Abstract
Nidogen 1 (NID1) is a glycoprotein found in basement membranes involved in cross-linking collagen IV and laminin. The role of NID in breast cancer has only been evaluated in a small number of studies and the findings of these studies have been inconsistent. Our previous work revealed that highly tumorigenic murine mammary tumor cells express high levels of Nid1 while weakly tumorigenic mammary tumor cells express low levels of Nid1. To investigate Nid1, two stable knockdown lines were created, and Nid1 knockdown was confirmed at both the mRNA and protein level. Nid1 knockdown significantly reduced cell proliferation and migration/invasion and these reductions in proliferation and migration/invasion could be rescued by conditioned media containing NID1 protein. The reduced migration/invasion observed in the Nid1 knockdown cells was not associated with significant alterations in the epithelial gene Cdh1 or the mesenchymal genes Snai1, Snai2, Twist1, Twist2, Zeb1 and Zeb2. Therefore, suppression of Nid1 expression reduces proliferation and migration/invasion in claudin-low murine mammary tumor cells.
Keywords: breast cancer, nidogen 1, migration, invasion, claudin-low, proliferation
Introduction
The basement membrane is a highly specialized extracellular matrix (ECM) that is located around all cell monolayers and multi-layered epithelia within the body (1–4). Its functions include structural support, regulation of cell signalling behaviors and providing a protective barrier. The main components of basement membranes include collagen IV, laminin, perlecan and nidogen. Of these components, it is known that collagen IV and laminin can self-assemble into superstructures individually, which plays a vital role in the stability of BMs. Perlecan and nidogen (NID) influence the structural integrity through connecting the collagen IV and laminin networks, increasing stability (1–4). NID, also known as entactin, mimics the function of perlecan by connecting the superstructures of collagen IV and laminin (4). Collagen IV contains a triple helical domain that fuses to the G2 domain of NID, while the γ-3 chain of laminin binds to the G3 domain of NID, effectively linking the two components (1,2,4).
Only a small number of studies have investigated the impact of NID1 in cancer with only 3 studies evaluating breast cancer (5–7). A study by Alečković et al (5) performed a secretome analysis of lung metastatic sublines of the human breast cancer cell line MDA-MB-231 and identified NID1 as potential regulator of metastasis. The importance of NID1 in lung cancer metastasis was further examined in MDA-MB-231 overexpressing NID1. Overexpression of NID1 increased lung metastases 10-fold and reduced survival in an experimental metastasis assay as well as promoting invasion and migration in a transwell assay (5). These investigators also showed that NID1 expression in breast cancer patients was associated with lung metastasis and poor patient prognosis (5). However, a recent study by Ferraro et al (7) showed the opposite; secretion of NID1 from endothelial cells inhibited the migration of the human breast cancer cell line SK-BR-3. Therefore, the impact of NID1 on breast cancer migration is inconclusive. The third study evaluating the function of NID1 in breast cancer found that a disintegrin and metalloproteinase with thrombospondin repeats 1 (ADAMTS1) can cleave NID1 inhibiting its function. ADAMTS1 levels were shown to be reduced in breast cancer samples and this was associated with reduced NID1 proteolysis and increased deposition of NID1 into tumor blood vessels (6).
A previous study from our lab looking at miR-200 levels found that re-expression of the miR-200b/200a/429 cluster impaired tumor growth and metastasis (8). RNA-sequencing revealed that Nid1 was the most significantly reduced gene in miR-200 transfected cells compared to control cells (8). Given that a higher Nid1 expression was associated with increased tumorigenicity and metastatic capacity this study evaluated the impact of Nid1 on mammary tumor cell proliferation and migration/invasion.
Materials and methods
Cell lines
RJ423 cells have been previously described (8–11). To reduce Nid1 expression, RJ423 cells were transfected with four unique shRNAs targeting Nid1 (cat. no. TL501474; Origene Technologies Inc.) or a control shRNA (cat. no. TR30021; Origene Technologies Inc.). Stable clones were selected using 5 µg/ml puromycin (InvivoGen). Two knockdown lines (RJ423shNid1A and RJ423shNid1D) were identified as having the greatest Nid1 mRNA knockdown relative to a control line (RJ423con). All cells were cultured as described in (8). RJ423shNid1A, RJ423shNid1D and RJ423con cells were maintained in 5 µg/ml puromycin.
RNA isolation and RT-qPCR
RNA isolation and RT-qPCR were performed as previously described (8). Primers for Nid1 (qMmuCID0018281), Cdh1 (qMmuCED0044197), Vim (qMmuCID0005527), Snai1 (qMmuCID0024342), Snai2 (qMmuCED0046072), Twist1 (qMmuCED0004065), Twist2 (qMmuCID0009652), Zeb1 (qMmuCID0009095), Zeb2 (qMmuCID0014662) and Hprt (qMmuCID0005679) were purchased from Bio-Rad Laboratories (Canada) Ltd. Gene expression was quantified and normalized to Hprt using the normalized expression (ΔΔCq) (12) mode in CFX-Manager 3.1 software [Bio-Rad Laboratories (Canada) Ltd.].
Collection of conditioned media
Cells were seeded in 10 mm culture dishes at a density of 2×106 cells per dish overnight in fully supplemented media. The next day, the fully supplemented media was removed, the cells were washed twice with phosphate-buffered saline (PBS) and 10 ml of serum-free media containing sodium pyruvate, glutamine and antibiotic-antimycotic was added to each dish. Twenty-four hours later, conditioned media was collected and either used immediately or stored at −20°C.
Western blotting
Western blotting was performed as previously described (13). NID1 protein was detected using a primary antibody against NID1 (cat. no. NBP1-97701; Novus Biologicals) at a 1:200 dilution while β-actin was detected using a 1:5,000 dilution of a primary antibody against β-actin (cat. no. 8457; Cell Signaling Technologies, Inc.). The appropriate HRP-linked secondary antibody (Cell Signaling Technology, Inc.) was used at a 1:2,000 dilution.
Cell proliferation
Immunofluorescence for phospho-histone H3 (PH3) was used to determine cell proliferation as previously described (8). For the conditioned media experiments, cells were allowed to attach to the coverslips overnight in fully supplemented media. The next day, the fully supplemented media was removed, the cells washed twice with PBS, and then the cells were cultured in conditioned media for 24 h prior to assessing proliferation.
Cell apoptosis
Apoptosis was assessed using the Annexin V-FITC Apoptosis Detection kit (eBioscience) according to the manufacturer's protocol. A BD Accuri C6 flow cytometer (BD Biosciences) was used to measure fluorescence at a flow rate less than 400 events/second and analyzed using the Accuri C6 software (BD Biosciences).
Transwell invasion chamber assay
Transwell assays were performed as previously described (8) with minor alterations. Since Matrigel contains NID1, the Cultrex® 24 Well Collagen IV Cell Invasion Assay kit (cat. no. 3458-024-K) was used for the transwell inserts. The amount of cell migration was analyzed using ImageJ software (National Institutes of Health). To determine whether NID1 served as a chemoattractant, cells were cultured in serum-free media in the upper chamber with conditioned media from the various cell lines in the lower chamber. To determine whether NID1 directly interacted with cells to alter migration, cells were cultured in conditioned media in the upper chamber and fully supplemented media was placed in the lower chamber.
Statistical analysis
Statistical analysis was conducted using one-way ANOVA followed by Tukey's test using GraphPad Prizm 8 (GraphPad Software, Inc.). Statistical significance (P<0.05) is indicated by asterisks.
Results
Nid1 shRNA reduces NID1 protein levels in conditioned media
Our previous worked revealed that re-expressing the miR-200b/200a/429 cluster in murine mammary tumor cells (RJ423-200ba429) suppressed tumor growth and metastasis in vivo (8). RNA sequencing of the RJ423-200ba429 cells revealed that Nid1 was the most significant, downregulated gene (Nid1 expression was decreased 3,821-fold, P-value 3.5×10−268, GSE113162). Consistent with this finding, Nid1 was expression was reduced 6,654-fold (P-value 3.5×10−240) in the poorly tumorigenic, non-metastatic murine mammary tumor cell line RJ345 compared to the highly tumorigenic, highly metastatic murine mammary tumor cell line RJ423. Since both RJ345 cells and RJ423-200ba429 cells are poorly tumorigenic, non-metastatic (8) and express almost no Nid1 (Table I) this study examined whether reducing Nid1 in RJ423 cells could negatively impact proliferation and/or migration.
Table I.
Characteristics of murine mammary tumor cells.
| Cell lines | miR-200 expression | Nid1 expression | Tumorigenicity | Metastatic |
|---|---|---|---|---|
| RJ345 | High | Low | Low | No |
| RJ423 | Low | High | High | Yes |
| RJ423-200ba429 | High | Low | Low | No |
| RJ423EV | Low | High | High | Yes |
miR-200, microRNA-200; Nid1, nidogen 1.
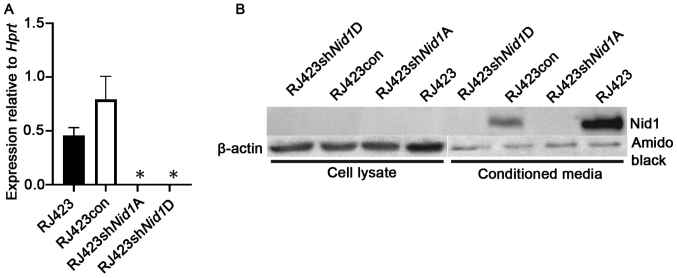
To reduce the expression of Nid1, RJ423 cells were stably transfected with 4 unique shRNA constructs (A-D) targeting mouse Nid1 or a control shRNA construct. The two most effective Nid1 shRNA constructs, shNid1A and shNid1D, reduced. Nid1 mRNA levels over 99% compared to RJ423 cells stably expressing the control shRNA construct (RJ423con; Fig. 1A).
Figure 1.
Nid1 levels in cells stably expressing Nid1 shRNA. (A) Expression levels of Nid1 mRNA relative to Hprt in parental RJ423, RJ423con, RJ423Nid1A and RJ423shNid1D cells. The bars represent the mean value and the error bars represent the SEM. *Indicates significant difference (P<0.05) from RJ423con cells (n=6). (B) Western blot analysis of NID1 protein from cell lysates and conditioned media from RJ423, RJ423con, RJ423shNid1A and RJ423shNid1D cells. β-actin was used as a loading control for the cell lysates while amido black was used to ensure similar loading for the conditioned media samples. Nid1, nidogen 1; Hprt, hypoxanthine phosphoribosyltransferase; con, control; sh, short hairpin RNA.
To confirm NID1 was suppressed at the protein level western blotting was performed on cell lysates from RJ423, RJ423con, RJ423shNid1A and RJ423shNid1D cells. The NID1 antibody failed to detect NID1 protein in any of the cell lysates (Fig. 1B). Since NID1 is a secreted protein conditioned media was collected from all four cell lines. As shown in Fig. 1B, NID1 was detected in the condition media from RJ423 cells and RJ423con cells but not from either RJ423shNid1A or RJ423shNid1D cells.
Nid1 knockdown inhibits proliferation but not apoptosis
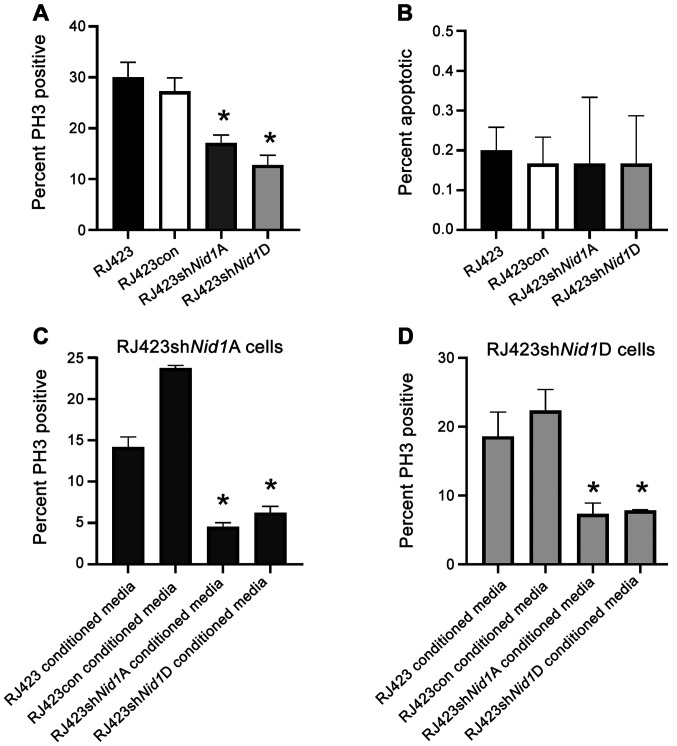
The impact of Nid1 suppression on cell proliferation was assessed using PH3 immunofluorescence. Both Nid1 shRNA constructs significantly reduced proliferation compared to the control cell line, RJ423con (Fig. 2A). The impact of Nid1 suppression on apoptosis was evaluated using Annexin V flow cytometry. RJ423, RJ423con, RJ4223shNid1A and RJ423shNid1D cells all had very low levels of apoptosis (<1%) and this level of basal apoptosis when cultured in fully supplemented media (Fig. 2B). Since there was no significant difference in apoptosis this cell response was not further evaluated.
Figure 2.
Impact of NID1 conditioned media on cell proliferation and apoptosis. (A) Percentage of PH3-positive cells in parental RJ423, RJ423con, RJ423shNid1A and RJ423shNid1D cells. (B) Percentage of apoptotic cells in RJ423, RJ423con, RJ423Nid1A and RJ423Nid1D cells. Percentage of PH3-positive cells from (C) RJ423shNid1A and (D) RJ423shNid1D cells treated with conditioned media from RJ423, RJ423con, RJ423shNid1A or RJ423shNid1D cells for 24 h. *P<0.05 vs. RJ423con cells (n=3). Nid1, nidogen 1; con, control; sh, short hairpin RNA; PH3, phospho-histone H3.
To confirm the decrease in proliferation resulted from Nid1 suppression, RJ423shNid1A and RJ423shNid1D cells were cultured in the presence of conditioned media from RJ423 cells, RJ423con cells, their own conditioned media and conditioned media from the other knockdown line. It was anticipated that conditioned media from RJ423 or RJ423con cells would increase proliferation in the Nid1 knockdown cells as both RJ423 and RJ423con cells secrete NID1 into the media. As shown in Fig. 2C and D proliferation of RJ423shNid1A and RJ423shNid1D cells was significantly increased when the cells were grown in conditioned media from RJ423con cells compared to when these cells were grown in conditioned media from either Nid1 knockdown cell line. Proliferation of RJ423shNid1A cells was also significantly increased when the cells were grown in conditioned media from RJ423 cells compared to when these cells were grown in conditioned media from either Nid1 knockdown cell line while the proliferation in the RJ423shNid1D cells was close to being significant when these cells were grown in condition media from RJ423 cells compared to condition media from RJ423shNid1A (P=0.058) or RJ423shNid1D (P=0.081) cells.
Nid1 knockdown impairs cell migration
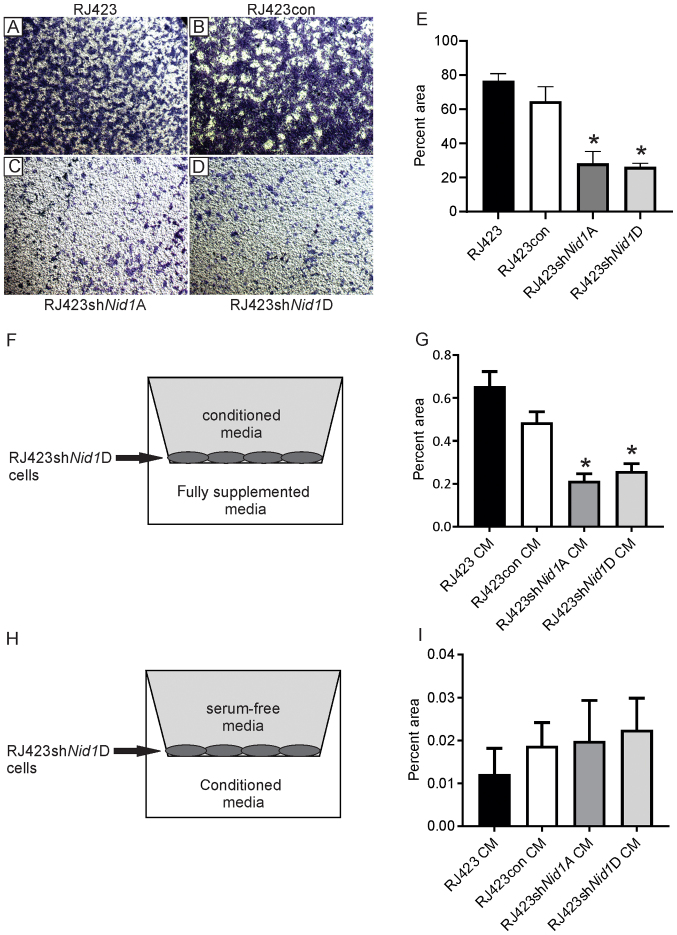
Next, migration and invasion were assessed using a transwell assay. The transwell inserts were coated with collagen IV rather than Matrigel as Matrigel contains NID1. Fig. 3A-D shows representative images of the stained cells that invaded and migrated through the collagen IV coated inserts. Quantification of these images (Fig. 3E) revealed that RJ423shNid1A and RJ423shNid1D cells were significantly less effective at invading and migrating than either the RJ423 or RJ423con cells.
Figure 3.
Impact of NID1 conditioned media on cell migration. Representative images of toluidine blue stained cells on the bottom of collagen-coated transwell inserts for (A) parental RJ423, (B) RJ423con, (C) RJ423shNid1A and (D) RJ423shNid1D cells (magnification, ×4). (E) Quantification of the percent area occupied by parental RJ423, RJ423con, RJ423shNid1A and RJ423shNid1D cells. Migration and invasion of RJ423shNid1D cells when conditioned media from RJ423, RJ423con, RJ423shNid1A or RJ423shNid1D cells were placed (F and G) in the upper chamber of the well with the cells or (H and I) in the lower chamber. (F and H) Schematic indicating the location of the conditioned media and cells. (G and I) Quantitative data from these experiments. *P<0.05 vs. RJ423con cells (n=3). Nid1, nidogen 1; con, control; sh, short hairpin RNA; CM, conditioned media.
To confirm that migration and invasion was dependent on NID1, migration of RJ423shNid1D cells was assessed using conditioned media from RJ423, RJ423con, RJ423shNid1A or RJ423shNid1D cells in the upper chamber and fully supplemented media in the lower chamber (Fig. 3F). The presence of conditioned media from RJ423 and RJ423con cells significantly increased invasion and migration of RJ423shNid1D cells compared the conditioned media from either RJ423shNid1A or RJ423shNid1D cells (Fig. 3G).
To assess whether NID1 served as a chemoattractant, RJ423shNid1D cells were grown in the upper chamber in serum-free media and conditioned media from RJ423, RJ423con, RJ423shNid1A or RJ423shNid1D cells was placed in the lower chamber (Fig. 3H). As shown in Fig. 3I, RJ423shNid1A cells invaded and migrated very poorly independent of the conditioned media used as a chemoattractant and there were no significant differences observed.
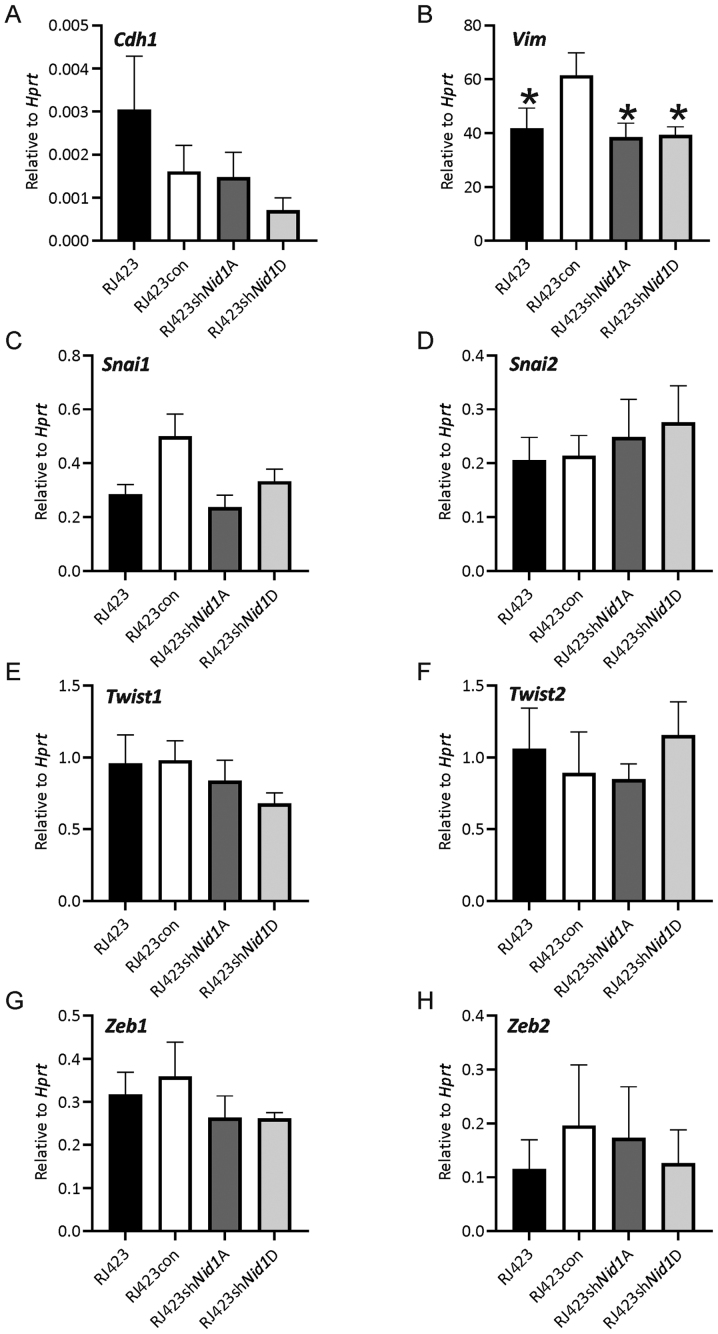
Enhanced migration and invasion are often associated with increased expression of mesenchymal genes and loss of epithelial genes in a process known as epithelial to mesenchymal transition (EMT). It was anticipated that one or more mesenchymal genes would be expressed at significantly reduced levels or an increase in expression of the epithelial gene, Cdh1, would be observed in the Nid1 knockdown lines compared to RJ423con and RJ423 parental cells. The only gene significantly altered was Vim in the Nid1 knockdown lines, however Vim expression in the Nid1 knockdown lines was almost identical to the parental RJ423 cells (Fig. 4).
Figure 4.
Expression levels of epithelial and mesenchymal genes following Nid1 knockdown. mRNA expression levels of (A) Cdh1, (B) Vim, (C) Snai1, (D) Snai2, (E) Twist1, (F) Twist2, (G) Zeb1 or (H) Zeb2 relative to Hprt. *P<0.05 vs. RJ423con cells (n=5). Nid1, nidogen 1; Hprt, hypoxanthine phosphoribosyltransferase; con, control; sh, short hairpin RNA.
Discussion
Our previous work looking at highly tumorigenic and metastatic murine mammary tumor cells compared to weakly tumorigenic, non-metastatic tumor cells identified Nid1 as a potential promoter of tumor development and metastasis (8). RJ423 cells are murine mammary tumor cells with characteristics of human claudin-low breast cancer that express high levels of Nid1. RJ423 cells rapidly form tumors when injected into the mammary fat pad of mice and induce lung metastases in mice following tail vein injection (8).
Suppression of Nid1 expression in RJ423 cells significantly reduced cell proliferation and this decrease in proliferation was rescued by incubating cells in conditioned media containing NID1. While it is possible that the conditioned media from RJ423 and RJ423con cells contain proteins other than NID1, presumably the conditioned media from RJ423con and RJ423shNid1A/RJ423shNid1D cells should be highly similar as the only difference between these cells was the stable knockdown of Nid1. While treating the cells with recombinant NID1 would confirm the impact of NID1 on proliferation, commercially available, full-length recombinant murine NID1 could not be found. In a study by Alečković et al (5), proliferation of MDA-MB-231 cells overexpressing NID1 cultured in vitro was not examined. However, this group did assess tumor size in vivo and found that overexpression of NID1 in MDA-MB-231 did not increase primary tumor growth. No other study investigated the impact of NID1 on breast cancer cell proliferation in vitro.
NID1 did not impact basal levels of apoptosis as RJ423con, RJ423shNid1A, and RJ423shNid1D cells. As no study has investigated the impact of NID1 on breast cancer cell apoptosis this is the first study showing that a reduction of NID1 does not increase mammary tumor cell apoptosis.
Reducing NID1 levels significantly impaired migration and invasion through collagen IV coated membranes and migration/invasion could be rescued when conditioned media containing NID1 was placed in the upper chamber with the RJ423shNid1D cells but not when the conditioned media containing NID1 was placed in the lower chamber. These findings suggest that NID1 is interacting directly with the cancer cells and not acting as a chemoattractant. It is not clear how NID1 alters cell migration and invasion, but it could be through binding to receptors on the cell and influencing signaling. NID1 has been reported to bind to integrins αvβ3 and α3β1 (14–16) and RNA sequencing by our lab has shown the presence of Itga3, Itgb3, and Itgb1 but not Itgav in RJ423 cells (GSE113162). Therefore, NID1 is presumably signaling through integrin α3β1 or another receptor that has yet to be identified. The observation that NID1 promotes migration and invasion is consistent with the findings of Alečković et al (5) who showed that MDA-MB-231 cells overexpressing NID1 increased migration and invasion in vitro and lung metastatic colonization in vivo. However, Ferraro et al (7) found that NID1 secreted by endothelial cells inhibited invasion a migration of the human breast cancer cell line, SK-BR-3. The main difference between the study by Ferraro et al (7) and our study and the study by Alečković et al (5) was that while the Ferraro et al (7) study utilized breast cancer cells with HER2 overexpression while our study (9) and the study by Alečković et al (5) used tumor cells with characteristics of claudin-low breast cancer. Thus, NID1 may promote migration and invasion of claudin-low breast cancer cells but not in other breast cancer subtypes.
Enhanced invasion and migration are often accompanied by an increase in mesenchymal gene expression, reduced epithelial gene expression and a more spindled-shape morphology. Moreover, NID1 has been implicated in inducing EMT in colorectal cancer cells (17) and ovarian cancer cells (18) but has not been investigated in breast cancer. No consistent increase in mesenchymal gene expression or decrease in epithelial gene expression was observed in either RJ423shNid1A or RJ423shNid1D cells compared to RJ423con cells suggesting that NID1 cannot restore epithelial gene expression patterns in claudin-low mammary tumor cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by a CIHR operating grant (MOP-136970) and a CIHR project grant (PJT-162218) to RAM.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113162
Authors' contributions
RJ performed reverse transcription-quantitative PCR, western blotting for NID1, proliferation assays, apoptosis assays and Transwell migration assays. CJM performed reverse transcription-quantitative PCR on some of the samples for Nid1 and some of the epithelial and mesenchymal genes. RAM created the cell lines, edited the manuscript and ran the project. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jayadev R, Sherwood DR. Basement membranes. Curr Biol. 2017;27:R207–R211. doi: 10.1016/j.cub.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 2.LeBleu VS, Macdonald B, Kalluri R. Structure and function of basement membranes. Exp Biol Med (Maywood) 2007;232:1121–1129. doi: 10.3181/0703-MR-72. [DOI] [PubMed] [Google Scholar]
- 3.Chang J, Chaudhuri O. Beyond proteases: Basement membrane mechanics and cancer invasion. J Cell Biol. 2019;218:2456–2469. doi: 10.1083/jcb.201903066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yurchenco PD. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol. 2011;3:a004911. doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alečković M, Wei Y, LeRoy G, Sidoli S, Liu DD, Garcia BA, Kang Y. Identification of Nidogen 1 as a lung metastasis protein through secretome analysis. Genes Dev. 2017;31:1439–1455. doi: 10.1101/gad.301937.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martino-Echarri E, Fernández-Rodríguez R, Rodríguez-Baena FJ, Barrientos-Durán A, Torres-Collado AX, Plaza-Calonge Mdel C, Amador-Cubero S, Cortés J, Reynolds LE, Hodivala-Dilke KM, Rodríguez-Manzaneque JC. Contribution of ADAMTS1 as a tumor suppressor gene in human breast carcinoma. Linking its tumor inhibitory properties to its proteolytic activity on nidogen-1 and nidogen-2. Int J Cancer. 2013;133:2315–2324. doi: 10.1002/ijc.28271. [DOI] [PubMed] [Google Scholar]
- 7.Ferraro DA, Patella F, Zanivan S, Donato C, Aceto N, Giannotta M, Dejana E, Diepenbruck M, Christofori G, Buess M. Endothelial cell-derived nidogen-1 inhibits migration of SK-BR-3 breast cancer cells. BMC Cancer. 2019;19:312. doi: 10.1186/s12885-019-5521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watson KL, Jones RA, Bruce A, Moorehead RA. The miR-200b/200a/429 cluster prevents metastasis and induces dormancy in a murine claudin-low mammary tumor cell line. Exp Cell Res. 2018;369:17–26. doi: 10.1016/j.yexcr.2018.04.024. [DOI] [PubMed] [Google Scholar]
- 9.Jones R, Watson K, Bruce A, Nersesian S, Kitz J, Moorehead R. Re-expression of miR-200c suppresses proliferation, colony formation and in vivo tumor growth of murine claudin-low mammary tumor cells. Oncotarget. 2017;8:23727–23749. doi: 10.18632/oncotarget.15829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell CI, Moorehead RA. Mammary tumors that become independent of the type I insulin-like growth factor receptor express elevated levels of platelet-derived growth factor receptors. BMC Cancer. 2011;11:480. doi: 10.1186/1471-2407-11-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell CI, Thompson DE, Siwicky MD, Moorehead RA. Murine mammary tumor cells with a claudin-low genotype. Cancer Cell Int. 2011;11:28. doi: 10.1186/1475-2867-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Chorner PM, Moorehead RA. A-674563, a putative AKT1 inhibitor that also suppresses CDK2 activity, inhibits human NSCLC cell growth more effectively than the pan-AKT inhibitor, MK-2206. PLoS One. 2018;13:e0193344. doi: 10.1371/journal.pone.0193344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dedhar S, Jewell K, Rojiani M, Gray V. The receptor for the basement membrane glycoprotein entactin is the integrin alpha 3/beta 1. J Biol Chem. 1992;267:18908–18914. [PubMed] [Google Scholar]
- 15.Dong LJ, Hsieh JC, Chung AE. Two distinct cell attachment sites in entactin are revealed by amino acid substitutions and deletion of the RGD sequence in the cysteine-rich epidermal growth factor repeat 2. J Biol Chem. 1995;270:15838–15843. doi: 10.1074/jbc.270.26.15838. [DOI] [PubMed] [Google Scholar]
- 16.Yi XY, Wayner EA, Kim Y, Fish AJ. Adhesion of cultured human kidney mesangial cells to native entactin: Role of integrin receptors. Cell Adhes Commun. 1998;5:237–248. doi: 10.3109/15419069809040294. [DOI] [PubMed] [Google Scholar]
- 17.Rokavec M, Bouznad N, Hermeking H. Paracrine induction of epithelial-mesenchymal transition between colorectal cancer cells and its suppression by a p53/miR-192/215/NID1 axis. Cell Mol Gastroenterol Hepatol. 2019;7:783–802. doi: 10.1016/j.jcmgh.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Zhu Y, Fan X, Zhang C, Wang Y, Zhang L, Zhang H, Wen T, Zhang K, Huo X, et al. NID1, a new regulator of EMT required for metastasis and chemoresistance of ovarian cancer cells. Oncotarget. 2017;8:33110–33121. doi: 10.18632/oncotarget.16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113162