Abstract
Accumulating evidence has indicated that circular RNAs (circRNAs) serve crucial roles in the progression of a diverse range of different types of cancer, including osteosarcoma (OS). The present study determined the expression pattern and function of circRNA homeodomain interacting protein kinase 3 (circHIPK3), a novel circular RNA, in OS. It was revealed that circHIPK3 expression was upregulated in OS tissue samples and OS cell lines. A localization assay revealed that circHIPK3 was primarily located in the cytoplasm. Using loss-of-function proliferation and Transwell assays, the present study revealed that circHIPK3-knockdown suppressed OS cell proliferation, migration and invasion. Furthermore, the present study screened potential microRNAs that may interact with circHIPK3. It was revealed that microRNA-637 (miR-637) expression was downregulated in OS according to a Gene Expression Omnibus data analysis. In addition, the present study demonstrated that miR-637 expression was downregulated in OS cell lines. A fluorescence in situ hybridization assay revealed that both miR-637 and circHIPK3 were located in the cytoplasm. An in-depth mechanism investigation demonstrated that circHIPK3 expression was inversely correlated with miR-637 expression, and that circHIPK3 was a target of miR-637. In addition, it was revealed that histone deacetylase 4 (HDAC4) was another downstream target gene of miR-637, as demonstrated using a luciferase assay. It was revealed that miR-637 suppressed OS cell proliferation, migration and invasion via targeting of HDAC4. Finally, the present study demonstrated that circHIPK3 sponged miR-637 to promote HDAC4 expression and OS cell proliferation, migration and invasion. In conclusion, the present study uncovered the role of the circHIPK3/miR-637/HDAC4 axis in OS cell proliferation, migration and invasion. It was demonstrated that circHIPK3 promoted OS cell proliferation, migration and invasion by modulating miR-637/HDAC4 signaling.
Keywords: osteosarcoma, circular RNA HIPK3, microRNA-637, migration and invasion, proliferation
Introduction
Osteosarcoma (OS) is the most prevalent primary malignant bone tumor in young adolescents (1). According to an epidemiology study of osteosarcoma in 2009, the incidence rate of OS is ~4 cases per million individuals in young children and adolescents in the United States (2). The prognosis of OS is usually unfavorable due to its aggressive nature (3). There is a high rate of disability and mortality among patients with OS (4). Due to the lack of effective biomarkers for an early diagnosis of OS, the majority of patients with OS are diagnosed at an advanced stage at their first visit (5,6). Therefore, identification of novel targets for the molecular treatment of OS is urgently required.
Circular (circ) RNAs are a type of endogenously-expressed covalently closed RNA transcripts with limited protein coding ability. Increasing evidence has indicated that circRNAs are intensively associated with cancer progression and metastasis (7,8). circRNA homeodomain interacting protein kinase 3 (circHIPK3) was first identified by ribominus RNA sequencing data from six human normal tissues and seven types of human cancer (9). circHIPK3 acts as an oncogene in various types of malignant tumor (10–13). Zheng et al (9) reported that circHIPK3 increases the expression levels of miR-7-targeting proto-oncogenes (protein tyrosine kinase 2, insulin like growth factor 1 receptor, EGFR and YY1 transcription factor), and promotes colorectal cancer growth and metastasis. Chen et al (14) revealed that circHIPK3 serves as a miR-124 sponge and regulates aquaporin 3 (Gill blood group)-mediated proliferation and migration in hepatocellular carcinoma. To the best of our knowledge, the role of circHIPK3 in OS remains to be elucidated.
Histone deacetylase 4 (HDAC4) is a key member of class IIa HDACs, and HDAC4 performs a wide variety of functions. Additionally, HDAC4 is post-transcriptionally regulated by several microRNAs (miRNAs/miRs), such as miR-1, miR-29, miR-140, mir-155, miR-200a, miR-206 and miR-365, in various cells (15). Our previous study demonstrated that HDAC4 is a target of miR-140-5p, and HDAC4 was closely associated with proliferation and apoptosis in OS (16). Zeng et al (17) demonstrated that HDAC4 is upregulated in esophageal carcinoma (EC), and HDAC4 promotes migration of EC cells by enhancing the epithelial-to-mesenchymal transition. Currently, to the best of our knowledge, research on circRNAs and HDAC4 is limited.
The present study focused on the expression and function of circHIPK3 in OS. It was revealed that circHIPK3 served as an oncogene, and circHIPK3 promoted OS cell proliferation, migration and invasion through modulation of HDAC4 via sponging of microRNA 637 (miR-637).
Materials and methods
Patients and tissue samples
A total of 12 chondroma (8 males and 4 females; age range, 17–49 years; mean age, 28.4 years) and 12 OS (6 males and 6 females; age range, 6–16 years; mean age, 11.5 years) samples were obtained from Shengjing Hospital (Shenyang, China) during tumorectomy between September 2014 and February 2019. All tissue specimens were histologically diagnosed by two pathologists according to the criteria defined by the World Health Organization (18). Written informed signed consent was obtained from each patient or the parents of the patients who were minors for the use of tissue specimens in the present study. The Institute Research Medical Ethics Committee of Shengjing Hospital approved the present study.
Cell culture
The human osteoblast hFOB 1.19 cell line was cultured in DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.). The four human OS cell lines (HOS, MG-63, U2OS and SJSA) were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.). All cell lines were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Media were supplemented with 10% (v/v) FBS (Invitrogen; Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin and 100 mg/ml streptomycin (Baomanbio). hFOB 1.19 cells were maintained at 34°C with 5% CO2. OS cell lines were maintained in a humidified atmosphere containing 5% CO2 at 37°C.
GEO database reanalysis and bioinformatics RNA-RNA interaction prediction
The differentially expressed miRNA data in OS were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) GSE70415 (19) and GSE28423 (20). Differentially expressed miRNAs between 18 ccRCC tissues and 18 paired ccRCC normal tissues were reanalyzed using the online software GEO2R (21). The expression levels of miR-508-3p in 18 ccRCC tissues and 18 paired ccRCC normal tissues were also reanalyzed in GSE116251 (22). To predict the potential circRNA-miRNA and mRNA-miRNA interactions, circBank (23), RegRNA (version 2.0) (24) and circularRNA interactome (25) were used according to the corresponding instructions.
RNase R treatment
The stability of circHIPK3 was determined by an Rnase R assay as previously described (26). Briefly, 2 mg total RNA was incubated for 30 min at 37°C with or without 5 U/µg RNase R (Epicentre; Illumina, Inc.) and subsequently purified using an RNeasy MinElute Cleaning kit (Qiagen GmBH), followed by analysis by reverse transcription (RT)-PCR.
Actinomycin D assay
The actinomycin D assay was performed as previously described (26). HOS and U2OS cells were exposed to 2 µg/ml actinomycin D for 30 min (Sigma-Aldrich; Merck KGaA) at 37°C at different time points (4, 8, 12 and 24 h). Total RNA was extracted at different time points and the expression levels of circHIPK3 and linear HIPK3 mRNA were determined by RT-quantitative (q)PCR.
Reverse transcription-quantitative PCR (RT-qPCR)
RT-qPCR was performed as previously described (27), and the 2−∆∆Cq method was used for quantification (28). Total RNA from tissue specimens and cultured cells was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RT reactions were performed at 44°C for 1 h followed by 92°C for 10 min to synthesize cDNA with the PrimeScript RT Master Mix (Takara Bio, Inc.) from 500 ng RNA. Stem-loop reverse transcription reactions were performed for miR-637 using the TaqMan MicroRNA detection kit (Thermo Fisher Scientific, Inc.). The qPCR analyses were performed using SYBR Premix Ex Taq II (Takara Bio, Inc.). β-actin and U6 were employed as endogenous controls for mRNAs and miR-637, respectively. PCR amplification conditions were as follows: 2 min at 95°C for one cycle, followed by denaturation for 15 sec at 95°C and extension for 60 sec at 60°C for 38 cycles. Primer sequences are listed in Table I.
Table I.
Primer and oligonucleotide sequences used in the present study.
| Primers/oligonucleotides | Sequence (5′-3′) |
|---|---|
| circHIPK3 forward | TATGTTGGTGGATCCTGTTCGGCA |
| circHIPK3 reverse | TGGTGGGTAGACCAAGACTTGTGA |
| HIPK3 forward | TGGAGACTGGGGGAAGATGA |
| HIPK3 reverse | CACACTAACTGGCTGAGGGG |
| HDAC4 forward | TCAGACATCTTTGGGAAGGG |
| HDAC4 reverse | CAACCTCCATCTTGCCTTGT |
| GAPDH forward | GTCAAGGCTGAGAACGGGAA |
| GAPDH reverse | AAATGAGCCCCAGCCTTCTC |
| miR-637 forward | ACUGGGGGCUUUCGGGCUCUGCGU |
| miR-637 reverse | ACGCAGAGCCCGAAAGCCCCCAGU |
| U6 forward primer | CTCGCTTCGGCAGCACA |
| U6 reverse primer | AACGCTTCACGAATTTGCGT |
| sicircHIPK3_1 sense | UGGCCUCACAAGUCUUGGU |
| sicircHIPK3_1 antisense | ACCAAGACUUGUGAGGCCAUA |
| sicircHIPK3_2 sense | GGUACUACAGGUAUGGCCUTT |
| sicircHIPK3_2 antisense | AGGCCAUACCUGUAGUACCGA |
| siHDAC4 sense | GAAAAGGUUUUACAGCAAATT |
| siHDAC4 antisense | UUUGCUGUAAAACCUUUUCTG |
| miR-637 mimic | ACUGGGGGCUUUCGGGCUCUGCGU |
| miR-637 inhibitor | ACGCAGAGCCCGAAAGCCCCCAGU |
circ, circular RNA; HDAC4, histone deacetylase 4; HIPK3, homeodomain interacting protein kinase 3; miR-637, microRNA-637; si, small interfering RNA.
RNA fluorescence in situ hybridization (FISH)
FISH was performed as previously described (27). Specific probes targeting circHIPK3 or miR-637 were synthesized by Guangzhou RiboBio Co., Ltd. The RNA FISH assay was performed using a FISH kit (Guangzhou RiboBio Co., Ltd.) according to the manufacturer's protocols. In brief, HOS and U2OS cells were seeded onto glass coverslips (0.8×0.8 cm) and cultured to 80–95% confluence. The coverslips were rinsed twice with PBS, fixed with 4% paraformaldehyde for 10 min at room temperature and blocked with 5% BSA (Sigma-Aldrich; Merck KGaA) for 1 h at room temperature. Then, the coverslips were permeabilized with 0.3% Triton X-100 (Sigma-Aldrich; Merck KGaA) for 15 min and incubated in a hybridization solution containing 1 µM Cy3-labeled circHIPK3 probes and 1 µM Dig-labeled locked nucleic acid miR-637 probes (Guangzhou RiboBio Co., Ltd.) supplemented with 1% BSA in a humid chamber at 37°C overnight. The next day, the coverslips were rinsed with a solution of 0.1% Tween-20 (Sigma-Aldrich; Merck KGaA) in 4X sodium citrate buffer (SSC; Sigma-Aldrich; Merck KGaA) for 5 min, a solution of 0.1% Tween-20 in 2X SSC for 5 min and a solution of 0.1% Tween-20 in 1X SSC for 5 min at 42°C in dark. Lastly, the coverslips were washed three times with 1X PBS for 5 min at room temperature. The signals of circHIPK3 probes or miR-637 probes were detected using Cy5-Streptavidin (Thermo Fisher Scientific, Inc.) or a tyramide-conjugated Alexa 488 fluorochrome TSA kit (Thermo Fisher Scientific, Inc.). Nuclei were counterstained with 4,6-diamidino-2-phenylindole at room temperature for 10 min. The images were acquired on a Leica SP5 confocal microscope (Leica Micosystems GmbH).
Oligonucleotide transfection
miR-637 mimics and negative control (NC mimic), miR-637 inhibitors and negative control (NC inhibitor), effective siRNA oligonucleotides that targeted the splicing sites of circHIPK3 (designed using circPrimer; version 1.2.0.5) (29) or targeted HDAC4 and scrambled control siRNAs were synthesized by Guangzhou RiboBio Co., Ltd. When the OS cells reached 80% confluence, the oligonucleotides (100 nM) were transfected using Lipofectamine® RNAiMax (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After 48 h of transfection, the OS cells were used for further detection. The sequences of the oligonucleotides are listed in Table I.
Cell Counting Kit 8 (CCK8) assay
The CCK8 assay was performed as previously described (30). HOS and U2OS cells were seeded in 96-well plates (2×103 cells/well) in 200 µl culture medium and conditioned at 37°C with 5% CO2. At days 1, 2, 3, 4 and 5, 10 µl CCK8 solution (Dojindo Molecular Technologies, Inc.) was added into each well and incubated at 37°C for 2 h according to the manufacturer's protocol. The absorbance was measured at an optical density of 450 nm using a microplate reader (Bio-Rad Laboratories, Inc.). The experiments were performed in triplicate.
Transwell assay
The transwell assay was performed as previously described (31). First, upper chambers (Corning Inc.) were pre-coated with (for the invasion assay; precoating for 1 h at room temperature) or without (for the migration assay) 20 µg Matrigel (BD Biosciences). HOS and U2OS cells (at a density of 1×105) were then seeded into the upper chambers. Culture medium without serum and medium with 10% FBS was added into the upper and lower chambers, respectively. Following incubation at 37°C for 24 h, the migrated or invaded OS cells were fixed with 4% paraformaldehyde at room temperature for 30 min, stained with crystal violet at 25°C for 1 min and counted under a light microscope (Olympus Corporation).
Wound healing assay
The wound healing assay was performed as previously described (32). HOS and U2OS cells were seeded and transfected with target plasmids in 6-well plates supplemented with culture medium with 10% FBS, and allowed to reach 70–80% confluence. A 200-µl pipette tip was used to scratch the artificial wound across the diameter of the wells and the culture medium was changed to serum-free medium. Wound closure was observed at 0 and 24 h, and images were captured under a phase contrast light microscope (magnification, ×200) at 0 and 24 h after wound incision. Each experiment was repeated three times.
Western blot analysis
The procedure was performed as previously described (33). Total proteins were extracted using RIPA lysis buffer (Sigma-Aldrich; Merck KGaA) and subsequently quantified using a bicinchoninic acid protein assay kit (Santa Cruz Biotechnology, Inc.). Samples (20 µg/lane) were separated via 10% SDS-PAGE and transferred onto a PVDF membrane (Amresco, LLC). Membranes were blocked with 5% BSA (Sigma-Aldrich; Merck KGaA) for 1 h at room temperature. Primary antibodies against HDAC4 (dilution, 1:500; cat. no. ab234084; Abcam) and GAPDH (dilution, 1:500; cat. no. ab8245; Abcam) were incubated with the membranes at 4°C overnight. The subsequent day, the membranes were incubated with secondary antibody (goat anti-mouse immunoglobulin G HRP-conjugated; cat. no. ab205719; dilution, 1:2,000; Abcam). Signals of targeted proteins were detected using an ECL Western Blotting Substrate kit (cat. no. ab65623; Abcam) and bands were analyzed with ImageJ software version 2 (National Institutes of Health).
Dual-luciferase reporter assay
The dual-luciferase reporter assay was performed as previously described (16). Wild-type (WT) and mutant (MUT) circHIPK3 (LUC-circHIPK3-WT and LUC-circHIPK3-MUT) or HDAC4 (LUC-HDAC4-WT and LUC-HDAC4-MUT) reporter plasmids that contained WT or MUT miR-637 seed sequences were synthesized by Shanghai GenePharma Co., Ltd. The constructed reporter plasmids and miR-637 mimic were co-transfected into HOS and U2OS cells using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, luciferase activity was measured and compared with Renilla luciferase activity using the Dual-Luciferase Reporter Assay system (Promega Corporation) according to the manufacturer's protocol.
Statistical analysis
All experiments were performed in triplicate and all data from three independent experiments are presented as the mean ± standard deviation. Statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software, Inc.). Survival curves were estimated by the Kaplan-Meier method and a log-rank test was performed to assess overall survival rate of patients with OS. A receiver operating characteristic curve (ROC) analysis was used to evaluate the clinical value of circHIPK3. The relationship between circHIPK3 and miR-637 was assessed by Pearson's correlation analysis. For comparisons, unpaired Student's t-test, Mann-Whitney U test and one-way ANOVA followed by Dunnett's post hoc test were performed, as appropriate. P<0.05 was considered to indicate a statistically significant difference.
Results
circHIPK3 is relatively highly expressed in OS cells, and predominantly localized in the cytoplasm
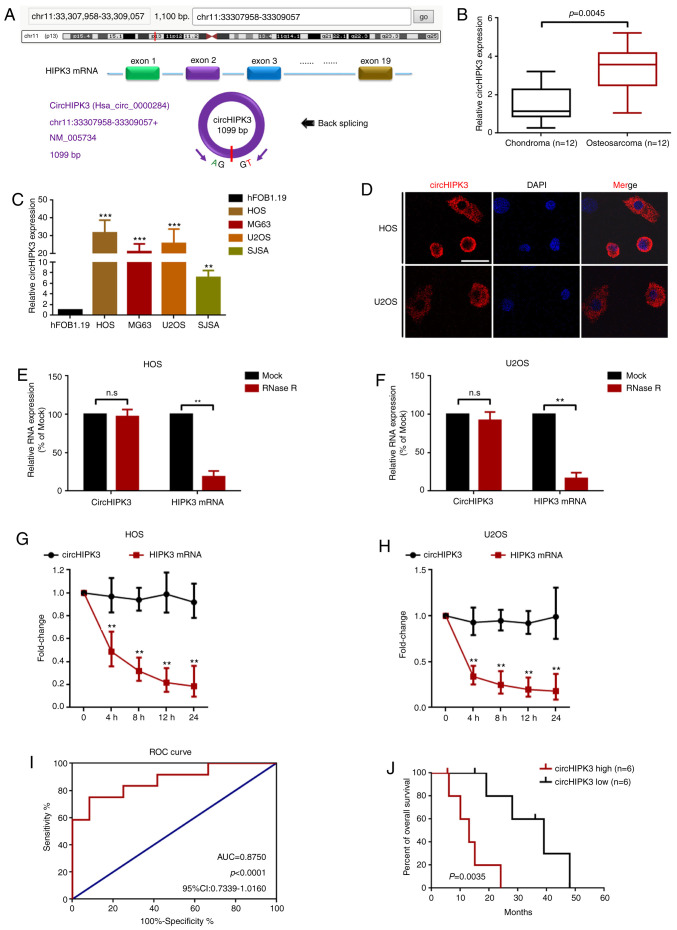
First, the present study demonstrated that circHIPK3 was derived from exon 2 of HIPK mRNA, as previously reported (26) (Fig. 1A and Fig. S1). The expression levels of circHIPK3 were then detected in 12 OS tissues and 12 paired chondroma tissues using RT-qPCR. As the data presented in Fig. 1B shows, circHIPK3 expression was significantly upregulated in OS tissues. In addition, the present study measured circHIPK3 expression in OS cells. RT-qPCR (Fig. 1C) demonstrated that circHIPK3 expression was upregulated in four OS cell lines (HOS, MG63, U2OS and SJSA) compared with in the normal osteoblast hFOB1.19 cell line. Furthermore, the present study revealed that circHIPK3 was mainly located in the cytoplasm of HOS and U2OS cells (Fig. 1D). A previous study considered circRNAs to be stable nucleic acids (34). Therefore, an RNase R assay was performed in the present study to evaluate the stability of circHIPK3. As shown in Fig. 1E and F, compared with the Mock group, RNase R led to a significant decrease in HIPK3 mRNA, while the expression of circHIPK3 was not markedly changed. Additionally, an actinomycin D assay indicated that circHIPK3 was more stable than the linear HIPK3 mRNA (Fig. 1G and H). Finally, the present study analyzed the clinical value of circHIPK3 in the collected cases. ROC curve and Kaplan-Meier analyses demonstrated that circHIPK3 had clinical value in OS (area under the curve, 0.8750; P<0.0001; Fig. 1I) and high circHIPK3 expression was closely associated with shorter overall survival of patients with OS (Fig. 1J).
Figure 1.
circHIPK3 is relatively highly expressed in OS cells, and predominantly localized in the cytoplasm. (A) circHIPK3 was derived from exon 2 of linear HIPK3 mRNA as illustrated. (B) circHIPK3 expression in 12 pairs of OS and chondroma tissue samples was determined using RT-qPCR. (C) circHIPK3 expression in four OS cell lines and in the normal osteoblast hFOB1.19 cell line was measured using RT-qPCR. **P<0.01 and ***P<0.001 vs. hFOB1.19. (D) circHIPK3 was mainly located in the cytoplasm according to a fluorescence in situ hybridization assay. Magnification, ×400; scale bar, 20 µm. An RNase R assay was performed to determine the stability of circHIPK3 in (E) HOS and (F) U2OS cells. **P<0.01. An actinomycin D assay was performed to check the stability of circHIPK3 in (G) HOS and (H) U2OS cells. **P<0.01. (I) Clinical values of circHIPK3 in OS were analyzed using an ROC curve (AUC, 0.8750; P<0.0001). (J) High expression levels of circHIPK3 were associated with shorter overall survival of patients with OS. n=6 for each group. P=0.0035. All data are presented as the mean ± SD of three independent experiments. AUC, area under the curve; circ, circular RNA; HIPK3, homeodomain interacting protein kinase 3; n.s, not significant (P>0.05); OS, osteosarcoma; ROC, receiver operating characteristic; RT-qPCR, reverse transcription-quantitative PCR.
Knockdown of circHIPK3 suppresses proliferation, migration and invasion in OS cells
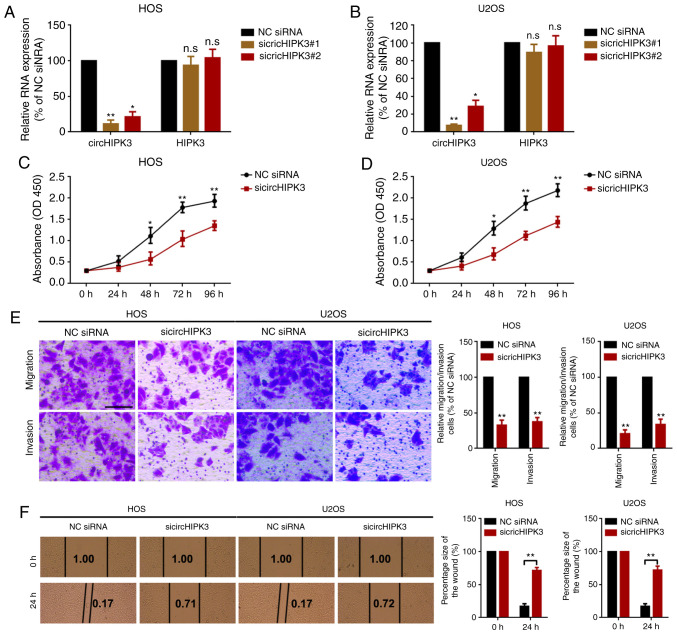
In order to investigate the role of circHIPK3 in OS, the present study performed loss-of-function assays. Two specific siRNAs that targeted the junction sites were designed to knock down circHIPK3 in OS cells. As demonstrated by RT-qPCR (Fig. 2A and B), sicircHIPK3#1 presented higher silencing efficacy. Therefore, this was selected as the RNA interference (RNAi) tool in the following RNAi experiments. Subsequently, the present study used a CCK8 assay to determine the proliferation ability of OS cells. As the data presented in Fig. 2C and D show, knockdown of circHIPK3 suppressed proliferation in HOS and U2OS cells. Finally, the present study performed a Transwell migration/invasion assay and a wound healing assay to detect the role of circHIPK3 in OS cell metastasis. As shown in the representative images in Fig. 2E and F, knockdown of circHIPK3 inhibited OS cell metastasis.
Figure 2.
Knockdown of circHIPK3 suppresses proliferation, migration and invasion in OS cells. Expression levels of circHIPK3 and HIPK3 mRNA in (A) HOS and (B) U2OS cells after transfection of specific circHIPK3 siRNAs were measured by reverse transcription-quantitative PCR. A Cell Counting Kit 8 assay was performed to investigate the proliferation ability of (C) HOS and (D) U2OS cells. (E) A Transwell assay was used to evaluate the metastatic ability of HOS and U2OS cells. Magnification, ×20; scale bar, 200 µm. (F) A wound healing assay was performed to determine the migration ability of HOS and U2OS cells. Magnification, ×4; scale bar, 500 µm. *P<0.05 and **P<0.01 vs. NC siRNA group. All data are presented as the mean ± SD of three independent experiments. circ, circular RNA; HIPK3, homeodomain interacting protein kinase 3; NC, negative control; n.s, not significant (P>0.05); OD, optical density; siRNA, small interfering RNA.
circHIPK3 sponges miR-637 in OS cells
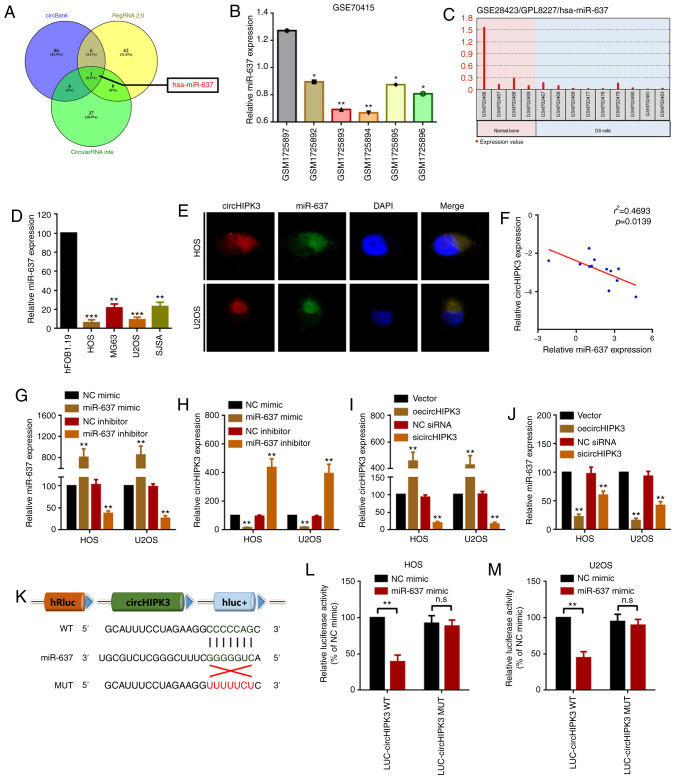
Accumulating evidence has indicated that circRNAs serve as miRNA sponges by harboring multiple miRNAs, and thereby function as miRNA inhibitors (7,35). The present study demonstrated that circHIPK3 was mainly located in the cytoplasm (Fig. 1D). Therefore, it was assumed that circHIPK2 may work via a similar mechanism in OS. The present study screened the miRNAs that may interact with circHIPK3 using the online tools circBank, RegRNA and circularRNA interactome. As demonstrated by a Venn diagram (Fig. 3A), only miR-637 was identified in all three databases. Therefore, the present study further investigated the expression levels of miR-637 in OS. By analyzing Gene Expression Omnibus (GEO) datasets GSE70415, it was revealed that miR-637 was expressed at low levels in OS cells (Fig. 3B). Using another non-coding RNA profiling array dataset (GSE28423), it was identified that miR-637 was expressed at low levels in 9 OS cell line compared with in 4 normal human bone samples (Fig. 3C). In the present study, it was revealed that miR-637 was downregulated in OS cell lines compared with in hFOB1.19 cells (Fig. 3D). Furthermore, using a FISH assay, it was revealed that both miR-637 and circHIPK3 were co-located in the cytoplasm (Fig. 3E). To further investigate the association between miR-637 and circHIPK3, the present study performed Spearman's correlation analysis. As shown in Fig. 3F, circHIPK3 expression was negatively correlated with miR-637 expression. In addition, it was demonstrated that upregulation and downregulation of miR-637 inversely regulated circHIPK3 expression (Fig. 3G and H). Conversely, miR-637 expression was inversely regulated by circHIPK3 upregulation and downregulation (Fig. 3I and J). These results indicated that miR-637 interacted with circHIPK3 in a reciprocal suppressive manner. Finally, a luciferase assay revealed that miR-637 targeted circHIPK3 via direct binding (Fig. 3K-M).
Figure 3.
circHIPK3 sponges miR-637 in OS cells. (A) Potential miRNAs that may interact with circHIPK3 were predicted by circBank (http://www.circbank.cn/), RegRNA 2.0 (http://regrna2.mbc.nctu.edu.tw/) and circularRNA interactome (https://circinteractome.nia.nih.gov/). miR-637 expression in GEO datasets (B) GSE70415 and (C) GSE28423 was analyzed using GEO2R. *P<0.05 and **P<0.01 vs. GSM1725897 group. (D) An RT-qPCR assay was used to determine the expression levels of miR-637 in OS cell lines. **P<0.01 and ***P<0.001 vs. hFOB1.19 group. (E) circHIPK3 and miR-637 were colocalized in the cytoplasm of HOS and U2OS cells as demonstrated by a fluorescence in situ hybridization assay. Magnification, ×400; scale bar, 20 µm. (F) Spearman's correlation analysis revealed that miR-637 expression was inversely correlated with circHIPK3 expression. (G) miR-637 and (H) circHIPK3 expression after different miR-637 interventions was investigated by RT-qPCR. (I) circHIPK3 and (J) miR-637 expression after different circHIPK3 interventions was measured by RT-qPCR. (K) Diagram of constructed luciferase reporter plasmids. miR-637 could directly target circHIPK3 in (L) HOS and (M) U2OS cells, as demonstrated by a luciferase assay. n.sP>0.05 and **P<0.01 vs. NC mimic group. All data are presented as the mean ± SD of three independent experiments. circ, circular RNA; GEO, Gene Expression Omnibus; HIPK3, homeodomain interacting protein kinase 3; LUC, luciferase; miR-637, microRNA-637; MUT, mutant; NC, negative control; n.s, not significant (P>0.05); oe, overexpression; OS, osteosarcoma; RT-qPCR, reverse transcription-quantitative PCR; siRNA, small interfering RNA; WT, wild-type.
miR-637 suppresses proliferation, migration and invasion partially via targeting of HDAC4 in OS cells
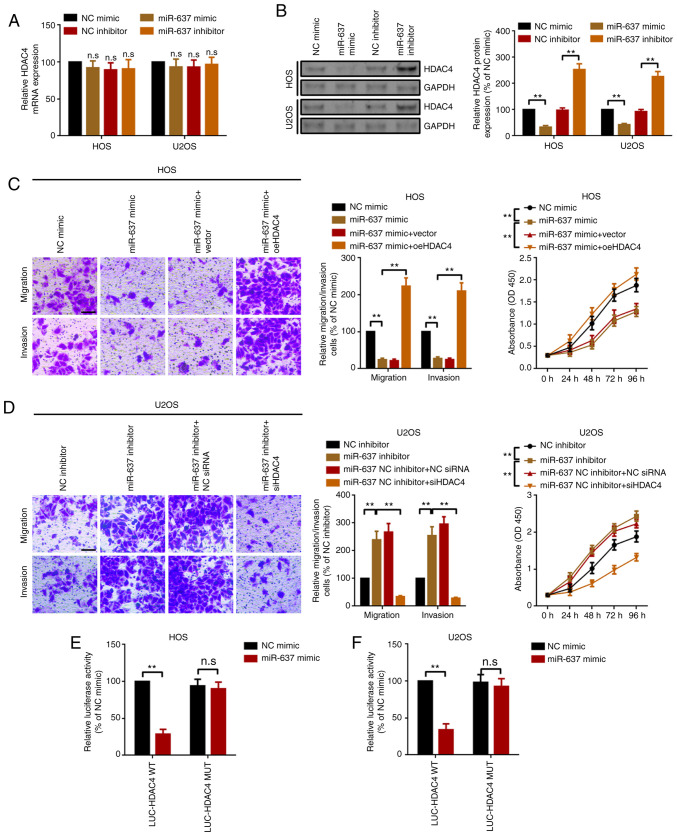
miRNAs are well-known to regulate their downstream genes via direct targeting. Our previous study indicated that HDAC4 serves as an oncogene in OS (16). The present study hypothesized that HDAC4 was also a target of miR-637. First, it was revealed that miR-637 had limited effects on the mRNA expression levels of HDAC4 (Fig. 4A). Furthermore, miR-637 inversely regulated HDAC4 protein expression (Fig. 4B), indicating that miR-637 regulated HDAC4 at the post-transcriptional level. HDAC4-specific small interfering RNAs (siHDAC4) and overexpression plasmids (oeHDAC4) were used to determine the role of HDAC4 in miR-637-mediated proliferation and motility changes. The silencing and overexpression efficacy of siHDAC4 and oeHDAC4 were demonstrated (Fig. S2). Secondly, the present study demonstrated that upregulation of miR-637 suppressed OS cell proliferation, migration and invasion, and the suppressive effect was reversed by upregulation of HDAC4 (transfection of HDAC4 overexpression plasmid oeHDAC4; Fig. 4C). Conversely, knockdown of miR-637 promoted OS cell proliferation, migration and invasion, and the effect was attenuated by knockdown of HDAC4 (transfection with siHDAC4; Fig. 4D). Subsequently, the present study demonstrated that HDAC4 was a downstream target of miR-637 using a luciferase assay (Fig. 4E and F). The aforementioned results indicated that miR-637 suppressed OS cell proliferation, migration and invasion partially by directly targeting HDAC4.
Figure 4.
miR-637 suppresses proliferation, migration and invasion partially via HDAC4 modulation in osteosarcoma cells. (A) HDAC4 mRNA expression in HOS and U2OS cells was measured by reverse transcription-quantitative PCR. (B) HDAC4 protein expression was investigated by western blotting. Metastatic and proliferation abilities of (C) HOS and (D) U2OS cells were determined by Transwell and Cell Counting Kit 8 assays. Magnification, ×20; scale bar, 200 µm. A luciferase assay was used to demonstrate the targeted binding effect between miR-637 and HDAC4 in (E) HOS and (F) U2OS cells. **P<0.01. All data are presented as the mean ± SD of three independent experiments. HDAC4, histone deacetylase 4; LUC, luciferase; miR-637, microRNA-637; MUT, mutant; NC, negative control; n.s, not significant (P>0.05); OD, optical density; oe, overexpression; siRNA, small interfering RNA; WT, wild-type.
circHIPK3 promotes proliferation, migration and invasion by modulating the miR-637/HDAC4 axis in OS
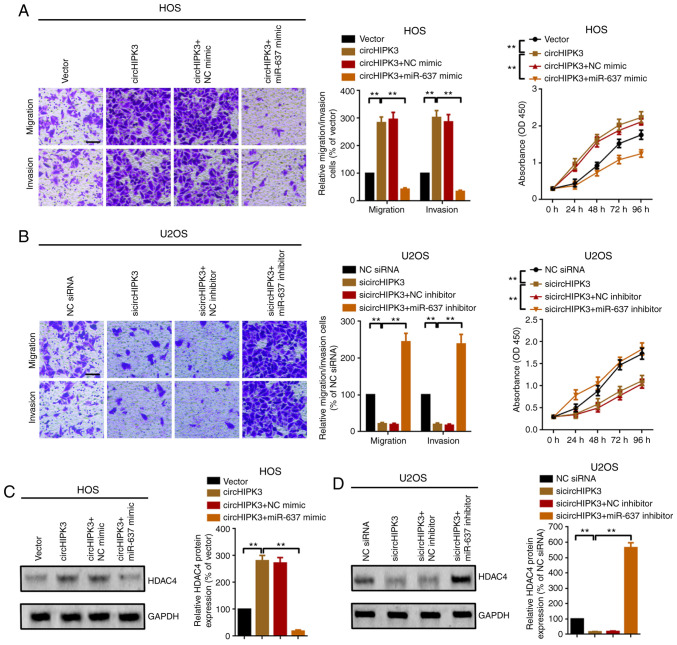
The present study investigated the association among circHIPK3, miR-637 and HDAC4. Transwell and CCK8 assays were performed to investigate the role of miR-637 in circHIPK3-induced proliferation, migration and invasion. As shown in Fig. 5A, overexpression of circHIPK3 promoted OS cell proliferation, migration and invasion. The facilitative effect was reversed by miR-637 mimics (upregulation of miR-637). Conversely, inhibition of circHIPK3 suppressed OS cell proliferation, migration and invasion, and the suppressive effect was attenuated by inhibition of miR-637 (transfection of miR-637 inhibitor; Fig. 5B). In addition, similar to the proliferation and metastatic assays, overexpression of circHIPK3 promoted HDAC4 protein expression, and the facilitative effect was attenuated by miR-637 (Fig. 5C). On the other hand, suppression of cirHIPK3 inhibited HDAC4 expression, while the suppressive effect was reversed by knockdown of miR-637 (Fig. 5D). In summary, all findings suggested that circHIPK3 promoted proliferation, migration and invasion via modulation of the miR-637/HDAC4 axis in OS.
Figure 5.
circHIPK3 promotes proliferation and metastasis via modulation of the miR-637/HDAC4 axis in osteosarcoma. Metastatic and proliferation abilities of (A) HOS and (B) U2OS cells were investigated using Transwell and Cell Counting Kit 8 assays. Magnification, ×20; scale bar, 200 µm. (C) HDAC4 protein expression after different circHIPK3 and miR-637 interventions was investigated by western blotting. **P<0.01. (D) Knockdown of circHIPK3 suppressed HDAC4 protein expression, while the suppressive effect was abolished by a further downregulation of miR-637, as determined by a western blotting assay. **P<0.01. All data are presented as the mean ± SD of three independent experiments. circ, circular RNA; HDAC4, histone deacetylase 4; HIPK3, homeodomain interacting protein kinase 3; miR-637, microRNA-637; NC, negative control; OD, optical density; n.s, not significant (P>0.05); siRNA, small interfering RNA.
Discussion
circRNAs are a type of RNA molecule with closed single-stranded structure. The formation of circRNAs primarily occurs through back-splicing of precursor mRNA or skipping events of thousands of genes in eukaryotes as covalently closed continuous loops (36). circRNAs function via multiple mechanisms, including modulation of transcription, miRNA sponging and associating with protein binding (37). circHIPK3 was first identified by ribominus RNA sequencing data from six human normal tissues and seven types of human cancer (9). circHIPK3 serves as an oncogene in various types of cancer, including gastric cancer, CRC, lung cancer and prostate cancer (38–41). In the present study, the expression and function of circHIPK3 in OS were determined. Using RT-qPCR, it was revealed that circHIPK3 expression was upregulated in OS tissue samples and in OS cells. In addition, the present study demonstrated that high levels of circHIPK3 were closely associated with shorter overall survival of patients with OS. Previous studies have demonstrated that circRNAs are abundantly enriched in the cytoplasm and serve as miRNAs sponges (42–44). Therefore, the present study performed a localization FISH assay to demonstrate the distribution of circHIPK3. A specific probe was designed to target circHIPK3, and it was revealed that circHIPK3 was prominently located in OS cell cytoplasm. Functionally, the present study revealed that circHIPK3 promoted OS cell proliferation, migration and invasion using loss-of-function CCK8 and Transwell assays.
circRNAs serve as miRNAs sponges by acting as competing endogenous RNAs (42). Since circHIPK3 was primarily located in the OS cell cytoplasm, the present study inferred that circHIPK3 may interact with certain miRNAs. miR-637 has been reported as a tumor suppressor in several types of cancer (45,46). In a melanoma-associated study, Zhang et al (47) revealed that overexpression of miR-637 suppresses proliferation and cell cycle G1-S transition, and induces apoptosis in melanoma cells. Du et al (45) reported that miR-637 is expressed at low levels in hepatoma, and miR-637 inhibits proliferation and invasion of hepatoma cells by targeted degradation of AKT1. By analyzing GEO datasets GSE70415 and GSE28423 and RT-qPCR, the present study demonstrated that miR-637 expression was downregulated in OS. Furthermore, it was revealed that both miR-637 and circHIPK3 were co-localized in the cytoplasm, and they interacted with each other in a reciprocal suppressive manner. These results demonstrated that miR-637 and circHIPK3 may interact on a biological basis. Furthermore, the results of the luciferase assay indicated that miR-637 could bind to circHIPK3 directly. Finally, the present study investigated the association between miR-637 and HDAC4, a well-known oncogene in various malignancies, including OS (15). In our previous study (16), the oncogenic role of HDAC4 in OS was observed. In the present study, it was revealed that HDAC4 was inversely regulated by miR-637 at the post-transcriptional level. In addition, the present study demonstrated that miR-637 targeted HDAC4 via direct binding. The functional Transwell and CCK8 assays suggested that miR-637 inhibited OS cell proliferation, migration and invasion partially via the HDAC4 signaling pathway.
In summary, the results of the present study suggested that circHIPK3 exerted a regulatory function by sponging miR-637 and promoting OS cell proliferation, migration and invasion partially via the HDAC4 signaling pathway. circHIPK3/miR-637/HDAC4 signaling may be a novel target in the treatment of OS.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by Key R&D Program of Liaoning Province (grant no. 20180550251).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GW conceived the experiments. YW, BL, MH and YS performed the experiments. ST analyzed the data. GW wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study and the associated experimental protocols (both human and animal experiments) were performed in compliance with ethical guidelines and approved by the Institute Research Medical Ethics Committee of the Shengjing Hospital of China Medical University, Shenyang, China (approval no. 2020PS051K). All chondroma and OS tissues were also used in accordance with the Declaration of Helsinki. Written informed consent was provided by each patient before the study. All patients agreed that the data from their samples could be used for experimental studies and paper presentations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sweetnam R. Osteosarcoma. Ann R Coll Surg Engl. 1969;44:38–58. [PMC free article] [PubMed] [Google Scholar]
- 2.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 3.Berner K, Johannesen TB, Berner A, Haugland HK, Bjerkehagen B, Bøhler PJ, Bruland ØS. Time-trends on incidence and survival in a nationwide and unselected cohort of patients with skeletal osteosarcoma. Acta Oncol. 2015;54:25–33. doi: 10.3109/0284186X.2014.923934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friebele JC, Peck J, Pan X, Abdel-Rasoul M, Mayerson JL. Osteosarcoma: A meta-analysis and review of the literature. Am J Orthop (Belle Mead NJ) 2015;44:547–553. [PubMed] [Google Scholar]
- 5.Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma. Curr Opin Pediatr. 2016;28:26–33. doi: 10.1097/MOP.0000000000000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zamborsky R, Kokavec M, Harsanyi S, Danisovic L. Identification of prognostic and predictive osteosarcoma biomarkers. Med Sci (Basel) 2019;7:28. doi: 10.3390/medsci7020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu N, Zhang J, Zhang LY, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22:3713–3718. doi: 10.26355/eurrev_201806_15250. [DOI] [PubMed] [Google Scholar]
- 11.Ke Z, Xie F, Zheng C, Chen D. CircHIPK3 promotes proliferation and invasion in nasopharyngeal carcinoma by abrogating miR-4288-induced ELF3 inhibition. J Cell Physiol. 2019;234:1699–1706. doi: 10.1002/jcp.27041. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F, Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu H, Chen Y, Jiang P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. 2018;506:455–462. doi: 10.1016/j.bbrc.2018.10.087. [DOI] [PubMed] [Google Scholar]
- 14.Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9:175. doi: 10.1038/s41419-017-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Qin G, Zhao TC. HDAC4: Mechanism of regulation and biological functions. Epigenomics. 2014;6:139–150. doi: 10.2217/epi.13.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, Qin B. Long noncoding RNA MALAT1 regulates HDAC4-mediated proliferation and apoptosis via decoying of miR-140-5p in osteosarcoma cells. Cancer Medicine. 2018;7:4584–4597. doi: 10.1002/cam4.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng LS, Yang XZ, Wen YF, Mail SJ, Wang MH, Zhang MY, Zheng XFS, Wang HY. Overexpressed HDAC4 is associated with poor survival and promotes tumor progression in esophageal carcinoma. Aging (Albany NY) 2016;8:1236–1249. doi: 10.18632/aging.100980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sissons HA. The WHO classification of bone tumors. Recent Results Cancer Res. 1976:104–108. doi: 10.1007/978-3-642-80997-2_8. [DOI] [PubMed] [Google Scholar]
- 19.Kawano M, Tanaka K, Itonaga I, Ikeda S, Iwasaki T, Tsumura H. microRNA-93 promotes cell proliferation via targeting of PTEN in osteosarcoma cells. J Exp Clin Cancer Res. 2015;34:76. doi: 10.1186/s13046-015-0192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Namløs HM, Meza-Zepeda LA, Barøy T, Østensen IHG, Kresse SH, Kuijjer ML, Serra M, Bürger H, Cleton-Jansen AM, Myklebost O. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One. 2012;7:e48086. doi: 10.1371/journal.pone.0048086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis S, Meltzer PS. GEOquery: A bridge between the gene expression omnibus (GEO) and bioconductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Ye Y, Chang DW, Lin SH, Huang M, Tannir NM, Matin S, Karam JA, Wood CG, Chen ZN, Wu X. Global and targeted miRNA expression profiling in clear cell renal cell carcinoma tissues potentially links miR-155-5p and miR-210-3p to both tumorigenesis and recurrence. Am J Pathol. 2018;188:2487–2496. doi: 10.1016/j.ajpath.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glažar P, Papavasileiou P, Rajewsky N. circBase: A database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(Suppl 2):S4. doi: 10.1186/1471-2105-14-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B, Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang C, Li B, Qin M, Yang C, He J, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10:3200. doi: 10.1038/s41467-019-11162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Zhong S, Wang J, Zhang Q, Xu H, Feng J. CircPrimer: A software for annotating circRNAs and determining the specificity of circRNA primers. BMC Bioinformatics. 2018;19:292. doi: 10.1186/s12859-018-2304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S, Zhang Y, Lu Z. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Lu Z, Wang N, Feng J, Zhang J, Luan L, Zhao W, Zeng X. Long noncoding RNA DANCR promotes colorectal cancer proliferation and metastasis via miR-577 sponging. Exp Mol Med. 2018;50:1–17. doi: 10.1038/s12276-018-0082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Zhang Y, Yang T, Zhao W, Wang N, Li P, Zeng X, Zhang W. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget. 2017;8:59417–59434. doi: 10.18632/oncotarget.19727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan Y, Wang Z, Qin B. A novel long noncoding RNA, LINC00483 promotes proliferation and metastasis via modulating of FMNL2 in CRC. Biochem Biophys Res Commun. 2019;509:441–447. doi: 10.1016/j.bbrc.2018.12.090. [DOI] [PubMed] [Google Scholar]
- 34.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 35.Arnaiz E, Sole C, Manterola L, Iparraguirre L, Otaegui D, Lawrie CH. CircRNAs and cancer: Biomarkers and master regulators. Semin Cancer Biol. 2019;58:90–99. doi: 10.1016/j.semcancer.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 37.Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi: 10.1016/j.omtn.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai C, Zhi Y, Wang K, Zhang P, Ji Z, Xie C, Sun F. CircHIPK3 overexpression accelerates the proliferation and invasion of prostate cancer cells through regulating miRNA-338-3p. OncoTargets Ther. 2019;12:3363–3372. doi: 10.2147/OTT.S196931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Li C, Liu X, Wang Y, Zhao R, Yang Y, Zheng X, Zhang Y, Zhang X. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine. 2019;48:277–288. doi: 10.1016/j.ebiom.2019.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X, Mao R, Su W, Yang X, Geng Q, Guo C, Wang Z, Wang J, Kresty LA, Beer DG, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy. 2020;16:659–671. doi: 10.1080/15548627.2019.1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu WG, Xu Q. Upregulation of circHIPK3 promotes the progression of gastric cancer via Wnt/β-catenin pathway and indicates a poor prognosis. Eur Rev Med Pharmacol Sci. 2019;23:7905–7912. doi: 10.26355/eurrev_201909_19004. [DOI] [PubMed] [Google Scholar]
- 42.Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui X, Wang J, Guo Z, Li M, Li M, Liu S, Liu H, Li W, Yin X, Tao J, Xu W. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer. 2018;17:123. doi: 10.1186/s12943-018-0877-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han TS, Hur K, Cho HS, Ban HS. Epigenetic associations between lncRNA/circRNA and miRNA in hepatocellular carcinoma. Cancers (Basel) 2020;12:E2622. doi: 10.3390/cancers12092622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du YM, Wang YB. MiR-637 inhibits proliferation and invasion of hepatoma cells by targeted degradation of AKT1. Eur Rev Med Pharmacol Sci. 2019;23:567–575. doi: 10.26355/eurrev_201901_16869. [DOI] [PubMed] [Google Scholar]
- 46.Li JX, Ding XM, Han S, Wang K, Jiao CY, Li XC. mir-637 inhibits the proliferation of cholangiocarcinoma cell QBC939 through interfering CTSB expression. Eur Rev Med Pharmacol Sci. 2018;22:1265–1276. doi: 10.26355/eurrev_201803_14467. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J, Liu WL, Zhang L, Ge R, He F, Gao TY, Tian Q, Mu X, Chen LH, Chen W, Li X. MiR-637 suppresses melanoma progression through directly targeting P-REX2a and inhibiting PTEN/AKT signaling pathway. Cell Mol Biol (Noisy-le-Grand) 2018;64:50–57. doi: 10.14715/cmb/2018.64.11.10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.