic options are available to patients with rare adult cancers versus more common cancers (60% vs 25%). The multiplicity of these cancers requires an international network for completing the description of somatic alterations in these cancers. Most available screening panels, developed for common solid cancers, often with limited genes interrogated, would most likely need to be adapted to include rare cancer specifics, leading to ‘customise’ screening strategy depending on the tumour type. Additionally, more clinical trials dedicated to rare cancers are needed.
The Arcagen project is aiming to bridge the gap and improve the knowledge of the molecular landscape of rare cancers by recruiting prospectively up to 1000 patients, using the EORTC SPECTA translational research platform. Going beyond the ‘molecular landscape’, it is also assessing actionability and matching treatment opportunities, in the light of the rapid evolution of the targeted treatment landscape and the need for infrastructures in healthcare systems to cope with the testing and treatment paradigm we are facing.
Acknowledgments
The authors would like to thank Lara Chalabreysse and Engjellush Shumeri, Hospices Civils de Lyon, CARDIOBIOTEC, France; Marie Karanian-Philippe, Isabelle Treilleux, Catherine Chassagne-Clement and Alexandra Meurgey, Centre Leon Berard, Lyon, France; Lieve Dirix, Arnaud Poncin, Marie-Sophie Robert and Stephane Lejeune, European Organization for Research and Treatment of Cancer, Brussels, Belgium.
Footnotes
Contributors: MM and JY-B set up the study, performed the analysis and wrote the manuscript. AS, MV, DJ, OH, RE, IR-C and NG performed the analysis and wrote the manuscript. VG, JF and SC wrote the manuscript.
Funding: This project is fully supported by an academic grant from Hoffmann-La Roche. This project was made possible thanks to the SPECTA platform supported by WBA and the EORTC, EURACAN (EC 739521) and LYRICAN networks (INCA-DGOS-INSERM 12563).
Competing interests: IR-C has Honoraria (self) from AbbVie, Agenus, Advaxis, BMS, PharmaMar, Genmab, Pfizer, AstraZeneca, Roche, GSK, MSD, Deciphera, Mersena, Merck Sereno, Novartis, Amgen, Tesaro and Clovis; honoraria (institution) from GSK, MSD, Roche and BMS; advisory/consulting fees from AbbVie, Agenus, Advaxis, BMS, PharmaMar, Genmab, Pfizer, AstraZeneca, Roche/Genentech, GSK, MSD, Deciphera, Mersena, Merck Sereno, Novartis, Amgen, Tesaro and Clovis; research grant/funding (self) from MSD, Roche and BMS; research grant/funding (institution) from MSD, Roche, BMS, Novartis, AstraZeneca and Merck Sereno and travel support from Roche, AstraZeneca and GSK. JF has Honoraria (self) from BMS, AstraZeneca, Roche, MSD, Merck Sereno; advisory/consulting fees from Servier, BMS, AstraZeneca, Innate Pharma, Roche/Genentech; research grant/funding (self) from AstraZeneca; research grant/funding (institution) from MSD, Roche, BMS, Novartis, AstraZeneca and Merck Sereno and travel support from BMS, MSD, AstraZeneca. J-YB has research support and honoraria from Roche Genentech. DJ, OH and RE are employees of FMI/Roche and Roche shareholders.
Patient consent for publication: Not required.
Ethics approval: The project was approved by the French Ethical Committee (CPP), under the number 2019-074B. All patients had provided at the time of sample collection written informed consent for future research and genomic analysis.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available on reasonable request. Data sharing follows the EORTC data sharing policy POL008.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Rare cancers Europe. Available: https://www.rarecancerseurope.org/
- 2.Gatta G, Ciccolallo L, Kunkler I, et al. Survival from rare cancer in adults: a population-based study. Lancet Oncol 2006;7:132–40. 10.1016/S1470-2045(05)70471-X [DOI] [PubMed] [Google Scholar]
- 3.Gatta G, Capocaccia R, Botta L, et al. Burden and centralised treatment in Europe of rare tumours: results of RARECAREnet-a population-based study. Lancet Oncol 2017;18:1022–39. 10.1016/S1470-2045(17)30445-X [DOI] [PubMed] [Google Scholar]
- 4.Gatta G, Capocaccia R, Trama A, et al. The burden of rare cancers in Europe. Adv Exp Med Biol 2010;686:285–303. 10.1007/978-90-481-9485-8_17 [DOI] [PubMed] [Google Scholar]
- 5.Blay J-Y, Coindre J-M, Ducimetière F, et al. The value of research collaborations and consortia in rare cancers. Lancet Oncol 2016;17:e62–9. 10.1016/S1470-2045(15)00388-5 [DOI] [PubMed] [Google Scholar]
- 6.Dufresne A, Brahmi M, Karanian M, et al. Using biology to guide the treatment of sarcomas and aggressive connective-tissue tumours. Nat Rev Clin Oncol 2018;15:443–58. 10.1038/s41571-018-0012-4 [DOI] [PubMed] [Google Scholar]
- 7.Diamandis P, Aldape K. World Health organization 2016 classification of central nervous system tumors. Neurol Clin 2018;36:439–47. 10.1016/j.ncl.2018.04.003 [DOI] [PubMed] [Google Scholar]
- 8.Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet 2018;392:432–46. 10.1016/S0140-6736(18)30990-5 [DOI] [PubMed] [Google Scholar]
- 9.Verweij J, Casali PG, Zalcberg J, et al. Progression-Free survival in gastrointestinal stromal tumours with high-dose imatinib: randomised trial. Lancet 2004;364:1127–34. 10.1016/S0140-6736(04)17098-0 [DOI] [PubMed] [Google Scholar]
- 10.Schlumberger M, Tahara M, Wirth LJ, et al. Lenvatinib versus placebo in radioiodine-refractory thyroid cancer. N Engl J Med 2015;372:621–30. 10.1056/NEJMoa1406470 [DOI] [PubMed] [Google Scholar]
- 11.Pestana RC, Sen S, Hobbs BP, et al. Histology-agnostic drug development - considering issues beyond the tissue. Nat Rev Clin Oncol 2020;17:555–68. 10.1038/s41571-020-0384-0 [DOI] [PubMed] [Google Scholar]
- 12.Covid-19 pandemic Management of patients with cancer : General Information. Available: http://euracan.ern-net.eu/
- 13.Engels EA. Epidemiology of thymoma and associated malignancies. J Thorac Oncol 2010;5:S260–5. 10.1097/JTO.0b013e3181f1f62d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubot C, Bernard V, Sablin MP, et al. Comprehensive genomic profiling of head and neck squamous cell carcinoma reveals FGFR1 amplifications and tumour genomic alterations burden as prognostic biomarkers of survival. Eur J Cancer 2018;91:47–55. 10.1016/j.ejca.2017.12.016 [DOI] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–82. 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer 2018;18:269–82. 10.1038/nrc.2018.11 [DOI] [PubMed] [Google Scholar]
- 17.Van Nieuwenhuysen E, Busschaert P, Neven P, et al. The genetic landscape of 87 ovarian germ cell tumors. Gynecol Oncol 2018;151:61–8. 10.1016/j.ygyno.2018.08.013 [DOI] [PubMed] [Google Scholar]
- 18.Winger BA, Cortopassi WA, Ruiz DG, et al. Atp-Competitive inhibitors midostaurin and avapritinib have distinct resistance profiles in exon 17–mutant kit. Cancer Res 2019. 10.1158/0008-5472.CAN-18-3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radovich M, Pickering CR, Felau I, et al. The integrated genomic landscape of thymic epithelial tumors. Cancer Cell 2018;33:244–58. 10.1016/j.ccell.2018.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girard N. Thymic epithelial tumours: from basic principles to individualised treatment strategies. Eur Respir Rev 2013;22:75–87. 10.1183/09059180.00007312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girard N. Targeted therapies for thymic malignancies. Thorac Surg Clin 2011. [DOI] [PubMed] [Google Scholar]
- 22.Girard N. Thymic tumors: relevant molecular data in the clinic. J Thorac Oncol 2010;5:S291–5. 10.1097/JTO.0b013e3181f209b9 [DOI] [PubMed] [Google Scholar]
- 23.Noeparast A, Teugels E, Giron P, et al. Non-V600 BRAF mutations recurrently found in lung cancer predict sensitivity to the combination of trametinib and dabrafenib. Oncotarget 2017;8:60094–108. 10.18632/oncotarget.11635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Our Proven Portfolio. That’s Our Foundation. Available: https://www.foundationmedicine.com/genomic-testing
- 25.Picard. Available: http://broadinstitute.github.io/picard/
- 26.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakken S, Fournous G, Vodák D, et al. Personal cancer genome reporter: variant interpretation report for precision oncology. Bioinformatics 2018;34:1778–80. 10.1093/bioinformatics/btx817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas B, Dobin A, Stransky N, et al. STAR-Fusion: fast and accurate fusion transcript detection from RNA-seq. bioRxiv 2017. 10.1101/120295 [DOI] [Google Scholar]
- 30.Nicorici D, Satalan M, Edgren H, et al. FusionCatcher - a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv 2014. 10.1101/011650 [DOI] [Google Scholar]
- 31.ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium Pan-Cancer analysis of whole genomes. Nature 2020;578 10.1038/s41586-020-1969-6 [DOI] [Google Scholar]
- 32.Kim J, Kim JH, Kang HG, et al. Integrated molecular characterization of adult soft tissue sarcoma for therapeutic targets. BMC Med Genet 2018;19:216. 10.1186/s12881-018-0722-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucchesi C, Khalifa E, Laizet Yec'han, et al. Targetable alterations in adult patients with soft-tissue sarcomas: insights for personalized therapy. JAMA Oncol 2018;4:1398–404. 10.1001/jamaoncol.2018.0723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coindre J-M. [New WHO classification of tumours of soft tissue and bone]. Ann Pathol 2012;32:S115–6. 10.1016/j.annpat.2012.07.006 [DOI] [PubMed] [Google Scholar]
- 35.Vilanova JC. Who classification of soft tissue tumors. Imaging Soft Tissue Tumors, 2017. [Google Scholar]
- 36.Cancer Genome Atlas Research Network. Electronic address: elizabeth.demicco@sinaihealthsystem.ca, Cancer Genome Atlas Research Network . Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell 2017;171:950–65. 10.1016/j.cell.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ray-Coquard I, Pautier P, Pignata S, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med 2019;381:2416–28. 10.1056/NEJMoa1911361 [DOI] [PubMed] [Google Scholar]
- 38.O'Connor JPB, Rose CJ, Waterton JC, et al. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res 2015;21:249–57. 10.1158/1078-0432.CCR-14-0990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Looijenga LHJ, Zafarana G, Grygalewicz B, et al. Role of gain of 12p in germ cell tumour development. APMIS 2003;111:161–73. 10.1034/j.1600-0463.2003.11101201.x [DOI] [PubMed] [Google Scholar]
- 40.Gadducci A, Guerrieri ME, Genazzani AR. New insights on the pathogenesis of ovarian carcinoma: molecular basis and clinical implications. Gynecol Endocrinol 2012;28:582–6. 10.3109/09513590.2011.649595 [DOI] [PubMed] [Google Scholar]
- 41.Fukumoto T, Zhang R, Bitler BG. Epigenetic inhibitors for the precision treatment of ARID1A-mutant ovarian cancers: what are the next steps? Expert Rev Precis Med Drug Dev 2018;3:233–6. 10.1080/23808993.2018.1503050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berns K, Caumanns JJ, Hijmans EM, et al. ARID1A mutation sensitizes most ovarian clear cell carcinomas to Bet inhibitors. Oncogene 2018;37:4611–25. 10.1038/s41388-018-0300-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito M, Fujiwara Y, Asao T, et al. The genomic and epigenomic landscape in thymic carcinoma. Carcinogenesis 2017;38:1084–91. 10.1093/carcin/bgx094 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
esmoopen-2020-001075supp001.pdf (43.9KB, pdf)
esmoopen-2020-001075supp002.pdf (289.4KB, pdf)
esmoopen-2020-001075supp003.pdf (243.9KB, pdf)