Abstract
B-cell lymphomas (BCLs) are malignant lymphoid tumours originating from the malignant proliferation and transformation of mature lymphocytes at various stages of differentiation and clonal expansion of the lymphatic and circulatory systems. Efforts to control or even eradicate BCLs are frequently hampered by the development of drug resistance. Autophagy is an evolutionarily conserved biological process of the energy metabolism. By degrading intracellular organelles and proteins, autophagy provides cells with biochemical reaction substrates for the maintenance of homeostasis under nutrient deprivation or other stressful conditions. Accumulating evidence indicates that autophagy plays an important role in chemotherapy resistance. S100A8 is an important member of the calcium-binding protein family that plays an important role in regulating tumour resistance to chemotherapy, while the specific molecular regulatory mechanisms remain unclear. In the present study, by employing three BCL cell lines (Daudi, SUDHL-4 and JeKo-1), it was demonstrated that BCL cells with a strong drug resistance also exhibited active autophagy. In addition, S100A8 was found to be crucial for regulating drug resistance and promoting autophagy in BCL cells. Interference of S100A8 significantly downregulated Bcl-2/adenovirus E1B 19-kDa protein-interacting protein 3 located in the mitochondria and endoplasmic reticulum to further inhibit autophagy. In addition, S100A8 interference markedly inhibited the formation of the BECN1-PI3KC3 complex and promoted B-cell lymphoma 2 expression, which collectively inhibited autophagy.
Keywords: S100A8, chemoresistance, autophagy, non-Hodgkin lymphoma
Introduction
Haematological malignancies comprise a group of heterogeneous tumours with prominent proliferative and self-replicating abilities (1–4). Despite advances in treatment, the quality of life of the patients following treatment is highly affected by chemotherapy resistance, which severely impedes the clinical treatment of B-cell lymphomas (BCLs) (5–7). BCLs comprise a heterogenous group of mature lymphoproliferative malignancies that are among the top 10 causes of cancer-related mortality, mainly occurring in the lymph nodes, spleen, thymus and other lymphoid organs (4,8,9). Almost all anti-lymphoma treatments are accompanied by chemotherapy resistance, including resistance to genotoxic agents, monoclonal antibodies, antibody-drug conjugates, targeted agents or multidrug combinations (5,10–12). Chemotherapy resistance has become the most important challenge to the successful treatment of BCLs.
Autophagy is an evolutionarily conserved biological process of the energy metabolism (13,14). By degrading intracellular organelles and proteins, autophagy provides cells with biochemical reaction substrates for the maintenance of homeostasis under nutrient deficiencies or other stress conditions (15,16). Autophagy has been confirmed to play a pivotal role in chemotherapy resistance (17). The inhibition of autophagy may enhance the cytotoxicity of chemotherapy drugs and the apoptosis of multiple malignant tumour cells, including BCL cells (18). Various studies have also demonstrated that multiple chemotherapeutic drugs increase autophagic flux, thereby enhancing drug resistance and promoting cell survival (19,20).
Autophagy-related 5 (ATG5) is a protein that, in humans, is encoded by the ATG5 gene located on chromosome 6. ATG5 acts as a proviral factor in the autophagic vesicle formation process. Conjugation with ATG12, through a ubiquitin-like conjugating system involving ATG7 as an E1-like activating enzyme and ATG10 as an E2-like conjugating enzyme, is crucial for the function of ATG5 (21,22). The ATG5-ATG12 conjugate acts as an E3-like enzyme, which is required for the lipidation of the ATG8 family of proteins and their association with the vesicle membranes (23,24). This ATG5-ATG12 conjugate is necessary for the conjugation of microtubule-associated protein 1A/1B-light chain 3 (LC3)-I and phosphatidylethanolamine to form LC3-II (23,25).
S100A8 is a multifunctional, calcium-binding protein that polymerizes with S100A9 to form calprotectin for the sequestration of metals, including iron, manganese and zinc, via chelation (26). S100A8 is part of the 22-member calcium-binding EF hand-containing S100 superfamily of proteins, which functions predominantly in the innate immune system (27). S100A8 is expressed in varying degrees in a variety of cells, and is abundant in myeloid cells (28). According to previous reports, S100A8 is mainly involved in cell proliferation and inflammation, and participates in the occurrence and development of various types of cancer (29). S100A8 has been shown to be associated with myeloid differentiation, autophagy, apoptosis and chemotherapy resistance (30–32).
The Bcl-2/adenovirus E1B 19-kDa protein-interacting protein 3 (BNIP3) is a BH3-only protein primarily localized to the mitochondria and endoplasmic reticulum (ER). BNIP3 is anchored to the outer mitochondrial and ER membrane via its C-terminus transmembrane domain, whereas the N-terminus faces the cytosol. BNIP3 induces cell death by perturbing mitochondrial function (33,34). The abnormal expression and function of BNIP3 may interfere with Ca2+ homeostasis and promote cell death (35,36). BNIP3 is also a potent inducer of autophagy in multiple types of cells (32,37).
The aim of the present study was to determine the role of S100A8 in the chemotherapy resistance of BCLs by investigating whether the resistance of BCL cells to Adriamycin (ADR) and vincristine (VCR) was positively correlated with autophagy, and whether S100A8 activates autophagy in BCL cells by promoting BNIP3, Beclin-1 (BECN1) and class III phosphatidylinositol-3-kinase (PI3KC3) expression, hoping to identify a novel target for the treatment of BCLs.
Materials and methods
Cell lines and cell culture
Daudi, SUDHL-4 and JeKo-1 cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI-1640 medium supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 2 mM glutamine in a humidified incubator with 5% CO2 and 95% air.
Cell viability assay
Cell viability was assessed by MTT assay. Daudi, SUDHL-4 and JeKo-1 cells were plated in 96-well plates at a density of 2×103 cells per well containing 100 µl culture medium. ADR (1 mg/ml) or VCR (1 mg/ml) was added to 96-well culture plates with 10 µl per well and incubated for 72 h, followed by incubation with MTT. The control cells were treated only with PBS prior to incubation with MTT. Dimethyl sulfoxide was used to dissolve the formazan. The relative cell viability was calculated by measuring the absorbance value/well at 570 nm and using the following equation: Mean optical density (OD) of treated cells/mean OD of control cells.
Lentivirus-mediated stable silencing of ATG5 and S100A8
The lentivirus particles of shATG5 LV-ATG5-RNAi (9513-1) were purchased from GENECHEM. The sequence for the synthesized shRNA targeting S100A8 was 5′-CCUGAAGAAAUUGCUAGAGTT-3′. The annealed oligonucleotide fragment was cloned into the lentivirus plasmid PLL3.7 (Addgene, Inc.) to establish the shRNA lentiviral vector. Recombinant lentiviruses expressing S100A8 shRNA or negative control (Shanghai GenePharma Co., Ltd.) were obtained by plasmid transformation. VSVG, pMDLg/pRRE, RSV-REV (Addgene, Inc.) and Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.) were used for lentivirus packaging in 293T cells. The supernatant of the 293T culture was collected and filtered using a 0.22-µm filter (EMD Millipore), followed by a concentration step. Medium containing lentivirus was stored at −80°C for future use.
Reverse transcription quantitative PCR (RT-qPCR) analysis
Total RNA was isolated using TRIzol® (Takara Bio, Inc.) and reverse-transcribed into cDNA using PrimeScript RT Master Mix (Takara Bio, Inc.). according to the manufacturer's instructions. RT-qPCR was performed using SYBR Green Real-time PCR Master Mix kit (Takara Bio, Inc.), according to the manufacturer's instructions. The following PCR conditions were used on the Light Cycler: 39 cycles at 95°C for 30 sec, 95°C for 5 sec, followed by 60°C for 30 sec in a 10-µl reaction volume. The relative expression was normalized to that of GAPDH, which was used as the internal control. The primers used were as follows: ATG5, 5′-CTCTGCAGTGGCTGAGTGAA-3′ forward and 5′-TCAATCTGTTGGCTGTGGGA-3′ reverse; S100A8, 5′-CCGAGCTGGAGAAAGCCTTG-3′ forward and 5′-AGGTCATCCCTGTAGACGGC-3′ reverse; GAPDH, 5′-AGAAGGCTGGGGCTCATTTG-3′ forward and 5′-AGGGGCCATCCACAGTCTTC-3′ reverse.
Western blotting (WB)
Cells were lysed in SDS lysis buffer (P0013G; Beyotime Institute of Biotechnology) containing 1% protease inhibitor PMSF (ST506; Beyotime Institute of Biotechnology). The extracted protein concentration was determined using the BCA protein assay kit (P0012S; Beyotime Institute of Biotechnology) and stored at −80°C. A total of 20 µg of each protein sample was separated by 12% SDS-PAGE and transferred to PVDF membranes (IPVH00010; EMD Millipore). Following blocking with 5% non-fat milk for 1 h at room temperature, the membranes were incubated with appropriate primary and secondary antibodies. Proteins were detected using an enhanced chemiluminescence kit (6305; Bio-Rad Laboratories, Inc.) under a gel electrophoresis imager (Bio-Rad Laboratories, Inc.). The primary antibodies for LC3 (source, rabbit; cat. no. ab192890; dilution, 1:1,000), P62 (cat. no. ab207305; source, rabbit; dilution, 1:2,000) and calnexin (source, rabbit; cat. no. ab13504; dilution, 1:2,000) were purchased from Abcam. The primary antibodies for B-cell lymphoma 2 (BCL2; cat. no. 12789-1-AP; source, rabbit; dilution, 1:500), BECN1 (cat. no. 11306-1-AP; source, rabbit; dilution, 1:500), PI3KC3 (cat. no. 20584-1-AP; source, rabbit; dilution, 1:100) and tubulin (cat. no. 10068-1-AP; source, rabbit; dilution, 1:2,000) were purchased from ProteinTech Group, Inc. The primary antibody for S100A8 (cat. no. HPA024372; source, rabbit; dilution, 1:500) was purchased from Merck KGaA. The primary antibodies for manganese-dependent superoxide dismutase 2 (cat. no. MAB3419; source, mouse; 0.5 µg/ml) and that for BNIP3 (cat. no. MAB4147; source, mouse; 0.5 µg/ml) were purchased from R&D. The secondary antibody for anti-rabbit (cat. no. SA00001-2; dilution, 1:2,000) was purchased from ProteinTech Group, Inc. The secondary antibody (anti-mouse; cat. no. SA00001-1; dilution, 1:2,000) was purchased from ProteinTech Group, Inc. The intensity of the western blot bands was quantified using ImageJ software (version 1.47; National Institutes of Health,). Only the blots with band intensity within the linear range were included.
Autophagy flux analysis of LC3 puncta
Adenovirus expressing mCherry-GFP-LC3 fusion protein (Ad-mCherry-GFP-LC3, C3011) was purchased from Beyotime Institute of Biotechnology. Cells were plated in 6-well plates and allowed to reach 50–70% confluence at the time of Ad-mCherry-GFP-LC3 transfection. Adenoviral infection was performed according to the manufacturer's instructions. The presence of mRFP-LC3 puncta indicated the autolysosomes in red fluorescent images.
Transmission electron microscopy (TEM) analysis of autophagosomes
Cells were harvested and centrifuged at 1,000 × g for 10 min at room temperature. The cell pellet was fixed with 4% glutaraldehyde for 24 h at 4°C and 1% osmium tetroxide for 2 h at 4°C. Thereafter, the cell pellet was dehydrated in a graded series of alcohol and acetone, followed by embedding in Epon 816 (Electron Microscopy Sciences). Ultrathin sections were cut by a Leica ultramicrotome (Leica Microsystems, Inc.) and stained with uranyl acetate for 4 min and followed by lead citrate for 2 min at room temperature. The exact thickness of the ultrathin section of the cell samples under the transmission electron microscope was 40 nm. TEM was conducted using a JEM-1400Plus transmission electron microscope (JEOL, Ltd.). The mean number of autophagosomes in tumour cells was counted and quantified.
Immunoprecipitation analysis
Cells were lysed at 4°C in ice-cold lysis buffer (Beyotime Institute of Biotechnology) and the cell pellet was prepared by centrifugation (12,000 × g for 10 min at room temperature). Immunoprecipitation was performed using anti-BECN1 antibody, which was captured with protein A+G agarose beads (Beyotime Institute of Biotechnology). The bead-bound proteins were then eluted by boiling in SDS sample buffer, subjected to PAGE and analyzed by WB.
Statistical analysis
Student's t-test was used when making comparisons in datasets containing two groups. One-way ANOVA test was used when making comparisons in datasets containing multiple groups, and Student-Newman-Keuls test was used as the post hoc test. All data were analyzed using GraphPad Prism software (version 8.3.1; GraphPad Software, Inc.) and presented as the mean ± SEM. *P<0.05, **P<0.01 and ***P<0.001 were considered to indicate statistically significant differences.
Results
Inhibition of autophagy significantly damages the drug resistance of BCL cells
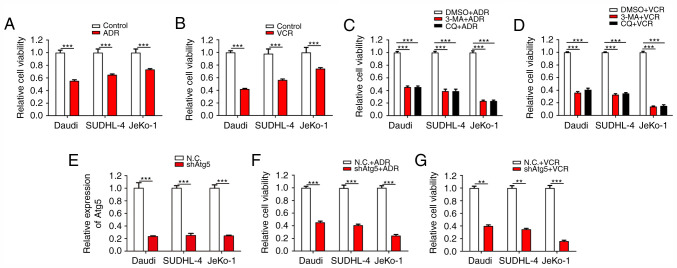
ADR and VCR are effective antitumour agents with a broad antitumour spectrum. They exert strong inhibitory effects on RNA synthesis and the proliferation of BCL cells. First, the inhibitory effects of ADR and VCR on the viability of different BCL cells, including Daudi (Burkitt lymphoma cells), SUDHL-4 (diffuse large B-cell lymphoma cells) and JeKo-1 cells (mantle cell lymphoma cells) were verified. As expected, both ADR and VCR notably decreased the viability of Daudi, SUDHL-4 and JeKo-1 cells (Fig. 1A and B). Among these three cell lines, JeKo-1 cells exhibited the strongest drug resistance against ADR and VCR treatments, followed by SUDHL-4 and Daudi cells. Considering the association between autophagy and drug resistance, it was then verified whether the inhibition of autophagy counteracted the resistance of BCL cells to ADR and VCR. The results demonstrated that, when the cells were treated with the synthetic autophagy inhibitors 3-methyladenine and chloroquine diphosphate salt combined with ADR and VCR, the viability of BCL cells, particularly JeKo-1 cells, was significantly inhibited (Fig. 1C and D). To further confirm the association between autophagy and drug resistance, lentivirus infection was conducted to inhibit autophagy via stable ATG5 interference in Daudi, SUDHL-4 and JeKo-1 cells (Fig. 1E). Cell viability assays revealed that Daudi-shATG5, SUDHL-4-shATG5 and Jeko-1-shATG5 cells were markedly more susceptible to ADR and VCR, as compared with their respective control cells (Fig. 1F and G). These results demonstrated that the resistance of BCL cells to ADR and VCR is partially dependent on autophagy.
Figure 1.
The inhibition of autophagy significantly reverses the drug resistance of BCL cells. (A and B) Daudi, SUDHL-4 and JeKo-1 cells were treated with ADR (1 µg/ml) or VCR (1 µg/ml) for 24 h. Cell viability was analyzed by the CCK-8 assay. (C and D) Cells were treated with 3-MA or CQ for 48 h, and then with ADR or VCR for an additional 24 h. Cell viability was analyzed by the CCK-8 assay. (E) RT-qPCR analysis was performed to confirm gene interference of ATG5 expression in BCL cells. (F and G) Cells were treated with ADR or VCR following ATG5 interference. Cell viability was analyzed by the CCK-8 assay. ***P<0.001 and **P<0.01. BCL, B-cell lymphoma; ADR, Adriamycin; CCK-8, Cell Counting Kit-8; 3-MA, 3-methyladenine; CQ, chloroquine diphosphate salt; VCR, vincristine; RT-qPCR, reverse transcription quantitative PCR.
The resistance of BCL cells to ADR and VCR is positively correlated with autophagy
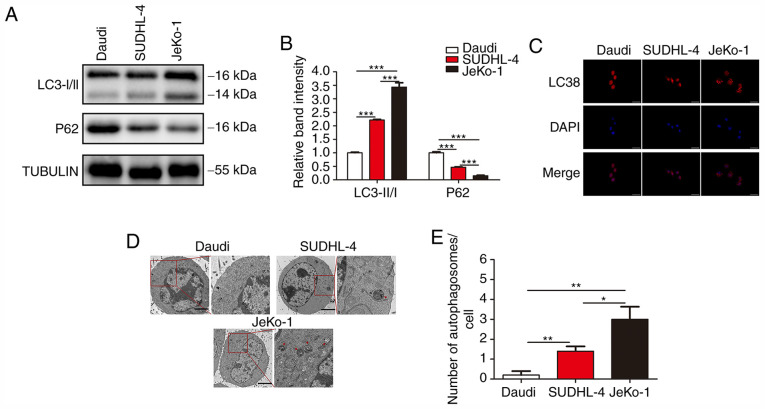
Since the drug resistance of BCL cells was found to be closely associated with autophagy and the drug resistance of BCL cells differed greatly, the autophagic activity in different BCL cells was examined. Three classic assays commonly used in autophagy research were conducted to characterize the status of autophagy: i) The protein level of LC3 and P62 was determined by WB; ii) mRFP-LC3 punta were examined by immunofluorescence; and iii) cytoplasmic accumulation of autophagosomes was detected by TEM. WB revealed that, among the three B-cell lymphoma cell lines, JeKo-1 cells exhibited the highest expression of LC3-II, followed by SUDHL-4 and Daudi cells; furthermore, Daudi cells exhibited the highest expression of P62, followed by SUDHL-4 and JeKo-1 cells (Fig. 2A and B). In the autophagy flux assays, there were markedly more mRFP-LC3 punta in the JeKo-1 cells compared with the Daudi and SUDHL-4 cells (Fig. 2C). The formation of autophagosomes and autolysosomes as analyzed by TEM was abundant in Jeko-1 cells and deficient in Daudi and SUDHL-4 cells (Fig. 2D and E). These results demonstrated that autophagy was activated in BCL cells exhibiting strong drug resistance.
Figure 2.
The resistance of BCL cells to ADR and VCR is associated with autophagy. (A and B) Western blotting for LC3-I, LC3-II and P62 in Daudi, SUDHL-4 and JeKo-1 cells. (C) Representative images of LC3-positive puncta in BCL cells. (D) Transmission electron microscopy of BCL cells. Autolysosomes are indicated by arrowheads. Scale bars, 5 µm. (E) Quantification of detectable autolysosomes in BCL cells. ***P<0.001, **P<0.01 and *P<0.05. BCL, B-cell lymphoma; ADR, Adriamycin; VCR, vincristine; LC3, microtubule-associated protein 1A/1B-light chain 3.
S100A8 expression is closely associated with the drug resistance of BCL cells
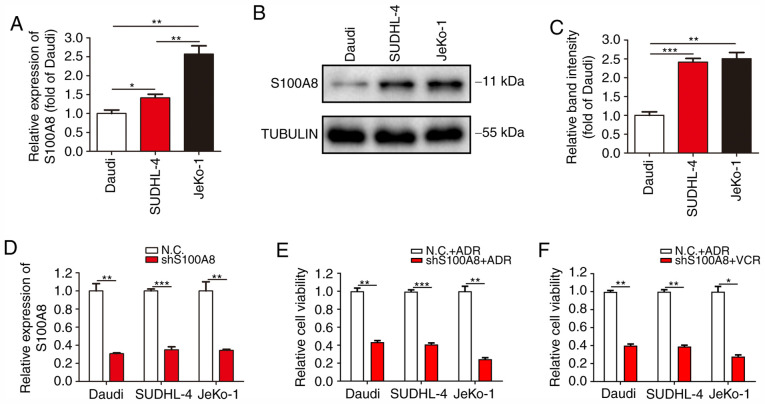
S100A8 is particularly highly expressed in myeloid cells and plays an important regulatory role in the differentiation, autophagy and chemotherapy resistance of myeloid cells. To verify whether the drug resistance and autophagic activity of Daudi, SUDHL-4 and JeKo-1 cells was associated with S100A8, the expression of S100A8 was detected in the three B-cell lymphoma cell lines. RT-qPCR analysis revealed that JeKo-1 cells exhibited the highest mRNA expression of S100A8, followed by SUDHL-4 and Daudi cells (Fig. 3A). WB revealed a markedly higher protein expression of S100A8 in JeKo-1 and SUDHL-4 cells, as compared with that in Daudi cells (Fig. 3B and C). To further confirm the association between S100A8 and drug resistance, lentivirus infection was conducted to inhibit S100A8 expression in Daudi, SUDHL-4 and JeKo-1 cells (Fig. 3D). Cell viability assay revealed that Daudi-shS100A8, SUDHL-4-shS100A8 and JeKo-1-shS100A8 cells were markedly more susceptible to ADR and VCR, as compared with their respective control cells (Fig. 3E and F). These results demonstrated that the resistance of BCL cells to ADR and VCR was closely associated with the expression of S100A8.
Figure 3.
S100A8 is closely associated with the drug resistance of BCL cells. (A) RT-qPCR analysis was performed to detect the mRNA expression of S100A8 in BCL cells. (B and C) Western blotting for S100A8 protein expression in BCL cells. (D) RT-qPCR analysis was performed to confirm gene interference in the S100A8 expression in BCL cells. (E and F) Cells were treated with ADR or VCR following S100A8 interference. Cell viability was analyzed by the Cell Counting Kit-8 assay. ***P<0.001, **P<0.01 and *P<0.05. BCL, B-cell lymphoma; RT-qPCR, reverse transcription quantitative PCR; ADR, Adriamycin; VCR, vincristine.
S100A8 augments autophagy in BCL cells
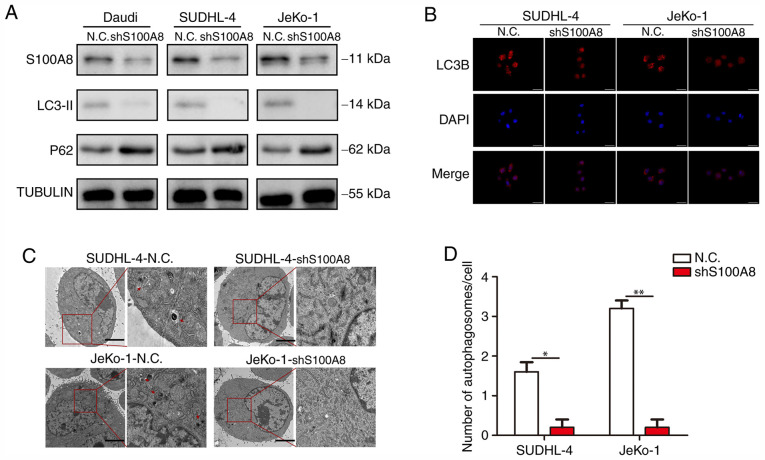
Our previous study demonstrated that the chemotherapy resistance of BCL cells was largely dependent on autophagy, and the stronger drug resistance of BCL cells was associated with a higher S100A8 expression. Next, an attempt was made to elucidate the association between S100A8 and autophagy in BCL cells. WB revealed that Daudi-shS100A8, SUDHL-4-shS100A8 and JeKo-1-shS100A8 cells exhibited a markedly lower LC3-II and higher P62 protein expression, as compared with their respective control cells (Fig. 4A). In the autophagy flux assay, there were markedly more mRFP-LC3 punta in SUDHL-4-shS100A8 and JeKo-1-shS100A8 cells, as compared with their respective control cells (Fig. 4B). Finally, the formation of autophagosomes and autolysosomes as analyzed by TEM was markedly deficient in BCL cells with S100A8 interference (Fig. 4C and D), indicating that S100A8 promoted autophagy in BCL cells.
Figure 4.
S100A8 increases autophagy in BCL cells. (A) Western blotting for LC3-II and P62 in BCL cells was performed following S100A8 interference. (B) Representative images of LC3-positive puncta in BCL cells following S100A8 interference. (C) Transmission electron microscopy of BCL cells following S100A8 interference. Autolysosomes are indicated by arrowheads. Scale bars, 5 µm. (D) Quantification of detectable autolysosomes in BCL cells following S100A8 interference. **P<0.01 and *P<0.05. BCL, B-cell lymphoma; LC3, microtubule-associated protein 1A/1B-light chain 3.
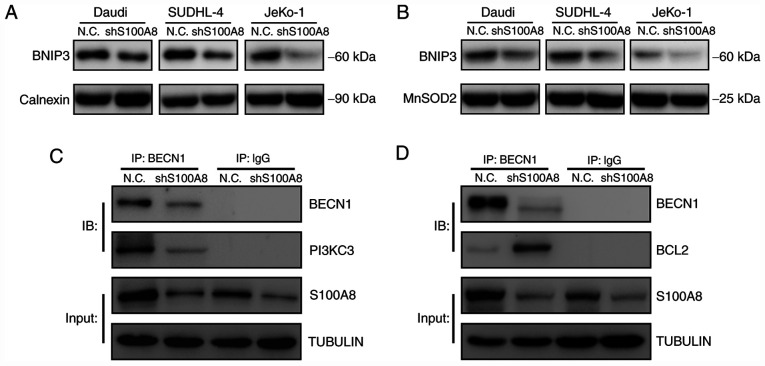
S100A8 activates autophagy by promoting BNIP3, BECN1 and PI3KC3 expression
Autophagy is an effective method used by cells to cope with environmental stressors, such as starvation, and is greatly involved in the normal physiological and pathological processes of the body. BNIP3 is anchored to the outer mitochondrial and ER membrane via its C-terminus transmembrane domain, and it can interact with LC3. The binding of BNIP3 to LC3 on the autophagosome is crucial for the removal of both ER (ER-phagy) and mitochondria (mitophagy) via autophagy. To explore the molecular mechanism underlying S100A8-mediated autophagy, the BNIP3 expression was tested in ER and mitochondria following S100A8 interference. The results demonstrated that S100A8 interference significantly decreased the BNIP3 protein expression, both in the ER and mitochondria, in Daudi, SUDHL-4 and JeKo-1 cells (Fig. 5A and B). Next the early autophagic signalling event of BECN1-PI3KC3 and BECN1-BCL2 complex formation was analyzed. The co-immunoprecipitation assay revealed that S100A8 interference significantly reduced the formation of the BECN1-PI3KC3 complex in JeKo-1 cells (Fig. 5C). Furthermore, S100A8 interference significantly increased the formation of the BECN1-BCL2 complex in JeKo-1 cells (Fig. 5D). Collectively, these results indicated that S100A8 may activate autophagy by promoting the expression of BNIP3 and the BECN1-PI3KC3 complex and inhibiting that of the BECN1-BCL2 complex.
Figure 5.
S100A8 promotes BNIP3, BECN1 and PI3KC3 expression. (A) WB for BNIP3 in the endoplasmic reticulum of BCL cells following S100A8 interference. (B) WB for BNIP3 in the mitochondria of BCL cells following S100A8 interference. Co-immunoprecipitation assay for (C) BECN1-PI3KC3 and (D) BECN1-BCL2 complexes in Jeko-1 cells following S100A8 interference. BNIP3, Bcl-2/adenovirus E1B 19-kDa protein-interacting protein 3; BECN1, Beclin-1; PI3KC3, class III phosphatidylinositol-3-kinase; WB, western blotting; BCL, B-cell lymphoma.
Discussion
The understanding of the occurrence and development of lymphomas, as well as the treatment of patients with lymphoma, has greatly improved (38,39). Chemotherapy is the main treatment strategy for almost all types of BCLs (40). However, drug resistance of lymphoma cells often renders chemotherapy ineffective and greatly shortens the long-term event-free survival of the patients. Therefore, overcoming the drug resistance of lymphoma cells is a major challenge for BCL treatment (5,40). In the present study, the BCL cell lines Daudi, SUDHL-4 and JeKo-1 were examined, which represent Burkitt's lymphoma, diffuse large B-cell lymphoma and mantle cell lymphoma, respectively, and account for 70–85% of all BCLs. Burkitt's lymphoma is strongly associated with infection by the oncogenic Epstein Barr virus (EBV). There is currently no research evidence on the association between EBV infection and chemoresistance enhancement by S100A8. It may be hypothesized that certain EBV proteins, such as EBNA1, EBNA3C and LMP1, may interact with S100A8, thereby affecting BECN1-PI3KC3 complex formation and eventually modifying chemoresistance by S100A8. Diffuse large B-cell lymphoma is the most prevalent non-Hodgkin lymphoma (NHL), accounting for 40% of all NHLs. Mantle cell lymphoma is the most genomically unstable lymphoma that is not sensitive to radiotherapy, chemotherapy or haematopoietic stem cell transplantation. However, T-cell or NK-cell lymphoma cells were not examined in the present study, which constitutes a limitation of the present study.
Rituximab binds the CD20 antigen on the surface of B lymphocytes, causing an enhanced immune elimination of malignant B cells. The present study did not involve immune response experiments in vivo but cellular chemoresistance experiments in vitro; therefore, the broad-spectrum chemotherapy drugs ADR and VCR were selected in the present study instead of rituximab.
Autophagy, also referred to as type II programmed cell death, is an evolutionarily conserved and strictly controlled metabolic process. Autophagy plays an important role in homeostasis and cell survival. Cancer cells have multiple responses to chemotherapy, from activating survival pathways to triggering cell death (41,42). Chemotherapy drugs can significantly increase the autophagic activity of BCL cells (43). Multiple studies have demonstrated that inhibition of autophagy enhances chemotherapy-induced BCL cell death (44–46). However, the specific molecular regulatory effects of S100A8 on autophagy and chemoresistance in BCL cells remain unclear. The present study indicated that the drug resistance of BCL cells was positively correlated with autophagic activity, and S100A8 was closely associated with the drug resistance of BCL cells by activating autophagy.
Prior to this study, S100A8 has been reported to promote autophagy in cancer cells through the cross-talk between mitochondria and lysosomes via reactive oxygen species (32), or through the activation of the autophagy initiation complex BECN1-PI3KC3 (30). In the present study, it was confirmed that S100A8 reduced the sensitivity of BCL cells to chemotherapy by maintaining BCL cell autophagy, thereby increasing the resistance of BCL cells to chemotherapy. The role of the S100A8 protein in pro-autophagic activities is mediated through the promotion of BECN1-PI3KC3 and BECN1-BCL2 complex formation (31). This study revealed that S100A8 stimulated BECN1-PI3KC3 complex formation as an early autophagic signalling event that significantly promoted autophagy. In addition, the dissociation of BCL2 from BECN1 was found to be an important mechanism involved in S100A8-activated autophagy.
LC3 is initially processed into its cytoplasmic form, LC3-I, then coupled with lipid phosphatidylethanolamine, generating its final LC3-II form (47). BNIP3 is a cell death-inducing factor of the Bcl-2 family of proteins, more precisely a member of the BH3-only subfamily. Proteins such as S100A8 in the BH3-only subfamily bind through a common BH3 domain, rather than the BH1 and BH2 domains as the other Bcl-2 family members. The binding of BNIP3 to LC3 is crucial for autophagosome formation (47,48). The present study demonstrated that S100A8 accelerated BNIP3 expression in ER and mitochondria, resulting in the promotion of autophagy.
In conclusion, the molecular mechanisms of S100A8-triggered autophagy and drug resistance of BCL cells was investigated in the present study. The involvement of the S1008-BNIP3/BECN1-PI3KC3 complex-autophagy-chemoresistance axis in the autophagic regulation of drug resistance in BCL cells was also confirmed. These findings may help overcome drug resistance in the treatment of BCLs.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Key Research Project from Science & Technology Department of Sichuan Province (grant no. 2019YFS0301); The Doctor Research Foundation of The Affiliated Hospital of Southwest Medical University (grant nos. 19032 and 19079); The Key Research Project from Health and Family Planning Commission of Sichuan Province (grant no. 18ZD014); The Applied Basic Research Project of Luzhou Science and Technology Bureau (grant no. 2019LZXNYDJ54); and the National Natural Science Foundation of China (grant no. 81450030).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
LZ performed the experiments and data analysis and wrote the manuscript. SZ, TZ, KY and XL participated in conducting this study. JT designed and oversaw this study, and contributed to the revision of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All institutional and national guidelines for the use of laboratory materials were followed.
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing interests.
References
- 1.Cruz-Rodriguez N, Combita AL, Zabaleta J. Epigenetics in hematological malignancies. Methods Mol Biol. 2018;1856:87–101. doi: 10.1007/978-1-4939-8751-1_5. [DOI] [PubMed] [Google Scholar]
- 2.Upadhyay VA, Fathi AT. Induction chemotherapy in acute myeloid leukaemia: Origins and emerging directions. Curr Opin Hematol. 2018;25:67–74. doi: 10.1097/MOH.0000000000000407. [DOI] [PubMed] [Google Scholar]
- 3.Stahl M, Kohrman N, Gore SD, Kim TK, Zeidan AM, Prebet T. Epigenetics in cancer: A hematological perspective. PLoS Genet. 2016;12:e1006193. doi: 10.1371/journal.pgen.1006193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz AD, Jaffe ES. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klener P, Klanova M. Drug resistance in non-Hodgkin lymphomas. Int J Mol Sci. 2020;21:2081. doi: 10.3390/ijms21062081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feugier P. A review of rituximab, the first anti-CD20 monoclonal antibody used in the treatment of B non-Hodgkin's lymphomas. Future Oncol. 2015;11:1327–1342. doi: 10.2217/fon.15.57. [DOI] [PubMed] [Google Scholar]
- 7.El-Galaly TC, Cheah CY, Kristensen D, Hutchison A, Hay K, Callréus T, Villa D. Potentials, challenges and future of chimeric antigen receptor T-cell therapy in non-Hodgkin lymphomas. Acta Oncol. 2020;59:766–774. doi: 10.1080/0284186X.2020.1741680. [DOI] [PubMed] [Google Scholar]
- 8.Swerdlow SH, Cook JR. As the world turns, evolving lymphoma classifications-past, present and future. Hum Pathol. 2020;95:55–77. doi: 10.1016/j.humpath.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 9.Jones SE. Non-Hodgkin lymphomas. JAMA. 1975;234:633–638. doi: 10.1001/jama.234.6.633. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Wang LX, Pang P, Twitty C, Fox BA, Aung S, Urba WJ, Hu HM. Cross-presentation of tumor associated antigens through tumor-derived autophagosomes. Autophagy. 2009;5:576–577. doi: 10.4161/auto.5.4.8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallo S, Gatti S, Sala V, Albano R, Costelli P, Casanova E, Comoglio PM, Crepaldi T. Agonist antibodies activating the Met receptor protect cardiomyoblasts from cobalt chloride-induced apoptosis and autophagy. Cell Death Dis. 2014;5:e1185. doi: 10.1038/cddis.2014.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu SF, Zheng B, Go M, Lau J, Spencer S, Raab H, Soriano R, Jhunjhunwala S, Cohen R, Caruso M, et al. A novel anti-CD22 anthracycline-based antibody-drug conjugate (ADC) that overcomes resistance to auristatin-based ADCs. Clin Cancer Res. 2015;21:3298–3306. doi: 10.1158/1078-0432.CCR-14-2035. [DOI] [PubMed] [Google Scholar]
- 13.Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 14.Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 15.Jardon MA, Rothe K, Bortnik S, Vezenkov L, Jiang X, Young RN, Lum JJ, Gorski SM. Autophagy: From structure to metabolism to therapeutic regulation. Autophagy. 2013;9:2180–2182. doi: 10.4161/auto.26378. [DOI] [PubMed] [Google Scholar]
- 16.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Zhou Y, Li Y, Yang L, Ma Y, Peng X, Yang S, Liu J, Li H. Autophagy: A novel mechanism of chemoresistance in cancers. Biomed Pharmacother. 2019;119:109415. doi: 10.1016/j.biopha.2019.109415. [DOI] [PubMed] [Google Scholar]
- 18.Piya S, Andreeff M, Borthakur G. Targeting autophagy to overcome chemoresistance in acute myleogenous leukemia. Autophagy. 2017;13:214–215. doi: 10.1080/15548627.2016.1245263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124:3307–3318. doi: 10.1002/cncr.31335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su LY, Luo R, Liu Q, Su JR, Yang LX, Ding YQ, Xu L, Yao YG. Atg5- and Atg7-dependent autophagy in dopaminergic neurons regulates cellular and behavioral responses to morphine. Autophagy. 2017;13:1496–1511. doi: 10.1080/15548627.2017.1332549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng W, Xie W, Yin D, Luo R, Liu M, Guo F. ATG5 and ATG7 induced autophagy interplays with UPR via PERK signaling. Cell Commun Signal. 2019;17:42. doi: 10.1186/s12964-019-0353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romanov J, Walczak M, Ibiricu I, Schüchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams RA, Woods KL, Juliano L, Mottram JC, Coombs GH. Characterization of unusual families of ATG8-like proteins and ATG12 in the protozoan parasite Leishmania major. Autophagy. 2009;5:159–172. doi: 10.4161/auto.5.2.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fletcher K, Ulferts R, Jacquin E, Veith T, Gammoh N, Arasteh JM, Mayer U, Carding SR, Wileman T, Beale R, Florey O. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 2018;37:e97840. doi: 10.15252/embj.201797840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruenster M, Vogl T, Roth J, Sperandio M. S100A8/A9: From basic science to clinical application. Pharmacol Ther. 2016;167:120–131. doi: 10.1016/j.pharmthera.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J. S100A8/A9 in inflammation. Front Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Anholts J, Kolbe U, Stegehuis-Kamp JA, Claas FHJ, Eikmans M. Calcium-binding proteins S100A8 and S100A9: Investigation of their immune regulatory effect in myeloid cells. Int J Mol Sci. 2018;19:1833. doi: 10.3390/ijms19071833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shabani F, Farasat A, Mahdavi M, Gheibi N. Calprotectin (S100A8/S100A9): A key protein between inflammation and cancer. Inflamm Res. 2018;67:801–812. doi: 10.1007/s00011-018-1173-4. [DOI] [PubMed] [Google Scholar]
- 30.Yang M, Zeng P, Kang R, Yu Y, Yang L, Tang D, Cao L. S100A8 contributes to drug resistance by promoting autophagy in leukemia cells. PLoS One. 2014;9:e97242. doi: 10.1371/journal.pone.0097242. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Yang L, Yang M, Zhang H, Wang Z, Yu Y, Xie M, Zhao M, Liu L, Cao L. S100A8-targeting siRNA enhances arsenic trioxide-induced myeloid leukemia cell death by down-regulating autophagy. Int J Mol Med. 2012;29:65–72. doi: 10.3892/ijmm.2011.806. [DOI] [PubMed] [Google Scholar]
- 32.Ghavami S, Eshragi M, Ande SR, Chazin WJ, Klonisch T, Halayko AJ, McNeill KD, Hashemi M, Kerkhoff C, Los M. S100A8/A9 induces autophagy and apoptosis via ROS-mediated cross-talk between mitochondria and lysosomes that involves BNIP3. Cell Res. 2010;20:314–331. doi: 10.1038/cr.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chourasia AH, Macleod KF. Tumor suppressor functions of BNIP3 and mitophagy. Autophagy. 2015;11:1937–1938. doi: 10.1080/15548627.2015.1085136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dhingra A, Jayas R, Afshar P, Guberman M, Maddaford G, Gerstein J, Lieberman B, Nepon H, Margulets V, Dhingra R, Kirshenbaum LA. Ellagic acid antagonizes Bnip3-mediated mitochondrial injury and necrotic cell death of cardiac myocytes. Free Radic Biol Med. 2017;112:411–422. doi: 10.1016/j.freeradbiomed.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Li L, Leavesley HW, Zhang X, Borowitz JL, Isom GE. Cyanide-induced apoptosis of dopaminergic cells is promoted by BNIP3 and Bax modulation of endoplasmic reticulum-mitochondrial Ca2+ levels. J Pharmacol Exp Ther. 2010;332:97–105. doi: 10.1124/jpet.109.159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson ÅB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Z, Chen C, Tang P, Zhang H, Yue J, Yu Z. BNIP3 induces apoptosis and protective autophagy under hypoxia in esophageal squamous cell carcinoma cell lines: BNIP3 regulates cell death. Dis Esophagus. 2017;30:1–8. doi: 10.1093/dote/dow025. [DOI] [PubMed] [Google Scholar]
- 38.Ferreri AJM. Therapy of primary CNS lymphoma: Role of intensity, radiation, and novel agents. Hematology Am Soc Hematol Educ Program. 2017;2017:565–577. doi: 10.1182/asheducation-2017.1.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witkowska M, Smolewski P, Robak T. Investigational therapies targeting CD37 for the treatment of B-cell lymphoid malignancies. Expert Opin Investig Drugs. 2018;27:171–177. doi: 10.1080/13543784.2018.1427730. [DOI] [PubMed] [Google Scholar]
- 40.Reagan PM, Barr PM. Chemotherapy-free treatment in non-Hodgkin lymphoma: A steep learning curve. Lancet Haematol. 2017;4:e152–e153. doi: 10.1016/S2352-3026(17)30040-6. [DOI] [PubMed] [Google Scholar]
- 41.Rivera Vargas T, Apetoh L. Danger signals: Chemotherapy enhancers? Immunol Rev. 2017;280:175–193. doi: 10.1111/imr.12581. [DOI] [PubMed] [Google Scholar]
- 42.Gunjur A. Short vs. long course adjuvant chemotherapy for colon cancer. Lancet Oncol. 2018;19:e236. doi: 10.1016/S1470-2045(18)30272-9. [DOI] [PubMed] [Google Scholar]
- 43.Gayle S, Landrette S, Beeharry N, Conrad C, Hernandez M, Beckett P, Ferguson SM, Xu T, Rothberg J, Lichenstein H. B-cell non-Hodgkin lymphoma: Selective vulnerability to PIKFYVE inhibition. Autophagy. 2017;13:1082–1083. doi: 10.1080/15548627.2017.1304871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng X, Li Y, Fan J, Zhao H, Xian Z, Sun Y, Wang Z, Wang S, Zhang G, Ju D. Recombinant human arginase induced caspase-dependent apoptosis and autophagy in non-Hodgkin's lymphoma cells. Cell Death Dis. 2013;4:e840. doi: 10.1038/cddis.2013.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan J, Zeng X, Li Y, Wang S, Wang Z, Sun Y, Gao H, Zhang G, Feng M, Ju D. Autophagy plays a critical role in ChLym-1-induced cytotoxicity of non-hodgkin's lymphoma cells. PLoS One. 2013;8:e72478. doi: 10.1371/journal.pone.0072478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu K, Sun XQ, Wang CQ, Gao TX, Sun P, Wang Y, Jiang WQ, Li ZM, Huang JJ. Metronomic combination chemotherapy using everolimus and etoposide for the treatment of non-Hodgkin lymphoma. Cancer Med. 2019;8:4688–4698. doi: 10.1002/cam4.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaaf MB, Keulers TG, Vooijs MA, Rouschop KM. LC3/GABARAP family proteins: Autophagy-(un)related functions. FASEB J. 2016;30:3961–3978. doi: 10.1096/fj.201600698R. [DOI] [PubMed] [Google Scholar]
- 48.Birgisdottir AB, Lamark T, Johansen T. The LIR motif-crucial for selective autophagy. J Cell Sci. 2013;126:3237–3247. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.