Abstract
Diamond-Blackfan anemia (DBA) is a rare hematopoietic disease characterized by a block in red cell differentiation. Most DBA cases are caused by mutations in ribosomal proteins and characterized by higher than normal activity of the tumor suppressor p53. Higher p53 activity is thought to contribute to DBA phenotypes by inducing apoptosis during red blood cell differentiation. Currently, there are few therapies available for DBA patients. We performed a chemical screen using zebrafish ribosomal small subunit protein 29 (rps29) mutant embryos that have a p53-dependent anemia and identified calmodulin inhibitors that rescued the phenotype. Our studies demonstrated that calmodulin inhibitors attenuated p53 protein amount and activity. Treatment with calmodulin inhibitors led to decreased p53 translation and accumulation but does not affect p53 stability. An FDA-approved calmodulin inhibitor, trifluoperazine, rescued hematopoietic phenotypes of DBA models in vivo in zebrafish and mouse models. In addition, trifluoperazine rescued these phenotypes in human CD34+ hematopoietic stem and progenitor cells. Erythroid differentiation was also improved in CD34+ cells isolated from a patient with DBA. This work uncovers a potential avenue of therapeutic development for patients with DBA.
One Sentence Summary:
Calmodulin inhibitors rescue erythropoiesis and p53-dependent phenotypes of Diamond-Blackfan anemia.
Introduction
Diamond-Blackfan anemia (DBA) is a congenital anemia that generally presents in young children(1). The primary symptom is anemia due to a block in erythroid differentiation. DBA is also associated with an elevated risk of craniofacial anomalies, short stature, thumb abnormalities, and an increased cancer predisposition(2). Ribosomal protein S19 (RPS19) was the first gene found mutated in patients with DBA(3). The sequencing of patient samples has identified mutations including deletions in 20 small and large subunit ribosomal proteins, estimated to explain the genetics of approximately 70% of patients with DBA(4, 5). Patients are heterozygous for these mutations and always maintain a wildtype copy of the affected ribosomal protein gene. In addition, a mutation in the ribosomal maturation factor TSR2 was described in one family(6). Mutations in erythroid-specific X-linked transcription factor GATA1 have also been identified in a subset of male patients with DBA who do not have ribosomal protein mutations(7). The genetics of DBA have strongly implicated the involvement of ribosomal proteins (RPs).
RP knockdown or mutation causes decreased ribosomal subunit assembly and aberrant processing of 18S or 28s rRNA(8-10). Stunted growth and homozygous lethality are observed in in vivo models of ribosomal protein mutants, including drosophila minutes(11), zebrafish(12), and mouse(13-15). Current evidence suggests that many effects of ribosomal protein mutations are p53-dependent(16-18). There are several mechanisms by which ribosomal protein deficiency can lead to p53 activation. Ribosomal protein deficiency leads to an increase of free ribosomal proteins, a subset of which bind MDM2 (mouse double minute 2 homolog) and putatively inhibit its p53 ubiquitination activity, leading to increased p53 protein in the context of ribosomal protein deficiency(19). TP53 translation can also be specifically upregulated as a result of ribosomal proteins or other factors that bind to untranslated regions (UTRs) of TP53 (20, 21). p53 activation has been shown to be a critical mediator of hematopoietic defects in many DBA models, including human CD34+ cells(22), mouse(16, 23), and zebrafish(24, 25). These studies are consistent with a role for p53 activation in the hematopoietic effects of ribosomal stress.
Standard DBA therapy includes regular blood transfusions and/or steroids(1). Steroids are thought to work by promoting proliferation of erythroid progenitor cells and are effective in DBA patient-derived cells and DBA mouse models(26-28). Although patients may undergo spontaneous remission, patients who remain on treatment have serious side effects such as iron overload and other complications. Currently, the only known cure for DBA is a hematopoietic stem cell transplant, which carries its own risks. Studies suggest that lenalidomide(14, 26), leucine(24, 29, 30), sotatercept(31, 32), and DYRK inhibition(33) may be useful therapies, and clinical trials with these drugs are completed or ongoing (lenalidomide – NCT01034592; leucine – NCT01362595; sotatercept – NCT01464164; DYRK inhibitors – NCT00443170). We also recently published results from a chemical screen in DBA patient-derived induced pluripotent stem cells (iPSCs), identifying autophagy as a therapeutic pathway in DBA(34). We previously characterized zebrafish rps29 mutants that have hematopoietic and endothelial defects(35). To identify potential therapeutics for DBA, we performed a chemical screen in rps29−/− zebrafish embryos and found several calmodulin (CaM) inhibitors that rescued the mutant phenotypes. The FDA-approved CaM inhibitor trifluoperazine (TFP) improved anemia in multiple in vitro and in vivo models of DBA, including patient-derived CD34+ cells, by reducing activation of p53 targets.
Results
Rps29−/− zebrafish embryos show increased p53 mRNA translation
We previously showed that rps29−/− zebrafish embryos have a defect in arterial specification, leading to decreased flk1 expression in the intersegmental vessels at 24 hours post fertilization (hpf) and decreased hematopoietic stem cells(35). Primitive erythropoiesis is also affected, as rps29−/− embryos have less hemoglobin. These embryos also have increased apoptosis, particularly in the head region, and die by five days post fertilization (dpf)(35). p53 pathways are activated in the rps29−/− embryo, and greater amounts of p53 mRNA are found in monosome and polysome fractions in the mutant (fig. S1A). Consistent with other models(16, 22-25), p53 mutation rescues all hematopoietic and apoptotic phenotypes(35). Also consistent with other studies of ribosomal protein mutants(8, 10), rps29−/− embryos have a defect in 40S ribosome subunit assembly formation, evidenced by a decrease in the 40S peak of the sedimentation profile, and an overall decrease in ribosome formation, as evidenced by a decrease in the 80S monosome peak (fig. S1B).
Chemical screen finds that calmodulin inhibitors rescue rps29−/− defects in zebrafish embryos
We performed a screen to identify chemicals that could reverse the endothelial and morphological defects of the rps29−/− mutant embryo (fig. S2A), because these readouts were more readily detectable in a high-throughput assay than the rescue of anemia. 600 bioactive chemicals were screened in duplicate. Rps29−/− embryos were treated starting at bud stage (10 hpf), scored for rescue of head morphology at 24 hpf, and subsequently fixed for in situ hybridization of flk1 and rps29. Embryos without rps29 staining (rps29−/− mutants) were scored for rescue of flk1 intersegmental vessel staining. Of the 600 compounds, 17 were validated as hits in the flk1 screen (table S1). One of the compounds identified in the screen to rescue flk1 expression was W-7, a naphthalenesulfonamide that inhibits calmodulin (CaM)(36) (Fig. 1A, table S2). We tested other naphthalenesulfonamides known to inhibit CaM, including A-7 and W-5(37), and they also rescued the vasculature defect (Fig. 1B, table S2). Several structurally dissimilar CaM inhibitors rescued flk1 expression, including CGS-9343B and members of the phenothiazine family such as trifluoperazine (TFP), as well as calcium channel blockers (Fig. 1B, fig. S2B). Taken together, the effects of these chemicals establish that multiple inhibitors of CaM improve flk1 expression in ribosomal protein deficiency. Of the 600 chemicals screened, only A-3 rescued the morphology of the rps29−/− mutant head (Fig. 1C, tables S1 and S2) with genotype validated by PCR. A-3 is a structural derivative of W-7 (fig. S2C) and a known CaM inhibitor(37). Treatment with A-3 or W-7 from 10 hpf also increased hemoglobin in the rps29 mutant embryos at 40 hpf (Fig. 1D, fig. S2D, table S2), establishing that calmodulin inhibition reverses the effects of RP deficiency in vivo in hematopoietic tissues.
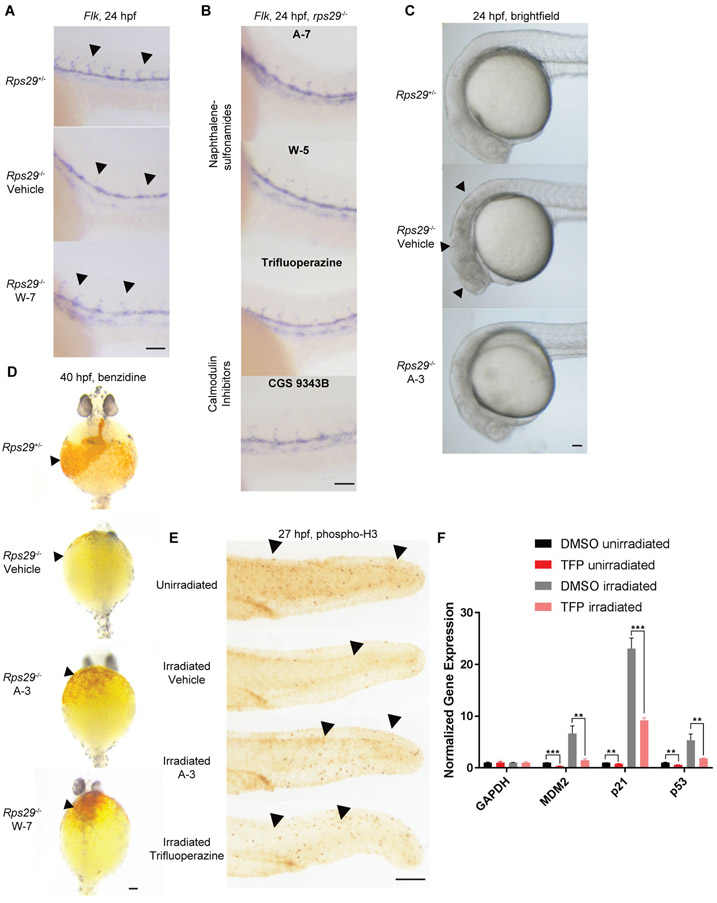
Fig. 1. Calmodulin inhibitors rescue rps29−/− defects and irradiation-induced phenotypes in zebrafish embryos.
A. Embryos from an rps29+/− incross were treated with DMSO (vehicle) or W-7 at 10 hpf and collected at 24 hpf for in situ hybridization of flk1. Arrowheads denote intersegmental vessels. Scale bar = 100 μm.
B. Rps29−/− embryos were treated with A-7, W-5, TFP, or CGS 9343B at 10 hpf and collected at 24 hpf for in situ hybridization of flk. Scale bar = 100 μm.
C. Embryos from an rps29+/− incross were treated with DMSO or A-3 at 10 hpf and imaged at 24 hpf. Arrowheads denote cell death in the head of the developing embryo, which looks dark and cloudy. Scale bar = 100 μm.
D. Embryos from an rps29+/− incross were treated with DMSO, A-3, or W-7 at 10 hpf and collected at 40 hpf for benzidine (o-dianisidine) staining of hemoglobinized cells. Arrowheads denote location of hemoglobin-positive cells (or lack thereof) around the yolk sac of the embryo. Scale bar = 100 μm.
E. Wildtype embryos were treated with DMSO, A-3, or TFP at 50% epiboly (5.25 hpf), irradiated at 10 Gy at 24 hpf, and collected for phospho-H3 staining at 25.5 hpf. Arrowheads denote phospho-H3 positive cells. Scale bar = 100 μm.
F. Wildtype embryos were treated with DMSO or TFP at 50% epiboly (5.25 hpf), irradiated at 10 Gy at 24 hpf, and collected for RNA isolation and qPCR at 25.5 hpf. *Student’s t-test -p < 0.01, *** p < 0.001.
We previously showed that defects in the rps29−/− embryo are mediated through p53(35, 38) and irradiation of zebrafish embryos activates p53 similarly to ribosomal protein deficiency(35), consistent with other studies demonstrating increased synthesis of p53 after irradiation(20). Irradiation of wildtype embryos at 24 hpf led to decreased cell proliferation as measured by phospho-H3 staining (Fig. 1E). We selected two structurally different CaM inhibitors to test: A-3, which was the most effective in the zebrafish model (Fig. 1D), and TFP, which is an FDA-approved antipsychotic. Both mitigated the decrease in proliferation induced by gamma-irradiation (Fig. 1E, fig. S2E, table S2). In zebrafish embryos treated with TFP or A-3, mRNA of p53 target genes p21, p53, and mdm2 was also decreased post-irradiation (Fig. 1F, fig. S3A). These data demonstrate that calmodulin inhibition attenuates p53 activity in the zebrafish embryo.
Calmodulin inhibitors decrease p53 activity in human cell models of DBA
We next validated whether calmodulin inhibitors also worked in human cell models of DBA. We first tested them in the A549 human cell line, which has wildtype p53, in which we transduced an shRNA for RPS19 or an shRNA for luciferase (Luc) as a control (Fig. 2A). As expected, RPS19 knockdown caused an increase in p53 protein. Treatment with A-3 or TFP modestly reduced the RPS19-induced increase of p53 protein compared to DMSO treatment and decreased p21 protein to the amount in control cells (Fig. 2A). RPS19 knockdown increased the expression of multiple p53 target genes by 5- to 15-fold, but it did not alter the expression of p53 itself (Fig. 2B). A-3 or TFP treatment reduced the expression of these target genes compared to DMSO treatment (Fig. 2B). Taken together, these data indicate that CaM inhibitors from two different classes reduce the activity of p53 in a RPS19-deficient cell line model. The remainder of the studies focused on only one drug candidate, TFP, because it is an FDA-approved drug with a favorable safety profile and over 60 years of use in patients(39).
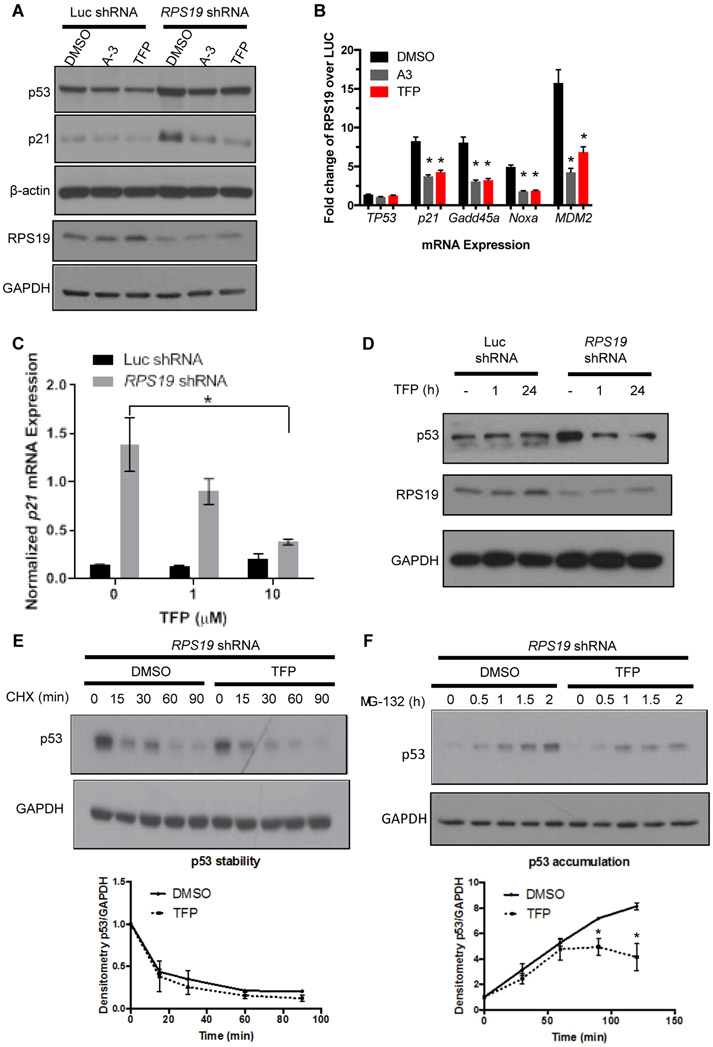
Fig. 2. TFP dose-dependently reduces p53 in RPS19-deficient human cells and primary human hematopoietic progenitors.
A. p53 and p21 protein, as measured by Western blot, in A549 cells with shRNA targeting luciferase or RPS19 and treated with DMSO, 50 μM A-3, or 20 μM TFP.
B. qPCR measuring expression of TP53, P21, GADD45A, NOXA, or MDM2 in A549 cells with shRNA targeting luciferase or RPS19 and treated with DMSO, A-3, or TFP. Student t-test, *p < 0.05 compared to DMSO control.
(C-D) CD34+ cells were transduced with RPS19 or control shRNA and selected for successfully transduced GFP+ cells.
C. qPCR measuring p21 mRNA in CD34+ cells treated with increasing doses of TFP. Student t-test, *p < 0.05.
D. Western blot measuring RPS19 and p53 proteins in CD34+ cells treated for increasing lengths of time with 10 μM TFP.
(E-F) CD34+ cells were infected with RPS19 shRNA and selected for GFP+ successfully transduced cells by FACS. Western blots of p53 and GAPDH were quantified by densitometry.
E. Cells were pre-treated with TFP for 15-30 minutes before treatment with 10 μM CHX for increasing lengths of time before lysates were collected for TP53 and GAPDH protein quantification.
F. Cells were pre-treated with TFP for 2 hours before treatment with 20 μM MG-132 for increasing lengths of time before lysates were collected for TP53 and GAPDH protein quantification. Two-way ANOVA, *p < 0.05.
To determine whether calmodulin inhibitors also worked in human blood cells, we isolated CD34+ cells from cord blood. In CD34+ cells, TFP dose-dependently reduced the expression of p21 (Fig. 2C), demonstrating that TFP can modulate p53 activity in the primary human in vitro DBA model. In the RPS19 shRNA CD34+ cells, TFP treatment restored the amount of p53 protein to that of Luc shRNA control within one hour of treatment (Fig. 2D). Treatment with the calcium chelator BAPTA also restored the amount of p53 protein in RPS19 shRNA cells to the amount observed in Luc shRNA control within one hour of treatment (fig. S3B).
Effects of TFP on translation result in decreased p53 accumulation
To investigate the mechanism by which TFP reduces p53 protein in the presence of RPS19-deficiency, we examined stability and accumulation of p53 using cycloheximide (CHX) and MG132, respectively. CD34+ cells transduced with RPS19 shRNA were pretreated with TFP or DMSO for 30 min and then treated with CHX for increasing amounts of time. Treatment with cycloheximide led to decreased p53 protein within 15 minutes, but Western blot analysis showed no difference in p53 protein stability in TFP- compared to DMSO-treated cells (Fig. 2E). This result suggested that TFP was not inducing p53 degradation or modulating the RP-MDM2-P53 axis. To investigate the effect of TFP on p53 accumulation, CD34+ cells transduced with RPS19 shRNA were pretreated with TFP or DMSO for 30 min and then treated with the proteasome inhibitor MG132 for increasing amounts of time. TFP reduced the accumulation of p53 compared to DMSO treatment (Fig. 2F). These data, in combination with the finding that TFP does not affect steady-state TP53 mRNA (Fig. 2B), demonstrate that TFP reduces the RP deficiency-induced synthesis of p53.
We next wanted to determine whether TFP affected p53 accumulation in other modes of p53 activation beyond RP deficiency. We hypothesized that TFP would also reduce p53 accumulation after gamma irradiation. To test this, CD34+ cells were irradiated with 10 Gy and treated with TFP for 1 hour before treatment with MG132 for increasing amounts of time. Similar to the effect observed in RPS19-deficient cells, TFP treatment also reduced p53 accumulation after irradiation (fig. S3C). The effect of TFP on p53 accumulation rather than stability suggests that TFP is acting at the level of translational regulation of p53.
The 5’ and 3’ UTRs of p53 were reported to play a role in its translational regulation(20, 21). Using a luciferase reporter construct with the luciferase gene flanked by p53 5’ and 3’ UTRs(20) (Fig. 3A), we observed that transduction of RPS19 shRNA or treatment with doxorubicin (positive control(40)) were both sufficient to increase luciferase signal, demonstrating that RP deficiency is sufficient for p53 induction via p53 UTRs (Fig. 3B). To determine if the p53 UTRs are sufficient for inhibition of p53 accumulation by TFP, cells were transduced with the shRNA for RPS19, sorted for GFP+ (marking successfully transduced cells), transfected with the UTR construct, and treated with DMSO or TFP. RPS19-knockdown cells treated with TFP had significantly lower luciferase signal compared to control-treated cells (p value < 0.001) (Fig. 3C). These data indicate that TFP reduces the translation of TP53 mRNA via the UTRs and independent of the coding sequence of TP53 mRNA.
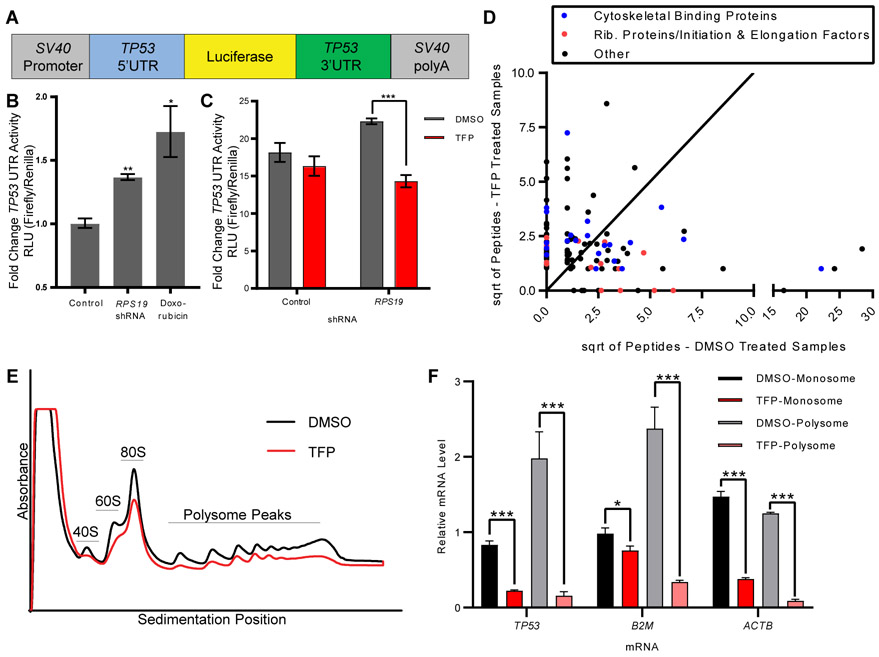
Fig. 3. TFP inhibition of translation is sufficient to decrease translation of p53.
A. Schematic of construct with TP53 UTRs and luciferase(20).
B. 293T cells were transduced with RPS19 shRNA or treated with doxorubicin before transfection with TP53 UTR luciferase construct and control Renilla luciferase construct. Y-axis shows relative luminescence units, representing the ratio of firefly luciferase (p53 UTR construct) to Renilla luciferase (control). Student t-test, *p < 0.05, **p <0.01.
C. 293T cells were transduced with RPS19 shRNA or control, treated with DMSO or TFP, and transfected with TP53 UTR luciferase construct and control Renilla luciferase construct. Y-axis shows relative luminescence units, representing the ratio of firefly luciferase (TP53 UTR construct) to Renilla luciferase (control). Student t-test, ***p < 0.001.
D. RNA Affinity Purification (RAP) identified proteins bound to TP53 mRNA in the presence or absence of TFP. Axes show the square roots of summed peptide counts from mass spectrometry. Each dot represents a protein observed bound to TP53 mRNA more than control mRNA.
E. CD34+ cells were transduced with shRNA against RPS19, sorted for successfully transduced GFP+ cells, and treated with DMSO or 10 μM TFP for 24 hours. Polysome profiles were generated, with the y-axis representing absorbance. Black line is the profile for DMSO-treated cells, red line is the profile for TFP-treated cells.
F. RNA was isolated from monosome and polysome fractions of treated cells, and TP53, β2M, and ACTβ mRNAs were measured by qPCR. Relative mRNA quantity represents Ct values normalized to each sample’s pool of all polysome fractions. Student’s t-test, *p < 0.05, *** p < 0.001.
To determine how TFP might be affecting the translation of TP53 mRNA, we performed RNA Affinity Purification (RAP) to identify proteins differentially bound to TP53 mRNA in the presence or absence of drug. Cells were irradiated at 10 Gy, then treated with DMSO or TFP for 2 hours, crosslinked, and subjected to pulldown with biotinylated DNA oligos specific to TP53 or LacZ (negative control) mRNAs. Recruited proteins were analyzed by mass spectrometry (data file S1), with the 125 proteins bound to TP53 mRNA over control pulldown highlighted in data file S1 and graphed in Fig. 3D. Of the 14 ribosomal proteins and elongation factors bound to TP53 mRNA (red dots in Fig. 3D), 9 were bound less in the presence of TFP, consistent with decreased translation of TP53 mRNA. In addition, annexins A1, A2, and A3 were all bound to p53 at lower amounts upon drug treatment (data file S1), consistent with literature suggesting a calcium-dependent association between the translation of TP53 mRNA and annexin A2(41).
We next generated polysome profiles of human CD34+ cells with or without TFP treatment to determine whether the effects of TFP were specific to p53 or more general translation effects, as has been suggested in other work demonstrating inhibition of translational activity by TFP(42). We transduced cells with RPS19 shRNA and treated with DMSO or TFP for 24 hours before assessing polysome profiles. Treatment with TFP caused a decrease in absorbance across the sedimentation fractions (Fig. 3E), consistent with a previously published report showing a similar effect upon calcium depletion(43). A decrease in absorbance across sedimentation fractions was also observed upon TFP treatment in irradiated and un-irradiated zebrafish embryos (fig. S4A). When assessing the amount of individual mRNAs in the polysome fractions, TP53 mRNA was decreased in the presence of TFP, as were β2M, β-actin, GAPDH, and EEF1A1 (Fig. 3F, fig. S4B). These data demonstrate that TFP treatment affects overall translation, and this is sufficient to decrease translation of TP53 and decrease TP53 transcriptional activity.
TFP improves anemia in both DBA mouse model and primary samples from patients with DBA
To determine if TFP could be a viable treatment for patients with DBA, we assessed the effect of TFP on erythropoiesis in a mouse model of DBA. We used a transgenic mouse model where the amount of Rps19 is controlled in the hematopoietic system using conditional expression of short hairpin RNA targeting Rps19, because the erythroid phenotype is reliable and comparable to what is observed in patients(23). Wildtype mice were irradiated and transplanted with bone marrow from mice harboring a knock-in allele that expresses a doxycycline-inducible Rps19 shRNA(23). Following engraftment of the transplanted cells, mice were administered doxycycline in drinking water and received an intraperitoneal (IP) injection of 5 mg/kg TFP every other day (Fig. 4A). After two weeks, blood was analyzed for red blood cell (RBC) counts and hemoglobin, and bone marrow was collected for qPCR analysis. In vehicle-treated mice, knockdown of Rps19 caused anemia and an induction of p53 target genes in hematopoietic tissues. Bone marrow collected from TFP-treated animals was assessed for reduction in p21 mRNA expression but there was no statistically significant difference due to one sample with high p21 expression (Fig. 4B). Both red blood cell counts and hemoglobin measures were significantly increased by TFP treatment in the presence of RPS19 knockdown (p = 0.0023 for red blood cells and 0.0022 for hemoglobin) (Fig. 4C, D). TFP showed no toxicity when the Rps19 knockdown was induced (fig. S5). In particular, mouse weight, which was decreased by doxycycline treatment and Rps19 knockdown, was restored to WT numbers, indicating that TFP-treated mice with Rps19 knockdown were healthier than vehicle-treated mice (fig. S5F). Notably, treatment with TFP in this model achieved therapeutic effects similar to those seen with dietary leucine treatment and dexamethasone treatment(27, 44).
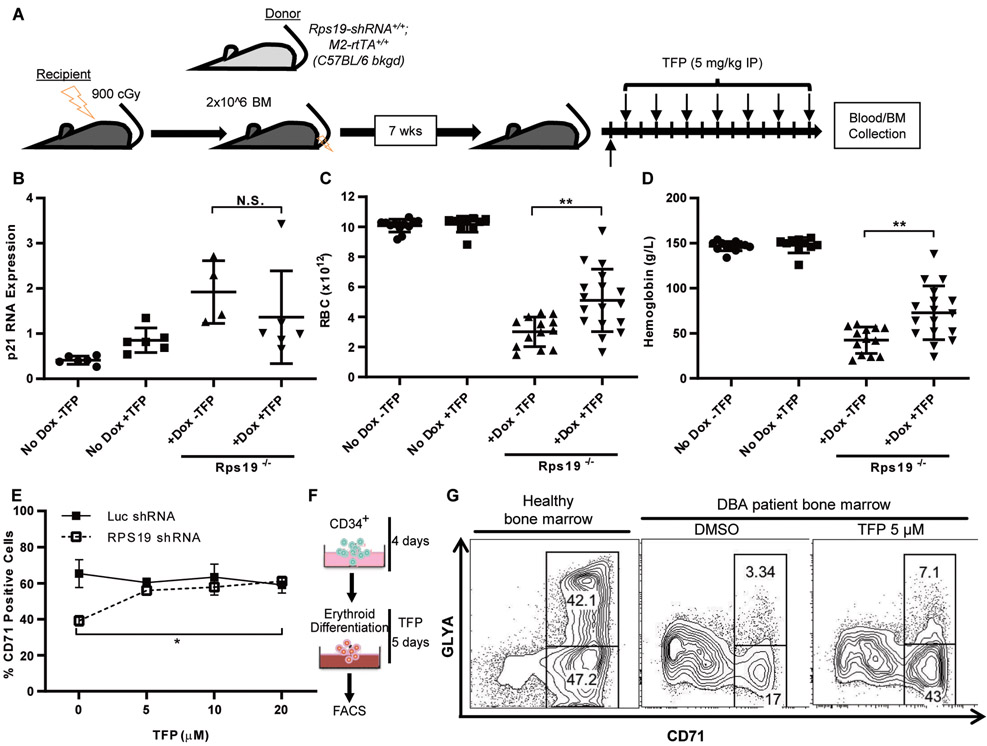
Fig. 4. TFP improves erythropoiesis in a DBA mouse model and primary cells from a patient with DBA.
A. Schematic of mouse transplantation and drug treatment. Unfractionated bone marrow from inducible Rps19 shRNA donor mice was transplanted into irradiated wildtype recipients. After engraftment, hairpin expression was induced with doxycycline and mice were treated with TFP or vehicle for two weeks.
B. RNA was isolated from bone marrow collected from recipient mice after two weeks of treatment.
(C-D) Peripheral blood samples from recipient mice were analyzed using a Hemavet hematology system for red blood cell counts (C) and hemoglobin (D). Student t-test, **p < 0.01.
E. CD34+ cells were expanded for four days prior to infection with shRNAs targeting luciferase or RPS19. Infected cells were selected with puromycin beginning on day 5, and drug was added to the medium beginning on day 7. Cells were processed for flow cytometry on day 10. Percentage of CD71+ cells, as measured by flow cytometry, in CD34+ cells treated with increasing doses of TFP. Two-way ANOVA, *p < 0.05.
F. Schematic of CD34+ HSPC differentiation. CD34+ cells were expanded for 4 days prior to addition of DMSO or TFP for 5 days.
G. CD34+ cells isolated from a DBA patient were expanded for four days prior to treatment with DMSO or TFP. Percentages of CD71+ and GLYA+ cells, as measured by flow cytometry, in treated cells. Left = healthy bone marrow, right = bone marrow from a patient with DBA, with and without TFP treatment.
To assess the effectiveness of TFP on the erythroid differentiation block in a human model of DBA, we used two-phase in vitro erythroid differentiation culture systems with primary human hematopoietic progenitors. We transduced cord blood-derived CD34+ progenitors with shRNAs for RPS19 or Luc during the expansion phase. After selecting for cells containing the shRNAs, the cells were placed in erythroid differentiation medium and cultured with TFP or DMSO. As expected, RPS19 knockdown decreased the percentage of transferrin receptor (CD71)-positive cells, demonstrating a block in differentiation (Fig. 4E). Treatment with TFP significantly increased the percentage of CD71+ cells to the amount observed in Luc shRNA control cells (p = 0.0066) (Fig. 4E).
Finally, we tested primary bone marrow CD34+ cells from a patient with DBA (UPN NCI-131) with an RPS19 R94X mutation (Fig. 4F). This patient’s cells produced fewer CD71+ and fewer GlyA+ cells than healthy CD34+ cells (Fig. 4G). Treatment with TFP led to improved erythroid differentiation compared to DMSO, with the percentage of CD71+ cells increasing to normal quantities, and of the percentage of GlyA+ cells doubling (Fig. 4G). DBA cells typically exhibit a differentiation block at the BFU-E (erythroid burst-forming unit) stage(26), and treatment with TFP improved the erythroid output of the cells, as measured by an increased percentages of CD71+ and CD71+GlyA+ erythroid progenitors. These data indicate that TFP improves erythropoiesis in multiple in vivo and in vitro models of DBA.
Discussion
Current therapies for DBA are suboptimal and ineffective for many patients, leaving a dire need for the development of new treatment options. Here, we present a chemical screen that found calmodulin inhibitors to rescue ribosomal protein mutation-mediated phenotypes in a zebrafish model of DBA. CaM inhibitors improved erythropoiesis and decreased activation of p53 transcriptional targets in both RP- and irradiation-induced phenotypes, and translated to human cell models as well. Lastly, we established that the FDA-approved compound TFP improves the erythroid defect in multiple DBA models and described a mechanism of TFP that reduces the irradiation- or ribosomal stress-induced accumulation of p53.
The effects of the compounds tested were reproducible across several species and disease models. Here, our studies spanned zebrafish, mouse, and human cell models and cells from patients with DBA. We recently published results of a chemical screen in iPS cells from patients with DBA (34). Five out of the top 22 hits in the iPS cell screen were modulators of CaM or calcium, including the naphthalenesulfonamide A-3. Overlap between these screens performed in two different organismal models of DBA suggests that calcium/CaM are the compound targets that might best translate to the clinic. In particular, the effect of TFP in the DBA mouse model was comparable to that of dexamethasone(27), one of the leading DBA treatments. In addition, CaM inhibitors were also effective when different small subunit ribosomal proteins were deficient, including rps29 in the zebrafish and RPS19 in human cell lines and primary human HSPCs. These data suggest that they would be effective in patients with DBA and different ribosomal protein mutations.
Current therapies for patients with DBA, including blood transfusions, corticosteroids, and HSC transplantation, have adverse effects. TFP and other phenothiazines are given to adults with schizophrenia and are well tolerated(39). One concern with reducing p53 activity in patients with DBA is that they are already at risk for certain cancers. However, we and others argue that it is the high amount of p53 in RP-deficient cells that causes cells to attenuate the p53 pathway, consistent with findings that zebrafish with ribosomal protein mutations have increased cancer incidence(45, 46). If TFP reduces the superactivation of p53 that results from the ribosomal protein deficiency, it should also decrease the constant proliferative inhibition of high p53 and reduce the risk of cells trying to circumvent high p53 activity. TFP has been used since the 1950s, and there is no observed increase in tumor incidence(47). We observed a decrease of WBC (white blood count) in wildtype mice treated with TFP, but not in the DBA mice. Decreased WBC has not been described in patients who take trifluoperazine. However, patients with schizophrenia treated with TFP and other phenothiazines have experienced other side effects associated with other antipsychotics(48) that may preclude its use as a therapy in children with DBA. For these reasons, TFP has entered Phase I/II clinical trials for adults in remission from DBA (ClinicalTrials.gov NCT03966053). In addition, we are also interested in identifying TFP derivatives that improve erythropoiesis in vivo but do not cross the blood-brain barrier.
If CaM inhibition is a successful means of p53 inhibition in DBA, these drugs may be beneficial to patients with other diseases with aberrant p53 activity. DBA is one of a group of diseases termed “ribosomopathies”, in which patients exhibit ribosome dysfunction caused by a mutation in or loss of a ribosomal protein or related gene. Ribosomopathies are nearly all thought to involve aberrant p53 activity(19), so reducing p53 activity could be therapeutic in all of these diseases. For example, patients with 5q- myelodysplastic syndrome (MDS) exhibit haploinsufficient loss of RPS14, which causes aberrant p53 activity and hematopoietic defects by a mechanism similar to that seen in patients with DBA(22, 49, 50). Patients with Shwachman-Bodian-Diamond syndrome, who have a mutation in a ribosome biogenesis gene, also exhibit hematopoietic symptoms (51). Patients with dyskeratosis congenita or Treacher Collins syndrome have mutations in genes involved in rRNA modification, which also induce p53 activation(52-54). Other bone marrow failure syndromes, including Fanconi’s anemia, are also thought to result from aberrant p53 activity (55). A therapy developed to target the p53 pathway could be effective in any ribosomopathy. Two patients with bone marrow failure have been identified with activating germline mutations in p53 itself(56). p53 activation is also thought to play a role in numerous other developmental syndromes, including CHARGE syndrome and p53-mediated microencephaly (reviewed in Bowen and Attardi(57). Targeting of the p53 pathway may be beneficial for these syndromes.
We also showed that TFP can inhibit p53 accumulation after irradiation. Activation of p53 is thought to be the cause of cytotoxicity for radiotherapy and chemotherapy(58, 59), and p53 inhibition by the chemical pifithrin has been explored to minimize the toxicities associated with cancer treatment(60). Further understanding of how calmodulin inhibition affects the p53 pathway may prove it a useful therapeutic approach for DBA and other diseases characterized by aberrant p53 activity.
Materials and Methods
Study design
The goal of this study was to identify therapeutics for DBA using a zebrafish model of the disease. Compounds were tested in two randomly assigned independent experiments of twenty embryos each, so approximately ten mutant embryos were scored per chemical, giving an 80% power with 0.1 Type 1 error rate and 0.53 minimum detectable effect. Screen scoring and all scoring was performed in a blinded fashion. One of the hit compounds classes, calmodulin inhibitors, was tested in independent in vitro and in vivo DBA models including primary patient-derived iPS cells, human CD34+ cells, zebrafish, and mouse. Mechanistic studies were performed in human cell lines and CD34+ cells. Drug effects were always measured relative to vehicle control. Data points were combined from independent biological replicates, and outliers were not excluded.
Embryo manipulation, screening, and chemical treatment
Fish were maintained under Boston Children’s Hospital Institutional Animal Care and Use Committee approved laboratory conditions. Studies were performed on AB wildtype strains and hi2903, an insertional mutant in the first intron of ribosomal protein s29 (rps29)(12). Gamma irradiation was performed on 24 hpf AB embryos, at one dose of 10 Gy. For the chemical treatments of AB wildtype fish, embryos were treated at 50% epiboly (5.25 hpf) before irradiation at 24 hpf. For all other chemical treatments, rps29+/− fish were incrossed, and embryos were collected for treatment at bud stage (10 hpf). Embryos were treated from bud to 24 or 48 hpf with compounds of known bioactivity. For screening, chemicals from two libraries were tested at 1:300 dilutions (in E3) from library stock: BIOMOL 480 (Enzo Life Sciences) and Sigma Lopac1280 (Sigma-Aldrich). The following chemicals were diluted in DMSO or water and tested in doses from 5-50 μg/mL: A-3 (Enzo Life Sciences), W-7 (Tocris Bioscience), A-5 (Tocris Bioscience), W-5 (Enzo Life Sciences), CGS-9343B (Tocris Bioscience), and TFP (Enzo Life Sciences). Nimodipine (Cayman Chemical) was diluted in DMSO and tested in doses from 1.7 to 17 μg/mL. YS-035 (Sigma-Aldrich) was diluted in water and used in doses from 0.8 to 8 μg/mL.
In situ hybridization, phospho-H3, and benzidine staining
Whole-mount in situ hybridization was performed as described(61). Antisense probes were synthesized from digested plasmid. Flk1 staining was counted as rescued if most of the intersegmental vessels had flk1 expression by ISH. Head morphology was counted as rescued if the mutant embryos could no longer be distinguished from wildtype embryos at 24 hpf. Benzidine staining was performed as previously described(62). Phospho-H3 antibody was used to identify proliferating cells as previously described(63).
Cell culture conditions and drug treatment
All cells were maintained at 37°C and 5% CO2. A549 cells (American Type Culture Collection) were cultured in F-12K medium (Gibco ThermoFisher) supplemented with 10% fetal calf serum (Gemini Bio) and 1% penicillin/streptomycin (Gibco ThermoFisher). Unless otherwise noted, drugs were added one day post-infection (described below), and medium with or without drug was changed daily for the course of the experiment. After 2-5 days of drug treatment, cells were trypsinized and collected for mRNA expression analysis or Western blotting.
293T cells (used for luciferase reporter assays, American Type Culture Collection) were cultured in Dulbecco's Modified Eagle Medium (DMEM, Gibco ThermoFisher) supplemented with 10% fetal calf serum and 1% penicillin/streptomycin. Cells were transfected with p53 UTR construct(41) and pRL-TK (Renilla luciferase reporter) using TransIT (MirusBio) following manufacturer’s instructions.
MCF-7 cells (used for RNA affinity purification, American Type Culture Collection) were cultured in Eagle’s Minimal Essential Medium (EMEM, Gibco ThermoFisher) supplemented with 10% fetal calf serum and 1% penicillin/streptomycin. Cells were irradiated at 10 Gy, treated with 10 μM TFP or DMSO, and collected 2 hours after treatment for RNA affinity purification.
Culture of CD34+ cells
CD34+ hematopoietic stem and progenitor cells (HSPCs) were purified from human umbilical cord blood and maintained in liquid culture. Two methods of culturing were used in this study:
1) Cells were cultured in medium supportive of erythroid differentiation [serum-free expansion medium (StemCell Technologies), 100 U/mL penicillin/streptomycin, 2 mM glutamine, 10 g/mL lipids (Sigma-Aldrich), 100 ng/mL stem cell factor (SCF), 10 ng/mL interleukin-3 (IL-3), and 0.5 U/mL erythropoietin (EPO)]. On day 7 of liquid culture, the concentration of EPO was increased to 3 U/mL. In experiments evaluating differentiation, 15 ng/mL granulocyte colony-stimulating factor (G-CSF) (Neupogen; Amgen) and 40 ng/mL FLT3 ligand were added to support myeloid differentiation as well. Cells were allowed to expand for 4 days prior to infection. After infection (see below), drug or vehicle was added to the medium. Cells were harvested for flow cytometric and/or gene expression analysis after 10 days of liquid culture.
2) For most experiments, CD34+ cell culture conditions were performed as described(34).
Lentiviral vectors and infection
Previously characterized lentiviral shRNAs in the pLKO.1 vector were obtained from the Broad Institute of Harvard and MIT(22). Lentivirus was produced in 293TL cells as described previously(64) and infected in the presence of 8 μg/mL polybrene (Sigma-Aldrich). Cells were selected 24 hours after infection in one of two ways, depending on the selection marker on the plasmid:
2 μg/mL puromycin (Sigma-Aldrich) for at least 48 hours prior to analysis
FACS (fluorescence-activated cell sorting) for GFP-positive cells as described in Doulatov et al.(34)
Polysome profiles
CD34+ cells were infected with RPS19 shRNA, sorted for GFP+ cells that were successfully transduced, and treated overnight with DMSO or 10 μM TFP. Equal numbers of cells (10 million) for each condition were then prepared for polysome fractionation as previously described(65). RNA was isolated and prepared for quantitative PCR as described below.
24 hpf wildtype (AB), rps29+/+ and rps+/−, embryos and rps29−/− embryos were prepared for polysome fractionation by dechorionation, treatment with 200 μg/mL cycloheximide for 10 min, removal of fish water, and freezing the embryos at −80C. Embryos at 50% epiboly were treated with DMSO or 50 μM TFP before 10 Gy irradiation at 24 hpf and subsequently collected for polysome fractionation at 25 hpf by dechorionation, treatment with 100 μg/mL cycloheximide for 10 min, removal of fish water, and freezing embryos at −80C.
Embryos were thawed and lysed at 4C in polysome lysis buffer [10 mM Tris-HCl pH 7.4, 5 mM MgCl2, 100 mM KCl, 1% Triton X-100, 2 mM DTT, 200 μg/mL cycloheximide, cOmplete EDTA-free protease inhibitor cocktail (Roche), and 500 U/mL RNasin Plus (Promega)] by gentle trituration through a 26G needle and incubation on ice for 30 min. The lysate was cleared of intact cells, nuclei, and mitochondria by centrifugation at 10,000g for 10 min at 4C. The supernatant was carefully layered onto an 11 mL 10-50% linear sucrose gradient made in 20 mM HEPES pH 7.4, 5 mM MgCl2, 100 mM KCl, 2 mM DTT, and 100 μg/mL cycloheximide. Lysates were ultracentrifuged at 35,000 RPM using a SW-41 Ti rotor at 4C for 2.5 hr. Gradients were analyzed and collected using a BioComp Poston Gradient Fractionator connected to a UV detector to monitor absorbance at 254 nm. Polysome profiles were normalized to the small ribosomal subunit.
RNA Affinity Purification (RAP)
Chemically treated and irradiated MCF-7 cells were subjected to RNA affinity purification (RAP) as previously described(66). After 2 hours of TFP or DMSO treatment (as negative control), cells were washed with PBS, trypsinized, and harvested into Falcon tubes. Approximately 40 15-cm plates were used for each treatment. Then cells were suspended into 35 ml cold PBS, crosslinked by addition of 8 ml 16% formaldehyde (final concentration, 3%), and kept in rotation for 30 minutes at room temperature. Excessive formaldehyde was quenched by addition of glycine to a final concentration of 0.25 M and incubation for 5 minutes at room temperature with shaking. Next, cells were centrifuged, washed twice with cold PBS, and cell pellets were flash frozen in liquid nitrogen and kept at −80°C before RAP. Cell pellets were lysed in lysis buffer containing 50 mM Tris-Cl pH 7.0, 10 mM EDTA, 1% SDS, 20 μl/ml RNaseOUT (Invitrogen), 2 mM PMSF, 2 mM benzamide, and 1x protease inhibitor cocktail (Roche). Lysates were passed through G20 and G26 syringe needles to dissociate cell aggregates, then sonicated for 5 cycles (30 seconds on, 30 seconds off). Sonicated lysates were centrifuged for 10 minutes at 4°C and 16000g, and cleared supernatants were collected for RAP. 3 ml of cleared lysates from each DMSO- or TFP-treated group were aliquoted into two 15-ml Falcon tubes (for LacZ and p53 probes) each containing 6 ml (2X) hybridization buffer [750 mM NaCl, 1% SDS, 50 mM Tris-Cl pH 7.0, 1 mM EDTA, 15% formamide, 20 μl/ml RNaseOUT (Invitrogen), 2 mM PMSF, 2 mM benzamide, and 1x protease inhibitor cocktail (Roche)]. 100 μl of pooled LacZ- or 5’-UTRL p53-specific probes were added to each Falcon tube (final concentration, 10 μM) and incubated in rotation at 37°C overnight. The following probes were used for RAP: p53 (GTGGCTCTAGACTTTTGAGA/3BioTEG/ and AATCCAGGGAAGCGTGTCAC/3BioTEG/); LacZ (CCAGTGAATCCGTAATCA/3BioTEG/, ATTAAGTTGGGTAACGCCAG/3BioTEG/, AATAATTCGCGTCTGGCCTT/3BioTEG/, and ATCTTCCAGATAACTGCCGT/3BioTEG/). Next, biotinylated DNA probes in complex with ribonucleoproteins were pooled down using streptavidin magnetic beads (Dynabeads MyOne Streptavidin C1; Invitrogen) and the beads were washed 5 times at room temperature with RAP wash buffer [2X SSC, 0.5% SDS, 2 mM PMSF, 2 mM benzamide, and 1x protease inhibitor cocktail (Roche)]. After the final wash, beads were incubated with protein loading buffer for 1 hour at 65°C for reverse-crosslinking, and then protein complexes were separated on Tris-Glycine polyacrylamide gels before mass spectrometry.
Mice
Mice expressing inducible Rps19 shRNA have been described previously(44). Wildtype C57BL/6 recipient mice were irradiated with 900 cGy and transplanted with 2x106 unfractionated bone marrow cells isolated from donor mice harboring two copies each of the Rps19 inducible hairpin construct and the doxycycline-responsive M2-rtTA element. Cells were allowed to engraft for 7 weeks prior to treatment. Beginning on day 1 of each experiment, mice were given doxycycline (2 mg/mL in drinking water, with 10 mg/mL sucrose) to induce the Rps19-deficiency phenotype. Starting on day 2, TFP (5 mg/kg) or saline vehicle was injected intraperitoneally every other day for two weeks. On day 15, peripheral blood was collected and analyzed for red blood cell counts and hemoglobin, and bone marrow was harvested for RNA isolation and qPCR analysis.
Quantitative PCR
Irradiated zebrafish embryos were collected and homogenized in Trizol (Invitrogen ThermoFisher) before RNA isolation and DNase treatment with the Direct-zol RNA miniprep kit (Zymo Research). cDNA was synthesized with equal amounts of RNA using SuperScript III First-Strand Synthesis (Invitrogen ThermoFisher) per manufacturer’s instructions. Real-time PCR was performed with SsoFast Evagreen Supermix (Biorad), and the delta-delta-Ct method was used for quantification, with normalization to GAPDH.
RNA was isolated from cells using the RNeasy kit according to manufacturer instructions (Qiagen). cDNA was synthesized with equal amounts of RNA using iScript cDNA synthesis kit (Biorad). Real-time PCR was performed with SsoFast Evagreen Supermix (Biorad), and gene expression was calculated relative to GAPDH according to methods previously described(67, 68).
For qPCR from polysome fractions, RNA was isolated using Trizol LS (Invitrogen ThermoFisher) following manufacturer’s instructions. RNA was treated with DNAse using the Turbo DNA-free kit (Invitrogen ThermoFisher). cDNA was synthesized with equal amounts of RNA using SuperScript III First-Strand Synthesis (Invitrogen ThermoFisher). Quantitative PCR was performed using PowerSYBR Green PCR mastermix (Applied Biosystems ThermoFisher). The delta-Ct method was used for quantification, with normalization to a pool of polysome fractions.
Western blots
Equal concentrations of protein lysates were run on 4-20% gradient gels (Invitrogen) and transferred using the iBlot Dry Blotting System (Invitrogen ThermoFisher) following manufacturer’s instructions. The following antibodies were used for Westerns: p53 – mouse monoclonal antibody DO-1 (Cell Signaling Technology), p21 – rabbit polyclonal monoclonal antibody 12D1 (Cell Signaling Technology), GAPDH – rabbit polyclonal antibody 14C10 (Cell Signaling Technology), RPS19 – rabbit monoclonal antibody EPR10423 (Abcam), and β-actin (C4) mouse monoclonal antibody (Santa Cruz Biotechnology).
Flow cytometry
For flow cytometry measuring cell surface markers, differentiated CD34+ cells were incubated for 30 minutes with PE-Cy5-conjugated anti-CD71 (BD Biosciences PharMingen). For flow cytometry in DBA patient-derived CD34+ cells, we followed the protocols described in Doulatov et al.(58). For Annexin V/7-AAD staining, PE Annexin V Apoptosis Kit I (BD Biosciences PharMingen) was used, following manufacturer instructions.
Statistics
For each zebrafish embryo experiment, p value is taken from binomial distribution calculations. All other p values were calculated using the Student’s T Test. * p<0.05, ** p<0.01, *** p<0.001.
Supplementary Material
Fig. S1. p53 is translated in higher amounts in a zebrafish model of Diamond-Blackfan anemia.
Fig. S2. Chemical screen identifies calmodulin inhibitors and calcium channel blockers.
Fig. S3. TFP and BAPTA decreases p53 accumulation.
Fig. S4. TFP treatment leads to decreased mRNAs in monosome and polysome fractions.
Fig. S5. TFP improves platelet counts and mouse weight in a mouse model of Diamond-Blackfan anemia.
Table S1. Summary of validated hits for screen phenotypes.
Table S2. Numbers of zebrafish embryos that respond to drug treatment with statistical analysis.
Data File S1. Proteins isolated by RNA affinity purification of TP53 mRNA were identified by mass spectrometry.
Acknowledgments
Funding: Studies were supported by National Heart, Lung, and Blood Institute (NHLBI) U01 HL100001 (to G.Q.D. and L.I.Z.), NHLBI U01 HL134812 (to G.Q.D. and L.I.Z) and the Henry and Marilyn Taub Foundation. This work was also supported by NHLBI 1F32HL124948-01 (to E.R.M.) and NSF Graduate Research Fellowships (to L.T.V and 174530 to A.N.S.). Funding for B.P.A. was provided in part by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics of the National Cancer Institute.
Footnotes
Competing Interests: A.M.T. and L.I.Z. are inventors on U.S. Patent No. 10420778 held by Boston Children’s Hospital that covers calmodulin inhibitors for the treatment of ribosomal disorders. E.R.M. and L.I.Z. are inventors on patent application No. 16/315,899 submitted by Boston Children’s Hospital that covers calmodulin inhibitors and other kinase inhibitors for the treatment of ribosomal disorders. S.D. and G.Q.D. are inventors on patent application No. 16/620,064 submitted by Boston Children’s Hospital for screening DBA patient-derived iPS models. E.R.M. has received consulting fees from CAMP4 Therapeutics and is a stockholder of CAMP4 Therapeutics. L.I.Z. is a founder and stockholder of Fate Therapeutics, CAMP4 Therapeutics, and Scholar Rock. He is a consultant for Celularity. R.I.G. is co-founder and scientific advisory board member of 28-7 Therapeutics. B.L.E. has received research funding from Celgene and Deerfield. He has received consulting fees from GRAIL, and he serves on the scientific advisory boards for and holds equity in Skyhawk Therapeutics and Exo Therapeutics. G.Q.D. has been an equity holder or received consulting fees from 28/7 Therapeutics and MPM Capital during the course of this study. All other authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are available in the main text or the supplementary materials. One DBA patient sample was provided to George Daley under a material transfer agreement with Blanche Alter at the National Cancer Institute.
References and notes
- 1.Vlachos A, Ball S, Dahl N, Alter BP, Sheth S, Ramenghi U, Meerpohl J, Karlsson S, Liu JM, Leblanc T, Paley C, Kang EM, Leder EJ, Atsidaftos E, Shimamura A, Bessler M, Glader B, Lipton JM, C. Participants of Sixth Annual Daniella Maria Arturi International Consensus, Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol 142, 859–876 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vlachos A, Rosenberg PS, Atsidaftos E, Alter BP, Lipton JM, Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Blood 119, 3815–3819 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D, Mohandas N, Carlsson B, Dahl N, The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet 21, 169–175 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Sakamoto KM, Narla A, Perspective on Diamond-Blackfan anemia: lessons from a rare congenital bone marrow failure syndrome. Leukemia 32, 249–251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulirsch JC, Verboon JM, Kazerounian S, Guo MH, Yuan D, Ludwig LS, Handsaker RE, Abdulhay NJ, Fiorini C, Genovese G, Lim ET, Cheng A, Cummings BB, Chao KR, Beggs AH, Genetti CA, Sieff CA, Newburger PE, Niewiadomska E, Matysiak M, Vlachos A, Lipton JM, Atsidaftos E, Glader B, Narla A, Gleizes PE, O'Donohue MF, Montel-Lehry N, Amor DJ, McCarroll SA, O'Donnell-Luria AH, Gupta N, Gabriel SB, MacArthur DG, Lander ES, Lek M, Da Costa L, Nathan DG, Korostelev AA, Do R, Sankaran VG, Gazda HT, The Genetic Landscape of Diamond-Blackfan Anemia. Am J Hum Genet 103, 930–947 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gripp KW, Curry C, Olney AH, Sandoval C, Fisher J, Chong JX, U. W. C. f. M. Genomics, Pilchman L, Sahraoui R, Stabley DL, Sol-Church K, Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am J Med Genet A 164A, 2240–2249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sankaran VG, Ghazvinian R, Do R, Thiru P, Vergilio JA, Beggs AH, Sieff CA, Orkin SH, Nathan DG, Lander ES, Gazda HT, Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest 122, 2439–2443 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, Leblanc T, Tchernia G, Da Costa L, Gleizes PE, Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood 109, 1275–1283 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gazda HT, Preti M, Sheen MR, O'Donohue MF, Vlachos A, Davies SM, Kattamis A, Doherty L, Landowski M, Buros C, Ghazvinian R, Sieff CA, Newburger PE, Niewiadomska E, Matysiak M, Glader B, Atsidaftos E, Lipton JM, Gleizes PE, Beggs AH, Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in diamond-blackfan anemia. Hum Mutat 33, 1037–1044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, Ellis SR, Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood 109, 980–986 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marygold SJ, Roote J, Reuter G, Lambertsson A, Ashburner M, Millburn GH, Harrison PM, Yu Z, Kenmochi N, Kaufman TC, Leevers SJ, Cook KR, The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol 8, R216 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N, Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A 101, 12792–12797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliver ER, Saunders TL, Tarle SA, Glaser T, Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development 131, 3907–3920 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keel SB, Phelps S, Sabo KM, O'Leary MN, Kirn-Safran CB, Abkowitz JL, Establishing Rps6 hemizygous mice as a model for studying how ribosomal protein haploinsufficiency impairs erythropoiesis. Exp Hematol 40, 290–294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgado-Palacin L, Varetti G, Llanos S, Gomez-Lopez G, Martinez D, Serrano M, Partial Loss of Rpl11 in Adult Mice Recapitulates Diamond-Blackfan Anemia and Promotes Lymphomagenesis. Cell Rep 13, 712–722 (2015). [DOI] [PubMed] [Google Scholar]
- 16.McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, Attardi LD, Barsh GS, Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet 40, 963–970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terzian T, Dumble M, Arbab F, Thaller C, Donehower LA, Lozano G, Justice MJ, Roop DR, Box NF, Rpl27a mutation in the sooty foot ataxia mouse phenocopies high p53 mouse models. J Pathol 224, 540–552 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watkins-Chow DE, Cooke J, Pidsley R, Edwards A, Slotkin R, Leeds KE, Mullen R, Baxter LL, Campbell TG, Salzer MC, Biondini L, Gibney G, Phan Dinh Tuy F, Chelly J, Morris HD, Riegler J, Lythgoe MF, Arkell RM, Loreni F, Flint J, Pavan WJ, Keays DA, Mutation of the diamond-blackfan anemia gene Rps7 in mouse results in morphological and neuroanatomical phenotypes. PLoS Genet 9, e1003094 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raiser DM, Narla A, Ebert BL, The emerging importance of ribosomal dysfunction in the pathogenesis of hematologic disorders. Leuk Lymphoma 55, 491–500 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Kastan MB, 5'-3'-UTR interactions regulate p53 mRNA translation and provide a target for modulating p53 induction after DNA damage. Genes Dev 24, 2146–2156 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takagi M, Absalon MJ, McLure KG, Kastan MB, Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123, 49–63 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, Wilson FH, Currie T, Khanna-Gupta A, Berliner N, Kutok JL, Ebert BL, Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood 117, 2567–2576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaako P, Flygare J, Olsson K, Quere R, Ehinger M, Henson A, Ellis S, Schambach A, Baum C, Richter J, Larsson J, Bryder D, Karlsson S, Mice with ribosomal protein S19 deficiency develop bone marrow failure and symptoms like patients with Diamond-Blackfan anemia. Blood 118, 6087–6096 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Ear J, Yang Z, Morimoto K, Zhang B, Lin S, Defects of protein production in erythroid cells revealed in a zebrafish Diamond-Blackfan anemia model for mutation in RPS19. Cell Death Dis 5, e1352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danilova N, Sakamoto KM, Lin S, Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. Br J Haematol 152, 217–228 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narla A, Dutt S, McAuley JR, Al-Shahrour F, Hurst S, McConkey M, Neuberg D, Ebert BL, Dexamethasone and lenalidomide have distinct functional effects on erythropoiesis. Blood 118, 2296–2304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sjogren SE, Siva K, Soneji S, George AJ, Winkler M, Jaako P, Wlodarski M, Karlsson S, Hannan RD, Flygare J, Glucocorticoids improve erythroid progenitor maintenance and dampen Trp53 response in a mouse model of Diamond-Blackfan anaemia. Br J Haematol 171, 517–529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Brien KA, Farrar JE, Vlachos A, Anderson SM, Tsujiura CA, Lichtenberg J, Blanc L, Atsidaftos E, Elkahloun A, An X, Ellis SR, Lipton JM, Bodine DM, Molecular convergence in ex vivo models of Diamond-Blackfan anemia. Blood 129, 3111–3120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pospisilova D, Cmejlova J, Hak J, Adam T, Cmejla R, Successful treatment of a Diamond-Blackfan anemia patient with amino acid leucine. Haematologica 92, e66–67 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Payne EM, Virgilio M, Narla A, Sun H, Levine M, Paw BH, Berliner N, Look AT, Ebert BL, Khanna-Gupta A, L-Leucine improves the anemia and developmental defects associated with Diamond-Blackfan anemia and del(5q) MDS by activating the mTOR pathway. Blood 120, 2214–2224 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ear J, Huang H, Wilson T, Tehrani Z, Lindgren A, Sung V, Laadem A, Daniel TO, Chopra R, Lin S, RAP-011 improves erythropoiesis in zebrafish model of Diamond-Blackfan anemia through antagonizing lefty1. Blood 126, 880–890 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Ge J, Apicella M, Mills JA, Garcon L, French DL, Weiss MJ, Bessler M, Mason PJ, Dysregulation of the Transforming Growth Factor beta Pathway in Induced Pluripotent Stem Cells Generated from Patients with Diamond Blackfan Anemia. PLoS One 10, e0134878 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siva K, Ek F, Chen J, Ghani Alattar A, Sigmundsson K, Olsson R, Wlodarski M, Lundback T, Flygare J, A Phenotypic Screening Assay Identifies Modulators of Diamond Blackfan Anemia. SLAS Discov 24, 304–313 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Doulatov S, Vo LT, Macari ER, Wahlster L, Kinney MA, Taylor AM, Barragan J, Gupta M, McGrath K, Lee HY, Humphries JM, DeVine A, Narla A, Alter BP, Beggs AH, Agarwal S, Ebert BL, Gazda HT, Lodish HF, Sieff CA, Schlaeger TM, Zon LI, Daley GQ, Drug discovery for Diamond-Blackfan anemia using reprogrammed hematopoietic progenitors. Sci Transl Med 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor AM, Humphries JM, White RM, Murphey RD, Burns CE, Zon LI, Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation. Exp Hematol 40, 228–237 e225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hidaka H, Yamaki T, Asano M, Totsuka T, Involvement of calcium in cyclic nucleotide metabolism in human vascular smooth muscle. Blood Vessels 15, 55–64 (1978). [DOI] [PubMed] [Google Scholar]
- 37.Inagaki M, Kawamoto S, Itoh H, Saitoh M, Hagiwara M, Takahashi J, Hidaka H, Naphthalenesulfonamides as calmodulin antagonists and protein kinase inhibitors. Mol Pharmacol 29, 577–581 (1986). [PubMed] [Google Scholar]
- 38.Ablain J, Durand EM, Yang S, Zhou Y, Zon LI, A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev Cell 32, 756–764 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch K, Mansi K, Haynes E, Adams CE, Sampson S, Furtado VA, Trifluoperazine versus placebo for schizophrenia. Cochrane Database Syst Rev, 1, CD010226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katoch A, George B, Iyyappan A, Khan D, Das S, Interplay between PTB and miR-1285 at the p53 3’UTR modulates the levels of p53 and its isoform Delta40p53alpha. Nucleic Acids Res 45, 10206–10217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharathchandra A, Lal R, Khan D, Das S, Annexin A2 and PSF proteins interact with p53 IRES and regulate translation of p53 mRNA. RNA Biol 9, 1429–1439 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Behnen P, Davis E, Delaney E, Frohm B, Bauer M, Cedervall T, O’Connell D, Akerfeldt KS, Linse S, Calcium-dependent interaction of calmodulin with human 80S ribosomes and polyribosomes. Biochemistry 51, 6718–6727 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Chin KV, Cade C, Brostrom CO, Galuska EM, Brostrom MA, Calcium-dependent regulation of protein synthesis at translational initiation in eukaryotic cells. J. Biol. Chem 262, 16509–16514 (1987) [PubMed] [Google Scholar]
- 44.Jaako P, Debnath S, Olsson K, Bryder D, Flygare J, Karlsson S, Dietary L-leucine improves the anemia in a mouse model for Diamond-Blackfan anemia. Blood 120, 2225–2228 (2012). [DOI] [PubMed] [Google Scholar]
- 45.MacInnes AW, Amsterdam A, Whittaker CA, Hopkins N, Lees JA, Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proc Natl Acad Sci U S A 105, 10408–10413 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai K, Amsterdam A, Farrington S, Bronson RT, Hopkins N, Lees JA, Many ribosomal protein mutations are associated with growth impairment and tumor predisposition in zebrafish. Dev Dyn 238, 76–85 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fond G, Macgregor A, Attal J, Larue A, Brittner M, Ducasse D, Capdevielle D, Antipsychotic drugs: pro-cancer or anti-cancer? A systematic review. Med Hypotheses 79, 38–42 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Marques LO, Lima MS, Soares BG, Trifluoperazine for schizophrenia. Cochrane Database Syst Rev, CD003545 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, Golub TR, Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 451, 335–339 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barlow JL, Drynan LF, Hewett DR, Holmes LR, Lorenzo-Abalde S, Lane AL, Jolin HE, Pannell R, Middleton AJ, Wong SH, Warren AJ, Wainscoat JS, Boultwood J, McKenzie AN, A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat Med 16, 59–66 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong CC, Traynor D, Basse N, Kay RR, Warren AJ, Defective ribosome assembly in Shwachman-Diamond syndrome. Blood 118, 4305–4312 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, Rey JP, Glynn EF, Ellington L, Du C, Dixon J, Dixon MJ, Trainor PA, Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med 14, 125–133 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pereboom TC, van Weele LJ, Bondt A, MacInnes AW, A zebrafish model of dyskeratosis congenita reveals hematopoietic stem cell formation failure resulting from ribosomal protein-mediated p53 stabilization. Blood 118, 5458–5465 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Calo E, Gu B, Bowen ME, Aryan F, Zalc A, Liang J, Flynn RA, Swigut T, Chang HY, Attardi LD, Wysocka J, Tissue-selective effects of nucleolar stress and rDNA damage in developmental disorders. Nature 554, 112–117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ceccaldi R, Parmar K, Mouly E, Delord M, Kim JM, Regairaz M, Pla M, Vasquez N, Zhang QS, Pondarre C, Peffault de Latour R, Gluckman E, Cavazzana-Calvo M, Leblanc T, Larghero J, Grompe M, Socie G, D'Andrea AD, Soulier J, Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell 11, 36–49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toki T, Yoshida K, Wang R, Nakamura S, Maekawa T, Goi K, Katoh MC, Mizuno S, Sugiyama F, Kanezaki R, Uechi T, Nakajima Y, Sato Y, Okuno Y, Sato-Otsubo A, Shiozawa Y, Kataoka K, Shiraishi Y, Sanada M, Chiba K, Tanaka H, Terui K, Sato T, Kamio T, Sakaguchi H, Ohga S, Kuramitsu M, Hamaguchi I, Ohara A, Kanno H, Miyano S, Kojima S, Ishiguro A, Sugita K, Kenmochi N, Takahashi S, Eto K, Ogawa S, Ito E, De Novo Mutations Activating Germline TP53 in an Inherited Bone-Marrow-Failure Syndrome. Am J Hum Genet 103, 440–447 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bowen ME, Attardi LD, The role of p53 in developmental syndromes. J Mol Cell Biol 11, 200–211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komarova EA, Gudkov AV, Chemoprotection from p53-dependent apoptosis: potential clinical applications of the p53 inhibitors. Biochem Pharmacol 62, 657–667 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Johnstone RW, Ruefli AA, Lowe SW, Apoptosis: a link between cancer genetics and chemotherapy. Cell 108, 153–164 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV, A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 285, 1733–1737 (1999). [DOI] [PubMed] [Google Scholar]
- 61.Thisse C, Thisse B, High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3, 59–69 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Paffett-Lugassy NN, Zon LI, Analysis of hematopoietic development in the zebrafish. Methods Mol Med 105, 171–198 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Murphey RD, Stern HM, Straub CT, Zon LI, A chemical genetic screen for cell cycle inhibitors in zebrafish embryos. Chem Biol Drug Des 68, 213–219 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE, A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Ludwig LS, Gazda HT, Eng JC, Eichhorn SW, Thiru P, Ghazvinian R, George TI, Gotlib JR, Beggs AH, Sieff CA, Lodish HF, Lander ES, Sankaran VG, Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med 20, 748–753 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chu C, Quinn J, Chang HY, Chromatin isolation by RNA purification (ChIRP). J Vis Exp, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larionov A, Krause A, Miller W, A standard curve based method for relative real time PCR data processing. BMC Bioinformatics 6, 62 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Macari ER, Lowrey CH, Induction of human fetal hemoglobin via the NRF2 antioxidant response signaling pathway. Blood 117, 5987–5997 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. p53 is translated in higher amounts in a zebrafish model of Diamond-Blackfan anemia.
Fig. S2. Chemical screen identifies calmodulin inhibitors and calcium channel blockers.
Fig. S3. TFP and BAPTA decreases p53 accumulation.
Fig. S4. TFP treatment leads to decreased mRNAs in monosome and polysome fractions.
Fig. S5. TFP improves platelet counts and mouse weight in a mouse model of Diamond-Blackfan anemia.
Table S1. Summary of validated hits for screen phenotypes.
Table S2. Numbers of zebrafish embryos that respond to drug treatment with statistical analysis.
Data File S1. Proteins isolated by RNA affinity purification of TP53 mRNA were identified by mass spectrometry.