ABSTRACT
In response to the invasion of microorganisms, plants actively balance their resources for growth and defence, thus ensuring their survival. The regulatory mechanisms underlying plant immunity and growth operate through complex networks, in which the brassinosteroid phytohormone is one of the central players. In the past decades, a growing number of studies have revealed a multi-layered crosstalk between brassinosteroid-mediated growth and plant immunity. In this Review, by means of the tango metaphor, we immerse ourselves into the intimate relationship between brassinosteroid and plant immune signalling pathways that is tailored by the lifestyle of the pathogen and modulated by other phytohormones. The plasma membrane is the unique stage where brassinosteroid and immune signals are dynamically integrated and where compartmentalization into nanodomains that host distinct protein consortia is crucial for the dance. Shared downstream signalling components and transcription factors relay the tango play to the nucleus to activate the plant defence response and other phytohormonal signalling pathways for the finale. Understanding how brassinosteroid and immune signalling pathways are integrated in plants will help develop strategies to minimize the growth–defence trade-off, a key challenge for crop improvement.
KEY WORDS: Plant innate immunity, Brassinosteroid, Hormone, Growth–defence trade-off, Phytohormone crosstalk, Biotic stress, Nanodomains
Summary: In this Review, we discuss molecular interactions between the brassinosteroid and immune signalling pathways, which contribute to the growth–defence trade-off in plants.
Introduction
As sessile organisms, plants are constantly exposed to numerous environmental stresses, including various pathogen attacks, throughout their life cycles. Therefore, accurate communication networks are required to monitor the surrounding environment and trigger prompt responses, thus ensuring the survival of the organism. Plants have evolved sophisticated defence strategies against potential pathogens that entail physical and chemical preformed barriers, such as waxy cuticles, lignified cell walls, trichomes, antimicrobial enzymes and secondary metabolites (Thordal-Christensen, 2003; Malinovsky et al., 2014). In addition, their refined innate immune system can detect invaders and generate manifold layers of defence responses upon an attack recognition (Fig. 1) (Jones and Dangl, 2006). The specific defence responses depend on the pathogen type. Based on their feeding preferences, pathogens are subdivided into three classes: (1) biotrophs that derive nutrients from living host cells, (2) necrotrophs that consume nutrients from dead or dying cells, and (3) hemibiotrophs that are at first biotrophic and become necrotrophic at a later stage (Glazebrook, 2005; Précigout et al., 2020).
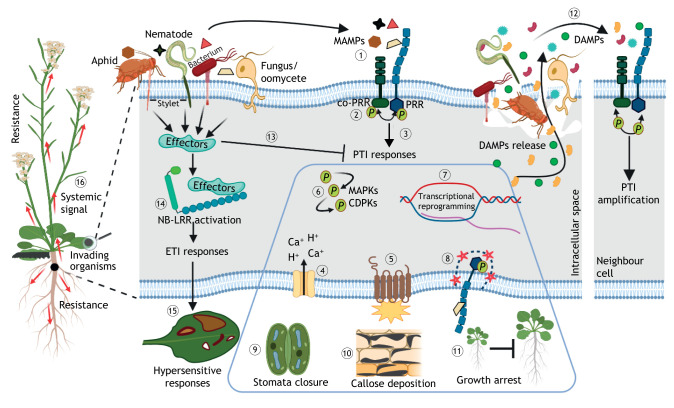
Fig. 1.
Plant defence mechanisms. Pathogens of different lifestyles, such as aphids, nematodes, bacteria and fungi, release microbe-associated molecular patterns (MAMPs) into the extracellular space of the plants they colonize. (1) MAMPs are recognized by the extracellular domain of pattern recognition receptors (PRRs). After ligand perception, (2) most of the known PRRs heterodimerize with co-receptors, followed by phosphorylation and activation of the intracellular kinases. (3) Then pattern-triggered immunity (PTI) responses (shown within the blue outline) are activated, including a set of very early responses (≤15 min) such as (4) rapid changes in the ion fluxes (H+ and Ca2+), (5) production of reactive oxygen species (ROS), (6) phosphorylation of MAPKs and Ca2+-dependent protein kinases (CDPKs). Subsequently (≤60 min), (7) transcription is reprogrammed in-depth to activate the expression of immune genes, and (8) activated PRRs are removed from the plasma membrane via endocytosis, allowing desensitization of the signal. Finally (hours to days), (9) stomatal closure, (10) callose deposition and (11) growth inhibition occur when the presence of the elicitor is maintained. (12) Host-derived elicitor molecules, designated damage-associated molecular patterns (DAMPs), are released upon pathogen perception or pathogen-induced cell damage, which are then recognized by PRRs of neighbouring cells for PTI amplification. (13) Pathogens deliver a suite of effector proteins into host cells through specialized structures, such as the aphid or the nematode stylet, the type III secretion system (T3SS) in bacteria and the haustorium in fungi, that target specific subcellular locations, where they can interfere with PTI and facilitate virulence. However, (14) intracellular nucleotide-binding domain leucine-rich repeat proteins (NB-LRRs) can recognize virulence effectors or effector-triggered perturbations of host structures, activating the effector-triggered immunity (ETI), (15) often accompanied by a hypersensitive response. (16) Long-lasting systemic signals are released from the infection sites. P circled in green indicates phosphorylation events. This figure was created with BioRender.
The plant innate immunity is activated once the preformed barriers are breached, and the invading microorganisms are detected by a two-tiered perception system (Fig. 1). The first tier or the pattern-triggered immunity (PTI) contributes mostly to the host basal defence against a broad range of pathogens. Microbe-associated molecular patterns (MAMPs) are recognized by plasma membrane (PM)-localized pattern recognition receptors (PRRs), which interact immediately with co-regulatory receptor kinases, followed by phosphorylation of intracellular kinase domains in both receptors and co-receptors (Schwessinger et al., 2011; Yu et al., 2017). Subsequently, a sequential set of protective responses are initiated, comprising early responses (seconds to minutes) that involve activation of mitogen-activated protein kinases (MAPKs) and Ca2+-dependent protein kinases (CDPKs), removal of activated receptors from the PM by endocytosis, transcriptional reprogramming, ion fluxes (H+ and Ca2+) and production of reactive oxygen species (ROS). These are followed by long-term responses (hours to days) that include stomatal closure, callose deposition and growth arrest (Yu et al., 2017) (Fig. 1). In addition, upon pathogen infection and cell injury, damage-associated molecular patterns (DAMPs) are released into the extracellular space, where they can then diffuse to neighbouring cells and amplify the PTI response (Yamaguchi and Huffaker, 2011; Hou et al., 2014; Yu et al., 2017; Ortiz-Morea and Reyes-Bermudez, 2019) (Fig. 1). Plants also use PRRs to identify nematodes, herbivorous insects and parasitic plants (Albert et al., 2020). All known PRRs are located in the PM and are either receptor kinases (RKs) or receptor-like proteins (RLPs). RKs consist of a ligand-binding ectodomain, a single-pass transmembrane domain and an intracellular kinase domain, whereas RLPs share the same overall structure but lack an intracellular kinase domain (Yu et al., 2017; He et al., 2018).
In general, successful pathogens have evolved diverse virulence effector molecules that are secreted into the host cells to suppress PTI and/or interfere with the host physiology for an effective colonization of their hosts (Fig. 1). These effectors are translocated into different compartments (e.g. PM, cytoplasm or nucleus) of the host cells, through specialized structures, such as the type III secretion system (T3SS) in bacteria, the haustorium in fungi, and the nematode or aphid stylet (Quentin et al., 2013; Choi et al., 2017; Rodriguez et al., 2017; Wang et al., 2019b). As counter defence, plants have developed a second tier of immunity, effector-triggered immunity (ETI). ETI is activated upon recognition of specific virulence effectors or of an effector-triggered perturbation of the host structures by intracellular immune receptors (Jones and Dangl, 2006). These receptors are mostly encoded by nucleotide-binding domain leucine-rich repeat proteins (NB-LRRs), also designated disease resistance (R) or NLR proteins (Jones and Dangl, 2006; Wang et al., 2019a) (Fig. 1). The responses activated by ETI are usually stronger and last longer than those by PTI, and are often accompanied by a localized programmed cell death, known as the hypersensitive response (HR) (Jones and Dangl, 2006). This strategy helps to contain any further spread of biotrophs and hemibiotrophs at their early stages.
Following the initial detection of the invading organisms and the activation of local responses, plants are capable of triggering long-lasting systemic signals through phytohormones to induce resistance in distal uninfected tissues, thus protecting the entire plant against a broad range of pathogens (Spoel and Dong, 2012) (Fig. 1). There are two branches of systemic immunity regulated by phytohormonal networks. On the one hand, systemic-acquired resistance (SAR), which depends mainly on salicylic acid (SA), mediates resistance against biotrophic and hemibiotrophic pathogens; on the other hand, induced systemic resistance (ISR), which is modulated synergistically by jasmonic acid (JA) and ethylene (ET), provides resistance against necrotrophic pathogens (Ton et al., 2002; Boyajyan et al., 2014). The SA and JA pathways mostly exhibit antagonistic relationships, where an elevated resistance against biotrophs is usually correlated with an increased susceptibility against necrotrophs, and vice versa (Robert-Seilaniantz et al., 2011). However, this is not always the case because synergistic interactions between SA and JA have been reported (Tamaoki et al., 2013; Liu et al., 2016). Although relatively little is known about the recognition mechanisms that trigger herbivory responses, it is believed that similar to pathogen recognition, plant cells are able to perceive danger signals derived from herbivores that allow them to respond defensively to insect attacks (Howe and Jander, 2008). The SA and JA pathways are also involved in the modulation of herbivory responses, more specifically in the production of toxins and defensive proteins that affect the attacking performance of the insect, and the emission of volatiles that attract predators and parasitoids to the invaders (Howe and Jander, 2008; Costarelli et al., 2020).
When autotrophic plants are attacked by pathogens, they must continuously balance the use of their resources between growth and defence, often in an opposite manner, hence defining a trade-off. Besides ET, JA and SA, other phytohormones, including auxin, gibberellins (GAs), abscisic acid (ABA) and cytokinins (CKs), play important roles in plant immunity. Currently, how these phytohormones are involved in immunity and how pathogens manipulate their respective signalling pathways during infections have been extensively reviewed from different perspectives (Shigenaga and Argueso, 2016; Berens et al., 2017; Bürger and Chory, 2019). An additional group of phytohormones, the brassinosteroids (BRs), also play a key role in the growth–defence trade-off (Lozano-Durán and Zipfel, 2015). BRs are polyhydroxylated steroidal phytohormones that have a major function in plant growth, development and responses to different environmental stresses (Nolan et al., 2020). Here, we review our current understanding of the crosstalk between BR signalling and plant immunity. We focus our discussion on the molecular interactions between signalling proteins regulating both pathways in the PM, cytoplasm and nucleus. Furthermore, we assess the connections with other phytohormones. Finally, we conclude on how the interactions between defence and BR signalling pathways regulate growth–defence trade-off in plants.
Diverse functions of BR phytohormones
Initially, BRs were described as growth-promoting phytohormones, although exogenous BRs can either promote or inhibit growth depending on their concentrations (Grove et al., 1979; Müssig et al., 2003). Genetic and biochemical approaches have established a pivotal role for BRs in many aspects of plant development and physiology by promoting: (1) cell elongation by alteration of the cytoskeleton dynamics (Wang et al., 2012; Ruan et al., 2018); (2) cell division through stimulation of cell cycle progression (González-García et al., 2011; Hacham et al., 2011); (3) xylem development (Kondo et al., 2014; Lee et al., 2019); (4) stomatal development (Gudesblat et al., 2012; Kim et al., 2012; Houbaert et al., 2018); (5) seed germination by activation of GA signalling and inactivation of ABA signalling (Steber and McCourt, 2001; Divi and Krishna, 2010; Kim et al., 2019); (6) plant reproduction through the regulation of floral transition, as well as male and female fertility (Vogler et al., 2014; Li and He, 2020); and (7) photomorphogenesis by regulation of the expression of photosynthesis and light-responsive genes (Song et al., 2009; Li and He, 2016). BRs have also been shown to function in plant responses to various abiotic stresses. Upon changes in temperature, BRs induce the expression of growth-promoting genes at elevated temperatures and that of cold-responsive genes in cold environments (Eremina et al., 2016; Li et al., 2017; Ibañez et al., 2018; Nolan et al., 2020). Furthermore, variations in salinity lead to the BR control of ET biosynthesis and signalling (Zhu et al., 2016; Planas-Riverola et al., 2019). In the context of low nutrient availability, BRs can also act as central regulators of the reprogramming of the root system architecture under this condition (Singh et al., 2018; Pandey et al., 2020). Finally, BR signalling activates stress-responsive genes, modulates ABA levels and promotes accumulation of osmoprotectant metabolites under drought conditions (Ye et al., 2017; Fàbregas et al., 2018; Planas-Riverola et al., 2019). Recently, BRs have also been linked to plant responses to pathogens (Albrecht et al., 2012; Lozano-Durán and Zipfel, 2015; Yu et al., 2018; Liao et al., 2020).
The BR signalling pathway
Tremendous progress has been made in elucidating the perception of BRs and their signalling mechanisms in the model plant Arabidopsis thaliana (comprehensively reviewed recently in Planas-Riverola et al., 2019; Kim and Russinova, 2020; Nolan et al., 2020). The PM-localized LRR RK BR INSENSITIVE1 (BRI1), which presents structural and activation similarities to known PRRs, initiates BR signalling upon direct recognition of the BR hormone in the apoplast (the space between the PM and the plant cell wall) (Hothorn et al., 2011; She et al., 2011). BRI1 requires a co-receptor, BRI1-ASSOCIATED KINASE1 (BAK1; also known as SOMATIC EMBRYOGENESIS RECEPTOR KINASE3; SERK3), and other SERKs (Li et al., 2002; Russinova et al., 2004; Sun et al., 2013; Bojar et al., 2014) (Figs 2 and 3). BR binding activates the intracellular kinase domain of BRI1 and induces the dissociation of the inhibitory proteins BRI1 KINASE INHIBITOR1 (BKI1) and BOTRYTIS-INDUCED KINASE1 (BIK1) (Wang and Chory, 2006; Lin et al., 2013). Subsequently, BRI1 activates BAK1 through phosphorylation (Wang et al., 2008), which releases BAK1 INTERACTING RECEPTOR-LIKE KINASE3 (BIR3) from BAK1 (Hohmann et al., 2018). The BRI1–BAK1 complex activates both BR SIGNALING KINASE1 (BSK1) (Tang et al., 2008) and CONSTITUTIVE DIFFERENTIAL GROWTH1 (CDG1) that, in turn, activate the phosphatase BRI1-SUPPRESSOR1 (BSU1) and its homologues, the BSU-like proteins (BSLs) (Kim et al., 2011). BSU1 dephosphorylates and inactivates the GSK3-like kinase, BR INSENSITIVE2 (BIN2), which subsequently undergoes proteasome degradation mediated by the F-box protein KINK SUPPRESSED IN BZR1-1D (KIB1) (Kim et al., 2011; Zhu et al., 2017). BIN2, a negative regulator of the BR signalling, inactivates two master transcription factors of the BR-dependent gene expression, BRASSINAZOLE-RESISTANT1 (BZR1) and BRI1-EMS-SUPPRESSOR1 (BES1, also known as BZR2) (Wang et al., 2002; Yin et al., 2002) through their phosphorylation and subsequent cytoplasmic retention due to the interaction with 14-3-3 proteins (Gampala et al., 2007). The BR-induced inactivation of BIN2 leads to the dephosphorylation of BZR1 and BES1 by PROTEIN PHOSPHATASE2A (PP2A); thereafter, both transcription factors translocate into the nucleus to induce a transcriptional reprogramming, either directly or through the interaction with other transcription factors (Wang et al., 2002; Yin et al., 2002; Tang et al., 2011; Oh et al., 2014). Recently, it has been shown that all transcription factors of the BZR1 family function redundantly in BR signalling (Chen et al., 2019). Nonetheless, additional studies are required to establish whether they follow the same activation mechanisms as previously described for BZR1 and BES1.
Fig. 2.
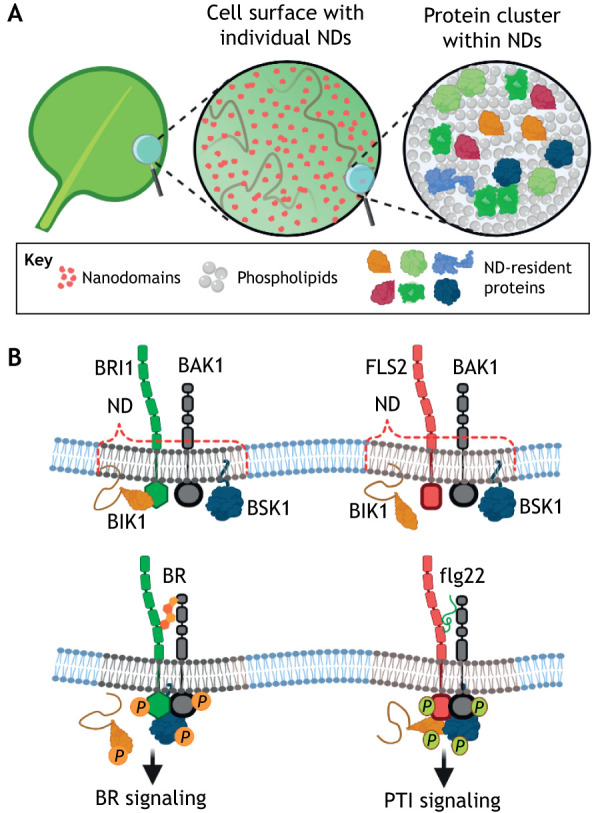
Crosstalk between brassinosteroid signalling and plant immunity in the plasma membrane. (A) The plasma membrane (PM) is compartmentalized into heterogeneously distributed specialized nanometer-scale platforms, referred to as nanodomains (NDs) that host distinct protein consortia. (B) BRI1 and FLS2, together with their signalling components, are segregated into different nanodomains (top). The spatial separation of BRI1 and FLS2, as well as other signalling components, maintains the signalling specificity upon ligand recognition, while independent pools of common signalling proteins are utilized. BR binding to BRI1 and the co-receptor BAK1 leads to the dissociation of BIK1 from BRI1 and triggers transphosphorylation between BRI1 and BAK1. The activated BRI1 and BAK1 receptor complex stimulates BSK1 (left bottom). In the presence of flg22, FLS2 interacts with BAK1, BIK1 and BSK1, followed by the phosphorylation of all the proteins (right bottom). P circled in orange indicates phosphorylation events triggered by BR, and P circled in green indicates phosphorylation events triggered by flg22. For convenience, not all known signalling components are shown. This figure was created with BioRender.
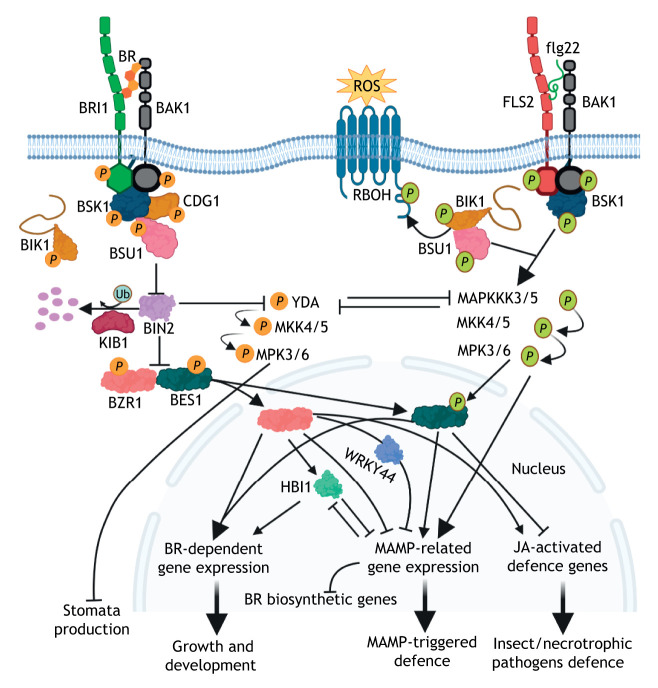
Fig. 3.
Intracellular crosstalk between brassinosteroid signalling and plant immunity. BAK1, BIK1, BSK1 and BSU1 are signalling components of both the BR–BRI1 and flg22–FLS2 ligand–receptor pairs. BR binding to BRI1 and BAK1 results in release of BIK1 from BRI1 and activation of the BRI1–BAK1 complex. BSK1 and CDG1 are activated through phosphorylation by the receptor complex. BSK1 or CDG1 activates BSU1, which, in turn, inactivates BIN2 by dephosphorylation and proteasomal degradation mediated by the F-box protein KIB1, allowing the dephosphorylated BZR1 and BES1 to enter the nucleus and induce BR-dependent gene expression, which modulates growth and development. After flg22 recognition, FLS2 and BAK1 bind and activate BIK1 and BSK1 through phosphorylation. Phosphorylated BIK1 dissociates from the FLS2–BAK1 complex and activates the respiratory burst oxidase protein D (RBOH), leading to a burst in ROS production, whereas BSK1 triggers downstream MAPK components. Notably, the MAPK pathway, which is mediated by MAPKKK3 or MAPKKK5 (MAPKKK3/5) during plant immunity, antagonizes the stomata developmental pathway that is mediated by YDA by competing for the downstream MAPK kinases (MKKs) upon BR perception. BZR1 and BES1 might play distinct roles in plant immunity. BR-activated BZR1 negatively affects immunity-related gene expression, directly, through HBI1 or in association with the WRKY transcription factor WRKY40. BZR1 also functions as a positive regulator of JA-activated defence genes that are associated with defence responses against insects and necrotrophic pathogens. In contrast, BES1, a direct substrate of MPK6, plays a positive role in microbe-associated molecular patterns (MAMP)-induced gene expression and antagonizes jasmonic acid (JA)-mediated plant defence responses. MAMP-triggered defence reduces the expression of the HBI1 gene and the BR biosynthetic genes. P circled in orange and green indicates phosphorylation events triggered by BR and flg22, respectively, and circled Ub denotes ubiquitylation. For convenience, not all known signalling components are shown. This figure was created with BioRender.
Crosstalk between BR signalling and plant immunity
BR signalling and plant immunity interact through complex regulatory networks that depend on pathogen feeding preferences, plant species and plant physiological status, which are often co-regulated with other phytohormones (Yu et al., 2018; Bürger and Chory, 2019; Liao et al., 2020). Moreover, different experimental approaches with either BR-related mutants or exogenous BRs revealed antagonistic or synergistic effects, or lack thereof, on plant immunity, leading to apparently opposing conclusions (Albrecht et al., 2012; Belkhadir et al., 2012; Miyaji et al., 2014; Lee et al., 2018; Liao et al., 2020). Here, we will discuss the increasing evidence that BR signalling and plant immunity interact in the PM and the intracellular space, depending on the different pathogen lifestyles. We will further examine the crosstalk of BRs with other phytohormones during the plant defence responses.
Crosstalk in the PM
The PM provides a protective barrier around the plant cell and acts as a communication interface between the outside environment and the cell interior. Receptor proteins, such as BRI1, which binds BRs to trigger BR-dependent responses, and PRRs, which recognize MAMPs and/or DAMPs to activate PTI, are embedded in this compartment (Russinova et al., 2004; Monaghan and Zipfel, 2012). Correct downstream signal transduction depends on the interactions of the activated receptors with additional regulatory proteins in the PM, which are often shared between the receptors. On the one hand, shared regulatory proteins might present a challenge for plant cells in triggering accurate responses, but, on the other hand, they might serve as hubs for signal integration and cross-regulation. Interestingly, BR PRR signalling pathways share regulatory components, including RKs, BAK1, BIR1, and the receptor-like cytoplasmic kinases (RLCKs), BIK1 and BSK1 (Gómez-Gómez and Boller, 2000; Li et al., 2002; Chinchilla et al., 2009; Lin et al., 2013; Bücherl et al., 2017).
BAK1 and other SERKs act as co-receptors of BRI1 (Gou et al., 2012; Sun et al., 2013) and of several PRRs; these include FLAGELLING SENSIN2 (FLS2), which recognizes the bacterial flagellin and the cognate peptide flg22 (Roux et al., 2011) (Figs 2 and 3), ELONGATION FACTOR-Tu RECEPTOR (EFR), which binds the bacterial elongation factor Tu and the cognate peptide elf18 (Roux et al., 2011), as well as PEP RECEPTOR1 (PEPR1) and PEPR2, which perceive a family of DAMP peptides (Peps) (Tang et al., 2015; Ortiz-Morea et al., 2016). BAK1 has been described as a positive regulator of BR and PTI signalling (Chinchilla et al., 2009; Roux et al., 2011), but its precise role in the crosstalk between BR hormones and PTI remains unknown.
Exogenous BRs and transgenic plants with constitutive BR signalling have revealed that flg22-induced outputs are negatively affected by BRs (Fig. 3), whereas the BR responses are not affected by FLS2 signalling, suggesting the existence of a unidirectional PTI regulation through BR perception (Albrecht et al., 2012; Belkhadir et al., 2012; Lozano-Durán et al., 2013). The effect of BRs on PTI responses has been proposed to operate through mechanisms both dependent and independent of BAK1 (Albrecht et al., 2012; Belkhadir et al., 2012).
Several observations support BAK1-dependent mechanisms. First, the callose deposition, a hallmark of PTI that strengthens the cell walls forming a physical barrier for pathogens (Yu et al., 2017), in plants overexpressing BRI1, was impaired by flg22, but not by the fungal MAMP chitin, which is recognized by a BAK1-independent pathway (Gimenez-Ibanez et al., 2009; Belkhadir et al., 2012). Second, the sensitivity to flg22 was recovered in BRI1-overproducing plants when the BAK1 dosage was increased by overexpression of BAK1–HA, indicating that the activated BRI1 might recruit BAK1 away from FLS2, thus affecting the flg22-triggered signalling in a BAK1-dependent manner (Belkhadir et al., 2012). However, later, BAK1–HA was shown to be partly functional in BR signalling and to possibly exert a dominant-negative effect on the endogenous BAK1 (Lozano-Durán et al., 2013), complicating the interpretation of the results. Finally, plants expressing a hyperactive BR receptor, BRI1sud1, which carries a G643E mutation that stabilizes the BR-bound state of the receptor (Santiago et al., 2013), presented an increased basal phosphorylation of FLS2, improved flg22-triggered responses and a flg22-dependent resistance to the hemibiotrophic bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pto DC3000) in a BAK1-dependent manner (Belkhadir et al., 2012). Hence, elevated flg22 responses activated by the hypermorphic BRI1sud1 might result from a cross-activation of FLS2 by BAK1, as a consequence of BRI1 hyperactivity (Lozano-Durán and Zipfel, 2015). However, the question is raised of why these responses do not occur in plants in which the BR signalling is activated either by BRI1 overexpression or by exogenous BRs (Belkhadir et al., 2012; Lozano-Durán and Zipfel, 2015).
BAK1-independent BR inhibition of PTI signalling has been supported by the observation that exogenous BRs are able to hinder the production of ROS, induced not only by flg22 but also by chitin, which does not require BAK1 (Albrecht et al., 2012). Moreover, the amount of BAK1 in a complex with FLS2 after co-treatment with BRs and flg22 remained unaffected, indicating that BAK1 is not rate limiting between the BRI1 and FLS2 pathways. Consistently, upon treatment with flg22, BAK1 complexed normally with FLS2 in plants overexpressing BRI1 (Lozano-Durán et al., 2013).
Recently, BRI1 and FLS2 have been found to be heterogeneously distributed into specialized nanometer-scale PM platforms, referred to as nanodomains, in which other PM-associated signalling components are also present, such as BAK1, BSK1 and BIK1 (Wang et al., 2015; Bücherl et al., 2017) (Fig. 2). Therefore, nanodomains could provide a scaffold for proteins to ensure physical separation of protein–protein interactions, thus allowing BRI1 and FLS2 to maintain differential signalling outputs by means of independent pools of shared proteins (Bücherl et al., 2017; Burkart and Stahl, 2017; Ott, 2017) (Fig. 2). Hence, activated BRI1 and PRRs would not compete for BAK1, because with their associated signalling components, they might be separated within the PM nanodomains even before binding of their ligands (Lozano-Durán and Zipfel, 2015). This assumption is supported by the fact that preformed BRI1–BAK1 complexes have been detected independently from the ligand-bound complexes in the PM nanodomains (Bücherl et al., 2013; Hutten et al., 2017). Although the presence of FLS2 and BAK1 in the same nanodomains has still to be demonstrated, the shared signalling components BIK1 and BSK1 cluster differentially with the FLS2 and BRI1 receptors in the PM nanodomain structures (Bücherl et al., 2017).
BIK1 has been shown to negatively regulate BR signalling through direct association with BRI1. After BR perception, BIK1 is phosphorylated by BRI1 causing its dissociation from the receptor (Lin et al., 2013). In contrast, BIK1 is required for signalling triggered by flg22, elf18 and Pep1, whereas bik1 mutants exhibit compromised resistance to infection with Pto DC3000 hrcC, a type III secretion mutant, suggesting that BIK1 positively controls the MAMP-induced immunity (Lu et al., 2010; Liu et al., 2013). Upon flg22 perception, BIK1 is phosphorylated by BAK1, and, inversely, BIK1 phosphorylates the FLS2–BAK1 complex (Lin et al., 2014). Subsequently, BIK1 dissociates from the complex and phosphorylates the NADPH oxidase respiratory burst oxidase protein D (RBOHD), which leads to a burst in ROS production (Kadota et al., 2014; Li et al., 2014) (Fig. 3). Recently, it has been reported that BIK1 activation and release from the PRR complex is induced by ligands and is dependent on monoubiquitylation (Ma et al., 2020). Interestingly, BIK1 was also found to play a negative role in immune responses that are mediated by the RLP-type immune receptors RLP23, RLP42 and aphid resistance (Lei et al., 2014; Wan et al., 2019). Taken together, these observations indicate that the functions of BIK1 in different receptor complexes are distinct and could lead to opposing signalling outputs.
BSK1, another RLCK, is a BRI1 substrate that positively regulates BR signalling (Tang et al., 2008) and, when associated with FLS2, positively controls the FLS2-activated defence (Tang et al., 2008; Shi et al., 2013) (Fig. 3). The bsk1 mutant displays an enhanced susceptibility to infections with virulent and avirulent strains of Pto DC3000, the fungal powdery mildew pathogen Golovinomyces cichoracearum and the oomycete Hyaloperonospora arabidopsidis (Shi et al., 2013). These phenotypes point to a convergent role of BSK1 in plant defence responses triggered by multiple pathogens, although whether BSK1 associates with other PRRs, in addition to FLS2, is unknown. BSK1 regulates plant immunity by phosphorylating MAPK KINASE KINASE5 (MAPKKK5), thus linking the signalling from the immune complex in the PM to the MAPK cascade (Yan et al., 2018). The BSK1 subfamily consists of 12 members, of which some play a redundant role in the BR signalling (Tang et al., 2008; Sreeramulu et al., 2013). BSK1, BSK3 and BSK5 are phosphorylated by BRI1 upon BR perception, and overexpression of any one of these proteins can partially suppress the dwarf phenotype of the weak bri1-5 mutant (Tang et al., 2008; Sreeramulu et al., 2013; Ren et al., 2019). Similarly, some BSK family members have been implicated in plant immunity control (Tang et al., 2008; Qi et al., 2011; Xu et al., 2014; Majhi et al., 2019). For example, BSK5 has recently been found to play a role in PTI responses mediated by multiple immune receptors, such as FLS2, EFR and PEPR1 (Majhi et al., 2019). BSK3 also interacts with multiple PRRs, but the functional relevance of these interactions remains unknown (Xu et al., 2014).
Intracellular crosstalk
Transmission of downstream BR signalling requires the activation of BSU1 or BSLs, through their phosphorylation by BSKs or CDG1 (Kim et al., 2011; Sreeramulu et al., 2013). BSU1 and BSLs positively regulate immune signalling, functioning upstream of the MAPK module (Park et al., 2019 preprint). The participation of BSU1 in plant immunity and in BR signalling is probably controlled by different phosphorylation patterns, interpreted as a phosphocode. BSU1 is phosphorylated on Ser251 by BIK1 in an flg22-dependent manner and on Ser764 by CDG1 upon BR perception (Kim et al., 2011; Park et al., 2019 preprint).
BIN2 and the two closely related GSK3-like kinases, BIN2-LIKE1 (BIL1) and BIL2, also regulate plant immunity, because impairment of their function, by either a GSK inhibitor or knockout mutants, reduced the levels of flg22- and chitin-induced ROS (Lozano-Durán et al., 2013). In addition, an inhibitory effect of BR signalling on immunity is supported by the dramatic increase of MAMP-induced MAPK activation in the BIN2 gain-of-function mutant, bin2-1 (Li and Nam, 2002). In this mutant, both the BR responses and the developmental signalling module, comprising MAPKKK YODA (YDA), MAPK KINASE4 or MAPK KINASE5 (referred to hereafter as MKK4/MKK5) and MAPK3 or MAPK6 (referred to hereafter as MPK3/MPK6) are blocked (Sun et al., 2018). The inhibitory effect of BR signalling on immunity might also be attributed to an antagonistic interaction between the immune signalling module, containing MAPKKK3 or MAPKKK5 (referred to hereafter as MAPKKK3/MAPKKK5), MKK4/MKK5 and MPK3/MPK6, and the developmental module (i.e. YDA–MKK4/MKK5–MPK3/MPK6), determined by the active state of BIN2 or of its homologues (Sun et al., 2018). For example, in the stomatal lineage, BIN2 and its homologues are inactivated by BRs, which then leads to activation of YDA, MKK4/MKK5 and MPK3/MPK6, and to subsequent inhibition of stomatal development (Kim et al., 2012; Khan et al., 2013). The developmental MAPK signalling module YDA–MKK4/MKK5–MPK3/MPK6 can plausibly compete for the shared MKK4/MKK5 proteins with the immune MAPK module MAPKKK3/MAPKKK5–MKK4/MKK5–MPK3/MPK6 (Sun et al., 2018) (Fig. 3). Moreover, another BIN2 homologue, ASKα, has been found to be rapidly induced under MAMP or DAMP perception and to positively regulate PTI responses through phosphorylation of glucose-6-phosphate dehydrogenase (G6PD), a key enzyme of the oxidative pentose phosphate pathway (Stampfl et al., 2016).
The transcription factors BZR1 and BES1, which control the majority of the BR-regulated genes, play essential roles in mediating plant immunity (Yu et al., 2011; Oh et al., 2014). Activation of BZR1 negatively controls early immune responses. For example, plants constitutively expressing active BZR1 versions exhibit an impaired flg22- or chitin-triggered signalling, a decreased flg22-induced resistance to Pto DC3000, and an enhanced susceptibility to the non-host strain Pseudomonas syringae pv. cilantro (Pci) 0788-9 (Lozano-Durán et al., 2013). BZR1 modulates these effects through the direct activation of the expression of a subset of WRKY transcription factors that negatively regulate MAMP-triggered ROS, including WRKY11, WRKY15 and WRKY18, and through the repression of defence genes by the formation of a protein complex with WRKY40 (Lozano-Durán et al., 2013) (Fig. 3). BZR1 also activates the basic helix-loop-helix transcription factor HOMOLOG OF BR ENHANCED EXPRESSION2 INTERACTING WITH INCREASED LEAF INCLINATION1 BINDING bHLH1 (HBI1), which stimulates BR biosynthetic genes to induce cell elongation and downregulates the expression of a subset of immune-related genes (Fan et al., 2014) (Fig. 3). Interestingly, the expression of HBI1, but not of BZR1, is inhibited by MAMPs (Fan et al., 2014), hinting at a negative and bidirectional crosstalk between PTI and BR signalling that differs from the initially postulated unidirectional regulation of PTI by BRs (Albrecht et al., 2012; Belkhadir et al., 2012). Bidirectional crosstalk between PTI and BR is further supported by the activation of PTI by flg22 or other MAMPs, resulting in the reduced expression of BR biosynthetic genes (Jiménez-Góngora et al., 2015) (Fig. 3).
In contrast to BZR1, BES1 has been shown to positively regulate plant immunity against bacterial pathogens, because the loss-of-function mutants bes1-1 and bes1-2 display a reduced flg22-induced expression of WRKY22 and FLG22-INDUCED RECEPTOR-LIKE KINASE1 (FRK1), both marker genes for PTI signalling and compromised resistance to Pto DC3000 (Kang et al., 2015). As a substrate of MPK6, BES1 phosphorylation is enhanced under MAMP perception (Kang et al., 2015). Similar to BSU1, the function of BES1 in plant immunity and development depends on phosphocodes. For instance, the two mutations S286A and S137A in BES1, which impair the MAMP-induced phosphorylation and fail to re-establish the Pto DC3000 resistance in the bes1-1 mutant, did not affect BR-mediated hypocotyl and root growth (Kang et al., 2015). Interestingly, the bes1-D gain-of-function mutant has an increased susceptibility to the necrotrophic fungi Alternaria brassicicola and Botrytis cinerea, and the insect herbivore Spodoptera exigua (Shin et al., 2016; Liao et al., 2020), thus indicating that BES1 can both positively or negatively regulate plant immunity and that this effect is associated with the pathogen lifestyle (Fig. 3). The negative effect of BES1 on immune responses to necrotrophs and herbivory is regulated by the suppression of the expression of defensins genes, such as PDF1.2a and PDF1.2b, and of the indole glucosinolate (GS) biosynthetic genes, respectively, acting in concert with JA (Shin et al., 2016; Liao et al., 2020).
Crosstalk of BRs with other phytohormones
BRs have been connected with JA, SA and GA in plant defence against different pathogens and insects. Recently, BRs have been reported to antagonize JA-activated plant defence against necrotrophic pathogens and herbivore insects through BES1 (Liao et al., 2020). The BR-activated BES1 interacts with the terminator region of PDF1.2a and PDF1.2b, suppressing their transcriptional activities and attenuating JA-induced responses against necrotrophic organisms (Liao et al., 2020). Moreover, the interaction of BES1 with the indolic GS-related MYB transcription factors MYB34, MYB51 and MYB122 represses the expression of the JA-induced indole-GS biosynthetic genes, including CYP79B3 and UGT74B, thereby diminishing the defence responses against the insect herbivore S. exigua (Liao et al., 2020). Although the bes1-D mutant is highly susceptible to S. exigua, and BRs regulate the GS biosynthesis through BZR1 and BES1 (Guo et al., 2013; Lee et al., 2018; Liao et al., 2020), the adverse effect of BRs on plant defence against insect attacks cannot be generalized. Experiments with the diamondback moth Plutella xylostella revealed that its larvae prefer to feed on bri1-5 mutant than on wild-type plants (Lee et al., 2018), and that the gain-of-function bzr1-1D plants show an increased resistance against thrip feeding, together with an enhanced expression of the JA-inducible gene VSP2 (Miyaji et al., 2014). Thus, the crosstalk between JA and BRs might play distinct roles in plant defence mediated by BZR1 and BES1 (Figs 3 and 4).
Fig. 4.
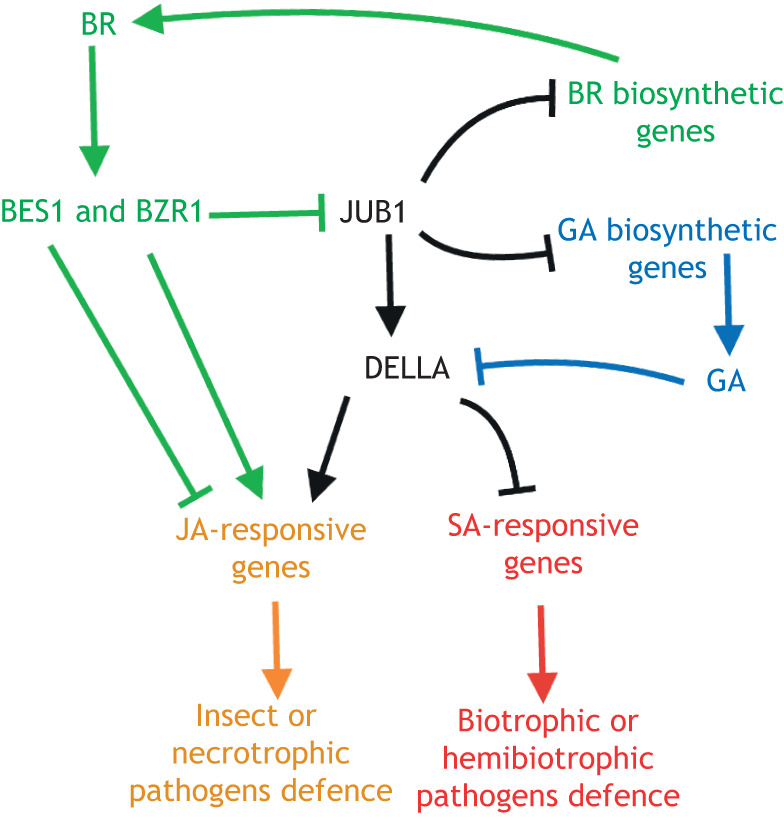
Signalling network integrating brassinosteroid, gibberellins, jasmonic acid and salicylic acid pathways during plant defence responses. In the presence of brassinosteroids (BRs), the transcription factors BES1 and BZR1 attenuate and stimulate jasmonic acid (JA)-induced responses against insects and necrotrophic pathogens, respectively. Activated BZR1 represses JUB1 expression, which directly suppresses genes involved in the biosynthesis of BR and gibberellin (GA), reducing their levels. JUB1 induces the accumulation and activation of DELLA proteins, which are, in turn, negatively affected by GA. DELLA proteins repress the expression of salicylic acid (SA) signalling genes but increase the expression of JA-mediated defence genes, leading to improved susceptibility to biotrophic and hemibiotrophic pathogens, and increased resistance to insects and necrotrophic pathogens, respectively. This figure was created with BioRender.
Interactions between GA and BRs also contribute to the modulation of plant responses against necrotrophic and biotrophic pathogens (Shahnejat-Bushehri et al., 2016a). The NAM, ATAF and CUC (NAC) transcription factor JUB1 directly represses the BR and GA biosynthetic genes, DWF4 and GA3OX1, respectively, decreasing BR and GA levels (Shahnejat-Bushehri et al., 2016a,b). Plants overexpressing JUB1 display an enhanced susceptibility to Pto DC3000 that is mainly attributed to an increased accumulation and activation of DELLA proteins (Shahnejat-Bushehri et al., 2016a). DELLA proteins suppress the expression of SA signalling genes but increase JA-mediated defence genes, improving the susceptibility of plants to biotrophic pathogens and increasing resistance against necrotrophic pathogens (Hou et al., 2010; Shahnejat-Bushehri et al., 2016a). Interestingly, BZR1 has been reported to repress JUB1, thus amplifying BR signalling through the release of the JUB1-mediated suppression of the BR biosynthesis and the formation of a negative-feedback loop (Shahnejat-Bushehri et al., 2016b). These results show that it is conceivable that JUB1, as a core regulatory module, induces DELLA accumulation, downregulates BR and upregulates JA levels to favour JA signalling (mainly associated to necrotrophic pathogen resistance) over that of SA (mainly associated to biotropic pathogen resistance) (Robert-Seilaniantz et al., 2011). Therefore, owing to reduced BR and GA levels, cell elongation will be restricted, while concomitantly the resistance against necrotrophic and biotrophic pathogens will be enhanced and attenuated, respectively (Fig. 4). The JUB1 signalling network could be fine-tuned by the interactions between BZR1 (or BES1) and particular DELLA proteins, resulting in opposite outputs. For example, on the one hand, the activity of REPRESSOR OF ga1-3 (RGA), a member of the DELLA family, is negatively affected by the activated BZR1 and BES1 (Li et al., 2012). On the other hand, the DELLA proteins RGA and GIBBERELLIN INSENSITIVE (GAI) inactivate the transcriptional activity of BZR1 by inhibiting its ability to bind to its targets (Gallego-Bartolomé et al., 2012; Bai et al., 2012). However, the relevance of these interactions with respect to plant immunity remains to be explored.
In addition, BRs antagonize GA- and SA-mediated immunity in rice (Oryza sativa) roots (De Vleesschauwer et al., 2012), suppress defence against root-knot nematodes by antagonizing the JA pathway (Nahar et al., 2013) and are involved in ET-induced pathogen resistance in Nicotiana benthamiana (Xiong et al., 2020). Therefore, the interaction between BRs and other phytohormones (JA, SA, GA and ET) seems to be a common feature of the modulation of growth and immune responses in the plant kingdom.
Taken together, the BR and plant immunity pathways are interconnected by sharing different signalling components in both the PM and the intracellular space (Figs 2 and 3). This relationship is in concert with the action of other phytohormones. Collectively, these interactions regulate the growth–defence trade-off as they allow plants to defend against pathogens but avoid growth-negative effects caused by overstimulation of immunity.
Concluding remarks and future perspectives
Over the past decades, remarkable progress has been made in understanding the mechanisms of BR and plant immune signalling individually. More recently, BRs have gained attention as important regulators of plant immunity and of growth–defence trade-off through remobilization of plant resources for growth and defence according to the specific type and duration of the biotic stresses. However, thus far, unravelling the action mechanisms of BRs during pathogen infections has been a challenging task due to the use of diverse experimental conditions and readouts.
BRs seemingly play negative and positive roles in plant defence that correlate with the function of the master transcription factors in BR signalling, BZR1 and BES1, as positive and negative regulators of plant immunity. Nevertheless, BR regulation of local and systemic plant defence responses against distinct types of invading organisms remains unclear. Unification of experimental conditions and readouts in future studies might help to obtain results that are more reliable. Research on the mechanisms, by which BZR1 and BES1 modulate downstream defence genes, would further advance our understanding on BR crosstalk with other phytohormones that control plant growth and immunity.
Although activation of PTI signalling by flg22 does not affect BR signalling, treatment with flg22 leads to quick and sustained repression of the BR biosynthetic genes (Jiménez-Góngora et al., 2015), possibly due to the existence of a bidirectional crosstalk between PTI and BR pathways. As the maintenance of BR homeostasis is critical for proper immune responses (Albrecht et al., 2012), the modulation of endogenous BR would help the plant to regulate its growth under a pathogen attack.
BRs and plant immunity interact at different cellular levels, of which the PM is a critical checkpoint prior to signal transduction to the cytoplasm and the nucleus. PM compartmentalization into nanodomains hosting distinct protein consortia is pivotal for the integration of BR signalling and plant immune responses. BRI1 and PRRs represent a suitable model to help us understand how biological systems link diverse external and developmental cues to elicit specific biological outcomes with a limited number of shared signalling modules. It would be interesting to visualize with high precision, through super-resolution microscopy, the nanodomain patterns of the receptors and their accessory proteins in different cell types during plant development and/or various pathogen attacks. As phosphocode-based regulation is crucial for the integration of BR and plant immune pathways, it is also worth investigating the phosphorylation-dependent function of other signalling components by the implementation of phosphoproteomics.
Recently, it has been reported that different root cell types activate immune responses according to their cell identity (Rich-Griffin et al., 2020) and that a tissue-specific distribution of BR signalling controls root growth (Vragović et al., 2015; Ackerman-Lavert and Savaldi-Goldstein, 2020). Considering these facts, it is plausible to hypothesize that a well-coordinated spatiotemporal control at the cell and tissue levels, determines the specificity within the complex immune and hormonal networks that modulate the growth–defence trade-off. With the advance of plant single-cell analysis, such as high-throughput single-cell RNA sequencing, it is expected that scientists will be able to decipher how this process is orchestrated in the coming years. Moreover, as the molecular mechanisms of BR and plant immune pathways each appear to be evolutionarily conserved in different plant species (Liu et al., 2017, Kim and Russinova, 2020), the knowledge acquired from model plants would be insightful when designing strategies for enhancing crop tolerance to biotic stresses while minimizing the compromised plant growth.
Acknowledgements
We thank Martine De Cock for help in preparing the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Our research was supported by the National Institutes of Health (NIH) (R01GM092893) and National Science Foundation (NSF) (MCB-1906060) to P.H. and the NIH (R01GM097247) and the Robert A. Welch Foundation (A-1795) to L.S. F.A.O.-M. is indebted to the PEW Latin American Fellows Program for a postdoctoral fellowship. Deposited in PMC for release after 12 months.
References
- Ackerman-Lavert M. and Savaldi-Goldstein S. (2020). Growth models from a brassinosteroid perspective. Curr. Opin. Plant Biol. 53, 90-97. 10.1016/j.pbi.2019.10.008 [DOI] [PubMed] [Google Scholar]
- Albert I., Hua C., Nürnberger T., Pruitt R. N. and Zhang L. (2020). Surface sensor systems in plant immunity. Plant Physiol. 182, 1582-1596. 10.1104/pp.19.01299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C., Boutrot F., Segonzac C., Schwessinger B., Gimenez-Ibanez S., Chinchilla D., Rathjen J. P., de Vries S. C. and Zipfel C. (2012). Brassinosteroids inhibit pathogen-associated molecular pattern–triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA 109, 303-308. 10.1073/pnas.1109921108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.-Y., Shang J.-X., Oh E., Fan M., Bai Y., Zentella R., Sun T.-P. and Wang Z.-Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14, 810-817. 10.1038/ncb2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y., Jaillais Y., Epple P., Balsemão-Pires E., Dangl J. L. and Chory J. (2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. USA 109, 297-302. 10.1073/pnas.1112840108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens M. L., Berry H. M., Mine A., Argueso C. T. and Tsuda K. (2017). Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 55, 401-425. 10.1146/annurev-phyto-080516-035544 [DOI] [PubMed] [Google Scholar]
- Bojar D., Martinez J., Santiago J., Rybin V., Bayliss R. and Hothorn M. (2014). Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 78, 31-43. 10.1111/tpj.12445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyajyan A., Devejyan H., Haykazyan V., Avetisyan G. and Khanoyan D. (2014). Molecular mechanisms and mediators of the immune response in plants. J. Plant Sci. 2, 23-30. [Google Scholar]
- Bücherl C. A., van Esse G. W., Kruis A., Luchtenberg J., Westphal A. H., Aker J., van Hoek A., Albrecht C., Borst J. W. and de Vries S. C. (2013). Visualization of BRI1 and BAK1(SERK3) membrane receptor heterooligomers during brassinosteroid signaling. Plant Physiol. 162, 1911-1925. 10.1104/pp.113.220152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücherl C. A., Jarsch I. K., Schudoma C., Segonzac C., Mbengue M., Robatzek S., MacLean D., Ott T. and Zipfel C. (2017). Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 6, e25114 10.7554/eLife.25114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger M. and Chory J. (2019). Stressed out about hormones: how plants orchestrate immunity. Cell Host Microbe 26, 163-172. 10.1016/j.chom.2019.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart R. C. and Stahl Y. (2017). Dynamic complexity: plant receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 40, 15-21. 10.1016/j.pbi.2017.06.016 [DOI] [PubMed] [Google Scholar]
- Chen L.-G., Gao Z., Zhao Z., Liu X., Li Y., Zhang Y., Liu X., Sun Y. and Tang W. (2019). BZR1 family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in Arabidopsis. Mol. Plant 12, 1408-1415. 10.1016/j.molp.2019.06.006 [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Shan L., He P., de Vries S. and Kemmerling B. (2009). One for all: the receptor-associated kinase BAK1. Trends Plant Sci. 14, 535-541. 10.1016/j.tplants.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Jayaraman J., Segonzac C., Park H.-J., Park H., Han S.-W. and Sohn K. H. (2017). Pseudomonas syringae pv. actinidiae type III effectors localized at multiple cellular compartments activate or suppress innate immune responses in Nicotiana benthamiana. Front. Plant Sci. 8, 2157 10.3389/fpls.2017.02157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costarelli A., Bianchet C., Ederli L., Salerno G., Piersanti S., Rebora M. and Pasqualini S. (2020). Salicylic acid induced by herbivore feeding antagonizes jasmonic acid mediated plant defenses against insect attack. Plant Signal. Behav. 15, 1704517 10.1080/15592324.2019.1704517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vleesschauwer D., Van Buyten E., Satoh K., Balidion J., Mauleon R., Choi I.-R., Vera-Cruz C., Kikuchi S. and Höfte M. (2012). Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 158, 1833-1846. 10.1104/pp.112.193672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divi U. K. and Krishna P. (2010). Overexpression of the brassinosteroid biosynthetic gene AtDWF4 in Arabidopsis seeds overcomes abscisic acid-induced inhibition of germination and increases cold tolerance in transgenic seedlings. J. Plant Growth Regul. 29, 385-393. 10.1007/s00344-010-9150-3 [DOI] [Google Scholar]
- Eremina M., Unterholzner S. J., Rathnayake A. I., Castellanos M., Khan M., Kugler K. G., May S. T., Mayer K. F. X., Rozhon W. and Poppenberger B. (2016). Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl. Acad. Sci. USA 113, E5982-E5991. 10.1073/pnas.1611477113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fàbregas N., Lozano-Elena F., Blasco-Escámez D., Tohge T., Martínez-Andújar C., Albacete A., Osorio S., Bustamante M., Riechmann J. L., Nomura T. et al. (2018). Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 9, 4680 10.1038/s41467-018-06861-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M., Bai M.-Y., Kim J.-G., Wang T., Oh E., Chen L., Park C. H., Son S.-H., Kim S.-K., Mudgett M. B. et al. (2014). The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern–triggered immunity in Arabidopsis. Plant Cell 26, 828-841. 10.1105/tpc.113.121111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E. G., Grau-Enguix F., Abbas M., Locascio A., Thomas S. G., Alabadí D. and Blázquez M. A. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 109, 13446-13451. 10.1073/pnas.1119992109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala S. S., Kim T.-W., He J.-X., Tang W., Deng Z., Bai M.-Y., Guan S., Lalonde S., Sun Y., Gendron J. M. et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13, 177-189. 10.1016/j.devcel.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Hann D. R., Ntoukakis V., Petutschnig E., Lipka V. and Rathjen J. P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423-429. 10.1016/j.cub.2009.01.054 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205-227. 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L. and Boller T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003-1011. 10.1016/S1097-2765(00)80265-8 [DOI] [PubMed] [Google Scholar]
- González-García M.-P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-García S., Russinova E. and Caño-Delgado A. I. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138, 849-859. 10.1242/dev.057331 [DOI] [PubMed] [Google Scholar]
- Gou X., Yin H., He K., Du J., Yi J., Xu S., Lin H., Clouse S. D. and Li J. (2012). Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 8, e1002452 10.1371/journal.pgen.1002452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove M. D., Spencer G. F., Rohwedder W. K., Mandava N., Worley J. F., Warthen J. D., Steffens G. L., Flippen-Anderson J. L. and Cook J. C. (1979). Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281, 216-217. 10.1038/281216a0 [DOI] [Google Scholar]
- Gudesblat G. E., Schneider-Pizoń J., Betti C., Mayerhofer J., Vanhoutte I., van Dongen W., Boeren S., Zhiponova M., de Vries S., Jonak C. et al. (2012). SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14, 548-554. 10.1038/ncb2471 [DOI] [PubMed] [Google Scholar]
- Guo R., Qian H., Shen W., Liu L., Zhang M., Cai C., Zhao Y., Qiao J. and Wang Q. (2013). BZR1 and BES1 participate in regulation of glucosinolate biosynthesis by brassinosteroids in Arabidopsis. J. Exp. Bot. 64, 2401-2412. 10.1093/jxb/ert094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y., Holland N., Butterfield C., Ubeda-Tomas S., Bennett M. J., Chory J. and Savaldi-Goldstein S. (2011). Brassinosteroid perception in the epidermis controls root meristem size. Development 138, 839-848. 10.1242/dev.061804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou J., Shan L. and Meng X. (2018). Plant cell surface receptor-mediated signaling – a common theme amid diversity. J. Cell Sci. 131, jcs209353 10.1242/jcs.209353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann U., Santiago J., Nicolet J., Olsson V., Spiga F. M., Hothorn L. A., Butenko M. A. and Hothorn M. (2018). Mechanistic basis for the activation of plant membrane receptor kinases by SERK-family coreceptors. Proc. Natl. Acad. Sci. USA 115, 3488-3493. 10.1073/pnas.1714972115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M., Belkhadir Y., Dreux M., Dabi T., Noel J. P., Wilson I. A. and Chory J. (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474, 467-471. 10.1038/nature10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Lee L. Y. C., Xia K., Yan Y. and Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19, 884-894. 10.1016/j.devcel.2010.10.024 [DOI] [PubMed] [Google Scholar]
- Hou S., Wang X., Chen D., Yang X., Wang M., Turrà D., Di Pietro A. and Zhang W. (2014). The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog. 10, e1004331 10.1371/journal.ppat.1004331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaert A., Zhang C., Tiwari M., Wang K., de Marcos Serrano A., Savatin D. V., Urs M. J., Zhiponova M. K., Gudesblat G. E., Vanhoutte I. et al. (2018). POLAR-guided signalling complex assembly and localization drive asymmetric cell division. Nature 563, 574-578. 10.1038/s41586-018-0714-x [DOI] [PubMed] [Google Scholar]
- Howe G. A. and Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41-46. 10.1146/annurev.arplant.59.032607.092825 [DOI] [PubMed] [Google Scholar]
- Hutten S. J., Hamers D. S., Aan den Toorn M., van Esse W., Nolles A., Bücherl C. A., de Vries S. C., Hohlbein J. and Borst J. W. (2017). Visualization of BRI1 and SERK3/BAK1 nanoclusters in Arabidopsis roots. PLoS ONE 12, e0169905 10.1371/journal.pone.0169905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañez C., Delker C., Martinez C., Bürstenbinder K., Janitza P., Lippmann R., Ludwig W., Sun H., James G. V., Klecker M. et al. (2018). Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Curr. Biol. 28, 303-310.e3. 10.1016/j.cub.2017.11.077 [DOI] [PubMed] [Google Scholar]
- Jiménez-Góngora T., Kim S.-K., Lozano-Durán R. and Zipfel C. (2015). Flg22-triggered immunity negatively regulates key BR biosynthetic genes. Front. Plant Sci. 6, 981 10.3389/fpls.2015.00981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D. G. and Dangl J. L. (2006). The plant immune system. Nature 444, 323-329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J. D. G., Shirasu K., Menke F., Jones A. et al. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54, 43-55. 10.1016/j.molcel.2014.02.021 [DOI] [PubMed] [Google Scholar]
- Kang S., Yang F., Li L., Chen H., Chen S. and Zhang J. (2015). The Arabidopsis transcription factor BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1 is a direct substrate of MITOGEN-ACTIVATED PROTEIN KINASE6 and regulates immunity. Plant Physiol. 167, 1076-1086. 10.1104/pp.114.250985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Rozhon W., Bigeard J., Pflieger D., Husar S., Pitzschke A., Teige M., Jonak C., Hirt H. and Poppenberger B. (2013). Brassinosteroid-regulated GSK3/Shaggy-like kinases phosphorylate mitogen-activated protein (MAP) kinase kinases, which control stomata development in Arabidopsis thaliana. J. Biol. Chem. 288, 7519-7527. 10.1074/jbc.M112.384453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.-J. and Russinova E. (2020). Brassinosteroid signalling. Curr. Biol. 30, R294-R298. 10.1016/j.cub.2020.02.011 [DOI] [PubMed] [Google Scholar]
- Kim T.-W., Guan S., Burlingame A. L. and Wang Z.-Y. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43, 561-571. 10.1016/j.molcel.2011.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-W., Michniewicz M., Bergmann D. C. and Wang Z.-Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419-422. 10.1038/nature10794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., Warpeha K. M. and Huber S. C. (2019). The brassinosteroid receptor kinase, BRI1, plays a role in seed germination and the release of dormancy by cold stratification. J. Plant Physiol. 241, 153031 10.1016/j.jplph.2019.153031 [DOI] [PubMed] [Google Scholar]
- Kondo Y., Ito T., Nakagami H., Hirakawa Y., Saito M., Tamaki T., Shirasu K. and Fukuda H. (2014). Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat. Commun. 5, 3504 10.1038/ncomms4504 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Lee J., Kim H. R., Chae W. B., Kim S.-J., Lim Y. P. and Oh M.-H. (2018). Brassinosteroids regulate glucosinolate biosynthesis in Arabidopsis thaliana. Physiol. Plant. 163, 450-458. 10.1111/ppl.12691 [DOI] [PubMed] [Google Scholar]
- Lee J., Han S., Lee H.-Y., Jeong B., Heo T.-Y., Hyun T. K., Kim K., Je B. I., Lee H., Shim D. et al. (2019). Brassinosteroids facilitate xylem differentiation and wood formation in tomato. Planta 249, 1391-1403. 10.1007/s00425-019-03094-6 [DOI] [PubMed] [Google Scholar]
- Lei J., Finlayson S. A., Salzman R. A., Shan L. and Zhu-Salzman K. (2014). BOTRYTIS-INDUCED KINASE1 modulates Arabidopsis resistance to green peach aphids via PHYTOALEXIN DEFICIENT4. Plant Physiol. 165, 1657-1670. 10.1104/pp.114.242206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.-F. and He J.-X. (2016). BZR1 interacts with HY5 to mediate brassinosteroid-and light-regulated cotyledon opening in Arabidopsis in darkness. Mol. Plant 9, 113-125. 10.1016/j.molp.2015.08.014 [DOI] [PubMed] [Google Scholar]
- Li Z. and He Y. (2020). Roles of brassinosteroids in plant reproduction. Int. J. Mol. Sci. 21, 872 10.3390/ijms21030872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. and Nam K. H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295, 1299-1301. 10.1126/science.1065769 [DOI] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K. A., Doke J. T., Tax F. E. and Walker J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213-222. 10.1016/S0092-8674(02)00812-7 [DOI] [PubMed] [Google Scholar]
- Li Q.-F., Wang C., Jiang L., Li S., Sun S. S. M. and He J.-X. (2012). An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 5, ra72 10.1126/scisignal.2002908 [DOI] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y. et al. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329-338. 10.1016/j.chom.2014.02.009 [DOI] [PubMed] [Google Scholar]
- Li H., Ye K., Shi Y., Cheng J., Zhang X. and Yang S. (2017). BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol. Plant 10, 545-559. 10.1016/j.molp.2017.01.004 [DOI] [PubMed] [Google Scholar]
- Liao K., Peng Y.-J., Yuan L.-B., Dai Y.-S., Chen Q.-F., Yu L.-J., Bai M.-Y., Zhang W.-Q., Xie L.-J. and Xiao S. (2020). Brassinosteroids antagonize jasmonate-activated plant defense responses through BRI1-EMS-SUPPRESSOR1 (BES1). Plant Physiol. 182, 1066-1082. 10.1104/pp.19.01220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Lu D., Gao X., Jiang S., Ma X., Wang Z., Mengiste T., He P. and Shan L. (2013). Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA 110, 12114-12119. 10.1073/pnas.1302154110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Li B., Lu D., Chen S., Zhu N., He P. and Shan L. (2014). Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc Natl Acad Sci. USA 111, 3632-3637. 10.1073/pnas.1318817111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wu Y., Yang F., Zhang Y., Chen S., Xie Q., Tian X. and Zhou J.-M. (2013). BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. USA 110, 6205-6210. 10.1073/pnas.1215543110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Sonbol F.-M., Huot B., Gu Y., Withers J., Mwimba M., Yao J., He S. Y. and Dong X. (2016). Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. commun. 7, 13099 10.1038/ncomms13099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.-L., Du L., Huang Y., Gao S.-M. and Yu M. (2017). Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 17, 47 10.1186/s12862-017-0891-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R. and Zipfel C. (2015). Trade-off between growth and immunity: role of brassinosteroids. Trends Plant Sci. 20, 12-19. 10.1016/j.tplants.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Lozano-Durán R., Macho A. P., Boutrot F., Segonzac C., Somssich I. E. and Zipfel C. (2013). The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLife 2, e00983 10.7554/eLife.00983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L. and He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107, 496-501. 10.1073/pnas.0909705107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Claus L. A. N., Leslie M. E., Tao K., Wu Z., Liu J., Yu X., Li B., Zhou J., Savatin D. V. et al. (2020). Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 581, 199-203. 10.1038/s41586-020-2210-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majhi B. B., Sreeramulu S. and Sessa G. (2019). BRASSINOSTEROID-SIGNALING KINASE5 associates with immune receptors and is required for immune responses. Plant Physiol. 180, 1166-1184. 10.1104/pp.18.01492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovsky F. G., Fangel J. U. and Willats W. G. T. (2014). The role of the cell wall in plant immunity. Front. Plant Sci. 5, 178 10.3389/fpls.2014.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaji T., Yamagami A., Kume N., Sakuta M., Osada H., Asami T., Arimoto Y. and Nakano T. (2014). Brassinosteroid-related transcription factor BIL1/BZR1 increases plant resistance to insect feeding. Biosci. Biotechnol. Biochem. 78, 960-968. 10.1080/09168451.2014.910093 [DOI] [PubMed] [Google Scholar]
- Monaghan J. and Zipfel C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349-357. 10.1016/j.pbi.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Müssig C., Shin G.-H. and Altmann T. (2003). Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 133, 1261-1271. 10.1104/pp.103.028662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar K., Kyndt T., Hause B., Höfte M. and Gheysen G. (2013). Brassinosteroids suppress rice defense against root-knot nematodes through antagonism with the jasmonate pathway. Mol. Plant-Microbe Interact. 26, 106-115. 10.1094/MPMI-05-12-0108-FI [DOI] [PubMed] [Google Scholar]
- Nolan T. M., Vukašinović N., Liu D., Russinova E. and Yin Y. (2020). Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32, 295-318. 10.1105/tpc.19.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.-Y., Ryu H., Hwang I. and Wang Z.-Y. (2014). TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat. Commun. 5, 4140 10.1038/ncomms5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Morea F. A., Savatin D. V., Dejonghe W., Kumar R., Luo Y., Adamowski M., Van den Begin J., Dressano K., Pereira de Oliveira G., Zhao X. et al. (2016). Danger-associated peptide signaling in Arabidopsis requires clathrin. Proc. Natl. Acad. Sci. USA 113, 11028-11033. 10.1073/pnas.1605588113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Morea F. A. and Reyes-Bermudez A. A. (2019). Endogenous peptides: key modulators of plant immunity. In Bioactive Molecules in Plant Defense (ed. Jogaiah S. and Abdelrahman M.), pp. 159-177. Cham, Switzerland: Springer. [Google Scholar]
- Ott T. (2017). Membrane nanodomains and microdomains in plant-microbe interactions. Curr. Opin. Plant Biol. 40, 82-88. 10.1016/j.pbi.2017.08.008 [DOI] [PubMed] [Google Scholar]
- Pandey A., Devi L. L. and Singh A. P. (2020). Emerging roles of brassinosteroid in nutrient foraging. Plant Science 296, 110474 10.1016/j.plantsci.2020.110474 [DOI] [PubMed] [Google Scholar]
- Park C. H., Youn J.-H., Xu S.-L., Kim J.-G., Bi Y., Xu N., Mudgett M. B., Kim S.-K., Kim T.-W. and Wang Z.-Y. (2019). BSU1 family phosphatases mediate Flagellin-FLS2 signaling through a specific phosphocode. bioRxiv, 685610 10.1101/685610. [Google Scholar]
- Planas-Riverola A., Gupta A., Betegón-Putze I., Bosch N., Ibañes M. and Caño-Delgado A. I. (2019). Brassinosteroid signaling in plant development and adaptation to stress. Development 146, dev151894 10.1242/dev.151894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Précigout P.-A., Claessen D., Makowski D. and Robert C. (2020). Does the latent period of leaf fungal pathogens reflect their trophic type? A meta-analysis of biotrophs, hemibiotrophs, and necrotrophs. Phytopathology 110, 345-361. 10.1094/PHYTO-04-19-0144-R [DOI] [PubMed] [Google Scholar]
- Qi Y., Tsuda K., Glazebrook J. and Katagiri F. (2011). Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol. Plant Pathol. 12, 702-708. 10.1111/j.1364-3703.2010.00704.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin M., Abad P. and Favery B. (2013). Plant parasitic nematode effectors target host defense and nuclear functions to establish feeding cells. Front. Plant Sci. 4, 53 10.3389/fpls.2013.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Willige B. C., Jaillais Y., Geng S., Park M. Y., Gray W. M. and Chory J. (2019). BRASSINOSTEROID-SIGNALING KINASE 3, a plasma membrane-associated scaffold protein involved in early brassinosteroid signaling. PLoS Genet. 15, e1007904 10.1371/journal.pgen.1007904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich-Griffin C., Eichmann R., Reitz M. U., Hermann S., Woolley-Allen K., Brown P. E., Wiwatdirekkul K., Esteban E., Pasha A., Kogel K.-H. et al. (2020). Regulation of cell type-specific immunity networks in Arabidopsis roots. Plant Cell 32, 2742-2762. 10.1105/tpc.20.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M. and Jones J. D. G. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317-343. 10.1146/annurev-phyto-073009-114447 [DOI] [PubMed] [Google Scholar]
- Rodriguez P. A., Escudero-Martinez C. and Bos J. I. B. (2017). An aphid effector targets trafficking protein VPS52 in a host-specific manner to promote virulence. Plant Physiol. 173, 1892-1903. 10.1104/pp.16.01458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., Holton N., Malinovsky F. G., Tör M., de Vries S. and Zipfel C. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2440-2455. 10.1105/tpc.111.084301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y., Halat L. S., Khan D., Jancowski S., Ambrose C., Belmonte M. F. and Wasteneys G. O. (2018). The microtubule-associated protein CLASP sustains cell proliferation through a brassinosteroid signaling negative feedback loop. Curr. Biol. 28, 2718-2729.e5. 10.1016/j.cub.2018.06.048 [DOI] [PubMed] [Google Scholar]
- Russinova E., Borst J.-W., Kwaaitaal M., Caño-Delgado A., Yin Y., Chory J. and de Vries S. C. (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16, 3216-3229. 10.1105/tpc.104.025387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J., Henzler C. and Hothorn M. (2013). Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341, 889-892. 10.1126/science.1242468 [DOI] [PubMed] [Google Scholar]
- Schwessinger B., Roux M., Kadota Y., Ntoukakis V., Sklenar J., Jones A. and Zipfel C. (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7, e1002046 10.1371/journal.pgen.1002046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnejat-Bushehri S., Nobmann B., Devi Allu A. and Balazadeh S. (2016a). JUB1 suppresses Pseudomonas syringae-induced defense responses through accumulation of DELLA proteins. Plant Signal. Behav. 11, e1181245 10.1080/15592324.2016.1181245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnejat-Bushehri S., Tarkowska D., Sakuraba Y. and Balazadeh S. (2016b). Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat. Plants 2, 16013 10.1038/nplants.2016.13 [DOI] [PubMed] [Google Scholar]
- She J., Han Z., Kim T.-W., Wang J., Cheng W., Chang J., Shi S., Wang J., Yang M., Wang Z.-Y. et al. (2011). Structural insight into brassinosteroid perception by BRI1. Nature 474, 472-476. 10.1038/nature10178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Shen Q., Qi Y., Yan H., Nie H., Chen Y., Zhao T., Katagiri F. and Tang D. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25, 1143-1157. 10.1105/tpc.112.107904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenaga A. M. and Argueso C. T. (2016). No hormone to rule them all: interactions of plant hormones during the responses of plants to pathogens. Semin. Cell Dev. Biol. 56, 174-189. 10.1016/j.semcdb.2016.06.005 [DOI] [PubMed] [Google Scholar]
- Shin S. Y., Chung H., Kim S. Y. and Nam K. H. (2016). BRI1-EMS-suppressor 1 gain-of-function mutant shows higher susceptibility to necrotrophic fungal infection. Biochem. Biophys. Res. Commun. 470, 864-869. 10.1016/j.bbrc.2016.01.128 [DOI] [PubMed] [Google Scholar]
- Singh A. P., Fridman Y., Holland N., Ackerman-Lavert M., Zananiri R., Jaillais Y., Henn A. and Savaldi-Goldstein S. (2018). Interdependent nutrient availability and steroid hormone signals facilitate root growth plasticity. Dev. Cell 46, 59-72.e4. 10.1016/j.devcel.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Song L., Zhou X.-Y., Li L., Xue L.-J., Yang X. and Xue H.-W. (2009). Genome-wide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis. Mol. Plant 2, 755-772. 10.1093/mp/ssp039 [DOI] [PubMed] [Google Scholar]
- Spoel S. H. and Dong X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89-100. 10.1038/nri3141 [DOI] [PubMed] [Google Scholar]
- Sreeramulu S., Mostizky Y., Sunitha S., Shani E., Nahum H., Salomon D., Hayun L. B., Gruetter C., Rauh D., Ori N. et al. (2013). BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 74, 905-919. 10.1111/tpj.12175 [DOI] [PubMed] [Google Scholar]
- Stampfl H., Fritz M., Dal Santo S. and Jonak C. (2016). The GSK3/Shaggy-like kinase ASKα contributes to pattern-triggered immunity. Plant Physiol. 171, 1366-1377. 10.1104/pp.15.01741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber C. M. and McCourt P. (2001). A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 125, 763-769. 10.1104/pp.125.2.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Han Z., Tang J., Hu Z., Chai C., Zhou B. and Chai J. (2013). Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 23, 1326-1329. 10.1038/cr.2013.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Nitta Y., Zhang Q., Wu D., Tian H., Lee J. S. and Zhang Y. (2018). Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep. 19, e45324 10.15252/embr.201745324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaoki D., Seo S., Yamada S., Kano A., Miyamoto A., Shishido H., Miyoshi S., Taniguchi S., Akimitsu K. and Gomi K. (2013). Jasmonic acid and salicylic acid activate a common defense system in rice. Plant Signal. Behav. 8, e24260 10.4161/psb.24260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.-W., Oses-Prieto J. A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A. L. and Wang Z.-Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321, 557-560. 10.1126/science.1156973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Yuan M., Wang R., Yang Y., Wang C., Oses-Prieto J. A., Kim T.-W., Zhou H.-W., Deng Z., Gampala S. S. et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13, 124-131. 10.1038/ncb2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Han Z., Sun Y., Zhang H., Gong X. and Chai J. (2015). Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res. 25, 110-120. 10.1038/cr.2014.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H. (2003). Fresh insights into processes of nonhost resistance. Curr. Opin. Plant Biol. 6, 351-357. 10.1016/S1369-5266(03)00063-3 [DOI] [PubMed] [Google Scholar]
- Ton J., Van Pelt J. A., Van Loon L. C. and Pieterse C. M. J. (2002). Differential effectiveness of salicylate-dependent and jasmonate/ethylene-dependent induced resistance in Arabidopsis. Mol. Plant-Microbe Interact. 15, 27-34. 10.1094/MPMI.2002.15.1.27 [DOI] [PubMed] [Google Scholar]
- Vogler F., Schmalzl C., Englhart M., Bircheneder M. and Sprunck S. (2014). Brassinosteroids promote Arabidopsis pollen germination and growth. Plant Reprod. 27, 153-167. 10.1007/s00497-014-0247-x [DOI] [PubMed] [Google Scholar]
- Vragović K., Sela A., Friedlander-Shani L., Fridman Y., Hacham Y., Holland N., Bartom E., Mockler T. C. and Savaldi-Goldstein S. (2015). Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. Proc. Natl. Acad. Sci. USA 112, 923-928. 10.1073/pnas.1417947112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W.-L., Zhang L., Pruitt R., Zaidem M., Brugman R., Ma X., Krol E., Perraki A., Kilian J., Grossmann G. et al. (2019). Comparing Arabidopsis receptor kinase and receptor protein-mediated immune signaling reveals BIK1-dependent differences. New Phytol. 221, 2080-2095. 10.1111/nph.15497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. and Chory J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313, 1118-1122. 10.1126/science.1127593 [DOI] [PubMed] [Google Scholar]
- Wang Z.-Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T. et al. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505-513. 10.1016/S1534-5807(02)00153-3 [DOI] [PubMed] [Google Scholar]
- Wang X., Kota U., He K., Blackburn K., Li J., Goshe M. B., Huber S. C. and Clouse S. D. (2008). Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15, 220-235. 10.1016/j.devcel.2008.06.011 [DOI] [PubMed] [Google Scholar]
- Wang X., Zhang J., Yuan M., Ehrhardt D. W., Wang Z. and Mao T. (2012). Arabidopsis MICROTUBULE DESTABILIZING PROTEIN40 is involved in brassinosteroid regulation of hypocotyl elongation. Plant Cell 24, 4012-4025. 10.1105/tpc.112.103838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Li H., Lv X., Chen T., Li R., Xue Y., Jiang J., Jin B., Baluška F., Šamaj J. et al. (2015). Spatiotemporal dynamics of the BRI1 receptor and its regulation by membrane microdomains in living Arabidopsis cells. Mol. Plant 8, 1334-1349. 10.1016/j.molp.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Wang J., Hu M., Wang J., Qi J., Han Z., Wang G., Qi Y., Wang H.-W., Zhou J.-M. and Chai J. (2019a). Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, eaav5870 10.1126/science.aav5870 [DOI] [PubMed] [Google Scholar]
- Wang S., McLellan H., Bukharova T., He Q., Murphy F., Shi J., Sun S., van Weymers P., Ren Y., Thilliez G. et al. (2019b). Phytophthora infestans RXLR effectors act in concert at diverse subcellular locations to enhance host colonization. J. Exp. Bot. 70, 343-356. 10.1093/jxb/ery360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., He R., Yang F., Zou L., Yi K., Lin H. and Zhang D. (2020). Brassinosteroids are involved in ethylene-induced Pst DC3000 resistance in Nicotiana benthamiana. Plant Biol. 22, 309-316. 10.1111/plb.13074 [DOI] [PubMed] [Google Scholar]
- Xu P., Xu S.-L., Li Z.-J., Tang W., Burlingame A. L. and Wang Z.-Y. (2014). A brassinosteroid-signaling kinase interacts with multiple receptor-like kinases in Arabidopsis. Mol. Plant 7, 441-444. 10.1093/mp/sst105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y. and Huffaker A. (2011). Endogenous peptide elicitors in higher plants. Curr. Opin. Plant Biol. 14, 351-357. 10.1016/j.pbi.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Yan H., Zhao Y., Shi H., Li J., Wang Y. and Tang D. (2018). BRASSINOSTEROID-SIGNALING KINASE1 phosphorylates MAPKKK5 to regulate immunity in Arabidopsis. Plant Physiol. 176, 2991-3002. 10.1104/pp.17.01757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Liu S., Tang B., Chen J., Xie Z., Nolan T. M., Jiang H., Guo H., Lin H.-Y., Li L. et al. (2017). RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 8, 14573 10.1038/ncomms14573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wang Z.-Y., Mora-Garcia S., Li J., Yoshida S., Asami T. and Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181-191. 10.1016/S0092-8674(02)00721-3 [DOI] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P. et al. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65, 634-646. 10.1111/j.1365-313X.2010.04449.x [DOI] [PubMed] [Google Scholar]
- Yu X., Feng B., He P. and Shan L. (2017). From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55, 109-137. 10.1146/annurev-phyto-080516-035649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M.-H., Zhao Z.-Z. and He J.-X. (2018). Brassinosteroid signaling in plant–microbe interactions. Int. J. Mol. Sci. 19, 4091 10.3390/ijms19124091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T., Deng X., Zhou X., Zhu L., Zou L., Li P., Zhang D. and Lin H. (2016). Ethylene and hydrogen peroxide are involved in brassinosteroid-induced salt tolerance in tomato. Sci. Rep. 6, 35392 10.1038/srep35392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.-Y., Li Y., Cao D.-M., Yang H., Oh E., Bi Y., Zhu S. and Wang Z.-Y. (2017). The F-box protein KIB1 mediates brassinosteroid-induced inactivation and degradation of GSK3-like kinases in Arabidopsis. Mol. Cell 66, 648-657.e4. 10.1016/j.molcel.2017.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]