Vaccines work primarily by eliciting antibodies, even when recovery from natural infection depends on cellular immunity. Large efforts have therefore been made to identify microbial antigens that elicit protective antibodies, but these endeavors have encountered major difficulties, as witnessed by the lack of vaccines against many pathogens. This review summarizes accumulating evidence that subdominant protein regions, i.e., surface-exposed regions that elicit relatively weak antibody responses, are of particular interest for vaccine development.
KEYWORDS: immune escape, Plasmodium falciparum, Streptococcus agalactiae, Streptococcus pyogenes, antibodies, dengue virus, immunodominance, influenza virus, subdominance, vaccine
SUMMARY
Vaccines work primarily by eliciting antibodies, even when recovery from natural infection depends on cellular immunity. Large efforts have therefore been made to identify microbial antigens that elicit protective antibodies, but these endeavors have encountered major difficulties, as witnessed by the lack of vaccines against many pathogens. This review summarizes accumulating evidence that subdominant protein regions, i.e., surface-exposed regions that elicit relatively weak antibody responses, are of particular interest for vaccine development. This concept may seem counterintuitive, but subdominance may represent an immune evasion mechanism, implying that the corresponding region potentially is a key target for protective immunity. Following a presentation of the concepts of immunodominance and subdominance, the review will present work on subdominant regions in several major human pathogens: the protozoan Plasmodium falciparum, two species of pathogenic streptococci, and the dengue and influenza viruses. Later sections are devoted to the molecular basis of subdominance, its potential role in immune evasion, and general implications for vaccine development. Special emphasis will be placed on the fact that a whole surface-exposed protein domain can be subdominant, as demonstrated for all of the pathogens described here. Overall, the available data indicate that subdominant protein regions are of much interest for vaccine development, not least in bacterial and protozoal systems, for which antibody subdominance remains largely unexplored.
INTRODUCTION
Vaccination has been described as the most effective medical intervention ever introduced (1, 2). While the eradication of smallpox may represent the greatest achievement of vaccinology so far, approximately 2.5 million deaths per year are currently prevented by vaccination against measles, polio, and other infectious diseases. Moreover, vaccination has allowed the eradication of rinderpest, a major veterinary disease affecting cattle and other animals (3). The importance of vaccination for human and animal health can therefore hardly be overestimated. On the other hand, efficacious vaccines are not available for several of the most important infectious agents, including emerging pathogens, emphasizing that novel approaches are needed for the identification of vaccine components (4).
Most vaccines act by eliciting protective antibodies, even when immunity from natural infection is mainly cell mediated (5–7). Accordingly, work in the field is focused on the identification of pathogen components, typically, surface proteins, that elicit protective antibodies. A number of different methods are currently employed for this purpose, and it is relevant to separately consider bacteria and protozoa on the one hand and viruses on the other.
For bacteria and protozoa, which have many surface proteins, two methods have attracted particular attention. In one method, genomic information is used to identify conserved surface proteins, which are screened for the ability to elicit protective antibodies, a procedure known as reverse vaccinology (8). In another method, antisera from infected patients are used to screen an array of proteins harbored by the pathogen, a procedure based on the assumption that a protein which elicits a good antibody response during natural infection is of interest for vaccine development (9–11). Both of these methods are aimed at the identification of surface proteins that elicit broadly protective antibody responses.
For viruses, the identification of a relevant protein is rarely a problem, since most pathogenic viruses have only one or a few surface proteins, but sequence variability and/or structural instability cause major difficulties. However, work in the virus field has been transformed by procedures allowing the screening of large collections of monoclonal antibodies (MAbs), which are studied for the presence of antibodies that bind to conserved epitopes and confer protective immunity, with the long-term aim to develop epitope-based vaccines (12–15).
Because the identification of vaccine components remains challenging, it is essential to evaluate novel methods. This review will summarize evidence that subdominant protein regions, i.e., regions that elicit a relatively weak antibody response during natural infection or after vaccination, may be of particular interest for vaccine development. It may seem counterintuitive that a region which elicits a weak antibody response would be attractive for vaccine development, but a weak response may represent an immune evasion mechanism (16–19), implying that the corresponding region is a key target for protective immunity. Thus, subdominant protein regions are potentially of much interest for vaccine development (20, 21).
The review will first consider the concepts of immunodominance and subdominance in antibody responses. The following sections will describe examples of subdominance in several major human pathogens, viz., the protozoan Plasmodium falciparum that causes malaria, two species of pathogenic streptococci, and the dengue and influenza viruses. These systems are presented because they provide clear examples of subdominance and together demonstrate the general interest of the subject. Influenza virus will be described in particular detail, because the detailed information available about that system offers unique insights into subdominance and its relevance for vaccine development. Later sections are devoted to the molecular basis of subdominance, its potential role in immune evasion, and implications for the development of novel vaccines.
IMMUNODOMINANCE AND SUBDOMINANCE IN ANTIBODY RESPONSES
More than 50 years ago, Michael Heidelberger proposed the term immunodominant to describe findings on antibody responses to bacterial polysaccharides (22). Heidelberger did not himself publish work on the subject, but his insights are acknowledged in the initial study (22) and mentioned in an obituary (23). As used in the early work, the term signifies that one part of an antigen elicits a stronger antibody response than another comparable part of the antigen (Fig. 1). Thus, the term implies that a purely quantitative comparison is made between the antibody responses to two surface-exposed parts of an antigen, without any inferences concerning the function of the antibodies or their targets. The comparison usually involves the use of standard laboratory techniques to determine the titer of antibodies to the two parts of the antigen. The concept of immunodominance may also be used to compare the response to two or more different molecules on a microbe (21), but such comparisons must be made with caution, since antigens may be expressed at different levels and at different times during an infection.
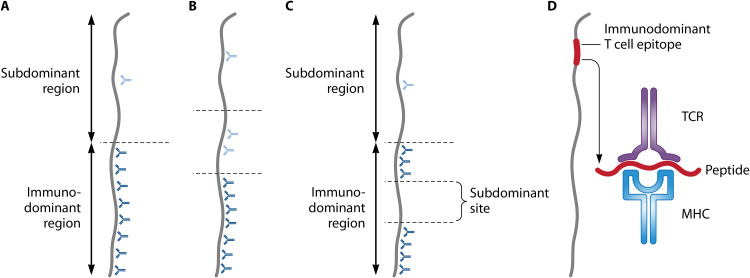
FIG 1.
Fundamentals of immunodominance and subdominance in antibody responses. Panels A to C refer to antibody responses, and for comparison, panel D shows immunodominance in T cell responses. The antigen shown here is a protein, and for simplicity, it is represented schematically as a rod. However, it should be noted that most epitopes recognized by an antibody are conformational. Note that antibody immunodominance is a quantitative concept, not a qualitative one. (A) In the simplest case, one region of a protein is immunodominant, implying that it elicits the quantitatively dominating antibody response, while another region is subdominant. Antibodies are represented by Y-forms, with those directed against subdominant and immunodominant regions shown in light blue and dark blue, respectively. (B) In some cases, a hierarchy of immunodominance may be discerned among different parts of a protein. (C) An immunodominant region may include a subdominant site, a situation that has attracted particular interest in viruses. (D) Immunodominance in T cell responses. In the adaptive immune response to a protein, major histocompatibility complex (MHC) molecules form complexes with peptides derived from the protein, and these complexes are presented for recognition by T cell receptors (TCRs). However, peptides that are efficiently presented for recognition represent only a small minority of all peptides that potentially can be derived from the protein. These rare peptides correspond to T cell epitopes that are immunodominant in the responding individual (28, 234). In the figure, the peptide is enlarged compared to the epitope.
Following the early observations, immunodominance in antibody responses attracted limited attention, although some studies of synthetic polypeptides and proteins reported interesting data (24, 25). A possible explanation for this development could be that studies of model proteins had resulted in a consensus that antibodies elicited by a native protein typically recognize antigenic sites all over the exposed surface (26). In contrast, studies of T cell responses showed that only certain sites in a protein are efficiently recognized, a phenomenon that similarly was referred to as immunodominance (26, 27). The intense interest in T cell responses (28, 29) has had the result that the term immunodominance is often used as a synonym for T cell immunodominance, but it should be noted that the molecular basis is very different in the two cases (Fig. 1). However, immunodominance implies that an immune response is limited to some determinants on an antigen, whether analyzed at the B or T cell level.
Immunodominance in antibody responses has recently attracted renewed attention because striking examples have been disclosed in microbial surface proteins, making the subject of interest for vaccine development (21). Given this development, it is relevant to reiterate that antibody immunodominance refers to a quantitative difference, not a qualitative one. Thus, the demonstration that one region in a microbial protein is immunodominant does not imply that it is a major target for protective antibodies, although that may be the case. Similarly, a key target for protective antibodies need not be immunodominant. There is considerable confusion in the literature, however, since much work is based on the assumption that a structure which elicits a relatively strong antibody response during an infection is of particular interest for vaccine development (9–11). To avoid misunderstandings, the term immunodominant should preferably be employed only in the quantitative sense, without reference to protection.
A protein region that is not immunodominant is now commonly referred to as subdominant, but the terms nonimmunodominant (17, 30–32), immunosubdominant (33), cryptic (34), and immunorecessive (35, 36) have also been used. In the simplest case, a protein can be divided into two regions, one of which is immunodominant and the other subdominant, but the situation can also be more complex, as when an immunodominant region includes a site that is subdominant (Fig. 1).
Before specific examples of subdominance are described, it is pertinent to consider some terms used in the text.
Terms Used To Designate Antibody Targets
Terms used in this review include epitope, site, domain, part, and region. An epitope, also called an antigenic determinant, is the structure recognized by one antibody (37). Epitopes are usually classified as continuous or conformational (discontinuous), describing that the amino acid residues in the epitope are contiguous or not in the peptide chain, but the classification is not unequivocal (37, 38). An antigenic site is defined by a set of antibodies that bind sufficiently close to compete for binding (39), but the term site is also used in a vaguer sense, to designate a limited part of an antigen. A domain is a protein segment that folds independently and typically retains its structure and function when studied in isolated form. The terms part and region are vaguer and often refer to large portions of a protein, e.g., amino- and carboxy-terminal parts. In addition to these terms, which have structural connotations, the description of an epitope may state whether it was identified with a MAb that confers protection against infection. Much work in the field is now based on the assumption that an epitope which is targeted by a protective MAb may be employed to develop an epitope-based vaccine (40).
PLASMODIUM FALCIPARUM: THE CS PROTEIN
Malaria and P. falciparum
With an estimated 228 million cases and 405,000 deaths in 2018, malaria remains a major cause of human disease (41). Most cases are caused by the protozoan P. falciparum, but other Plasmodium species also contribute to the global disease burden. Infection starts with the bite of an infected mosquito and deposition in the skin of approximately 10 to 100 sporozoites, the infective form of the parasite (42) (Fig. 2A). From the skin, the sporozoites slowly trickle into the bloodstream and reach the liver, where they infect hepatocytes (43). The subsequent blood stages, with infection of erythrocytes, cause the clinical signs of malaria. Species of Plasmodium pathogenic for animals pass through similar stages and provide invaluable models for the parasites that cause human infections. The only available malaria vaccine, designated RTS,S, provides partial protection by eliciting antibodies against the major surface protein of the sporozoite, the circumsporozoite protein (CSP) (44, 45). Blocking infection at the sporozoite stage is attractive, because control at this stage prevents clinical disease. Since CSP has distinct subdominant and immunodominant domains, it is of particular relevance for this review.
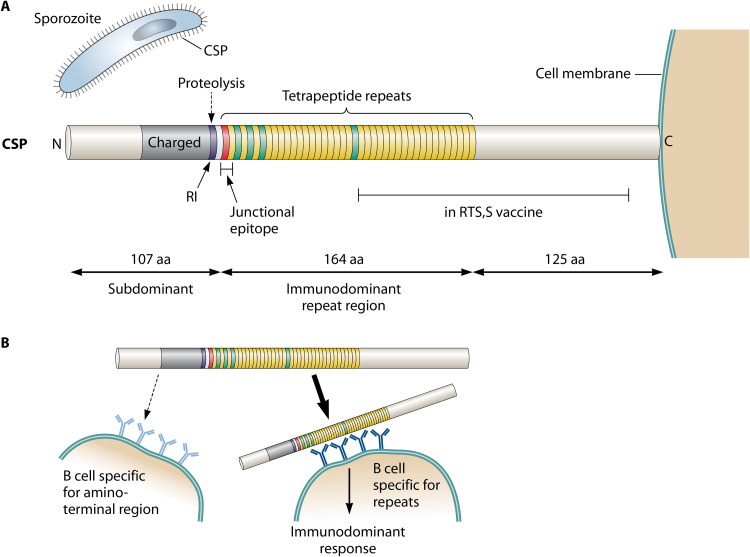
FIG 2.
Circumsporozoite protein (CSP) of P. falciparum and its subdominant amino-terminal domain. (A) The sporozoite of P. falciparum is covered by CSP, in which a central repeat region is surrounded by distinct amino-terminal and carboxy-terminal domains. The repeat region is mainly composed of NANP tetrapeptide sequences (yellow) but starts with a single NPDP sequence (red) and also contains a few NVDP repeats (green) (47). While the repeat region is immunodominant, the amino-terminal region, which also is exposed on the sporozoite surface, is subdominant. In contrast, the carboxy-terminal region may be largely hidden in the intact CSP molecule. The immunodominant repeat region is the key component of the malaria vaccine RTS,S, as indicated. During an infection, CSP is cleaved at the RI site in the amino-terminal domain. Antibodies that bind close to this site (61) or to a junctional epitope located at the beginning of the repeat region (66–68) may protect against infection and may share the ability to block proteolysis at the RI site. Little is known about the structure of the subdominant amino-terminal domain, but it is noteworthy that the part located close to the repeats is charged (47), while the first 50 amino acid (aa) residues include 7 tyrosine residues. (B) Model for immunodominance and subdominance in CSP. The repeats of CSP may promote multipoint and high-avidity binding to cognate B cells, making the repeat region immunodominant by diverting the protein from B cells recognizing the nonrepeated domains, in particular, the surface-exposed amino-terminal domain (56, 57). The resulting subdominance of the amino-terminal domain may favor microbial virulence by allowing the microbe to evade potently protective antibodies directed against that domain. The limited protection conferred by antibodies to the repeats would be the price the microbe pays to achieve this result.
Circumsporozoite Protein
CSP is required for the development of sporozoites in both mosquitos and humans (46) and has three distinct domains: an amino-terminal region, a central repeat region, and a carboxy-terminal region (47). The repeat region has attracted most interest, although little is known about its role during pathogenesis, because early work indicated that it can be a target for protective antibodies (48). In P. falciparum, the repeat region is composed of tetrapeptides, which vary between 37 and 44 in number (49). Almost all repeats have the sequence NANP, but a few repeats with related sequences are also present (Fig. 2A).
While the function of the repeat region remains unclear, specific functions have been ascribed to the amino- and carboxy-terminal domains. In particular, the available data support a model in which the carboxy-terminal domain has cell-binding properties but is masked by the amino-terminal domain, allowing the sporozoite to move through the skin (46). When the sporozoite reaches the liver, it binds to hepatocytes (46) and a sporozoite protease is activated (50), causing cleavage in the amino-terminal domain at (or close to) a site designated RI. The cleavage apparently causes exposure of the carboxy-terminal domain, which binds to a hepatocyte receptor (51) and thereby promotes cellular invasion (46, 50).
The central repeat region of CSP is immunodominant.
Classical work showed that immunization with irradiated sporozoites elicits protective immunity, which is mainly directed against CSP (48). Both antibodies and T cells contribute to the protection (52, 53), but the antibody response has attracted particular interest (48, 54, 55). Importantly, almost all antibodies elicited by CSP are directed against the repeat region, which therefore is immunodominant (48). The reason for the immunodominance of this region is not known, but the repeats may promote multipoint and high-avidity binding to cognate B cells, resulting in a strong response, while the responses to other parts of the protein become strongly limited (56, 57) (Fig. 2B). The very size of the repeat region and preferential exposure of this region on the sporozoite surface (58) may also contribute to its immunodominance. Because the repeat region of CSP is strikingly immunodominant (59, 60), it follows that the amino- and carboxy-terminal domains are subdominant. It is particularly noteworthy that the entire amino-terminal domain is subdominant, although it is exposed on the sporozoite surface (46, 50, 61). In contrast, the carboxy-terminal domain may be largely hidden (46, 62).
Attempts to develop a human malaria vaccine were focused on the immunodominant repeat region of CSP, and promising results were eventually obtained with the RTS,S vaccine, in which the potent adjuvant AS01 is combined with virus-like particles (formed by hepatitis B virus surface antigen) containing one part of CSP (44). The part of CSP included in RTS,S comprises 19 NANP repeat motifs and most of the carboxy-terminal region, which was added to provide T cell epitopes, but the amino-terminal domain was not included (Fig. 2A). Importantly, the development of the RTS,S vaccine provided evidence that a vaccine can protect against malaria and demonstrated that infections may be blocked at the sporozoite stage, but in a phase III trial, the protection was modest and short-lived (63), implying that an efficient malaria vaccine is not yet available.
Subdominant amino-terminal domain in CSP.
Early studies of the amino- and carboxy-terminal domains of CSP suggested that they are not suitable for vaccine development (64, 65), and vaccine work was focused on the immunodominant repeat region. While recent studies with MAbs support the notion that the carboxy-terminal domain is not a target for protective antibodies (62), interesting data have been reported for the amino-terminal domain. In particular, a mouse MAb that confers protection against experimental infection binds close to the RI site and prevents proteolytic cleavage of CSP (61), suggesting that proteolysis at this site plays a key role in pathogenesis. Moreover, some human MAbs that protect against infection bind to a “junctional epitope,” located at the beginning of the repeat region (45, 54, 66–68) (Fig. 2A). At least one of these human MAbs prevents proteolytic cleavage of CSP, possibly through a steric effect on the adjacent RI site (67), again focusing interest on the RI site as a target for protective antibodies. Similarly, simultaneous binding of multiple antirepeat antibodies to CSP results in the formation of a complex which may sterically hinder access of the protease to CSP (69). Thus, evidence is accumulating that the subdominant amino-terminal domain may be of interest for vaccine development. However, the properties of the mouse MAb described above are controversial (70), emphasizing that other MAbs to the amino-terminal domain should be characterized and studied with regard to protective ability. It would also be of much interest to know the three-dimensional (3D) structure of the amino-terminal domain and to know whether immunization with this domain in its native form elicits protective immunity. In this context, it is noteworthy that the repeat region of CSP was not effective as a vaccine component until it was presented on virus-like particles and was combined with a potent adjuvant (44), suggesting that it may be of value to employ a similar methodology in studies of the amino-terminal domain. Of note, the subdominance of the amino-terminal domain, when present in CSP, should not represent a problem for vaccine work, since this domain elicits a good antibody response when used in an isolated form (50, 71).
Vaccine development.
Because the protection conferred by RTS,S is modest, studies are in progress to modify this CSP-derived vaccine to make it more immunogenic (72). Given the data available for the amino-terminal domain of CSP, it will also be of considerable interest to evaluate whether this subdominant domain is suitable for vaccine development when used in an isolated form, i.e., in the absence of the immunodominant repeat region (61). Moreover, many P. falciparum proteins other than CSP are being evaluated as vaccine components (45, 73). Interestingly, studies of one polymorphic protein, the blood-stage antigen MSP2, suggest that a vaccine containing two variants of this protein may elicit functionally active antibodies directed against a conserved region which is subdominant in natural infections (74). Thus, subdominant protein regions could be of broad interest for the development of a malaria vaccine.
STREPTOCOCCAL SURFACE PROTEINS
Surface proteins of pathogenic streptococci provide clear examples of bacterial proteins in which a subdominant region is a target for protective antibodies. Like many surface proteins of Gram-positive bacteria (75–77), these proteins have elongated structure and repeat regions, providing a parallel to the CSP of P. falciparum.
Streptococcus agalactiae: the Rib and α Proteins
S. agalactiae, also known as group B streptococcus (GBS), is the major cause of life-threatening bacterial infections in the neonatal period (78). No vaccine is available. All strains of GBS express a surface protein belonging to the Alp family, in which the members have a unique amino-terminal domain and an extended region with long repeats (79–82). The most common members in this family are the Rib and α proteins (76) (Fig. 3A).
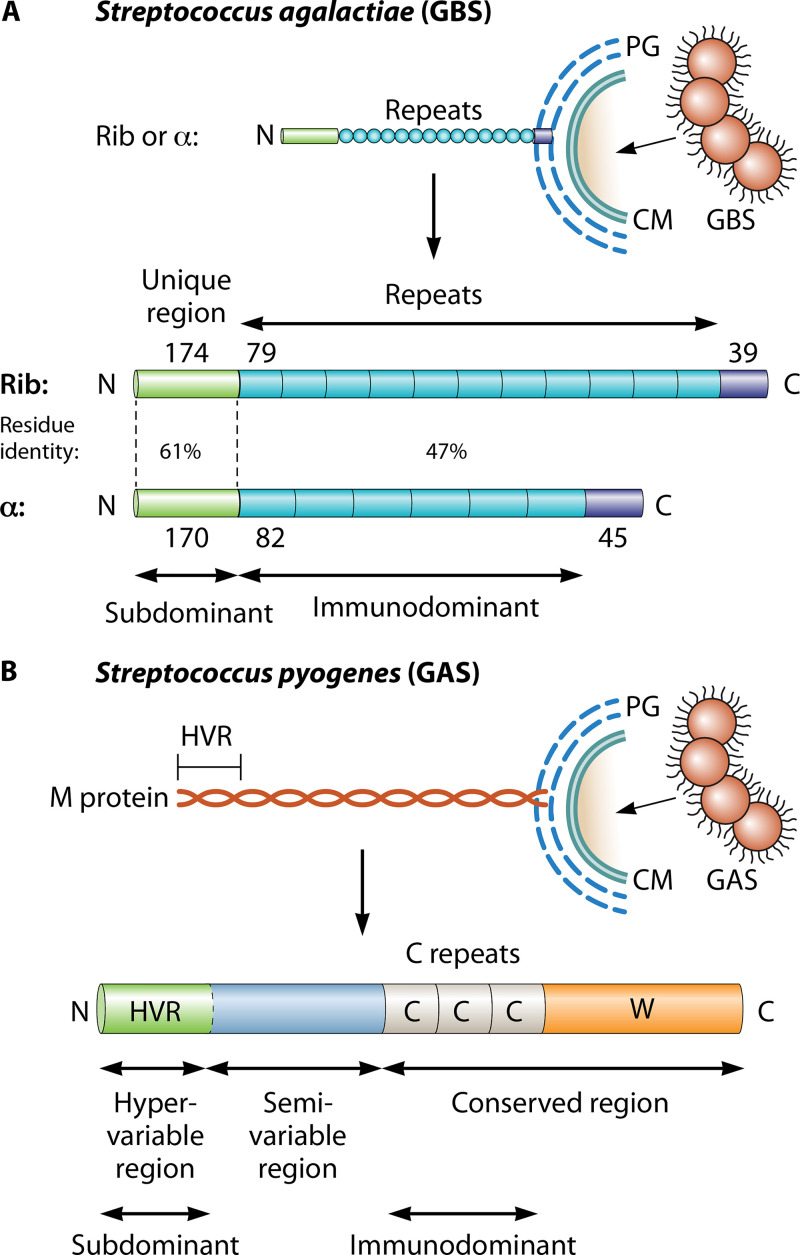
FIG 3.
Subdominant domains in streptococcal surface proteins. (A) Schematic of the surface-exposed forms of the Rib and α proteins of Streptococcus agalactiae (GBS) (79, 80). These two proteins are the most common members of a family of elongated and highly repetitive streptococcal proteins. Each protein has a surface-distal amino-terminal domain, a region with long repeats, and a short carboxy-terminal region that promotes covalent linkage to the bacterial peptidoglycan (PG) layer, located outside the cell membrane (CM). The number of amino acid residues in a region (or repeat) is indicated. While all repeats are completely identical within Rib or α, they vary in number among clinical isolates and are different in Rib and α but show 47% residue identity. The amino-terminal domains are strikingly subdominant but are targets for protective antibodies (17). (B) Schematic of the surface-exposed form of Streptococcus pyogenes M protein. All strains of S. pyogenes express an M protein, encoded by the emm gene (95). This fibrillar coiled-coil protein has an amino-terminal hypervariable region (HVR) of ∼50 to 100 amino acid residues, a conserved carboxy-terminal region that includes C repeats (each with 35 or 42 residues), and a wall-spanning region (W). The HVR exhibits extreme sequence divergence among strains but is stable within a strain and represents a distinct domain that in many (but not all) M proteins specifically binds a human complement regulator. The central part of M protein is also variable among strains and typically includes domains that bind human plasma proteins, e.g., fibrinogen or IgA (95). While the HVR is the major target for protective antibodies, it is strikingly subdominant (18).
The amino-terminal domains of Rib and α are subdominant.
Analysis of the immune response to Rib and α demonstrated that the repeat regions are immunodominant and targeted by >90% of the antibodies, while the entire amino-terminal domains are subdominant (17). It is not known why the repeat regions of the intact proteins are immunodominant, but multipoint binding of the repeats to cognate B cells may contribute, as proposed for the CSP protein of P. falciparum (see above), and the very size of the repeat region may also play a role.
Although the amino-terminal domains of Rib and α were strikingly subdominant when present in the intact proteins, a fusion protein derived from these domains elicited a better antibody response than a control protein derived from the repeats, was immunogenic even without adjuvants, and elicited good protective immunity (17). Thus, the subdominant amino-terminal regions of Rib and α are targeted by protective antibodies and are of interest for vaccine development. While antibodies to the repeats also have some protective ability (17, 83), providing a parallel to CSP, the protection afforded by these antibodies may, in certain situations, be reduced through the appearance of bacterial mutants with a decreased number of repeats (84).
Streptococcus pyogenes: the M Protein
The human pathogen S. pyogenes (group A streptococcus [GAS]) is best known as the cause of superficial infections of the skin and pharynx (“strep throat”) but also causes invasive infections and the autoimmune disease rheumatic fever, which together cause >500,000 deaths per year (85). A vaccine is not available. Among the virulence factors of S. pyogenes, particular interest has been focused on the surface-localized M protein (Fig. 3B), which blocks phagocytosis and was the first bacterial surface protein implicated in virulence (86).
M protein is a fibrillar coiled-coil protein (87, 88) with an amino-terminal hypervariable region (HVR) that varies extensively in sequence among circulating bacterial strains but not within a strain. Clinical isolates of S. pyogenes can therefore be classified into ∼240 M (emm) types, each of which has a unique HVR sequence, as determined at the protein or DNA level (89, 90). In contrast, the carboxy-terminal half of M protein contains relatively conserved C repeats (91) (Fig. 3B). The HVR plays an active role in virulence (92, 93) and is the target for type-specific antibodies that protect against infection (94, 95). In spite of the extreme sequence divergence among different HVRs, most of them bind a human complement regulator, a property retained by HVRs studied in isolated form (96–98). In particular, most HVRs share the ability to bind the plasma protein C4b-binding protein (C4BP) (96, 97), a finding explained by the demonstration that these C4BP-binding HVRs have similar 3D structures (99). Thus, all available evidence indicates that the HVR of M protein is a well-defined ligand-binding domain. Given this situation, it is attractive to hypothesize that the currently known HVRs have been selected during evolution as the most fit ones, a circumstance that would favor sequence conservation and may explain why the number of known types is relatively limited (18).
While type-specific anti-HVR antibodies can be prepared by the immunization of animals, it has remained unclear whether infection in humans results in type-specific immunity (100). This situation, and unexpected laboratory findings, led to the hypothesis that the HVR may be subdominant (18), although highly variable regions in microbial proteins are typically predicted to be immunodominant. While this hypothesis may seem paradoxical, it makes sense from an evolutionary point of view, since it should be advantageous for the bacterium if the HVR elicits a weak antibody response, allowing escape from protective antibodies.
The hypervariable region of M protein is a subdominant domain.
Support for the hypothesis that the HVR is subdominant came from extensive studies of two M proteins (18). As shown in these studies, the HVR is strikingly subdominant compared to other parts of the M protein, not only in immunized animals but also in S. pyogenes-infected humans. Further dissection of the antibody response demonstrated that it is mainly directed against the conserved C repeats (101).
The conclusion that the HVR of an M protein is subdominant is supported by several early reports, which indicated that type-specific antibodies, i.e., antibodies to the HVR, appear only slowly, if at all, in infected patients (102–104). Moreover, treatment of patients with penicillin prevents the appearance of type-specific antibodies, suggesting that an anti-HVR response requires extended antigen exposure (103, 104). In agreement with these early reports, a recent study of infected humans indicated that past infection may not prevent future infection, implying that many infections do not result in a protective anti-HVR response (100). While these different studies did not exclude that the entire M protein is weakly immunogenic in humans, the results can now be explained by the subdominance of the HVR. Further support for the subdominance of the HVR comes from a study of mouse MAbs, which indicated that only one of 19 MAbs elicited by an M protein was directed against the HVR (94). The molecular basis for the subdominance of the HVR is not known, but selective elimination by proteases might contribute (101).
The subdominance of the M protein HVR suggests that this domain is so important for virulence that evolution has favored the appearance of two mechanisms that allow escape from antibody attack on the HVR: sequence divergence and subdominance (18). This conclusion follows from the observation that antibodies to the HVR eventually accumulate during a prolonged infection (103, 104), probably making the host immune to infection with strains of the same M type (i.e., with the same HVR) as the infecting strain but not to strains of other M types. Thus, the sequence divergence among strains implies that S. pyogenes can escape preexisting antibodies elicited by a strain of a different M type (91). In contrast, the antibodies elicited during the course of a single infection cannot be evaded through sequence variation, since the HVR typically is genetically stable. However, the subdominance of the HVR should delay the appearance of protective antibodies, thereby prolonging the infection and favoring transmission to new hosts. An important corollary to these arguments is that sequence divergence and subdominance promote antibody escape at different stages of an infection (18).
While the HVR is subdominant when present in an intact M protein, peptides derived from HVRs elicit good antibody responses (105), a finding that forms the basis for attempts to develop a vaccine based on multivalent fusion proteins derived from HVRs (106). However, even such a vaccine may cover too few of the many circulating M types (107), and it is of some concern that an immune response to HVRs may contribute to the development of the autoimmune disease rheumatic fever (108). Thus, the subdominance of M protein HVRs is conceptually intriguing, but it remains uncertain whether these domains can be employed for vaccine development.
DENGUE VIRUS: THE E PROTEIN
Dengue virus, which is transmitted by mosquitos, causes infections that typically are asymptomatic or mild, but a life-threatening condition with hemorrhagic fever and/or shock develops in some patients (109, 110). The incidence of infections has increased during the last few decades, with worldwide effects on health and economy (109). Major efforts have therefore been made to develop a vaccine, work that has been complicated by the possibility that some antibodies may enhance disease, a phenomenon known as antibody-dependent enhancement (ADE). This situation emphasizes that detailed understanding of the antibody response to dengue virus is essential. Here, the presentation will be focused on the subdominance of an entire domain in the surface E protein.
Dengue Virus and Other Flaviviruses
Dengue virus is a member of the flavivirus family, a group of enveloped RNA viruses that also includes West Nile virus (WNV), yellow fever virus (YFV), tickborne encephalitis virus (TBEV), and Zika virus (111, 112). The surface of the virus is covered by dimers of the E protein, which has three distinct domains (111, 113) (Fig. 4A). Domain III has been implicated in the binding to host cell receptors, domain II promotes dimer formation and includes a fusion loop (FL) that triggers membrane fusion, while the centrally located domain I may have a structural role (111). During the formation of progeny virus, premature membrane fusion is prevented by binding of the FL in domain II to the viral protein prM (Fig. 4B). As described below, this interaction is of relevance for studies of the antibody response. Proteolytic degradation of prM into pr and M results in the release of pr and retention of the small M fragment, while FL becomes largely hidden within the E dimer (113–115). Incomplete processing may result in the formation of immature virions, on which uncleaved prM-FL complexes are exposed (113, 115), but it is unclear to what extent such virions are formed in vivo (116).
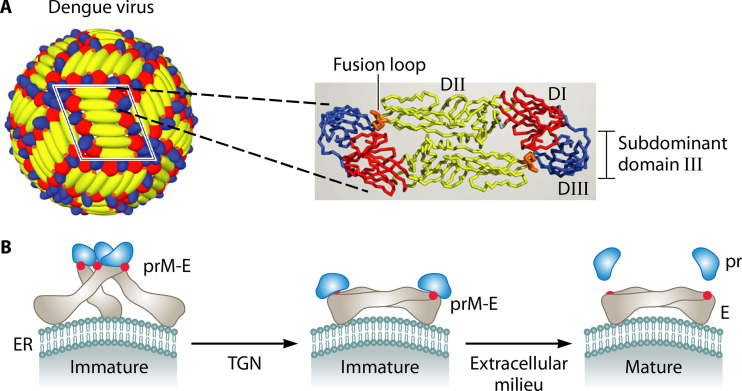
FIG 4.
E protein of dengue virus and its subdominant domain III. (A) Schematic of dengue virus and its surface E protein. The mature dengue virion (left) is covered by 30 rafts, each of which contains three antiparallel E protein dimers. One raft is framed in black, and a single dimer is shown as ribbons (right). The positions of the three domains (DI to DIII) are indicated for one E protein monomer. While domain III is subdominant, the fusion loop in domain II is part of an immunodominant site. Adapted from reference 113. (B) Protein maturation during dengue virus replication (114). In the endoplasmic reticulum (ER), the surface of the immature virus particle is covered by trimeric prM-E spikes, in which prM prevents premature membrane fusion promoted by the fusion loop (red spot) in the E protein. During transport through the acidic trans-Golgi network (TGN), the complex is rearranged and prM is then cleaved into pr and M. The small M fragment is retained in the viral membrane, with negligible surface exposure, while the pr fragment remains bound to the FL. When the virus particle is released from the cell, pr dissociates from the complex and the FL becomes largely hidden within the E dimer (113–115). Adapted from reference 114 with permission of AAAS.
Antibody Response to the E Protein: Immunodominant and Subdominant Sites
Dengue virus occurs in four serotypes, reflecting antigenic differences in the E protein. Infection with virus of one type usually elicits antibodies that confer lifelong and type-specific protective immunity. However, most of the antibodies elicited during an infection are not type-specific but cross-reacting and are poorly neutralizing and directed against the conserved FL in domain II and also against the prM protein (113, 117). Thus, the FL represents an immunodominant site on the E protein while other sites, which are subdominant, elicit protective responses (113, 117). The immunodominant response to FL and prM may be elicited by FL-prM complexes exposed on immature virions and by FL epitopes transiently exposed through “breathing” of E dimers on mature virions (113, 115). The restricted accessibility of the FL on mature virions, and the absence of prM, can explain the limited ability of antibodies to FL and prM to neutralize infectious virions.
While a primary infection mainly elicits poorly neutralizing antibodies directed against immunodominant sites, studies of MAbs have identified several targets for antibodies that have good neutralizing ability. These potent antibodies recognize a variety of subdominant sites on the E protein, all of which are of interest for vaccine development. One group of potent antibodies is those conferring type-specific protective immunity after natural infection. These antibodies are mainly directed against quaternary epitopes, which are formed by E proteins on the virion surface and include amino acid residues located in two or all three of the domains (118–120). Potent antibodies in a second group are broadly neutralizing and directed against highly conserved quaternary epitopes, designated E dimer epitopes (EDEs), which are located at the E dimer interface and span a valley that contains the FL (121–123). While EDE epitopes apparently elicit a limited response during a primary dengue infection, which confers type-specific immunity, they may elicit stronger responses during secondary infections, which result in protective responses with greater breadth (124, 125). Because the EDE epitopes are conserved, they are of considerable interest as targets for vaccine-induced antibodies. Potent antibodies in a third group are directed against sites in domain III, the whole of which is subdominant, as outlined below.
Subdominant domain III: a target for protective antibodies.
Early work in the mouse showed that many strongly neutralizing and type-specific MAbs were directed against domain III of the E protein (126). It therefore seemed possible that the type-specific neutralizing antibodies elicited during human infections are directed against domain III. However, studies of polyclonal human antibodies, elicited during natural infection, indicated that antibodies specific for domain III are not essential for neutralization or protection (127, 128) and constitute only a small fraction of the total response to the E protein (127, 129). Thus, the entire domain III is subdominant and is not important as a target for protective immunity during natural infection. Similarly, domain III is subdominant in the E protein homologs of the flaviviruses WNV, TBEV, and YFV, as demonstrated by studies of polyclonal human antisera (130–132). Of note, some collections of MAbs have compositions that provide little support for the conclusion that domain III is subdominant (133, 134), but the available data are not contradictory, since collections of MAbs may not be representative of the antibodies present in polyclonal antisera (135). The molecular basis for the subdominance of domain III is not known, but the strong responses to FL and prM may contribute by limiting responses to other sites.
Antibody-Dependent Enhancement
It is unclear why only a minority of all infections with dengue virus result in serious disease (110), but a prevailing model posits that preexisting cross-reactive antibodies play a major role by causing ADE. According to this model, certain preexisting antibodies do not neutralize the virus but bind to a virus particle and promote internalization into Fc receptor-bearing cells, in which the virus replicates (113, 136–138). Particular interest has been focused on the strong antibody responses elicited by the FL in the E protein and by the prM protein (113, 117), implying that a vaccine must not elicit such antibodies. While the triggering of ADE may be prevented if a balanced and long-lasting neutralizing response is elicited against all four serotypes, the problems involved in eliciting such a balanced response have been described as formidable (117). Given this situation, it is of interest that ADE may not be triggered by a vaccine based on the subdominant domain III (see below).
Dengue Vaccines
Work on dengue vaccines has mainly been focused on tetravalent formulations of live attenuated viruses, but problems encountered with such vaccines have stimulated interest in subunit vaccines based on the E protein (113, 117). One interesting approach involves the use of modified E protein dimers, which retain ability to display the subdominant and conserved EDE epitopes but have been stabilized to limit breathing, thereby reducing the formation of anti-FL antibodies that may promote ADE (123). Another approach is focused on the subdominant domain III, which does not include the FL and is a target for potent MAbs (31, 126, 133). It has remained unclear whether derivatives of domain III are suitable as vaccine components (139), but encouraging results were recently obtained with a vaccine based on virus-like particles (VLPs) formed by hepatitis B virus (HBV) S protein (140, 141), the classical system also used to develop the RTS,S malaria vaccine. This dengue vaccine contained a fusion protein derived from the HBV S protein and domain III of all four dengue serotypes. The antibodies elicited by this vaccine were broadly neutralizing and protected against infection in a model of passive vaccination. Moreover, studies in an in vivo model suggested that the vaccine may not trigger ADE (140). Thus, it may be possible to develop a safe dengue vaccine based on the subdominant domain III of the E protein.
INFLUENZA VIRUS: HEMAGGLUTININ
Influenza and Influenza Virus
Influenza remains one of the globally most important infectious diseases, with seasonal disease causing ∼500,000 deaths annually and the emergence of a pandemic representing a constant threat (142). This situation has led to intense efforts to develop good vaccines, resulting in extraordinary insights into protein structure, mechanisms of pathogenesis, and immune responses. The holy grail in the field is the development of a universal vaccine that protects against most if not all strains.
Influenza virus is an enveloped RNA virus that comes in four serological types, of which types A and B cause epidemics in humans. The type A virus, IAV, is clinically most important and is the only type causing pandemics. The surface of the virus is covered with spikes of two kinds, representing the glycoproteins hemagglutinin (HA) and neuraminidase (NA). The major surface protein is HA, which has two key roles: binding to sialic acid receptors and triggering of membrane fusion (143–145). An intact HA molecule is a trimer of identical subunits, each of which comprises a globular head, a largely α-helical stem, and a membrane anchor, with the receptor binding site (RBS) located in a shallow hydrophobic pocket at the top of the head (Fig. 5A). While the head varies extensively in sequence among strains, the stem is largely conserved. On the basis of antigenic properties and sequences, HA and NA are classified into subtypes (18 for HA and 11 for NA), with virus strains given designations such as H1N1 and H3N2, the two subtypes that have dominated during the last century. Moreover, HAs can be divided into two distinct groups, with H1 and H3 belonging to groups 1 and 2, respectively. Subtypes are further classified into strains, with new strains appearing every year, requiring the development of seasonal vaccines that often have suboptimal properties. In contrast, a pandemic may be associated with the rare emergence of a new subtype, which has acquired its HA from an animal strain of IAV.
FIG 5.
Hemagglutinin (HA) of influenza virus: subdominant sites as targets for protective antibodies. (A) Models of influenza virus and hemagglutinin (HA). (Left) In a viral particle, the glycoproteins HA and neuraminidase (NA) are anchored in a lipid envelope surrounding the core, which contains the eight RNA segments of the genome. (Right) Top and side views of an HA trimer of subtype H1, showing the immunodominant head and the subdominant stem (149). The location of the five highly variable and immunodominant sites in the head are indicated (Sa, blue; Sb, gold; Ca1, purple; Ca2, orange; Cb, red). Right panel adapted from reference 149 with permission from Springer Nature. (B) Cartoon showing the location of conserved sites (red) that are targets for protective antibodies in HA. These sites are of at least five types and include the RBS, a partially occluded site located at the monomer interface in the head, other conserved sites in the head, and two sites in the stem, one of which is occluded on native HA, i.e., not accessible to antibodies. Except for this occluded site in the stem, which has unique properties, the various sites may be described as subdominant. Of note, the whole head is variable and immunodominant compared to the stem, but it includes subdominant sites that are conserved and protective. In contrast, the entire stem is subdominant but contains protective sites. Panels C to F, and the corresponding legends, describe procedures that may allow an antibody response to be targeted to a subdominant site. (C) A stem-only construct (“mini-HA”), derived from the subdominant stem of H1 HA, elicits broadly protective antibodies (191). Figure based on PDB 5CJQ and produced with VMD (235). Of note, the structure of mini-HA differs slightly from that for the stem of intact HA, as it adopts a more open splayed conformation (191). (D) Use of chimeric HA proteins to focus responses on the subdominant stem (33, 196). (Left) Adults have preexisting antibodies to the head (top) and the stem (bottom) of H1 HA, although the response to the stem is very limited. (Middle) Immunization with a chimeric H8/1 protein may elicit a memory response to the H1 stem but only a primary response to the “exotic” H8 head. (Right) Boosting with an H5/1 chimera may further boost the response to the H1 stem while again eliciting a primary response to the “exotic” head. As in Fig. 1, antibodies directed against subdominant and immunodominant regions are shown in light blue and dark blue, respectively. (E) Schematic representation of “breathing” in an HA molecule, resulting in the exposure of a site in the head that is located at the monomer interface and is a target for protective antibodies (157, 159, 160). Formation of antibodies to this site may be favored by immunization with hyperglycosylated HA (197). (F) Use of mosaic nanoparticles to focus antibody responses on sites in the HA head that are subdominant and conserved (198). In this procedure, nanoparticles were covered with monomeric HA heads. (Left) A nanoparticle covered with heads from a single virus strain will elicit antibodies directed almost exclusively against strain-specific immunodominant sites, which promote very strong binding to cognate B cells, thereby diverting the antigen from B cells recognizing any other site(s). (Right) Use of a mosaic nanoparticle covered with several different heads will reduce the local concentration of strain-specific immunodominant sites, potentially favoring a response to a conserved and shared subdominant site, which may promote binding of relatively high avidity to cognate B cells.
Antibody Response to HA: Immunodominant Head, Subdominant Stem
The resolution of an IAV infection depends on the appearance of protective antibodies, most of which are directed against the HA head and prevent binding of the virus to the sialic acid receptor (144, 146). The immunity is typically strain specific, reflecting the sequence variability in the head. The large majority of all antibodies elicited by HA, whether protective or not, are also directed against the head, which therefore is immunodominant in the classical sense of the term, while the stem is subdominant (147–149). Antibodies to NA also contribute to protection (150) but will not be considered here.
The selective pressure exerted by protective antibodies directed against HA favors antigenic drift and shift, the two classical mechanisms causing sequence variability (142, 151). Over the years, antigenic drift in a subtype results in extensive sequence variability in several regions of the head (152, 153), but the RBS remains highly conserved, reflecting its limited ability to change without losing its ability to bind sialic acid (154). Nevertheless, many of the antibodies elicited during an infection bind to the RBS, prompting the question of why these antibodies do not confer broad protection. The explanation for this apparent paradox is that an antibody to the conserved RBS typically has a footprint that is much larger than the RBS, resulting in binding not only to the RBS but also to variable loops located outside the RBS, making the antibodies specific for one strain (154, 155). Other strain-specific antibodies only bind to variable sites and act by sterically blocking binding to the receptor (154). Moreover, some rare MAbs have a minimal footprint and bind almost exclusively to the RBS, allowing them to neutralize many, if not most, strains of IAV (153).
Here, it is of relevance to consider how antibodies to HA are evaluated for their ability to inhibit infection. The most important method involves the use of an animal model, in which passively administered antibodies are tested for their ability to confer protective immunity. However, two in vitro surrogate methods are in widespread use, neutralization and hemagglutination inhibition (HAI). While neutralization determines the ability of antibodies to block virus infection of cells, HAI measures the ability to interfere with binding to the sialic acid receptor. All three methods are valuable, but it is essential to note that they may give different results (13, 156–161). In particular, certain antibodies require interaction with Fc receptors for the ability to confer in vivo protection (156) and may lack activity in neutralization tests (157–161).
Immunodominant and subdominant sites within the HA head.
Pioneering studies of H1 and H3 strains indicated that five highly variable sites in the HA head are major targets for neutralizing antibodies (162, 163) as shown for H1 HA in Fig. 5A. Similarly, the majority of all antibodies (whether neutralizing or not) elicited during an infection are directed against these five sites, as shown for the H1 protein (148), confirming that these sites indeed are immunodominant in the quantitative sense of the term, compared to other sites in HA. The five sites are largely composed of protruding loops, a characteristic that probably contributes to their immunodominance and may allow extensive sequence variability without effects on the overall structure of the head (144).
While the five immunodominant sites were identified in strains for which glycosylation of the HA head is limited, additional glycan groups are typically present in seasonal strains, potentially modulating the antibody response (164–166). Moreover, differential glycosylation may explain some of the differences between the H1 and H3 systems in the location of the five classical sites (167). However, factors other than glycosylation are probably also important in determining the strength of the antibody response to the five sites. Extensive attempts have recently been made to identify a hierarchy of immunodominance among these sites (21, 149, 168), but the situation is highly complex, and the factors that determine the strength of the response to a given site remain unclear.
In contrast to the strain-specific immunity elicited by the variable and immunodominant sites, evidence has accumulated that certain conserved sites in the head can elicit broadly protective MAbs (Fig. 5B). Because immunity typically is strain specific, these conserved sites apparently elicit antibodies only rarely during an infection, implying that they may be described as subdominant. These conserved sites in the head include broadly conserved sites that are partially occluded and located at the interface between two HA monomers (157, 159, 160, 169) (see below). Other protective sites are exposed on the HA head and are conserved within one serotype (170–174). Moreover, certain rare antibodies with a small footprint specifically bind to the conserved RBS, as noted above (153).
The HA stem: a subdominant domain targeted by protective antibodies.
Work on influenza immunity was for many years focused on the HA head, reflecting the importance of the immunodominant sites as targets for strain-specific protective antibodies. Although an early study of a mouse MAb suggested that the stem may also be a target for protective antibodies (175), this part of HA attracted only limited attention until human protective MAbs were described 15 years later (176–180). However, intense efforts are now made to develop vaccines based on the stem (see below).
An occluded site in the stem.
Further evidence that the stem is of interest as a target for protective immunity was reported in a recent study describing a novel class of protective MAbs that show broad reactivity and bind to a site in the stem (161). While previously identified anti-stem MAbs bind to epitopes exposed on intact HA (178, 181–183), MAbs in the new class apparently do not bind to native HA, implying that the binding site is occluded on free virus particles (Fig. 5B). Although the properties of this occluded site do not conform to the classical definition of a subdominant site, it is of relevance to consider it here, since it elicits a poor antibody response after influenza vaccination (184) but nevertheless is a target for protective antibodies. This relatively conserved site is probably exposed at some stage of the infectious process, allowing access of antibodies that protect against infection (161). These antibodies do not neutralize the virus in vitro but confer protection by interacting with Fc receptors on effector cells. Interestingly, the occluded epitope may be exposed on the postfusion form of HA that appears when the virus is exposed to low pH during cellular invasion, suggesting that this form of HA may elicit broadly protective antibodies (161, 185). Since the interface site in the head is also partially occluded (see above), it now seems possible that occluded sites in HA are of general interest as targets for antibodies that protect against influenza (185). Of note, studies of the E protein in flaviviruses have similarly indicated that sites, which are at least partially occluded, may be targets for protective antibodies (115, 186–188).
Vaccine Development: Focus on Subdominant Targets in HA
Conserved sites in the stem or head of HA are currently attracting much interest as potential targets for broadly protective antibodies induced by a vaccine (146, 185, 189, 190) (Fig. 5B). Except for the occluded site in the stem, all of these sites may be described as subdominant, although the term should be used with caution, because different sites may vary with regard to size and surface exposure. Particular interest has so far been focused on the possibility that the stem may be employed to develop a universal vaccine.
For the stem, protein engineering has allowed the expression in mammalian cells of vaccine candidates with promising properties (191, 192). One of these antigens, designated “mini-HA” (191, 193), can be viewed as a soluble form of the trimeric stem (Fig. 5C), while another antigen was prepared by immobilizing stems on ferritin nanoparticles (192, 194). Both of these antigens elicited good protective antibody responses in animal models, indicating that the HA stem is not inherently weakly immunogenic. While the initial studies reported protection against viruses in group 1, work is in progress to develop stem-only vaccines that protect against viruses in both groups (194). A stem-only immunogen that elicits protective immunity has also been expressed in bacteria (195), but the absence of glycan groups in this protein may affect its stability and antigenic properties (164, 194).
To focus responses on the stem is also the aim of a method employing sequential immunizations with chimeric HAs (33, 196) (Fig. 5D). In these chimeras, the stem domain of a common HA is combined with an “exotic” head domain, to which the recipient has not been exposed. The method requires that some antibodies have previously been elicited against the stem. After immunization with a chimera, the stem will elicit a memory response, while the head will only elicit a primary response. An advantage with this strategy is that vaccines can be prepared by standard procedures, but the immunization schedules may become complex (33).
For the HA head, the recently described sites at the trimer interface are of considerable interest for vaccine development (157, 159, 160) (Fig. 5E). While these highly conserved sites are partially occluded on the virus particle, they are apparently exposed transiently through breathing of HA trimers, especially on the surface of infected cells (159, 160, 169). Intriguingly, different MAbs against these sites may be directed against either of the complementary HA surfaces exposed through breathing (160). For vaccine development, it is of interest that antibody responses to these interface sites may be favored by immunization with a hyperglycosylated HA, in which the added glycan groups limit the response to immunodominant sites (197).
An ingenious procedure to focus antibody responses on subdominant sites in the HA head involves the use of nanoparticles covered with a mosaic of immobilized monomeric HA heads (198) (Fig. 5F). This method is based on the prediction that a nanoparticle covered with identical HA heads will elicit antibodies that are directed almost exclusively to strain-specific immunodominant sites, while exposure of different heads on a mosaic nanoparticle may enhance the formation of antibodies to a shared subdominant site. The effects were relatively limited in the initial studies, but this method may have much potential.
Overall, the various methods that focus antibody responses on subdominant HA sites (Fig. 5) now offer much hope for the development of a universal influenza vaccine. Moreover, the occluded site in the stem is also of considerable interest for vaccine development.
MOLECULAR BASIS OF ANTIBODY SUBDOMINANCE
General Aspects
Little is yet known about the molecular mechanisms that make one surface-exposed region of a microbial protein subdominant, but a variety of processes can probably contribute. Factors such as B cell precursor frequency (199), mode of immunization (149, 200), antigen dose (201, 202), and local antigen concentration (203) may contribute, but most interest has been focused on the properties of the antigen (180), the aspect considered here.
If the subdominance of a protein region allows a pathogen to evade protective antibodies, as discussed in this review, the question may be raised as to why the whole protein has not evolved to become weakly immunogenic. The answer could be that it is most important for the pathogen to evade antibodies to a region targeted by protective antibodies, while the other regions may even contribute to the selective subdominance of one region, in particular, by acting as decoys. Moreover, it is not simple to envisage mechanisms which would limit the antibody response to a whole protein without interfering with its function, although such a situation has been described for the Env protein of HIV-1, which has unique properties (204). Accordingly, the examples considered below all refer to situations where only one region of a protein elicits a weak antibody response, i.e., is subdominant.
Tandem repeat structures in proteins and bacterial pili.
Many surface proteins in bacteria and protozoa have a region with tandem repeats, which is located next to a unique domain (75–77, 205). Studies of several such proteins have shown that the unique domain is subdominant (Fig. 2 and 3), a result that may at least partly be explained by multipoint and high-avidity binding of the repeats to cognate B cells (56, 57) (Fig. 2B). Of note, this model implies that the unique region may elicit a good response if separated from the repeats (17). A formally similar mechanism could affect the antibody response to bacterial pili, which typically are composed of a unique tip adhesin and a rod built from repeating protein units (206). Studies of type 1 pili in Escherichia coli have indeed indicated that the tip adhesin is subdominant in intact pili but elicits a good response when administered in an isolated form (207).
Physicochemical properties.
The presence of a strongly immunogenic region in an antigen may cause selective binding to specific B cells, making an adjacent region subdominant. Thus, the outcome would be similar to that caused by tandem repeats. It follows that the isolated subdominant region may be employed as a vaccine component, as described for domain III in the E protein of dengue virus and the stem of influenza virus HA (Fig. 4 and 5). In another scenario, the breathing of a protein may result in limited responses to epitopes that are exposed only transiently (Fig. 5E). In that case, it may be possible to enhance the antibody response through modifications of the protein that focus the response on the breathing sites (197).
Sensitivity to proteolytic attack.
A protein region may be eliminated through proteolytic attack at an early stage of an infection, potentially resulting in subdominance. Possible examples include the amino-terminal regions of P. falciparum CSP (61) and streptococcal M protein (101) (Fig. 2A and 3B). In this case, a mutant protein that is not cleaved might elicit a stronger antibody response to the subdominant region.
Binding of a host protein.
Many surface proteins of pathogenic bacteria specifically bind a human plasma protein (75–77, 95, 208). Little is known about the effects of these interactions on antibody responses, but studies of a Neisseria meningitidis protein indicate that binding of the human plasma protein factor H limits antibody responses to the binding site (209, 210). Thus, the binding of a host ligand may contribute to subdominance. In this case, immunization with a mutant protein, which does not bind the host ligand, may enhance the formation of protective antibodies (209, 210).
Glycosylation.
Glycosylation of a protein site may limit the formation of antibodies and reduce the binding of preexisting antibodies (164, 211, 212). Thus, glycosylation may confer subdominance to a protein site or even a large region (213). On the other hand, extensive glycosylation of one region may redirect the antibody response to subdominant and broadly protective epitopes in other parts, a finding of relevance to vaccine development (197, 214).
Other possible mechanisms.
If a protein region is poorly accessible for B cells, it may elicit a weak response, even if it is accessible to free antibodies, a situation that could contribute to the subdominance of the HA stem (215). Subdominance may also result from tolerance, reflecting a similarity between the subdominant part of an antigen and a host structure (216). This aspect is potentially problematic for vaccine development, because the induction of antibodies to a subdominant site might induce autoimmunity if the site is subdominant because of tolerance (216, 217).
IS SUBDOMINANCE A MECHANISM OF IMMUNE ESCAPE?
Evolutionary Arguments
For a pathogen, it should be advantageous if a protein region, which is a target for protective antibodies, elicits a subdominant antibody response during an infection. This argument suggests that subdominance has evolved as a mechanism of immune evasion, although its molecular basis may vary among different systems. Support for this notion comes from classical Darwinian arguments (19) and from the well-documented ability of pathogens to evade immune attack through a variety of mechanisms (218). Thus, subdominance may be interpreted in the light of Dobzhansky’s classical dictum that “Nothing in biology makes sense except in the light of evolution” (219). However, it is possible that subdominance is unrelated to immune evasion in at least some cases.
Comparison with antigenic variation.
Assuming that subdominance plays a substantial role in antibody escape, it is of interest to make a comparison with antigenic variation, which arguably is the most important mechanism by which pathogens escape antibodies (220, 221) (Fig. 6). This comparison is particularly relevant, because both antigenic variation and subdominance are found in a variety of systems, suggesting that not only antigenic variation but also subdominance may be of general importance.
FIG 6.
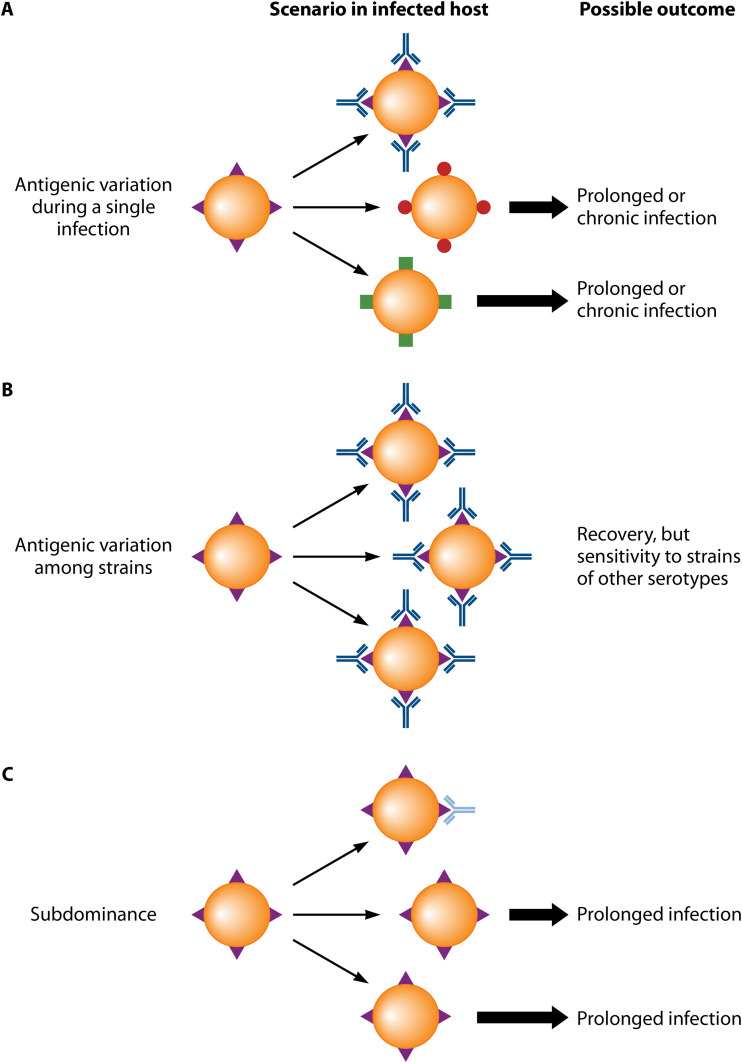
Antibody escape through antigenic variation or subdominance: a comparison. Schematic comparing the well-known role of antigenic variation and the potential role of subdominance in immune escape. Of note, antigenic variation comes in two forms, as indicated below. (A) Antigenic variation during the course of a single infection. Protein variants are indicated in different colors. Microbes expressing new variants escape antibody attack and can grow (thick arrows) because they are not recognized by antibodies elicited by an earlier variant. This situation may result in the establishment of a prolonged infection, during which new antigenic variants are repeatedly selected for. (B) Antigenic variation among strains. In this case, an antibody response to strain-specific determinants typically results in recovery and immunity, but the host may subsequently be infected by a variant strain circulating in the population. The hemagglutinin (HA) of influenza virus offers a classical example. (C) Subdominance may promote immune escape by strongly limiting the formation of antibodies to a protective site. As a result, an infection may be prolonged.
Antigenic variation implies that a pathogen evades host immunity through structural variation in a surface component, and it typically results from extensive sequence variability in a protein (151, 220, 221). For many pathogens, the sequence variability is generated during a single infection, allowing the microbe to escape newly formed antibodies in the infected host, resulting in a prolonged infection (Fig. 6A). For other pathogens, such as influenza virus, new mutants appear too rarely to allow immune escape within a single host, allowing a protective antibody response to be elicited against the antigenic variant expressed by the infecting strain. However, the host may subsequently be infected with a strain expressing a different variant (Fig. 6B). In either case, antigenic variation is characterized by changes in the quality, but not necessarily in the quantity, of protective antibodies. In contrast, subdominance would promote immune escape by reducing the quantity of antibodies, a situation that could favor the establishment of a prolonged infection, such as the antigenic variation that occurs during a single infection (Fig. 6C). Of note, a combination of antigenic variation and subdominance may result in particularly efficient antibody escape (18).
SUBDOMINANCE AND VACCINE DEVELOPMENT: GENERAL COMMENTS
The subdominant regions described in this review were recognized during the characterization of antibody responses to microbial proteins and were not specifically searched for. On the other hand, evidence is now accumulating that subdominant regions may be of general interest for vaccine development. It may then be argued that some vaccine studies could benefit from being primarily focused on the identification of subdominant regions (or sites), with subsequent evaluation of those targets as possible vaccine components. This argument raises two questions: how to identify a subdominant region or site in a given protein, and how to focus the analysis on relevant proteins.
Methods To Identify Subdominant Regions or Sites in Microbial Proteins
In discussing this problem, it is instructive to separately focus on elongated and globular proteins.
Elongated surface proteins are common in Gram-positive bacterial pathogens, such as staphylococci and streptococci, and are typically composed of a series of distinct domains (75–77, 82, 95), a feature that may simplify the identification of a subdominant region. This argument is based on the fact that a whole protein domain may be subdominant, as described in this review. Accordingly, the different domains in a protein may be directly compared for their ability to elicit antibodies. Specifically, a polyclonal antiserum elicited by the intact protein may be tested for reactivity with each of the isolated domains, potentially allowing the identification of one domain that reacts weakly and therefore may be subdominant. To verify this preliminary conclusion, an inhibition analysis must be performed, demonstrating that the other domains inhibit most or all of the antibody reactivity with the intact protein (17, 18, 101). Interestingly, surface proteins with multiple distinct domains are common not only in Gram-positive bacterial pathogens but also in the protozoan Plasmodium falciparum (222), implying that similar analysis may be performed for those proteins, as described in an early study of CSP (60). Moreover, some viral proteins are composed of at least two distinct domains, allowing antibody responses to those domains to be compared, as described above for the E protein of dengue virus and for the HA of influenza virus.
For globular proteins (or domains), such as the surface proteins of many viruses and Gram-negative bacteria, the location of antibody binding sites usually cannot be analyzed with protein fragments or peptides, since epitopes typically are conformational (223). However, pioneering work on the HA of influenza virus demonstrated that immunodominant sites could be identified through the characterization of the binding sites of many MAbs (162, 163). These sites in influenza virus were initially identified as targets for neutralizing antibodies, but as noted above, it is now known that they are immunodominant also in the quantitative sense of the term (148). With the caveat that collections of MAbs may not have properties representative of polyclonal sera (39, 135), this demanding method provides information on immunodominant sites and, by inference, also on subdominant sites. Much of the present work on viruses indeed employs large collections of MAbs, aiming at the identification of rare MAbs that bind to previously unknown epitopes of interest for vaccine development (13, 14, 169, 224).
Interestingly, recent studies indicate that electron microscopy may also be used for epitope mapping (225, 226). This semiquantitative method has relatively low resolution but can give important information on the location of immunodominant sites and therefore also about subdominant sites. While the method has so far been used only for viruses, it should be of interest to apply it in a variety of systems.
In discussing methodology, it should be noted that the identification of a surface-exposed protein region as subdominant is, by necessity, based on laboratory findings, which indicate that the antibody response to one region is relatively weak or even negligible. It is conceivable, however, that antibodies are present but not detected in laboratory tests, an aspect that should be kept in mind during studies of subdominance. Antibodies may, for example, go undetected in tests with a purified protein, because they are directed at quaternary epitopes present only on the surface of the pathogen (117, 120, 224). Problems associated with the use of pure antigens may in some cases be avoided by performing the analysis with whole microbes, but it is noteworthy that the antigenic properties of a microbial surface protein may not be the same in the laboratory as in the infected host (116).
Focusing the Analysis on Relevant Proteins
To limit the amount of work needed to identify subdominant regions or sites, it is obviously essential to focus the analysis on relevant proteins. The selection of a protein to study will not be a problem for viruses, which typically have one or very few surface proteins, but as noted above, the subsequent identification of subdominant sites or epitopes in a viral protein may be very demanding and may require the characterization of large numbers of MAbs. Moreover, the problem of how to selectively elicit a polyclonal antibody response against a subdominant site is challenging and is currently attracting intense interest (15, 117, 227–229).
For bacteria and protozoa, the identification of a protein in which a subdominant region is of interest for vaccine work will require methodologies different from those used for viruses. These cellular microbes typically have numerous surface proteins, limiting the possibility to screen collections of MAbs elicited by the pathogen, because most MAbs may be directed against surface structures that are of limited interest as vaccine components. Accordingly, screenings with MAbs have so far found little application in work with bacteria and protozoa, although collections of MAbs were recently employed in pioneering studies allowing the identification of a novel protective epitope on the circumsporozoite of P. falciparum (66, 67). In contrast, much work on the identification of vaccine components in bacteria and protozoa has employed reverse vaccinology (8, 230) or the screening of protein arrays with patient sera (9–11). Notwithstanding the interest of these methods, it should be noted that they are focused on the identification of intact proteins, not subdominant regions.
How can a subdominant region of interest for vaccine work be identified among the many surface proteins expressed by a bacterial or protozoal pathogen? It is probably unrealistic to analyze all surface proteins for the presence of subdominant regions, but a possible way forward could involve studies of a few well-characterized virulence factors. The data summarized in this review provide support for this proposal, since the proteins described here had been extensively studied long before it was demonstrated that they have subdominant regions. Thus, it is possible that even well-characterized surface proteins may have subdominant regions that are of interest for vaccine development. Indeed, many surface proteins in bacteria and protozoa have been studied in detail with regard to structure and function, while knowledge about their antigenic properties is limited (75–77, 222).
It is beyond the scope of this review to consider problems associated with vaccine development, but it should be noted that potential difficulties include the induction of autoimmunity (216, 217), limited duration of the protective response, effects of preexisting immunity (231, 232), and antibody-dependent enhancement of disease (138).
CONCLUDING REMARKS
As summarized in this review, accumulating data now indicate that subdominant regions are of interest for vaccine development. Here, it is of interest to consider three aspects that are of general interest. First, subdominance may characterize a whole surface-exposed protein domain, as demonstrated for all of the systems described in this review. This feature may facilitate vaccine development, since it should be much simpler to focus antibody responses on an intact domain than on a subdominant site that cannot be studied in an isolated form. Second, studies of subdominant regions are of interest, even for proteins known to have an immunodominant region that elicits protective antibodies. This conclusion follows from the observation that the antibodies elicited by the immunodominant region may not have optimal properties. For example, the immunodominant head of influenza virus HA elicits strain-specific protective antibodies, while the subdominant stem elicits broadly protective antibodies. Third, subdominance is not correlated with sequence conservation. An example is provided by dengue virus, for which the three domains in the E protein show similar variability among different strains (233), while domain III stands out as being subdominant. This situation may seem paradoxical but is not surprising, since the extent of sequence variation in a region probably depends on many factors, including the potency and concentration of antibodies to that region.
In summary, a subdominant region in a microbial protein may represent an Achilles’ heel that can be targeted by vaccine-induced antibodies. While subdominant regions have recently attracted much interest in studies of viruses, in particular, influenza virus, little information is available about bacterial and protozoal pathogens. Thus, it will be especially important to analyze how common subdominant regions are in surface proteins of those infectious agents. It will also be essential to gain insights into the molecular basis of subdominance, since such knowledge may favor the development of optimal vaccines and may provide insights of general interest about the role of subdominance in microbial immune evasion.
ACKNOWLEDGMENTS
Outstanding contributions to the studies of streptococci and immune responses were made by the following colleagues: Thomas Areschoug, Karin Berggård, Fredric Carlsson, Mattias Gustafsson, Eskil Johnsson, Bodil Kristensen, Jonas Lannergård, Anna Norrby-Teglund, Jenny Persson, Charlotta Sandin, Lars Stenberg, Margaretha Stålhammar-Carlemalm, Anette Thern, Johan Waldemarsson, and Maria Wästfelt. I also thank Erik Lindahl and Peter Rådström. Skillful help with the figures was provided by Katarina Jandér.
I declare no conflicts of interest.
REFERENCES
- 1.Rodrigues CMC, Pinto MV, Sadarangani M, Plotkin SA. 2017. Whither vaccines? J Infect 74 Suppl 1:S2–S9. doi: 10.1016/S0163-4453(17)30184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappuoli R, Pizza M, Del Giudice G, De Gregorio E. 2014. Vaccines, new opportunities for a new society. Proc Natl Acad Sci U S A 111:12288–12293. doi: 10.1073/pnas.1402981111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mariner JC, House JA, Mebus CA, Sollod AE, Chibeu D, Jones BA, Roeder PL, Admassu B, van 't Klooster GG. 2012. Rinderpest eradication: appropriate technology and social innovations. Science 337:1309–1312. doi: 10.1126/science.1223805. [DOI] [PubMed] [Google Scholar]
- 4.Koff WC, Burton DR, Johnson PR, Walker BD, King CR, Nabel GJ, Ahmed R, Bhan MK, Plotkin SA. 2013. Accelerating next-generation vaccine development for global disease prevention. Science 340:1232910. doi: 10.1126/science.1232910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadevall A, Pirofski LA. 2003. Exploiting the redundancy in the immune system: vaccines can mediate protection by eliciting 'unnatural' immunity. J Exp Med 197:1401–1404. doi: 10.1084/jem.20030637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambert PH, Liu M, Siegrist CA. 2005. Can successful vaccines teach us how to induce efficient protective immune responses? Nat Med 11:S54–S62. doi: 10.1038/nm1216. [DOI] [PubMed] [Google Scholar]
- 7.Plotkin SA. 2010. Correlates of protection induced by vaccination. Clin Vaccine Immunol 17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delany I, Rappuoli R, Seib KL. 2013. Vaccines, reverse vaccinology, and bacterial pathogenesis. Cold Spring Harb Perspect Med 3:a012476. doi: 10.1101/cshperspect.a012476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Etz H, Minh DB, Henics T, Dryla A, Winkler B, Triska C, Boyd AP, Sollner J, Schmidt W, von Ahsen U, Buschle M, Gill SR, Kolonay J, Khalak H, Fraser CM, von Gabain A, Nagy E, Meinke A. 2002. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc Natl Acad Sci U S A 99:6573–6578. doi: 10.1073/pnas.092569199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SJ, Liang L, Juarez S, Nanton MR, Gondwe EN, Msefula CL, Kayala MA, Necchi F, Heath JN, Hart P, Tsolis RM, Heyderman RS, MacLennan CA, Felgner PL, Davies DH, McSorley SJ. 2012. Identification of a common immune signature in murine and human systemic salmonellosis. Proc Natl Acad Sci U S A 109:4998–5003. doi: 10.1073/pnas.1111413109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hviid L. 2019. Looking for needles in the plasmodial haystack. mSphere 4:e00146-19. doi: 10.1128/mSphere.00146-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burton DR, Poignard P, Stanfield RL, Wilson IA. 2012. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corti D, Lanzavecchia A. 2013. Broadly neutralizing antiviral antibodies. Annu Rev Immunol 31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 14.Crowe JE., Jr 2017. Principles of broad and potent antiviral human antibodies: insights for vaccine design. Cell Host Microbe 22:193–206. doi: 10.1016/j.chom.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sesterhenn F, Yang C, Bonet J, Cramer JT, Wen X, Wang Y, Chiang CI, Abriata LA, Kucharska I, Castoro G, Vollers SS, Galloux M, Dheilly E, Rosset S, Corthésy P, Georgeon S, Villard M, Richard CA, Descamps D, Delgado T, Oricchio E, Rameix-Welti MA, Más V, Ervin S, Eléouët JF, Riffault S, Bates JT, Julien JP, Li Y, Jardetzky T, Krey T, Correia BE. 2020. De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science 368:eaay5051. doi: 10.1126/science.aay5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burton DR, Parren PW. 2000. Vaccines and the induction of functional antibodies: time to look beyond the molecules of natural infection? Nat Med 6:123–125. doi: 10.1038/72200. [DOI] [PubMed] [Google Scholar]
- 17.Stålhammar-Carlemalm M, Waldemarsson J, Johnsson E, Areschoug T, Lindahl G. 2007. Nonimmunodominant regions are effective as building blocks in a streptococcal fusion protein vaccine. Cell Host Microbe 2:427–434. doi: 10.1016/j.chom.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Lannergård J, Gustafsson MC, Waldemarsson J, Norrby-Teglund A, Stålhammar-Carlemalm M, Lindahl G. 2011. The hypervariable region of Streptococcus pyogenes M protein escapes antibody attack by antigenic variation and weak immunogenicity. Cell Host Microbe 10:147–157. doi: 10.1016/j.chom.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Little TJ, Allen JE, Babayan SA, Matthews KR, Colegrave N. 2012. Harnessing evolutionary biology to combat infectious disease. Nat Med 18:217–220. doi: 10.1038/nm.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nabel GJ. 2013. Designing tomorrow’s vaccines. N Engl J Med 368:551–560. doi: 10.1056/NEJMra1204186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Angeletti D, Yewdell JW. 2018. Understanding and manipulating viral immunity: antibody immunodominance enters center stage. Trends Immunol 39:549–561. doi: 10.1016/j.it.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Lüderitz O, Staub AM, Westphal O. 1966. Immunochemistry of O and R antigens of Salmonella and related Enterobacteriaceae. Bacteriol Rev 30:192–255. doi: 10.1128/MMBR.30.1.192-255.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabat EA. 1992. Michael Heidelberger April 29, 1888-June 25, 1991. J Immunol 148:301–307. [PubMed] [Google Scholar]
- 24.Sela M. 1969. Antigenicity: some molecular aspects. Science 166:1365–1374. doi: 10.1126/science.166.3911.1365. [DOI] [PubMed] [Google Scholar]
- 25.Wicker LS, Benjamin CD, Miller A, Sercarz EE. 1984. Immunodominant protein epitopes. II. The primary antibody response to hen egg white lysozyme requires and focuses upon a unique N-terminal epitope. Eur J Immunol 14:447–453. doi: 10.1002/eji.1830140512. [DOI] [PubMed] [Google Scholar]
- 26.Berzofsky JA. 1988. Immunodominance in T lymphocyte recognition. Immunol Lett 18:83–92. doi: 10.1016/0165-2478(88)90046-6. [DOI] [PubMed] [Google Scholar]
- 27.Sercarz EE, Lehmann PV, Ametani A, Benichou G, Miller A, Moudgil K. 1993. Dominance and crypticity of T cell antigenic determinants. Annu Rev Immunol 11:729–766. doi: 10.1146/annurev.iy.11.040193.003501. [DOI] [PubMed] [Google Scholar]
- 28.Yewdell JW. 2006. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity 25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Kim A, Sadegh-Nasseri S. 2015. Determinants of immunodominance for CD4 T cells. Curr Opin Immunol 34:9–15. doi: 10.1016/j.coi.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obeid OE, Steward MW. 1994. The potential of immunization with synthetic peptides to overcome the immunosuppressive effect of maternal anti-measles virus antibodies in young mice. Immunology 82:16–21. [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson LN, Tharakaraman K, Rowley KJ, Costa VV, Chan KR, Wong YH, Ong LC, Tan HC, Koch T, Cain D, Kirloskar R, Viswanathan K, Liew CW, Tissire H, Ramakrishnan B, Myette JR, Babcock GJ, Sasisekharan V, Alonso S, Chen J, Lescar J, Shriver Z, Ooi EE, Sasisekharan R. 2015. Structure-guided design of an anti-dengue antibody directed to a non-immunodominant epitope. Cell 162:493–504. doi: 10.1016/j.cell.2015.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tas JM, Mesin L, Pasqual G, Targ S, Jacobsen JT, Mano YM, Chen CS, Weill JC, Reynaud CA, Browne EP, Meyer-Hermann M, Victora GD. 2016. Visualizing antibody affinity maturation in germinal centers. Science 351:1048–1054. doi: 10.1126/science.aad3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nachbagauer R, Palese P. 2018. Development of next generation hemagglutinin-based broadly protective influenza virus vaccines. Curr Opin Immunol 53:51–57. doi: 10.1016/j.coi.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Oscherwitz J, Yu F, Jacobs JL, Liu TH, Johnson PR, Cease KB. 2009. Synthetic peptide vaccine targeting a cryptic neutralizing epitope in domain 2 of Bacillus anthracis protective antigen. Infect Immun 77:3380–3388. doi: 10.1128/IAI.00358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothbard JB, Fernandez R, Schoolnik GK. 1984. Strain-specific and common epitopes of gonococcal pili. J Exp Med 160:208–221. doi: 10.1084/jem.160.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, Connell MJ, Stevens E, Schroeter A, Chen M, Macpherson S, Serra AM, Adachi Y, Holmes MA, Li Y, Klevit RE, Graham BS, Wyatt RT, Baker D, Strong RK, Crowe JE Jr, Johnson PR, Schief WR. 2014. Proof of principle for epitope-focused vaccine design. Nature 507:201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Regenmortel MH. 2009. What is a B-cell epitope? Methods Mol Biol 524:3–20. doi: 10.1007/978-1-59745-450-6_1. [DOI] [PubMed] [Google Scholar]
- 38.Barlow DJ, Edwards MS, Thornton JM. 1986. Continuous and discontinuous protein antigenic determinants. Nature 322:747–748. doi: 10.1038/322747a0. [DOI] [PubMed] [Google Scholar]
- 39.Crowe JE. 2019. Antibody determinants of influenza immunity. J Infect Dis 219:S21–S29. doi: 10.1093/infdis/jiz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sesterhenn F, Bonet J, Correia BE. 2018. Structure-based immunogen design - leading the way to the new age of precision vaccines. Curr Opin Struct Biol 51:163–169. doi: 10.1016/j.sbi.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.World Health Organization. 2019. WHO world malaria report 2019. https://www.who.int/publications-detail/world-malaria-report-2019. Accessed 1 May 2020.
- 42.Medica DL, Sinnis P. 2005. Quantitative dynamics of Plasmodium yoelii sporozoite transmission by infected anopheline mosquitoes. Infect Immun 73:4363–4369. doi: 10.1128/IAI.73.7.4363-4369.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flores-Garcia Y, Nasir G, Hopp CS, Munoz C, Balaban AE, Zavala F, Sinnis P. 2018. Antibody-mediated protection against Plasmodium sporozoites begins at the dermal inoculation site. mBio 9:e02194-18. doi: 10.1128/mBio.02194-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen J, Nussenzweig V, Nussenzweig R, Vekemans J, Leach A. 2010. From the circumsporozoite protein to the RTS, S/AS candidate vaccine. Hum Vaccin 6:90–96. doi: 10.4161/hv.6.1.9677. [DOI] [PubMed] [Google Scholar]
- 45.Cockburn IA, Seder RA. 2018. Malaria prevention: from immunological concepts to effective vaccines and protective antibodies. Nat Immunol 19:1199–1211. doi: 10.1038/s41590-018-0228-6. [DOI] [PubMed] [Google Scholar]
- 46.Coppi A, Natarajan R, Pradel G, Bennett BL, James ER, Roggero MA, Corradin G, Persson C, Tewari R, Sinnis P. 2011. The malaria circumsporozoite protein has two functional domains, each with distinct roles as sporozoites journey from mosquito to mammalian host. J Exp Med 208:341–356. doi: 10.1084/jem.20101488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dame JB, Williams JL, McCutchan TF, Weber JL, Wirtz RA, Hockmeyer WT, Maloy WL, Haynes JD, Schneider I, Roberts D, Sanders GS, Reddy EP, Diggs CL, Miller LH. 1984. Structure of the gene encoding the immunodominant surface antigen on the sporozoite of the human malaria parasite Plasmodium falciparum. Science 225:593–599. doi: 10.1126/science.6204383. [DOI] [PubMed] [Google Scholar]
- 48.Nussenzweig V, Nussenzweig RS. 1989. Rationale for the development of an engineered sporozoite malaria vaccine. Adv Immunol 45:283–334. doi: 10.1016/s0065-2776(08)60695-1. [DOI] [PubMed] [Google Scholar]
- 49.Neafsey DE, Juraska M, Bedford T, Benkeser D, Valim C, Griggs A, Lievens M, Abdulla S, Adjei S, Agbenyega T, Agnandji ST, Aide P, Anderson S, Ansong D, Aponte JJ, Asante KP, Bejon P, Birkett AJ, Bruls M, Connolly KM, D'Alessandro U, Dobano C, Gesase S, Greenwood B, Grimsby J, Tinto H, Hamel MJ, Hoffman I, Kamthunzi P, Kariuki S, Kremsner PG, Leach A, Lell B, Lennon NJ, Lusingu J, Marsh K, Martinson F, Molel JT, Moss EL, Njuguna P, Ockenhouse CF, Ogutu BR, Otieno W, Otieno L, Otieno K, Owusu-Agyei S, Park DJ, Pelle K, Robbins D, Russ C, et al. . 2015. Genetic diversity and protective efficacy of the RTS,S/AS01 malaria vaccine. N Engl J Med 373:2025–2037. doi: 10.1056/NEJMoa1505819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coppi A, Pinzon-Ortiz C, Hutter C, Sinnis P. 2005. The Plasmodium circumsporozoite protein is proteolytically processed during cell invasion. J Exp Med 201:27–33. doi: 10.1084/jem.20040989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerami C, Frevert U, Sinnis P, Takacs B, Clavijo P, Santos MJ, Nussenzweig V. 1992. The basolateral domain of the hepatocyte plasma membrane bears receptors for the circumsporozoite protein of Plasmodium falciparum sporozoites. Cell 70:1021–1033. doi: 10.1016/0092-8674(92)90251-7. [DOI] [PubMed] [Google Scholar]
- 52.Overstreet MG, Cockburn IA, Chen YC, Zavala F. 2008. Protective CD8 T cells against Plasmodium liver stages: immunobiology of an 'unnatural' immune response. Immunol Rev 225:272–283. doi: 10.1111/j.1600-065X.2008.00671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dups JN, Pepper M, Cockburn IA. 2014. Antibody and B cell responses to Plasmodium sporozoites. Front Microbiol 5:625. doi: 10.3389/fmicb.2014.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan J, Piccoli L, Lanzavecchia A. 2019. The antibody response to Plasmodium falciparum: cues for vaccine design and the discovery of receptor-based antibodies. Annu Rev Immunol 37:225–246. doi: 10.1146/annurev-immunol-042617-053301. [DOI] [PubMed] [Google Scholar]
- 55.Julien JP, Wardemann H. 2019. Antibodies against Plasmodium falciparum malaria at the molecular level. Nat Rev Immunol 19:761–775. doi: 10.1038/s41577-019-0209-5. [DOI] [PubMed] [Google Scholar]
- 56.Kemp DJ, Coppel RL, Anders RF. 1987. Repetitive proteins and genes of malaria. Annu Rev Microbiol 41:181–208. doi: 10.1146/annurev.mi.41.100187.001145. [DOI] [PubMed] [Google Scholar]
- 57.Schofield L. 1991. On the function of repetitive domains in protein antigens of Plasmodium and other eukaryotic parasites. Parasitol Today 7:99–105. doi: 10.1016/0169-4758(91)90166-l. [DOI] [PubMed] [Google Scholar]
- 58.Patra AP, Sharma S, Ainavarapu SR. 2017. Force spectroscopy of the Plasmodium falciparum vaccine candidate circumsporozoite protein suggests a mechanically pliable repeat region. J Biol Chem 292:2110–2119. doi: 10.1074/jbc.M116.754796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zavala F, Cochrane AH, Nardin EH, Nussenzweig RS, Nussenzweig V. 1983. Circumsporozoite proteins of malaria parasites contain a single immunodominant region with two or more identical epitopes. J Exp Med 157:1947–1957. doi: 10.1084/jem.157.6.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zavala F, Tam JP, Hollingdale MR, Cochrane AH, Quakyi I, Nussenzweig RS, Nussenzweig V. 1985. Rationale for development of a synthetic vaccine against Plasmodium falciparum malaria. Science 228:1436–1440. doi: 10.1126/science.2409595. [DOI] [PubMed] [Google Scholar]
- 61.Espinosa DA, Gutierrez GM, Rojas-Lopez M, Noe AR, Shi L, Tse SW, Sinnis P, Zavala F. 2015. Proteolytic cleavage of the Plasmodium falciparum circumsporozoite protein is a target of protective antibodies. J Infect Dis 212:1111–1119. doi: 10.1093/infdis/jiv154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scally SW, Murugan R, Bosch A, Triller G, Costa G, Mordmuller B, Kremsner PG, Sim BKL, Hoffman SL, Levashina EA, Wardemann H, Julien JP. 2018. Rare PfCSP C-terminal antibodies induced by live sporozoite vaccination are ineffective against malaria infection. J Exp Med 215:63–75. doi: 10.1084/jem.20170869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.RTS,S Clinical Trials Partnership. 2015. Efficacy and safety of RTS,S/AS01 malaria vaccine with or without a booster dose in infants and children in Africa: final results of a phase 3, individually randomised, controlled trial. Lancet 386:31–45. doi: 10.1016/S0140-6736(15)60721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ballou WR, Rothbard J, Wirtz RA, Gordon DM, Williams JS, Gore RW, Schneider I, Hollingdale MR, Beaudoin RL, Maloy WL, Miller LH, Hockmeyer WT. 1985. Immunogenicity of synthetic peptides from circumsporozoite protein of Plasmodium falciparum. Science 228:996–999. doi: 10.1126/science.2988126. [DOI] [PubMed] [Google Scholar]
- 65.Heppner DG, Gordon DM, Gross M, Wellde B, Leitner W, Krzych U, Schneider I, Wirtz RA, Richards RL, Trofa A, Hall T, Sadoff JC, Boerger P, Alving CR, Sylvester DR, Porter TG, Ballou WR. 1996. Safety, immunogenicity, and efficacy of Plasmodium falciparum repeatless circumsporozoite protein vaccine encapsulated in liposomes. J Infect Dis 174:361–366. doi: 10.1093/infdis/174.2.361. [DOI] [PubMed] [Google Scholar]
- 66.Tan J, Sack BK, Oyen D, Zenklusen I, Piccoli L, Barbieri S, Foglierini M, Fregni CS, Marcandalli J, Jongo S, Abdulla S, Perez L, Corradin G, Varani L, Sallusto F, Sim BKL, Hoffman SL, Kappe SHI, Daubenberger C, Wilson IA, Lanzavecchia A. 2018. A public antibody lineage that potently inhibits malaria infection through dual binding to the circumsporozoite protein. Nat Med 24:401–407. doi: 10.1038/nm.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kisalu NK, Idris AH, Weidle C, Flores-Garcia Y, Flynn BJ, Sack BK, Murphy S, Schon A, Freire E, Francica JR, Miller AB, Gregory J, March S, Liao HX, Haynes BF, Wiehe K, Trama AM, Saunders KO, Gladden MA, Monroe A, Bonsignori M, Kanekiyo M, Wheatley AK, McDermott AB, Farney SK, Chuang GY, Zhang B, Kc N, Chakravarty S, Kwong PD, Sinnis P, Bhatia SN, Kappe SHI, Sim BKL, Hoffman SL, Zavala F, Pancera M, Seder RA. 2018. A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite. Nat Med 24:408–416. doi: 10.1038/nm.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oyen D, Torres JL, Aoto PC, Flores-Garcia Y, Binter S, Pholcharee T, Carroll S, Reponen S, Wash R, Liang Q, Lemiale F, Locke E, Bradley A, King CR, Emerling D, Kellam P, Zavala F, Ward AB, Wilson IA. 2020. Structure and mechanism of monoclonal antibody binding to the junctional epitope of Plasmodium falciparum circumsporozoite protein. PLoS Pathog 16:e1008373. doi: 10.1371/journal.ppat.1008373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fisher CR, Sutton HJ, Kaczmarski JA, McNamara HA, Clifton B, Mitchell J, Cai Y, Dups JN, D'Arcy NJ, Singh M, Chuah A, Peat TS, Jackson CJ, Cockburn IA. 2017. T-dependent B cell responses to Plasmodium induce antibodies that form a high-avidity multivalent complex with the circumsporozoite protein. PLoS Pathog 13:e1006469. doi: 10.1371/journal.ppat.1006469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thai E, Costa G, Weyrich A, Murugan R, Oyen D, Flores-Garcia Y, Prieto K, Bosch A, Valleriani A, Wu NC, Pholcharee T, Scally SW, Wilson IA, Wardemann H, Julien JP, Levashina EA. 2020. A high-affinity antibody against the CSP N-terminal domain lacks Plasmodium falciparum inhibitory activity. J Exp Med 217:e20200061. doi: 10.1084/jem.20200061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bongfen SE, Ntsama PM, Offner S, Smith T, Felger I, Tanner M, Alonso P, Nebie I, Romero JF, Silvie O, Torgler R, Corradin G. 2009. The N-terminal domain of Plasmodium falciparum circumsporozoite protein represents a target of protective immunity. Vaccine 27:328–335. doi: 10.1016/j.vaccine.2008.09.097. [DOI] [PubMed] [Google Scholar]
- 72.Collins KA, Snaith R, Cottingham MG, Gilbert SC, Hill AVS. 2017. Enhancing protective immunity to malaria with a highly immunogenic virus-like particle vaccine. Sci Rep 7:46621. doi: 10.1038/srep46621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Draper SJ, Sack BK, King CR, Nielsen CM, Rayner JC, Higgins MK, Long CA, Seder RA. 2018. Malaria vaccines: recent advances and new horizons. Cell Host Microbe 24:43–56. doi: 10.1016/j.chom.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng G, Boyle MJ, Cross N, Chan JA, Reiling L, Osier F, Stanisic DI, Mueller I, Anders RF, McCarthy JS, Richards JS, Beeson JG. 2018. Human immunization with a polymorphic malaria vaccine candidate induced antibodies to conserved epitopes that promote functional antibodies to multiple parasite strains. J Infect Dis 218:35–43. doi: 10.1093/infdis/jiy170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Navarre WW, Schneewind O. 1999. Surface proteins of Gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev 63:174–229. doi: 10.1128/MMBR.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lindahl G, Stalhammar-Carlemalm M, Areschoug T. 2005. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin Microbiol Rev 18:102–127. doi: 10.1128/CMR.18.1.102-127.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Foster TJ. 2019. Surface proteins of Staphylococcus aureus. Microbiol Spectr 7:GPP3-0046-2018. doi: 10.1128/microbiolspec.GPP3-0046-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seale AC, Bianchi-Jassir F, Russell NJ, Kohli-Lynch M, Tann CJ, Hall J, Madrid L, Blencowe H, Cousens S, Baker CJ, Bartlett L, Cutland C, Gravett MG, Heath PT, Ip M, Le Doare K, Madhi SA, Rubens CE, Saha SK, Schrag SJ, Sobanjo-Ter Meulen A, Vekemans J, Lawn JE. 2017. Estimates of the burden of group B streptococcal disease worldwide for pregnant women, stillbirths, and children. Clin Infect Dis 65:S200–S219. doi: 10.1093/cid/cix664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Michel JL, Madoff LC, Olson K, Kling DE, Kasper DL, Ausubel FM. 1992. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc Natl Acad Sci U S A 89:10060–10064. doi: 10.1073/pnas.89.21.10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wästfelt M, Stâlhammar-Carlemalm M, Delisse AM, Cabezon T, Lindahl G. 1996. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J Biol Chem 271:18892–18897. doi: 10.1074/jbc.271.31.18892. [DOI] [PubMed] [Google Scholar]
- 81.Auperin TC, Bolduc GR, Baron MJ, Heroux A, Filman DJ, Madoff LC, Hogle JM. 2005. Crystal structure of the N-terminal domain of the group B streptococcus alpha C protein. J Biol Chem 280:18245–18252. doi: 10.1074/jbc.M412391200. [DOI] [PubMed] [Google Scholar]
- 82.Whelan F, Lafita A, Griffiths SC, Cooper REM, Whittingham JL, Turkenburg JP, Manfield IW, St John AN, Paci E, Bateman A, Potts JR. 2019. Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection. Proc Natl Acad Sci U S A 116:26540–26548. doi: 10.1073/pnas.1911776116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kling DE, Gravekamp C, Madoff LC, Michel JL. 1997. Characterization of two distinct opsonic and protective epitopes within the alpha C protein of the group B Streptococcus. Infect Immun 65:1462–1467. doi: 10.1128/IAI.65.4.1462-1467.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Madoff LC, Michel JL, Gong EW, Kling DE, Kasper DL. 1996. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc Natl Acad Sci U S A 93:4131–4136. doi: 10.1073/pnas.93.9.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect Dis 5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 86.Lancefield RC, Todd EW. 1928. Antigenic differences between matt hemolytic streptococci and their glossy variants. J Exp Med 48:769–790. doi: 10.1084/jem.48.6.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Manjula BN, Fischetti VA. 1980. Tropomyosin-like seven residue periodicity in three immunologically distinct streptococcal M proteins and its implications for the antiphagocytic property of the molecule. J Exp Med 151:695–708. doi: 10.1084/jem.151.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Phillips GN Jr, Flicker PF, Cohen C, Manjula BN, Fischetti VA. 1981. Streptococcal M protein: alpha-helical coiled-coil structure and arrangement on the cell surface. Proc Natl Acad Sci U S A 78:4689–4693. doi: 10.1073/pnas.78.8.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Manjula BN, Acharya AS, Fairwell T, Fischetti VA. 1986. Antigenic domains of the streptococcal Pep M5 protein. Localization of epitopes crossreactive with type 6 M protein and identification of a hypervariable region of the M molecule. J Exp Med 163:129–138. doi: 10.1084/jem.163.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bessen DE, Smeesters PR, Beall BW. 2018. Molecular epidemiology, ecology, and evolution of group A streptococci. Microbiol Spectr 6:CPP3-0009-2018. doi: 10.1128/microbiolspec.CPP3-0009-2018. [DOI] [PubMed] [Google Scholar]
- 91.Fischetti VA. 1989. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev 2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carlsson F, Berggård K, Stålhammar-Carlemalm M, Lindahl G. 2003. Evasion of phagocytosis through cooperation between two ligand-binding regions in Streptococcus pyogenes M protein. J Exp Med 198:1057–1068. doi: 10.1084/jem.20030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Waldemarsson J, Stålhammar-Carlemalm M, Sandin C, Castellino FJ, Lindahl G. 2009. Functional dissection of Streptococcus pyogenes M5 protein: the hypervariable region is essential for virulence. PLoS One 4:e7279. doi: 10.1371/journal.pone.0007279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jones KF, Fischetti VA. 1988. The importance of the location of antibody binding on the M6 protein for opsonization and phagocytosis of group A M6 streptococci. J Exp Med 167:1114–1123. doi: 10.1084/jem.167.3.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smeesters PR, McMillan DJ, Sriprakash KS. 2010. The streptococcal M protein: a highly versatile molecule. Trends Microbiol 18:275–282. doi: 10.1016/j.tim.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 96.Morfeldt E, Berggård K, Persson J, Drakenberg T, Johnsson E, Lindahl E, Linse S, Lindahl G. 2001. Isolated hypervariable regions derived from streptococcal M proteins specifically bind human C4b-binding protein: implications for antigenic variation. J Immunol 167:3870–3877. doi: 10.4049/jimmunol.167.7.3870. [DOI] [PubMed] [Google Scholar]
- 97.Persson J, Beall B, Linse S, Lindahl G. 2006. Extreme sequence divergence but conserved ligand-binding specificity in Streptococcus pyogenes M protein. PLoS Pathog 2:e47. doi: 10.1371/journal.ppat.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gustafsson MC, Lannergård J, Nilsson OR, Kristensen BM, Olsen JE, Harris CL, Ufret-Vincenty RL, Stålhammar-Carlemalm M, Lindahl G. 2013. Factor H binds to the hypervariable region of many Streptococcus pyogenes M proteins but does not promote phagocytosis resistance or acute virulence. PLoS Pathog 9:e1003323. doi: 10.1371/journal.ppat.1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Buffalo CZ, Bahn-Suh AJ, Hirakis SP, Biswas T, Amaro RE, Nizet V, Ghosh P. 2016. Conserved patterns hidden within group A Streptococcus M protein hypervariability recognize human C4b-binding protein. Nat Microbiol 1:16155. doi: 10.1038/nmicrobiol.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Campbell PT, Tong SYC, Geard N, Davies MR, Worthing KA, Lacey JA, Smeesters PR, Batzloff MR, Kado J, Jenney AWJ, McVernon J, Steer AC. 2020. Longitudinal analysis of group A Streptococcus emm types and emm clusters in a high-prevalence setting: relationship between past and future infections. J Infect Dis 221:1429–1437. doi: 10.1093/infdis/jiz615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lannergård J, Kristensen BM, Gustafsson MC, Persson JJ, Norrby-Teglund A, Stålhammar-Carlemalm M, Lindahl G. 2015. Sequence variability is correlated with weak immunogenicity in Streptococcus pyogenes M protein. Microbiologyopen 4:774–789. doi: 10.1002/mbo3.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kuttner AG, Lenert TF. 1944. The occurrence of bacteriostatic properties in the blood of patients after recovery from streptococcal pharyngitis. J Clin Invest 23:151–161. doi: 10.1172/JCI101478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Denny FW Jr, Perry WD, Wannamaker LW. 1957. Type-specific streptococcal antibody. J Clin Invest 36:1092–1100. doi: 10.1172/JCI103504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Siegel AC, Johnson EE, Stollerman GH. 1961. Controlled studies of streptococcal pharyngitis in a pediatric population—behavior of the type-specific immune response. N Engl J Med 265:566–571. doi: 10.1056/NEJM196109212651202. [DOI] [Google Scholar]
- 105.Beachey EH, Seyer JM, Dale JB. 1987. Protective immunogenicity and T lymphocyte specificity of a trivalent hybrid peptide containing NH2-terminal sequences of types 5, 6, and 24 M proteins synthesized in tandem. J Exp Med 166:647–656. doi: 10.1084/jem.166.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pastural E, McNeil SA, MacKinnon-Cameron D, Ye L, Langley JM, Stewart R, Martin LH, Hurley GJ, Salehi S, Penfound TA, Halperin S, Dale JB. 2020. Safety and immunogenicity of a 30-valent M protein-based group A streptococcal vaccine in healthy adult volunteers: a randomized, controlled phase I study. Vaccine 38:1384–1392. doi: 10.1016/j.vaccine.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 107.Giffard PM, Tong SYC, Holt DC, Ralph AP, Currie BJ. 2019. Concerns for efficacy of a 30-valent M-protein-based Streptococcus pyogenes vaccine in regions with high rates of rheumatic heart disease. PLoS Negl Trop Dis 13:e0007511. doi: 10.1371/journal.pntd.0007511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gray LA, D'Antoine HA, Tong SYC, McKinnon M, Bessarab D, Brown N, Remenyi B, Steer A, Syn G, Blackwell JM, Inouye M, Carapetis JR. 2017. Genome-wide analysis of genetic risk factors for rheumatic heart disease in aboriginal Australians provides support for pathogenic molecular mimicry. J Infect Dis 216:1460–1470. doi: 10.1093/infdis/jix497. [DOI] [PubMed] [Google Scholar]
- 109.Diamond MS, Pierson TC. 2015. Molecular insight into dengue virus pathogenesis and its implications for disease control. Cell 162:488–492. doi: 10.1016/j.cell.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grange L, Simon-Loriere E, Sakuntabhai A, Gresh L, Paul R, Harris E. 2014. Epidemiological risk factors associated with high global frequency of inapparent dengue virus infections. Front Immunol 5:280. doi: 10.3389/fimmu.2014.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mukhopadhyay S, Kuhn RJ, Rossmann MG. 2005. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol 3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 112.Hasan SS, Sevvana M, Kuhn RJ, Rossmann MG. 2018. Structural biology of Zika virus and other flaviviruses. Nat Struct Mol Biol 25:13–20. doi: 10.1038/s41594-017-0010-8. [DOI] [PubMed] [Google Scholar]
- 113.Rey FA, Stiasny K, Vaney MC, Dellarole M, Heinz FX. 2018. The bright and the dark side of human antibody responses to flaviviruses: lessons for vaccine design. EMBO Rep 19:206–224. doi: 10.15252/embr.201745302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 115.Dowd KA, Pierson TC. 2018. The many faces of a dynamic virion: implications of viral breathing on flavivirus biology and immunogenicity. Annu Rev Virol 5:185–207. doi: 10.1146/annurev-virology-092917-043300. [DOI] [PubMed] [Google Scholar]
- 116.Raut R, Corbett KS, Tennekoon RN, Premawansa S, Wijewickrama A, Premawansa G, Mieczkowski P, Ruckert C, Ebel GD, De Silva AD, de Silva AM. 2019. Dengue type 1 viruses circulating in humans are highly infectious and poorly neutralized by human antibodies. Proc Natl Acad Sci U S A 116:227–232. doi: 10.1073/pnas.1812055115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Slon Campos JL, Mongkolsapaya J, Screaton GR. 2018. The immune response against flaviviruses. Nat Immunol 19:1189–1198. doi: 10.1038/s41590-018-0210-3. [DOI] [PubMed] [Google Scholar]
- 118.de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WM, White LJ, Diamond MS, Baric RS, Crowe JE Jr, de Silva AM. 2012. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci U S A 109:7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Teoh EP, Kukkaro P, Teo EW, Lim AP, Tan TT, Yip A, Schul W, Aung M, Kostyuchenko VA, Leo YS, Chan SH, Smith KG, Chan AH, Zou G, Ooi EE, Kemeny DM, Tan GK, Ng JK, Ng ML, Alonso S, Fisher D, Shi PY, Hanson BJ, Lok SM, MacAry PA. 2012. The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci Transl Med 4:139ra83. doi: 10.1126/scitranslmed.3003888. [DOI] [PubMed] [Google Scholar]
- 120.Gallichotte EN, Baric RS, de Silva AM. 2018. The molecular specificity of the human antibody response to dengue virus infections. Adv Exp Med Biol 1062:63–76. doi: 10.1007/978-981-10-8727-1_5. [DOI] [PubMed] [Google Scholar]
- 121.Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney MC, Kikuti CM, Navarro Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A, Girard-Blanc C, Petres S, Shepard WE, Despres P, Arenzana-Seisdedos F, Dussart P, Mongkolsapaya J, Screaton GR, Rey FA. 2015. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 520:109–113. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- 122.Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NTH, Duangchinda T, Grimes JM, Tsai WY, Lai CY, Wang WK, Malasit P, Farrar J, Simmons CP, Zhou ZH, Rey FA, Mongkolsapaya J, Screaton GR. 2015. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol 16:170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rouvinski A, Dejnirattisai W, Guardado-Calvo P, Vaney MC, Sharma A, Duquerroy S, Supasa P, Wongwiwat W, Haouz A, Barba-Spaeth G, Mongkolsapaya J, Rey FA, Screaton GR. 2017. Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope. Nat Commun 8:15411. doi: 10.1038/ncomms15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tsai WY, Lai CY, Wu YC, Lin HE, Edwards C, Jumnainsong A, Kliks S, Halstead S, Mongkolsapaya J, Screaton GR, Wang WK. 2013. High-avidity and potently neutralizing cross-reactive human monoclonal antibodies derived from secondary dengue virus infection. J Virol 87:12562–12575. doi: 10.1128/JVI.00871-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Coloma J, Harris E. 2015. Broad and strong: the ultimate antibody to dengue virus. Nat Immunol 16:135–137. doi: 10.1038/ni.3081. [DOI] [PubMed] [Google Scholar]
- 126.Pierson TC, Fremont DH, Kuhn RJ, Diamond MS. 2008. Structural insights into the mechanisms of antibody-mediated neutralization of flavivirus infection: implications for vaccine development. Cell Host Microbe 4:229–238. doi: 10.1016/j.chom.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wahala WM, Kraus AA, Haymore LB, Accavitti-Loper MA, de Silva AM. 2009. Dengue virus neutralization by human immune sera: role of envelope protein domain III-reactive antibody. Virology 392:103–113. doi: 10.1016/j.virol.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wahala WM, Huang C, Butrapet S, White LJ, de Silva AM. 2012. Recombinant dengue type 2 viruses with altered E protein domain III epitopes are efficiently neutralized by human immune sera. J Virol 86:4019–4023. doi: 10.1128/JVI.06871-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lai CY, Tsai WY, Lin SR, Kao CL, Hu HP, King CC, Wu HC, Chang GJ, Wang WK. 2008. Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross-reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J Virol 82:6631–6643. doi: 10.1128/JVI.00316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Oliphant T, Nybakken GE, Austin SK, Xu Q, Bramson J, Loeb M, Throsby M, Fremont DH, Pierson TC, Diamond MS. 2007. Induction of epitope-specific neutralizing antibodies against West Nile virus. J Virol 81:11828–11839. doi: 10.1128/JVI.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jarmer J, Zlatkovic J, Tsouchnikas G, Vratskikh O, Strauß J, Aberle JH, Chmelik V, Kundi M, Stiasny K, Heinz FX. 2014. Variation of the specificity of the human antibody responses after tick-borne encephalitis virus infection and vaccination. J Virol 88:13845–13857. doi: 10.1128/JVI.02086-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vratskikh O, Stiasny K, Zlatkovic J, Tsouchnikas G, Jarmer J, Karrer U, Roggendorf M, Roggendorf H, Allwinn R, Heinz FX. 2013. Dissection of antibody specificities induced by yellow fever vaccination. PLoS Pathog 9:e1003458. doi: 10.1371/journal.ppat.1003458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, Rey FA, Varani L, Whitehead SS, Diamond MS, Harris E, Lanzavecchia A, Sallusto F. 2010. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Smith SA, de Alwis R, Kose N, Durbin AP, Whitehead SS, de Silva AM, Crowe JE Jr. 2013. Human monoclonal antibodies derived from memory B cells following live attenuated dengue virus vaccination or natural infection exhibit similar characteristics. J Infect Dis 207:1898–1908. doi: 10.1093/infdis/jit119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sajadi MM, Dashti A, Rikhtegaran Tehrani Z, Tolbert WD, Seaman MS, Ouyang X, Gohain N, Pazgier M, Kim D, Cavet G, Yared J, Redfield RR, Lewis GK, DeVico AL. 2018. Identification of near-pan-neutralizing antibodies against HIV-1 by deconvolution of plasma humoral responses. Cell 173:1783.e14–1795.e14. doi: 10.1016/j.cell.2018.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Halstead SB. 2003. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res 60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- 137.Katzelnick LC, Gresh L, Halloran ME, Mercado JC, Kuan G, Gordon A, Balmaseda A, Harris E. 2017. Antibody-dependent enhancement of severe dengue disease in humans. Science 358:929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.St John AL, Rathore APS. 2019. Adaptive immune responses to primary and secondary dengue virus infections. Nat Rev Immunol 19:218–230. doi: 10.1038/s41577-019-0123-x. [DOI] [PubMed] [Google Scholar]
- 139.Frei JC, Wirchnianski AS, Govero J, Vergnolle O, Dowd KA, Pierson TC, Kielian M, Girvin ME, Diamond MS, Lai JR. 2018. Engineered dengue virus domain III proteins elicit cross-neutralizing antibody responses in mice. J Virol 92:e01023-18. doi: 10.1128/JVI.01023-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ramasamy V, Arora U, Shukla R, Poddar A, Shanmugam RK, White LJ, Mattocks MM, Raut R, Perween A, Tyagi P, de Silva AM, Bhaumik SK, Kaja MK, Villinger F, Ahmed R, Johnston RE, Swaminathan S, Khanna N. 2018. A tetravalent virus-like particle vaccine designed to display domain III of dengue envelope proteins induces multi-serotype neutralizing antibodies in mice and macaques which confer protection against antibody dependent enhancement in AG129 mice. PLoS Negl Trop Dis 12:e0006191. doi: 10.1371/journal.pntd.0006191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shukla R, Ramasamy V, Rajpoot RK, Arora U, Poddar A, Ahuja R, Beesetti H, Swaminathan S, Khanna N. 2019. Next generation designer virus-like particle vaccines for dengue. Expert Rev Vaccines 18:105–117. doi: 10.1080/14760584.2019.1562909. [DOI] [PubMed] [Google Scholar]
- 142.Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, Palese P, Shaw ML, Treanor J, Webster RG, Garcia-Sastre A. 2018. Influenza. Nat Rev Dis Primers 4:3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wilson IA, Cox NJ. 1990. Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol 8:737–771. doi: 10.1146/annurev.iy.08.040190.003513. [DOI] [PubMed] [Google Scholar]
- 144.Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 145.Gamblin SJ, Vachieri SG, Xiong X, Zhang J, Martin SR, Skehel JJ. 8 June 2020. Hemagglutinin structure and activities. Cold Spring Harb Perspect Med doi: 10.1101/cshperspect.a038638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Krammer F. 2019. The human antibody response to influenza A virus infection and vaccination. Nat Rev Immunol 19:383–397. doi: 10.1038/s41577-019-0143-6. [DOI] [PubMed] [Google Scholar]
- 147.Sui J, Sheehan J, Hwang WC, Bankston LA, Burchett SK, Huang CY, Liddington RC, Beigel JH, Marasco WA. 2011. Wide prevalence of heterosubtypic broadly neutralizing human anti-influenza A antibodies. Clin Infect Dis 52:1003–1009. doi: 10.1093/cid/cir121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Altman MO, Bennink JR, Yewdell JW, Herrin BR. 2015. Lamprey VLRB response to influenza virus supports universal rules of immunogenicity and antigenicity. Elife 4:e07467. doi: 10.7554/eLife.07467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Angeletti D, Gibbs JS, Angel M, Kosik I, Hickman HD, Frank GM, Das SR, Wheatley AK, Prabhakaran M, Leggat DJ, McDermott AB, Yewdell JW. 2017. Defining B cell immunodominance to viruses. Nat Immunol 18:456–463. doi: 10.1038/ni.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, Lee J, Wan H, Rojas KT, Kirkpatrick E, Henry C, Palm AE, Stamper CT, Lan LY, Topham DJ, Treanor J, Wrammert J, Ahmed R, Eichelberger MC, Georgiou G, Krammer F, Wilson PC. 2018. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 173:417.e10–429.e10. doi: 10.1016/j.cell.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Webster RG, Laver WG. 1971. Antigenic variation in influenza virus. Biology and chemistry. Prog Med Virol 13:271–338. [PubMed] [Google Scholar]
- 152.Lee JM, Eguia R, Zost SJ, Choudhary S, Wilson PC, Bedford T, Stevens-Ayers T, Boeckh M, Hurt AC, Lakdawala SS, Hensley SE, Bloom JD. 2019. Mapping person-to-person variation in viral mutations that escape polyclonal serum targeting influenza hemagglutinin. Elife 8:e49324. doi: 10.7554/eLife.49324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wu NC, Wilson IA. 2017. A perspective on the structural and functional constraints for immune evasion: insights from influenza virus. J Mol Biol 429:2694–2709. doi: 10.1016/j.jmb.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Knossow M, Skehel JJ. 2006. Variation and infectivity neutralization in influenza. Immunology 119:1–7. doi: 10.1111/j.1365-2567.2006.02421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bizebard T, Gigant B, Rigolet P, Rasmussen B, Diat O, Bosecke P, Wharton SA, Skehel JJ, Knossow M. 1995. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature 376:92–94. doi: 10.1038/376092a0. [DOI] [PubMed] [Google Scholar]
- 156.DiLillo DJ, Palese P, Wilson PC, Ravetch JV. 2016. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest 126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lee J, Boutz DR, Chromikova V, Joyce MG, Vollmers C, Leung K, Horton AP, DeKosky BJ, Lee CH, Lavinder JJ, Murrin EM, Chrysostomou C, Hoi KH, Tsybovsky Y, Thomas PV, Druz A, Zhang B, Zhang Y, Wang L, Kong WP, Park D, Popova LI, Dekker CL, Davis MM, Carter CE, Ross TM, Ellington AD, Wilson PC, Marcotte EM, Mascola JR, Ippolito GC, Krammer F, Quake SR, Kwong PD, Georgiou G. 2016. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat Med 22:1456–1464. doi: 10.1038/nm.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Henry Dunand CJ, Leon PE, Huang M, Choi A, Chromikova V, Ho IY, Tan GS, Cruz J, Hirsh A, Zheng NY, Mullarkey CE, Ennis FA, Terajima M, Treanor JJ, Topham DJ, Subbarao K, Palese P, Krammer F, Wilson PC. 2016. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe 19:800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bangaru S, Lang S, Schotsaert M, Vanderven HA, Zhu X, Kose N, Bombardi R, Finn JA, Kent SJ, Gilchuk P, Gilchuk I, Turner HL, Garcia-Sastre A, Li S, Ward AB, Wilson IA, Crowe JE Jr. 2019. A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell 177:1136.e18–1152.e18. doi: 10.1016/j.cell.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Watanabe A, McCarthy KR, Kuraoka M, Schmidt AG, Adachi Y, Onodera T, Tonouchi K, Caradonna TM, Bajic G, Song S, McGee CE, Sempowski GD, Feng F, Urick P, Kepler TB, Takahashi Y, Harrison SC, Kelsoe G. 2019. Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell 177:1124.e16–1135.e16. doi: 10.1016/j.cell.2019.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Adachi Y, Tonouchi K, Nithichanon A, Kuraoka M, Watanabe A, Shinnakasu R, Asanuma H, Ainai A, Ohmi Y, Yamamoto T, Ishii KJ, Hasegawa H, Takeyama H, Lertmemongkolchai G, Kurosaki T, Ato M, Kelsoe G, Takahashi Y. 2019. Exposure of an occluded hemagglutinin epitope drives selection of a class of cross-protective influenza antibodies. Nat Commun 10:3883. doi: 10.1038/s41467-019-11821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 163.Wiley DC, Wilson IA, Skehel JJ. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 164.Tate MD, Job ER, Deng YM, Gunalan V, Maurer-Stroh S, Reading PC. 2014. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 6:1294–1316. doi: 10.3390/v6031294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wei CJ, Boyington JC, Dai K, Houser KV, Pearce MB, Kong WP, Yang ZY, Tumpey TM, Nabel GJ. 2010. Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci Transl Med 2:24ra21. doi: 10.1126/scitranslmed.3000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Medina RA, Stertz S, Manicassamy B, Zimmermann P, Sun X, Albrecht RA, Uusi-Kerttula H, Zagordi O, Belshe RB, Frey SE, Tumpey TM, Garcia-Sastre A. 2013. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci Transl Med 5:187ra70. doi: 10.1126/scitranslmed.3005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Das SR, Puigbò P, Hensley SE, Hurt DE, Bennink JR, Yewdell JW. 2010. Glycosylation focuses sequence variation in the influenza A virus H1 hemagglutinin globular domain. PLoS Pathog 6:e1001211. doi: 10.1371/journal.ppat.1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu STH, Behzadi MA, Sun W, Freyn AW, Liu WC, Broecker F, Albrecht RA, Bouvier NM, Simon V, Nachbagauer R, Krammer F, Palese P. 2018. Antigenic sites in influenza H1 hemagglutinin display species-specific immunodominance. J Clin Invest 128:4992–4996. doi: 10.1172/JCI122895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wu Y, Gao GF. 2019. “Breathing” hemagglutinin reveals cryptic epitopes for universal influenza vaccine design. Cell 177:1086–1088. doi: 10.1016/j.cell.2019.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhu X, Guo YH, Jiang T, Wang YD, Chan KH, Li XF, Yu W, McBride R, Paulson JC, Yuen KY, Qin CF, Che XY, Wilson IA. 2013. A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J Virol 87:12619–12635. doi: 10.1128/JVI.01577-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Benjamin E, Wang W, McAuliffe JM, Palmer-Hill FJ, Kallewaard NL, Chen Z, Suzich JA, Blair WS, Jin H, Zhu Q. 2014. A broadly neutralizing human monoclonal antibody directed against a novel conserved epitope on the influenza virus H3 hemagglutinin globular head. J Virol 88:6743–6750. doi: 10.1128/JVI.03562-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Thornburg NJ, Zhang H, Bangaru S, Sapparapu G, Kose N, Lampley RM, Bombardi RG, Yu Y, Graham S, Branchizio A, Yoder SM, Rock MT, Creech CB, Edwards KM, Lee D, Li S, Wilson IA, García-Sastre A, Albrecht RA, Crowe JE Jr. 2016. H7N9 influenza virus neutralizing antibodies that possess few somatic mutations. J Clin Invest 126:1482–1494. doi: 10.1172/JCI85317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nogales A, Piepenbrink MS, Wang J, Ortega S, Basu M, Fucile CF, Treanor JJ, Rosenberg AF, Zand MS, Keefer MC, Martinez-Sobrido L, Kobie JJ. 2018. A highly potent and broadly neutralizing H1 influenza-specific human monoclonal antibody. Sci Rep 8:4374. doi: 10.1038/s41598-018-22307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bangaru S, Zhang H, Gilchuk IM, Voss TG, Irving RP, Gilchuk P, Matta P, Zhu X, Lang S, Nieusma T, Richt JA, Albrecht RA, Vanderven HA, Bombardi R, Kent SJ, Ward AB, Wilson IA, Crowe JE Jr. 2018. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat Commun 9:2669. doi: 10.1038/s41467-018-04704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Okuno Y, Isegawa Y, Sasao F, Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67:2552–2558. doi: 10.1128/JVI.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, Goudsmit J. 2008. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lee PS, Wilson IA. 2015. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr Top Microbiol Immunol 386:323–341. doi: 10.1007/82_2014_413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Angeletti D, Yewdell JW. 2018. Is it possible to develop a “universal” influenza virus vaccine? Outflanking antibody immunodominance on the road to universal influenza vaccination. Cold Spring Harb Perspect Biol 10:a028852. doi: 10.1101/cshperspect.a028852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 182.Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, Wachter-Rosati L, Palmer-Hill FJ, Yuan AQ, Walker PA, Vorlaender MK, Bianchi S, Guarino B, De Marco A, Vanzetta F, Agatic G, Foglierini M, Pinna D, Fernandez-Rodriguez B, Fruehwirth A, Silacci C, Ogrodowicz RW, Martin SR, Sallusto F, Suzich JA, Lanzavecchia A, Zhu Q, Gamblin SJ, Skehel JJ. 2016. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell 166:596–608. doi: 10.1016/j.cell.2016.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Joyce MG, Wheatley AK, Thomas PV, Chuang GY, Soto C, Bailer RT, Druz A, Georgiev IS, Gillespie RA, Kanekiyo M, Kong WP, Leung K, Narpala SN, Prabhakaran MS, Yang ES, Zhang B, Zhang Y, Asokan M, Boyington JC, Bylund T, Darko S, Lees CR, Ransier A, Shen CH, Wang L, Whittle JR, Wu X, Yassine HM, Santos C, Matsuoka Y, Tsybovsky Y, Baxa U, NISC Comparative Sequencing Program, Mullikin JC, Subbarao K, Douek DC, Graham BS, Koup RA, Ledgerwood JE, Roederer M, Shapiro L, Kwong PD, Mascola JR, McDermott AB. 2016. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell 166:609–623. doi: 10.1016/j.cell.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang TT, Tan GS, Hai R, Pica N, Ngai L, Ekiert DC, Wilson IA, García-Sastre A, Moran TM, Palese P. 2010. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci U S A 107:18979–18984. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kuraoka M, Adachi Y, Takahashi Y. 2020. Hide and seek: interplay between influenza viruses and B cells. Int Immunol 32:605–611. doi: 10.1093/intimm/dxaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J Virol 80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, Diamond MS, Kuhn RJ, Rossmann MG. 2008. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol 15:312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- 188.Li J, Watterson D, Chang CW, Che XY, Li XQ, Ericsson DJ, Qiu LW, Cai JP, Chen J, Fry SR, Cheung STM, Cooper MA, Young PR, Kobe B. 2018. Structural and functional characterization of a cross-reactive dengue virus neutralizing antibody that recognizes a cryptic epitope. Structure 26:51.e4–59.e4. doi: 10.1016/j.str.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 189.Wei CJ, Crank MC, Shiver J, Graham BS, Mascola JR, Nabel GJ. 2020. Next-generation influenza vaccines: opportunities and challenges. Nat Rev Drug Discov 19:239–252. doi: 10.1038/s41573-019-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mathew NR, Angeletti D. 2019. Recombinant influenza vaccines: saviors to overcome immunodominance. Front Immunol 10:2997. doi: 10.3389/fimmu.2019.02997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, de Man M, Ding Z, Apetri A, Kukrer B, Sneekes-Vriese E, Tomkiewicz D, Laursen NS, Lee PS, Zakrzewska A, Dekking L, Tolboom J, Tettero L, van Meerten S, Yu W, Koudstaal W, Goudsmit J, Ward AB, Meijberg W, Wilson IA, Radosevic K. 2015. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 192.Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, Lingwood D, Moin SM, Andersen H, Okuno Y, Rao SS, Harris AK, Kwong PD, Mascola JR, Nabel GJ, Graham BS. 2015. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21:1065–1070. doi: 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 193.van der Lubbe JEM, Verspuij JWA, Huizingh J, Schmit-Tillemans SPR, Tolboom J, Dekking L, Kwaks T, Brandenburg B, Meijberg W, Zahn RC, Roozendaal R, Kuipers H. 2018. Mini-HA is superior to full length hemagglutinin immunization in inducing stem-specific antibodies and protection against group 1 influenza virus challenges in mice. Front Immunol 9:2350. doi: 10.3389/fimmu.2018.02350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Corbett KS, Moin SM, Yassine HM, Cagigi A, Kanekiyo M, Boyoglu-Barnum S, Myers SI, Tsybovsky Y, Wheatley AK, Schramm CA, Gillespie RA, Shi W, Wang L, Zhang Y, Andrews SF, Joyce MG, Crank MC, Douek DC, McDermott AB, Mascola JR, Graham BS, Boyington JC. 2019. Design of nanoparticulate group 2 influenza virus hemagglutinin stem antigens that activate unmutated ancestor B cell receptors of broadly neutralizing antibody lineages. mBio 10:e02810-18. doi: 10.1128/mBio.02810-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mallajosyula VV, Citron M, Ferrara F, Lu X, Callahan C, Heidecker GJ, Sarma SP, Flynn JA, Temperton NJ, Liang X, Varadarajan R. 2014. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci U S A 111:E2514–E2523. doi: 10.1073/pnas.1402766111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bernstein DI, Guptill J, Naficy A, Nachbagauer R, Berlanda-Scorza F, Feser J, Wilson PC, Solorzano A, Van der Wielen M, Walter EB, Albrecht RA, Buschle KN, Chen YQ, Claeys C, Dickey M, Dugan HL, Ermler ME, Freeman D, Gao M, Gast C, Guthmiller JJ, Hai R, Henry C, Lan LY, McNeal M, Palm AE, Shaw DG, Stamper CT, Sun W, Sutton V, Tepora ME, Wahid R, Wenzel H, Wohlbold TJ, Innis BL, Garcia-Sastre A, Palese P, Krammer F. 2020. Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates: interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect Dis 20:80–91. doi: 10.1016/S1473-3099(19)30393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bajic G, Maron MJ, Adachi Y, Onodera T, McCarthy KR, McGee CE, Sempowski GD, Takahashi Y, Kelsoe G, Kuraoka M, Schmidt AG. 2019. Influenza antigen engineering focuses immune responses to a subdominant but broadly protective viral epitope. Cell Host Microbe 25:827.e6–835.e6. doi: 10.1016/j.chom.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kanekiyo M, Joyce MG, Gillespie RA, Gallagher JR, Andrews SF, Yassine HM, Wheatley AK, Fisher BE, Ambrozak DR, Creanga A, Leung K, Yang ES, Boyoglu-Barnum S, Georgiev IS, Tsybovsky Y, Prabhakaran MS, Andersen H, Kong WP, Baxa U, Zephir KL, Ledgerwood JE, Koup RA, Kwong PD, Harris AK, McDermott AB, Mascola JR, Graham BS. 2019. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat Immunol 20:362–372. doi: 10.1038/s41590-018-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, Schief WR, Nemazee D, Crotty S. 2018. Precursor frequency and affinity determine B cell competitive fitness in germinal centers, tested with germline-targeting HIV vaccine immunogens. Immunity 48:133.e6–146.e6. doi: 10.1016/j.immuni.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Cirelli KM, Carnathan DG, Nogal B, Martin JT, Rodriguez OL, Upadhyay AA, Enemuo CA, Gebru EH, Choe Y, Viviano F, Nakao C, Pauthner MG, Reiss S, Cottrell CA, Smith ML, Bastidas R, Gibson W, Wolabaugh AN, Melo MB, Cossette B, Kumar V, Patel NB, Tokatlian T, Menis S, Kulp DW, Burton DR, Murrell B, Schief WR, Bosinger SE, Ward AB, Watson CT, Silvestri G, Irvine DJ, Crotty S. 2019. Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell 177:1153.e28–1171.e28. doi: 10.1016/j.cell.2019.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Keitel WA, Atmar RL, Nino D, Cate TR, Couch RB. 2008. Increasing doses of an inactivated influenza A/H1N1 vaccine induce increasing levels of cross-reacting antibody to subsequent, antigenically different, variants. J Infect Dis 198:1016–1018. doi: 10.1086/591465. [DOI] [PubMed] [Google Scholar]
- 202.Atmar RL, Keitel WA. 2020. Searching for improved flu vaccines - the time is now. J Infect Dis 221:1–4. doi: 10.1093/infdis/jiz545. [DOI] [PubMed] [Google Scholar]
- 203.Angeletti D, Kosik I, Santos JJS, Yewdell WT, Boudreau CM, Mallajosyula VVA, Mankowski MC, Chambers M, Prabhakaran M, Hickman HD, McDermott AB, Alter G, Chaudhuri J, Yewdell JW. 2019. Outflanking immunodominance to target subdominant broadly neutralizing epitopes. Proc Natl Acad Sci U S A 116:13474–13479. doi: 10.1073/pnas.1816300116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Klasse PJ, Ozorowski G, Sanders RW, Moore JP. 2020. Env exceptionalism: why are HIV-1 Env glycoproteins atypical immunogens? Cell Host Microbe 27:507–518. doi: 10.1016/j.chom.2020.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Davies HM, Nofal SD, McLaughlin EJ, Osborne AR. 2017. Repetitive sequences in malaria parasite proteins. FEMS Microbiol Rev 41:923–940. doi: 10.1093/femsre/fux046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Proft T, Baker EN. 2009. Pili in Gram-negative and Gram-positive bacteria - structure, assembly and their role in disease. Cell Mol Life Sci 66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner JS, Burlein J, Barren P, Koenig S, Leath S, Jones CH, Hultgren SJ. 1997. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 208.Kronvall G, Simmons A, Myhre EB, Jonsson S. 1979. Specific absorption of human serum albumin, immunoglobulin A, and immunoglobulin G with selected strains of group A and G streptococci. Infect Immun 25:1–10. doi: 10.1128/IAI.25.1.1-10.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Granoff DM, Giuntini S, Gowans FA, Lujan E, Sharkey K, Beernink PT. 2016. Enhanced protective antibody to a mutant meningococcal factor H-binding protein with low-factor H binding. JCI Insight 1:e88907. doi: 10.1172/jci.insight.88907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Granoff DM, Ram S, Beernink PT. 2013. Does binding of complement factor H to the meningococcal vaccine antigen, factor H binding protein, decrease protective serum antibody responses? Clin Vaccine Immunol 20:1099–1107. doi: 10.1128/CVI.00260-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Pentiah K, Lees WD, Moss DS, Shepherd AJ. 2015. N-Linked glycans on influenza A H3N2 hemagglutinin constrain binding of host antibodies, but shielding is limited. Glycobiology 25:124–132. doi: 10.1093/glycob/cwu097. [DOI] [PubMed] [Google Scholar]
- 212.Watanabe Y, Bowden TA, Wilson IA, Crispin M. 2019. Exploitation of glycosylation in enveloped virus pathobiology. Biochim Biophys Acta Gen Subj 1863:1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Zhou T, Zheng A, Baxa U, Chuang GY, Georgiev IS, Kong R, O'Dell S, Shahzad-Ul-Hussan S, Shen CH, Tsybovsky Y, Bailer RT, Gift SK, Louder MK, McKee K, Rawi R, Stevenson CH, Stewart-Jones GBE, Taft JD, Waltari E, Yang Y, Zhang B, Shivatare SS, Shivatare VS, Lee CD, Wu CY, NISC Comparative Sequencing Program, Mullikin JC, Bewley CA, Burton DR, Polonis VR, Shapiro L, Wong CH, Mascola JR, Kwong PD, Wu X. 2018. A neutralizing antibody recognizing primarily N-linked glycan targets the silent face of the HIV envelope. Immunity 48:500.e6–513.e6. doi: 10.1016/j.immuni.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Eggink D, Goff PH, Palese P. 2014. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J Virol 88:699–704. doi: 10.1128/JVI.02608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Andrews SF, Graham BS, Mascola JR, McDermott AB. 2018. Is it possible to develop a “universal” influenza virus vaccine? Immunogenetic considerations underlying B-cell biology in the development of a pan-subtype influenza A vaccine targeting the hemagglutinin stem. Cold Spring Harb Perspect Biol 10:a029413. doi: 10.1101/cshperspect.a029413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Finney J, Watanabe A, Kelsoe G, Kuraoka M. 2019. Minding the gap: the impact of B-cell tolerance on the microbial antibody repertoire. Immunol Rev 292:24–36. doi: 10.1111/imr.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Bajic G, van der Poel CE, Kuraoka M, Schmidt AG, Carroll MC, Kelsoe G, Harrison SC. 2019. Autoreactivity profiles of influenza hemagglutinin broadly neutralizing antibodies. Sci Rep 9:3492. doi: 10.1038/s41598-019-40175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Finlay BB, McFadden G. 2006. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 219.Dobzhansky T. 1973. Nothing in biology makes sense except in the light of evolution. Am Biol Teach 35:125–129. doi: 10.2307/4444260. [DOI] [Google Scholar]
- 220.Borst P. 1991. Molecular genetics of antigenic variation. Immunol Today 12:A29–33. doi: 10.1016/S0167-5699(05)80009-X. [DOI] [PubMed] [Google Scholar]
- 221.Lipsitch M, O'Hagan JJ. 2007. Patterns of antigenic diversity and the mechanisms that maintain them. J R Soc Interface 4:787–802. doi: 10.1098/rsif.2007.0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wahlgren M, Goel S, Akhouri RR. 2017. Variant surface antigens of Plasmodium falciparum and their roles in severe malaria. Nat Rev Microbiol 15:479–491. doi: 10.1038/nrmicro.2017.47. [DOI] [PubMed] [Google Scholar]
- 223.Laver WG, Air GM, Webster RG, Smith-Gill SJ. 1990. Epitopes on protein antigens: misconceptions and realities. Cell 61:553–556. doi: 10.1016/0092-8674(90)90464-p. [DOI] [PubMed] [Google Scholar]
- 224.Sok D, Burton DR. 2018. Recent progress in broadly neutralizing antibodies to HIV. Nat Immunol 19:1179–1188. doi: 10.1038/s41590-018-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Bianchi M, Turner HL, Nogal B, Cottrell CA, Oyen D, Pauthner M, Bastidas R, Nedellec R, McCoy LE, Wilson IA, Burton DR, Ward AB, Hangartner L. 2018. Electron-microscopy-based epitope mapping defines specificities of polyclonal antibodies elicited during HIV-1 BG505 envelope trimer immunization. Immunity 49:288.e8–300.e8. doi: 10.1016/j.immuni.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Nogal B, Bianchi M, Cottrell CA, Kirchdoerfer RN, Sewall LM, Turner HL, Zhao F, Sok D, Burton DR, Hangartner L, Ward AB. 2020. Mapping polyclonal antibody responses in non-human primates vaccinated with HIV Env trimer subunit vaccines. Cell Rep 30:3755.e7–3765.e7. doi: 10.1016/j.celrep.2020.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Burton DR. 2019. Advancing an HIV vaccine; advancing vaccinology. Nat Rev Immunol 19:77–78. doi: 10.1038/s41577-018-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Graham BS, Gilman MSA, McLellan JS. 2019. Structure-based vaccine antigen design. Annu Rev Med 70:91–104. doi: 10.1146/annurev-med-121217-094234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Weidenbacher PA, Kim PS. 2019. Protect, modify, deprotect (PMD): a strategy for creating vaccines to elicit antibodies targeting a specific epitope. Proc Natl Acad Sci U S A 116:9947–9952. doi: 10.1073/pnas.1822062116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, Galeotti CL, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood DW, Jeffries AC, Saunders NJ, Granoff DM, Venter JC, Moxon ER, Grandi G, Rappuoli R. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 231.Guthmiller JJ, Wilson PC. 2018. Harnessing immune history to combat influenza viruses. Curr Opin Immunol 53:187–195. doi: 10.1016/j.coi.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 232.Yewdell JW, Santos JJS. 21 January 2020. Original antigenic sin: how original? How sinful? Cold Spring Harb Perspect Med doi: 10.1101/cshperspect.a038786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Modis Y, Ogata S, Clements D, Harrison SC. 2005. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol 79:1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Blum JS, Wearsch PA, Cresswell P. 2013. Pathways of antigen processing. Annu Rev Immunol 31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]