Abstract
The tumor microenvironment in hepatocellular carcinoma can be classified into cellular and non-cellular components. Myeloid-derived suppressor cells (MDSCs) are cellular components of this microenvironment that serve an important role in the progression of hepatocellular carcinoma. Fibrinogen-like protein 2 (FGL2) has been demonstrated to promote tumor progression by regulating cellular components of the tumor microenvironment in various types of malignant tumor. The present study aimed to determine the expression of FGL2 in hepatocellular carcinoma and its effect on the tumor microenvironment in order to determine novel targets for liver cancer treatment. Immunohistochemistry and reverse transcription quantitative PCR were performed to determine the expression level of FGL2 and the correlation with surface markers of human MDSCs in hepatocellular carcinoma. Furthermore, a mouse hepatocellular carcinoma cell line overexpressing FGL2 was established by stable transfection of a lentivirus expressing FGL2. In addition, fresh bone marrow cells extracted from mouse femurs were in vitro cultured using conditioned medium derived from the cell line overexpressing FGL2. An orthotopic hepatocellular carcinoma mouse model was also established. The results demonstrated that FGL2 expression level in hepatocellular carcinoma tissues was closely associated with tumor size. FGL2 level was positively correlated with the expression level of the MDSC surface markers CD11b and CD33 in hepatocellular carcinoma. The in vitro results demonstrated that FGL2 could maintain the undifferentiated state of bone marrow cells, therefore promoting MDSC accumulation. Furthermore, in the orthotopic hepatocellular carcinoma mouse model, we observed that overexpression of FGL2 could promote tumor growth and significantly increase the number of MDSCs in the tumors and spleen. Taken together, these findings suggested that FGL2 may promote hepatocellular carcinoma tumor growth by promoting the accumulation of MDSCs in the tumor microenvironment.
Keywords: hepatocellular carcinoma, fibrinogen-like protein 2, myeloid-derived suppressor cells, orthotopic hepatocellular carcinoma model
Introduction
Primary liver cancer is a common malignant tumor in China (1). Liver cancer can be divided into three types as follows: Hepatocellular carcinoma, bile duct cell carcinoma and a mixture of both types. Hepatocellular carcinoma represents 70–90% of all primary liver cancers worldwide. Primary liver cancer is the second leading cause of tumor-associated mortality worldwide and in China. The main cause is chronic infection of hepatitis B virus (HBV) and hepatitis C virus (HCV) (2).
The tumor microenvironment is the major site for tumor cell proliferation and differentiation. Previous studies have demonstrated that the tumor microenvironment significantly affects tumorigenesis and development, metastasis, and response to immunotherapy and chemotherapy (3,4). The hepatocellular carcinoma tumor microenvironment is divided into cellular and non-cellular components (5). The cellular components of this microenvironment include liver sinusoidal endothelial cells and hepatic stellate cells (6,7). In addition, infiltrating immune cells are present in hepatocellular carcinoma, and mainly include neutrophils, lymphocytes, tumor-associated macrophages, dendritic cells and myeloid-derived suppressor cells (8–10).
Myeloid-derived suppressor cells (MDSCs) form a group of heterogeneous cells composed of bone marrow progenitor cells and immature bone marrow cells. These cells are precursors of dendritic cells, granulocytes and macrophages. In healthy individuals, immature bone marrow cells produced by the bone marrow can rapidly differentiate into mature dendritic cells, macrophages and granulocytes (11). However, under certain pathological conditions, such as cancer, infection, inflammation, sepsis, trauma, transplantation or autoimmune diseases, the differentiation of immature bone marrow cells into mature bone marrow cells is blocked (12,13). This phenomenon eventually leads to the expansion of MDSCs.
In humans, MDSCs express CD11b and CD33 on their surface, whereas in mouse, MDSCs express CD11b and glucocorticoid receptor 1 Gr-1, which includes two types, Ly6G and Ly6C (14,15). MDSCs express high levels of the immunosuppressive molecule arginase 1 (Arg-1) and inducible nitric oxide synthase (iNOS) that exert immunosuppressive functions in humans and mice (16,17). MDSCs are one of the main components of the tumor microenvironment. They migrate to peripheral lymphoid organs, such as the spleen, to infiltrate into the tumor, contributing therefore to the formation of the tumor microenvironment (18,19).
Fibrinogen-like protein 2 (FGL2) comprises two forms, a membrane-associated form and a secreted form (20,21). The cleavage of the amino-terminal hydrophobic segment results in the secreted form of FGL2 that has immunoregulatory functions. The carboxyl terminus of FGL2 contains a fibrinogen-related domain (FRED) that is similar to the β and γ chains of fibrinogen (22). This domain has been demonstrated to have immunoregulatory effects (23). The immunoregulatory function of FGL2 serves crucial roles in certain diseases, including viral hepatitis, transplant rejection and cancers (24). Previous studies have reported that FGL2 is involved in the promotion of tumor progression by regulating the cellular components of the tumor microenvironment in some types of cancer, including renal clear cell carcinoma, gliomas and lung cancer (25–27). However, the expression of FGL2 in hepatocellular carcinomas and its effect on the tumor microenvironment remain unknown.
Materials and methods
Clinical samples and clinicopathological data
A total of 40 pairs of hepatocellular carcinoma cancer and peritumor fresh and FFPE tissue samples were collected between October 2017 and December 2017 from patients who were pathologically diagnosed with hepatocellular carcinoma at The First Hospital of China Medical University. The clinicopathological characteristics of patients are presented in Table I. The age of the patients ranged from 39 to 72 years, and the mean age was 54 years. All clinical samples were collected after obtaining written informed consent from patients. The study was approved by the Ethics Committee of the China Medical University.
Table I.
Association between FGL2 expression level and the clinicopathological characteristics of patients with hepatocellular carcinoma.
| Clinicopathological characteristics | Number of patients | Low/medium expression of FGL2 | High expression of FGL2 | P-value |
|---|---|---|---|---|
| Total cases | 40 | 23 | 17 | |
| Age, years | ||||
| ≤53 | 22 | 11 | 11 | 0.348 |
| >53 | 18 | 12 | 6 | |
| Sex | ||||
| Male | 32 | 20 | 12 | 0.250 |
| Female | 8 | 3 | 5 | |
| Tumor size, cm | ||||
| ≤5 | 23 | 17 | 6 | 0.024a |
| >5 | 17 | 6 | 11 | |
| Number of tumors | ||||
| Single | 28 | 16 | 12 | 1.000 |
| Multiple | 12 | 7 | 5 | |
| HBsAg expression | ||||
| + | 30 | 16 | 14 | 0.471 |
| – | 10 | 7 | 3 | |
| Cirrhosis | ||||
| + | 32 | 19 | 13 | 0.702 |
| – | 8 | 4 | 4 | |
| AFP level, ng/dl | ||||
| ≤200 | 19 | 13 | 6 | 0.216 |
| >200 | 21 | 10 | 11 | |
| TNM stage | ||||
| I | 23 | 14 | 9 | 0.749 |
| II–IV | 17 | 9 | 8 | |
P<0.05. +, positive; -, negative; AFP, alpha-fetoprotein; HBsAg, hepatitis B surface antigen; TNM, tumor-node-metastasis.
Immunohistochemistry
Serial pathological tissue sections (5 µm-thick) were dewaxed using xylene and rinsed with gradient ethanol (100, 95, 90, 80 and 70%, 5 min each). Antigen retrieval was performed using citrate buffer at high pressure. Sections were incubated with rabbit anti-FGL2 polyclonal antibody (1:100; cat. no. ab198029; Abcam) overnight at 4°C, washed with PBS and incubated with secondary antibody (goat anti-rabbit; ready to use; MaxVision; cat. no. KIT-5004) for 30 min at room temperature. Streptavidin-peroxidase solution (ready to use; MaxVision; cat. no. KIT-5004) was then added and incubated for 30 min at room temperature. Sections were color developed using DAB, stained with hematoxylin for 5 min at room temperature and dried. Transparent sections were neutral resin counterstained and imaged using a microscope (magnification, ×200; Nikon Eclipse Ti-S; Nikon Corporation). The expression level of FGL2 was evaluated using the Engers scoring method (28). In addition, tumor tissue cases were divided into high and low/medium groups according to the median expression level using the Engers scoring method.
Reverse transcription quantitative (RT-q) PCR
TRIZOL (Invitrogen; Thermo Fisher Scientific, Inc.) were used to extract total RNA from human hepatocellular carcinoma tissues. cDNA was generated using 5× Prime Script RT Master Mix (Takara Bio, Inc.) on PCR System 9700 (GeneAmp) under the following conditions: 37°C for 15 min, 85°C for 5 sec and 4°C. The sequences of the primers were as follows: FGL2, forward, 5′-ACAAAGGTGTCCGTAATGGG-3′ and reverse, 5′-GTTTGAAGGAGGACTTGTAGCC-3′; CD11b, forward, 5′-CTGTTTACCTGTTTCACGGAAC-3′ and reverse, 5′-GATTGCCTTGACTCTCAGTACT-3′; and CD33, forward, 5′-CGATCTTCTCCTGGTTGTCAG-3′ and reverse, 5′-GATGGTTCTCTCCGCGTAGTCAC-3′. Human GAPDH Endogenous Reference Genes Primers (cat. no. B662104; Sangon Biotech Co., Ltd.) and Mouse GAPDH Endogenous Reference Genes Primers (cat. no. B662304; Sangon Biotech Co, Ltd.) were used as the internal controls. The mRNA expression was determined using TB Green Premix Ex TaqII (Takara Bio, Inc.) on Applied Biosystems® 7500 (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the following thermocycling conditions: 95°C for 30 sec, followed by 40 cycles of 95°C for 3 sec and 60°C for 30 sec. The relative expression levels were normalized to endogenous control and were expressed as 2−ΔCt or 2−ΔΔCt (29).
Cell line experiments
The C57BL/6 mouse-derived Hepa1-6 hepatocellular carcinoma cell line was purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Cells were cultured in DMEM medium (cat. no. SH30243.01; HyClone; Cytiva) supplemented with 10% FBS (cat. no. VS500T; Ausbian) and penicillin-streptomycin (100 U/ml penicillin and 100 µg/ml streptomycin; Gibco; Thermo Fisher Scientific, Inc.; cat. no. 15070063) and placed at 37°C in a humidified incubator containing 5% CO2. Cells were transfected with LV5-FGL2 [overexpressing (OE) FGLV2] and LV5-Negative Control (NC) lentivirus (Shanghai GenePharma Co., Ltd.) using standard protocols. The transfection efficiency was verified in 293T cells. The MOI of Hepa1-6 was 100, and 1×108 lentivirus were used to transfect 1×106 Hepa1-6 cells for 24 h. After 48 h of transfection, light and green fluorescence microscopy (magnification ×4; Nikon Eclipse Ti-S) images were taken to verify the proportion of transfected cells. After 1 week of transfection, the mRNA and protein expression of FGL2 was tested by RT-qPCR, western blotting and ELISA. Transfected cells were subsequently used for further experiments. To generate conditioned media (CM) for MDSC culture, ~1×106 Hepa1-6 cells were cultured in serum-free DMEM medium for 48 h. CM was then collected, centrifuged at 300 × g at 37°C and for 5 min to remove any debris and was freshly used.
Western blotting and ELISA assay
Western blotting was performed to detect the FGL2 expression in Hepa1-6 cells transfected with LV5-FGL2 and LV5-NC.
Hepa1-6 cells were washed with ice-cold PBS and lysed by RIPA (Beyotime Institute of Biotechnology). Protein concentration was determined using BCA Protein Assay Kit (Beyotime Institute of Biotechnology). Proteins (25 µg per lane) were separated by 10% SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked by 5% skimmed milk dissolved in TBST for 60 min at room temperature and were incubated with rabbit anti-FGL2 polyclonal (1:900; cat. no. ab198029; Abcam) or anti-GAPDH (1:10,000; cat. no. ab8245; Abcam) antibodies overnight at 4°C. Membranes were washed with TBST and incubated with the secondary antibodies goat anti-rabbit IgG (1:50,000; cat. no. ab205718; Abcam) or goat anti-mouse IgG (1:20,000; cat. no. ab205719; Abcam) for 60 min at room temperature. Enhanced chemiluminescence reagent (Pierce; Thermo Fisher Scientific, Inc.) was used to detect the signal on the membrane.
ELISA assay was performed to detect the FGL2 expression in the CM of Hepa1-6 cells transfected with LV5-FGL2 and LV5-NC. After centrifugation of the CM to remove cell debris as aforementioned, FGL2 expression level in CM was measured using an ELISA kit (cat. no. CSB-EL008654MO; Cusabio Biotech Co., Ltd.) according to the manufacturers protocol.
Isolation of bone marrow cells from mice
A total of 8 male C57BL/6 mice (5–6 weeks old) were purchased from the Experimental Animal Center of China Medical University. Animal experiments were approved by the Laboratory Animal Management and Use Committee (approval no. 2020016). Mice were euthanized by cervical dislocation by trained certified people. Mice were placed in 75% alcohol for 1–2 min and were positioned on a sterile bench. Skin and muscles were excised, the epiphysis was cut at both ends of the femur and bone marrow was extracted. Briefly, 1 ml of DMEM was aspirated into a syringe and inserted into the bone marrow cavity. The bone marrow cavity was rinsed to wash out the bone marrow tissues that was filtered using a 200-mesh nylon strainer (Beijing Solarbio Science & Technology Co., Ltd.) to obtain fresh bone marrow cells. After centrifugation (300 × g; 4°C; 5 min), the fresh bone marrow cells were cultured in CM collected from cell line culture.
Flow cytometry
Two flow cytometry tubes containing single-cell suspensions (from bone marrow cells, tumor or spleen) in cell staining buffer (CSB; cat. no. 00-4222-57; Invitrogen; Thermo Fisher Scientific, Inc.) were prepared per sample, one serving as the negative control. Each tube contained ~1×106 cells, and 400 µl CSB solution was added to the tube 1 that was stored on ice. Mouse Fc Block (2 µl; BD Biosciences) was added to the tube 2 of each sample. Then, APC rat anti-mouse CD11b (1 µl; BD Pharmingen; BD Biosciences; cat. no. 557657) and PE rat anti-mouse Ly-6G and Ly-6C (1 µl; BD Pharmingen; BD Biosciences; cat. no. 561084) were added to the tube 2 of each sample. After incubation on ice for 20 min in the dark, cells were washed twice with CSB solution, diluted in 500 µl CSB and analyzed by flow cytometry (BD FACSAria III; BD Biosciences) and data were analyzed by Flowjo (v10.0.7; BD Biosciences).
Establishment of the hepatocellular carcinoma orthotopic mouse model
FGL2-OE Hepa1-6 cells and NC Hepa1-6 cells were harvested, washed twice with PBS, resuspended in PBS and counted. Cells were diluted in PBS to the density of 5×107/ml. A total of 12 C57BL/6 mice (age, 5–6 weeks) purchased from the Experimental Animal Center of China Medical University were anesthetized using an intraperitoneal injection of 1% sodium pentobarbital (60 mg/kg). Using sterile ophthalmic scissors, the abdomen was opened using a 1 cm incision under the xiphoid. Tumor cell suspension (FGL2-OE Hepa1-6 cells or NC Hepa1-6 cells; n=6 mice per group; 20 µl, ~1×106 cells) was then injected at a 20° angle from the liver surface into the liver parenchyma of the left lobe. The abdomen was closed layer by layer using surgical suture. Mice were placed in the incubator immediately after the operation and had free access to food and water for 2 weeks (temperature 20–22°C, humidity 40–60% and 12/12 h light/dark cycle, before the euthanasia by cervical dislocation. Animal health and behavior were monitored twice a day. No unplanned death occurred before the humane endpoints. Tumor size and volume were measured with a caliper using the following formula: Length × width × height × 0.52 (mm3).
Preparation of single-cell suspensions
A total of 12 tumor-bearing C57BL/6 mice (6 per group) were euthanized by cervical dislocation. For tumor single-cell suspension, liver tumors in situ were excised and washed with PBS. Tumors were cut into small pieces and washed with PBS. Tumor tissues were then digested with 0.1% collagenase solution in a 37°C water bath for 30 min. Cell suspensions were filtered using a 200-mesh nylon strainer, washed twice with PBS and resuspended into CSB solution.
For spleen single-cell suspension, spleens were excised and placed in a 24-well plate containing 2 ml PBS. Tissues were crushed using a 1 ml syringe core and filtered with a 200-mesh nylon strainer. Erythrocytes were lysed using erythrocyte-lysing solution, and the cell suspension was washed twice with PBS solution and resuspended with CSB solution.
Statistical analysis
SPSS 22.0 (IBM Corp.) was used for statistical analysis. χ2 test was used for clinical-pathological data analysis, linear Pearson correlation coefficient was used for correlation analysis, and unpaired t-test was used to compare means between groups. P<0.05 was considered to indicate a statistically significant difference.
Results
FGL2 expression level in hepatocellular carcinoma and analysis of clinicopathological characteristics
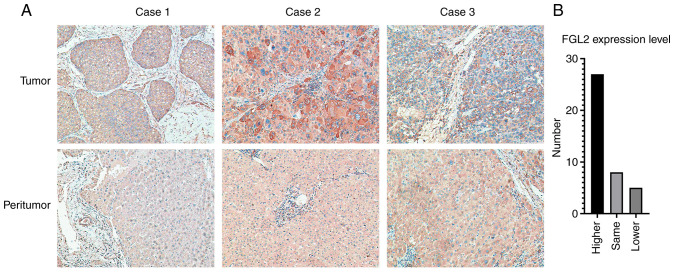
Immunohistochemistry was used to determine the expression level of FGL2 in 40 pairs of hepatocellular carcinoma tumor tissues and peritumor tissue samples. In tumor tissues, FGL2 was mainly expressed in the cancer nest and cytoplasm. In peritumor tissues, FGL2 is mainly expressed in hepatocytes and in some vascular endothelial cells (Fig. 1A). Among the 40 pairs of samples, 27 pairs of tumor tissues had higher expression level of FGL2 relative to peritumor tissues, eight pairs of tumor tissues expressed the same level of FGL2 relative to peritumor tissues, whereas five pairs of tumor tissues had lower FGL2 expression level relative to peritumor tissues (Fig. 1B). Furthermore, the results from association analysis between FGL2 expression level and the clinicopathological characteristics of patients demonstrated that FGL2 expression level was only associated with hepatocellular carcinoma tumor size (P<0.05; Table I).
Figure 1.
FGL2 expression in tumor and peritumor tissues from patients with hepatocellular carcinoma. (A) FGL2 expression in tumor tissues was higher compared with their corresponding matched peritumor tissues (three representative tumor/peritumor sample pairs; magnification, ×200). (B) Number of patients with higher, same or lower expression level of FGL2. FGL2, fibrinogen-like protein 2.
Positive correlation between FGL2 expression and expression of MDSC surface markers in hepatocellular carcinoma
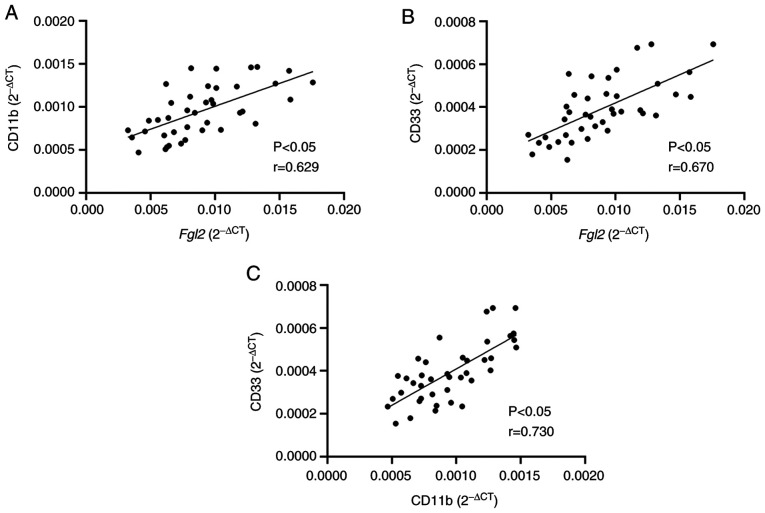
RT-qPCR was performed to determine the mRNA expression of FGL2 and of CD11b and CD33, which are surface markers of human MDSCs, in the hepatocellular carcinoma tumor tissue samples. Linear Pearson correlation coefficient was used to determine the correlation between the expression levels of FGL2 and CD11b/CD33. The results demonstrated that FGL2 expression level was positively correlated with CD11b and CD33 expression levels (r=0.629 and r=0.670, respectively; P<0.05; Fig. 2A). In addition, a positive correlation between the expression levels of CD11b and CD33 was observed (r=0.730; P<0.05; Fig. 2B). These findings suggested that FGL2 expression level may be correlated with the number of MDSCs in the tumor microenvironment of hepatocellular carcinoma.
Figure 2.
Correlation analysis between the expression level of FGL2 and surface markers of myeloid-derived suppressor cells in hepatocellular carcinoma tissues. (A and B) Linear Pearsons correlation analysis between FGL2 expression levels and (A) CD11b and (B) CD33 in 40 hepatocellular carcinoma tumor samples. (C) Linear Pearsons correlation analysis between CD11b and CD33. FGL2, fibrinogen-like protein 2.
Conditioned media from FGL2-OE Hepa1-6 cells increases the in vitro proportion of MDSCs in mouse bone marrow cells
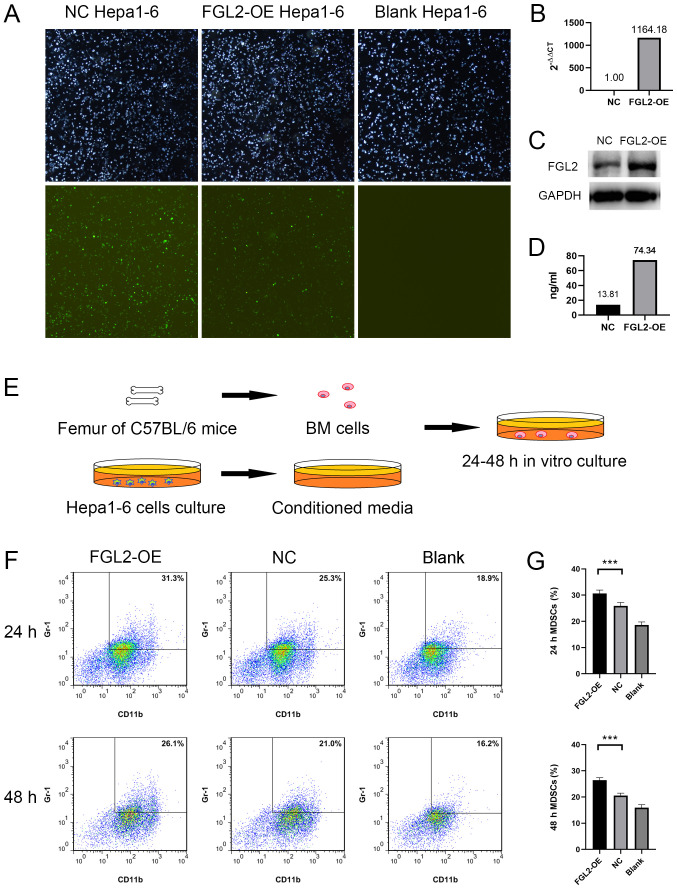
FGL2-OE Hepa1-6 and NC Hepa1-6 cell lines were established by lentivirus transfection. Light and green fluorescence microscopy images demonstrated the transfection efficiency (Fig. 3A). One week post LV5-FGL2 lentivirus transfection, FGL2 mRNA expression level in FGL2-OE Hepa1-6 cells was significantly upregulated (Fig. 3B). FGL2 protein expression was increased in FGL2-OE Hepa1-6 cells and in the culture supernatants compared with NC Hepa1-6 cells (Fig. 3C and D). Bone marrow cells harvested from the femurs of C57BL/6 mice were cultured in vitro using CM from either FGL2-OE Hepa1-6 or NC Hepa1-6 cells for 24–48 h (Fig. 3E). Bone marrow cells cultured in serum-free DMEM medium were used as control. Flow cytometry was used to determine the proportion of CD11b+/Gr-1+ cells in the three groups. The results demonstrated that the proportions of CD11b+/Gr-1+ cells in bone marrow cells cultured using FGL2-OE Hepa1-6 CM for 24 and 48 h were significantly higher compared with cells cultured in NC Hepa1-6 CM (n=8, P<0.001; t-test; Fig. 3F and G).
Figure 3.
Conditioned media from FGL2-OE Hepa1-6 cells increased the proportion of MDSCs in mouse BM cells. (A) Hepa1-6 cells stably transfected with LV5-FGL2 or LV5-NC lentivirus (48 h post-transfection). Light and green fluorescence microscopy images (magnification, ×4). (B) One week post LV5-FGL2 lentivirus transfection, FGL2 mRNA expression level in FGL2-OE Hepa1-6 cells was upregulated 1168 fold compared with NC Hepa1-6 cells according to reverse transcription quantitative PCR experiment. (C) FGL2 protein expression was increased in FGL2-OE Hepa1-6 cells compared with NC Hepa1-6 cells according to western blotting experiment. (D) FGL2 protein level was significantly higher in FGL2-OE Hepa1-6 cell culture supernatants compared with NC Hepa1-6 cell supernatants according to ELISA assay. (E) BM cells extracted from C57BL/6 mice femurs were cultured for 24–48 h using conditioned media from FGL2-OE Hepa1-6 cell, NC Hepa1-6 cell or conventional medium. (F and G) Proportion of CD11b+/Gr-1+ cells in BM cells cultured in FGL2-OE Hepa1-6 conditioned media for 24 and 48 h was higher compared with BM cells cultured in NC Hepa1-6 conditioned media (n=8). ***P<0.001. BM, bone marrow; FGL2, fibrinogen-like protein 2; MDSCs, myeloid-derived suppressor cells; NC, negative control; OE, overexpressing.
FGL2-OE Hepa1-6 cells promote tumor growth and MDSC accumulation in tumors and spleen
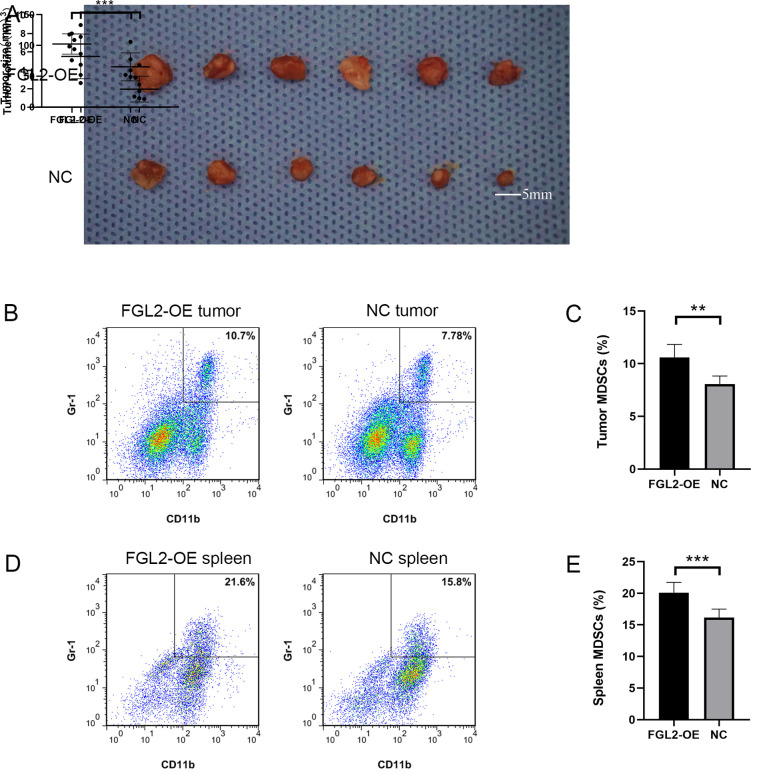
The FGL2-OE Hepa1-6 and NC Hepa1-6 cell lines were used to establish an orthotopic hepatocellular carcinoma model in C57BL/6 mice. Two weeks post-inoculation, the size and volume of the tumors in the FGL2-OE Hepa1-6 group were significantly larger compared with tumors in the NC Hepa1-6 group (Fig. 4A). The diameter of the tumors in the FGL2-OE Hepa1-6 group ranged from 5.1 mm to 8.0 mm, and the diameter of the tumors in the NC Hepa1-6 group ranged from 3.2 mm to 7.1 mm. Single-cell suspensions from the harvested tumors were used to determine the proportion of CD11b+/Gr-1+ cells by flow cytometry. The results demonstrated that the proportion of CD11b+/Gr-1+ cells in the tumors of the FGL2-OE Hepa1-6 group was significantly higher compared with the NC Hepa1-6 group (n=6; P<0.001; t-test; Fig. 4B and C). In addition, the proportion of CD11b+/Gr-1+ cells in the spleen of FGL2-OE Hepa1-6 tumor-bearing C57BL/6 mice was significantly higher compared with mice in the NC Hepa1-6 group (n=6; P<0.001; t-test; Fig. 4D and E).
Figure 4.
Overexpression of FGL2 promotes tumor growth through the accumulation of MDSCs in mice tumors and spleen. (A) Tumor size and volume in the FGL2-OE Hepa1-6 orthotopic hepatocellular carcinoma model group was significantly higher compared with tumors in the NC Hepa1-6 group. (B and C) Proportion of CD11b+/Gr-1+ cells in the tumors of mice in the FGL2-OE Hepa1-6 orthotopic hepatocellular carcinoma model group was higher compared with mice in the NC Hepa1-6 group (n=6). (D and E) Proportion of CD11b+/Gr-1+ cells in the spleen of mice in the FGL2-OE Hepa1-6 orthotopic hepatocellular carcinoma model group was higher compared with spleen in the NC Hepa1-6 group (n=6). *P<0.05, **P<0.01 and ***P<0.001. FGL2, fibrinogen-like protein 2; MDSCs, myeloid-derived suppressor cells; NC, negative control; OE, overexpressing.
Discussion
Liver cancer is the second leading cause of cancer-associated mortality worldwide and in China (1,2). Most primary liver cancers are hepatocellular carcinomas (2). FGL2 has been reported to play a role in promoting tumor progression by regulating the cellular components of the tumor microenvironment (25–27). In the present study, the expression level of FGL2 in hepatocellular carcinomas was associated with tumor size. It was therefore hypothesized that FGL2 may promote tumor progression by affecting certain cellular components in the tumor microenvironment. The present study demonstrated a positive correlation between the expression level of FGL2 and surface markers of MDSCs in hepatocellular carcinomas. These results suggested that FGL2 may promote tumor progression by promoting the accumulation of MDSCs in hepatocellular carcinoma. Immunohistochemistry will be performed in the future to determine CD11b and CD33 expression in human hepatocellular carcinoma tissues.
MDSCs express high levels of the immunosuppressive molecules Arg-1 and iNOS, which can significantly inhibit the function of CD4+ and CD8+ T lymphocytes (30) and inhibit NK cell activation (31), in addition to suppressing antitumor immune response through direct and indirect mechanisms (32). MDSCs are some of the main components of the tumor microenvironment, and can also migrate to peripheral lymphoid organs, such as the spleen (33). Previous studies reported that the suppression of T cells in peripheral lymphoid organs by MDSCs requires direct cell contact (19,34). Furthermore, the number of MDSCs in the peripheral blood and tumors of patients with hepatocellular carcinoma is significantly higher compared with healthy people (35) and is associated with tumor stage, size and Child-Pugh classification (36). In addition, MDSCs presence has been associated with poor overall survival and early relapse in patients with hepatocellular carcinoma (37–40).
In the present study, a mouse hepatocellular carcinoma cell line overexpressing FGL2 was established by lentivirus stable transfection. Furthermore, fresh bone marrow cells harvested from the femurs of C57BL/6 mice were cultured in vitro using conditioned media from hepatocellular carcinoma cells overexpressing FGL2. The results demonstrated that FGL2 could maintain the undifferentiated state of bone marrow cells in vitro, thereby promoting the accumulation of MDSCs. In vivo, the overexpression of FGL2 could promote tumor growth. In addition, MDSC levels in the tumors and spleen of mouse overexpressing FGL2 were significantly increased. It was therefore hypothesized that FGL2 may promote the accumulation of MDSCs to stimulate tumor growth. The original low expression of FGL2 in mouse hepatocellular carcinoma cell line makes it not suitable for silencing FGL2. Therefore, FGL2 was only overexpressed in Hepa1-6 cells. FGL2 knockdown in mice will be performed in the future. Previous studies have demonstrated that FGL2 could specifically bind to FcγR receptors, including FcγRIIB (41–43). FcγRIIB is a transmembrane protein that is abundantly expressed on the surface of myeloid cells (44,45). FGL2 in hepatocellular carcinoma may help maintaining the undifferentiated state of bone marrow cells directly via FcγRIIB, and therefore promote the accumulation of MDSCs required for tumor growth. FGL2 may also upregulate the expression of several cytokines, including tumor growth factor β and interleukin 6 (46,47), in the human hepatocellular carcinoma tumor microenvironment in order to promote the maintenance and recruitment of MDSCs. FGL2 may also induce the accumulation of MDSCs by direct and indirect mechanisms. Further investigation is required to test this hypothesis.
The liver is exposed to large amounts of antigen from the gastrointestinal tract via the portal vein blood flow. To prevent continuous immune stimulation and autoimmune damage induced by antigen exposure, the liver undergoes innate and adaptive immune responses and develops an intrinsic tolerance mechanism (48,49). The immune characteristics of the tumor microenvironment, the immune checkpoint molecules and the prognosis differ between patients with hepatocellular carcinoma (50,51). Immunotherapy for hepatocellular carcinoma has been rapidly developed due to the crucial role that immunosuppressive cells play in anti-tumor immune response (52). Recently, immunotherapy has become the first-line treatment method to treat solid tumors. This includes anti-cytotoxic T lymphocyte antigen 4 (CTLA-4) and anti-programmed cell death 1 (PD-1/PD-L1) monoclonal antibodies (53,54). Targeted therapy for tumor-associated macrophages has rapidly progressed in the recent years (55), and targeted therapy for MDSCs shows great potential (56). Since the accumulation of MDSCs in tumors could increase resistance to anti-CTLA-4/anti-PD-1 therapy, combined targeted therapy with anti-MDSCs may significantly increase the efficacy of this therapy (57). Because of the unique immunosuppressive microenvironment of the liver and hepatocellular carcinoma insensitivity to traditional chemotherapy and radiotherapy, precise cell therapy and associated cytokines targeted to the immunosuppressive microenvironment may represent a successful therapeutic strategy for the treatment of hepatocellular carcinoma. Further investigation is required to better understand the role of FGL2 on the tumor microenvironment of hepatocellular carcinomas.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural Science Foundation of China (grant no. 81373173).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors contributions
BQL performed the main experimental work and wrote the manuscript. ZYB performed in vitro experiments. JYZ contributed to animal experiments. HL performed the important experimental design work and contributed to the writing and revising of the manuscript. All authors read and approved the final version.
Ethics approval and consent to participate
All clinical samples were collected after obtaining written informed consent from patients. The study was approved by the Ethics Committee of the China Medical University. Animal experiments were approved by the Laboratory Animal Management and Use Committee of the China Medical University (approval no. 2020016).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.An L, Zeng HM, Zheng RS, Zhang SW, Sun KX, Zou XN, Chen R, Wang SM, Gu XY, Wei WW, He J. Liver cancer epidemiology in China, 2015. Zhonghua Zhong Liu Za Zhi. 2019;41:721–727. doi: 10.3760/cma.j.issn.0253-3766.2019.10.001. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Eggert T, Greten TF. Tumor regulation of the tissue environment in the liver. Pharmacol Ther. 2017;173:47–57. doi: 10.1016/j.pharmthera.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017;387:61–68. doi: 10.1016/j.canlet.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 5.Sevic I, Spinelli FM, Cantero MJ, Reszegi A, Kovalszky I, García MG, Alaniz L. The role of the tumor microenvironment in the development and progression of hepatocellular carcinoma. In: Tirnitz-Parker J, editor. Hepatocellular Carcinoma [Internet] Codon Publications; Brisbane (AU): 2019. Chapter 2. [DOI] [PubMed] [Google Scholar]
- 6.Yin Z, Dong C, Jiang K, Xu Z, Li R, Guo K, Shao S, Wang L. Heterogeneity of cancer-associated fibroblasts and roles in the progression, prognosis, and therapy of hepatocellular carcinoma. J Hematol Oncol. 2019;12:101. doi: 10.1186/s13045-019-0782-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas H. LSEC stretch promotes fibrosis during hepatic vascular congestion. Nat Rev Gastroenterol Hepatol. 2019;16:262–263. doi: 10.1038/s41575-019-0146-6. [DOI] [PubMed] [Google Scholar]
- 8.Tesi RJ. MDSC; the most important cell you have never heard of. Trends Pharmacol Sci. 2019;40:4–7. doi: 10.1016/j.tips.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 9.Tahmasebi Birgani M, Carloni V. Tumor microenvironment, a paradigm in hepatocellular carcinoma progression and therapy. Int J Mol Sci. 2017;18:405. doi: 10.3390/ijms18020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novikova MV, Khromova NV, Kopnin PB. Components of the hepatocellular carcinoma microenvironment and their role in tumor progression. Biochemistry (Mosc) 2017;82:861–873. doi: 10.1134/S0006297917080016. [DOI] [PubMed] [Google Scholar]
- 11.Condamine T, Mastio J, Gabrilovich DI. Transcriptional regulation of myeloid-derived suppressor cells. J Leukoc Biol. 2015;98:913–922. doi: 10.1189/jlb.4RI0515-204R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porembka MR, Mitchem JB, Belt BA, Hsieh CS, Lee HM, Herndon J, Gillanders WE, Linehan DC, Goedegebuure P. Pancreatic adenocarcinoma induces bone marrow mobilization of myeloid-derived suppressor cells which promote primary tumor growth. Cancer Immunol Immunother. 2012;61:1373–1385. doi: 10.1007/s00262-011-1178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinha P, Okoro C, Foell D, Freeze HH, Ostrand-Rosenberg S, Srikrishna G. Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J Immunol. 2008;181:4666–4675. doi: 10.4049/jimmunol.181.7.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer. 2013;13:739–752. doi: 10.1038/nrc3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ugel S, De Sanctis F, Mandruzzato S, Bronte V. Tumor-induced myeloid deviation: When myeloid-derived suppressor cells meet tumor-associated macrophages. J Clin Invest. 2015;125:3365–3376. doi: 10.1172/JCI80006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergenfelz C, Leandersson K. The generation and identity of human myeloid-derived suppressor cells. Front Oncol. 2020;10:109. doi: 10.3389/fonc.2020.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsieh CC, Hung CH, Lu L, Qian S. Hepatic immune tolerance induced by hepatic stellate cells. World J Gastroenterol. 2015;21:11887–11892. doi: 10.3748/wjg.v21.i42.11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Fang F, Jiao H, Zheng X, Huang L, Yi X, Zhao W. Activated hepatic stellate cells regulate MDSC migration through the SDF-1/CXCR4 axis in an orthotopic mouse model of hepatocellular carcinoma. Cancer Immunol Immunother. 2019;68:1959–1969. doi: 10.1007/s00262-019-02414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37:208–220. doi: 10.1016/j.it.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuwaraj S, Ding J, Liu M, Marsden PA, Levy GA. Genomic characterization, localization, and functional expression of FGL2, the human gene encoding fibroleukin: A novel human procoagulant. Genomics. 2001;71:330–338. doi: 10.1006/geno.2000.6444. [DOI] [PubMed] [Google Scholar]
- 21.Marazzi S, Blum S, Hartmann R, Gundersen D, Schreyer M, Argraves S, von Fliedner V, Pytela R, Rüegg C. Characterization of human fibroleukin, a fibrinogen-like protein secreted by T lymphocytes. J Immunol. 1998;161:138–147. [PubMed] [Google Scholar]
- 22.Yang G, Hooper WC. Physiological functions and clinical implications of fibrinogen-like 2: A review. World J Clin Infect Dis. 2013;3:37–46. doi: 10.5495/wjcid.v3.i3.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan CW, Kay LS, Khadaroo RG, Chan MW, Lakatoo S, Young KJ, Zhang L, Gorczynski RM, Cattral M, Rotstein O, Levy GA. Soluble fibrinogen-like protein 2/fibroleukin exhibits immunosuppressive properties: Suppressing T cell proliferation and inhibiting maturation of bone marrow-derived dendritic cells. J Immunol. 2003;170:4036–4044. doi: 10.4049/jimmunol.170.8.4036. [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Yang PS, Zhu T, Manuel J, Zhang J, He W, Shalev I, Zhang L, Cybulsky MI, Grant DR, et al. Characterization of fibrinogen-like protein 2 (FGL2): Monomeric FGL2 has enhanced immunosuppressive activity in comparison to oligomeric FGL2. Int J Biochem Cell Biol. 2013;45:408–418. doi: 10.1016/j.biocel.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Tang M, Cao X, Li P, Zhang K, Li Y, Zheng QY, Li GQ, Chen J, Xu GL, Zhang KQ. Increased expression of fibrinogen-like protein 2 is associated with poor prognosis in patients with clear cell renal cell carcinoma. Sci Rep. 2017;7:12676. doi: 10.1038/s41598-017-13149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan J, Kong LY, Hu J, Gabrusiewicz K, Dibra D, Xia X, Heimberger AB, Li S. FGL2 as a multimodality regulator of tumor-mediated immune suppression and therapeutic target in gliomas. J Natl Cancer Inst. 2015;107:djv137. doi: 10.1093/jnci/djv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Y, Zhang L, Zha H, Yang F, Hu C, Chen L, Guo B, Zhu B. Stroma-derived fibrinogen-like protein 2 activates cancer-associated fibroblasts to promote tumor growth in lung cancer. Int J Biol Sci. 2017;13:804–814. doi: 10.7150/ijbs.19398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engers R, Mueller M, Walter A, Collard JG, Willers R, Gabbert HE. Prognostic relevance of Tiam1 protein expression in prostate carcinomas. Br J Cancer. 2006;95:1081–1086. doi: 10.1038/sj.bjc.6603385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 30.Vetsika EK, Koukos A, Kotsakis A. Myeloid-derived suppressor cells: Major figures that shape the immunosuppressive and angiogenic network in cancer. Cells. 2019;8:1647. doi: 10.3390/cells8121647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Law AMK, Valdes-Mora F, Gallego-Ortega D. Myeloid-derived suppressor cells as a therapeutic target for cancer. Cells. 2020;9:561. doi: 10.3390/cells9030561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian X, Shen H, Li Z, Wang T, Wang S. Tumor-derived exosomes, myeloid-derived suppressor cells, and tumor microenvironment. J Hematol Oncol. 2019;12:84. doi: 10.1186/s13045-019-0772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andreu P, Johansson M, Affara NI, Pucci F, Tan T, Junankar S, Korets L, Lam J, Tawfik D, DeNardo DG, et al. FcRgamma activation regulates inflammation-associated squamous carcinogenesis. Cancer Cell. 2010;17:121–134. doi: 10.1016/j.ccr.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoechst B, Ormandy LA, Ballmaier M, Lehner F, Krüger C, Manns MP, Greten TF, Korangy F. A new population of myeloid-derived suppressor cells in hepatocellular carcinoma patients induces CD4+CD25+Foxp3+ T cells. Gastroenterology. 2008;135:234–243. doi: 10.1053/j.gastro.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 36.Wang D, An G, Xie S, Yao Y, Feng G. The clinical and prognostic significance of CD14(+) HLA-DR(−/low) myeloid-derived suppressor cells in hepatocellular carcinoma patients receiving radiotherapy. Tumour Biol. 2016;37:10427–10433. doi: 10.1007/s13277-016-4916-2. [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, Ma X, Zhu C, Liu L, Wang G, Yuan X. The role of myeloid-derived suppressor cells in patients with solid tumors: A meta-analysis. PLoS One. 2016;11:e0164514. doi: 10.1371/journal.pone.0164514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao XH, Tian L, Wu J, Ma XL, Zhang CY, Zhou Y, Sun YF, Hu B, Qiu SJ, Zhou J, et al. Circulating CD14+ HLA-DR−/low myeloid-derived suppressor cells predicted early recurrence of hepatocellular carcinoma after surgery. Hepatol Res. 2017;47:1061–1071. doi: 10.1111/hepr.12831. [DOI] [PubMed] [Google Scholar]
- 39.Mizukoshi E, Yamashita T, Arai K, Terashima T, Kitahara M, Nakagawa H, Iida N, Fushimi K, Kaneko S. Myeloid-derived suppressor cells correlate with patient outcomes in hepatic arterial infusion chemotherapy for hepatocellular carcinoma. Cancer Immunol Immunother. 2016;65:715–725. doi: 10.1007/s00262-016-1837-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Fu X, Li T, Yan H. The prognostic value of myeloid derived suppressor cell level in hepatocellular carcinoma: A systematic review and meta-analysis. PLoS One. 2019;14:e0225327. doi: 10.1371/journal.pone.0225327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu H, Shalev I, Manuel J, He W, Leung E, Crookshank J, Liu MF, Diao J, Cattral M, Clark DA, et al. The FGL2-FcgammaRIIB pathway: A novel mechanism leading to immunosuppression. Eur J Immunol. 2008;38:3114–3126. doi: 10.1002/eji.200838338. [DOI] [PubMed] [Google Scholar]
- 42.Morris AB, Farley CR, Pinelli DF, Adams LE, Cragg MS, Boss JM, Scharer CD, Fribourg M, Cravedi P, Heeger PS, Ford ML. Signaling through the inhibitory Fc receptor FcgammaRIIB induces CD8+T cell apoptosis to limit T cell immunity. Immunity. 2020;52:136–150.e6. doi: 10.1016/j.immuni.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selzner N, Liu H, Boehnert MU, Adeyi OA, Shalev I, Bartczak AM, Xue-Zhong M, Manuel J, Rotstein OD, McGilvray ID, et al. FGL2/fibroleukin mediates hepatic reperfusion injury by induction of sinusoidal endothelial cell and hepatocyte apoptosis in mice. J Hepatol. 2012;56:153–159. doi: 10.1016/j.jhep.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 44.Takai T, Li M, Sylvestre D, Clynes R, Ravetch JV. FcR gamma chain deletion results in pleiotrophic effector cell defects. Cell. 1994;76:519–529. doi: 10.1016/0092-8674(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 45.Ravetch JV, Bolland S. IgG Fc receptors. Annu Rev Immunol. 2001;19:275–290. doi: 10.1146/annurev.immunol.19.1.275. [DOI] [PubMed] [Google Scholar]
- 46.Pan G, Zhao Z, Tang C, Ding L, Li Z, Zheng D, Zong L, Wu Z. Soluble fibrinogen-like protein 2 ameliorates acute rejection of liver transplantation in rat via inducing Kupffer cells M2 polarization. Cancer Med. 2018;7:3168–3177. doi: 10.1002/cam4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin SJ, Liu Y, Deng SH, Liao LH, Lin TL, Ning Q, Luo XP. Neuroprotective effects of activated protein C on intrauterine inflammation-induced neonatal white matter injury are associated with the downregulation of fibrinogen-like protein 2/fibroleukin prothrombinase and the inhibition of pro-inflammatory cytokine expression. Int J Mol Med. 2015;35:1199–1212. doi: 10.3892/ijmm.2015.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buonaguro L, Mauriello A, Cavalluzzo B, Petrizzo A, Tagliamonte M. Immunotherapy in hepatocellular carcinoma. Ann Hepatol. 2019;18:291–297. doi: 10.1016/j.aohep.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 49.Brown ZJ, Greten TF. Hoshida Y. Immune Therapies. Cham (CH): Humana Press; 2019. Hepatocellular carcinoma: Translational precision medicine approaches (Internet) Chapter 12. [PubMed] [Google Scholar]
- 50.Ma L, Hernandez MO, Zhao Y, Mehta M, Tran B, Kelly M, Rae Z, Hernandez JM, Davis JL, Martin SP, et al. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell. 2019;36:418–430.e6. doi: 10.1016/j.ccell.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W, Wang H, Ma Z, Zhang J, Ou-Yang W, Qi Y, Liu J. Multi-omics analysis of microenvironment characteristics and immune escape mechanisms of hepatocellular carcinoma. Front Oncol. 2019;9:1019. doi: 10.3389/fonc.2019.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu C, Rong D, Zhang B, Zheng W, Wang X, Chen Z, Tang W. Current perspectives on the immunosuppressive tumor microenvironment in hepatocellular carcinoma: Challenges and opportunities. Mol Cancer. 2019;18:130. doi: 10.1186/s12943-019-1047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao H, Chen W, Dai Y, Richardson JJ, Guo J, Yuan K, Zeng Y, Xie K. Expression of programmed cell death-ligands in hepatocellular carcinoma: Correlation with immune microenvironment and survival outcomes. Front Oncol. 2019;9:883. doi: 10.3389/fonc.2019.00883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hilmi M, Vienot A, Rousseau B, Neuzillet C. Immune therapy for liver cancers. Cancers (Basel) 2019;12:77. doi: 10.3390/cancers12010077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Heide D, Weiskirchen R, Bansal R. Therapeutic targeting of hepatic macrophages for the treatment of liver diseases. Front Immunol. 2019;10:2852. doi: 10.3389/fimmu.2019.02852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu LC, Chang CJ, Hsu CH. Targeting myeloid-derived suppressor cells in the treatment of hepatocellular carcinoma: Current state and future perspectives. J Hepatocell Carcinoma. 2019;6:71–84. doi: 10.2147/JHC.S159693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chesney JA, Mitchell RA, Yaddanapudi K. Myeloid-derived suppressor cells-a new therapeutic target to overcome resistance to cancer immunotherapy. J Leukoc Biol. 2017;102:727–740. doi: 10.1189/jlb.5VMR1116-458RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.