ABSTRACT
In sexually reproducing metazoans, spermatogenesis is the process by which uncommitted germ cells give rise to haploid sperm. Work in model systems has revealed mechanisms controlling commitment to the sperm fate, but how this fate is subsequently executed remains less clear. While studying the well-established role of the conserved nuclear hormone receptor transcription factor, NHR-23/NR1F1, in regulating C. elegans molting, we discovered that NHR-23/NR1F1 is also constitutively expressed in developing primary spermatocytes and is a critical regulator of spermatogenesis. In this novel role, NHR-23/NR1F1 functions downstream of the canonical sex-determination pathway. Degron-mediated depletion of NHR-23/NR1F1 within hermaphrodite or male germlines causes sterility due to an absence of functional sperm, as depleted animals produce arrested primary spermatocytes rather than haploid sperm. These spermatocytes arrest in prometaphase I and fail to either progress to anaphase or attempt spermatid-residual body partitioning. They make sperm-specific membranous organelles but fail to assemble their major sperm protein into fibrous bodies. NHR-23/NR1F1 appears to function independently of the known SPE-44 gene regulatory network, revealing the existence of an NHR-23/NR1F1-mediated module that regulates the spermatogenesis program.
KEY WORDS: C. elegans, Spermatogenesis, NHR-23, Nuclear hormone receptor, Meiosis, Auxin-inducible degron
Summary: NHR-23, a well-characterized regulator of C. elegans molting, is identified as a critical regulator of spermatogenesis in both hermaphrodite and male animals.
INTRODUCTION
Transcription factors perform diverse roles in the development of specialized cell types. Many work synergistically in gene regulatory networks to execute complex programs of cell differentiation (Drapek et al., 2017; Levine and Davidson, 2005; Medwig-Kinney et al., 2020). These networks coordinate the expression of enzymes, modifiers and structural proteins that characterize a specific cell type. One challenge of studying transcription factors is that they often function with distinct co-regulators in multiple developmental contexts. For example, in the nematode Caenorhabditis elegans, the GATA-1 transcription factor ELT-1 is essential for epidermal cell fates during embryogenesis (Page et al., 1997) but later regulates the expression of multiple genes required for spermatogenesis (del Castillo-Olivares et al., 2009). In knockout or null mutants, if disruption of the first developmental event results in embryonic or larval lethality, later functions will be missed. Traditionally, this challenge has been addressed using conditional mutants, mRNA depletion by RNA interference or genetic mosaics. A powerful new addition to this investigational toolbox is the auxin-inducible degron (AID) system for the conditional disruption of protein function (Nishimura et al., 2009; Zhang et al., 2015). In this system, a gene of interest tagged with an AID sequence is co-expressed with a TIR1 F-box protein driven by a tissue-specific promoter. In the presence of the plant hormone auxin, the TIR1-containing E3 ubiquitin ligase polyubiquitylates the AID sequence, leading to degradation of the AID-tagged protein. This conditional protein depletion system is ideal for characterizing the distinct functions of an essential, multi-functional transcription factor.
One motivation of developing the AID system in C. elegans was to study events in meiosis required for gametogenesis (Zhang et al., 2015). During gametogenesis, stem cell precursors enact a developmental program producing highly specialized haploid sperm or oocytes. C. elegans is a powerful model for studying gametogenesis as hermaphrodites produce a limited number of sperm before switching exclusively to producing oocytes, whereas males produce sperm continuously (Ellis and Schedl, 2007) (Fig. 1A,B). Extensive studies of C. elegans sex determination (Barton and Kimble, 1990; Ellis and Schedl, 2007) identified the transcription factor TRA-1, a homolog of GLI and cubitus interruptus, as the key regulator of somatic sex determination and the spermatocyte/oocyte decision (Hodgkin, 1987; Schedl et al., 1989; Zarkower and Hodgkin, 1992). Within the germline, TRA-1 promotes oogenesis and inhibits spermatogenesis by repressing expression of two germline-specific, RNA-binding proteins (FOG-1 and FOG-3) (Chen and Ellis, 2000; Jin et al., 2001). A different RNA-binding translational repressor, PUF-8, maintains sperm fate (Subramaniam and Seydoux, 2003).
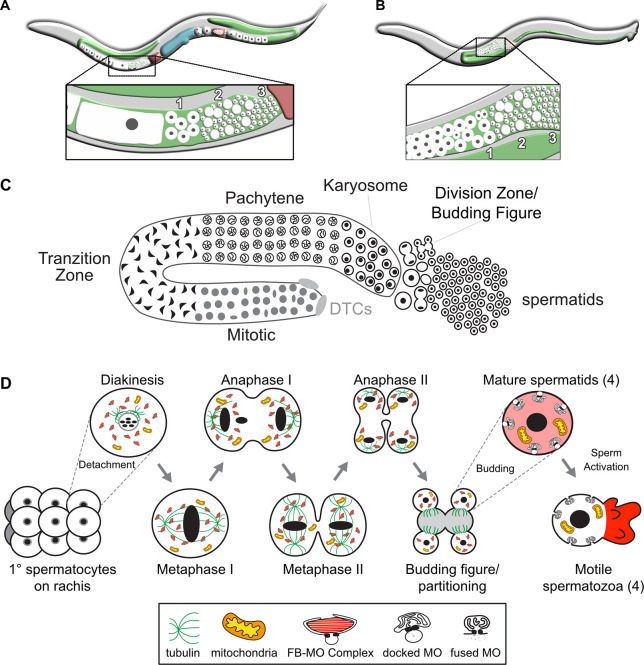
Fig. 1.
Overview of C. elegans spermatogenesis. (A,B) Cartoons depicting a young adult C. elegans hermaphrodite (A) and male (B), and their respective germlines. The hermaphrodite germline (A) is transitioning from spermatogenesis to oogenesis. The enlarged views highlight the linear arrangement of the primary spermatocytes (1), residual bodies (RBs) (2) and mature haploid spermatids (3). (C) Stylized cartoon of a surface view of the male germline highlighting its overall linear organization. Mitotic proliferation of the germline stem cells is maintained by two somatic distal tip cells (DTCs) that form the germ cell niche. The early events of meiotic prophase, including homolog pairing and formation of the synaptonemal complex, occur in the transition zone. Following an extended pachytene stage, spermatocytes enter a karyosome stage before mature spermatocytes detach from the syncytial germline and divide meiotically. The first meiotic division is often incomplete, leaving secondary spermatocytes linked by a cytoplasmic connection. Following anaphase II, the spermatocytes morph into budding figures that split into residual bodies and haploid spermatids. (D) Details of the meiotic divisions and post-meiotic partitioning event. Once spermatocytes detach from the germline syncytium, they pass through a brief diakinesis stage before undergoing nuclear envelope breakdown and initiating meiotic divisions. During the post-meiotic partitioning event, microtubules become acentrosomal and localize to the developing residual body (Winter et al., 2017). Components retained in the spermatids include fibrous body-membranous organelles (FB-MO), mitochondria, chromatin and centrioles. Components discarded within the RB include the tubulin, actin, endoplasmic reticulum and ribosomes of the cell; mature sperm are thus both transcriptionally and translationally inactive. Following separation from the RB, FBs disassemble and release unpolymerized MSP and the MOs dock with the plasma membrane. Males store sperm in this inactive spermatid state. During spermatid activation, MOs fuse with the plasma membrane and unpolymerized MSP localizes to the pseudopod, where it forms fibers that are required for spermatozoon motility.
Beyond the initial sperm fate decision, the subsequent control of sperm differentiation remains poorly understood. Understanding the gene networks that regulate sperm differentiation offers an inroad into this question. Many C. elegans germline expressed genes primarily rely on mRNA 3′ untranslated regions (UTRs), not promoters, to ensure expression at the correct time and location during germline development (Merritt et al., 2008). In contrast, spermatocyte promoters provide spatiotemporal control of gene expression, making transcription factors direct regulators of sperm differentiation (Merritt et al., 2008). The C. elegans transcription factor SPE-44 is widely distributed on autosomes of developing spermatocytes and directly regulates other transcription factors, e.g. elt-1, as well as genes whose expression is required for sperm function, such as kinases and structural components (Kulkarni et al., 2012). Which other transcription factors contribute to gene expression regulation in C. elegans spermatogenesis and to what degree they control similar or disparate sets of genes remains unknown.
Spermatogenesis is a complex cellular process involving a host of dynamic subcellular events coordinated by a large number of genes. Because of its linear organization, the full developmental sequence of spermatogenesis can be analyzed in individual gonads (Fig. 1C, Chu and Shakes, 2013). Undifferentiated germ cells proliferate mitotically at the distal end while mature primary spermatocytes divide meiotically at the proximal end (Fig. 1C,D). The commitment to sperm fate occurs as undifferentiated germ cells exit mitosis and initiate meiotic homolog pairing. Transcription of genes required for spermatogenesis and translation of most sperm proteins occurs within an extended pachytene zone. During the subsequent karyosome stage, global transcription ceases as chromosomes detach from the nuclear envelope and coalesce into a central mass (Shakes et al., 2009). Although transcription stops, translation and assembly of sperm-specific complexes continue (Chu and Shakes, 2013; Shakes et al., 2009). Throughout meiotic prophase, each individual spermatocyte is linked to a shared central rachis via a syncytial connection (Fig. 1D). Following the karyosome stage, primary spermatocytes detach from the rachis before undergoing the meiotic divisions. Post-meiotic development is limited to a post-meiotic partitioning event. Following anaphase II, haploid spermatids bud from a central residual body (Fig. 1D). Then during sperm activation, these sperm become motile by extending a pseudopod (Chu and Shakes, 2013; Ward et al., 1981). Despite having large collections of mutants affecting the processes of sperm differentiation (Nishimura and L'Hernault, 2010), how these events are coordinated is not well understood.
C. elegans NHR-23/NR1F1 (hereafter referred to as NHR-23) is a nuclear hormone receptor (NHR) transcription factor previously shown to be involved in embryonic epidermal development (Kostrouchova et al., 1998), larval molting (Kostrouchova et al., 1998, 2001) and diet-induced developmental acceleration (Macneil et al., 2013). The mammalian orthologs (RORs/NR1F) regulate circadian rhythms (Akashi and Takumi, 2005; André et al., 1998; Sato et al., 2004) and the insect ortholog (DHR3) regulates metamorphosis (Kageyama et al., 1997; Lam et al., 1997; White et al., 1997). Loss or reduction of nhr-23 activity leads to embryonic lethality, larval arrest or larval lethality (Kostrouchova et al., 1998, 2001; Kouns et al., 2011). Lethality following nhr-23 inactivation occurs early in development, masking an unanticipated later role for NHR-23 during spermatogenesis, which we describe here. Our results are the first to show that nhr-23 is expressed within developing spermatocytes and functions downstream of the canonical sex-determination pathway. Depletion of NHR-23 protein results in sperm-specific sterility. Affected spermatocytes arrest in meiosis I prometaphase and exhibit developmental defects in biogenesis of an essential sperm-specific organelle. NHR-23 promotes spermatogenesis in a pathway that is distinct from that of SPE-44. Our work provides new insights into how transcription factors coordinately execute the spermatogenesis program.
RESULTS
NHR-23:GFP is expressed in sperm-producing germlines
To explore potential functions of NHR-23 beyond molting, we wanted to know where and when NHR-23 is expressed. To address this issue, we used CRISPR/Cas9-mediated genome editing to introduce a multifunctional GFP::AID*::3×FLAG sequence into the endogenous nhr-23 locus, tagging all known isoforms. AID* is a minimal, 44 amino acid auxin-inducible degron sequence. Consistent with previous promoter reporters and immunolabeling (Kostrouchova et al., 1998), NHR-23::GFP expression was observed in epidermal cell nuclei of 1.5-stage embryos, and in hypodermal and seam cells of developing larvae (unpublished data). Somatic cell expression persisted in L4 larvae, specifically in hypodermal cells of the head, vulval precursor cells of hermaphrodites, and hypodermal and tail cells of males (Fig. 2A). Notably, NHR-23::GFP was also expressed in the sperm-producing germlines of males and L4 hermaphrodites, but not in hermaphrodites producing only oocytes (Fig. 2B). Previous studies missed this spermatocyte expression as transgenes are frequently silenced in the germline (Kelly et al., 1997) and immunolabeling was limited to embryos (Kostrouchova et al., 1998, 2001).
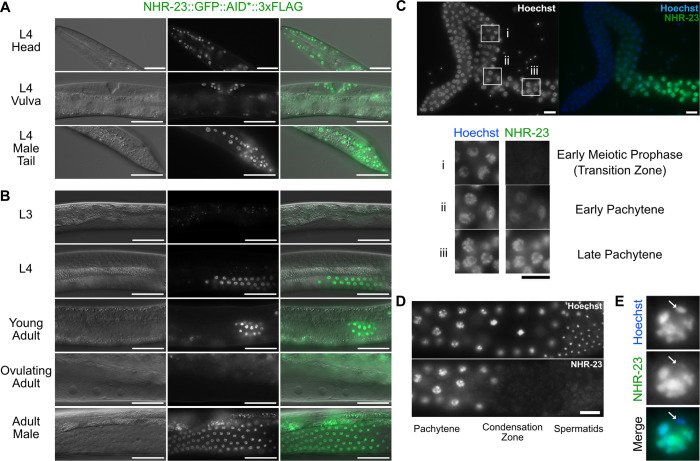
Fig. 2.
NHR-23::GFP::AID*::3xFLAG is expressed in somatic cells throughout development and in spermatogenic germlines. A strain carrying GFP::AID*::3xFLAG knocked-in to the endogenous nhr-23 gene to produce a C-terminal translational fusion to all known nhr-23 isoforms was used to monitor endogenous NHR-23 expression. (A) Representative DIC and GFP images of NHR-23::GFP::AID*::3xFLAG L4 larvae, specifically hypodermal cells of a hermaphrodite head, vulval precursor cells and seam/hypodermal cells of the male tail. (B) Representative DIC and GFP images of NHR-23::GFP::AID*::3xFLAG in pachytene cells of the germline in L3, L4, young adult, ovulating adult and adult male worms. (C) Fluorescent images of NHR-23::GFP::AID*::3xFLAG in a dissected adult male germline. (i-iii) Cells in early meiotic prophase (transition zone) (i), and in early (ii) and late (iii) pachytene. (D) Representative fluorescent images of NHR-23::GFP::AID*::3xFLAG in a late pachytene male germline. (E) Representative image of a late pachytene nucleus expressing NHR-23::GFP::AID*::3xFLAG. The location of the X chromosome is indicated with a white arrow. Scale bars: 40 µm in A,B; 10 µm in C-E. A minimum of 12 P0 animals were analyzed in A-C. Nuclei are visualized using Hoechst stain in C-E.
In males (and sperm-producing hermaphrodites), NHR-23::GFP was first observed in early pachytene spermatocytes (Fig. 2C), increased in intensity through late pachytene (Fig. 2B,C) and became undetectable during late meiotic prophase with onset of chromatin condensation (Fig. 2D). NHR-23 was undetectable in meiotically dividing spermatocytes or mature spermatids. This overall pattern is similar to that reported for SPE-44 (Kulkarni et al., 2012). Also like SPE-44, NHR-23::GFP labeled most chromosomes along their entire length yet failed to label a region of DNA in each pachytene stage spermatocyte (Fig. 2E). Presumably NHR-23 is not labeling the X chromosome, which is transcriptionally silent during spermatocyte meiosis (Kelly et al., 2002).
Auxin-induced NHR-23-depletion during or before spermatogenesis causes infertility in males and hermaphrodites
Because both nhr-23 null mutants and RNAi knockdowns of L1 larvae arrest as early larvae (Kostrouchova et al., 1998, 2001), previous studies failed to assess potential NHR-23 functions in the germline. Conversely, RNAi treatment of L3/L4 hermaphrodites or adult males failed to detectably deplete NHR-23 in the germline (J.M.R. and J.D.W., unpublished). We therefore turned to auxin-inducible degradation (Nishimura et al., 2009; Zhang et al., 2015), which allows for tissue-specific temporally regulated depletion of proteins. Our multifunctional NHR-23 knock-in construct contained the AID* sequence necessary for auxin- and TIR-dependent depletion of target proteins. A separate TIR1 transgene included the strong mex-5 promoter (Schubert et al., 2000), which would allow NHR-23-depletion primarily in the germline. Hermaphrodites expressing either mex-5::TIR1 or nhr-23::GFP::AID*::3xFLAG grew to adulthood and were fully fertile whether grown on control or 4 mM auxin media (Fig. 3A). Hermaphrodites expressing both constructs were fully fertile on control media but produced no progeny when grown on media with 4 mM auxin. Crossing NHR-23-depleted hermaphrodites to wild-type males restored fertility (Fig. 3B). These results suggest NHR-23 is necessary for sperm production in hermaphrodites.
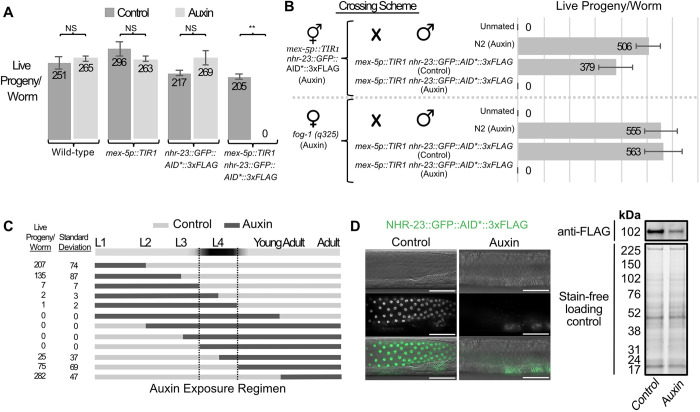
Fig. 3.
NHR-23-depletion in the germline during periods of spermatogenesis causes infertility in hermaphrodites and males. (A) Average number of live progeny produced by wild-type, mex-5p::TIR1, nhr-23::GFP::AID*::3xFLAG or mex-5p::TIR1 nhr-23::GFP::AID*::3xFLAG hermaphrodites grown from L1 through adulthood on MYOB media with or without 4 mM auxin. Student's t-test was performed for each genotype comparing brood sizes on control media versus 4 mM auxin. P>0.01 was considered non-significant (NS). **P<0.00001. n=12 for each brood size. Data are mean±s.e.m. (B) Average number of live progeny produced by mex-5p::TIR1 nhr-23::GFP::AID*::3xFLAG or fog-1(q325) hermaphrodites grown from L1 on MYOB media with 4 mM auxin. Hermaphrodites were either unmated or crossed to males of the indicated genotype; males were grown from L1 to adulthood on control or 4 mM auxin media (n=12). Data are mean±s.e.m. (C) Average number of live progeny produced by mex-5p::TIR1 nhr-23::GFP::AID*::3xFLAG hermaphrodites shifted on or off 4 mM auxin at different points in development. Dark horizontal bands represent growth on media with 4 mM auxin and light horizontal bands represent growth on control media lacking auxin. n=12 for each condition. (D) DIC and fluorescent images of mex-5p::TIR1 nhr-23::GFP::AID*::3xFLAG adult male germlines after animals were grown from L1 on MYOB media±4 mM auxin. NHR-23-depletion results in minimally detectable GFP signal by fluorescence microscopy, but FLAG signal remains detectable in western blots. Scale bars: 40 μm. Anti-FLAG immunoblot analyses of lysates are from synchronized male mex-5p::TIR1 nhr-23::GFP::AID*::3xFLAG animals grown on control or 4 mM auxin media. Marker size (in kDa) is provided. Predicted size of the fusion protein is 98.7 kDa, based on the NHR-23 isoform expressed in adults in a Nanopore direct RNA-sequencing dataset (Roach et al., 2020). Stain-free analysis, which visualizes total protein on the membrane is provided as a loading control.
We next carried out a set of experiments to determine when during development NHR-23 is necessary for fertility. Synchronized mex-5::TIR, nhr-23::GFP::AID*::3xFLAG hermaphrodites were placed on control or 4 mM auxin media (Fig. 3C). Hermaphrodites were shifted from control media to auxin, or vice versa. A short auxin exposure early in larval development (L1-L2) or continuous exposure after young adulthood had little or no effect on hermaphrodite self-fertility. The auxin-sensitive period was the L4 larval stage, with auxin exposure causing partially penetrant or complete self-infertility. Self-infertility was more pronounced in ‘shift-off’ treatments, presumably due to the reported slow recovery of AID-tagged proteins following auxin removal (Zhang et al., 2015). Western blotting experiments revealed ∼70% depletion of NHR-23 (Fig. 3D; Fig. S2). As we specifically depleted NHR-23::GFP in the germline in adult males, it is possible that somatic expression below the detection threshold of our fluorescent imaging contributes to the residual NHR-23::GFP detected following auxin treatment.
Although spermatogenesis is similar in C. elegans males and hermaphrodites, sex-specific differences do exist (L'Hernault et al., 1988; Minniti et al., 1996; Nance et al., 1999, 2000; Shakes and Ward, 1989). To test whether NHR-23 also functioned during male spermatogenesis, we repeated the crossing experiment from Fig. 3B using mex-5::TIR1, nhr-23::GFP::AID*::3xFLAG males. Males grown on control media sired progeny when crossed with either NHR-23-depleted hermaphrodites or feminized but otherwise wild-type fog-1 ‘females’, but males grown on 4 mM auxin did not sire any progeny (Fig. 3B). We confirmed that NHR-23-depleted males were mating both by observation and assessing the transfer of seminal fluid while mating with spe-8 hermaphrodites (Shakes and Ward, 1989) (Fig. S3). Sperm activation defects preclude self-fertility in spe-8 hermaphrodites, but their sperm can be transactivated by male seminal fluid. Non-celibate, NHR-23-depleted adult males mated with spe-8 hermaphrodites produced no outcross progeny, but some matings induced spe-8 self-progeny. Unexpectedly, some crosses set up with L4 males yielded both self- and outcross progeny, suggesting that incomplete NHR-23 degradation allows the early production of functional sperm.
NHR-23 acts following the sperm/oocyte decision
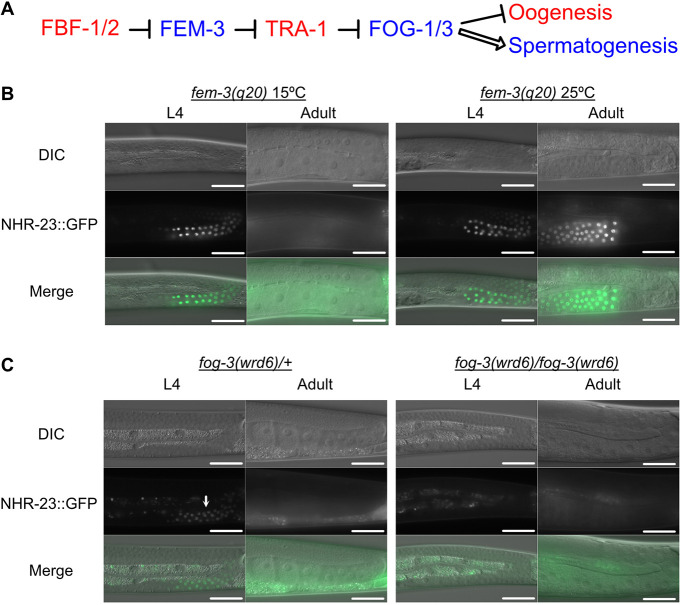
The infertility phenotype could suggest that NHR-23 was either a spermatocyte/oocyte cell fate determinant or responsible for executing the sperm fate program. To place NHR-23 in relation to known sex determination factors (Ellis and Schedl, 2007), we performed epistasis experiments (Fig. 4A). We crossed nhr-23::GFP::AID*::3xFLAG into a temperature-sensitive (ts) gain-of-function fem-3(q20) allele genetic background (Barton et al., 1987). At 15°C, hermaphrodites produce both sperm and oocytes, and NHR-23 expression was limited to the L4 stage (Fig. 4B). At 25°C, the masculinized germline produced only sperm, and NHR-23 expression persisted throughout adulthood. These results indicate that NHR-23 functions downstream of FEM-3. Next, we tested fog-3 (feminization of germline), which, along with fog-1, is believed to act as the terminal, germline-specific selector for sperm fate. As fog-3 is close to nhr-23 on chromosome I, we directly engineered a putative null allele of fog-3 in an nhr-23::GFP::BioTag::AID*::3xFLAG strain (wrd1 allele) by introducing a premature termination codon followed by a frameshift at the fog-3 locus (wrd6 allele). Because this experiment used a nhr-23 strain distinct from the wrd8 allele used elsewhere in this study, we verified that germline-specific NHR-23 depletion in a sun-1p::TIR1; nhr-23(wrd1[GFP::BioTag::AID*::3xFLAG]) strain also caused sterility (Fig. S1). Importantly, this result demonstrates an equivalent phenotype with two distinct AID*-tagged nhr-23 alleles and TIR1 transgenes being driven by two different germline-specific promoters (mex-5p versus sun-1p) (Fig. 3A; Fig. S2). In fog-3(wrd6) heterozygotes, NHR-23::GFP was expressed specifically in L4 hermaphrodites (Fig. 4C). In fog-3(wrd6) homozygotes, germline feminization is accompanied by a complete loss of NHR-23::GFP expression in L4 animals. These results show NHR-23 operates downstream of the known sex-determination pathway, after spermatocyte cell fate has been decided.
Fig. 4.
nhr-23 is downstream of the germline sex-determination pathway. (A) Simplified schematic of the C. elegans sex-determination pathway and their effect on gamete fate. The wild-type function of the factors in blue is to promote spermatogenesis, whereas the factors in red promote oogenesis. (B) DIC and GFP images of mex-5p::TIR1 nhr-23::GFP::AID*:: 3xFLAG; fem-3(q20) L4 and adult hermaphrodites grown from L1 onwards at permissive (15°C) or restrictive (25°C) temperatures. (C) DIC and GFP images of mex-5p::TIR1 nhr-23::GFP::AID*::3xFLAG; fog-3(wrd6) L4 and adult hermaphrodites. The white arrow indicates NHR-23::GFP::AID*::3xFLAG expression in late pachytene cells of a fog-3(wrd6) heterozygous hermaphrodite germline. Scale bars: 40 µm.
Germline depletion of NHR-23 results in spermatocyte arrest
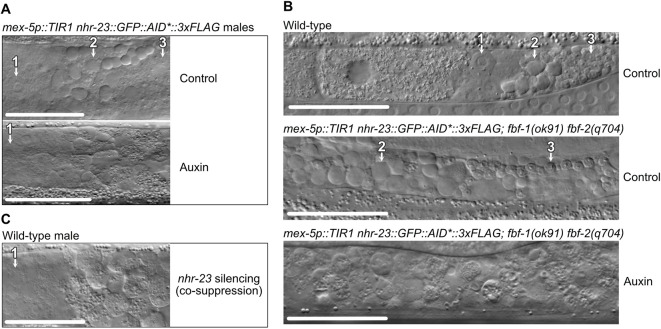
To better understand the nature of sperm-related fertility defects, we used DIC optics to examine gonads of control and NHR-23-depleted animals. The proximal gonad of control males contained a linear sequence of: (1) spermatocytes completing meiotic prophase; (2) a discrete zone of meiotically dividing spermatocytes, budding figures, and residual bodies; and, finally, (3) a zone of tightly packed spermatids with their small, highly refractive chromatin mass (Figs 1B and 5A). NHR-23-depleted germlines produced spermatocytes that progressed normally through meiotic prophase. However, the rest of the gonad filled with what appeared to be primary spermatocytes. Haploid spermatids were notably absent. Depletion of NHR-23 in masculinized germlines of fbf-1(ok91) fbf-2(q704) hermaphrodites yielded similar defects (Fig. 5B), indicating NHR-23-depletion during either male or hermaphrodite spermatogenesis results in a spermatocyte arrest phenotype.
Fig. 5.
NHR-23 is necessary for sperm development. (A) DIC images of the proximal germlines of mex-5p::TIR1, nhr-23::GFP::AID*::3xFLAG young adult males grown from L1 onwards on control or 4 mM auxin media. (B) DIC images of wild-type and masculinized mex-5p::TIR1, nhr-23::GFP::AID*::3xFLAG; fbf-1(ok91) fbf-2(q704) hermaphrodites grown from L1 onwards on control or 4 mM auxin media. (1) Primary spermatocytes have a flat morphology with a large nucleus. (2) Residual bodies appear as highly refractile raised button-like structures. (3) Spermatids are readily discernible through their characteristic small refractive nuclei. (C) DIC image of the proximal germline in a wild-type young adult male F1 progeny from a P0 animal injected with a mex-5p::nhr-23 cDNA co-suppression construct, which promotes an RNAi-like depletion of targeted mRNA. Scale bars: 40 µm.
To independently confirm our NHR-23 depletion results, we depleted nhr-23 mRNA in the germline using transgene-mediated co-suppression, which is similar to RNAi (Dernburg et al., 2000). This method yielded viable but infertile male animals with the same spermatogenesis defects as their auxin-mediated depletion counterparts, confirming the phenotypes are due to NHR-23-depletion (Fig. 5C).
NHR-23-depleted primary spermatocytes arrest in a metaphase I-like state
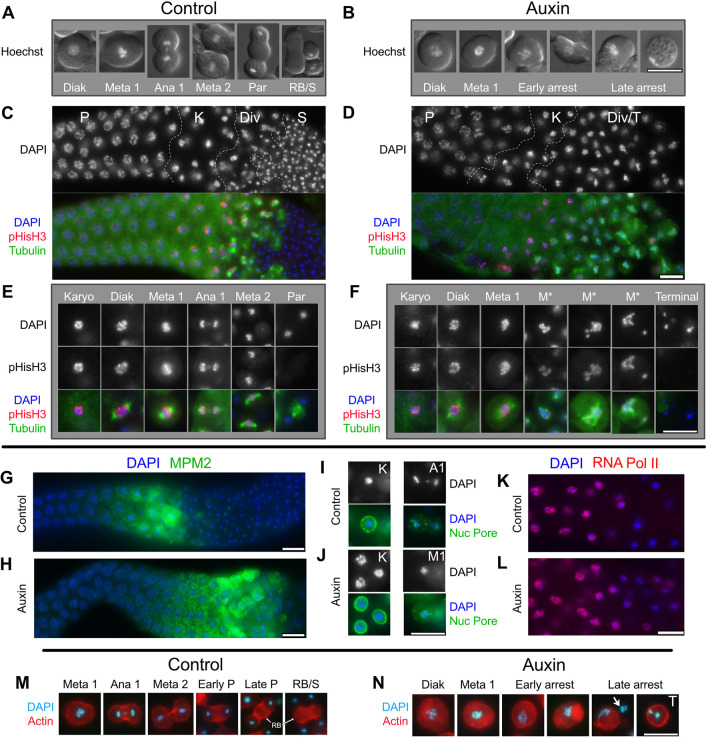
To determine which stages of spermatogenesis are affected by NHR-23 depletion, we assessed nuclear morphology in male germlines. Hoechst staining of DNA in control male germlines revealed the expected stages: (1) diakinesis stage (Diak) spermatocytes with intact nuclear envelopes; (2) meiotically dividing spermatocytes; and (3) cells undergoing the post-meiotic partitioning (Par) event that yields a central residual body (RB) and four haploid spermatids (Figs 1 and 6A). NHR-23-depleted male germlines included diakinesis spermatocytes and spermatocytes with chromosome patterns, suggesting they were arresting in prometaphase or metaphase I (Fig. 6B). Notably, these germlines lacked secondary spermatocytes, budding figures or spermatids (Figs 1D and 6B). In many cells, chromosomes were spread out relative to the initial metaphase clusters (Fig. 6B, early arrest), but there was no evidence of actual anaphase chromosome segregation. Nuclear envelope breakdown was independently confirmed by labeling fixed gonads with nuclear pore antibodies (Fig. 6I,J). These data suggested that spermatocyte development was aberrant, with an arrest in prometaphase or metaphase I.
Fig. 6.
Metaphase I-like arrest in NHR-23-depleted spermatocytes. (A,B) Live spermatocytes ordered according to stage. Differential interference contrast (DIC) images of cells were overlaid by epifluorescence images of their Hoechst stained nuclei. (C-J) Isolated and fixed male gonads, and individual spermatocytes labeled with DAPI (blue) and indicated antibodies. (C,D) Gonad images show the proximal gonad from late pachytene (P) to either haploid spermatids (S) or terminal arrest (T) co-labeled with antibodies against α-tubulin (green) and phosphorylated histone H3 (ser10) (red). Controls (nhr-23::AID* without auxin or him-5 with auxin) in C; nhr-23::AID* with auxin in D. (E,F) Higher magnification images of individual spermatocytes. (G,H) Isolated control (G) and NHR-23-depleted (H) proximal male gonads (pachytene and later meiotic stages) co-labeled with antibodies against MPM2 (green), which binds diverse mitotic and meiotic phosphorylated proteins. (I,J) Staged spermatocytes co-labeled with anti-nuclear pore protein (green) in control (I) and NHR-23-depleted (J) males. (K,L) Control (K) and NHR-23-depleted (L) gonads (pachytene through karyosome) co-labeled with anti-phospho-RNA polymerase II CTD repeat (red) show switching off of global transcription in karyosome spermatocytes. (M,N) Aldehyde-fixed and staged spermatocytes with DNA pseudo-colored in cyan (DAPI) and actin microfilaments labeled with rhodamine. Arrow in N indicates the chromatin of an adjacent lysed cell. P, pachytene; K, karyosome; Div, meiotic divisions; Diak, diakinesis; Meta 1/M1, metaphase I; M*, aberrant metaphase I; Ana1/A1, anaphase I; Meta 2, metaphase II; Par, post-meiotic partitioning; RB, residual body; S, haploid spermatids; T, terminal-stage spermatocyte. Scale bars: 10 µm.
To determine how well NHR-23-depleted spermatocytes were entering or exiting M-phase, isolated male gonads were first co-labeled with DAPI, anti-tubulin and anti-phospho-histone H3 serine 10 [pHisH3(ser10)], a proxy for activity of the cell cycle regulator Aurora kinase (Hsu et al., 2000) (Fig. 6C-F). The most obvious NHR-23-depletion phenotype was an increase in the meiotic division zone (Div); large numbers of detached spermatocytes containing metaphase-like chromosomes with associated microtubule spindles (Fig. 6D). Recently detached spermatocytes had relatively normal diakinesis and metaphase I spindles, but metaphase chromosome alignment was impaired. Slightly older spermatocytes had more dispersed chromosomes (M*) and multipolar asters with four or more asters (Fig. 6F). In control germlines, anti-pHisH3(ser10) labeled chromosomes through both meiotic divisions and only became undetectable in post-meiotic budding figures (Fig. 6E). In NHR-23-depleted germlines, pHisH3 labeling persisted in arrested, metaphase-like cells (M*; Fig. 6F). The very oldest spermatocytes exhibited a distinct terminal/late arrest phenotype; these spermatocytes lacked microtubule spindles and their chromosomes had aggregated into one or more tight masses and failed to label with anti-pHis3(ser10) (Fig. 6F). To assess CDK-cyclin B activity, gonads were labeled for a phospho-epitope primarily found in proteins phosphorylated at inception of mitosis or meiosis (MPM2). In control spermatocytes (Fig. 6G), MPM2 labeling turned on during late meiotic prophase (karyosome stage), remained high through meiotic divisions and then dropped abruptly during post-meiotic partitioning. In NHR-23-depleted spermatocytes (Fig. 6H), MPM2 labeling remained high in metaphase-arrested spermatocytes, only dropping in terminal arrest spermatocytes. Anti-nuclear pore antibodies confirmed that nuclear envelope breakdown occurred in a timely fashion in both control and NHR-23-depleted spermatocytes (Fig. 6I,J). Together, these results suggest that the cell cycle regulators of M-phase entry are unaffected by NHR-23 depletion. However, depleted spermatocytes remain in a prolonged prometaphase state, develop multipolar spindles and never progress to anaphase. M-phase markers do eventually turn off, but only in late, terminal stage spermatocytes.
We hypothesized that the observed meiotic defects might arise from earlier defects during the poorly understood karyosome stage (Fig. 1) (Shakes et al., 2009). In control gonads labeled with an antibody against active RNA polymerase II, entry into the karyosome stage was marked by abrupt cessation of global transcription (Fig. 6K) (Shakes et al., 2009). In NHR-23-depleted gonads, switching off global transcription was both delayed and more gradual (Fig. 6L). In a complementary fashion, the switching on of pHisH3(ser10) labeling, which corresponds to chromosome condensation, was also delayed and more gradual (Fig. 6D,F). Closer examination revealed that, during the karyosome stage, the chromosomes in both control and depleted spermatocytes properly detached from the nuclear envelope, but the compaction of chromosomes into a single tight mass was more limited in NHR-23-depleted spermatocytes (Fig. 6I,J, karyosome nuclei labeled ‘K’). Such aberrations in chromosome compaction during karyosome stage might contribute to subsequent defects in meiotic chromosome segregation as well as the prolonged M-phase arrest, if such defects trigger a cell cycle checkpoint.
NHR-23-depleted spermatocytes have defects in both cytokinesis and post-meiotic partitioning
In previous studies, a metaphase I arrest of the cell cycle was not sufficient to block other aspects of sperm development. Notably, spermatocytes lacking a functional anaphase-promoting complex arrest in metaphase I but still undergo post-meiotic partitioning to generate motile, albeit anucleate, sperm (Golden et al., 2000; Sadler and Shakes, 2000). Although NHR-23-depleted germlines do not make mature spermatids, we wondered whether they nevertheless attempted aspects of normal, post-meiotic partitioning and residual body formation, which is mediated in part by actin microfilaments (Hu et al., 2019; Winter et al., 2017). In control spermatocytes, actin localization to the cortex was uniform in metaphase and enriched in cortical rings during anaphase (Fig. 6M). In partitioning stage spermatocytes, actin localized in a central band and ultimately degraded with the residual body (RB; Fig. 6M). In NHR-23-depleted spermatocytes, actin patterns remained largely cortical in both metaphase-arrested and late terminally arrested spermatocytes (Fig. 6N). Consistent with our DIC analysis, NHR-23-depleted spermatocytes showed no evidence of cytokinesis or residual body formation.
NHR-23 is required for biogenesis of sperm-specific fibrous body-membranous organelle (FB-MO) complexes
Among C. elegans mutants with spermatogenesis defects, several exhibit a spermatocyte-arrest phenotype. Four of these (spe-4, spe-5, spe-6 and wee-1.3) exhibit a vacuolated phenotype by DIC that is very similar to NHR-23-depleted spermatocytes (Fig. 7A). Other spermatocyte arrest mutants (spe-39, spe-44) do not exhibit the same vacuolated phenotype (Fig. 7A) (Kulkarni et al., 2012; Zhu and L'Hernault, 2003). In previous studies, transmission electron microscopy found these vacuoles are swollen, distended membranous organelles (MOs) (Lamitina and L'Hernault, 2002; L'Hernault and Arduengo, 1992; Machaca and L'Hernault, 1997; Varkey et al., 1993). To determine whether vacuoles in NHR-23-depleted spermatocytes were MOs, we crossed a PEEL-1::GFP transgene into our mex-5p::TIR1, nhr-23::GFP::AID*::3xFLAG strain. PEEL-1 is the toxin component of a toxin-antidote selfish genetic element (Seidel et al., 2008) that also serves as a useful MO marker (Seidel et al., 2011). In control primary spermatocytes and budding figures, PEEL-1::GFP marked internal membranes (Figs 1 and 7B). In mature spermatids, PEEL-1::GFP exhibited a bright punctate pattern corresponding to plasma membrane-docked MOs. In NHR-23-depleted spermatocytes, PEEL-1::GFP colocalized with vacuoles in the corresponding DIC images (Fig. 7B), indicating that MOs form but then vacuolate in terminal arrest spermatocytes.
Fig. 7.
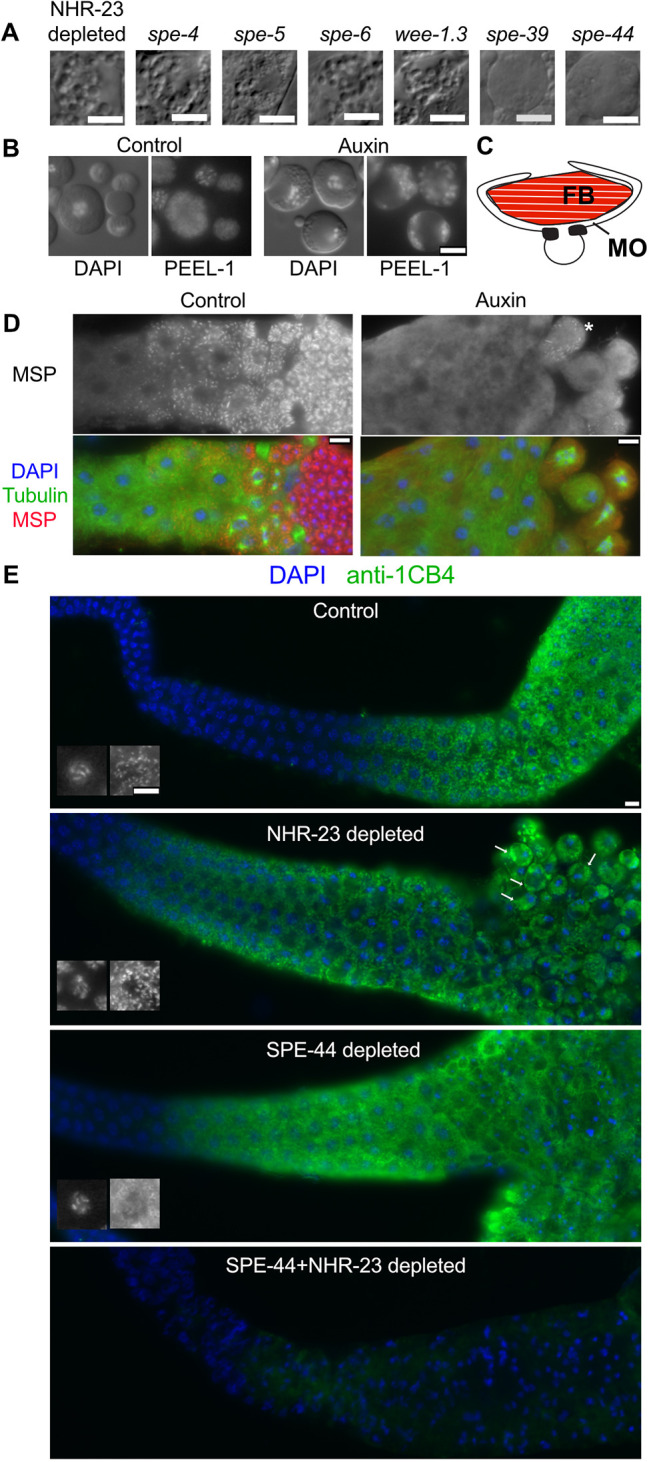
FB-MO defects in NHR-23 spermatocytes and synergy with SPE-44. (A) DIC images of arrested spermatocytes from spermatogenesis-defective mutants. (B) Control and NHR-23-depleted spermatocytes visualized by DIC/Hoechst (left) and the MO marker PEEL::GFP (right). (C) Schematic of an FB-MO complex showing an MSP-enriched fibrous body (FB) enveloped within arm-like extensions of the Golgi-derived membranous organelle (MO). (D) Proximal gonads from control (left) and NHR-23-depleted (right) males co-labeled with DAPI and antibodies against tubulin and MSP. Asterisk indicates a single spermatocyte in the NHR-23-depleted gonad with MSP polymers. (E) Gonads from animals with indicated protein depletions co-labeled with DAPI and the anti-MO antibody 1CB4. Black and white insets show DAPI staining (left) and 1CB4-stained MOs (right) in pachytene stage spermatocytes and highlight defects in SPE-44 spermatocytes. Arrows indicate clumping of 1CB4-stained bodies following NHR-23 depletion. Scale bars: 5 µm.
In developing spermatocytes, MOs serve as sites where major sperm protein (MSP) assembles into fibrous bodies (FBs) (Fig. 7C). MSP is a nematode-specific protein that drives the actin-like cell motility of spermatozoa while also serving as a signaling molecule for oocyte maturation (Miller et al., 2001). Not surprisingly, MO defects are often associated with either abnormalities in FB assembly, as in spe-5 and spe-39 mutants (Machaca and L'Hernault, 1997; Zhu and L'Hernault, 2003), or a complete failure in FB formation, as in spe-6 and spe-44 (Kulkarni et al., 2012; Varkey et al., 1993). In wild-type animals, MSP is first expressed in mid-pachytene spermatocytes and, by the end of the karyosome stage, individual spermatocytes fill with discrete uniformly sized FBs (Fig. 7D) (Chu and Shakes, 2013). During post-meiotic partitioning, FB-MO complexes facilitate MSP delivery to spermatids. In NHR-23-depleted spermatocytes, MSP was expressed, but in most spermatocytes it remained dispersed throughout the cytoplasm (Fig. 7D). NHR-23-depleted germlines sometimes included individual spermatocytes with MSP assemblages (Fig. 7D, asterisk) that were more variable in shape and size than normal FBs.
To investigate potential defects in FB-MO biogenesis, gonads were immunolabeled with 1CB4, an antibody labeling glycosylated MO proteins (Okamoto and Thomson, 1985). In control gonads, 1CB4 labeled discrete cytoplasmic organelles starting in mid-pachytene spermatocytes (Fig. 7E, inset). This pattern persisted until the spermatid stage, when individual MOs docked with the plasma membrane (Figs 1D and 7E). In NHR-23-depleted germlines, the 1CB4 pattern in pachytene spermatocytes was similar to controls (Fig. 7E, inset). However, in arrested primary spermatocytes, MOs aggregated into clumps adjacent to the plasma membrane, suggesting a defect in either MO docking or later stages of MO morphogenesis.
Of the spermatocyte arrest mutants shown in Fig. 7A, only the spe-44 gene encodes an early transcriptional regulator (Kulkarni et al., 2012). Like NHR-23-depleted spermatocytes, spe-44 spermatocytes exhibit defects in MSP assembly and meiotic progression (Kulkarni et al., 2012). However, spe-44 spermatocytes arrest in the second rather than the first meiotic division, and arrested spermatocytes do not fill with distended MOs (Fig. 7A). We confirmed that SPE-44 depletion causes sterility, as previously reported (Fig. S4A) (Kasimatis et al., 2018). To determine whether NHR-23 and SPE-44 play similar or distinct roles in spermatogenesis, we compared 1CB4 patterns as a proxy for their role in FB-MO biogenesis. In SPE-44-depleted pachytene spermatocytes, the 1CB4 pattern within developing spermatocytes did not form discrete structures (Fig. 7E, inset), matching the published pattern for spe-44(ok1400) mutants (Kulkarni et al., 2012). As this spe-44 pattern is distinct from NHR-23-depletion defects, we also examined doubly depleted males which typically exhibited a more severe synthetic phenotype (Fig. 7E).
NHR-23 and SPE-44 independently regulate spermatogenesis
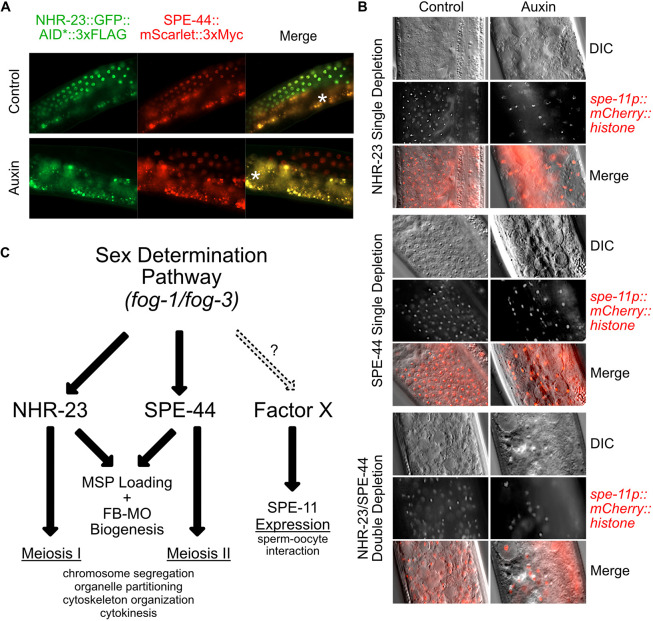
The synthetic FB-MO phenotype observed following NHR-23+SPE-44 double depletion raised the possibility that these regulators could function in distinct pathways. In spe-44(ok1400) mutants, levels of nhr-23 mRNA do not differ significantly from wild type (Kulkarni et al., 2012). To test whether SPE-44 might function downstream of NHR-23, we generated a strain carrying an mScarlet::3xMyc cassette inserted into the 3′ end of spe-44 by CRISPR/Cas9-mediated genome editing. There is only a single spe-44 isoform and our strain results in a C-terminal translational fusion. We confirmed that the tag did not compromise SPE-44 function, as animals with the mScarlet::3xMyc knock-in had a wild-type brood size (Fig. S4B), and SPE-44::mScarlet::3xMyc localization was similar to previous reports (Fig. 8A) (Kulkarni et al., 2012). On control media, spermatocytes in strains carrying both spe-44::mScarlet::3xMyc and mex-5::TIR1, nhr-23::GFP::AID*::3xFLAG exhibited an almost complete overlap in expression for SPE-44 and NHR-23 (Fig. 8A). When grown on auxin, depletion of NHR-23 did not affect the level or expression pattern of SPE-44::mScarlet::3xMyc. These data indicate NHR-23 regulates neither SPE-44 expression nor its localization.
Fig. 8.
Control of transcription in spermatogenesis is facilitated by multiple factors with divergent targets. (A) Fluorescent images of NHR-23::GFP::AID*::3xFLAG and spe-44::mScarlet::3xMyc. Worms containing both translational fusions as well as the mex-5p::TIR1 driver were grown from L1 onwards on control or 4 mM auxin media. The asterisks indicate gut autofluorescence. (B) Representative DIC and fluorescent images of spe-11p::mCherry::H2B in adult males expressing mex-5p::TIR1 in the germline. Worms individually expressed NHR-23::AID*::3xFLAG or SPE-44::AID*::3xFLAG, or both, were grown from L1 until the young adult stage on control or 4 mM auxin media. (C) Model depicting the coordinated control of gene expression prior to and during the stages of spermatogenesis. NHR-23 and SPE-44 control distinct sets of genes to promote the events of meiosis I and II, respectively. They redundantly promote MSP loading and FB-MO biogenesis. A third pathway controlled by yet unidentified factors promotes the expression of genes involved in the sperm-oocyte interaction, such as spe-11.
These data raised the possibility that C. elegans spermatogenesis is coordinated by multiple transcriptional regulatory pathways. If so, then spermatogenesis-specific genes like spe-11 that are known not to be regulated by SPE-44 may instead be NHR-23 target genes. We acquired a strain in which the spe-11 promoter drives expression of an mCherry::Histone H2B::unc-54 3′UTR reporter (Frøkjaer-Jensen et al., 2008). We crossed AID* alleles of nhr-23 (Zhang et al., 2015) and spe-44 (Kasimatis et al., 2018) lacking a fluorescent protein fusion, as well as a mex-5p::TIR1 allele, into this spe-11 reporter strain. In animals grown on control media, the spe-11 promoter drove mCherry::histone expression, starting in pachytene spermatocytes with the histone fusion protein persisting in spermatids (Fig. 8B) (Frøkjaer-Jensen et al., 2008). Following individual depletion of either NHR-23 or SPE-44, expressed mCherry fusion protein persisted in meiotically arrested nuclei, despite the spermatocyte defects and their meiotic arrest in meiosis I or II, respectively. To test whether NHR-23 and SPE-44 redundantly regulated spe-11, we created a spe-11 reporter strain that would allow simultaneous germline-specific depletion of NHR-23 and SPE-44. Co-depletion of NHR-23 and SPE-44 resulted in germline defects so severe that it was difficult to identify individual cells within the proximal gonad. Yet reporter expression was still detectable in nuclei (Fig. 8B). Together these results suggest not only that NHR-23 and SPE-44 function in independent pathways, but also that spe-11 is regulated either by factors upstream of NHR-23 and SPE-44 or by an additional, yet to be identified, transcriptional regulatory pathway (Fig. 8C).
DISCUSSION
C. elegans spermatogenesis offers a powerful model with which to define how gene-regulatory networks implement and coordinate the cell cycle and developmental programs. In this study, we identify NHR-23 as a critical regulator of spermatogenesis in both hermaphrodite and male animals.
NHR-23 is expressed in primary spermatocytes and acts in the L4 stage in hermaphrodites. NHR-23 functions downstream of the sex determination pathway (Fig. 4), and inactivation in hermaphrodite germlines causes a loss of mature spermatids (Figs 3–5). However, cells recognizable as primary spermatocytes developed (Figs 5–7) and progressed through meiotic prophase (Fig. 6). Numerous cell cycle markers indicated that affected spermatocytes entered but did not complete meiosis. Instead, the fully condensed chromosomes clustered within a disorganized and ultimately multipolar microtubule array (Fig. 6). Although microtubules appeared to associate with the chromatin, there was no evidence of even attempted anaphase segregation (Fig. 6). Similarly, we never observed cytokinesis or spermatid budding divisions. The oldest cells contained terminal chromatin masses, which might represent either a variation of normal sperm chromatin remodeling or apoptotic compaction (Fig. 6). In addition to these meiotic roles, NHR-23 is necessary for proper FB-MO biogenesis. Golgi-derived MOs developed in meiotic prophase spermatocytes, yet MSP failed to assemble into associated FBs and the morphology of MOs in arrested spermatocytes was clearly abnormal (Fig. 7).
Multiple gene regulatory networks might control C. elegans spermatogenesis
The meiotic phenotypes following NHR-23-depletion are similar to, but distinct from, those caused by inactivation of spe-44, a critical transcriptional regulator of spermatogenesis (Fig. 8C). In both NHR-23-depletion and spe-44 mutants, meiotic prophase is largely unaffected except that chromatin compaction is delayed during the karyosome stage (Kulkarni et al., 2012). spe-44 spermatocytes also fail to assemble MSP into FBs (Kulkarni et al., 2012). Both NHR-23 and SPE-44 proteins are first expressed in developing spermatocytes and have similar mRNA expression profiles in a spatiotemporal analysis of germline expression (Tzur et al., 2018). NHR-23 and SPE-44 both function downstream of the canonical sex-determination pathway (Fig. 4) (Kulkarni et al., 2012). Although both are essential to complete the spermatocyte meiotic divisions, NHR-23 is necessary for the first meiotic division (Fig. 6), whereas SPE-44 is necessary for the second meiotic division (Fig. 8C) (Kulkarni et al., 2012). Their microtubule spindle patterns also differ; in NHR-23-depleted spermatocytes, the microtubule asters remain close to the chromosomes and cell center (Fig. 6D,F), whereas those in spe-44 mutants move to the cell cortex (Kulkarni et al., 2012). SPE-44 regulates ELT-1 (del Castillo-Olivares et al., 2009; Kulkarni et al., 2012), which promotes MSP gene expression in conjunction with a histone methyltransferase, SET-17 (Engert et al., 2018). However, SPE-44 does not regulate nhr-23 (Kulkarni et al., 2012), nor does NHR-23-depletion affect SPE-44 expression (Fig. 8A), consistent with these two transcription factors regulating distinct pathways. Co-depletion of NHR-23 and SPE-44 led to additive phenotypes such as reduction of the MO marker 1CB4 (Fig. 7E) and more severe germline morphology defects (Fig. 8B). NHR-23-depleted spermatocytes developed distended MOs, similar to those of several sperm function mutants [e.g. spe-4(lf), spe-6(lf) and wee-1.3(gf)] (Lamitina and L'Hernault, 2002; L'Hernault et al., 1988; Muhlrad and Ward, 2002; Varkey et al., 1993). spe-44 does not regulate these genes (Kulkarni et al., 2012) and does not exhibit this mutant phenotype (Kulkarni et al., 2012) (Fig. 7A). These results suggest that NHR-23 and SPE-44 function in separate pathways to regulate distinct cellular processes.
Additional undiscovered gene regulatory pathways are likely to help execute the C. elegans spermatogenesis program. spe-11p::mCherry::H2B reporter activity was unaffected by NHR-23, SPE-44 or NHR-23+SPE-44 depletions. These data could suggest the presence of three or more distinct pathways controlling sperm morphogenesis in C. elegans or, alternatively, that spe-11 is directly regulated by a factor upstream of both NHR-23 and SPE-44 (Fig. 8C). Notably, SPE-44 only accounts for one-quarter of sperm-expressed genes (Kulkarni et al., 2012). As we further characterize NHR-23 and SPE-44, it will be informative to compare their function with spermatogenesis regulators in other systems. In Drosophila, BAM promotes differentiation of spermatogonia into spermatocytes (McKearin and Spradling, 1990), while the sperm-specific transcriptional program is promoted by two complexes: tMAC and tTAF (Beall et al., 2007; Laktionov et al., 2018; Metcalf and Wassarman, 2007). In mice, retinoic acid receptor signaling promotes both spermatogonia differentiation and meiotic entry (Chung et al., 2004; Gely-Pernot et al., 2012, 2015), and MYBL1 coordinates the expression of four downstream transcription factors (CREM-τ, TRF2, RFX2 and Sox30) (Bolcun-Filas et al., 2011; Horvath et al., 2009; Li et al., 2013; Zhang et al., 2018). Zhang et al. (2018) suggest that tree-like regulatory cascades would be more efficient than a network enriched in nodes. Spermatogenesis is a rapidly evolving process. Defining the regulatory architecture across a range of model organisms will shed insight into conserved features controlling sperm morphogenesis, as well as distinctive features that have evolved separately in mammals, insects, nematodes and other organisms (White-Cooper and Bausek, 2010).
Circadian rhythm regulators in spermatogenic tissues
Before this study, the best characterized role for nhr-23 in C. elegans was as a key regulator of molting (Frand et al., 2005; Kostrouchova et al., 1998, 2001; Kouns et al., 2011; Patel and Frand, 2018 preprint). Like many molting factors, nhr-23 mRNA levels oscillate over the course of each larval stage (Gissendanner et al., 2004; Hendriks et al., 2014; Kostrouchova et al., 2001). Yet in sperm-producing germlines, NHR-23 is expressed constitutively, suggesting a radically different mode of regulation in soma versus germ tissues. Interestingly, many core mammalian circadian rhythm regulators also oscillate in the soma but exhibit constitutive, non-oscillatory expression in the testes (Alvarez et al., 2008; Kang et al., 2010; Kennaway et al., 2012; Morse et al., 2003). Although the non-circadian roles of these factors in spermatogenesis have not been deeply explored, male mice that are null for the clock gene Bmal1 (Arntl) have fertility defects and are difficult to mate (Alvarez et al., 2008), and depletion of the NHR-23 homolog RORα/NR1F1 in rat Sertoli cells reduces sperm count (Mandal et al., 2018). It remains unclear how these timers have been co-opted to promote spermatogenesis. Our work could provide an entry point into understanding both the function of these circadian rhythm factors during spermatogenesis and how their expression is altered from oscillating to constitutive expression in the testes.
Future perspectives
Most known spermatogenesis regulators have been identified in forward genetic screens and candidate-based testing of male germline-enriched factors. These approaches have been incredibly powerful but would miss factors, such as NHR-23, that also have earlier essential roles in embryogenesis and molting. Similarly, as NHR-23 is maternally loaded, it was not identified by focusing on male-enriched transcriptional regulators (Ebbing et al., 2018; Kulkarni et al., 2012; Reinke et al., 2004; Tzur et al., 2018). High-throughput GFP::AID* tagging of NHR-23-regulated genes could provide an exciting avenue for identifying novel regulators of spermatogenesis. New datasets that detail mRNA expression with respect to position in the germline can also be analyzed to identify transcriptional regulators expressed before, coincident with, and after the NHR-23/SPE-44 zone of expression. The mammalian homologs of NHR-23 are regulated by several sterol ligands (Kallen et al., 2002, 2004; Soroosh et al., 2014; Wang et al., 2010), raising the exciting possibility that hormonal signaling through NHR-23 coordinates spermatogenic and somatic development events. Going forward, determining factors that either regulate or are regulated by NHR-23 and SPE-44 will deepen our understanding of how the NHR-23 node controls spermatogenesis and how gene regulatory networks control the complex cellular events underpinning sperm morphogenesis.
MATERIALS AND METHODS
Strains and culture
C. elegans were cultured as originally described (Brenner, 1974), except worms were grown on MYOB media instead of NGM. MYOB agar was made as previously described (Church et al., 1995).
Strains used in this study
- JDW5
nhr-23(wrd1[GFP^BioTag::AID*::3xFLAG]) I; ieSi38 [Psun-1::TIR1::mRuby::sun-1 3′UTR, cb-unc-119(+)] IV
- JDW26
nhr-23(wrd1[GFP^BioTag::AID*::3xFLAG]) I/hT2[bli-4(e937) let-?(q782) qIs48] (I;III) ; ieSi38 [Psun-1::TIR1::mRuby::sun-1 3′UTR, cb-unc-119(+)] IV
- JDW29
nhr-23(wrd8[nhr-23::GFP^degron::3xFLAG]) I
- JDW32
fog-3(wrd6[fog-3 N-term STOP with frameshift]), nhr-23(wrd1[GFP^BioTag::AID*::3xFLAG]) I/hT2[bli-4(e937) let-?(q782) qIs48] (I;III); ieSi38 [Psun-1::TIR1::mRuby::sun-1 3′UTR, cb-unc-119(+)] IV
- JDW40
wrdSi11(mex-5p::TIR1:F2A:mTagBFP2:tbb-2 3′UTR), nhr-23(wrd8[nhr-23::GFP^AID*::3xFLAG]) I
- JDW45
wrdSi12(mex-5p::TIR1:F2A:mTagBFP2:tbb-2 3′UTR), nhr-23(wrd8[nhr-23::GFP^AID*::3xFLAG]) I; fem-3(q20) IV
- JDW47
wrdSi13(mex-5p::TIR1:F2A:mTagBFP2:tbb-2 3′UTR), nhr-23(wrd8[nhr-23::GFP^AID*::3xFLAG]) I; fbf-1(ok91), fbf-2(q704)/mIn1 [dpy-10(e128) mIs14] II
- JDW86
wrdSi15(mex-5p::TIR1:F2A:mTagBFP2:tbb-2 3′UTR) I; spe-44(fx110[spe-44::AID*]) IV; him-5(e1490) V
- JDW87
wrdSi16(mex-5p::TIR1:F2A:mTagBFP2:tbb-2 3′UTR), nhr-23(wrd8[nhr-23::GFP^AID*::3xFLAG]) I; him-5(e1490) V
- JDW89
wrdSi18(mex-5p::TIR1:F2A:mTagBFP2:tbb-2 3′UTR) I
- JDW92
wrdSi19(mex-5p::TIR1:F2A:mTagBFP2:tbb-2 3′UTR), nhr-23(kry61[nhr-23::AID*::TEV::3xFLAG]) I; him-5(e1490) V
- JDW101
spe-44(wrd20[spe-44::30 amino acid linker::mScarlet^3xMyc])
- JDW146
wrdSi19(mex-5p::TIR1:F2A:mTagBFP2:tbb-2 3′UTR), nhr-23(kry61[nhr-23::AID*::TEV::3xFLAG]) I; spe-44(fx110[spe-44::AID*]) IV; him-5(e1490) V
- KRY87
nhr-23(kry61(nhr-23::AID*::TEV::3xFLAG)) I (Zhang et al., 2015)
Strains provided by the Caenorhabditis Genetics Center
N2 (Bristol)
- BA676
spe-6(hc92) unc-32(e189) III; eDp6 (III;f)
- BA714
sDf5/spe-4(hc78) I
- BA763
spe-5(hc93) I; sDp2 (I;f)
- DR466
him-5(e1490) V
- DS179
spe-8(hc50) I; him-8 (e1489) IV
- PX627
fxIs1 I; spe-44(fx110[spe-44::degron]) IV (Kasimatis et al., 2018)
- SL753
spe-39(tx12) V/nT1 [unc-?(n754) let-?] (IV;V)
- SL940
wee-1.3(q89eb94) unc-4(e120)/mIn1 [dpy-10(e128) mIs14] II
Other strains
oxSi1091 [Pmex-5::Cas9(smu-2 introns) unc-119+] II; unc-119(ed3) III (EG9615) was a gift from Erik Jorgensen (University of Utah/HHMI, USA) and is currently unpublished.
Immunocytochemistry
Intact gonads were obtained by dissection of individual males in 5-10 µl of egg buffer (Edgar, 1995) on ColorFrost Plus slides (Fisher Scientific, 12-550) coated with poly-L-lysine (Sigma Aldrich, P8290). Samples were freeze-cracked in liquid nitrogen. Sperm spreads to analyze detached spermatocytes and spermatids were obtained by applying slight pressure to the coverslip before freeze-cracking. Most samples were fixed overnight in −20°C methanol. Specimen preparation and antibody labeling followed established protocols (Shakes et al., 2009). Primary antibodies included: 1:100 Alexa Fluor 488-conjugated anti-α-tubulin (mouse monoclonal DM1A, Millipore-Sigma, 16-323), 1:200 rabbit anti-phosphorylated histone H3 (S10) (Millipore, 06-570, lot 2649123), 1:100 MPM-2 mouse monoclonal (EMD Millipore, 05-368), 1:200 anti-nuclear pore antibody Mab414 (Abcam, ab24609), 1:250 anti-RNA polymerase II CTD repeat Ser2 (Abcam, ab5095), 1:200 rabbit anti-SPE-44 (Kulkarni et al., 2012), 1:15,000 G3197 rabbit anti-MSP polyclonal (Kosinski, 2005; a gift from David Greenstein, University of Minnesota, Minneapolis, MN, USA ) and 1:40 1CB4 monoclonal (Okamoto and Thomson, 1985; a gift from Steve L'Hernault, Emory University, Atlanta, GA, USA). All samples were incubated with primary antibodies for 2 h at room temperature except MPM-2 samples, which were incubated overnight at 4°C. Affinity-purified secondary antibodies included 1:300 Alexa Fluor Plus 555 goat anti-rabbit IgG (Invitrogen, A32732) and 1:100 Alexa Fluor 488 goat anti-mouse IgG (H+L) (Jackson ImmunoResearch, 115-545-146).
Final slides were mounted with DAPI containing Fluoro Gel II mounting medium (Electron Microscopy Sciences #50-246-93). Images were acquired under differential interference contrast or epifluorescence using an Olympus BX60 microscope equipped with a QImaging EXi Aqua CCD camera. Photos were taken, merged and exported for analysis using the program iVision. In some cases, the levels adjust function in Adobe Photoshop was used to spread the data containing regions of the image across the full range of tonalities. When the experiment was designed to compare levels of fluorescence between control and experimental samples, images from the same experimental preparation were captured with the same exposure and any post-experimental processing was carried out identically. However, when the critical imaging information was to distinguish between the structures of chromatin, microtubule array or 1CB4 staining structures, the images were optimized for the individual cells.
For actin staining, slides with dissected gonads were fixed for 10 min in 4% fresh paraformaldehyde (ThermoFisher Scientific) in 1×PBS. Samples were quenched in 1 M glycine (ThermoFisher Scientific) in PBS (ThermoFisher Scientific, BP3994) for at least 5 min, permeabilized in 0.1% Triton-X-100 (ThermoFisher Scientific, BP151) in PBS for 5 min, then dip washed in 1× PBS. Slides were incubated with rhodamine-conjugated phalloidin (Invitrogen, #R415) diluted 1:100 in 1× PBS for 15 min in the dark before washing three times in PBS for 5 min each. Slides were mounted as above. For DIC/Hoechst preparations, males were dissected in egg buffer with 100 mg/ml Hoechst 33342 (Sigma Aldrich, 94403) on non-plus slides and immediately imaged according to Edgar (1995).
CRISPR/Cas9 genome editing
A thorough description of the plasmids and oligonucleotides described in this section is provided in Tables S1 and S2, respectively. nhr-23(wrd1[GFP^BioTag::AID*::3xFLAG]) was generated using a pJW1598 repair template, based on the previously described set of self-excising cassette (SEC) vectors for genome editing. Briefly, nhr-23 5′ and 3′ homology arms were PCR amplified from N2 genomic DNA and Gibson cloned into a AvrII+SpeI double-digested pJW1592 vector (Ashley et al., 2020 preprint) as described by Dickinson et al. (2015). N2 animals were injected and knock-ins were recovered as previously described (Dickinson et al., 2015). A previously described Cas9+sgRNA plasmid (pJW1254) (Zhang et al., 2015) targeting the 3′ end of nhr-23 was used to generate the knock-in.
nhr-23(wrd8[nhr-23::GFP^AID*::3xFLAG]) was generated with the self-excising cassette (SEC method) (Dickinson et al., 2015) using a pJW1725 (nhr-23:GFP:SEC:degron:3XFLAG with nhr-23 homology arms, U6p::sgRNA) repair template. The repair template (pJW1725) was constructed using SapTrap (Schwartz and Jorgensen, 2016) cloning with an SEC selection block (Dickinson et al., 2018). The 5′ homology arm was PCR amplified from a gBlock (IDT) and contained silent mutations to inactivate the PAM and two SapI restriction sites. The 3′ homology arm was PCR amplified from N2 genomic DNA. The 5′ and 3′ homology arm PCRs were cloned into pCR-Blunt II-TOPO using a Zero Blunt TOPO PCR Cloning Kit (Invitrogen, 450245) to generate pJW1776 and pJW1781, respectively. pJW1725 was assembled through SapTrap cloning using pDD379 (backbone), pMLS287 (flexible linker), pDD372 (GFP), pDD363 (SEC), pJW1659 (linker::AID*::3xFLAG), pJW1776 (5′ homology arm), pJW1781 (3′ homology arm) and annealed oligos 3488+3489 (nhr-23 targeting sgRNA) (Tables S1 and S2). The plasmid was injected into EG9615, which stably expresses Cas9, and knock-in animals were recovered as described (Dickinson et al., 2015). The strain was outcrossed two times to remove the mex-5p::Cas9 allele and the SEC was excised to make JDW29.
spe-44(wrd20[spe-44::30 amino acid linker::mScarlet^3xMyc]) was generated using SapTrap cloning with an SEC cassette, similar to the approach described above. Oligos 3916+3917 were annealed and Sap Trap cloned (Schwartz and Jorgensen, 2016) into pJW1838 (Ashley et al., 2020 preprint) to generate pJW1872(U6p::sgRNA (F+E) targeting spe-44). A pJW1876 repair template [30 amino acid flexible linker::mScarlet^SEC (Lox511I)^3xMyc with spe-44 homology arms] has been previously described (Ashley et al., 2020 preprint). The plasmid was injected into EG9615 and knock-in animals were recovered as described previously (Dickinson et al., 2015). The strain was outcrossed twice to remove the mex-5p::Cas9 allele and the SEC was excised to make JDW101.
wrdSi10 was generated with the self-excising cassette method (Dickinson et al., 2015) and a recently described TIR::F2A::mTagBFP2 construct (Ashley et al., 2020 preprint). pJW1357 (mex-5p::TIR1:F2A:mTagBFP2) (Ashley et al., 2020 preprint) and pCZGY2747 (eft-3p::Cas9; LGI ttTi4348 sgRNA vector) were injected into JDW29 to make JDW39. wrd10 was subsequently outcrossed to N2 animals once to generate JDW84, then heat-shocked at 34°C for 2-3 h to excise the SEC to make JDW89. These two strains carrying a mex-5p::TIR1:F2A:mTagBFP2 with and without the SEC, respectively, are part of our recent AID toolkit and are available through the Caenorhabditis Genetics Center.
wrdSi12, wrdSi13 and wrdSi16 were made by crossing JDW39 to JK816 [fem-3(q20) IV], JK3107 [fbf-1(ok91) fbf-2(q704)/mIn1 [dpy-10(e128) mIs14] II] and DR466 [him-5(e1490) V], respectively. wrdSi19 was made by crossing KRY87 [nhr-23[kry61(nhr-23::AID*-TEV-3xFLAG)]] I to JDW83 [wrdSi10(mex-5p::TIR1:F2A:mTagBFP2:tbb-2 3′UTR+SEC, I:-5.32); him-5(e1490) V]. The Rol phenotype produced by expression of the dominant sqt-1 allele within the SEC was used as a genetic marker to follow the insertion. Once the resultant strains were established, the SEC was excised by heat shock for 2-3 h at 34°C in wrdSi12, wrdSi13, wrdSi16 and wrdSi19, thereby making JDW45, JDW47, JDW87 and JDW92, respectively.
JDW32 [fog-3(wrd6)] was made by ribonucleoprotein-based co-CRISPR injection into JDW29 (Arribere et al., 2014; Paix et al., 2015). sgRNAs were generated by in vitro transcription of PCR templates, as described by Leonetti et al. (2016). JDW29 animals were injected with 3.8 µM dpy-10 sgRNA, 25.0 µM fog-3 sgRNA, 15.1 µM Cas9 protein, 0.4 µM dpy-10 repair oligo and 1.5 µM fog-3 repair oligo. The sequence of the sgRNAs and repair oligos is provided in Table S3. The insertion was targeted to the middle of exon 2 in the fog-3 locus and contained an EcoRI site to facilitate genotyping, followed by an in-frame stop codon. An additional base was inserted after the stop codon to produce a frameshift. F1 rollers were selected and screened by PCR and restriction digestion as described previously (Ward, 2015).
Western blot
Gravid JDW87 adults were allowed to lay eggs for 1 h on MYOB with and without 4 mM auxin (indole 3-acetic acid, Alfa Aesar, AAA1055622), then removed. Progeny were allowed to grow until L4+1 day at 20°C. 100 hand-picked males were collected, washed three times in M9 with 0.05% gelatin (VWR, 97062-620) and resuspended in 30 µl M9+gelatin. Worms were freeze/thawed three times in liquid nitrogen and 10 µl Laemmli sample buffer with 10% BME mixed in. Samples were boiled for 10 min at 95°C, centrifuged for 5 min at full speed and the supernatant was removed to a new tube. Proteins were separated on a SDS-PAGE Mini-PROTEAN TGX 4-15% gradient gel (BioRad, 456-1086) along a 1:1 mix of Amersham ECL Rainbow Molecular Weight Markers (95040-114) and Precision Plus Protein Unstained Standards (1610363). Proteins were transferred to a PVDF membrane using a Trans-Blot Turbo Transfer System. Membrane was washed in TBST and blocked in TBST+5% milk (VWR 10128-600) for 1 h at room temperature. Membrane was incubated in TBST+1:1000 anti-FLAG-M2-HRP (Sigma, 8592) at 4°C overnight while rolling, washed five times in TBST and incubated in 10 ml Supersignal West Femto Maximum Sensitivity Substrate (Pierce, 34095) for 5 min. Separated proteins on the gel, transferred proteins on the PVDF and the final blot were imaged using the ‘chemi high-resolution’ setting on a Bio-Rad ChemiDoc MP System. For depletion quantitation, the blot images were taken to avoid pixel saturation, and the number of pixels in each band were quantified using a densitometry feature in ImageLab software (Bio-Rad). Percent depletion was calculated by: [(pixels in control band−pixels in depletion band)/(pixels in depletion band)]×100%.
Auxin depletion
Control media consisted of MYOB agar+0.25% ethanol. Auxin media was made by dissolving indole 3-acetic acid in 100% ethanol to 1.6 M and then mixing it into melted MYOB agar at 55°C to a final concentration of 4 mM prior to pouring plates. Temperature of the media was monitored with a Lasergrip 1080 infrared thermometer gun (Etekcity). For live-progeny counts, gravid adults were allowed to lay F1 eggs for 1 h on control or auxin media and then removed. F1 progeny grew at 20°C until L4, then were individually placed onto equivalent media and allowed to lay eggs for 24 h (n=12 per condition). These worms were transferred to a new and equivalent set of media each day for 3 days and F2 eggs were counted. Once the F2 progeny of these worms grew to L4, they were counted. For matings, F1 hermaphrodite progeny were grown to L4, then individually picked to plates and either left alone or mated with males (picked as L4s) for 24 h at 20°C. The hermaphrodite animal was then transferred to new plates daily over 4 days. The eggs laid on each plate were counted after removing the parent. Viable progeny were quantified when the F1s reached L4 or adult stages (2-3 days post egg laying). To determine the auxin-sensitive period of development, F1 progeny eggs were laid on MYOB media+ethanol with or without 4 mM auxin, allowed to grow until the appropriate stage and then transferred to the opposite plate-type for the duration of their development. Once F1 progeny reached L4+1 day, hermaphrodites were transferred to fresh plates daily over 4 days and F2 viability was scored as above. For cell morphology and immunohistochemistry, worms were grown from the late L1 stage on 4 mM auxin media.
Co-suppression
nhr-23 cDNA was PCR amplified and Gateway cloned (Invitrogen) into pDONR221 to produce pJW258. The mex-5 promoter was amplified from C. elegans N2 genomic DNA and an nhr-23 cDNA was amplified from pJW258. PCR stitching was used to create a chimeric mex-5p::nhr-23 cDNA PCR product, as previously described (Dernburg et al., 2000). This product (50 ng/µl) was injected into JDW45 [fem-3(q20)] hermaphrodites along with 10 ng/µl pCFJ90 [Pmyo-2::mCherry::unc-54utr], a pharyngeal-expressed co-injection marker. Worms were incubated at 25°C to masculinize the germlines of the subsequent F1 progeny. myo-2p::mCherry-positive F1 young adults were then imaged for defective/arrested spermatocytes.
Mating/transactivation
In a variant of a transactivation experiment (Shakes and Ward, 1989), single spe-8 L4 hermaphrodites were crossed to three NHR-23-depleted or control males at room temperature (22°C). Controls included unmated spe-8 hermaphrodites as well as crossing to him-5 males or males containing the NHR-23 auxin construct that were grown on non-auxin plates. After 24 h, the males were removed. The mated spe-8 hermaphrodites were transferred to fresh plates daily, and the size of each brood was counted. For the crosses involving NHR-23-depleted males, L4 hermaphrodite progeny were assessed for self-fertility as an indicator of genotype (infertile SPE self-progeny resulting from transactivation or fertile spe/+ outcross progeny). For the crosses involving NHR-23-depleted males, one set of crosses was performed with young adult males (L4+24 h) to potentially deplete the males of their earliest produced sperm.
Statistical analyses
Statistical tests and numbers of animals analyzed are detailed in figure legends.
Supplementary Material
Acknowledgements
We thank Diana Chu and Penny Sadler for their critical reading of the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.M.R., R.M.-M., D.C.S., J.D.W.; Methodology: J.M.R., K.N.M., D.C.S., J.D.W.; Validation: J.M.R., R.M.-M., D.C.S., J.D.W.; Formal analysis: J.M.R., A.L.A., R.M.-M., D.C.S., J.D.W.; Investigation: J.M.R., A.L.A., K.N.M., R.M.-M., H.N.S., G.A.A., L.C.J., K.A.S., D.C.S., J.D.W.; Resources: J.M.R., D.C.S., J.D.W.; Data curation: J.M.R., A.L.A., K.N.M., D.C.S., J.D.W.; Writing - original draft: J.M.R., D.C.S., J.D.W.; Writing - review & editing: J.M.R., K.N.M., R.M.-M., D.C.S., J.D.W.; Visualization: J.M.R., D.C.S., J.D.W.; Supervision: J.M.R., D.C.S., J.D.W.; Project administration: D.C.S., J.D.W.; Funding acquisition: D.C.S., J.D.W.
Funding
This work was funded by the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) [R00GM107345 to J.D.W. and R15GM096309 to D.C.S.] and the National Science Foundation (NSF) Division of Molecular and Cellular Biosciences (CAREER award 1942922). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.193862.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.193862.reviewer-comments.pdf
References
- Akashi M. and Takumi T. (2005). The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 12, 441-448. 10.1038/nsmb925 [DOI] [PubMed] [Google Scholar]
- Alvarez J. D., Hansen A., Ord T., Bebas P., Chappell P. E., Giebultowicz J. M., Williams C., Moss S. and Sehgal A. (2008). The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice. J. Biol. Rhythms 23, 26-36. 10.1177/0748730407311254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- André E., Conquet F., Steinmayr M., Stratton S. C., Porciatti V. and Becker-André M. (1998). Disruption of retinoid-related orphan receptor β changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J. 17, 3867-3877. 10.1093/emboj/17.14.3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X. H., Artiles K. L., Hartman P. S. and Fire A. Z. (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837-846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley G., Duong T., Levenson M. T., Martinez M. A. Q., Hibshman J. D., Saeger H. N., Doonan R., Palmisano N. J., Martinez-Mendez R., Davidson B. et al. (2020). Expanding the Caenorhabditis elegans auxin-inducible degron system toolkit with internal expression and degradation controls and improved modular constructs for CRISPR/Cas9-mediated genome editing. bioRxiv. 10.1101/2020.05.12.090217 [DOI] [Google Scholar]
- Barton M. K. and Kimble J. (1990). fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics 125, 29-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M. K., Schedl T. B. and Kimble J. (1987). Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics 115, 107-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall E. L., Lewis P. W., Bell M., Rocha M., Jones D. L. and Botchan M. R. (2007). Discovery of tMAC: a Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B. Genes Dev. 21, 904-919. 10.1101/gad.1516607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E., Bannister L. A., Barash A., Schimenti K. J., Hartford S. A., Eppig J. J., Handel M. A., Shen L. and Schimenti J. C. (2011). A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development 138, 3319-3330. 10.1242/dev.067645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics77, 71-94. [DOI] [PMC free article] [PubMed]
- Chen P. and Ellis R. E. (2000). TRA-1A regulates transcription of fog-3, which controls germ cell fate in C. elegans. Development 127, 3119-3129. [DOI] [PubMed] [Google Scholar]
- Chu D. S. and Shakes D. C. (2013). Spermatogenesis. In Germ Cell Development in C. elegans (ed. Schedl T.), pp. 171-203. New York, NY: Springer New York. [Google Scholar]
- Chung S. S. W., Sung W., Wang X. and Wolgemuth D. J. (2004). Retinoic acid receptor alpha is required for synchronization of spermatogenic cycles and its absence results in progressive breakdown of the spermatogenic process. Dev. Dyn. 230, 754-766. 10.1002/dvdy.20083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church D. L., Guan K. L. and Lambie E. J. (1995). Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development 121, 2525-2535. [DOI] [PubMed] [Google Scholar]
- del Castillo-Olivares A., Kulkarni M. and Smith H. E. (2009). Regulation of sperm gene expression by the GATA factor ELT-1. Dev. Biol. 333, 397-408. 10.1016/j.ydbio.2009.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., Zalevsky J., Colaiácovo M. P. and Villeneuve A. M. (2000). Transgene-mediated cosuppression in the C. elegans germ line. Genes Dev. 14, 1578-1583. [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Pani A. M., Heppert J. K., Higgins C. D. and Goldstein B. (2015). Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200, 1035-1049. 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Slabodnick M. M., Chen A. H. and Goldstein B. (2018). SapTrap assembly of repair templates for Cas9-triggered homologous recombination with a self-excising cassette. microPublication Biology. 10.17912/W2KT0N. [DOI] [PMC free article] [PubMed]
- Drapek C., Sparks E. E. and Benfey P. N. (2017). Uncovering gene regulatory networks controlling plant cell differentiation. Trends Genet. 33, 529-539. 10.1016/j.tig.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbing A., Vértesy Á., Betist M. C., Spanjaard B., Junker J. P., Berezikov E., Van Oudenaarden A. and Korswagen H. C. (2018). Spatial transcriptomics of C. elegans males and hermaphrodites identifies sex-specific differences in gene expression patterns. Dev. Cell 47, 801-813.e6. 10.1016/j.devcel.2018.10.016 [DOI] [PubMed] [Google Scholar]
- Edgar L. G. (1995). Blastomere culture and analysis. In Methods in Cell Biology (ed. Epstein H. F. and Shakes D. C.), pp. 303-321. Academic Press. [DOI] [PubMed] [Google Scholar]
- Ellis R. and Schedl T. (2007). Sex determination in the germ line. WormBook (ed. The C. elegans Research Community). 10.1895/wormbook.1.82.2 [Google Scholar]
- Engert C. G., Droste R., Van Oudenaarden A. and Horvitz H. R. (2018). A Caenorhabditis elegans protein with a PRDM9-like SET domain localizes to chromatin-associated foci and promotes spermatocyte gene expression, sperm production and fertility. PLoS Genet. 14, e1007295 10.1371/journal.pgen.1007295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frand A. R., Russel S. and Ruvkun G. (2005). Functional genomic analysis of C. elegans molting. PLoS Biol. 3, e312 10.1371/journal.pbio.0030312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., Olesen S.-P., Grunnet M. and Jorgensen E. M. (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375-1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gely-Pernot A., Raverdeau M., Célébi C., Dennefeld C., Feret B., Klopfenstein M., Yoshida S., Ghyselinck N. B. and Mark M. (2012). Spermatogonia differentiation requires retinoic acid receptor γ. Endocrinology 153, 438-449. 10.1210/en.2011-1102 [DOI] [PubMed] [Google Scholar]
- Gely-Pernot A., Raverdeau M., Teletin M., Vernet N., Féret B., Klopfenstein M., Dennefeld C., Davidson I., Benoit G., Mark M. et al. (2015). Retinoic acid receptors control spermatogonia cell-fate and induce expression of the SALL4A transcription factor. PLoS Genet. 11, e1005501 10.1371/journal.pgen.1005501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissendanner C. R., Crossgrove K., Kraus K. A., Maina C. V. and Sluder A. E. (2004). Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev. Biol. 266, 399-416. 10.1016/j.ydbio.2003.10.014 [DOI] [PubMed] [Google Scholar]
- Golden A., Sadler P. L., Wallenfang M. R., Schumacher J. M., Hamill D. R., Bates G., Bowerman B., Seydoux G. and Shakes D. C. (2000). Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151, 1469-1482. 10.1083/jcb.151.7.1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks G.-J., Gaidatzis D., Aeschimann F. and Großhans H. (2014). Extensive oscillatory gene expression during C. elegans larval development. Mol. Cell 53, 380-392. 10.1016/j.molcel.2013.12.013 [DOI] [PubMed] [Google Scholar]
- Hodgkin J. (1987). A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes Dev. 1, 731-745. 10.1101/gad.1.7.731 [DOI] [PubMed] [Google Scholar]
- Horvath G. C., Kistler M. K. and Kistler W. S. (2009). RFX2 is a candidate downstream amplifier of A-MYB regulation in mouse spermatogenesis. BMC Dev. Biol. 9, 63 10.1186/1471-213X-9-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.-Y., Sun Z.-W., Li X., Reuben M., Tatchell K., Bishop D. K., Grushcow J. M., Brame C. J., Caldwell J. A., Hunt D. F. et al. (2000). Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279-291. 10.1016/S0092-8674(00)00034-9 [DOI] [PubMed] [Google Scholar]
- Hu J., Cheng S., Wang H., Li X., Liu S., Wu M., Liu Y. and Wang X. (2019). Distinct roles of two myosins in C. elegans spermatid differentiation. PLoS Biol. 17, e3000211 10.1371/journal.pbio.3000211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S.-W., Kimble J. and Ellis R. E. (2001). Regulation of cell fate in Caenorhabditis elegans by a novel cytoplasmic polyadenylation element binding protein. Dev. Biol. 229, 537-553. 10.1006/dbio.2000.9993 [DOI] [PubMed] [Google Scholar]
- Kageyama Y., Masuda S., Hirose S. and Ueda H. (1997). Temporal regulation of the mid-prepupal gene FTZ-F1: DHR3 early late gene product is one of the plural positive regulators. Genes Cells 2, 559-569. 10.1046/j.1365-2443.1997.1460344.x [DOI] [PubMed] [Google Scholar]
- Kallen J. A., Schlaeppi J.-M., Bitsch F., Geisse S., Geiser M., Delhon I. and Fournier B. (2002). X-ray structure of the hRORα LBD at 1.63 Å: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORα. Structure 10, 1697-1707. 10.1016/S0969-2126(02)00912-7 [DOI] [PubMed] [Google Scholar]
- Kallen J., Schlaeppi J.-M., Bitsch F., Delhon I. and Fournier B. (2004). Crystal structure of the human RORα Ligand binding domain in complex with cholesterol sulfate at 2.2 Å. J. Biol. Chem. 279, 14033-14038. 10.1074/jbc.M400302200 [DOI] [PubMed] [Google Scholar]
- Kang T.-H., Lindsey-Boltz L. A., Reardon J. T. and Sancar A. (2010). Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc. Natl Acad. Sci. USA 107, 4890-4895. 10.1073/pnas.0915085107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimatis K. R., Moerdyk-Schauwecker M. J. and Phillips P. C. (2018). Auxin-mediated sterility induction system for longevity and mating studies in Caenorhabditis elegans. G3 (Bethesda) 8, 2655-2662. 10.1534/g3.118.200278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Xu S., Montgomery M. K. and Fire A. (1997). Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics146, 227-238. [DOI] [PMC free article] [PubMed]
- Kelly W. G., Schaner C. E., Dernburg A. F., Lee M.-H., Kim S. K., Villeneuve A. M. and Reinke V. (2002). X-chromosome silencing in the germline of C. elegans. Development 129, 479-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennaway D. J., Boden M. J. and Varcoe T. J. (2012). Circadian rhythms and fertility. Mol. Cell. Endocrinol. 349, 56-61. 10.1016/j.mce.2011.08.013 [DOI] [PubMed] [Google Scholar]
- Kosinski M. (2005). C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development 132, 3357-3369. 10.1242/dev.01916 [DOI] [PubMed] [Google Scholar]
- Kostrouchova M., Krause M., Kostrouch Z. and Rall J. E. (1998). CHR3: a Caenorhabditis elegans orphan nuclear hormone receptor required for proper epidermal development and molting. Development 125, 1617-1626. [DOI] [PubMed] [Google Scholar]
- Kostrouchova M., Krause M., Kostrouch Z. and Rall J. E. (2001). Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 98, 7360-7365. 10.1073/pnas.131171898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouns N. A., Nakielna J., Behensky F., Krause M. W., Kostrouch Z. and Kostrouchova M. (2011). NHR-23 dependent collagen and hedgehog-related genes required for molting. Biochem. Biophys. Res. Commun. 413, 515-520. 10.1016/j.bbrc.2011.08.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni M., Shakes D. C., Guevel K. and Smith H. E. (2012). SPE-44 implements sperm cell fate. PLoS Genet. 8, e1002678 10.1371/journal.pgen.1002678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laktionov P. P., Maksimov D. A., Romanov S. E., Antoshina P. A., Posukh O. V., White-Cooper H., Koryakov D. E. and Belyakin S. N. (2018). Genome-wide analysis of gene regulation mechanisms during Drosophila spermatogenesis. Epigenet. Chromatin 11, 1-15. 10.1186/s13072-018-0183-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam G. T., Jiang C. and Thummel C. S. (1997). Coordination of larval and prepupal gene expression by the DHR3 orphan receptor during Drosophila metamorphosis. Development 124, 1757-1769. [DOI] [PubMed] [Google Scholar]
- Lamitina S. T. and L'Hernault S. W. (2002). Dominant mutations in the Caenorhabditis elegans Myt1 ortholog wee-1.3 reveal a novel domain that controls M-phase entry during spermatogenesis. Development 129, 5009-5018. [DOI] [PubMed] [Google Scholar]
- Leonetti M. D., Sekine S., Kamiyama D., Weissman J. S., Huang B. (2016). A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc. Natl. Acad. Sci. USA113, E3501-3508. 10.1073/pnas.1606731113 [DOI]
- Levine M. and Davidson E. H. (2005). Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 102, 4936-4942. 10.1073/pnas.0408031102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault S. W. and Arduengo P. M. (1992). Mutation of a putative sperm membrane protein in Caenorhabditis elegans prevents sperm differentiation but not its associated meiotic divisions. J. Cell Biol. 119, 55-68. 10.1083/jcb.119.1.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault S. W., Shakes D. C. and Ward S. (1988). Developmental genetics of chromosome I spermatogenesis-defective mutants in the nematode Caenorhabditis elegans. Genetics 120, 435-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Z., Roy C. K., Dong X., Bolcun-Filas E., Wang J., Han B. W., Xu J., Moore M. J., Schimenti J. C., Weng Z. et al. (2013). An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell 50, 67-81. 10.1016/j.molcel.2013.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machaca K. and L'Hernault S. W. (1997). The Caenorhabditis elegans spe-5 gene is required for morphogenesis of a sperm-specific organelle and is associated with an inherent cold-sensitive phenotype. Genetics 146, 567-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macneil L. T., Watson E., Arda H. E., Zhu L. J. and Walhout A. J. M. (2013). Diet-Induced Developmental Acceleration Independent of TOR and Insulin in C. elegans. Cell 153, 240-252. 10.1016/j.cell.2013.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal K., Sarkar R. K., Sen Sharma S., Jain A. and Majumdar S. S. (2018). Sertoli cell specific knockdown of RAR-related orphan receptor (ROR) alpha at puberty reduces sperm count in rats. Gene 641, 18-24. 10.1016/j.gene.2017.10.032 [DOI] [PubMed] [Google Scholar]
- McKearin D. M. and Spradling A. C. (1990). bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 4, 2242-2251. 10.1101/gad.4.12b.2242 [DOI] [PubMed] [Google Scholar]
- Medwig-Kinney T. N., Smith J. J., Palmisano N. J., Tank S., Zhang W. and Matus D. Q. (2020). A developmental gene regulatory network for C. elegans anchor cell invasion. Development 147, dev185850 10.1242/dev.185850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D. and Seydoux G. (2008). 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18, 1476-1482. 10.1016/j.cub.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf C. E. and Wassarman D. A. (2007). Nucleolar colocalization of TAF1 and testis-specific TAFs during Drosophila spermatogenesis. Dev. Dyn. 236, 2836-2843. 10.1002/dvdy.21294 [DOI] [PubMed] [Google Scholar]
- Miller M. A., Nguyen V. Q., Lee M.-H., Kosinski M., Schedl T., Caprioli R. M. and Greenstein D. (2001). A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291, 2144-2147. 10.1126/science.1057586 [DOI] [PubMed] [Google Scholar]
- Minniti A. N., Sadler C. and Ward S. (1996). Genetic and molecular analysis of spe-27, a gene required for spermiogenesis in Caenorhabditis elegans hermaphrodites. Genetics 143, 213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D., Cermakian N., Brancorsini S., Parvinen M. and Sassone-Corsi P. (2003). No circadian rhythms in testis: Period1 expression is clock independent and developmentally regulated in the mouse. Mol. Endocrinol. 17, 141-151. 10.1210/me.2002-0184 [DOI] [PubMed] [Google Scholar]
- Muhlrad P. J. and Ward S. (2002). Spermiogenesis initiation in Caenorhabditis elegans involves a casein kinase 1 encoded by the spe-6 gene. Genetics 161, 143-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J., Minniti A. N., Sadler C. and Ward S. (1999). spe-12 encodes a sperm cell surface protein that promotes spermiogenesis in Caenorhabditis elegans. Genetics 152, 209-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J., Davis E. B. and Ward S. (2000). spe-29 encodes a small predicted membrane protein required for the initiation of sperm activation in Caenorhabditis elegans. Genetics 156, 1623-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H. and L'Hernault S. W. (2010). Spermatogenesis-defective (spe) mutants of the nematode Caenorhabditis elegans provide clues to solve the puzzle of male germline functions during reproduction. Dev. Dyn. 239, 1502-1514. 10.1002/dvdy.22271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Fukagawa T., Takisawa H., Kakimoto T. and Kanemaki M. (2009). An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Meth. 6, 917-922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Okamoto H. and Thomson J. N. (1985). Monoclonal antibodies which distinguish certain classes of neuronal and supporting cells in the nervous tissue of the nematode Caenorhabditis elegans. J. Neurosci. 5, 643-653. 10.1523/JNEUROSCI.05-03-00643.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page B. D., Zhang W., Steward K., Blumenthal T. and Priess J. R. (1997). ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes Dev.. 11, 1651-1661. 10.1101/gad.11.13.1651 [DOI] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D. and Seydoux G. (2015). High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201, 47-54. 10.1534/genetics.115.179382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. and Frand A. R. (2018). A regulatory loop between the retinoid-related orphan nuclear receptor NHR-23 and let-7 family microRNAs modulates the C. elegans molting cycle. bioRxiv 10.1101/506261 [DOI] [Google Scholar]
- Reinke V., Gil I. S., Ward S. and Kazmer K. (2004). Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131, 311-323. 10.1242/dev.00914 [DOI] [PubMed] [Google Scholar]
- Roach N. P., Sadowski N., Alessi A. F., Timp W., Taylor J. and Kim J. K. (2020). The full-length transcriptome of C. elegans using direct RNA sequencing. Genome Res. 30, 299-312. 10.1101/gr.251314.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler P. L. and Shakes D. C. (2000). Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo. Development 127, 355-366. [DOI] [PubMed] [Google Scholar]
- Sato T. K., Panda S., Miraglia L. J., Reyes T. M., Rudic R. D., McNamara P., Naik K. A., FitzGerald G. A., Kay S. A. and Hogenesch J. B. (2004). A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43, 527-537. 10.1016/j.neuron.2004.07.018 [DOI] [PubMed] [Google Scholar]
- Schedl T., Graham P. L., Barton M. K. and Kimble J. (1989). Analysis of the role of tra-1 in germline sex determination in the nematode Caenorhabditis elegans. Genetics 123, 755-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert C. M., Lin R., de Vries C. J., Plasterk R. H. A. and Priess J. R. (2000). MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol. Cell 5, 671-682. 10.1016/S1097-2765(00)80246-4 [DOI] [PubMed] [Google Scholar]
- Schwartz M. L. and Jorgensen E. M. (2016). SapTrap, a toolkit for high-throughput CRISPR/Cas9 gene modification in Caenorhabditis elegans. Genetics 202, 1277-1288. 10.1534/genetics.115.184275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Ailion M., Li J., van Oudenaarden A., Rockman M. V. and Kruglyak L. A. (2011). Novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biol.9, e1001115. 10.1371/journal.pbio.1001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Rockman M. V. and Kruglyak L. (2008). Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science 319, 589-594. 10.1126/science.1151107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes D. C. and Ward S. (1989). Initiation of spermiogenesis in C. elegans: a pharmacological and genetic analysis. Dev. Biol. 134, 189-200. 10.1016/0012-1606(89)90088-2 [DOI] [PubMed] [Google Scholar]
- Shakes D. C., Wu J.-C., Sadler P. L., Laprade K., Moore L. L., Noritake A. and Chu D. S. (2009). Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet. 5, e1000611 10.1371/journal.pgen.1000611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroosh P., Wu J., Xue X., Song J., Sutton S. W., Sablad M., Yu J., Nelen M. I., Liu X., Castro G. et al. (2014). Oxysterols are agonist ligands of RORγt and drive Th17 cell differentiation. Proc. Natl Acad. Sci. USA 111, 12163-12168. 10.1073/pnas.1322807111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K. and Seydoux G. (2003). Dedifferentiation of primary spermatocytes into germ cell tumors in C. elegans lacking the Pumilio-like protein PUF-8. Curr. Biol. 13, 134-139. 10.1016/S0960-9822(03)00005-8 [DOI] [PubMed] [Google Scholar]
- Tzur Y. B., Winter E., Gao J., Hashimshony T., Yanai I. and Colaiácovo M. P. (2018). Spatiotemporal gene expression analysis of the Caenorhabditis elegans germline uncovers a syncytial expression switch. Genetics 210, 587-605. 10.1534/genetics.118.301315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkey J. P., Jansma P. L., Minniti A. N. and Ward S. (1993). The Caenorhabditis elegans spe-6 gene is required for major sperm protein assembly and shows second site non-complementation with an unlinked deficiency. Genetics 133, 79-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kumar N., Crumbley C., Griffin P. R. and Burris T. P. (2010). A second class of nuclear receptors for oxysterols: Regulation of RORα and RORγ activity by 24S-hydroxycholesterol (cerebrosterol). Biochim. Biophys. Acta 1801, 917-923. 10.1016/j.bbalip.2010.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. D. (2015). Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics 199, 363-377. 10.1534/genetics.114.172361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., Argon Y. and Nelson G. A. (1981). Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J. Cell Biol. 91, 26-44. 10.1083/jcb.91.1.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K. P., Hurban P., Watanabe T. and Hogness D. S. (1997). Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science 276, 114-117. 10.1126/science.276.5309.114 [DOI] [PubMed] [Google Scholar]
- White-Cooper H. and Bausek N. (2010). Evolution and spermatogenesis. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 365, 1465-1480. 10.1098/rstb.2009.0323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter E. S., Schwarz A., Fabig G., Feldman J. L., Pires-daSilva A., Müller-Reichert T., Sadler P. L. and Shakes D. C. (2017). Cytoskeletal variations in an asymmetric cell division support diversity in nematode sperm size and sex ratios. Development 144, 3253-3263. 10.1242/dev.153841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D. and Hodgkin J. (1992). Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell 70, 237-249. 10.1016/0092-8674(92)90099-X [DOI] [PubMed] [Google Scholar]
- Zhang L., Ward J. D., Cheng Z. and Dernburg A. F. (2015). The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374-4384. 10.1242/dev.129635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Xie D., Lin X., Ma L., Chen J., Zhang D., Wang Y., Duo S., Feng Y., Zheng C. et al. (2018). The transcription factor SOX30 is a key regulator of mouse spermiogenesis. Development 145, dev164723 10.1242/dev.164723 [DOI] [PubMed] [Google Scholar]
- Zhu G.-D. and L'Hernault S. W. (2003). The Caenorhabditis elegans spe-39 gene is required for intracellular membrane reorganization during spermatogenesis. Genetics 165, 145-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.