Abstract
Rhodopsin, a prototypical G-protein-coupled receptor, is responsible for scoptic vision at low-light levels. Although rhodopsin’s photoactivation cascade is well understood, it remains unclear how lipid and zinc binding to the receptor are coupled. Using native mass spectrometry, we developed a novel data analysis strategy to deconvolve zinc and lipid bound to the proteoforms of rhodopsin and investigated the allosteric interaction between lipids and zinc binding. We discovered that phosphatidylcholine bound to rhodopsin with a greater affinity than phosphatidylserine or phosphatidylethanolamine, and that binding of all lipids was influenced by zinc but with different effects. In contrast, zinc binding was relatively unperturbed by lipids. Overall, these data reveal that lipid binding can be strongly and differentially influenced by metal ions.
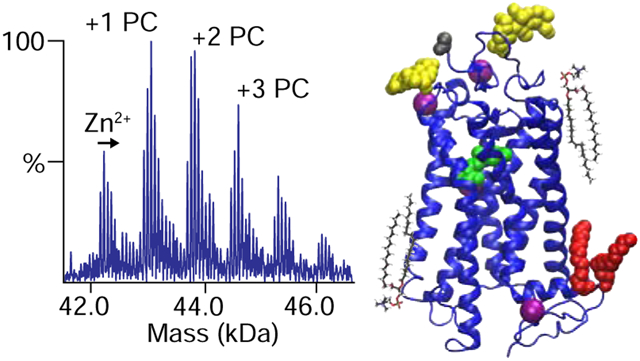
Graphical Abstract
Introduction
Rhodopsin is an essential protein in vision and plays a central role in autosomal dominant retinitis pigmentosa, and congenital stationary night blindness.1,2 Rhodopsin is the G-protein-coupled receptor (GPCR) found in the disc membranes of the rod outer segments (ROS) of the eye. It is a sensitive photoreceptor that absorbs visible light around 500 nm, allowing for vision in low-light level or scoptic conditions.3 Upon photon absorption, rhodopsin’s retinal chromophore photoisomerizes from the 11-cis to all-trans form.4 After activation, rhodopsin undergoes further conformational changes that result in short-lived intermediate products. The photocycle then reaches an equilibrium between the inactive metarhodopsin I (MI) and the active metarhodopsin II state (MII), before decaying by hydrolysis into free retinal and chromophore-free opsin.5
The activity of rhodopsin is modulated both by its lipid and aqueous environments.6 The ROS membranes are concentrated in phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) lipids.7 Changes in the lipid environment as the disc membrane matures cause differences in membrane properties, such as curvature, that can affect the activity of rhodopsin.8,9,10 Furthermore, reconstitution into synthetic lipid membranes has shown that the MI-MII equilibrium is affected by both the lipid head groups and hydrocarbon tails.11 , However, individual lipid interactions with rhodopsin12 have not been as well studied as bulk membrane effects.13
In addition to interacting with lipids, rhodopsin also binds zinc in the form of Zn2+. The retina contains some of the highest concentrations of zinc in the body.14 Altered zinc concentrations can contribute to age-related diseases, vision loss, and cataracts.15 Rhodopsin has three proposed zinc ion binding sites, one within the transmembrane region and two on the intradiscal side.14 The transmembrane site involving residues Glu122 and His211 has been suggested to be a high affinity binding site.16 The intradiscal binding sites involving Glu201/Gln279 and Glu197/His195 are lower affinity and are believed to destabilize rhodopsin, but may be non-specific.17,14 In addition to specific zinc binding regions, there may be other non-specific binding sites. Zinc binding has clear effects on rhodopsin stability and activity,18 but it is not clear how zinc and lipid binding are related.
To explore the allostery between lipid and zinc interactions and their effects on rhodopsin activity, we examined the influence of different lipids and different zinc concentrations on dark state rhodopsin with native mass spectrometry (MS). Native MS preserves noncovalent complexes for mass analysis. Because bound zinc and lipids have distinct mass shifts, native MS enabled us to simultaneously quantify the amount of bound lipid and zinc. To remove interferences between proteoforms of rhodopsin and zinc bound states, we developed a novel double deconvolution data analysis approach. Using these new MS methods, we discovered that zinc and lipid binding are interrelated, and the effects of zinc binding differ between lipids.
Experimental Methods
Materials.
Ammonium acetate, zinc acetate, 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate hydrate (CHAPS), and dodecyl trimethylammonium bromide (DTAB) were purchased from Sigma Aldrich. Ethylenediaminetetraacetic acid (EDTA) was purchased from Fisher. Hydroxyapatite was purchased from Bio-Rad Laboratories. Tetraethylene glycol monooctyl ether (C8E4) was purchased from Anatrace. 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) were obtained from Avanti Polar Lipids.
Protein Purification.
Rhodopsin was purified from frozen bovine retinas (W.L. Lawson Co., Omaha, NE) according to previous protocols.19 Purification was performed under dim red-light conditions and on ice or at 4 °C except where noted.6 To purify ROS membranes, retinas were thawed at 4 °C overnight, and then homogenized under a direct flow of argon gas. The homogenate was clarified by centrifugation at 2,600 ×g for 20 min prior to a second round of homogenization and centrifugation. The supernatants were pooled and centrifuged at 8,000 ×g for 1 hr after dilution in a 10 mM Tris-acetate buffer. The pellet was then introduced into a discontinuous sucrose step gradient of densities of 1.15, 1.13, 1.11, and 1.10 g/mL. Sucrose gradients were centrifuged in polyallomer tubes using a swinging bucket rotor at 113,000 ×g for 1 hr. The band at the interface of the 1.11 g/mL and the 1.13 g/mL layers corresponded to the ROS membranes and was collected by syringe. The collected rod disk membrane carpets were washed threefold, each time by diluting with two volumes of double-distilled water, and centrifuged at 48,000 ×g for 30 min to isolate the pelleted membranes from the aqueous supernatant.
To solubilize rhodopsin, the membranes were incubated at a concentration of 8–12 mg/mL in a solution of 100 mM DTAB containing 15 mM sodium phosphate at pH 6.8 for 1 hr on ice. Samples were centrifuged again at 48,000 ×g for 30 min, and the pellets were discarded. The supernatant was then loaded into a hydroxyapatite column equilibrated with a 100 mM DTAB and 15 mM sodium phosphate buffer at pH 6.8. The column was poured with around 30 mL of hydroxyapatite per 50 mg rhodopsin. Rhodopsin was eluted with a linear gradient of 0–0.5 M NaCl in detergent buffer at a flow rate of 0.6 mL/min. Fractions of 2 mL were collected, and their concentrations were measured using UV/visible spectroscopy. Those fractions with an A280/A500 ratio below 2.0 were combined together and concentrated using a 30 kDa centrifugal filter to a concentration of 10 mg/mL and exchanged into a 30 mM CHAPS and 15 mM sodium phosphate buffer (pH 6.8) for storage.6
Purified rhodopsin was further buffer exchanged from the CHAPS buffer into an MS-compatible buffer containing 200 mM ammonium acetate and 0.5% (v/v) C8E4 detergent by size exclusion chromatography (SEC) on a Superdex 200 Increase 10/300 column (GE Healthcare). For EDTA-treated samples, 0.1 mM EDTA at pH 7.8 was added prior to SEC and incubated for 10 min. The rhodopsin concentration was determined to be 1.3 μM using the A280 absorbance and a molar absorptivity of 40,600 M−1cm−1.6 Purified rhodopsin was then divided into 10 μL aliquots, flash frozen, and stored at −80 °C.
Lipid and Mass Spectrometry Sample Preparation.
Lipid solutions were prepared by first dissolving the lipids in chloroform. Phosphate analysis was used to determine the concentration of the lipid stock solution.20 Lipids were then dried overnight in a vacuum and solubilized in ammonium acetate buffer with C8E4 to a concentration of 155 μM that was 5 times the working concentration. The solutions were then further diluted using the rhodopsin sample to obtain a 10:1 lipid to rhodopsin molar ratio (an absolute lipid concentration of 13.0 μM). Zinc acetate was dissolved in ultrapure water to achieve stock solutions that were 5 times the working concentrations of 20, 250, and 750 ppm. Solutions were mixed using 6 μL of rhodopsin, 2 μL of lipid stock, and 2 μL of zinc stock solution. For the control solution, the lipid and zinc were replaced with C8E4 detergent in ammonium acetate. Samples were mixed and incubated in the dark for 10 min before introduction to the MS.
Mass Spectrometry.
Native electrospray ionization (ESI) was performed using borosilicate needles pulled inhouse with a P-1000 micropipette puller (Sutter Instrument, Novato, CA). Native MS was performed using a Q Exactive HF quadrupole-Orbitrap instrument equipped with Ultra-High Mass Range (UHMR) modifications (Thermo Fisher Scientific, Bremen). Instrumental parameters were used as previously published, including a spray voltage of 1.5 kV and 25–50 V of desolvation voltage in the insource trapping region to remove detergent.21,22 All samples were analyzed in the dark for one minute of analysis. Mass spectra were collected for triplicate mixtures, and error bars are shown as the standard deviation from the three replicates, except for POPC at 750 ppm, which was done in duplicate.
Mass Spectrometry Data Analysis.
Analysis of native mass spectra was performed using UniDec23 and MetaUniDec software.21 The mass range was set between 40–49 kDa with a charge range of 5–25. Deconvolved spectra showed a holo-rhodopsin peak at 41,983 Da that included several post-translational modifications (PTM), including palmitoylation, acetylation, and glycosylation.24
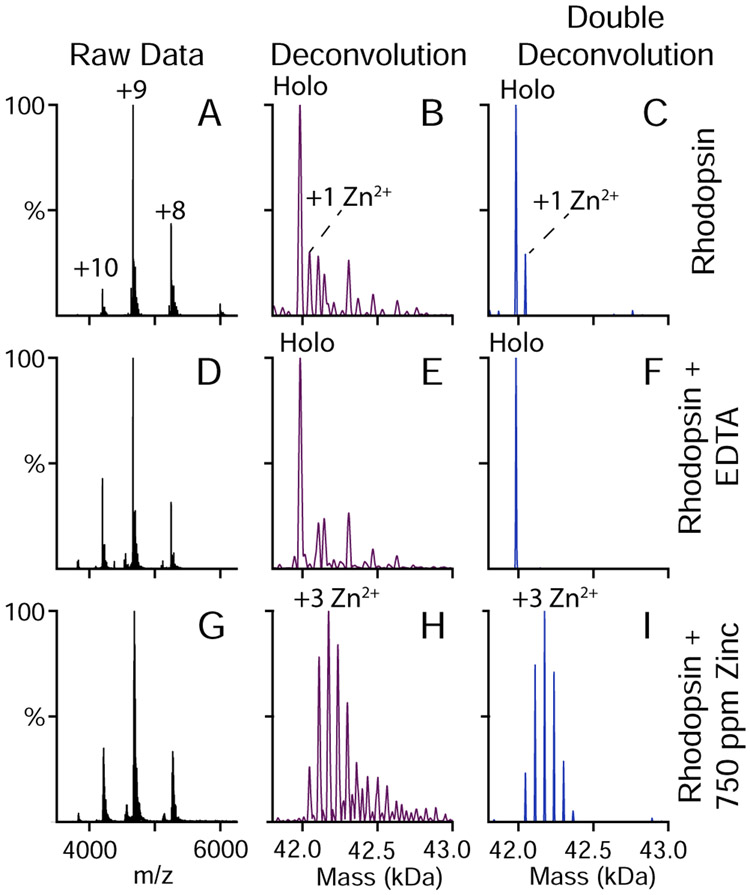
Though relatively simple, the different glycosylation proteoforms present in holo-rhodopsin limited the quantification of bound zinc because some peaks for different proteoforms with different numbers of bound zinc overlapped. To aid in quantitation, we developed a custom script to take the initial deconvolution results from UniDec and perform a second deconvolution with a conventional Richardson-Lucy algorithm (Figure 1).25,26 Here, a deconvolved zero-charge mass spectrum of EDTA-treated rhodopsin with no zinc or lipid added (Figure 1E) was used as the deconvolution kernel/template or point spread function. Using this double deconvolution approach, the contributions from all proteoforms were combined into a single peak, resulting in spectra with unique peak assignments and few artifacts. As shown in Figure 1, this approach significantly improved the quantification of bound species. A similar approach was previously shown by Klassen and coworkers using a sliding window subtractive method, which required more manual guidance.27
Figure 1.
Native mass spectra of rhodopsin (A, D, G), zero-charge mass spectra deconvolved by UniDec (B, E, H), and doubly deconvolved zero-charge mass spectra subjected to Richardson-Lucy deconvolution (C, F, I). Data are for rhodopsin under standard purification conditions (A, B, C), rhodopsin purified with EDTA added to remove bound zinc (D, E, F), and rhodopsin with 750 ppm zinc added (G, H, I).
The amount of lipid and zinc binding to rhodopsin was quantified by extracting the intensities of peaks in the doubly deconvolved spectra for all possible combinations of bound lipid and zinc using the largest peak height within a ±10 Da window. The intensities of each zinc bound state were then summed for all lipid bound states to obtain the relative amounts of bound zinc. The average numbers of bound zinc ions per rhodopsin were then calculated weighted by their relative intensities summed across all lipid bound states. The average amount of bound lipid was calculated in a similar manner by summing across all zinc bound states and calculating the weighted average.
Results and Discussion
Rhodopsin Proteoforms.
To understand the relationship between zinc and lipid binding, we performed native MS of rhodopsin solubilized in micelles of C8E4 nonionic detergent. Native MS uses nondenaturing ionization to preserve noncovalent complexes for mass analysis, which allows simultaneous and independent detection of multiple different bound ligands.28,29 Rhodopsin was purified from bovine retinas and exchanged into C8E4 detergent micelles using SEC. All steps were performed under dark conditions with a small amount of red light to prevent photoactivation of rhodopsin.
First, we measured the native mass spectrum of dark-state rhodopsin in the absence of added zinc or lipid. We discovered that rhodopsin was a monomer in C8E4 detergent at these concentrations. No higher ordered oligomers were observed. Under standard purification conditions, some zinc ions remained bound to rhodopsin (Figure 1A-C), indicating tight zinc interactions that persisted through multiple steps of purification. To remove the residual zinc, we added EDTA prior to the SEC buffer exchange, which stripped the remaining zinc from the complex (Figure 1D-F).
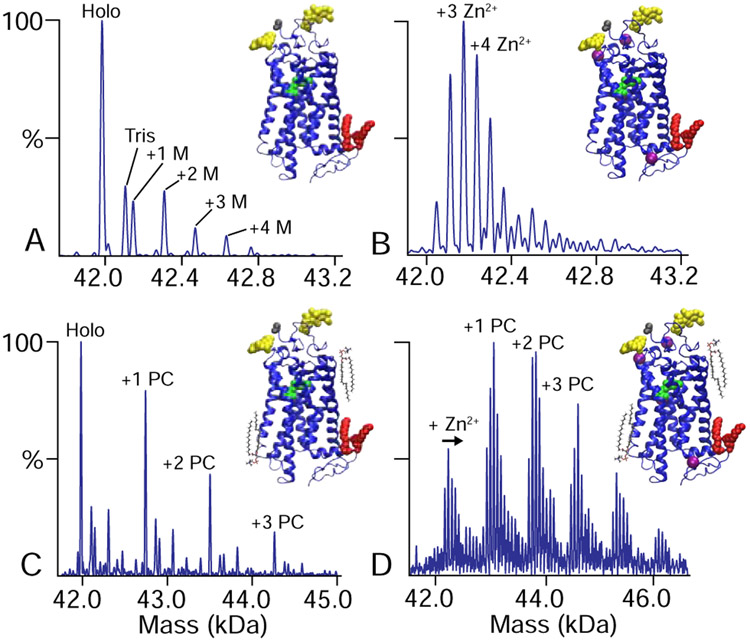
Significantly, the zinc-free mass spectrum (Figure 2A) showed a relatively simple pattern of post-translational modifications (PTMs). The most abundant mass was the rhodopsin protein with the expected post-translational modifications: two glycans, two palmitoylations, N-terminal acetylation, and retinylation.24,30 Additional peaks were observed for one, two, three, or four additional mannose (M) units added to the glycans, which matched prior glycomics results.20 Apart from the expected PTMs, an extra peak was observed at +121 Da that was not conclusively assigned to any covalent or noncovalent adduct, but it may correspond to residual Tris buffer from the purification process.
Figure 2.
Deconvolved zero-charge mass spectra of zinc-free rhodopsin (A) with added (B) zinc, (C) PC lipids, and (D) both PC and zinc. Added zinc concentrations were 750 ppm in B and D. Rhodopsin structures are shown with glycans in yellow, retinal in green, palmitoyl groups in red, zinc in magenta, and lipids in black.
Native MS Reveals Zinc Binding to Rhodopsin.
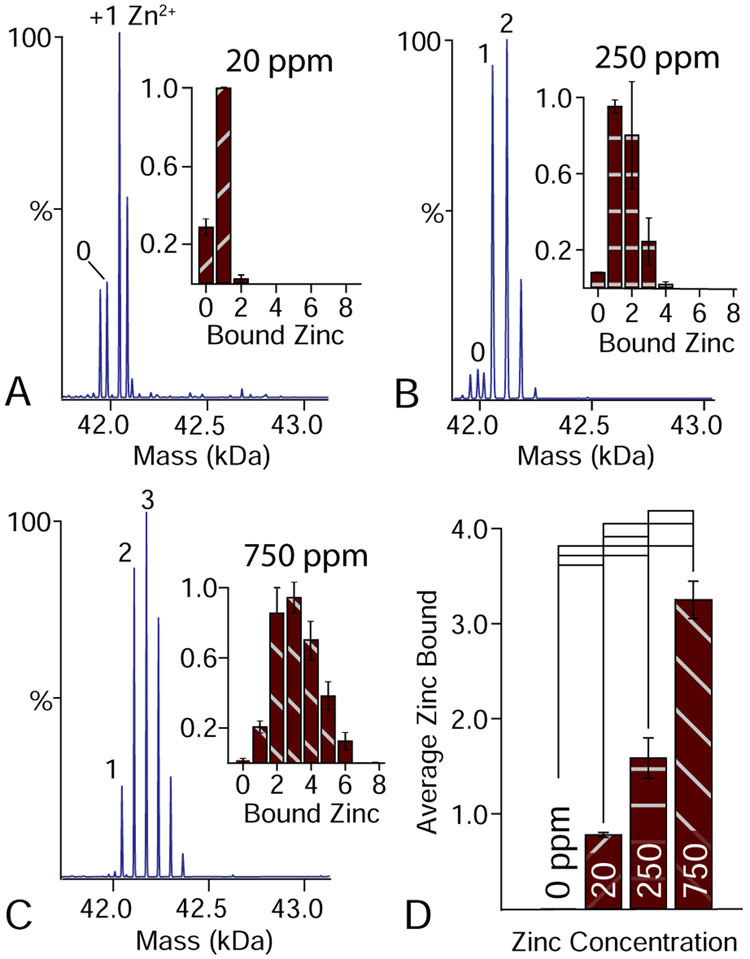
Because zinc remains tightly associated with rhodopsin during purification, we next measured the binding of exogenous zinc to rhodopsin. Retinas have some of the highest zinc levels in the body, with concentrations around 500–600 ppm.31 To bracket the physiological zinc concentration, we added zinc acetate at 20, 250, and 750 ppm. Using native MS, we then collected dark-state mass spectra of rhodopsin with bound zinc (Figure 2B). Each bound zinc ion created a new peak series with an additional 63 Da in mass. When double deconvolution is applied (Figure 1I), the known PTMs are computationally removed and only bound zinc is observed. From the double deconvolved spectra, we quantified the average number of zinc ions bound as a function of its concentration. A clear trend was observed with more zinc bound at higher concentrations (Figure 3). Interestingly, rhodopsin with 20 ppm zinc had 0.84±0.03 bound zinc ions while 250 ppm only showed a moderate increase to 1.58±0.18 bound zinc ions. Further increasing the concentration to 750 ppm increased binding to 3.25±0.15. Overall, these data are consistent with one higher affinity binding site and two with lower affinity bind sites.12 Additional bound states at higher concentrations are likely nonspecific, as has previously been discussed.18
Figure 3.
Doubly deconvolved zero-charge mass spectra of rhodopsin with varying concentrations of bound zinc (A–C). Insets quantify the numbers of zinc ions bound. Extra peaks for 20 ppm result from partial degradation. (D) Average number of bound zinc ions at different concentrations of added zinc. Significant differences at the 95% confidence level are indicated by black lines.
Lipid-Rhodopsin Interactions Depend on Zinc Binding.
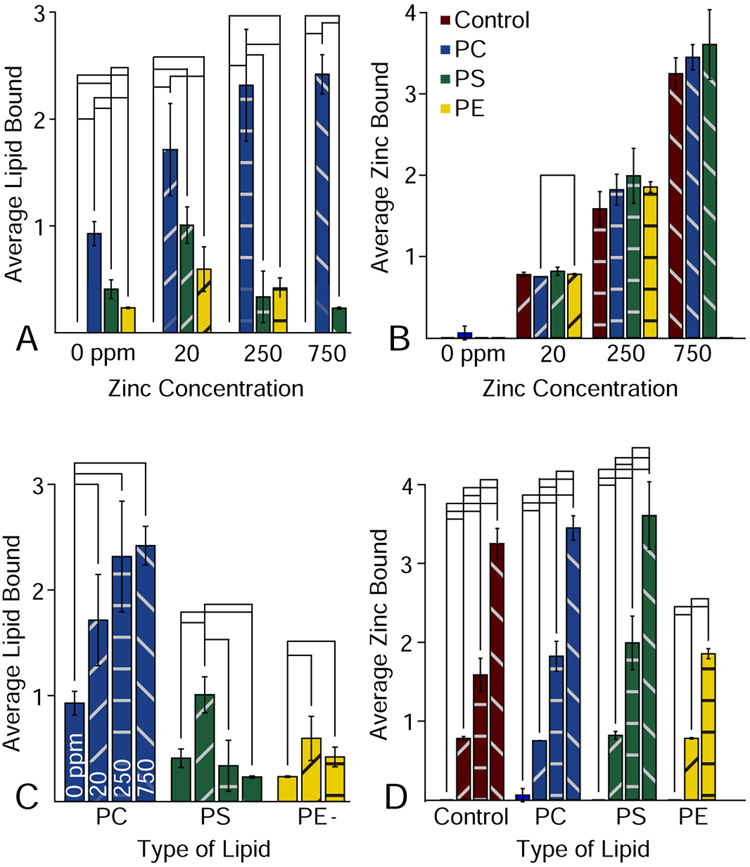
We next sought to investigate the binding of lipids to rhodopsin by native MS. Rhodopsin is known to be sensitive to the lipid bilayer environment, and changes in the disc membrane as the rod outer segments matures play a physiological role in modulating rhodopsin activity.32,33 Here, we added POPC, POPS, and POPE lipids solubilized in C8E4 detergent at a 10:1 molar ratio to rhodopsin, because these are the most common lipid head groups in retinal membranes.34 Bound lipids were easily detected as new peaks in the mass spectrum that were shifted by the mass of the lipid (Figure 2C). Similar to zinc, we quantified the average number of bound lipid molecules. At the same concentration, more POPC molecules were bound than POPE and POPS, which showed comparable levels of binding (Figure 4A). The greater association of PC versus PE and PS in the dark state is in line with its role in stabilizing rhodopsin upon light-activation to large-scale conformational changes.
Figure 4.
Average number of lipids bound to rhodopsin grouped by zinc concentration (A) and lipid type (C). The average amount of zinc binding grouped by zinc concentration (B) and lipid type (D). Significant differences at the 95% confidence level are marked by black lines.
To explore how lipid binding altered zinc binding and vice versa, we added the same zinc concentrations to each of the lipid bound samples. Lipids generally had no statistically significant impact on the amount of bound zinc (Figure 4B). In contrast, the effects of zinc on lipid binding varied dramatically between lipids (Figure 4C). POPC showed an overall trend of higher lipid binding with higher zinc concentrations. However, POPS first showed an initial rise in lipid binding from 0 to 20 ppm zinc, but additional zinc beyond this level caused a decrease in lipid binding. Because PS lipids are capable of chelating divalent cations, it may be that zinc forms a bridge between PS and anionic residues on rhodopsin at low concentrations but induces lipid clustering at higher concentrations. Finally, POPE showed initially higher levels of lipid binding with increased zinc but was not stable at 750 ppm zinc.
Together, these data reveal that zinc and lipid binding are interrelated in complex ways that depend on the identity of the lipid. Zinc binding is largely unaffected by lipids, but lipid binding was significantly affected by zinc. However, the precise effects of zinc on lipid binding depended on both the lipid type and the zinc concentration. Overall, the effects of metals and other divalent cations may represent important modulators of lipid interactions that have been understudied. Future studies will perform two-component titrations to quantify allostery in zinc and lipid binding, but lipid binding results, especially from POPS, reveal that conventional KD models may be insufficient to capture the complex interplay of metals and lipids on the surface of membrane proteins
Conclusion
Enabled by novel data analysis methods, we used the unique power of native MS to simultaneously monitor zinc and lipid binding to rhodopsin. We discovered that zinc binding is largely independent of lipids, but lipid binding can be significantly influenced by zinc in a manner that depends on the concentration of zinc and type of lipid. These data reveal that divalent cations and metals may play an important and largely unexplored role in membrane protein-lipid interactions.
Highlights.
Native MS was used to study lipid and zinc binding to bovine rhodopsin.
Novel double deconvolution data processing was introduced.
Zinc binding was relatively unperturbed by lipid binding.
Lipid binding was significantly influenced by zinc in different ways.
ACKNOWLEDGMENT
The authors thank Deseree Reid for assistance in sample analysis. This work was funded by the Bisgrove Scholar Award from Science Foundation Arizona and the National Institute of General Medical Sciences and National Institutes of Health under Award Number R35 GM128624 to M.T.M. Additional funding was provided by National Science Foundation awards CHE 1904125 and MCB 11817862 to M.F.B. and National Institutes of Health awards EY012049 and EY02604 to M.F.B. Support of S.M.D.C. was by a Technology Research Initiative Fund predoctoral fellowship from the Arizona Board of Regents. S.D.E.F. was partially supported by a Goldwater research scholarship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- (1).Tam BM; Qazalbash A; Lee HC; Moritz OL The Dependence of Retinal Degeneration Caused by the Rhodopsin P23H Mutation on Light Exposure and Vitamin a Deprivation. Biochem. Mol. Biol 2010, 51 (3), 1327–1334. [DOI] [PubMed] [Google Scholar]
- (2).Rakoczy EP; Kiel C; McKeone R; Stricher F; Serrano L Analysis of Disease-Linked Rhodopsin Mutations Based on Structure, Function, and Protein Stability Calculations. J. Mol. Biol 2011, 405 (2), 584–606. [DOI] [PubMed] [Google Scholar]
- (3).Latorraca NR; Venkatakrishnan AJ; Dror RO GPCR Dynamics: Structures in Motion. Chem. Rev 2017, 117 (1), 139–155. [DOI] [PubMed] [Google Scholar]
- (4).Nathans J Rhodopsin : Structure, Function, and Genetics. Biochemistry 1992, 31 (21), 4923–4931. [DOI] [PubMed] [Google Scholar]
- (5).Struts AV; Salgado GFJJ; Martínez-Mayorga K; Brown MF Retinal Dynamics Underlie Its Switch from Inverse Agonist to Agonist during Rhodopsin Activation. Nat. Struct. Mol. Biol 2011, 18 (3), [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Brown MF Membrane Protein Structure and Dynamics; 2012; Vol. 914. [Google Scholar]
- (7).Turcu Francisca E. Reyes, Ventii Karen H., W. K. D. Role of Membrane Integrity on G Protein-Coupled Receptors: Rhodopsin Stability and Function. Annu Rev Biochem 2010, 50 (3), 363–397. [Google Scholar]
- (8).Albert A; Alexander D; Boesze-battaglia K Cholesterol in the Rod Outer Segment : A Complex Role in a “ Simple ” System. Chem. Phys. Lipids 2016, 199, 94–105. [DOI] [PubMed] [Google Scholar]
- (9).Soubias O; Teague WE; Hines KG; Gawrisch K The Role of Membrane Curvature Elastic Stress for Function of Rhodopsin-like G Protein-Coupled Receptors. Biochimie 2015, 107 (A), 28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Soubias O; Teague WE; Hines KG; Gawrisch K Rhodopsin/Lipid Hydrophobic Matching - Rhodopsin Oligomerization and Function. Biophys. J 2015, 108 (5), 1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Gibson NJ; Brown MF Lipid Headgroup and Acyl Chain Composition Modulate the MI–MII Equilibrium of Rhodopsin in Recombinant Membranes. Biochemistry 1993, 32 (9), 2438–2454. [DOI] [PubMed] [Google Scholar]
- (12).Salas-Estrada LA; Leioatts N; Romo TD; Grossfield A Lipids Alter Rhodopsin Function via Ligand-like and Solvent-like Interactions. Biophys. J 2018, 114 (2), 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Watts A; Volotovski ID; Volotovski ID Rhodopsin-Lipid Associations in Bovine Rod Outer Segment Membranes. Identification of Immobilized Lipid by Spin-Labels. Biochemistry 1979, 18 (22), 5006–5013.. [DOI] [PubMed] [Google Scholar]
- (14).Toledo D; Cordomi A; Proietti MG; Benfatto M; Del Valle LJ; Pérez JJ; Garriga P; Sepulcre F Structural Characterization of a Zinc High-Affinity Binding Site in Rhodopsin. Photochem. Photobiol 2009, 85 (2), 479–484. [DOI] [PubMed] [Google Scholar]
- (15).Handa JT; Cano M; Wang L; Datta S; Liu T Lipids, Oxidized Lipids, Oxidation-Specific Epitopes, and Age-Related Macular Degeneration. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 2017, 1862 (4), 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Stojanovic A; Stitham J; Hwa J Critical Role of Transmembrane Segment Zinc Binding in the Structure and Function of Rhodopsin. J. Biol. Chem 2004, 279 (34), 35932–35941. [DOI] [PubMed] [Google Scholar]
- (17).Gleim S; Stojanovic A; Arehart E; Byington D; Hwa J Conserved Rhodopsin Intradiscal Structural Motifs Mediate Stabilization: Effects of Zinc. Biochemistry 2009, 48 (8), 1793–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Park PSH; Sapra KT; Koliński M; Filipek S; Palczewski K; Muller DJ Stabilizing Effect of Zn2+ in Native Bovine Rhodopsin. J. Biol. Chem 2007, 282 (15), 11377–11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Botelho AV; Gibson NJ; Thurmond RL; Wang Y; Brown MF Conformational Energetics of Rhodopsin Modulated by Nonlamellar-Forming Lipids. Biochemistry 2002, 41 (20), 6354–6368. [DOI] [PubMed] [Google Scholar]
- (20).Chifflet S; Torriglia A; Chiesa R; Tolosa S A Method for the Determination of Inorganic Phosphate in the Presence of Labile Organic Phosphate and High Concentrations of Protein: Application to Lens ATPases. Anal. Biochem 1988, 168 (1), 1–4. [DOI] [PubMed] [Google Scholar]
- (21).Reid DJ; Diesing JM; Miller MA; Perry SM; Wales JA; Montfort WR; Marty MT MetaUniDec: High-Throughput Deconvolution of Native Mass Spectra. J. Am. Soc. Mass Spectrom 2019, 30 (1), 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Reid DJ; Keener JE; Wheeler AP; Zambrano DE; Diesing JM; Reinhardt-Szyba M; Makarov A; Marty MT Engineering Nanodisc Scaffold Proteins for Native Mass Spectrometry. Anal. Chem 2017, 89 (21), 11189–11192. [DOI] [PubMed] [Google Scholar]
- (23).Marty MT; Baldwin AJ; Marklund EG; Hochberg GKA; Benesch JLP; Robinson CV Bayesian Deconvolution of Mass and Ion Mobility Spectra: From Binary Interactions to Polydisperse Ensembles. Anal. Chem 2015, 87 (8), 4370–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Murray AR; Fliesler SJ; Al-Ubaidi MR Rhodopsin: The Functional Significance of Asn-Linked Glycosylation and Other Post-Translational Modifications. Ophthalmic Genet. 2009, 30 (3), 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lucy LB An Iterative Technique for the Rectification of Observed Distributions. Astron. J 1974, 79 (6). [Google Scholar]
- (26).Richardson WH Bayesian-Based Iterative Method of Image Restoration. J. Opt. Soc. Am 1972, 62 (1), 55. [Google Scholar]
- (27).Kitov PI; Han L; Kitova EN; Klassen JS Sliding Window Adduct Removal Method (SWARM) for Enhanced Electrospray Ionization Mass Spectrometry Binding Data. J. Am. Soc. Mass Spectrom 2019, 30 (8), 1446–1454. [DOI] [PubMed] [Google Scholar]
- (28).Mehmood S; Allison TM; Robinson CV Mass Spectrometry of Protein Complexes: From Origins to Applications. Annu. Rev. Phys. Chem 2015, 66 (1), 453–474. [DOI] [PubMed] [Google Scholar]
- (29).Leney AC; Heck AJR Native Mass Spectrometry: What Is in the Name? J. Am. Soc. Mass Spectrom 2017, 28 (1), 5–13. [DOI] [PubMed] [Google Scholar]
- (30).Whitelegge JP; Gundersen CB; Faull KF Electrospray-Ionization Mass Spectrometry of Intact Intrinsic Membrane Proteins. Protein Sci. 1998, 7 (6),. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Galin MA; Nano HD; Hall T Ocular Zinc Concentration. Invest. Ophthalmol 1962, 1, 142–148. [PubMed] [Google Scholar]
- (32).Tainer JA; Roberts VA; Getzoff ED Protein Metal-Binding Sites. Curr Opin Biotechnol 1992, 3(4), 378–387. [DOI] [PubMed] [Google Scholar]
- (33).Sakmar TP; Huber T Rhodopsin. Encycl. Neurosci 2010, 1271 (8), 365–372. [Google Scholar]
- (34).Cartailler J-P; Luecke H X-Ray Crystallographic Analysis of Lipid-Protein Interactions in the Bacteriorhodopsin Purple Membrane. Annu. Rev. Biophys. Biomol. Struct 2003, 32 (1), 285–310. [DOI] [PubMed] [Google Scholar]
- (35).Acar N; Berdeaux O; Grégoire S; Cabaret S; Martine L; Gain P; Thuret G; Creuzot-Garcher CP; Bron AM; Bretillon L Lipid Composition of the Human Eye: Are Red Blood Cells a Good Mirror of Retinal and Optic Nerve Fatty Acids? PLoS One 2012, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]