Abstract
Long non-coding RNA growth arrest specific 5 (GAS5) exerts inhibitory effects through the modulation of several target microRNAs (miRs) in cancer. However, its potential roles and underlying relationship during colorectal cancer (CRC) progression are unclear. Therefore, we explored the role of the negative feedback loop formed by the GAS5/miR-34a axis and mammalian target of rapamycin/sirtuin 1 (mTOR/SIRT1) pathway on macroautophagy and apoptosis in CRC. Expression of GAS5, miR-34a, SIRT1 and mTOR in CRC patients and cell lines was detected by quantitative reverse transcription polymerase chain reaction. Online bioinformatic analysis was used to predict the downstream miRs of GAS5. Luciferase assay and western blotting were performed to demonstrate miR-34a as a downstream target gene of GAS5 in CRC cells. The effects of the GAS5/miR-34a axis on apoptosis, macroautophagy, and the mTOR/SIRT1 pathway were assessed by flow cytometry, transmission electron microscopy and western blotting, respectively. Our results suggested that GAS5 was downregulated and acted as a molecular sponge of miR-34a during CRC progression. miR-34a participated in regulating GAS5-suppressed CRC cell macroautophagy and induced apoptosis through the mTOR/SIRT1 pathway. GAS5-mediated macroautophagy was maintained in an equilibrium state that might have a protective effect on CRC cell apoptosis. The mTOR signaling pathway suppressed GAS5 expression and formed a negative regulation feedback loop with miR-34a in CRC cells. Our results suggested that the GAS5/miR-34a/SIRT1/mTOR negative regulatory feedback loop mediated CRC cell macroautophagy, and maintained the cells in an autonomous equilibrium state, but not excessive activation state, which functions as a strong antiapoptotic phenotype during human CRC progression.
Keywords: colorectal cancer, lncRNA GAS5, macroautophagy, miR-34a, negative feedback regulatory loop
Introduction
Colorectal cancer (CRC) is a common digestive tract tumor. Despite advances in surgical resection combined with chemotherapy and radiotherapy, the median survival rate of CRC patients remains low and it is still a serious threat to human health (1). Although multifactorial etiology is linked with high susceptibility to CRC development, including genetic factors and unhealthy dietary lifestyle (2), the exact molecular mechanisms underlying human CRC development are far from clear.
Long non-coding RNAs (lncRNAs) are ncRNAs that play important roles including regulation of gene expression and tumorigenesis (3). Growth arrest specific 5 (GAS5) is a newly discovered lncRNA, which is encoded by a poorly conserved gene mapped to chromosome 1q25.1. The gene consists of 12 exons and 11 introns from which 29 transcripts are produced from alternative splicing, many of which contain retained introns (4). Several studies have shown that GAS5 has a critical tumor-suppressive effect during progression of prostate cancer, renal cancer, ovarian cancer, cervical cancer as well as CRC (5–10). Additionally, accumulating evidence has confirmed that GAS5 binds to microRNAs (miRs), such as miR-21, miR-196A, miR-205, miR-222 and miR-103, sponging their inhibitory effect on the target genes to perform tumor-suppressive roles (5,11–15). It is important to note that our previous studies have verified that miR-34a acts as a tumor-suppressor gene in human CRC (16,17) and our bioinformatic analyses have revealed the presence of a common binding site for miR-34a and GAS5. These results have broadened our exploration of the interaction between GAS5 and miR-34a in the progression of CRC.
Recently, the roles of macroautophagy regulated by ncRNAs in the malignant progression of human cancers have been investigated. Our previous studies demonstrated that miR-34a-regulated macroautophagy enhanced the sensitivity of oxaliplatin and induced CRC cell apoptosis (16,17). GAS5 performs key regulatory roles in macroautophagy in response to external stimuli in various types cells (18–20). Although increasing evidence has focused on the association between macroautophagy regulated by miR-34a and GAS5 in malignant progression of human cancers, as well as other pathological processes, to our knowledge, the precise roles of GAS5/miR-34a axis-regulated macroautophagy and its effect on CRC progression requires further investigation.
In the present study, we demonstrated that GAS5 acted as a tumor-suppressive factor in progression of human CRC by regulating the miR-34a/SIRT1 axis. miR-34a participated in mediating GAS5-mediated suppression of CRC cell macroautophagy and induced apoptosis via the mammalian target of rapamycin/sirtuin 1 (mTOR/SIRT1) pathway. Moreover, GAS5-regulated macroautophagy maintained cells in an equilibrium state that protected against CRC cell apoptosis. GAS5/miR-34a/SIRT1/mTOR formed a negative regulatory feedback loop that might explain the macroautophagy striking in the relatively activated balance in CRC cells.
Materials and methods
Patients and tissue samples
The present study protocol was approved by the Institutional Research Ethics Committee of Harbin Medical University (KY2018-208). All patients gave signed informed consent according to our institutional guidelines. We obtained 75 CRC samples and their paired adjacent normal colon tissues from patients (median age, 64 years; range, 49–79 years) immediately after surgical resection between March 2018 and March 2019 at the Second Affiliated Hospital of Harbin Medical University. The samples were stored at −80°C. The tissues were reviewed by a pathologist and classified according to the 7th Tumor-Node-Metastasis (TNM) classification of the International Union against Cancer (21). Information concerning the clinical characteristics was collected from medical records.
Animal model of azoxymethane (AOM)-induced CRC
Thirty-six male Wistar rats (40–60 g, aged 3 weeks) purchased from the Animal Center of the Second Affiliated Hospital of Harbin Medical University were housed with free access to sterile food and water under a standard 12-h light/dark cycle and controlled temperature (22±2°C) and humidity (55±5%). The health and behavior of the rats were monitored every 2 days. Animal experiments were carried out in strict accordance with the Harbin Medical University Institutional Animal Care and Use Committee (KY2018-208). The rats were randomly divided into three groups of 12. Once weekly, for 6 weeks, animals in the control group received an equal volume of saline, while animals in the AOM groups received AOM (15 mg/kg i.p.). Rats in the AOM1 group were sacrificed at 12 weeks after the first AOM treatment, while rats in the AOM2 group were sacrificed at 24 weeks after the first AOM treatment. Animals were sacrificed by anesthesia overdose with i.p. injection of sodium pentobarbital (Nembutal; 200 mg/kg). The colon tissue samples were obtained from each rat after decapitation (17).
Cell culture and treatment
The normal colon epithelial cell line FHC and human CRC cell lines HT29, HCT116, SW480 and SW620 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and routinely cultured as previously described (17). All the cell lines used in the present study were authenticated by short tandem repeat (STR) profiling. The GAS5 plasmid for GAS5 overexpression and the hsa-miR-34a mimics or hsa-miR-34a inhibitor were synthesized by GenePharma. The CRC cell lines HT29 and SW480 were transfected with either 200 nM pcDNA3.1-GAS5 or 100 nM miR-34a mimics (or miR-34a inhibitors) for 48 h. The corresponding negative control was performed in all experiments. The mTOR siRNA (50 nM, 48 h) was purchased from Santa Cruz Biotechnology, Inc. Transient transfection of the siRNA or miRNA was conducted using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). A potent mTOR activator, MYH1485 (2 nM, 12 h; Sigma-Aldrich; Merck KGaA), was used to enhance mTOR expression, which in turn inhibited macroautophagy flux. Rapamycin (10 nM, 24 h; Cell Signaling Technology, Inc.), an macroautophagy promoter, was used to further activate CRC cell macroautophagy as previously described (17).
Binding sites prediction and luciferase assay
The putative binding sites between GAS5 and miR-34a were predicted by online software DIANA-LncBase v2 (http://www.microrna.gr/LncBase). Luciferase activity was measured using a Dual-Luciferase Reporter Assay system (Promega Corp.), and promoter activity was expressed as the ratio of firefly luciferase activity to Renilla luciferase activity as previously described (17).
Real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent and total miRNAs were extracted using mirVana miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc.). cDNA was synthesized from 2 µg total miRNAs using the High Capacity cDNA Reverse Transcription kit (Ambion; Thermo Fisher Scientific, Inc.). Expression levels of miRNAs and mRNAs were assessed with real-time RT-qPCR using the Power SYBR Green and a 7500 Sequence Detection System (Thermo Fisher Scientific, Inc.) as previously described (17). The names and the primer sequences of the detected genes are listed in Table I. Changes in mRNAs and miRNAs were quantified using the 2−∆∆Cq method (22) and β-actin mRNA and U6 small nuclear RNA were used as references.
Table I.
Primers used for RT-qPCR.
| Primer name | Primer sequence: 5′-3′ |
|---|---|
| Forward GAS5 | CTTGCCTGGACCAGCTTAAT |
| Reverse GAS5 | CAAGCCGACTCTCCATACCT |
| Forward mTOR | TCCGAGAGATGAGTCAAGAGG |
| Reverse mTOR | CACCTTCCACTCCTATGAGGC |
| Forward SIRT1 | CCCAGAACATAGACACGCTGGA |
| Reverse SIRT1 | ATCAGCTGGGCACCTAGGACA |
| Forward β-actin | ATGTTGAGACCTTCAACACC |
| Reverse β-actin | AGGTAGTCAGTCAGGTCCCGGCC |
| Forward miR-34a | TGGTGTCGTGGAGTCG |
| Reverse miR-34a | GGCATCTCTCGCTTCATCTT |
| Forward U6 | CTCGCTTCGGCAGCACA |
| Reverse U6 | AACGCTTCACGAATTTGCGT |
Western blotting
Total protein from each experimental group was lysed in RIPA buffer (Beyotime Institute of Biotechnology). The BCA kit (Beyotime Institute of Biotechnology) was used to estimate the concentration of total protein. Total protein (20–40 µg) was separated by 10–15% SDS-PAGE and blotted on nitrocellulose membranes (Amersham Pharmacia Biotech). Membranes were blocked with blocking buffer (Beyotime Institute of Biotechnology) for 2 h at room temperature, and then the membranes were probed with antibodies against mTOR, phospho (p)-mTOR, SIRT1, MAPLC3β, Beclin1, Bcl-2, Bax, matrix metalloproteinase (MMP)2, MMP9 and β-actin. Enhanced chemiluminescence detection reagents of donkey anti-mouse IgG Alexa Fluor 680 or donkey anti-rabbit IgG Alexa Fluor 680 were incubated with the membranes for 12 h at 4°C and visualized with an Odyssey Infrared Imaging system (LI-COR Biosciences). Densitometry was performed using an Alpha Imager 2200 (Applied Biosystems; Thermo Fisher Scientific, Inc). β-actin was used as a normalization control. The names and manufacturers of the detected genes are listed in Table II.
Table II.
Antibodies used for western blot analysis.
| Antibody | Product/cat. no. (manufacturer) | Species | Dilution |
|---|---|---|---|
| mTOR | sc-517464 (Santa Cruz Biotechnology, Inc., USA) | Mouse | 1:200 |
| p-mTOR | sc-293089 (Santa Cruz Biotechnology, Inc., USA) | Mouse | 1:200 |
| SIRT1 | sc-135792 (Santa Cruz Biotechnology, Inc., USA) | Mouse | 1:500 |
| LC3 | sc-28266 (Santa Cruz Biotechnology, Inc., USA) | Rabbit | 1:500 |
| Beclin1 | sc-11427 (Santa Cruz Biotechnology, Inc., USA) | Rabbit | 1:500 |
| Bax | sc-526 (Santa Cruz Biotechnology, Inc., USA) | Mouse | 1:200 |
| Bcl-2 | sc-7382 (Santa Cruz Biotechnology, Inc., USA) | Mouse | 1:200 |
| MMP2 | sc-13594 (Santa Cruz Biotechnology, Inc., USA) | Mouse | 1:500 |
| MMP9 | sc-393859 (Santa Cruz Biotechnology, Inc., USA) | Mouse | 1:500 |
| β-actin | sc-69879 (Santa Cruz Biotechnology, Inc., USA) | Mouse | 1:500 |
| Anti-rabbit IgG, Alexa Fluor 680 | A10043 (Thermo Fisher Scientific, Inc., USA) | Donkey | 1:5,000 |
| Anti-mouse IgG, Alexa Fluor 680 | A32788 (Thermo Fisher Scientific, Inc., USA) | Donkey | 1:5,000 |
Cell migration and invasion assays
The invasive potential of cells was measured in 6.5-µm Transwell chambers with 8.0-µm Pore Polycarbonate Membrane Insert (Corning Inc.). The filter of the top chamber was Matrigel-coated with 50 µl diluted Matrigel and incubated at 37°C for 2 h. The lower chambers were filled with 600 µl Dulbecco's modified Eagle's medium (DMEM) containing 5% fetal bovine serum (FBS) as chemoattractant for a further 24 h. Cells were serum-free-starved overnight, harvested, and suspended in migration medium (DMEM with 0.5% bovine serum albumin). A suspension of 5,000 CRC cells in 100 µl migration medium was added to each top chamber. After the cells were incubated for 16 h, the non-invading cells that remained on the upper surface were removed with a cotton swab. The invasive cells on the lower surface of the membrane insert were fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.2% Triton X-100 at room temperature for 15 min, and stained with 0.1% crystal violet for 5 min. The number of cells on the lower surface, which had invaded through the membrane, was counted under a light microscope in five random fields at a magnification of ×100. The procedure for the Transwell migration assays was the same as the Transwell invasion assay except that the filter of the top chamber was not coated with Matrigel.
Evaluation of macroautophagy and apoptosis by flow cytometry
CRC cell macroautophagy and apoptosis were evaluated by flow cytometry as previously described (17). CRC cells (104) were washed with PBS and then incubated with 0.05 mmol/l mono-dansylcadaverine (MDC; Sigma-Aldrich; Merck KGaA) at 37°C for 45 min. The cells were washed three times with PBS and analyzed by flow cytometry immediately (BD Biosciences). Apoptosis was detected with flow cytometry using Annexin V-fluoroisothiocyanate (FITC)/propidium iodide (PI) kit (Becton-Dickinson). After treatment, the cells from each group were washed three times with PBS and stained with Annexin V-FITC/PI in the dark. The rate of apoptosis was quantified by flow cytometry (BD Biosciences).
Transmission electron microscopy
CRC cells were harvested and fixed with 2.5% glutaraldehyde at 4°C for 2 h and then in 1% osmic acid PBS at pH 7.4, at 37°C for 2 h. After dehydration and embedding, the ultrathin sections were prepared on uncoated copper grids with an Ultrotome (Leica, Reichert Ultracuts) and stained with uranyl acetate and lead citrate. Images were recorded under a transmission electron microscope (JEM 1230; Geol) (17).
Statistical analysis
Statistical analysis was carried out by SPSS version 21 (IBM Corp.) or GraphPad Prism version 5.0 (GraphPad Software, Inc.). Results are presented as the mean ± SEM. Comparisons of quantitative data between two groups were performed using Student's two-tailed t-tests, and among three or more groups were performed by one-way ANOVA test followed by Tukey's test. Correlation analysis between GAS5, miR-34a, SIRT1 and mTOR mRNA expression levels was examined by Pearson's rank correlation analysis. The Chi-square test analysis was used to calculate the P-values in Table III. P<0.05 was assigned to indicate a significant difference.
Table III.
Association between GAS5 expression levels and clinicopathological features in the CRC patients (N=75).
| GAS5 | ||||
|---|---|---|---|---|
| Characteristics | n | Low (n=37) | High (n=38) | P-value |
| Age (years) | 0.786 | |||
| <60 | 35 | 18 | 17 | |
| ≥60 | 40 | 19 | 21 | |
| Sex | 0.125 | |||
| Male | 35 | 17 | 18 | |
| Female | 40 | 20 | 20 | |
| Tumor size (cm) | 0.062 | |||
| <5 | 44 | 26 | 18 | |
| ≥5 | 31 | 11 | 20 | |
| Differentiation | 0.170 | |||
| Well/moderate | 29 | 3 | 26 | |
| Poor | 46 | 34 | 12 | |
| Lymphatic node metastasis | 0.016 | |||
| Present | 36 | 33 | 3 | |
| Absent | 39 | 4 | 35 | |
| Clinical stages | 0.022 | |||
| I/II | 38 | 3 | 35 | |
| III/IV | 37 | 34 | 3 | |
| CA19-9 level (U/ml) | 0.690 | |||
| <37 | 38 | 19 | 19 | |
| ≥37 | 37 | 18 | 19 | |
| CEA level (ng/ml) | 0.081 | |||
| <5 | 23 | 3 | 20 | |
| ≥5 | 52 | 34 | 18 | |
The median value of GAS5 expression levels in CRC patients is 0.8699. P<0.05 indicates a statistically significant difference. CRC, colorectal cancer; GAS5, growth arrest specific 5; CA19-9, cancer antigen 19-9; CEA, carcinoembryonic antigen. Significant P-values are displayed in bold print.
Results
Expression of GAS5, miR-34a, SIRT1 and mTOR in human CRC tissues, cell lines and rat colon tissues
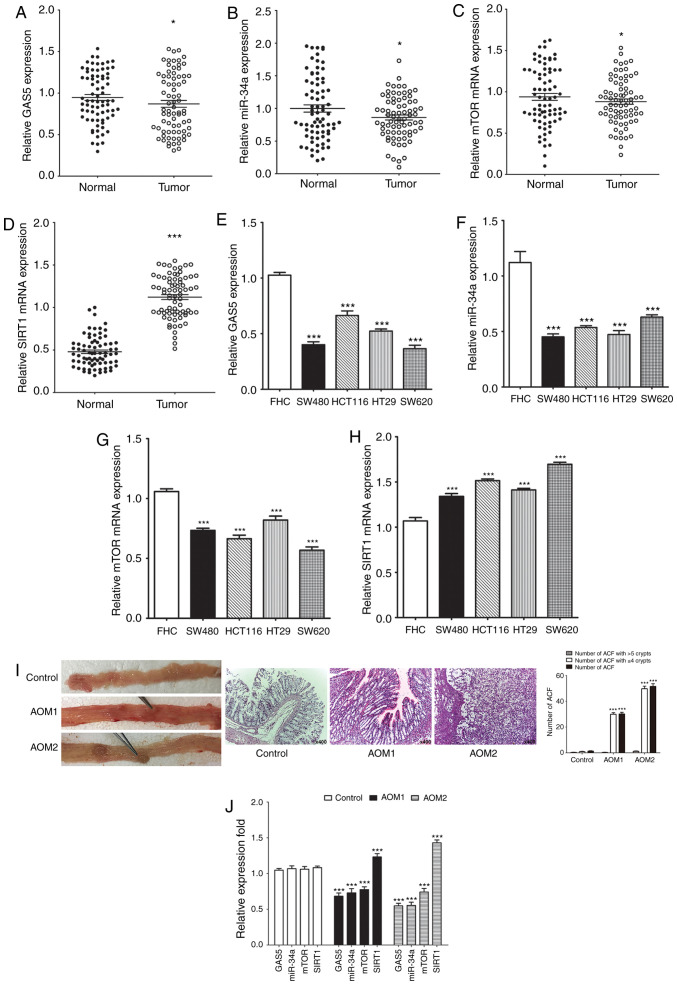
We initially identified GAS5, miR-34a, SIRT1 and mTOR expression in human CRC specimens to understand better the underlying complex mechanisms of human CRC. Expression levels of GAS5, miR-34a and mTOR were significantly decreased (Fig. 1A-C) while those of SIRT1 mRNA were significantly upregulated (Fig. 1D) in most of the CRC tissues compared to the paired adjacent non-tumor tissues. All the CRC cells displayed significantly lower expression levels of GAS5, miR-34a and mTOR, and SIRT1 mRNA was significantly higher when compared to these levels in the FHC cells (Fig. 1E-H). The AOM-induced CRC rat model was used to determine the potential mechanism of GAS5 in vivo. All rats treated with AOM developed polyps and preneoplastic aberrant crypt foci (ACF) (Fig. 1I). The number of ACF with ≤4 crypts and total number of ACF in the entire colon of the rats were significantly different. There was no significant difference in the number of ACF with >5 crypts in the entire colon of the rats. Consistent with the results in vitro, expression of GAS5, miR-34a and mTOR was dramatically decreased, while expression of SIRT1 was significantly upregulated in the colon tissues from AOM-treated rats compared to the control group (Fig. 1J). These results showed that the GAS5/miR-34a axis is involved in inhibition of CRC malignant behavior through the mTOR/SIRT1 pathway in vitro and in vivo.
Figure 1.
Expression of GAS5, miR-34a, SIRT1 and mTOR in human CRC tissues, cell lines and rats colon tissues. (A-D) RT-qPCR was performed and revealed downregulation of GAS5, miR-34a and mTOR mRNA expression and upregulation of SIRT1 mRNA expression in 75 CRC samples compared to their paired adjacent nontumor tissues. (E-H) RT-qPCR revealed downregulation of GAS5, miR-34a and mTOR mRNA expression and upregulation of SIRT1 mRNA expression in HT29, HCT116, SW480 and SW620 CRC cell lines compared to FHC cells. (I) All rats treated with AOM developed polyps and ACF. Numbers of ACF in the entire colon of the rats were quantified. (J) Expression of GAS5, miR-34a, SIRT1 and mTOR in the colon samples of rats was determined by RT-qPCR. *P<0.05 and ***P<0.001 means a significant difference vs. the adjacent nontumor tissues, FHC cells or the control group. CRC, colorectal cancer; GAS5, growth arrest specific 5; miR/miRNA, microRNA; SIRT1, sirtuin 1; mTOR, mammalian target of rapamycin.
Association between expression of GAS5, miR-34a, SIRT1 and mTOR in human CRC tissues and the clinicopathological significance of GAS5 expression in CRC patients
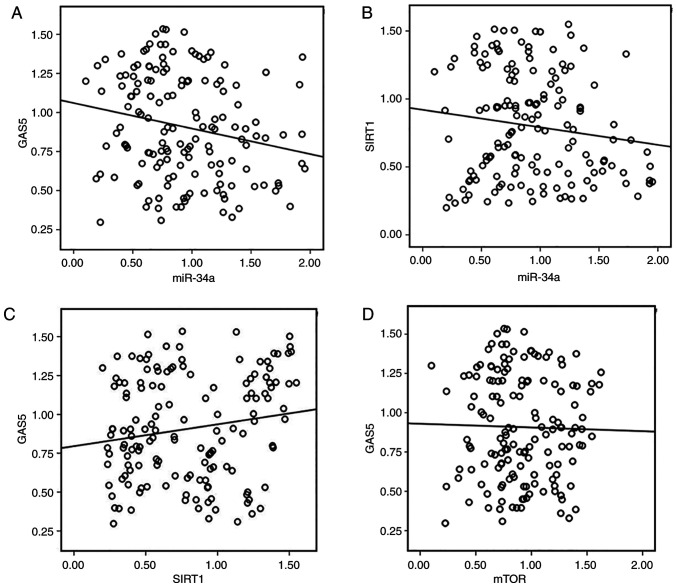
To evaluate further the clinical value and underlying mechanism of the GAS5/miR-34a/SIRT1 axis in CRC patients, we analyzed the correlation between expression of GAS5, miR-34a, SIRT1 and mTOR in human CRC tissues. Although the correlation between GAS5 and miR-34a (R=0.209, P=0.010, Fig. 2A), miR-34a and SIRT1 (R=−0.182, P=0.026, Fig. 2B) was weakly inverse the correlation was significant. Expression of GAS5 and SIRT1 was positively correlated in most of the tissues (R=0.163, P=0.046, Fig. 2C). However, our data indicated a mild inverse correlation between GAS5 and mTOR expression in CRC but the correlation was not significant (R=0.032, P=0.700, Fig. 2D). The 75 CRC patients were divided into two groups according to the median value (0.8699) of GAS5 mRNA expression in CRC tissues to evaluate further the association between expression of GAS5 and clinicopathological characteristics of CRC (Table III). The data indicated that GAS5 expression was inversely associated with samples with lymphatic metastasis and advanced clinical stages. However, age, sex, tumor size, poor cell differentiation, and high expression of carcinoembryonic antigen (CEA) and carbohydrate antigen (CA)19-9 had no association with GAS5 mRNA expression.
Figure 2.
Correlation between expression of GAS5, miR-34a, SIRT1 and mTOR in human CRC tissues. (A) Inverse correlation between GAS5 and miR-34a expression was determined by Spearman's rank correlation analysis (R=−0.209, P=0.010). (B) Inverse correlation between miR-34a and SIRT1 expression was determined by Spearman's rank correlation analysis (R=−0.182, P=0.026). (C) Expression of GAS5 and SIRT1 was positively correlated in the experimental tissues (R=0.163, P=0.046). (D) A mild inverse but not significant correlation between GAS5 and mTOR expression in human colon and CRC tissues (R=−0.032, P=0.700). CRC, colorectal cancer; GAS5, growth arrest specific 5; miR/miRNA, microRNA; SIRT1, sirtuin 1; mTOR, mammalian target of rapamycin.
GAS5 acts as a molecular sponge of miR-34a in CRC cells
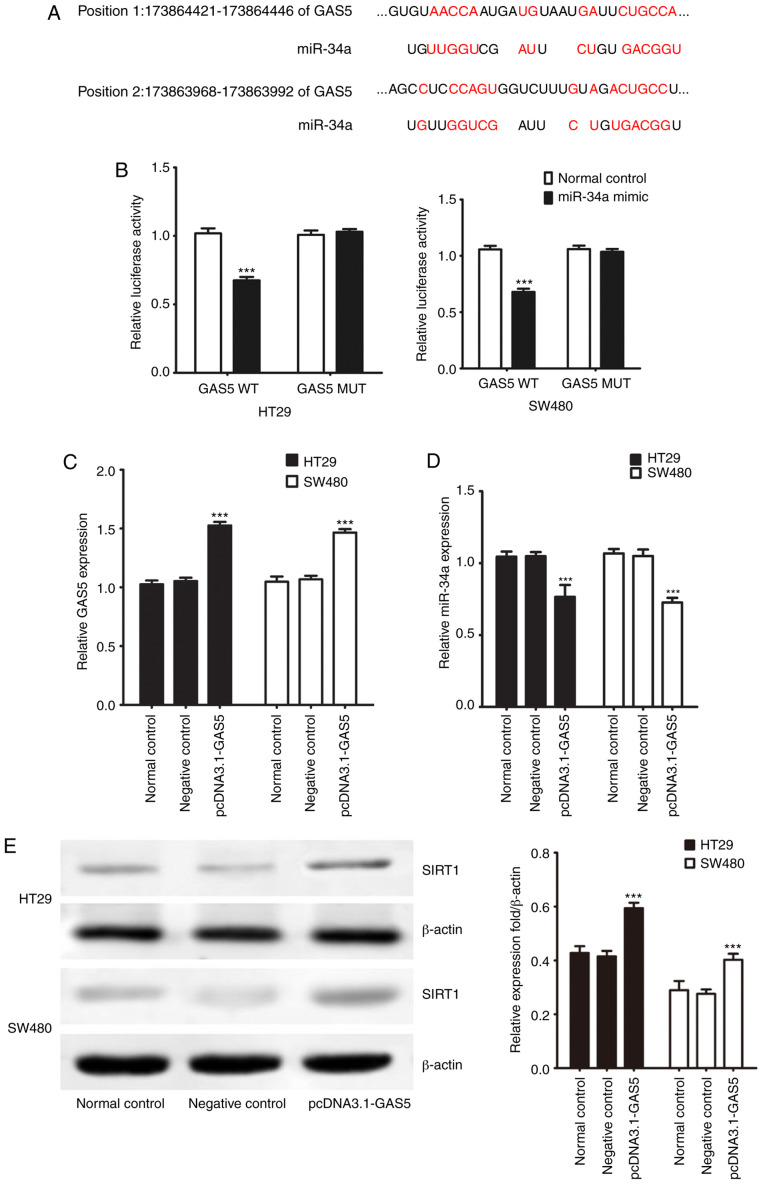
lncRNAs act as molecular sponges of miRNAs to exert their regulatory functions. Online bioinformatic analysis using the DIANA Tools suggested the presence of a common binding site for miR-34a and GAS5 (Fig. 3A). We previously found that SIRT1 is one of the target genes of miR-34a in CRC cells (17). Here, GAS5 WT/MUT (wild-type/mutant) was cloned into the double luciferase reporter gene vector and cotransfected into CRC cell lines with miR-34a mimics and mimic negative control to verify the targeting effect of GAS5 on miR-34a. miR-34a mimic transfection decreased luciferase activity compared to the negative control group (Fig. 3B). Additionally, after transfection with the mutated 3′ untranslated region (UTR) of the GAS5 gene, the luciferase activities did not differ significantly between the miR-34a mimics and negative control groups. Transfection of pcDNA3.1-GAS5 increased endogenous GAS5 expression but decreased miR-34a expression in the CRC cells (Fig. 3C and D). Western blotting showed that transfection with pcDNA3.1-GAS5 significantly promoted SIRT1 protein expression in the CRC cells (Fig. 3E). Thus, the results indicated that GAS5 negatively regulated expression of miR-34a at the post-transcriptional level in CRC cells. Since our correlation analysis of clinical specimens indicated that lncRNA GAS5 was related to lymph node metastasis, we evaluated cell invasion and migration activities in vitro. Protein expression of MMP2 and MMP9 was not significantly influenced by overexpression of GAS5 in CRC cells compared to the control group (Fig. S1). HT29 and SW480 cells were transfected with pcDNA3.1-GAS5 to evaluate cell invasion and migration activities using a cell invasion assay in Transwell chambers. Transfection with pcDNA3.1-GAS5 had no significant effect on cell migration and invasive capacity in the CRC cells (Fig. S2).
Figure 3.
GAS5 acts as a molecular sponge of miR-34a in CRC HT29 and SW480 cells. (A) Data from DIANA Tools showed a common binding site for GAS5 and miR-34a. (B) Luciferase reporter assay showed a reduction in luciferase activity of wild-type (WT) GAS5 3′ UTR following transfection with miR-34a mimic in CRC cells. (C) RT-qPCR analysis of expression of GAS5 in CRC cells after pcDNA3.1-GAS5 transfection. (D) RT-qPCR analysis of expression of miR-34a in CRC cells after pcDNA3.1-GAS5 transfection. (E) SIRT1 protein expression was examined by western blotting. ***P<0.001 indicates a significant difference vs. the normal control group. CRC, colorectal cancer; GAS5, growth arrest specific 5; miR/miRNA, microRNA; SIRT1, sirtuin 1.
miR-34a participates in regulating GAS5-mediated suppression of CRC cell macroautophagy and induces apoptosis through the mTOR/SIRT1 pathway
Previous studies have demonstrated that both miR-34a and GAS5 act as key regulators of macroautophagy and apoptosis (16–20). We investigated whether miR-34a was involved in mediating GAS5-regulated macroautophagy and apoptosis in CRC cells. Our data indicated that the number of MDC-positive cells (Fig. 4A) and autophagosomes (Fig. 4B) in pcDNA3.1-GAS5 treatment and miR-34a mimics transfection groups were significantly decreased compared to the controls. Western blotting showed lower expression of LC3-II, Beclin I and Bcl-2, and higher expression of SIRT1, p-mTOR and Bax (Fig. 4D) in the pcDNA3.1-GAS5 and miR-34a mimic transfection groups. Similarly, the apoptotic rate was significantly increased in CRC cells following transfection with pcDNA3.1-GAS5 and miR-34a mimics (Fig. 4C). Macroautophagy was enhanced but apoptosis was not further induced in HT29 and SW480 cell lines by miR-34a inhibitor transfection compared to the control group. Treatment with pcDNA3.1-GAS5 in combination with miR-34a mimics had no significant effect on p-mTOR expression, macroautophagy flux, or apoptosis, but impaired SIRT1 expression compared with pcDNA3.1-GAS5 treatment alone in the CRC cells (Fig. 4). Compared with pcDNA3.1-GAS5 treatment alone, the induction of p-mTOR expression and apoptosis and inhibitory effect on macroautophagy flux were impaired by pcDNA3.1-GAS5 treatment in combination with miR-34a inhibitor in CRC cells. This was shown by more MDC-positive cells (Fig. 4A) and autophagosomes (Fig. 4B), increased expression of LC3-II, Beclin I and Bcl2, decreased expression of Bax (Fig. 4D), and apoptotic rate detected by flow cytometry (Fig. 4C). Notably, no significant change was found in SIRT1 expression level between pcDNA3.1-GAS5 and pcDNA3.1-GAS5 treatment in combination with miR-34a inhibitor. These data indicated that the miR-34a/SIRT1 axis at least partly participated in mediating GAS5-suppressed macroautophagy and induced apoptosis through activation of the mTOR pathway in CRC cells.
Figure 4.
miR-34a participates in regulating GAS5-mediated suppression of CRC cell macroautophagy and induces apoptosis through the mTOR/SIRT1 pathway. (A) CRC HT29 and SW480 cells were transfected with pcDNA3.1-GAS5 with or without miR-34a mimics and miR-34a inhibitor. Flow cytometry detected the number of MDC-positive cells to evaluate macroautophagy levels. (B) Autophagic vacuole formation was shown by transmission electron microscopy of CRC cell lines. The arrows indicate the autophagosomes (scale bar, 1 µm). (C) Analysis of apoptosis by flow cytometry in HT29 and SW480 cells. (D) Western blotting was performed to detect p-mTOR, mTOR, SIRT1 and macroautophagy- and apoptosis-related proteins in CRC cells. ***P<0.001 indicates a significant difference vs. the control group; ###P<0.001 means a significant difference vs. the pcDNA3.1-GAS5 treatment group. MDC, mono-dansylcadaverine; CRC, colorectal cancer; GAS5, growth arrest specific 5; miR/miRNA, microRNA; SIRT1, sirtuin 1; mTOR, mammalian target of rapamycin; LC3, microtubule-associated protein light chain 3; p-, phosphorylated.
Activation of GAS5-suppressed macroautophagy is involved in promoting apoptosis in CRC cells
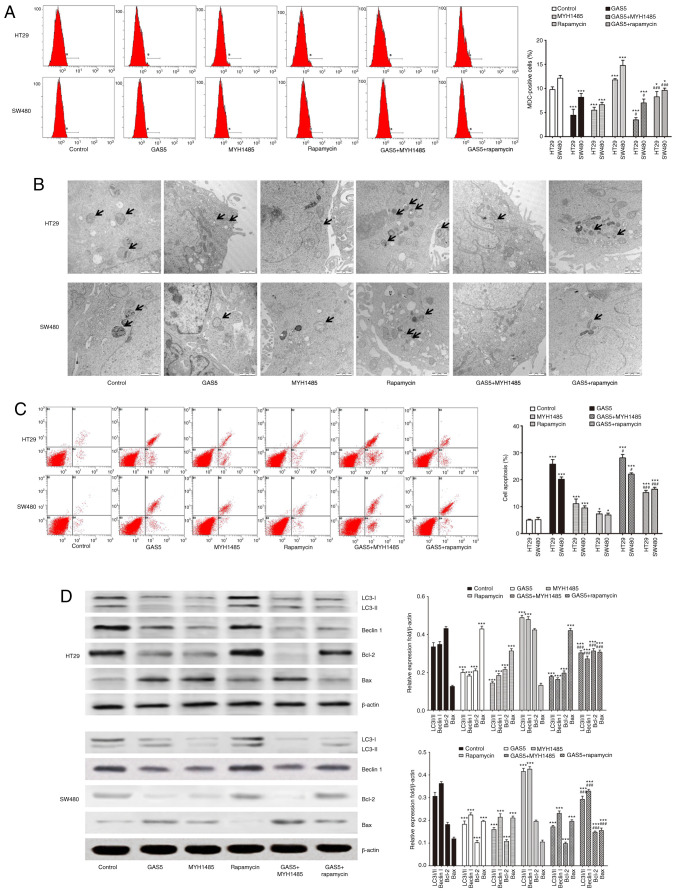
To address whether GAS5 regulated macroautophagy-modulated apoptosis in CRC cells, we transfected CRC cells with pcDNA3.1-GAS5 with or without MYH1485 and rapamycin treatment. pcDNA3.1-GAS5 transfection inhibited macroautophagy and induced apoptosis in CRC cells, as shown by decreased MDC-positive cells (Fig. 5A) and autophagosomes (Fig. 5B); lower expression of LC3-II, Beclin I and Bcl-2; and higher expression of Bax (Fig. 5D) as well as apoptosis detected by flow cytometry (Fig. 5C). MYH1485 treatment dramatically suppressed macroautophagy and induced apoptosis of CRC cells, while rapamycin further activated macroautophagy and protected CRC cells against apoptosis, as shown by more MDC-positive cells (Fig. 5A) and autophagosomes (Fig. 5B); and higher expression of macroautophagy-related protein and Bax; and lower expression of Bcl-2 (Fig. 5D) as well as decreased apoptosis detected by flow cytometry (Fig. 5C). Additionally, MYH1485 in combination with pcDNA3.1-GAS5 transfection enhanced the inhibitory effect on macroautophagy and induction of apoptosis in CRC cells, while rapamycin in combination with pcDNA3.1-GAS5 transfection attenuated the inhibitory effect on macroautophagy and induction of apoptosis in CRC cells compared with pcDNA3.1-GAS5 treatment alone (Fig. 5). Thus, the results strongly suggested that GAS5-mediated macroautophagy maintained cells in an equilibrium state that might have a protective effect on CRC cell apoptosis.
Figure 5.
Activation of GAS5-mediated suppression of macroautophagy is involved in promoting apoptosis in CRC HT29 and SW480 cells. (A) CRC cells were treated with pcDNA3.1-GAS5 with or without MYH1485 and rapamycin, and then flow cytometry was used to detect the number of MDC-positive cells to evaluate macroautophagy levels. (B) Macroautophagy vacuole formation is shown by transmission electron microscopy of CRC cell lines. The arrows indicate the autophagosomes (scale bar, 1 µm). (C) Apoptosis was analyzed by flow cytometry in CRC cells. (D) Western blotting shows expression of LC3, Beclin1, Bcl-2 and Bax in CRC cells. Quantification was performed. *P<0.05, ***P<0.001 means a significant difference vs. the control group; #P<0.05, ###P<0.001 means a significant difference vs. the pcDNA3.1-GAS5 treatment group. MDC, mono-dansylcadaverine; GAS5, growth arrest specific 5; CRC, colorectal cancer; LC3, microtubule-associated protein light chain 3.
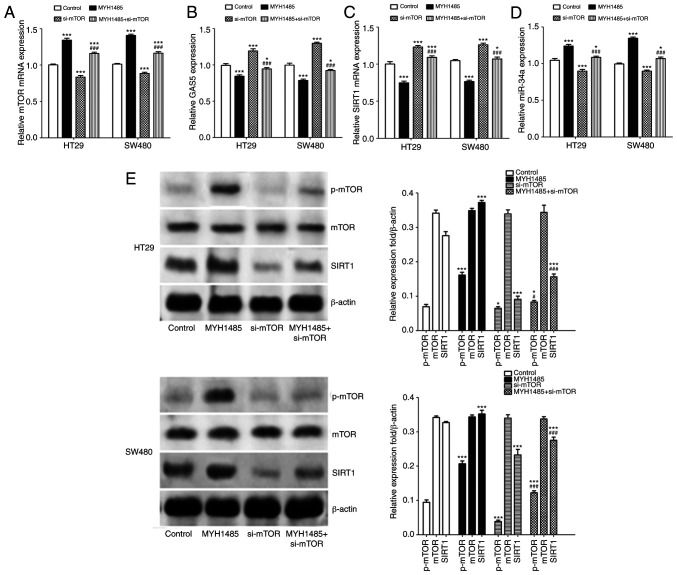
mTOR/SIRT1 pathway inhibits expression of GAS5 and forms a negative regulation feedback loop with miR-34a in CRC cells
The above results indicated that miR-34a participates in regulating GAS5-mediated macroautophagy and maintained CRC cells in an equilibrium state through the mTOR/SIRT1 pathway. More importantly, it has been reported that SIRT1 enhances mTOR expression in different conditions (23), and under conditions in which mTOR activity is high, GAS5 translation is promoted due to the presence of the 5′-TOR sequence. Conversely, when mTOR activity is low, this in turn results in accumulation of GAS5 transcripts (24). Thus, we speculated that a negative regulation feedback loop might be involved in mediating GAS5-regulated macroautophagy in CRC cells. To confirm our hypothesis, we examined the effect of activation of mTOR by MYH1485 or inhibition of mTOR by transfection of mTOR siRNAs on GAS5, miR-34a and SIRT1 expression in CRC cells. Compared with the control group, MYH1485 significantly elevated mTOR mRNA and protein expression in CRC cells, while transfection of mTOR siRNAs decreased expression of mTOR mRNA and protein. Additionally, when treated with MYH1485 and mTOR siRNAs simultaneously, mTOR expression was upregulated compared to transfection with mTOR siRNAs alone (Fig. 6A and E). As shown in Fig 6B and C, after treatment with MYH1485, GAS5 and SIRT1 mRNA expression was significantly downregulated and miR-34a expression was upregulated (Fig. 6D), while converse results were obtained when HT29 and SW480 cells were transfected with mTOR siRNAs. When treated with MYH1485 and mTOR siRNAs simultaneously, promotion of expression of GAS5 and SIRT1 and inhibition of expression of miR-34a were impaired compared to transfection with mTOR siRNAs alone (Fig. 6B, C and E). Based on these results and together with the inhibitory effect of pcDNA3.1-GAS5 transfection on miR-34a and the promotive effect on SIRT1 and mTOR expression in CRC cells, we suggest that a GAS5/miR-34a/mTOR/SIRT1 negative regulatory feedback loop in CRC cells is involved in mediating CRC progression.
Figure 6.
mTOR/SIRT1 pathway inhibits expression of GAS5 and forms a negative regulatory feedback loop with miR-34a in CRC HT29 and SW480 cells. (A-D) CRC cells were treated with MYH1485 with or without si-mTOR transfection, and then RT-qPCR was used to analysis mTOR mRNA, GAS5, SIRT1 mRNA and miR-34a expression. (E) Western blotting shows expression of p-mTOR, mTOR and SIRT1 in CRC cells. Quantification of the results was performed. *P<0.05, ***P<0.001 means a significant difference vs. the control group; #P<0.05, ###P<0.001 means a significant difference vs. the si-mTOR treatment group. GAS5, growth arrest specific 5; CRC, colorectal cancer; SIRT1, sirtuin 1; mTOR, mammalian target of rapamycin; p-, phosphorylated.
Discussion
Most studies have demonstrated that long non-coding RNA (lncRNA) growth arrest specific 5 (GAS5) is overexpressed in growth-arrested cells, as its name implies. This is further considered in the slowest dividing cells in the body as opposed to its lowest levels in other rapidly dividing cells, the most important of which are cancer cells (25). GAS5 performs a tumor-suppressor role in human cancer (5–15). However, separate studies have indicated that GAS5 promotes proliferation, migration and invasion in esophageal cancer and hepatocellular carcinoma (26,27). In human colorectal cancer (CRC), recent studies have verified that GAS5 contributes to lymphatic metastasis (10). There is increasing evidence for significant changes in miRNA expression profiles in cancer cells when GAS5 is activated under different conditions (5,11–15) and most of our previous studies were concerned with the tumor-suppressive gene miR-34a during progression of human gastrointestinal cancer (16,17). Thus, here we selected miR-34a as the candidate gene to investigate further the mechanism of GAS5 in the progression of CRC. We revealed that both GAS5 and miR-34a expression levels were aberrantly decreased in most, but not all human CRC tissues. In line with a previous study (10), our present results revealed that all the CRC cells had significantly lower levels of GAS5 and miR-34a compared to FHC cells. HT29 and SW480 cells expressed medium expression levels of GAS5, thus, these two cell lines were selected to explore further the role of GAS5 in the progression of CRC. The AOM-induced CRC rat model was used to evaluate the inhibitory effect of GAS5 on malignancy in vivo. We found that AOM intervention markedly induced the formation of colon tumors and blocked expression of GAS5 and miR-34a in colon tissues of rats. Our analysis of clinicopathological characteristics of CRC patients indicated that decreased expression of GAS5 was inversely correlated with lymphatic metastasis and advanced clinical stage. A crucial step in CRC invasion is the degradation of basement membrane, which is catalyzed by proteolytic enzymes, such as matix metalloproteinases (MMPs) (28). Thus, in the present study, we detected protein expression of MMP2 and MMP9 in CRC cells, and Transwell migration and invasion assays were performed to evaluated migration and invasion of CRC cells exposed to forced upregulation of GAS5. We verified that forced upregulation of GAS5 inhibited expression of miR-34a in CRC cells. However, no significant difference in MMP2 and MMP9 protein expression and no significant effect on cell migration and invasion were found by forced upregulation of GAS5 in HT29 and SW480 cells. Although expression of GAS5 and miR-34a was markedly decreased in most human CRC tissues in the present study, expression of GAS5 and miR-34a showed a significant weakly inverse correlation. The inconsistent results observed between the in vivo and in vitro models in our study could have many possible explanations. First, we measured the expression of GAS5 in tumor tissues obtained directly from CRC patients. The clinical-feature specificity of CRC may antagonize or interfere with the effect of GAS5 on CRC malignant behavior and miR-34a expression (29). Moreover, CRC cell lines are highly essential for functional molecular analysis. Yet, they may be different from primary tumors, which possibly through building up new mutations attempt to adjust their artificial environment (30). Such mutations could easily alter cellular responses and regulatory mechanisms, and thereby affect the expression of GAS5 and miR-34a. Second, miR-34a could be a non-specific molecule which can be regulated by internal stimuli (other regulatory molecules or polymorphisms, oxidative molecules, other associated disease states and tumor stage) and external stimuli (cellular response to environmental exposures, including chemotherapy and food) (4). Third, miR-34a acts as a target for many lncRNAs (31). We speculate that other unique regulatory networks participate in the development of CRC and regulate the expression of miR-34a. Every signal study focuses on one or a few targets and infers a tumor-suppressor or an oncogenic function based on its effect on the studied signaling pathway. However, miR-34a is a small molecule among the larger network, and the function of miR-34a could easily differ according to countless variables (4). These speculations need further investigation in the future. The final but the most important aspect is that the GAS5/miR-34a/SIRT1/mTOR negative regulatory feedback loop may partly explain why the expression of GAS5 and miR-34a was markedly decreased in most human CRC tissues, but their expression levels showed a significant weak inverse correlation. The above results suggest that GAS5 is involved in suppressing CRC malignant behavior at least partly through mediating miR-34a expression.
Sirtuin 1 (SIRT1) is a class III nuclear deacetylase that can inactivate the p53 pathway (32). Our previous study confirmed that miR-34a suppresses progression of CRC via binding to the site within the 3′ UTR of SIRT1 to silence SIRT1 mRNA (17). Regarding mammalian targets of rapamycin(mTOR), it was found to be involved in regulating macroautophagy activity independent of its enzymatic function based on cumulative evidence. Additionally, it has been reported that SIRT1 can activate the mTOR signaling pathway under different conditions (23). However, some studies have indicated that SIRT1 acts as a negative regulator of mTOR (33). In the present study, SIRT1 expression was significantly increased in most CRC tissues, all CRC cell lines, and AOM-treated colon tissues of rats, whereas mTOR expression showed opposite trends. Most importantly, we found a significant inverse correlation between miR-34a and SIRT1, and a significant positive correlation between GAS5 and SIRT1. Although GAS5 and mTOR revealed an inverse correlation, the result was not significant. In vitro, we found that forced upregulation of miR-34a had no significant effect on p-mTOR expression but impaired SIRT1 expression upon treatment with pcDNA3.1-GAS5 in CRC cells. Moreover, exogenous inhibition of miR-34a reduced the induction of p-mTOR expression but caused no significant change in SIRT1 expression after pcDNA3.1-GAS5 treatment in CRC cells. We speculate that the expression pattern of individuals, the clinical-feature specificity, or the different target genes involved in the regulatory network of CRC might be involved in antagonizing or interfering with expression of the mTOR/SIRT1 pathway, which remains to be elucidated in the future. The luciferase reporter results and clinical studies confirmed that activation of GAS5 inhibition of malignancy might be partly through regulating miR-34a in combination with suppressing SIRT1 expression, while the mTOR signaling pathway is more likely downregulated during progression of CRC.
Mounting evidence shows the inhibitory effects on macroautophagy and proliferation when GAS5 is activated in several types of human malignancies (18–20). Our own and other studies have indicated that miR-34a inhibits CRC cell macroautophagy and induces apoptosis through different signaling pathway in different conditions (16,17,34). We know that the mTOR pathway acts as a classical negative regulator of macroautophagy, and miR-34a was selected as the candidate gene to investigate the mechanism of GAS5 in the progression of CRC. Thus, in our present study, we investigated whether the miR-34a/SIRT1 axis is involved in mediating GAS5-regulated macroautophagy and apoptosis through the mTOR signaling pathway in CRC cells. We found that overexpression of GAS5 in combination with promotion of miR-34a expression had no significant effect on macroautophagy flux or apoptosis in CRC cells, but the inhibitory effect of macroautophagy and induction of apoptosis were impaired by overexpression of GAS5 in combination with silencing miR-34a in CRC cells compared to overexpression of GAS5 alone. Microtubule-associated protein light chain 3 (LC3) is a soluble protein that is distributed ubiquitously in mammalian tissues and cultured cells. A cytosolic form of LC3 (LC3-I) is conjugated to phosphatidylethanolamine to form LC3-phosphatidylethanolamine conjugate (LC3-II), which is recruited to autophagosomal membranes. At the same time, LC3-II in autolysosomal lumen is degraded. Thus, lysosomal turnover of the autophagosomal marker LC3-II reflects macroautophagy activity (35). Moreover, Beclin 1 is a central scaffold protein that assembles components for promoting or inhibiting macroautophagy. Binding of Beclin I with Bcl-2 family and (or) class III phosphatidylinositol 3-kinase (PI3KC3) has been shown to play a vital role during the formation of mammalian autophagosomes in response to external stimuli. Thus, detecting LC3 and Beclin I by immunoblotting or immunofluorescence has become a reliable method for monitoring macroautophagy-related processes in different conditions (36). For the detection of apoptosis, it is established that cancer cells reprogram the Bcl-2 family interaction network that regulates mitochondrial apoptosis. Overexpression of antiapoptotic Bcl-2 family members such as Bcl-2 and Bcl-XL bind and neutralize the BH3 death domains of the activated proapoptotic Bcl-2 members, such as Bax (37). Cells deficient in Bax become less sensitive to various apoptotic stimuli and become resistant when Bax is deleted. Selective inhibitors of antiapoptotic Bcl-2 proteins are effective inducers of apoptosis because they release BH3-only proteins from the antiapoptotic Bcl-2 proteins to activate Bax (38). Thus, protein expression of Bcl-2 and Bax was measured in the present study by western blotting. In another method to analyze apoptosis in vitro, we used Annexin V-FITC, a protein fluorophore with high affinity to phosphatidylserines during the early stage of apoptosis, and PI staining, red fluorescence to detect dead cells, which are used as a standard protocol for measuring late apoptotic cells and dead cells (39). Caspases also have been used as an apoptosis-specific target. In particular, caspase-3- and caspase-7-specifc cleavable peptide substrates have been extensively used as caspase-cleavable imaging probes for apoptosis imaging for monitoring of caspase activity in tumor cells (40). The activity of caspases regulated by GAS5 in CRC cells remains to be elucidated in the future. As we noted previously, GAS5 exerts its molecular effects by targeting multiple genes and signaling pathways. The antitumor effects and potential mechanisms underlying GAS5-mediated macroautophagy or miRNAs might be regulated by complicated signaling pathways (18–20). A limitation of the present study might be that our data only partly suggested that miR-34a participated in mediating GAS5 suppression of macroautophagy and induction of apoptosis in CRC cells through inhibiting SIRT1 expression and activating the mTOR signaling pathway, and more studies are needed in the future.
It has been established that macroautophagy has a dual role in regulating apoptosis of cancer cells (41). Although increasing evidence has focused on the association between macroautophagy and progression of malignant tumors, it is far from being clarified. The present study indicated that MYH1485 suppressed CRC cell macroautophagy and induced apoptosis, but rapamycin treatment caused the inverse trend. More importantly, in combination with activation of GAS5, MYH1485 enhanced the inhibitory effect on macroautophagy and promotion of apoptosis, while rapamycin in combination with activation of GAS5 attenuated the inhibitory effect on macroautophagy and induction of apoptosis in CRC cells compared to upregulation of GAS5 alone. These results showed that GAS5-regulated macroautophagy developed a steady activation state that exerted an antiapoptotic effect during CRC progression. Thus, we thought that negative feedback regulation between GAS5 and its target pathways might be involved in mediating macroautophagy activation, but not excessive activation, which might be induced by apoptosis of CRC cells.
A negative feedback regulatory loop might not only amplify a response but also control a self-sustained mode that is autonomous from the original stimuli (42). Recently, there has been a focus on positive or negative feedback regulations between lncRNAs and their target miRNAs (43). SIRT1 has been reported to activate the mTOR pathway directly in different conditions (23), and notably, GAS5 is one of the downstream targets that is negatively regulated by the mTOR pathway (24). Thus, we examined the effect of mTOR activation or silencing on the GAS5/miR-34a axis and SIRT1 expression in CRC cells. Our data indicated that silencing mTOR expression resulted in upregulation of GAS5 and SIRT1 expression and downregulation of miR-34a expression, while converse results were obtained when the mTOR pathway was activated in CRC cells. Based on these results and together with the inhibitory effect of exogenous overexpression of GAS5 on miR-34a and promotive effect on SIRT1 and mTOR expression in CRC cells, we proposed a GAS5/miR-34a/mTOR/SIRT1 negative regulatory feedback loop in CRC cells that mediates CRC progression. Our present study suggests that after deficient expression of GAS5, the miR-34a/SIRT1 axis is activated and in turn promotes mTOR pathway phosphorylation, which results in accumulation of GAS5 transcripts. The GAS5/miR-34a/SIRT1/mTOR negative regulatory feedback loop might partly explain why the CRC cell macroautophagy was in an autonomously relative equilibrium state, but not excessive activation state, which function as a strong anti-apoptosis phenotype during human CRC progression.
In summary, we demonstrated that GAS5 acts as a tumor-suppressive factor in progression of human CRC, and exogenous overexpression of GAS5 inhibits tumorigenesis, at least in part regulated through the miR-34a/SIRT1 axis. miR-34a participates in regulating GAS5-suppressed CRC cell macroautophagy and induces apoptosis through the mTOR/SIRT1 pathway. GAS5-mediated regulation of macroautophagy maintains CRC cells in an equilibrium state that protects against apoptosis. GAS5/miR-34a/SIRT1/mTOR forms a negative regulatory feedback loop that provides new insight into the mechanisms of CRC progression and the influence of macroautophagy on the malignant behavior of CRC through external factors. Macroautophagy may be a potential method to screen out relevant target molecules and provide a reliable basis for the clinical diagnosis and treatment of CRC.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CRC
colorectal cancer
- lncRNA
long non-coding RNA
- GAS5
growth arrest specific 5
- miR/miRNA
microRNA
- MMP
matrix metalloproteinase
- SIRT1
sirtuin 1
- UTR
untranslated region
- AOM
azoxymethane
- PBS
phosphate-buffered saline
- MDC
mono-dansylcadaverine
- RT-qPCR
quantitative reverse transcription polymerase chain reaction
- NC
negative control
- mTOR
mammalian target of rapamycin
- CA
carbohydrate antigen
- CEA
carcinoembryonic antigen
Funding
The present study was supported by The Science Foundation of the Health Commission of Heilongjiang Province (2018346).
Availability of data and materials
The data used in the present study are available from the corresponding author upon reasonable request.
Authors' contributions
PFQ designed the experiments. HGZ and YW performed the experiments. ZXZ analyzed the data. PFQ and FJW carried out the literature review, drafted the manuscript and revised it critically for important intellectual content. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All patients provided written informed consent according to our institutional guidelines and the study protocol was approved by the Institutional Review Board of Harbin Medical University. Animal experiments were carried out in strict accordance with the Harbin Medical University Institutional Animal Care and Use Committee (KY2018-208).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.Doubeni CA, Corley DA, Quinn VP, Jensen CD, Zauber AG, Goodman M, Johnson JR, Mehta SJ, Becerra TA, Zhao WK, et al. Effectiveness of screening colonoscopy in reducing the risk of death from right and left colon cancer: A large community-based study. Gut. 2018;67:291–298. doi: 10.1136/gutjnl-2016-312712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibb EA, Brown CJ, Lam W L. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toraih EA, Alghamdi SA, El-Wazir A, Hosny MM, Hussein MH, Khashana MS, Fawzy MS. Dual biomarkers long non-coding RNA GAS5 and microRNA-34a co-expression signature in common solid tumors. PLoS One. 2018;13:e0198231. doi: 10.1371/journal.pone.0198231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xue D, Zhou C, Lu H, Xu R, Xu X, He X. lncRNA GAS5 inhibits proliferation and progression of prostate cancer by targeting miR-103 through AKT/mTOR signaling pathway. Tumor Biol. 2016 Oct 14; doi: 10.1007/s13277-016-5429-8. (Epub ahead of print). doi: 10.1007/s13277-016-5429-8. [DOI] [PubMed] [Google Scholar]
- 6.Liu L, Pang X, Shang W, Xie H, Feng Y, Feng G. Long non-coding RNA GAS5 sensitizes renal cell carcinoma to sorafenib via miR-21/SOX5 pathway. Cell Cycle. 2018;18:257–263. doi: 10.1080/15384101.2018.1475826. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Gao J, Liu M, Zou Y, Mao M, Shen T, Zhang C, Song S, Sun M, Zhang S, Wang B, et al. Long non-coding RNA growth arrest-specific transcript 5 is involved in ovarian cancer cell apoptosis through the mitochondria-mediated apoptosis pathway. Oncol Rep. 2015;34:3212–3221. doi: 10.3892/or.2015.4318. [DOI] [PubMed] [Google Scholar]
- 8.Wen Q, Liu Y, Lyu H, Xu X, Wu Q, Liu N, Yin Q, Li J, Sheng X. Long noncoding RNA GAS5, which acts as a tumor suppressor via microRNA 21, regulates cisplatin resistance expression in cervical cancer. Int J Gynecol Cancer. 2017;27:1096–1108. doi: 10.1097/IGC.0000000000001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye K, Wang S, Zhang H, Han H, Ma B, Nan W. Long noncoding RNA GAS5 suppresses cell growth and epithelial-mesenchymal transition in osteosarcoma by regulating the miR-221/ARHI pathway. J Cell Biochem. 2017;118:4772–4781. doi: 10.1002/jcb.26145. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Y, Song D, Xiao K, Yang C, Ding Y, Deng W, Tong S. lncRNA GAS5 contributes to lymphatic metastasis in colorectal cancer. Oncotarget. 2016;7:83727–83734. doi: 10.18632/oncotarget.13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, Wu F, Mo YY. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W, Hong L, Xu X, Wang Q, Huang J, Jiang L. lncRNA GAS5 suppresses the tumorigenesis of cervical cancer by downregulating miR-196a and miR-205. Tumour Biol. 2016;39:101042831771131. doi: 10.1177/1010428317711315. [DOI] [PubMed] [Google Scholar]
- 13.Guo C, Song WQ, Sun P, Jin L, Dai HY. lncRNA-GAS5 induces PTEN expression through inhibiting miR-103 in endometrial cancer cells. J Biomed Sci. 2015;22:100. doi: 10.1186/s12929-015-0213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao ZQ, Wang JF, Chen DH, Ma XS, Wu Y, Tang Z, Dang XW. Long non-coding RNA GAS5 suppresses pancreatic cancer metastasis through modulating miR-32-5p/PTEN axis. Cell Biosci. 2017;7:66. doi: 10.1186/s13578-017-0192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Gu J, Lu H. The GAS5/miR-222 Axis regulates proliferation of gastric cancer cells through the PTEN/Akt/mTOR Pathway. Dig Dis Sci. 2017;62:3426–3437. doi: 10.1007/s10620-017-4831-4. [DOI] [PubMed] [Google Scholar]
- 16.Sun C, Wang FJ, Zhang HG, Xu XZ, Jia RC, Yao L, Qiao PF. miR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-β/Smad4 pathway. World J Gastroenterol. 2017;23:1816–1827. doi: 10.3748/wjg.v23.i10.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiao PF, Yao L, Zeng ZL. Catalpol-mediated microRNA-34a suppresses autophagy and malignancy by regulating SIRT1 in colorectal cancer. Oncol Rep. 2020;43:1053–1066. doi: 10.3892/or.2020.7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu J, Wang Y, Wang X, Zhou D, Wang X, Zhou M, He Z. Effect of the lncRNA GAS5-MiR-23a-ATG3 axis in regulating autophagy in patients with breast cancer. Cell Physiol Biochem. 2018;48:194–207. doi: 10.1159/000491718. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Huang C, He Y, Sang Z, Liu G, Dai H. Knockdown of Long non-coding RNA GAS5 increases miR-23a by targeting ATG3 involved in autophagy and cell viability. Cell Physiol Biochem. 2018;48:1723–1734. doi: 10.1159/000492300. [DOI] [PubMed] [Google Scholar]
- 20.Zhang N, Yang GQ, Shao XM, Wei L. GAS5 modulated autophagy is a mechanism modulating cisplatin sensitivity in NSCLC cells. Eur Rev Med Pharmacol. 2016;20:2271–2277. [PubMed] [Google Scholar]
- 21.Sobin L, Gospodarowicz M, Wittekind C, editors. TNM Classification of Malignant Tumors. https://media.wiley.com/product_data/coverImage300/60/14443589/1444358960.jpg UICC International Union Against Cancer. (7th edition) 2009
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Zhang T, Du X, Zhao L, He M, Lin L, Guo C, Zhang X, Han J, Yan H, Huang K, et al. SIRT1 facilitates primordial follicle recruitment independent of deacetylase activity through directly modulating Akt1 and mTOR transcription. FASEB J. 2019;33:14703–14716. doi: 10.1096/fj.201900782R. [DOI] [PubMed] [Google Scholar]
- 24.Pickard MR, Williams GT. Molecular and cellular mechanisms of action of tumour suppressor GAS5 lncRNA. Genes (Basel) 2015;6:484–499. doi: 10.3390/genes6030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/S0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 26.Li W, Zhao W, Lu Z, Zhang W, Yang X. Long noncoding RNA GAS5 promotes proliferation, migration, and invasion by regulation of miR-301a in esophageal cancer. Oncol Res. 2018;26:1285–1294. doi: 10.3727/096504018X15166193231711. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Tao R, Hu S, Wang S, Zhou X, Zhang Q, Wang C, Zhao X, Zhou W, Zhang S, Li C, et al. Association between indel polymorphism in the promoter region of lncRNA GAS5 and the risk of hepatocellular carcinoma. Carcinogenesis. 2015;36:1136–1143. doi: 10.1093/carcin/bgv099. [DOI] [PubMed] [Google Scholar]
- 28.Jia YL, Shi L, Zhou JN, Fu CJ, Chen L, Yuan HF, Wang YF, Yan XL, Xu YC, Zeng Q, et al. Epimorphin promotes human hepatocellular carcinoma invasion and metastasis through activation of focal adhesion kinase/extracellular signal-regulated kinase/matrix metalloproteinase-9 axis. Hepatology. 2011;54:1808–1818. doi: 10.1002/hep.24562. [DOI] [PubMed] [Google Scholar]
- 29.Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, Klaus S, Snyman H, Hannon GJ, Bork P, Arendt D. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463:1084–1088. doi: 10.1038/nature08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borrell B. How accurate are cancer cell lines? Nature. 2010;463:858. doi: 10.1038/463858a. [DOI] [PubMed] [Google Scholar]
- 31.Li C, Liu T, Zhang Y, Li Q, Jin LK. lncRNA-ZDHHC8P1 promotes the progression and metastasis of colorectal cancer by targeting miR-34a. Eur Rev Med Pharmacol Sci. 2019;23:1476–1486. doi: 10.26355/eurrev_201902_17105. [DOI] [PubMed] [Google Scholar]
- 32.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 33.Romeo-Guitart D, Leiva-Rodriguez T, Forés J, Casas C. Improved motor nerve regeneration by SIRT1/Hif1a-mediated autophagy. Cells. 2019;8:1354. doi: 10.3390/cells8111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu K, Huang J, Xie M, Yu Y, Zhu S, Kang R, Cao L, Tang D, Duan X. miR-34a regulates autophagy and apoptosis by targeting HMGB1 in the retinoblastoma cell. Autophagy. 2014;10:442–452. doi: 10.4161/auto.27418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 36.Fujiwara N, Usui T, Ohama T, Sato K. Regulation of beclin 1 protein phosphorylation and autophagy by protein phosphatase 2A (PP2A) and death-associated protein kinase 3. J Biol Chem. 2016;13:10858–10866. doi: 10.1074/jbc.M115.704908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letai AG. Diagnosing and exploiting cancer's addiction to blocks in apoptosis. Nat Rev Cancer. 2008;8:121–132. doi: 10.1038/nrc2297. [DOI] [PubMed] [Google Scholar]
- 38.Reyna DE, Garner TP, Lopez A, Kopp F, Choudhary GS, Sridharan A, Narayanagari SR, Mitchell K, Dong B, Bartholdy BA, et al. Direct activation of BAX by BTSA1 overcomes apoptosis resistance in acute myeloid leukemia. Cancer Cell. 2017;32:490–505.e10. doi: 10.1016/j.ccell.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shim MK, Yoon HY, Lee S, Jo MK, Park J, Kim JH, Jeong SY, Kwon IC, Kim K. Caspase-3/-7-specific metabolic precursor for bioorthogonal tracking of tumor apoptosis. Sci Rep. 2017;7:16635. doi: 10.1038/s41598-017-16653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brentnall M, Rodriguez-Menocal L, De Guevara RL, Cepero E, Boise LH. Caspase-9, caspase-3 and caspase-7 have distinct roles during intrinsic apoptosis. BMC Cell Biol. 2013;14:32. doi: 10.1186/1471-2121-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maiuri M, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 42.Misso G, Zarone MR, Lombardi A, Grimaldi A, Cossu AM, Ferri C, Russo M, Vuoso DC, Luce A, Kawasaki H, et al. miR-125b Upregulates miR-34a and sequentially activates stress adaption and cell death mechanisms in multiple myeloma. Mol Ther Nucleic Acids. 2019;16:391–406. doi: 10.1016/j.omtn.2019.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian F, Wang J, Zhan Z, Yang J. lncRNA SNHG7/miR-34a-5p/SYVN1 axis plays a vital role in proliferation, apoptosis and autophagy in osteoarthritis. Biol Res. 2020;53:9. doi: 10.1186/s40659-020-00275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in the present study are available from the corresponding author upon reasonable request.