ABSTRACT
Accurate measurements of cell morphology and behaviour are fundamentally important for understanding how disease, molecules and drugs affect cell function in vivo. Here, by using muscle stem cell (muSC) responses to injury in zebrafish as our biological paradigm, we established a ‘ground truth’ for muSC behaviour. This revealed that segmentation and tracking algorithms from commonly used programs are error-prone, leading us to develop a fast semi-automated image analysis pipeline that allows user-defined parameters for segmentation and correction of cell tracking. Cell Tracking Profiler (CTP) is a package that runs two existing programs, HK Means and Phagosight within the Icy image analysis suite, to enable user-managed cell tracking from 3D time-lapse datasets to provide measures of cell shape and movement. We demonstrate how CTP can be used to reveal changes to cell behaviour of muSCs in response to manipulation of the cell cytoskeleton by small-molecule inhibitors. CTP and the associated tools we have developed for analysis of outputs thus provide a powerful framework for analysing complex cell behaviour in vivo from 4D datasets that are not amenable to straightforward analysis.
KEY WORDS: Muscle, Zebrafish, Cell tracking, Segmentation, In vivo imaging, Phagosight, Icy, Imaris
Summary: Analysis of cell shape and movement from 3D time-lapses is currently very challenging. Here, we develop Cell Tracking Profiler and use it to examine RhoA and myosin II control of muscle stem cell behaviour during zebrafish regeneration.
INTRODUCTION
Analysis of cell shape and behaviour are becoming increasingly important for understanding how cells respond to drugs or molecules. As a consequence it is crucial to be able to measure parameters of cell shape and movement, and then to interpret the meaning of such changes. Much progress has been made in creating workflows optimised for analysing cells cultured in vitro in 2D environments. It is becoming increasingly clear, however, that cells behave quite differently in 3D environments or in vivo. Many of the image analysis tools designed for investigating cell behaviour in 2D are not suitable for analysing such complex data. To characterise cell behaviour from such datasets without extensive distortion of the primary data represents a fundamental problem in image analysis, as cells in vivo often show extensive changes in their shape, move rapidly and can be hard to discriminate from adjacent cells if in close proximity (Driscoll and Danuser, 2015).
A number of programs are available for automated analysis of cell behaviour from 3D datasets. These include commercial programs such as Imaris (Bitplane AG) and AMIRA (FEI, Inc.) and open source programs such as ImageJ/Fiji (Collins, 2007; Schindelin et al., 2012), Icy (de Chaumont et al., 2012), Phagosight (Henry et al., 2013) and tTy/qTfy (Hilsenbeck et al., 2016). These programs use a variety of algorithms to segment and track cells, ranging from user-driven annotation to automated processes with minimal user intervention.
All image analysis programs display similar limitations when attempting to segment 3D datasets in which cells display a uniform labelling including: (1) an inability to accurately identify the boundaries of cells, (2) problems in coping with diverse cell sizes and shapes and (3) being able to identify discrete boundaries between adjacent cells. These issues become important when tracking cells, as the segmentation efficiency will dictate how well a cell can be tracked (Nketia et al., 2017). Furthermore, commercially available software packages do not make their code available, limiting options for users to modify how images are processed.
Several studies have compared software packages for their effectiveness in cell segmentation or tracking (Maška et al., 2014; Ulman et al., 2017). Performance of most software packages has been poor when analysing complex in vivo 3D imaging data, as these rely on specific labels (nuclear or membrane), do not accurately segment cells in 3D or require computationally expensive methods for segmenting. Given the importance of cell behaviour for effective regeneration, we performed a benchmark study of several available programs (Imaris, Icy and Phagosight) to evaluate their ability to accurately describe migratory muscle stem cells (muSCs) in the myotome of regenerating zebrafish (Knappe et al., 2015; Roy et al., 2017). We found that a fundamental limitation of the programs we tested was the segmentation efficiency and ease of correcting inappropriate tracking. To facilitate analysis of 3D time-lapse data we have therefore designed Cell Tracking Profiler (CTP), an image analysis tool that functions within the freely available Icy package (de Chaumont et al., 2012). A key strength of this tool is that it allows user input to control segmentation and correct tracking of cells. Using a variety of statistical tools, we evaluated how outputs from CTP can be analysed to identify parameters of cell shape and movement. Our comparisons against other image analysis packages reveal that CTP offers a powerful and intuitive user-led tool that can provide accurate measures of complex cell behaviour. As an example, we used CTP to identify specific changes to muSC shape and movement in response to inhibition of RhoA kinase proteins (ROCK) and myosin II.
Our development of this workflow provides the basis for a medium-throughput approach to analyse time-lapsed datasets from a variety of biological systems.
RESULTS
Morphological changes in muSCs during migration to sites of injury
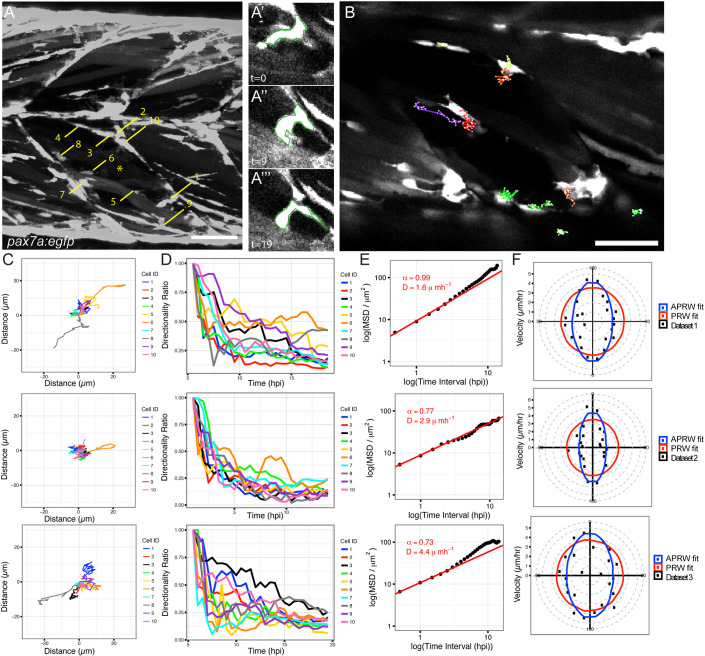
We wished to measure cell shape and behaviour of muSCs, a complex and migratory cell type, in order to determine how they are regulated during tissue repair. Using a zebrafish injury model in which muSCs are labelled with green fluorescent protein (GFP) driven by the pax7a promoter (pax7a:egfp) (Knappe et al., 2015), we aimed to segment and track cells from time-lapse datasets as accurately as possible. Larvae expressing the pax7a:egfp transgene were injured in the ventral 12th myotome at 5 days post fertilisation (dpf) using a sharpened tungsten needle. Larvae were immobilised in agarose and the response of GFP+ muSCs to injury was visualised by time-lapse confocal microscopy. As is common for many transgenic animals, GFP is distributed throughout the cytoplasm of muSCs in pax7a:egfp larvae, making it difficult to automatically detect cell boundaries using standard thresholding approaches. To therefore establish a ‘ground truth’ for cell shape, a manual segmentation of selected cells was performed by hand from three ‘baseline’ datasets (datasets 1–3). These differ in that images for datasets 1 and 2 were acquired from 5 h after injury, whereas dataset 2 commenced 0.5 h after injury and the z-interval for datasets 1 and 2 was 1 µm, and for dataset 3 was 5 µm. Segmentation of 10 representative cells from each dataset at three time-points from the time-lapse (representing 30 datapoints) revealed a diversity of muSC sizes and shapes within each individual dataset (Fig. 1A). By examining the temporal profile of muSCs in the injured myotome, it was apparent that shape and size for each cell changed over time in a non-uniform manner.
Fig. 1.
Characterisation of muSC behaviour from ground truth datasets. Muscle stem cells in transgenic pax7a:egfp zebrafish larvae show dynamic shape changes as they respond to muscle injury in a time-lapsed movie (dataset 1, acquisition interval 30 min). Manual segmentation of 10 cells (numbered in A) in an injured myotome (*) was performed at three time-points (time-point 0, 9, 19). Cell 1 changes shape over time (A′,A″,A‴). Selected cells were manually tracked over time using MTrackJ (B). Tracks were mapped to a common origin (C) for 10 selected cells from datasets 1–3 and the directionality ratio plotted (D). The MSD was used to calculate D (diffusion constant of cells) and plotted as a logged value against the log of time (E). A line of best fit (red) was generated, from which the α parameter was calculated. Cell movement was fitted against models of persistent random walk (PRW) and anisotropic PRW (APRW), with goodness of fit of the data to either model described by an R-squared value (F). Scale bars: 50 µm.
To define how muSCs migrate towards a muscle injury, we established a ground truth by tracking the selected 10 muSCs using the ImageJ manual tracker plugin MTrackJ (Fig. 1B; Movies 1 and 2). Each cell was tracked and parameters for duration, point speed and directionality were measured (Fig. 1C; Table 1). To determine whether muSCs responding to injury showed rapid movement, we evaluated instantaneous speed. This is a calculation of the speed of cell movement in-between time-points, and can be used to infer how fast cells move regardless of their directionality. Cells showed considerable variability in their behaviour, and no clear trend was discernible. We then tested whether muSCs showed persistence in their movement after injury by plotting the directionality ratio relative to time (Gorelik and Gautreau, 2014). A feature of this measure is that values decay from 1 as a function of their trajectory relative to their initial trajectory, such that cells showing a more-persistent movement will show values closer to 1 over time. Variability in cell behaviour of all three datasets was apparent, with some cells showing high persistence lasting for some time (e.g. cells 6 and 9 in dataset 1, and cell 3 in dataset 3), whereas others showed a rapid drop in persistence, for example, cell 1 in dataset 1 (Fig. 1D). We calculated the directional autocorrelation of cells to assess whether cell shape was related to the direction of cell migration (Gorelik and Gautreau, 2014), but observed no clear trend over time. An analysis of cell trajectories by determining the mean square displacement (MSD) indicated cell movement was sub-diffusive with alpha parameter values calculated as <1 and a diffusion constant (D)>1 (Fig. 1E). MSD assumes cell movement fits a model of persistent random walk (PRW), but recent descriptions of cell behaviour in 3D have suggested that cells move in an anisotropic mode (Wu et al., 2015). We therefore evaluated whether the angular velocity of muSCs fitted better to a model of PRW compared to an anisotropic persistent random walk (APRW, Fig. 1F). In all 3 datasets, we observed that the velocity of the tracked cells showed a better fit to the APRW model than to a PRW model (Table 1).
Table 1.
Evaluation of models of cell movement to explain behaviour of the 10 selected ground truth cells in datasets 1–3
Cell shape affects automated segmentation efficiency
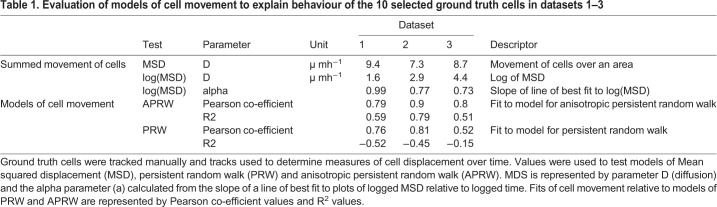
To explore the potential and limits of automated segmentation to identify and measure muSCs, we compared three programs that have been used to analyse migratory cell behaviour: Imaris (Bitplane AG), Icy (de Chaumont et al., 2012) and Phagosight (Henry et al., 2013). Our criteria for choosing these programs was that they are commonly used for analysing time-lapse imaging data and all employ preset criteria to some degree that can affect the image segmentation and tracking. We first evaluated how well each program could segment muSCs cells from datasets 1–3 relative to the ground truth obtained by manual segmentation. The criteria used to compare efficiency of these segmentation processes were: (1) how many cells were identified at three specified time points, (2) how many of a pre-defined population of selected cells of interest at each time point could be identified, and (3) a comparison of the segmented cell volume to the ground truth volume expressed as a ratio.
Cell segmentation was performed using Imaris, Icy and Phagosight with default settings as described in the Materials and Methods section. Dataset 1 had 276 objects cumulatively over the three time-points; Icy identified 324 objects, Phagosight identified 224 and Imaris identified 144 objects. Dataset 2 had 254 objects; Icy identified 290, Phagosight identified 190 and Imaris identified 648 objects. Dataset 3 had 410 objects; Icy identified 499 objects, Phagosight identified 249 objects and Imaris identified 636 objects (Fig. 2A). Relative to the number of objects identified from the ground truth across all three datasets Icy identified ∼18% more objects, Phagosight identified ∼30% fewer objects and Imaris identified ∼52% more objects. Icy therefore identified the number of objects from all the datasets that was closest to that in the ground truth.
Fig. 2.
Evaluation of segmentation accuracy by Imaris, Icy and Phagosight relative to a ground truth. The number of segmented objects by Imaris (purple), Icy (green) and Phagosight (cyan) were compared to a manually defined ground truth (dark blue) from datasets 1–3 (A). The volumes of the 10 segmented objects from dataset 1 were compared relative to a ground truth by plotting average values with standard deviation at 5, 9.5 and 14.5 h post injury (hpi) for each method (B). The effectiveness of each method for detecting the 10 cells at three time points (representing 30 data points) were scored as good (blue), none (red) or false (green) for each dataset (C–E). The volumes of the selected 30 objects segmented by each method were compared by star plots in which the outputs of the three programs were normalised to the ground truth (blue) for each dataset (F–H).
A comparison of the volumes of the segmented objects revealed that Phagosight segmented cells with a wider spread of volumes compared to Icy and Imaris (Fig. 2B). We noted that Phagosight often identified cells in close proximity as single objects when compared against the ground truth, which suggests this is why fewer objects are identified when compared to Icy or Imaris.
We then assessed how well each program could identify the 10 cells of interest that we had defined by manual annotation from each dataset at each of three time points (30 objects). Datasets 1–3 were segmented using optimal parameters (Fig. S1). These parameters were defined by comparing the number of objects segmented relative to the number of objects in the ground truth and whether detection was good (a single cell), false detection (cell detected, but not resolved as a single object or merged with another cell) or not detected. Key criteria for selecting optimal segmentation parameters were: (1) the number of objects segmented should be as close to the ground truth as possible, (2) that there should be a minimal number of false detections of the selected 30 objects, and (3) as many cells as possible should be detected from the 30 selected objects (Table S1). Based on these criteria, we identified parameters of segmentation that minimised the difference between actual number of objects (the ground truth) and the detected number of objects. The difference in the number of detected objects compared to the ground truth with these optimal parameters was 204 objects for Imaris, 58 for Icy and 92 for Phagosight, although each program showed distinct differences in their ability to segment the cells from the individual datasets (Fig. 2C–E). Using optimal segmentation parameter values Icy had a slightly higher false detection rate than Imaris (2/90 compared to 1/90) but was better than Phagosight (15/90). Imaris had the lowest number of missing detections (4/90) and highest number of good detections overall (85/90) compared to Icy (missing, 11/90; good, 77/90) or Phagosight (missing, 13/90; good, 62/90).
To evaluate the accuracy of segmentation by Icy, Imaris and Phagosight, we compared volumes of objects against volumes obtained from the manually defined ground truth. Focusing on the 10 cells of interest at one time point, we noted that overall, all three programs failed to segment the entire cell volume relative to manual segmentation (Fig. 2F–H). We compared the calculated cell volume relative to the ground truth using a mixed effects multiple level linear regression analysis. Comparing the cumulative values of the segmented objects revealed that all three methods significantly underestimated cell volume compared to the ground truth (Fig. S2). Differences in the cumulative volumes for objects segmented by each program were apparent for each dataset.
Analysing cell migration
In order to accurately describe cell behaviour during tissue repair, it is necessary to track them accurately through a 4D space. Given the time-intensive nature of manually annotating and tracking cells an automated method is necessary. Icy, Imaris and Phagosight use different approaches to track cells – the Spot Tracking tool in Icy evaluates past and futures frames to predict the position of a cell over time; Phagosight employs a keyhole algorithm that makes predictions about cell movement from past and future frames in a restricted model of directional movement (Reyes-Aldasoro et al., 2008); and Imaris offers several proprietary algorithms for tracking cells through the Surfaces tool.
To evaluate the accuracy of Imaris, Icy and Phagosight in tracking muSCs, we first compared outputs from these programs to the ground state obtained by manually tracking cells. Tracking for all three programs was performed using an optimally segmented labelled image for each dataset generated by the plugin Hierarchical K clustering of Means (HK Means) in Icy (Fig. 3A–C). These acted as the reference dataset to launch the tracking process with the different software programs and so allow a direct comparison between them. Tracking by Icy was performed using ‘Spot Tracker’, by Imaris using ‘Surfaces with the option ‘track over time’ and by Phagosight using the ‘keyholetracker.m’ script using default values.
Fig. 3.
Evaluation of the tracking accuracy of Imaris, Icy and Phagosight for ground truth datasets. Tracking efficiency of Imaris (A), Icy (B) and Phagosight (C) was evaluated by analysing cell movement of cells that were segmented by HK Means to ensure consistency. Tracks for cells are shown relative to cell position at the beginning of the time-lapse (A–C). Tracks for the 10 selected cells used for assessing segmentation efficiency were compared against a ground truth generated by manual tracking. Scoring of tracking was performed by counting the number of track breaks, loss or inappropriate links for each program for all three datasets (D). Accuracy of tracking for each program was determined by calculating alpha, beta and Jaccard coefficients of fit (E) relative to the ground truth. Scale bars: 50 µm.
Tracking efficiency by Imaris, Icy and Phagosight was then assessed by a manual scoring of how well they could identify tracks for the 10 cells of interest segmented by HK Means at each time-point. Effectiveness of tracking was scored by whether a segmented cell was linked between time points (+1), if this was inappropriate (i.e. to another cell, −2) or was broken between subsequent time points (−1). Our rationale for allocating a high penalty for inappropriate tracking was that this would introduce more variability to values of cell movement compared to that caused by missing tracks. Comparing the programs for their ability to track cells reveals that on average, Icy was able to generate longer and fewer tracks than Imaris or Phagosight. The performance of Imaris was nearly as good as Icy and was better than Phagosight (Fig. 3D).
As an alternative method for calculating accuracy of cell tracking by each program, we also used the Track Performance Tool. This compares tracking of cells from the same dataset by performing a pairwise comparison to a reference dataset (Chenouard et al., 2014). Using the manually tracked data as the reference, alpha, beta and Jaccard similarity coefficients of fit were obtained for each dataset (Fig. 3E). These coefficients describe localisation and tracking errors when tracks are compared against the ground truth (alpha) and factoring in non-paired (spurious) tracks (beta); the Jaccard similarity coefficient describes the fit of the tracking data only. Using these coefficients, Imaris was observed to be the most effective at tracking cells relative to the ground truth for dataset 1 (Movie 1) and 2, whereas Icy performed better for dataset 3 (Movie 2).
Cell Tracking Profiler – a semi-automated package for segmenting and tracking cells
In order to optimally segment and track cells in a semi-automated manner, we built a Java-based package that allowed tuneable segmentation and fast, accurate tracking together with an ability to manipulate and correct tracks. Outputs from Cell Tracking Profiler (CTP) include parameters of cell shape (volume, surface, sphericity and convexity) and movement (displacement, distance, directionality and instantaneous speed) for each cell over time (Fig. 4). To facilitate post-processing of data we wrote a MATLAB script, CTP2R, to process outputs from CTP and to select regions of interest (ROIs) to analyse. CTP2R calculates mean squared displacement and directional autocorrelation of cells for one or two user-defined regions of interest (ROIs) and writes values for each cell into a spreadsheet that can be opened and plotted in R software.
Fig. 4.
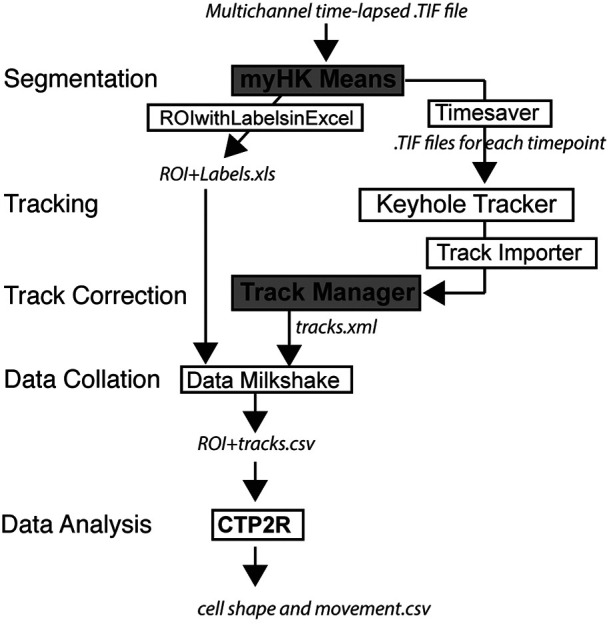
CTP forms a practical workflow for managing cell segmentation and tracking and can feed outputs into a variety of tools for statistical analysis. CTP runs in Icy under ‘plugins’. Users define segmentation parameters for processing of datasets by HK-Means. Outputs, including images and measurements of cells were saved by ‘Time-Saver’ into two files. Objects are tracked by the Keyhole Tracker program in MATLAB Runtime then imported into the Icy plugin Track Manager for editing. The Milkshake plugin then collates information from tracking with values of cell shape (from file ROI+Labels) to produce a tabulated sheet of data. This can be analysed using a MATLAB script, CTP2R, which can normalise data to time-point 0 (t0), generate values of directional autocorrelation, mean squared displacement (MSD) and select a region of interest (ROI), and average parameter values over time. Output files in CTP are shown in italics, modified Icy plugins are shown in a grey box, new scripts and plugins are shown in boxes, and plugins shown in bold indicate user input is required.
Our criteria for building CTP were the need for: (1) speed of segmentation and tracking, (2) ability to modify cell tracks, (3) linking of cell shape and movement parameters, and (4) to be freely available code on a robust platform. We used HK Means from Icy for segmentation, as we observed that the number of objects segmented was most similar to the ground truth in our evaluation of the three test datasets (Fig. 2A). Although Imaris and Icy were effective at cell tracking, the code for Imaris is not available and the Icy tool, Spot Tracker, cannot take the files generated by HK Means as input. Based on our requirements for an imaging pipeline that allowed user input for defining segmentation and for correction of tracks, we therefore decided to build CTP to run from Icy in a Java environment and use the keyhole tracking algorithm from Phagosight for cell tracking. CTP was built by modifying existing modules already present in Icy (HK Means, ROI with Labels and Track Manager), by writing new plugins (ROIwithLabelstoExcel, Timesaver and Milkshake) and modifying the MATLAB program Phagosight to run ‘Keyhole Tracker’ in Java using MATLAB Runtime. It incorporates a version of HK Means (myHKMeans) that was modified to rapidly segment objects cells from three separate channels with user input to define optimal values suitable for the range of cell shape. Outputs are a spreadsheet with measures of segmented object shape corresponding to a label (generated by ROIwithLabelstoExcel) and .TIF files for each time point (generated by Timesaver). Segmented images are imported by Timesaver into a modified version of Phagosight utilising MATLAB Runtime to run within Java by Keyhole Tracker. Outputs are imported into the Icy plugin Track Manager tool and corrected manually. Cell positions and labels from corrected files in Track Manager are generated by a Detection class, and the Milkshake plugin will then combine these together with measures of cell shape into a spreadsheet that contains all values of cell shape and movement for each cell (Fig. 4).
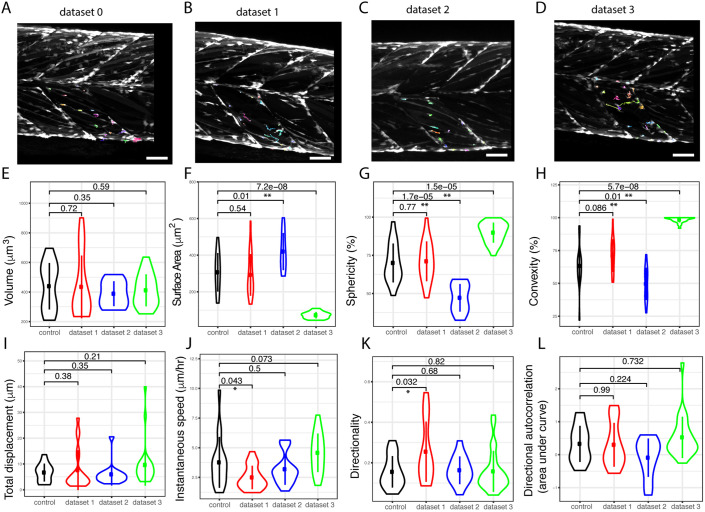
Analysis of muSC behaviour using CTP
We investigated the ability of CTP to segment and track all cells in datasets 1-3 relative to an uninjured control animal (dataset 0). In order to determine whether there were significant changes to cell behaviour and shape in response to injury we selected regions of interest using CTP2R. This allowed us to compare values for cells in injured myotomes relative to those in uninjured myotomes (Fig. 5A–D). There were significant differences in parameters of shape (surface area, sphericity, convexity) between cells in injured myotomes from datasets 2 and 3, but not dataset 1 relative to the uninjured control animal (Fig. 5E–H). However, the differences of cell shape observed between datasets 2 and 3 relative to dataset 0 were opposite, with cells in dataset 2 showing a larger surface area, but lower convexity and sphericity, whereas cells in dataset 3 had a smaller surface area but were more convex and spherical. Dataset 1 showed no significant differences in cell shape between cells in injured myotomes relative to dataset 0. Cells in datasets 2 and 3 showed no difference in their measures of movement (displacement, instantaneous speed, directionality and autocorrelation) relative to dataset 0. In contrast, cells in dataset 1 showed a lower instantaneous speed but more directional movement than cells in dataset 0 (Fig. 5I–L).
Fig. 5.
Tracking and processing of datasets 0, 1, 2 and 3 from injured larvae using CTP. Time-lapse images from injured (datasets 1–3) and an uninjured larva (control, dataset 0) (A–D) were processed by CTP using optimal parameters for segmentation and tracks corrected using Track Manager. Cells in the injured myotome (dataset 0, n=84; dataset 1, n=97; dataset 2, n=90; dataset 3, n=137) were selected using CTP2R (coloured tracks) and measures of shape (cell volume, surface area, sphericity and convexity; E–H) and movement (total displacement, instantaneous speed and directionality; I–K) are plotted as Violin plots (with median and interquartile range indicated). Directional autocorrelation was compared between datasets by measuring the area from beneath the curve of plotted values (L). Significance of difference between datasets for each parameter was calculated using a one-way ANOVA and pairwise comparisons performed using a Tukey posthoc test with significance denoted as P<0.05 (*) or P<0.01 (**). Scale bars: 50 μm.
In order to determine whether CTP was able to extract sufficient detail of cell shape and movement to reveal the effect of a perturbation, we analysed the behaviour of cells exposed to RhoA kinase (ROCK) inhibitors in 5 dpf larvae (Fig. 6A–C). At this stage, muscle progenitor cells expressing the pax7a:egfp transgene undergo extensive rearrangement as the myotome develops (Hollway et al., 2007; Roy et al., 2017). A comparison of cell shape and movement revealed clear differences between animals treated with DMSO vehicle control (dataset 4; Movie 3) relative to those treated with 15 µM Y-27632 (dataset 5; Movie 4) or 10 µM ROCKOUT (dataset 6; Table S2). Measures of cell shape (volume, sphericity, convexity and roundness) were significantly altered by inhibition of ROCK (P<0.01, Fig. 6D; Table S2). Cells with inhibited ROCK activity had a lower volume and surface area compared to DMSO-treated control animals, and were also more spherical and had higher convexity. Several parameters of movement (instantaneous speed, displacement, directionality and directional autocorrelation; P<0.001) were strongly affected by inhibition of ROCK using Y-27632 (P<0.02). Movement was less affected by exposure to ROCKOUT, as only directionality and directional autocorrelation were significantly altered compared to control animals (P<0.05, Fig. 6E; Table S2). This likely reflects the lower efficacy of ROCKOUT for inhibiting ROCK (Yarrow et al., 2005). ROCK inhibition by Y-27632 therefore resulted in a lower migratory speed and reduced directional movement of muSCs.
Fig. 6.
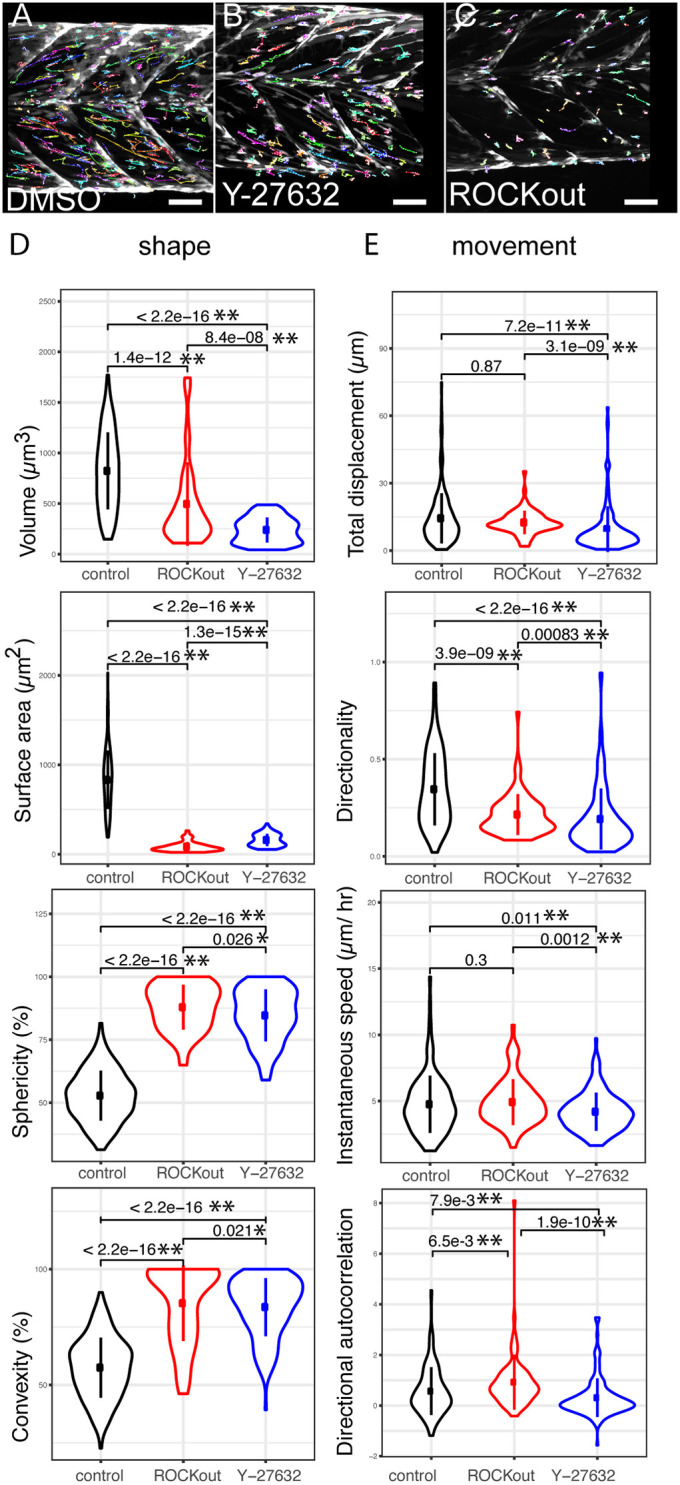
Outputs from CTP from analyses of datasets generated from animals exposed to DMSO and small-molecule inhibitors of ROCK activity. GFP+ cells (dataset 4, n=283; dataset 5, n=39; dataset 6, n=78) were tracked (coloured lines) from time-lapses of pax7a:egfp larvae exposed to DMSO (A, dataset 4), Y-27632 (B, dataset 5) or ROCKOUT (C, dataset 6). Violin plots (with median and interquartile range indicated) of cell shape (D) and movement (E) were generated from CTP outputs after correction of tracking and significance of differences tested by Kruskal–Wallis tests and pairwise comparisons performed with a Dunn post-hoc test. Significant differences between conditions are shown with P<0.05 (*) and P<0.01 (**) indicated. Scale bars: 50 µm.
RhoA signalling is a known regulator of myosin II contractility and hence can control cell migration. We wanted to understand which aspects of cell migration are myosin II dependent in the context of injury, and so compared muSC behaviour between injured and uninjured myotomes in the presence or absence of 10 µM blebbistatin (datasets 7–12, Fig. 7A,B; Movie 5). Larvae at 7 dpf were injured in the ventral 12th myotome, and muSCs in the injured and adjacent uninjured myotome imaged by time-lapsed microscopy. We found that muSC shape (volume, surface area, convexity and sphericity) in uninjured myotomes was mostly unaffected by blebbistatin treatment (datasets 10–12) compared to muSCs in untreated control larvae (datasets 7–9, P>0.05), except for roundness, which was slightly increased (P<0.05, compare blue plots in Fig. 7C; Table S3, comparison of conditions 0–2). Distance travelled and instantaneous speed of muSCs were reduced by blebbistatin in uninjured myotomes compared to untreated control larvae (P<0.05), but directionality and directional autocorrelation were unaffected (Fig. 7D; Table S3, comparison of conditions 0–2). No significant difference in cell shape was identified when comparing segmented cells from injured myotomes relative to uninjured myotomes in untreated control animals (P>0.05, Table S3, comparison of conditions 0–1, datasets 7–9). However, there was an increased speed, displacement and directionality of muSCs in injured myotomes relative to uninjured control myotomes, as expected (P<0.01, Table S3).
Fig. 7.
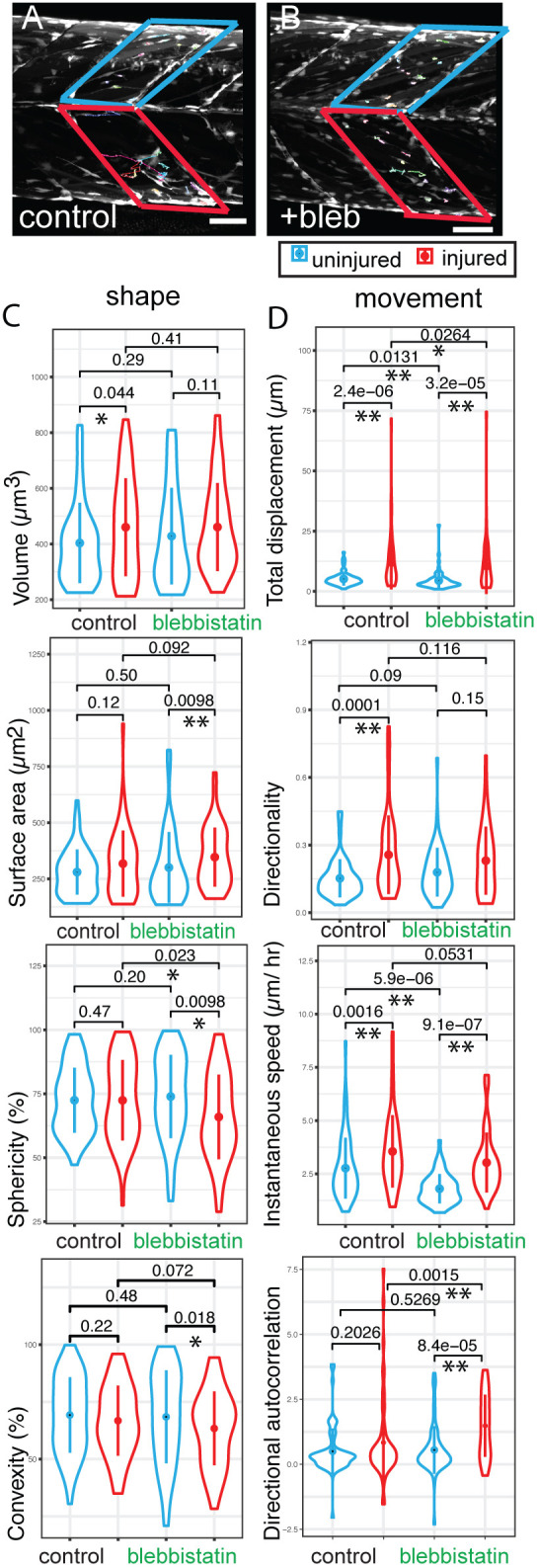
Outputs from CTP from analyses of datasets generated from 7 dpf pax7a:egfp larvae exposed to the myosin II inhibitor blebbistatin. Tracking of GFP+ cells in injured (red box) and uninjured (blue box) myotomes of pax7a:egfp larvae in the absence (A, dataset 7, n=34; dataset 8, n=47; dataset 9, n=48) or presence of 10 µM blebbistatin (B, dataset 10, n=38; dataset 11, n=31; dataset 12, n=36) was performed (n=3 animals for each condition with injured and uninjured myotomes in each animal). Violin plots (with median and interquartile range indicated) of shape (C) and movement (D) were generated from CTP outputs for cells in injured myotomes (red) and adjacent uninjured myotomes (blue) in larvae exposed to blebbistatin or water. Significance of differences in parameters of shape and movement were evaluated using Kruskal–Wallis tests and pairwise comparisons tested by Dunn post-hoc tests. Significant differences between conditions are shown with P<0.05 (*) and P<0.01(**) indicated. Scale bars: 50 µm.
When comparing muSCs responding to injury in the presence (datasets 10–12) or absence of blebbistatin (datasets 7–9), the only parameter of cell shape that was affected was sphericity, which was reduced compared to that in untreated control larvae (P<0.05, Table S3, comparison of conditions 1–3). Displacement of cells in injured myotomes was reduced by blebbistatin treatment (P<0.05, Table S3, comparison of conditions 1–3) although values of directional autocorrelation were higher in the presence of blebbistatin (P<0.01, Table S3).
DISCUSSION
A fundamental limitation for analysing cell behaviour from in vivo or 3D tissue engineering constructs is the software used for extracting measures of cell shape and movement. Although a variety of packages exist for this purpose, they either rely on features for cell segmentation and tracking that may not always be available (such as nuclear or membrane labelling) or require the user to adjust a number of different parameters to find the ‘best segmentation’, which is based on the perception of the user. We required a solution for analysing complex 4D datasets using the expression of GFP in muscle stem cells during regeneration in zebrafish larvae. These cells make numerous contacts with each other, have differing levels of fluorescence and show highly variable changes in shape over time. Our evaluation of several commonly used packages for cell tracking (Icy, Imaris and Phagosight) revealed differing abilities of the segmentation and tracking algorithms utilised by each package to analyse time lapses of muSCs responding to injury. In order to analyse these datasets and allow user defined guidance for cell segmentation and tracking, we have therefore developed a package that runs in the freely available Java based Icy platform. Cell Tracking Profiler (CTP) allows the user to identify the best conditions for accurate segmentation of cells, to track them over time and then manually correct this tracking data.
CTP was designed as a flexible package for analysing cell behaviour from complex 4D datasets in which the ability to discriminate between adjacent objects is limited, such as in transgenic animals expressing GFP or brightfield images. Extracting information about cell shape and tracking them from such datasets is often problematic with existing analysis tools due to the number of variables that can be adjusted to modify outcomes. Our solution was to create a semi-automated solution. CTP is a robust package that offers the user the ability to adjust the segmentation process in a reproducible manner, and then adjust and validate the tracking information manually after the tracking process is complete. A MATLAB script then selects regions of interest to focus on and extract multiple measures of cell shape and movement for subsequent statistical analysis.
Several packages have been described for analysing 3D live-cell imaging data, including plugins that run in ImageJ/Fiji or that are written in a variety of programming languages (Driscoll and Danuser, 2015; Hilsenbeck et al., 2016; Rajasekaran et al., 2016). The accuracy of many automated cell tracking packages can be evaluated by comparing automatically segmented and tracked data against a ground truth. We did this by generating a ground truth for a number of datasets in which we counted the number of cells and segmented a selection of these by hand at defined intervals. We found that there is high variability in how well cells are segmented and their volume when comparing several different packages. This was compounded by differences between datasets, which were acquired with different z-intervals and time, further increasing variability.
The human eye is a powerful tool for determining whether segmentation of cells is accurate. One important requirement of CTP was for it to identify objects from different datasets using the same optimal segmentation parameters and so reduce variability when comparing across conditions. We have therefore designed CTP to permit the user to set the optimal parameters for segmenting cells using Hierarchical K clustering of Means (HK Means). HK Means uses an unsupervised clustering method for identifying objects based on a defined number of objects classes and the minimum and maximum size of the objects, thus allowing considerable flexibility for defining how segmentation occurs (Dufour et al., 2008).
Cell tracking is a challenging problem that operates best when cells are clearly segmented and individual cells show a spatial overlap between adjacent time frames. Rapid movement of cells in random directions, overlap with neighbouring cells or large shape changes can all affect the ability of commonly applied methods for tracking cells. The keyhole method algorithm we have used for tracking muSCs in CTP relates movement of cells between adjacent time frames using a model of constrained movement relative to the previous position (Reyes-Aldasoro et al., 2008). This method does not require overlap between objects in adjacent time points and has been used successfully for tracking rapidly migrating neutrophils and macrophages in zebrafish (Henry et al., 2013; Paredes et al., 2018). By moving outputs to the Track Manager of Icy, CTP allows the user to correct inappropriate tracking by examining tracks by eye. Outputs are then fed to the post-processing tool CTP2R, which extracts multiple measures of cell shape and movement as well as calculating the mean squared displacement and directional autocorrelation of cell movement.
Our major criteria for creating this analysis pipeline was that it worked relatively rapidly and allowed user-driven segmentation and tracking. We acknowledge that there are methods that enable superior segmentation or tracking as recently highlighted in the latest Cell Tracking Challenge (Maška et al., 2014; Ulman et al., 2017). However, comparisons between CTP and such programs are problematic as the ground truth datasets used for the Cell Tracking Challenge did not have cells with such complex morphology or behaviour as were present in our example datasets. Additionally, CTP represents a flexible, user-driven package, unlike many other available tools, and can easily be adjusted as needed to run within Icy, thus providing a relatively easy to use tool for biologists. Recent developments in machine learning tools for cell segmentation and tracking have greatly expanded the opportunity for biologists to analyse 4D imaging data (Kan, 2017). Examples include Ilastik, the trainable Weka running in Fiji and variations of U-Net (Arganda-Carreras et al., 2017; Berg et al., 2019; Hesamian et al., 2019). Training of machine learning algorithms or networks by flagging features for classification is a powerful method for improving the accuracy of segmentation and tracking. Caveats with such methods include a dependence on the datasets or features used for training, which may not capture heterogeneity or be non-representative for the structures of interest. This can bias the trained algorithm or network, and so result in poor or inappropriate segmentation for datasets that show high variability in cell shape relative to the training datasets (Hesamian et al., 2019; Kan, 2017). One advantage of using intensity-based methods and defining a range of cell sizes for image segmentation, as employed by HK Means, is that the user can define these parameters without needing to select the most appropriate datasets for training. It also is much less computationally intensive to run HK Means-based segmentation compared to machine learning-based methods, making it attractive for users who do not have access to powerful image analysis workstations.
Our purpose for creating CTP was to allow us to characterise the dynamics of resident stem cells in response to muscle injury in vivo. Our analyses of muSC behaviour revealed that alpha values for Mean Squared Displacement were less than one. This has been described as indicative of a sub-diffusive movement found in cells within confined environments (Luzhansky et al., 2018). Additionally, we noted that a model of anisotropic persistent random walk (APRW) was better for explaining muSCs movement than a persistent random walk model (PRW) commonly used for explaining cell movement in vitro (Gorelik and Gautreau, 2014). This implies that the mode of muSC movement is constrained during regeneration. Muscle is a densely packed tissue, composed of myofibres, connective cells, blood vessels and axons. In the myotome of zebrafish larvae, there is a dense packing of myofibres in a stereotyped orientation that would restrict movement of the muSCs as they respond to injury. MuSCs migrate along and between myofibres in mammals (Baghdadi et al., 2018; Webster et al., 2016). We and others have noted that migration of muSCs in zebrafish occurs in a similarly restricted manner, with migratory cells using myofibres as a guide (Gurevich et al., 2016; Knappe et al., 2015; Pipalia et al., 2016; Seger et al., 2011). Although this has been described from observations, our results are the first quantitative measures describing this behaviour. Our finding that the movement of muSCs is constrained by the architecture of the muscle tissue is relevant for considering how changes to this architecture can affect cell migration, such as in fibrosis or changes to the extracellular matrix.
RhoA signalling is an important regulator of cell shape and migration for a wide variety of cell types. We found that inhibition of the RhoA kinase ROCK by use of the potent inhibitor Y-27632 results in muSCs assuming a more rounded shape, and showing a reduced directionality and speed. Although significant, ROCK inhibitor-induced changes to cell speed were not extreme, which suggests the speed of migrating muSC was not strongly affected due to loss of RhoA signalling. Inhibition of actinomyosin function is associated with cell rounding due a loss of resistance to osmotic pressure (Stewart et al., 2011). As RhoA signalling is required for myosin II phosphorylation and actin bundling, the inhibition of ROCK could therefore have caused impaired cortical myosin contraction and cell protrusivity in muSCs. ROCK phosphorylates myosin II, and myosin II-dependent functions have been described to include regulation of actin flow and stabilisation of focal adhesions, which is critical for mesenchymal cell migration (Aguilar-Cuenca et al., 2014). We noted that migration speed and cell displacement is impaired by blebbistatin treatment, but could not see obvious changes to shape except for a small reduction in sphericity. There are several potential reasons why we did not see changes to cell shape upon blebbistatin treatment but did in in response to ROCK inhibitors. This could reflect differential requirements for ROCK in controlling cell behaviour by modifying localised actinomyosin contractility, regulating formin during actin bundling, or controlling myosin and actin flow (Königs et al., 2014; Ridley, 2015; Ruprecht et al., 2015). Alternatively, it could be due to myosin II-independent cell movements based on actin flow (Lomakin et al., 2015). Many cells are able to switch their mode of cell migration to myosin II-independent modes in both 3D artificial and in vivo environments (Liu et al., 2015; Ruprecht et al., 2015), and potentially muSCs show a similar plasticity. Finally, it is possible that we were not able to discriminate changes to cell movement as the effects of blebbistatin treatment were only transient. Blebbistatin can form aggregates when diluted in water and is photolabile, so it is possible that over the course of the time-lapse, inhibition of myosin II was variable, so affecting our ability to discriminate changes to cell shape and movement (Várkuti et al., 2016).
The above caveat highlights an important consideration when analysing descriptions of cell shape and movement using standard statistical methods, such as ANOVA or Kruskal–Wallis tests. As each measurement of a cell is related to other measurements taken of that same cell over time, any statistical model attempting to discriminate differences due to an independent variable, such as a drug, should consider the time dependence of changes to a parameter. Furthermore, many parameters of shape and movement will be related to each other, implying that any change to one parameter should involve change to others. Ideally this relationship would be included in a statistical model that can then discriminate between changes to multiple parameters of cell shape and movement in a time-dependent manner.
In summary, we show that by creating a user-defined semi-automated process for dataset segmentation and cell tracking, we can obtain biological signatures for cell responses to specific molecular manipulations. CTP runs in Icy, an open source platform, providing great scope for further development and integration with other analysis suites and provides an easy to use tool for biologists aiming to quantify cell behaviour from 4D datasets.
MATERIALS AND METHODS
Zebrafish husbandry and imaging
Transgenic 5 or 7 days post fertilisation (dpf) Tg[pax7a:egfp]; pfeffer zebrafish (Danio rerio) larvae were injured as previously described (Knappe et al., 2015) and/or treated with small-molecule inhibitors. Images were acquired at 1 µm (datasets 0–2, 4, 6 and 7–12) or 5 µm (datasets 3 and 5) intervals every 20 min using a Nikon C1 confocal microscope equipped with an Argon-ion 488 nm laser and a 20× water-dipping objective (NA 0.7) under a constant temperature of 28.5°C.
All animal work was performed according to local and Home Office regulations under project license PBC5F9B13 with reference to the ARRIVE guidelines for use of animals in scientific experiments (Kilkenny et al., 2010).
Image analysis
Time-lapse image sequences were processed by ImageJ to remove movement artefacts using the plugin ‘Correct for 3D drift’ (http://fiji.sc/Correct_3D_drift) and, if necessary, cropped to focus only on relevant regions. No other manipulations were performed to the images prior to analysis. Datasets that were considered representative were selected for analysis and were obtained from larvae showing no obvious signs of ill health. Parameters used for Imaris 8.0 (Bitplane AG), Icy 1.7.3 (de Chaumont et al., 2012) and Phagosight (Henry et al., 2013) were defined by comparing to the ground truth. The ground truth dataset was obtained by: (1) cells were counted by Cell Counter (Image J/Fiji) at three time points for each time-lapse; (2) ROIs were defined for 10 cells per dataset by drawing around the cell in each slice of the z-stack, then merging to generate a 3D object; (3) the selected 10 cells were tracked over time using MTrackJ (Image J/Fiji). Optimal values for segmentation were identified by: (1) counting the number of objects identified, (2) scoring the identified objects against the manually defined subset of 10 cells at three different points in the time-lapse, and (3) evaluating the size of the segmented objects relative to the manually annotated ground truth. Parameter values that best identified cells from the ground truth as discrete objects were selected as optimal.
Cell movement was scored by comparing automatically generated values relative to the manual ground truth. A ground truth was established by tracking the selected 10 cells for each dataset using MTrackJ (Image J/Fiji). Cell tracking was performed using Surfaces in Imaris, Spot Tracker in Icy and by employing a keyhole tracking algorithm in Phagosight. Measures of tracking used for calculating efficiency were: (1) the number of tracks identified, (2) the length of continuous tracks, and (3) the number of times a track was interrupted, lost or falsely connected. Optimal parameter values were defined as those that: (1) resulted in the longest contiguous tracks for cells defined in the ground truth, and (2) resulted in the fewest number of false connections.
Determination of the segmentation and tracking efficiency of Imaris, Icy and Phagosight were performed by calculating alpha, beta and Jaccard co-efficients using the Track Analysis tool in Track Manager (Chenouard et al., 2014).
Software development
Cell Tracking Profiler (CTP) was generated using the Protocols tool in Icy. It is a semi-automated tool allowing segmentation and tracking of cells from 4D datasets. Compiled versions of CTP for Mac OS X and Windows XP are available from GitHub that allow easy installation within Icy (https://github.com/KnightLabKCL/CellTrackingProfiler). Icy-specific tools are also available from the plugins menu in Icy through option ‘caroff’.
CTP incorporates a modified version of the Icy segmentation plugin HK-Means Keyhole Tracker [the tracking algorithm from Phagosight was modified to run in Java through a MATLAB Runtime (Henry et al., 2013)] and scripts for extracting information about cell shape, position and movement.
Manual correction of tracked cells is then achieved using the Icy Track Manager plugin before the corrected cell tracks are linked to information about cell shape by the Milkshake tool. The output is a tabulated file that can be opened by spreadsheet managers, such as Excel (Microsoft Corporation) or imported into other image analysis programs. This file describes several parameters of cell shape and movement, including volume, surface area, convexity, sphericity, x, y and z position, distance, displacement, speed for each segmented cell in the dataset for each channel.
Specifics of how CTP was generated
Segmentation
HK-means was modified to access a temporal sequence in Protocols, which is used during the building of the Cell Tracking Profiler protocol. Functions ‘ROI Statistics’, ‘Workbook to File’ and ‘Select file’ were implemented from within Protocols to generate an Excel file, ‘ROI.xls’, in which ROI measurements are generated that correspond to each cell. In the same manner, the labelled sequence from HK-Means is used to extract the labels for each cell into file ‘labels.xls’. To merge ROI.xls and labels.xls an Icy plugin was written in Java called ‘ROI with Labels to Excel’. This plugin looks for the coordinates of cells to obtain the label of each cell, represented by a ROI and generates an output file called ‘ROI+labels.xls’, containing all the information about the ROIs.
To save the labelled image generated by HK-Means in a format readable by the reading functions of Phagosight, the function Time Saver was written. This is a JavaScript file that generates a folder of TIFs with each TIF representing a single labelled time point. In each saved image, the background is identified with the value of 0 and each segmented cell corresponds to a unique label value that also acts as an identifier.
Tracking
The MATLAB program Phagosight (Henry et al., 2013) was modified to accept the input from Time Saver as follows:
The initialisation process was adapted to read the TIF files of the folder obtained from Time Saver and to generate all the necessary folders and files required by Phagosight to perform the tracking. This initialisation process was implemented in a MATLAB function called createFolders.
The segmentation process using the Otsu algorithm was removed so the modified version of Phagosight will perform the tracking directly on the labelled TIFs imported by Time Saver and adapted to MATLAB format using createFolders.
A new function, trackingProcess, was written to perform the tracking process of Phagosight. This was modified from the previously published Phagosight code by removing the initial segmentation step.
The function exportTracksToICY was written to save the tracks in a format readable by the Icy plugin Track Manager. This function has been modified to read and write the cell labels from the segmentation process with its corresponding track in an XML file.
To automate the tracking process a function called keyholeTracker was implemented to launch the creation of the folders, the tracking process, and the generation of the output file simultaneously.
To be able to run this workflow through Icy, a compiled version of the keyholeTracker was generated using the MATLAB Compiler tool. The compiled keyholeTracker was implemented as an Icy plugin, called Keyhole Tracker, thus allowing the launch of the modified version of Phagosight from within Icy (see Fig. 4)
A Detection class was written to import information from the Keyhole Tracker plugin output, in particular, the file of tracks including the coordinates of each cell over time and its label into the Icy plugin Track Manager. This makes it possible to read and save the values for distinct labels (representing cells) from Track Manager after modification/correction of the tracks by the user.
To obtain all available measurements of cell shape and movement from the analysed dataset an Icy plugin called Milkshake was created. This is a Java plugin that generates an Excel file (Microsoft) with individual worksheets describing parameters of cell shape, intensity, displacement and speed and a folder of .csv files with each file representing an individual cell. To generate these files, Milkshake requires two input files: the files of measurements of each ROI with associated labels generated by CellTrackingProfiler (Channel0_ROI+labels.xls), and the files of tracks generated at the end of the tracking process or saved by users after correction of tracks (Channel0_tracks.xml file). Milkshake can be launched manually, or using the DataMilkshake protocol created to run as an Icy plugin in Protocols.
Post-analysis processing
CTP2R is a post-processing application for selection of data to reformat for statistical analysis. CTP2R was written in MATLAB and compiled as a standalone executable using the MATLAB compiler. The script allows for selection of specific ROIs and additionally calculates directional autocorrelation and the mean squared displacement for each cell (equations from Gorelik and Gautreau, 2014). This post-processed data is then reformatted into a long, single sheet format for easy plotting using the ggplot2 library in R software. Normalisation of data was performed by comparing each value to the value for that parameter at t=0 for the time-lapse in question.
To generate a single value to represent the directional autocorrelation for each cell, the area under the curve was calculated within CTP2R using the trapezoidal rule. Tests of fit to models of isotropic or anisotropic persistent random walk (PRW or APRW, respectively) were performed using a MATLAB script as previously described in Wu et al. (2015).
Statistical analysis
Cell volumes identified by Phagosight, Icy and Imaris were compared against the ground truth for each of the selected 10 cells at each time-point. A multi-level mixed effects linear regression analysis was used to predict the average cell volume. The 95% confidence intervals give an idea of the certainty around the calculated averages of cell volume and their overlap can be used to determine significance of difference between conditions.
Analysis of cell shape and movement produced by CTP2R were performed using R software and plotted using the ggplot2 library. Distribution of data was evaluated for normality by Shapiro–Wilks tests. Statistical tests for differences of cell shape and movement were performed by one-way ANOVA, Wilcoxon-Mann–Whitney tests or Kruskal–Wallis tests as appropriate, dependent on distribution of the data.
Supplementary Material
Acknowledgments
We thank Pia Cumine and Stefanie Knappe for help with generation of imaging data, Alexis Gautreau for advice on DiPer, Brian Stramer for helpful discussions and Ron de Bruin for assistance with Excel macros.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: C.C.R.-A., A.D., R.K.; Methodology: C.M., L.C., J.A.S.-L., C.C.R.-A., A.V., F.d.C., A.D., R.K.; Software: C.M., L.C., J.A.S.-L., C.C.R.-A., F.d.C., A.D., S.D.; Validation: C.M.; Formal analysis: C.M., L.C., A.V., F.W.; Investigation: C.M., L.C., A.V., R.K.; Resources: S.D., J.-C.O.-M., R.K.; Data curation: C.M., S.D.; Writing - original draft: C.M., L.C., C.C.R.-A., J.-C.O.-M., R.K.; Writing - review & editing: R.K.; Supervision: J.-C.O.-M., R.K.; Project administration: C.C.R.-A., R.K.; Funding acquisition: C.C.R.-A., J.-C.O.-M., R.K.
Funding
L.C. was funded through the Erasmus+ Training scheme. R.K. was funded by grants from the Biotechnology and Biological Sciences Research Council (BBSRC; BB/P002390/1) and the Wellcome Trust (202852/Z/16/Z, 108111/Z/15/Z). J.A.S.-L. is funded by a City University doctoral scholarship. C.C.R.-A. is partly funded by grants from the Leverhulme Trust (RPG-2017-054) and the Australian Research Council (DP170102235). This work was partially supported by grants from the Agence Nationale de la Recherche Labex IBEID (ANR-10-LABX-62-IBEID), France-BioImaging infrastructure (ANR-10-INBS-04) and the program PIA INCEPTION (ANR-16-CONV-0005). Deposited in PMC for release after 6 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.241422.supplemental
References
- Aguilar-Cuenca R., Juanes-García A. and Vicente-Manzanares M. (2014). Myosin II in mechanotransduction: master and commander of cell migration, morphogenesis, and cancer. Cell. Mol. Life Sci. 71, 479-492. 10.1007/s00018-013-1439-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arganda-Carreras I., Kaynig V., Rueden C., Eliceiri K. W., Schindelin J., Cardona A. and Sebastian Seung H. (2017). Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33, 2424-2426. 10.1093/bioinformatics/btx180 [DOI] [PubMed] [Google Scholar]
- Baghdadi M. B., Firmino J., Soni K., Evano B., Di Girolamo D., Mourikis P., Castel D. and Tajbakhsh S. (2018). Notch-induced miR-708 antagonizes satellite cell migration and maintains quiescence. Cell Stem Cell 23, 859-868.e5. 10.1016/j.stem.2018.09.017 [DOI] [PubMed] [Google Scholar]
- Berg S., Kutra D., Kroeger T., Straehle C. N., Kausler B. X., Haubold C., Schiegg M., Ales J., Beier T., Rudy M. et al. (2019). ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226-1232. 10.1038/s41592-019-0582-9 [DOI] [PubMed] [Google Scholar]
- Chenouard N., Smal I., de Chaumont F., Maška M., Sbalzarini I. F., Gong Y., Cardinale J., Carthel C., Coraluppi S., Winter M. et al. (2014). Objective comparison of particle tracking methods. Nat. Methods 11, 281-289. 10.1038/nmeth.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T. J. (2007). ImageJ for microscopy. BioTechniques 43, S25-S30. 10.2144/000112517 [DOI] [PubMed] [Google Scholar]
- de Chaumont F., Dallongeville S., Chenouard N., Hervé N., Pop S., Provoost T., Meas-Yedid V., Pankajakshan P., Lecomte T., Le Montagner Y. et al. (2012). Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods 9, 690-696. 10.1038/nmeth.2075 [DOI] [PubMed] [Google Scholar]
- Driscoll M. K. and Danuser G. (2015). Quantifying modes of 3D cell migration. Trends Cell Biol. 25, 749-759. 10.1016/j.tcb.2015.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour A., Meas-Yedid V., Grassart A. and Olivo-Marin J.-C. (2008). Automated quantification of cell endocytosis using active contours and wavelets. 19th International Conference on Pattern Recognition, Tampa, FL, 2008, pp. 1-4. 10.1109/ICPR.2008.4761748 [DOI]
- Gorelik R. and Gautreau A. (2014). Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc. 9, 1931-1943. 10.1038/nprot.2014.131 [DOI] [PubMed] [Google Scholar]
- Gurevich D. B., Nguyen P. D., Siegel A. L., Ehrlich O. V., Sonntag C., Phan J. M. N., Berger S., Ratnayake D., Hersey L., Berger J. et al. (2016). Asymmetric division of clonal muscle stem cells coordinates muscle regeneration in vivo. Science 353, aad9969 10.1126/science.aad9969 [DOI] [PubMed] [Google Scholar]
- Henry K. M., Pase L., Ramos-Lopez C. F., Lieschke G. J., Renshaw S. A. and Reyes-Aldasoro C. C. (2013). PhagoSight: an open-source MATLAB® package for the analysis of fluorescent neutrophil and macrophage migration in a zebrafish model. PLoS ONE 8, e72636 10.1371/journal.pone.0072636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesamian M. H., Jia W., He X. and Kennedy P. (2019). Deep learning techniques for medical image segmentation: achievements and challenges. J. Digit Imaging 32, 582-596. 10.1007/s10278-019-00227-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilsenbeck O., Schwarzfischer M., Skylaki S., Schauberger B., Hoppe P. S., Loeffler D., Kokkaliaris K. D., Hastreiter S., Skylaki E., Filipczyk A. et al. (2016). Software tools for single-cell tracking and quantification of cellular and molecular properties. Nat. Biotechnol. 34, 703-706. 10.1038/nbt.3626 [DOI] [PubMed] [Google Scholar]
- Hollway G. E., Bryson-Richardson R. J., Berger S., Cole N. J., Hall T. E. and Currie P. D. (2007). Whole-somite rotation generates muscle progenitor cell compartments in the developing zebrafish embryo. Dev. Cell 12, 207-219. 10.1016/j.devcel.2007.01.001 [DOI] [PubMed] [Google Scholar]
- Kan A. (2017). Machine learning applications in cell image analysis. Immunol. Cell Biol. 95, 525-530. 10.1038/icb.2017.16 [DOI] [PubMed] [Google Scholar]
- Kilkenny C., Browne W. J., Cuthill I. C., Emerson M. and Altman D. G. (2010). Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 8, e1000412 10.1371/journal.pbio.1000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe S., Zammit P. S. and Knight R. D. (2015). A population of Pax7-expressing muscle progenitor cells show differential responses to muscle injury dependent on developmental stage and injury extent. Front. Aging Neurosci. 7, 161 10.3389/fnagi.2015.00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Königs V., Jennings R., Vogl T., Horsthemke M., Bachg A. C., Xu Y., Grobe K., Brakebusch C., Schwab A., Bähler M. et al. (2014). Mouse macrophages completely lacking Rho subfamily GTPases (RhoA, RhoB, and RhoC) have severe lamellipodial retraction defects, but robust chemotactic navigation and altered motility. J. Biol. Chem. 289, 30772-30784. 10.1074/jbc.M114.563270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-J., Le Berre M., Lautenschlaeger F., Maiuri P., Callan-Jones A., Heuzé M., Takaki T., Voituriez R. and Piel M. (2015). Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell 160, 659-672. 10.1016/j.cell.2015.01.007 [DOI] [PubMed] [Google Scholar]
- Lomakin A. J., Lee K.-C., Han S. J., Bui D. A., Davidson M., Mogilner A. and Danuser G. (2015). Competition for actin between two distinct F-actin networks defines a bistable switch for cell polarization. Nat. Cell Biol. 17, 1435-1445. 10.1038/ncb3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzhansky I. D., Schwartz A. D., Cohen J. D., MacMunn J. P., Barney L. E., Jansen L. E. and Peyton S. R. (2018). Anomalously diffusing and persistently migrating cells in 2D and 3D culture environments. APL Bioeng 2, 026112 10.1063/1.5019196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maška M., Ulman V., Svoboda D., Matula P., Matula P., Ederra C., Urbiola A., España T., Venkatesan S., Balak D. M. W. et al. (2014). A benchmark for comparison of cell tracking algorithms. Bioinformatics 30, 1609-1617. 10.1093/bioinformatics/btu080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nketia T. A., Sailem H., Rohde G., Machiraju R. and Rittscher J. (2017). Analysis of live cell images: methods, tools and opportunities. Methods 115, 65-79. 10.1016/j.ymeth.2017.02.007 [DOI] [PubMed] [Google Scholar]
- Paredes A. D., Benavidez D., Cheng J., Mangos S., Donoghue M. and Bartholomew A. (2018). An automated quantitative image analysis pipeline of in vivo oxidative stress and macrophage kinetics. J. Biol. Methods 5, e101 10.14440/jbm.2018.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipalia T. G., Koth J., Roy S. D., Hammond C. L., Kawakami K. and Hughes S. M. (2016). Cellular dynamics of regeneration reveals role of two distinct Pax7 stem cell populations in larval zebrafish muscle repair. Dis. Model. Mech. 9, 671-684. 10.1242/dmm.022251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekaran B., Uriu K., Valentin G., Tinevez J.-Y. and Oates A. C. (2016). Object segmentation and ground truth in 3D embryonic imaging. PLoS ONE 11, e0150853 10.1371/journal.pone.0150853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Aldasoro C. C., Akerman S. and Tozer G. M. (2008). Measuring the velocity of fluorescently labelled red blood cells with a keyhole tracking algorithm. J. Microsc. 229, 162-173. 10.1111/j.1365-2818.2007.01877.x [DOI] [PubMed] [Google Scholar]
- Ridley A. J. (2015). Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol. 36, 103-112. 10.1016/j.ceb.2015.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. D., Williams V. C., Pipalia T. G., Li K., Hammond C. L., Knappe S., Knight R. D. and Hughes S. M. (2017). Myotome adaptability confers developmental robustness to somitic myogenesis in response to fibre number alteration. Dev. Biol. 431, 321-335. 10.1016/j.ydbio.2017.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht V., Wieser S., Callan-Jones A., Smutny M., Morita H., Sako K., Barone V., Ritsch-Marte M., Sixt M., Voituriez R. et al. (2015). Cortical contractility triggers a stochastic switch to fast amoeboid cell motility. Cell 160, 673-685. 10.1016/j.cell.2015.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger C., Hargrave M., Wang X., Chai R. J., Elworthy S. and Ingham P. W. (2011). Analysis of Pax7 expressing myogenic cells in zebrafish muscle development, injury, and models of disease. Dev. Dyn. 240, 2440-2451. 10.1002/dvdy.22745 [DOI] [PubMed] [Google Scholar]
- Stewart M. P., Helenius J., Toyoda Y., Ramanathan S. P., Muller D. J. and Hyman A. A. (2011). Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature 469, 226-230. 10.1038/nature09642 [DOI] [PubMed] [Google Scholar]
- Ulman V., Maška M., Magnusson K. E. G., Ronneberger O., Haubold C., Harder N., Matula P., Matula P., Svoboda D., Radojevic M. et al. (2017). An objective comparison of cell-tracking algorithms. Nat. Methods 14, 1141-1152. 10.1038/nmeth.4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várkuti B. H., Képiró M., Horváth I. A., Végner L., Ráti S., Zsigmond A., Hegyi G., Lenkei Z., Varga M. and Málnási-Csizmadia A. (2016). A highly soluble, non-phototoxic, non-fluorescent blebbistatin derivative. Sci. Rep. 6, 26141 10.1038/srep26141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M. T., Manor U., Lippincott-Schwartz J. and Fan C.-M. (2016). Intravital imaging reveals ghost fibers as architectural units guiding myogenic progenitors during regeneration. Cell Stem Cell 18, 243-252. 10.1016/j.stem.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P.-H., Giri A. and Wirtz D. (2015). Statistical analysis of cell migration in 3D using the anisotropic persistent random walk model. Nat. Protoc. 10, 517-527. 10.1038/nprot.2015.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarrow J. C., Totsukawa G., Charras G. T. and Mitchison T. J. (2005). Screening for cell migration inhibitors via automated microscopy reveals a Rho-kinase inhibitor. Chem. Biol. 12, 385-395. 10.1016/j.chembiol.2005.01.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.