ABSTRACT
The germline genome is guarded against invading foreign genetic elements by small RNA-dependent gene-silencing pathways. Components of these pathways localize to, or form distinct aggregates in the vicinity of, germ granules. These components and their dynamics in and out of granules are currently being intensively studied. Here, we report the identification of PLP-1, a Caenorhabditis elegans protein related to the human single-stranded nucleic acid-binding protein Pur-alpha, as a component of germ granules in C. elegans. We show that PLP-1 is essential for silencing different types of transgenes in the germ line and for suppressing the expression of several endogenous genes controlled by the germline gene-silencing pathways. Our results reveal that PLP-1 functions downstream of small RNA biogenesis during initiation of gene silencing. Based on these results and the earlier findings that Pur-alpha proteins interact with both RNA and protein, we propose that PLP-1 couples certain RNAs with their protein partners in the silencing complex. PLP-1 orthologs localized on RNA granules may similarly contribute to germline gene silencing in other organisms.
KEY WORDS: Pur-alpha, puf-8, P granules, piRNA, Major sperm proteins
Summary: PLP-1, the C. elegans ortholog of the conserved Pur-alpha proteins, functions downstream of small RNA biogenesis during initiation of gene silencing in the germ line.
INTRODUCTION
Maintenance of germline genome integrity is crucial for faithful genetic inheritance across generations. This task is accomplished by germline-specific genome surveillance mechanisms that recognize ‘self’ and ‘non-self’ nucleic acid sequences. These mechanisms protect the expression of endogenous genes that normally function in the germ line, but can silence the expression of foreign genetic elements, including artificially-introduced transgenes, and restrict the expression of certain endogenous loci in the germ line.
Silencing of some transposons, transgenes, certain endogenous loci and the sequence-specific silencing triggered by experimentally-introduced dsRNA (RNAi) all involve three major steps: generation of primary small interfering RNAs (primary siRNAs); amplification of the initial signal, which involves the production of secondary siRNAs in many organisms; and systemic disruption of target expression by cleavage of the target RNA, although other modes of regulation are also employed (for a review see Billi et al., 2014). The different silencing pathways rely on distinct proteins, but also share certain components, particularly in the latter steps. For example, in Caenorhabditis elegans, the mutator proteins are essential for transposon silencing and contribute to the RNAi response (Ketting et al., 1999; Phillips et al., 2012). In the RNAi pathway, exogenous dsRNAs are cleaved by the ribonuclease Dicer into primary siRNAs, which then associate with and guide the Argonaute (AGO) protein RDE-1 to target sequences (Bernstein et al., 2001; Ketting et al., 2001; Parrish and Fire, 2001; Yigit et al., 2006), whereas in the pathway that silences transgenes and endogenous genes, endogenously-encoded small RNAs, called piRNAs, guide the Piwi AGO protein PRG-1 to target sequences (Bagijn et al., 2012; Lee et al., 2012).
Once at the target, the primary siRNA-AGO complex recruits the RNA-dependent RNA polymerase (RdRP) system, which generates secondary siRNAs using the target sequence as template (Pak and Fire, 2007; Sijen et al., 2001). In C. elegans, many of the secondary siRNAs are 22-nucleotides (nt) long and contain a triphosphorylated guanosine at the 5′ end (22G-RNAs) (Gent et al., 2010; Pak and Fire, 2007). The 22G-RNAs associate with either a distinct AGO, CSR-1, or one of 12 worm-specific AGOs (called WAGOs), which guide small RNAs to target sequences (Gerson-Gurwitz et al., 2016; Gu et al., 2009; Guang et al., 2008). In addition to AGOs, several proteins aid RdRP recruitment. These include the Dicer-related RNA helicase DRH-3 and the Tudor proteins ERI-5 and EKL-1 (Gu et al., 2009; Thivierge et al., 2012). In addition, the mutator proteins MUT-2/RDE-3, MUT-7 and MUT-16 also apparently participate directly or indirectly in the biogenesis/stability of 22G-RNAs (Phillips et al., 2012). MUT-2/RDE-3 is a ribonucleotidyltransferase that adds uridine-guanosine dinucleotide (pUG) repeats to the targets. The pUG tails not only facilitate RdRP recruitment, but convert such pUG-tailed RNAs into templates for RdRPs (Shukla et al., 2020).
Many proteins mentioned above have been observed to localize to germ granules (Batista et al., 2008; Claycomb et al., 2009; Conine et al., 2010; Gu et al., 2009). These granules, known as P granules in C. elegans, are aggregates of RNAs and proteins (Eddy, 1975; Strome and Wood, 1982). P granules, like the other RNA granules, are biomolecular condensates having physical properties characteristic of liquid droplets (Brangwynne et al., 2009). Their components are important in the transport, stability and translational regulation of certain germline mRNAs, and organize gene-silencing pathways in the germ line (Dodson and Kennedy, 2019; Lev et al., 2019; Sheth et al., 2010; Spike et al., 2008; Updike and Strome, 2010; Voronina and Seydoux, 2010). Recent findings reveal distinct subregions adjacent to where the core P granule components, such as PGL-1, localize. It emerges that the components in these subregions have distinct functions and assemble independently of the core P granule components. For example, the mutator proteins, nucleated by MUT-16, form distinct foci adjacent to P granules. The formation and stability of these mutator foci do not require the known P granule components (Phillips et al., 2012). Similarly, ZNFX-1, which functions in the transgene-silencing pathway, and WAGO-4 form distinct liquid-like condensates, called Z granules, sandwiched between P granules and mutator foci (Ishidate et al., 2018; Wan et al., 2018). What other components, if any, are present in these structures and how the various components coalesce into physically and functionally distinct subregions are poorly understood. Importantly, how being part of such assemblies helps the individual components to function in gene silencing is not clear.
Here, we identify PLP-1 (Pur-alpha-like protein-1), the C. elegans ortholog of the conserved Pur-alpha proteins, as a new component of the germline gene-silencing machinery. PLP-1 localizes to P granules, and its loss causes temperature-sensitive sterility. Our experiments reveal a role for PLP-1 in silencing transgenes, repetitive DNA arrays and piRNA sensors, and in mounting a robust RNAi response. Furthermore, we find that PLP-1 modulates the expression of several germline-enriched genes that have previously been shown to be controlled by the silencing pathways.
RESULTS
Simultaneous loss of PLP-1 and PUF-8 disrupts oogenesis
The conserved RNA-binding protein PUF-8 is a key regulator of germ cell development in C. elegans. However, even null alleles of puf-8 cause only temperature-sensitive sterility (Subramaniam and Seydoux, 2003). Exploiting this feature, previous work in our laboratory isolated a number of mutant alleles that displayed constitutive sterility when in double mutant combination with the puf-8 mutation (Maheshwari et al., 2016; Vaid et al., 2013). While mapping one such new mutant allele, we discovered that the RNAi-mediated depletion of PLP-1, which shares sequence similarity with the human protein Pur-alpha (purine-rich element binding protein-alpha; Bergemann et al., 1992), made puf-8(−) worms sterile at 20°C, a temperature at which they were otherwise fertile. To examine this sterility phenotype further, we used the ok2155 allele of plp-1, which is a 1107 bp deletion that deletes the first two exons and part of the promoter region, and constructed a puf-8(ok302); plp-1(ok2155) double mutant strain (hereafter referred to as puf-8; plp-1) and examined the germ line systematically (Table S1). When raised at 20°C, all the puf-8 and plp-1 single mutants were fertile (n=548 and 500, respectively). By contrast, ∼99% of the puf-8; plp-1 double mutants were sterile (n=358), which reveals a potential redundancy between puf-8 and plp-1 in their germline function.
The C. elegans genome encodes a paralog of PLP-1, named PLP-2; these proteins share a high degree of amino acid sequence similarity (Fig. S1). To test whether PLP-2 could functionally substitute PLP-1, we generated plp-1(ok2155); plp-2(ok2153). The ok2153 allele deletes the entire coding sequence of plp-2. Similar to plp-1 and plp-2 single mutants that were all fertile (n=200), all the plp-1; plp-2 double mutants were also fertile (n=200). Moreover, in contrast to the sterility observed in puf-8; plp-1 worms, none of the puf-8; plp-2 worms were sterile (n=200). These results indicate that plp-2 does not functionally overlap with plp-1 in the germ line.
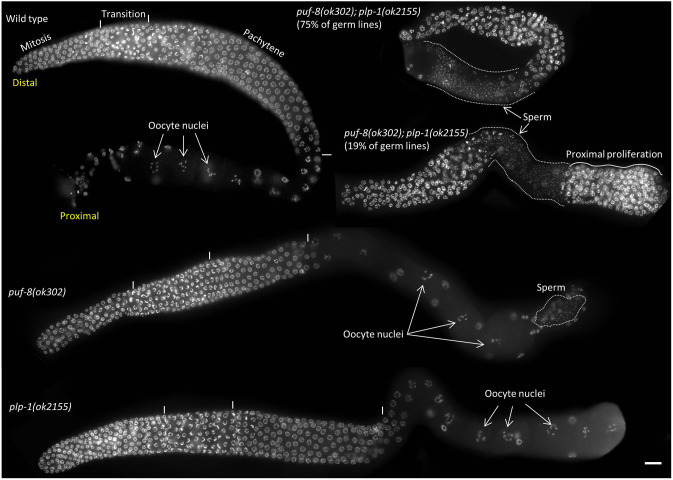
To more precisely determine the puf-8; plp-1 defect, we examined germ cell nuclei at different stages of development in animals grown at 20°C. For this, we visualized the chromatin morphology of germ cells, which is unique to each developmental stage, by staining with the DNA-binding dye DAPI. Germ cells in C. elegans hermaphrodites develop within two hairpin-shaped tubes, called the gonadal arms. Both gonadal arms are closed at one end, known as the distal end, and open at the other end, known as the proximal end. Both gonadal arms open into a common uterus at their proximal ends. The germline stem cells (GSCs) are present at the distalmost part; they progress through the various stages of meiosis and gametogenesis as they migrate proximally. All germ cells present within a gonadal arm are usually referred to as the germ line. As observed in wild-type germ lines, germ cell nuclei of all developmental stages, including the two types of gametes, were present in both puf-8 and plp-1 single mutant germ lines. By contrast, at least four kinds of defects could be observed in puf-8; plp-1 double mutant hermaphrodite germ lines: ∼75% of them did not have any oocytes and had only sperm at the proximal end; 19% had proliferating cells at the proximal end, reminiscent of the germ cell tumors observed in puf-8 mutant worms raised at 25°C (Subramaniam and Seydoux, 2003); ∼4% had neither type of gametes; and the remaining 2% had both oocytes and sperm (n=712) (Fig. 1). In addition, these germ lines were shorter and had fewer germ cells in the distal part when compared to wild type and the two single mutants. However, no obvious defects were observed in puf-8; plp-1 male germ lines when compared to puf-8 mutants (Fig. S2).
Fig. 1.
puf-8; plp-1 double mutant germ lines do not produce oocytes. Germ lines extruded from 1-day-old adults and stained with DAPI are shown. Approximate boundaries of different developmental stages are indicated by vertical lines and labeled above the wild-type germ line. Although oocyte nuclei can be seen in wild-type and the two single mutant germ lines, they are absent in the puf-8; plp-1 double mutant. The double mutant germ lines are relatively shorter, and the chromatin morphologies characteristic of the different developmental stages are not discernible in them. In some of these double mutant germ lines, proliferating germ cells are seen in the proximal region. Scale bar: 20 µm.
In C. elegans hermaphrodites, absence of oocytes can result from defects in either the spermatogenesis-to-oogenesis switch or oocyte development. To distinguish between these two possibilities, we feminized the germ lines using a fem-3 loss-of-function mutation [fem-3(e1996)], which eliminates spermatogenesis without affecting oogenesis (Ahringer and Kimble, 1991; Barton et al., 1987; Hodgkin, 1986). As expected, fem-3, puf-8; fem-3 and plp-1 fem-3 germ lines produced only oocytes, whereas no gametes were seen in puf-8; plp-1 fem-3 triple mutant germ lines (Fig. S3). Similarly, 95% of puf-8; plp-1 germ lines (n=260) did not produce any gametes when depleted of FOG-2, another protein required for spermatogenesis (Schedl and Kimble, 1988). These observations point to a role for both puf-8 and plp-1 in oocyte development, and show that at least one of these two genes must be active for normal oogenesis.
Simultaneous loss of PUF-8 and PLP-1 had only a marginal effect on germ cell proliferation. The number of metaphase nuclei, detected by immunostaining with anti-phosphohistone H3 antibodies, was slightly reduced in the mitotic zone of puf-8; plp-1 germ lines (Fig. S4). However, the excessive proliferation of germ cells caused by gain-of-function mutations in glp-1, a gene required for GSC maintenance (Pepper et al., 2003), was unaffected by the loss of puf-8 and plp-1. Excessive proliferation has also been observed in gld-2; gld-1 double mutants; these two genes function in a redundant fashion to promote meiotic entry (Kadyk and Kimble, 1998). Even here, loss of PUF-8 and PLP-1 had no effect: we did not see any reduction in the size of the proliferating germ cell population in gld-2; gld-1; puf-8; plp-1 quadruple mutants when compared with that in gld-2; gld-1 double mutants (Fig. S5). Both PUF-8 and PLP-1 could be dispensed with for meiotic entry as well: the expression pattern of HIM-3, a synaptonemal complex protein that is often used as a meiotic marker (Zetka et al., 1999), was not altered in puf-8; plp-1 germ lines (Fig. S4).
The plp-1 single mutant worms display a temperature-sensitive sterility phenotype
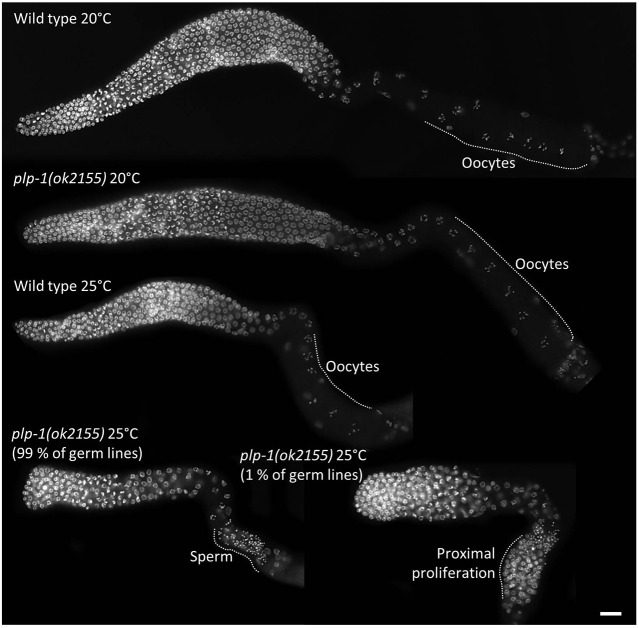
We wished to test whether plp-1 single mutants, like puf-8 single mutants, displayed any temperature-sensitive phenotype. For this, we shifted embryos collected from wild-type and plp-1(ok2155) hermaphrodites maintained at 20°C to 25°C and allowed them to hatch and develop to adulthood at 25°C. Embryos of both genotypes hatched and reached the adult stage normally. However, whereas all wild-type embryos developed into fertile adults (n=114), none of the plp-1 adults were fertile (n=248; Table S2). A similar observation has been made by another independent study that was published while this work was in progress (Lalani et al., 2014). A closer look at germ cells following DAPI staining revealed that the majority of plp-1 germ lines did not produce oocytes, whereas a few of them developed germ cell tumors at the proximal end at about 72 h after hatching. Even these few did not have any oocytes (Fig. 2). However, a few oocytes could be seen in 63% of plp-1 germ lines a day later (2-day-old adults; n=162). These oocytes were presumably defective, because a few of them appeared to have undergone endomitotic reduplication. None of the plp-1 worms grown at 25°C produced embryos, even by 120 h after hatching (n=68). These results reveal an essential function for plp-1 in oocyte development at elevated temperatures.
Fig. 2.
plp-1 mutant animals display temperature-sensitive oogenesis defects. Extruded germ lines of wild-type and plp-1 mutant hermaphrodites grown at the indicated temperatures were stained with DAPI. Similar to the puf-8; plp-1 double mutant (Fig. 1), plp-1 single mutants grown at 25°C do not produce oocytes, and some of them develop proximal tumors. Scale bar: 20 µm.
PLP-1 is essential in males for mating
Our attempts to generate plp-1(ok2155) males uncovered a role for PLP-1 in the mating process. We obtained plp-1(ok2155/+) heterozygous males by mating plp-1(ok2155/ok2155) hermaphrodites with wild-type males, and backcrossed them with plp-1(ok2155/ok2155) hermaphrodites. The resulting male progeny were backcrossed again, and this process was repeated for several successive generations. Genotyping of the male progeny by PCR revealed the presence of plp-1(ok2155/ok2155) homozygous males among the progeny. However, ∼50% of male progeny were heterozygous even after ten successive backcrosses. We then tested the ability of these males to mate by setting up 24 single-male crosses with fem-3(e1996) hermaphrodites, which, as mentioned above, do not produce any sperm but produce normal oocytes. We genotyped the male parents of these crosses after allowing sufficient time for mating (48 h). Whereas 12 out of 16 plp-1(ok2155/+) heterozygous males sired progeny, none of the eight plp-1(ok2155/ok2155) homozygous males sired progeny (Table S3). The fem-3(e1996) hermaphrodites used in these crosses did produce progeny when subsequently mated with wild-type males, indicating that these hermaphrodites were not defective in producing oocytes or mating. We inspected the germ lines of males that failed to sire progeny by staining with DAPI and found that the sperm production in these males was normal. However, the plp-1 males did not show normal mating behavior: unlike wild-type males, plp-1 males did not stop forward locomotion upon encountering a hermaphrodite, or go around the hermaphrodite in search of the vulva. Consistently, we did not find any sperm in the uterus of hermaphrodites mated with these males. Thus, the failure of plp-1 males to sire progeny most likely resulted from some somatic defect that compromised their normal mating behavior.
PLP-1 localizes to P granules throughout the germ line and to similar perinuclear granules in some somatic cells
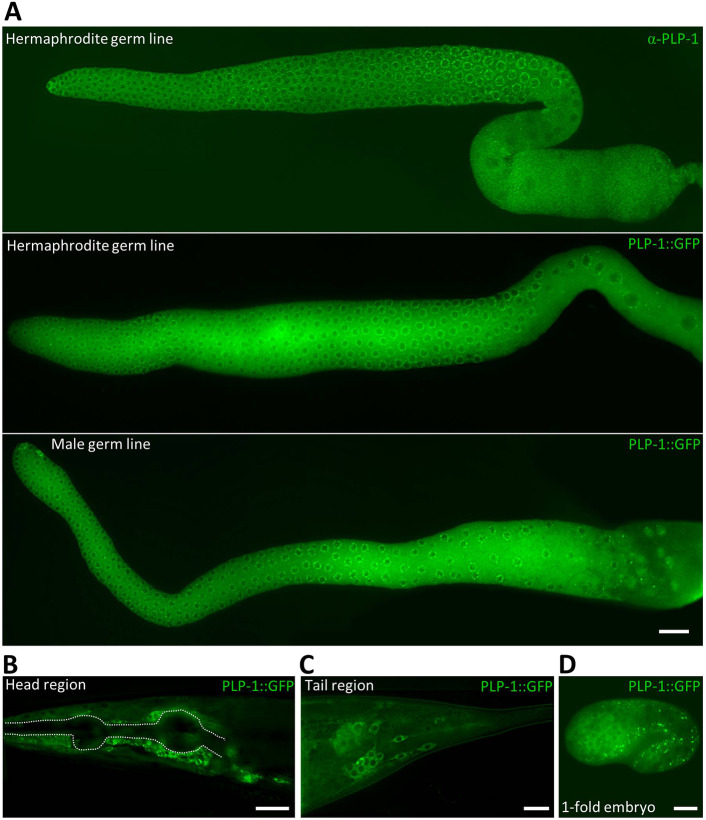
To begin to understand how PLP-1 functions in the germ line, we determined its expression pattern using both PLP-1-specific antibodies and an integrated transgene that expresses PLP-1::GFP under the control of the plp-1 promoter and 3′ UTR. This transgene restored fertility in puf-8; plp-1 mutants, indicating that the GFP fusion did not interfere with PLP-1 activity (88% of 209 animals were fertile). As shown in Fig. 3A, PLP-1 was present throughout the germ line, with prominent concentration on granules arranged in what appeared as perinuclear circles. Distribution of PLP-1 in the core cytoplasm was more readily noticeable in unfixed, live germ lines expressing the plp-1::gfp transgene. Similar distribution patterns were observed in both hermaphrodite and male germ lines. In worms expressing the plp-1::gfp transgene, we noticed GFP fluorescence in several cells surrounding the pharyngeal bulbs (Fig. 3B) and in cells in the tail region (Fig. 3C). Even in these somatic cells, PLP-1::GFP was concentrated on perinuclear granules, in addition to being present in the cytoplasm. Both the plp-1::gfp transgene and anti-PLP-1 antibodies revealed the presence of PLP-1 in the cytoplasm of both somatic and germline blastomeres throughout embryonic development (Fig. 3D, Fig. 4B; Fig. S6). Similar localization patterns were observed using the mCherry tag as well, and in worms in which endogenous PLP-1 was tagged with GFP (Fig. S7).
Fig. 3.
Distribution patterns of PLP-1. (A) Extruded germ lines are shown. Top: hermaphrodite germ line fixed and immunostained with anti-PLP-1 antibodies. Middle: germ line from a live hermaphrodite carrying the Pplp-1::plp-1::gfp::plp-1 3′ UTR transgene. Bottom: germline from a live male carrying the same transgene as above. PLP-1::GFP in the core cytoplasm is noticeable in the transgenic germ lines, whereas the perinuclear puncta of PLP-1/PLP-1::GFP are visible in all three. (B-D) PLP-1::GFP is expressed in somatic cells around the pharynx (B), tail region (C) and late-stage embryo (D). Perinuclear PLP-1::GFP puncta are seen in some of these somatic cells as well. Dotted line in B outlines the pharynx. Scale bars: 20 µm (A,B); 10 µm (C,D).
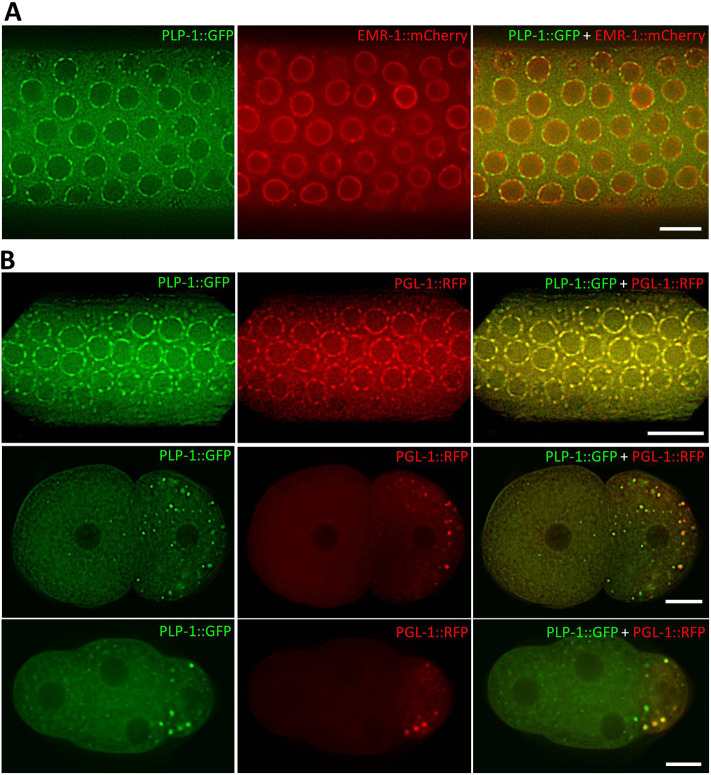
Fig. 4.
PLP-1 localizes to perinuclear P granules in the germ line and to P granules and unknown granules in embryos. (A) A section of germ line from a hermaphrodite showing the expression of PLP-1::GFP and the nuclear envelope marker EMR-1::mCherry. These fusion proteins are expressed by the transgenes Pplp-1::plp-1::gfp::plp-1 3′ UTR and Ppie-1::mCherry::emr-1::hip-1 3′ UTR, respectively. As can been seen in the merged image on the right, the PLP-1::GFP puncta surround the nuclear membrane, indicating that these puncta are cytoplasmic. (B) PLP-1::GFP and the P granule marker PGL-1::RFP colocalize in the germ line (top) and in two-cell (middle) and four-cell embryos (bottom). Note: Not all PLP-1::GFP puncta colocalize with the P granules in embryos, and a few smaller and fainter PLP-1::GFP granules are visible in the somatic blastomeres as well. Scale bars: 10 µm.
We confirmed that the PLP-1 circles were indeed perinuclear by using the nuclear membrane marker, EMR-1::mCherry (Fig. 4A) (Pushpa et al., 2013). To determine whether PLP-1 is a component of P granules, we generated double transgenic lines expressing both PLP-1::GFP and an RFP-tagged version of PGL-1, which is a core component of P granules (Kawasaki et al., 1998; Wolke et al., 2007). We found that PLP-1::GFP indeed colocalized with PGL-1::RFP in both adults and embryos (Fig. 4B). Intriguingly, unlike in adult germ lines, not all PLP-1::GFP granules colocalized with PGL-1::RFP in embryos. Localization of PLP-1 on perinuclear granules in the germ line could be detected in anti-PLP-1-immunostained embryos as well (Fig. S6).
Localization of PLP-1 to P granules in embryos has been observed previously (Witze et al., 2009). However, in contrast to this earlier study, which found PLP-1 mostly in the nuclei of embryos (Witze et al., 2009), we did not observe any detectable nuclear localization of PLP-1 in the transgenic reporter lines described above or in germ lines and embryos immunostained with PLP-1-specific antibodies. We think differences in antibody specificities and immunostaining conditions are the probable causes of the variation between our observations and those previously published.
PLP-1 is essential for silencing transgenes and piRNA targets in the germ line, and for a robust RNAi response
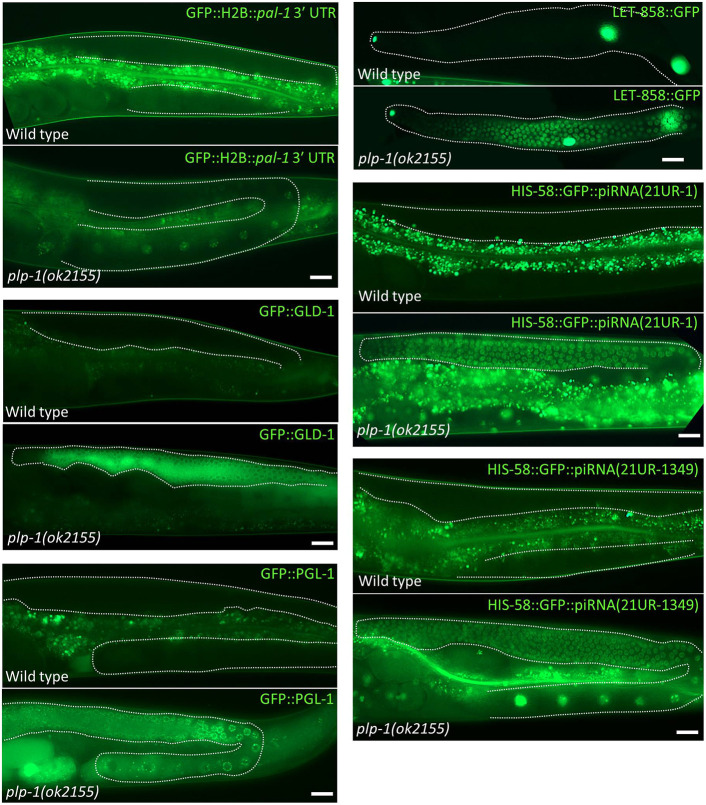
PUF-8 regulates the translation of several mRNAs in the germ line by acting via their 3′ UTR sequences (Maheshwari et al., 2016; Mainpal et al., 2011; Vaid et al., 2013). Transgenic reporter strains suitable for assaying 3′ UTR activity in the germ line are available in our laboratory for several potential targets of PUF-8. The redundancy between puf-8 and plp-1 described above prompted us to screen these reporter strains to test whether PLP-1 affected the reporter expression in any of them. In the case of the gfp::H2B::pal-1 3′ UTR transgene, GFP::H2B expression was seen in oocytes, which is similar to the endogenous PAL-1 expression pattern, but only in worms grown at 25°C (Mainpal et al., 2011). By contrast, when this transgene was introduced into plp-1 worms, GFP::H2B could be readily detected in oocytes even when the worms were grown at 20°C (Fig. 5). Except for this change, we did not see any other alterations in the expression pattern. We reasoned that the pal-1 3′ UTR transgene might be silenced in the germ line at 20°C, and this silencing might be dependent on PLP-1. To test this, we introduced two other integrated transgenes, namely gfp::gld-1 and gfp::pgl-1, into the plp-1(ok2155) genetic background. These two transgenes have been previously shown to be silenced in the germline at 20°C (Cheeks et al., 2004; Merritt et al., 2008). Consistent with our reasoning, both these transgenes remained active in plp-1 germ lines for at least 100 generations at 20°C (we did not observe beyond this), whereas they became silent within three generations in wild-type germ lines (Fig. 5).
Fig. 5.
Integrated and extrachromosomal array transgenes, and piRNA sensors, are not silenced in plp-1 germ lines. GFP fluorescence images of live wild-type and plp-1(ok2155) hermaphrodites are shown. GFP::H2B::pal-1 3′ UTR, GFP::GLD-1 and GFP::PGL-1 are expressed by integrated, single-copy transgenes (left). LET-858::GFP is expressed by the extrachromosomal array ccEx7271 carrying the let-858::gfp transgene; in these extruded germ lines, GFP fluorescence can be seen in three somatic nuclei – the distal tip cell nucleus at the extreme left and two other larger nuclei at the right – in both genotypes, whereas it is visible in germline nuclei only in the plp-1 mutant (right, top). Expression of HIS-58, which is a histone H2B, fused to GFP by integrated transgenes containing the target sequences of piRNA(21UR-1) (right, middle) or piRNA(21UR-1349) (right, bottom) in the 3′ UTR. The germ line is outlined (dotted white lines) in all images. Non-specific autofluorescence of gut granules is seen in some images. Scale bars: 20 µm.
Extrachromosomal DNA arrays, consisting of multiple tandem repeats, are readily silenced in wild-type germ lines at 20°C. To test whether PLP-1 is involved in this process as well, we checked the effect of plp-1 mutation on the expression of the ccEx7271 extrachromosomal array, which expresses GFP under control of the let-858 promoter and has been shown to be silenced in the germ line (Kelly et al., 1997; Tabara et al., 1999b). As expected, the ccEx7271 array became silent in the first generation at 20°C in control germ lines. By contrast, it remained active for at least 30 generations in the plp-1(ok2155) genetic background (Fig. 5).
Next, we compared the expression of two different piRNA sensors in wild-type and plp-1 germ lines. Both these piRNA sensors have been shown to faithfully reflect the status of piRNA-mediated gene silencing in the germ line (Bagijn et al., 2012). As shown in Fig. 5, both the piRNA sensors were silenced in wild-type germ lines by the third generation, but remained active in plp-1 germ lines even at the 90th generation, demonstrating that PLP-1 is also essential for piRNA-mediated silencing.
Mutants defective in silencing of foreign genetic elements in the germ line have been shown to be resistant to RNAi (Tabara et al., 1999b). Therefore, we targeted three different genes, namely mex-3, pos-1 and spn-4, by RNAi and compared the levels of RNAi penetrance between wild-type and plp-1 worms. These three genes were chosen because depletion of proteins encoded by them results in fully-penetrant embryonic lethality (Huang et al., 2002; Tabara et al., 1999a). The penetrance of mex-3(RNAi) did not vary significantly between wild-type and plp-1 worms (Table S4). However, ∼12% of pos-1(RNAi) and ∼15% of spn-4(RNAi) embryos of plp-1 worms were viable (n=1368 and 1590, respectively), whereas only ∼1.2% of pos-1(RNAi) and ∼1.4% of spn-4(RNAi) embryos of wild-type worms were viable (n=1901 and 1805, respectively), revealing a slight reduction in the ability of plp-1 worms to elicit an RNAi response. Thus, although RNAi-mediated gene silencing per se does not depend on plp-1, its robustness does appear to require plp-1.
PLP-1 participates in the initiation of germline gene silencing
Some factors, such as PRG-1, initiate transgene silencing in the germ line, whereas others, such as MUT-2/RDE-3 and MUT-7, maintain the silenced state (Lee et al., 2012; Shirayama et al., 2012). This functional distinction among the components of silencing pathways prompted us to determine whether PLP-1 functions during the initiation and/or maintenance of silencing for the gfp::pgl-1 transgene, ccEx7271 extrachromosomal array and piRNA sensor-1 that we tested in experiments described in the preceding section. When introduced from strains in which these were already silenced, all three sequences remained silent in the plp-1 mutant background, even after ten generations. By contrast, they became active in the first generation itself when introduced into the rde-3 mutant background (Fig. 6). Taken together, these observations and results indicate that PLP-1 only functions during the initiation step of transgene silencing, and is dispensable for the maintenance step for the three sequences tested here.
Fig. 6.
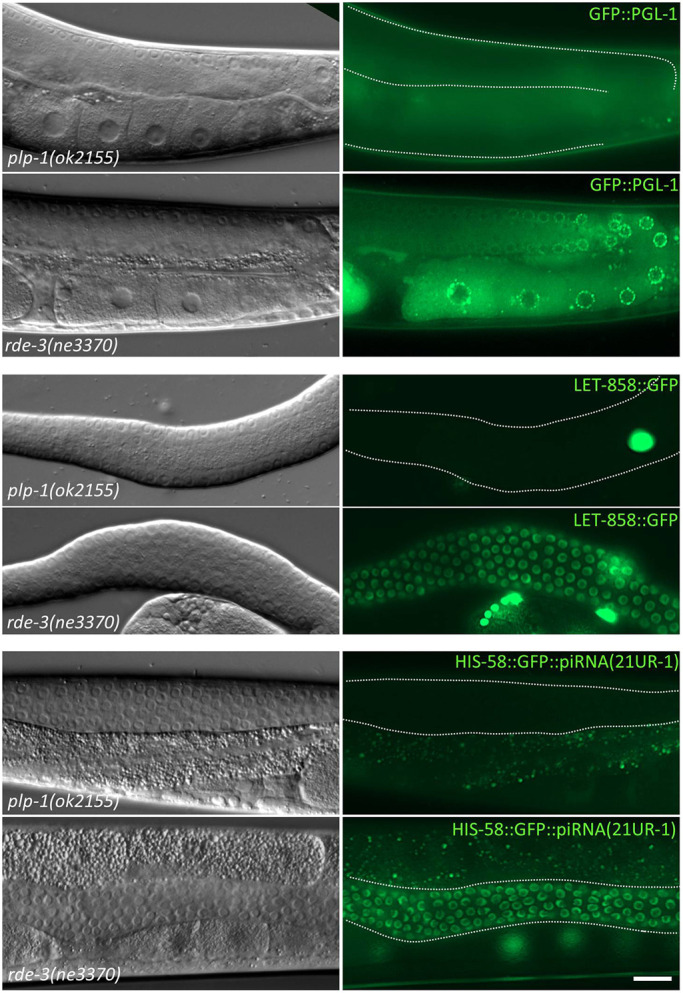
Already silenced transgenes are not activated in plp-1 mutants. GFP fluorescence (right) and the corresponding differential interference contrast images (left) of the indicated genotypes are shown. Only part of the germ line, outlined by dotted white lines for the plp-1 worms, is visible in all images. Already silenced gfp::pgl-1 transgene, ccEx7271 extrachromosomal array and piRNA sensor-1 [piRNA(21UR-1)] are not expressed when introduced into plp-1 germ lines, but are activated in rde-3 germ lines. Scale bar: 20 µm.
PLP-1 is not essential for small RNA biogenesis
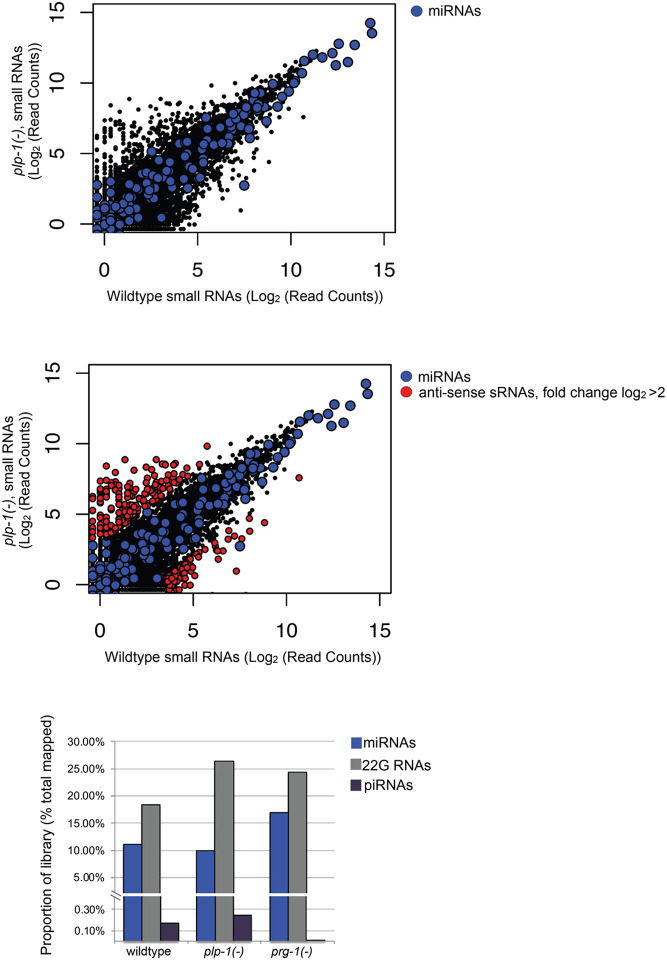
Germline gene silencing depends on the generation of small RNAs. In some silencing-defective mutants, the biogenesis of small RNAs is disrupted. For instance, piRNAs are not produced in prg-1 mutants (Batista et al., 2008). Therefore, we profiled small RNA transcripts using high-throughput sequencing and compared the levels of different classes of small RNAs among wild-type (positive control), prg-1 (negative control) and plp-1 worms. Although there were slight changes in relative contributions of the different classes, the levels of transcripts of all classes were comparable between wild-type and plp-1 worms. By contrast, consistent with the previous findings, piRNAs were absent in prg-1 mutants assayed in parallel (Fig. 7, Table S5). Based on a cutoff of 4-fold, 312 mRNAs had differential levels of small RNAs in plp-1 and wild-type worms. However, they were not enriched for any specific class of small RNAs. Furthermore, despite the involvement plp-1 in the silencing of piRNA sensors (Fig. 5), levels of piRNAs were not reduced in plp-1 mutants. These results indicate that plp-1 is not essential for the biogenesis of small RNAs, and suggest that it may function at a step downstream of small RNA synthesis in the silencing pathways.
Fig. 7.
Biogenesis of small RNAs is not substantially affected in plp-1 mutants. Top: small antisense RNA expression in young adult wild-type and plp-1(−) animals. Counts are normalized to total RNAs that aligned to the cell genome. miRNA counts are overlaid in blue. Middle: small antisense RNA expression in young adult wild-type and plp-1(−) animals. Counts are normalized to total RNAs that aligned to the cell genome. miRNA counts are overlaid in blue. Red-colored dots indicate protein-coding genes with at least 50 mapped reads and 4-fold more or less reads in plp-1(−) animals relative to the number of reads in wild-type animals (212 and 76 genes, respectively). Bottom: bar graph indicates the proportion of the total library that are miRNAs, 22G antisense small RNAs or annotated piRNAs for the indicated strains. Counts are normalized to total RNAs that aligned to the cell genome. See Materials and Methods, and Table S5 for more details.
Loss of PLP-1 and disruption of germline gene-silencing pathways affect expression of the same set of endogenous germline genes
Disruption of germline gene-silencing pathways leads to an increase in the expression levels of several endogenous genes. For example, the levels of more than 2000 endogenous mRNAs are significantly higher in prg-1 mutants than in wild-type worms (Bagijn et al., 2012; Lee et al., 2012; Reed et al., 2020). Similarly, the transcription of ∼1600 genes has been shown to be upregulated in mut-16 mutants (Phillips et al., 2012; Reed et al., 2020). This prompted us to investigate whether the expression patterns of endogenous genes are similarly altered in plp-1 mutants. We generated transcriptome profiles for both wild-type and plp-1 mutant genotypes using high-throughput sequencing and compared the two. This comparison identified 1367 genes as differentially expressed between wild-type and plp-1 animals with a cutoff value of ≥2-fold difference; of these, 780 were upregulated and 587 were downregulated in plp-1 (Table S6). RNA from hermaphrodites grown at 20°C was used for transcriptome profiling, and as shown in Fig. 1, the germline architecture of plp-1 mutants grown at 20°C did not display any noticeable defects and was comparable with that of wild type. Thus, the observed differential expression unlikely resulted from changes in the number or kind of cells in the germ line.
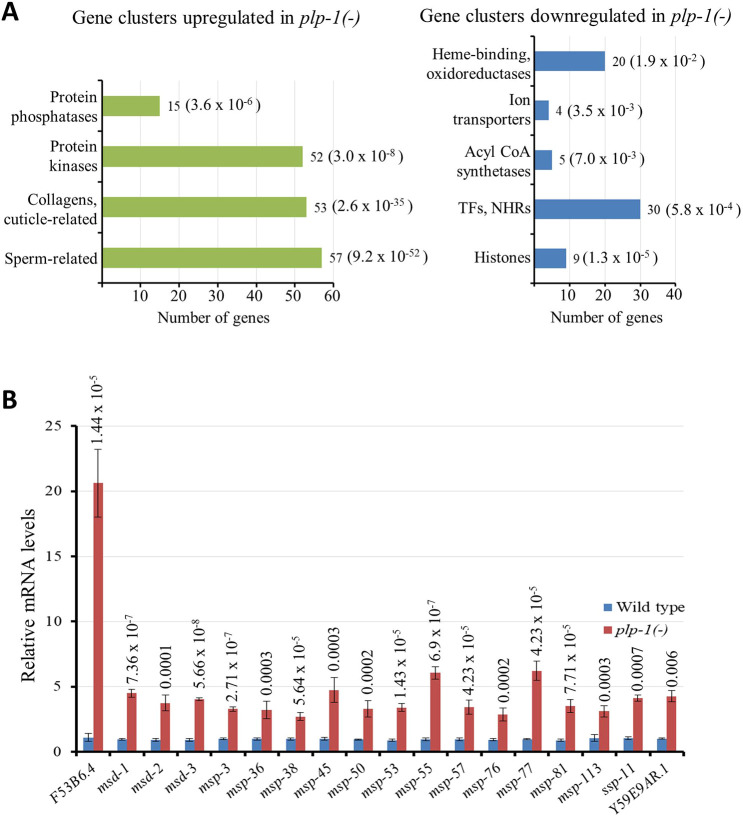
Although genes belonging to diverse classes – classified based on potential or confirmed biochemical functions – were represented in both upregulated and downregulated groups, three classes stood out: there were 57 sperm-related genes, 53 collagen and other cuticle-related genes and 52 genes encoding protein kinases among the genes upregulated in plp-1 mutants (Fig. 8A). By contrast, there were only one sperm-related and nine cuticle-related genes in the downregulated group. On the other hand, genes encoding transcription factors and nuclear hormone receptors (30 genes), heme-binding proteins and oxidoreductases (20 genes) and histones (nine genes) were more represented in the downregulated group. We performed quantitative reverse transcription-polymerase chain reaction (qRT-PCR) for 18 sperm-related genes in the upregulated group, and found that the transcript levels of all 18 were indeed higher in plp-1 mutants when compared with the levels in wild-type animals (Fig. 8B). Furthermore, we compared the transcript abundance between soma and germ line for five of the above 18 genes. All five were enriched in the germ line, and their upregulation in plp-1 mutants primarily resulted from an increase in the germ line, rather than soma (Fig. S8).
Fig. 8.
Sperm-related genes and collagen and other cuticle-related genes are significantly upregulated in plp-1 mutants. (A) Bar graphs compare the groups of genes for which mRNA levels are significantly higher (left) or lower (right) in plp-1 mutants than in wild-type animals. The P-values for the enrichment are indicated in parentheses (Fisher's exact test used). The lists of genes are presented in Tables S7,S8. The mRNA levels were measured using high-throughput transcriptome sequencing. Total processed and mapped reads ranged from 43,557,204 to 60,518,575 among the six samples [three biological replicates each of plp-1(−) and wild type]. See Materials and Methods and Table S6 for more details. (B) Validation of the sequencing results. Transcript levels of 18 sperm-related genes, which were identified as upregulated by transcriptome sequencing, were quantitated using qRT-PCR. Error bars represent s.d. The P-values, indicated above the bars, were calculated by Student's t-test (paired, two-tailed).
Many endogenous genes upregulated in prg-1 mutants have also been found to be upregulated in mut-16 mutants (Reed et al., 2020). These common gene expression changes probably result from the disruption of gene-silencing pathways in these two mutants. Because plp-1 mutants also displayed gene-silencing defects, we expected a significant overlap among the sets of genes upregulated in prg-1, mut-16 and plp-1 mutants. This indeed was the case: 44% of the 780 genes upregulated in plp-1 were also in the class shown to be upregulated in mut-16 (P<9.6×10−176), and this number increased to 57% when plp-1 and prg-1 sets were compared (P<8.8×10−203). We found 316 genes – a little over 40% of the 780 genes – to be common to all three sets (Fig. 9).
Fig. 9.
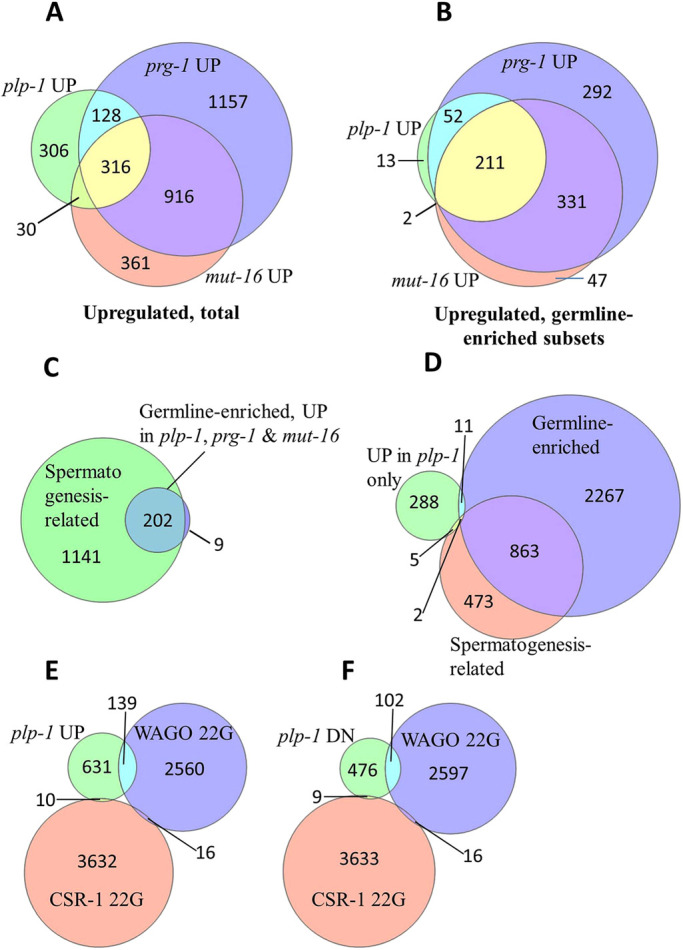
Meta-analysis of transcriptome datasets. (A-F) Proportional Venn diagram representations show (A) significant overlap of genes upregulated in plp-1, prg-1 and mut-16 mutants (P<8.8×10−203 for the overlap between plp-1 and prg-1; P<9.6×10−176 for the overlap between plp-1 and mut-16). (B) The overlap is substantially higher among the upregulated genes that are more abundantly expressed in the germ line (germline-enriched). (C) Spermatogenesis-related genes dominate (95%) among germline-enriched genes that are upregulated in all three mutants. (D) Majority of genes upregulated only in plp-1 are non-germline-enriched. (E,F) Whereas targets of WAGO-class 22G-RNAs are essentially equally represented in both plp-1(−)-upregulated (plp-1 UP) and plp-1(−)-downregulated (plp-1 DN) groups, targets of CSR-1-22G-RNAs are almost excluded from these two groups. See Table S6 for lists of genes represented in the Venn diagrams shown here. Numbers of genes in each set are indicated in the diagrams. prg-1 and mut-16 datasets are from Reed et al. (2020); germline-enriched and spermatogenesis-related datasets are from Reinke et al. (2004); and lists of targets of WAGO- and CSR-1-22G-RNAs are from Lee et al. (2012).
About 34% of genes upregulated in prg-1 and 36% of genes upregulated in mut-16 mutants belong to the ‘germline enriched’ category as defined by Reed et al. (2020) and Reinke et al. (2004). We noticed a similar trend with plp-1 as well: 36% of genes upregulated in plp-1 mutants belong to this category. Strikingly, when only the germline-enriched subsets were compared, 95% of genes upregulated in plp-1 mutants have been found to be upregulated in prg-1 mutants as well (P<0.000e+00); this overlap is 77% between plp-1 and mut-16 mutants (P<3.6×10−281). Overall, 211 germline-enriched genes were found to be upregulated in all three mutants, and 96% of these (202/211) are spermatogenesis-related, indicating that the same components that silence transgenes and other foreign genetic elements in the germ line might also control the expression of spermatogenesis-related genes (Fig. 9). In mutants such as mut-16 and set-2, and in germ lines depleted of P granules, some soma-enriched genes are misexpressed in the germ line (Knutson et al., 2017; Robert et al., 2014; Rogers and Phillips, 2020). We did not find significant overlap between these soma-enriched gene sets and the set of genes upregulated in plp-1 mutants (Table S6). Furthermore, only 22 of the 780 genes upregulated in plp-1 belong to the soma-enriched category. Conversely, the 306 genes upregulated in plp-1, but not in prg-1 or mut-16, were more or less excluded from the germline-enriched class (Fig. 9). We randomly chose five of the 306 genes and validated their differential expression using qRT-PCR, and found two of them to be upregulated in plp-1 mutant germ lines (Fig. S9). Thus, in the germ line, plp-1 may not play a major role in suppressing somatic genes, although it may suppress a few non-germline-enriched genes.
In C. elegans, some small RNAs, called WAGO-22G-RNAs, associate with WAGOs, whereas others associate with CSR-1. In prg-1 mutants, mRNAs targeted by WAGO-22G-RNAs, but not by the CSR-1-22G RNAs, have been found to be upregulated (Lee et al., 2012). We examined the set of genes differentially expressed in plp-1 mutants and found that the targets of WAGO-22G-RNAs were more or less equally represented in both upregulated and downregulated groups, although neither group was enriched for these targets. By contrast, CSR-1-22G-RNA targets were almost completely excluded from both groups (Fig. 9). Thus, plp-1 does not appear to be engaged directly in the CSR-1 pathways.
DISCUSSION
We have identified PLP-1 as a component of the germline gene-silencing pathway. We have shown that PLP-1 functions in silencing extrachromosomal arrays, integrated transgenes and piRNA sensors. Like many AGOs, PLP-1 localizes to P granules, and loss of PLP-1 causes temperature-sensitive germline defects, much like some of the other silencing pathway mutations. Furthermore, our work reveals a remarkable overlap among the sets of germline-related genes upregulated in plp-1 mutants and two other mutants, namely prg-1 and mut-16 mutants, which are known to disrupt gene-silencing pathways.
We identified plp-1 based on its genetic interaction with puf-8: both puf-8 and plp-1 single mutants are fertile at 20°C, but the puf-8; plp-1 double mutants are sterile at this temperature. The encoded proteins of these two genes do not share sequence or structural similarity. PUF-8 is a PUF family protein containing the conserved PUF RNA-binding domain (Wickens et al., 2002). Like many other PUF proteins, PUF-8 regulates translation by binding to specific 3′ UTR sequences of its target mRNAs (Maheshwari et al., 2016; Mainpal et al., 2011; Vaid et al., 2013). On the other hand, PLP-1 shares sequence similarity with Pur repeat-containing proteins such as the human Pur-alpha (Fig. S10) (Daniel and Johnson, 2018). The Pur proteins bind both single-stranded DNA and RNA through their Pur repeats, which are well conserved from bacteria to humans (Bergemann et al., 1992; Chepenik et al., 1998). The Pur repeat – named after the human ortholog, which binds purine-rich single-stranded DNA – shares neither sequence nor structural similarity with the PUF domain. Although Pur-alpha has been implicated in translational regulation (Gallia et al., 2001), it does not seem to regulate translation the way PUF proteins do, that is, by binding to the 3′ UTR of specific target mRNAs. Consistent with this notion, none of the 3′ UTR reporter fusions regulated by PUF-8 showed any change in expression when introduced into plp-1 mutants (L.T., R.V. and K.S., unpublished). Therefore, we do not think PUF-8 and PLP-1 substitute each other biochemically. However, there are some notable similarities: both proteins potentially bind RNA and protein, and localize to P granules (Ariz et al., 2009; this study). Both puf-8 and plp-1 single mutants display temperature-sensitive sterility phenotype (Subramaniam and Seydoux, 2003; this study). In addition, PUF-8 and the mammalian ortholog of PLP-1 are implicated in mRNA transport (Johnson et al., 2006; Kanai et al., 2004; Pushpa et al., 2013). Thus, PUF-8 and PLP-1 may control gene expression at the RNA level, but through different biochemical mechanisms. For example, PLP-1 may control the levels of certain germline mRNAs, while PUF-8 may regulate their translation. In such a scenario, it is not difficult to imagine how increased mRNA levels, when combined with a lack of translational control, could have damaging consequences, whereas either one alone might be inconsequential. Alternatively, PUF-8 and PLP-1 may contribute to P granule integrity such that their simultaneous absence, but not the absence of either one, perturbs the integrity of P granules sufficiently enough to alter the stability and/or translation of mRNAs associated with these granules. Such a perturbation may also happen under temperature stress (25°C) in the absence of either of these two proteins, given the liquid-droplet-like nature of P granules. A third possibility is that PUF-8 facilitates the P granule association of PLP-1, or vice versa, but we did not see any alteration in the localization of either protein in the absence of the other (L.T., R.V. and K.S., unpublished).
Germline gene-silencing pathway components such as PRG-1 and mutator proteins are involved in the formation of small RNAs. Whereas PRG-1 is essential for the accumulation of piRNAs, mutator proteins such as MUT-16 contribute to silencing through their facilitation of siRNA amplification (Batista et al., 2008; Phillips et al., 2012). By contrast, our results reveal that PLP-1 is not essential for small RNA accumulation (Fig. 7). Thus, PLP-1 must function downstream of small RNA biogenesis in the silencing pathways. How does PLP-1 contribute to gene silencing in the germ line? Although PLP-1 is not essential for survival of C. elegans, its mouse ortholog is essential for survival (Khalili et al., 2003). The Pur proteins participate in a variety of cellular functions, such as transcription, translation, activation of Cdk9 and mRNA transport (Darbinian et al., 2001; Gallia et al., 2001; Johnson et al., 2006; Kanai et al., 2004; Zhang et al., 2005). It has been proposed that the remarkable versatility of Pur proteins emerges from their ability to interact with both single-stranded nucleic acids and other proteins. During the course of evolution, the Pur proteins seem to have retained their ability to bind single-stranded nucleic acids by conserving the amino acid sequence of Pur repeats, which interact with nucleic acids. At the same time, these proteins seem to have adapted themselves to diverging cellular needs by interacting with new protein partners, probably facilitated by the diversification of the C-terminal region involved in protein-protein interactions (Daniel and Johnson, 2018). We think this flexibility most likely enables PLP-1 to couple, or strengthen the interaction among, some of the proteins and RNAs in the germline gene-silencing machinery. The notion that PLP-1 may facilitate RNA-protein interactions in P granule-like structures might explain its role in the somatic cells as well, where again we find PLP-1 on granules. Identifying the protein partners of PLP-1 will help test this idea and provide mechanistic insights into its function.
An earlier study implicated PLP-1 in the transcriptional modulation of the endoderm-specific transcription factor END-1 (Witze et al., 2009). It is conceivable that PLP-1 regulates transcription, at least of some genes, in the germ line as well. Misexpression of these genes in the absence of PLP-1 can potentially interfere with the germline gene-silencing machinery, and thus account for the silencing defects of plp-1 mutants. One paradox that remains in need of explanation is why PLP-1 localizes primarily on cytoplasmic granules in the germ line and soma. Although we cannot formally rule out the presence of PLP-1 in nuclei, results from three independent types of experiments, described here, demonstrated the cytoplasmic, perinuclear granules to be the major sites of PLP-1 localization in both the tissues. This localization pattern of PLP-1 and its involvement in germline gene-silencing are strikingly similar to that of many other silencing pathway components. Therefore, we favor a model in which the predominant function of PLP-1 is in P granules and similar RNA granules in the soma, where it facilitates RNA-protein interactions. This model may be useful to explain the functions of Pur proteins found on RNA granules in other organisms, such as Drosophila and mammals (Aumiller et al., 2012; Kanai et al., 2004).
MATERIALS AND METHODS
Strain maintenance and genetic crosses
All C. elegans strains used in this study were maintained as per established protocols (Brenner, 1974) at 20°C, unless mentioned otherwise. Strains of different genotypes used in this study were constructed using standard genetic crosses. Briefly, strains carrying the integrated transgenes, GFP::H2B::pal-1 3′ UTR, GFP::PGL-1 and GFP::GLD-1, and the ccEx7271 extrachromosomal array were maintained in an active state at 25°C. They were mated with plp-1(+/−) males at 25°C; the progeny were shifted to 20°C and cloned. The transgenes and the ok2155 allele of plp-1 were detected using single-worm PCRs, and animals homozygous for both the transgene and ok2155 were obtained by further cloning and single-worm PCRs (Fig. S11). A similar strategy was employed for the piRNA sensor 21UR-1349. The piRNA sensor 21UR-1 was transferred from the prg-1(−) background (strain SX1288), in which it is active, into the plp-1 mutant. Maintenance at 20°C of strains carrying the transgenes, extrachromosomal array and piRNA sensors mentioned above for several generations led to complete silencing. Such silenced transgenes – their presence was detected by PCR – were then introduced into plp-1 and rde-3 mutants through crosses set up at 20°C. All strains used in this study are listed in Table S9. Marc Ridyard and Eric Miska (The Wellcome Trust/Cancer Research UK Gurdon Institute, Cambridge, UK) gifted the piRNA sensor strains SX1286, SX1287, SX1288 and SX2110; and Meetu Seth and Craig Mello (University of Massachusetts Medical School, Worcester, MA, USA) gifted the strain WM250.
RNA interference
Coding sequences of target genes were PCR-amplified from cDNA and inserted at the EcoRV site of pSV2 vector (Mainpal et al., 2011) by the TA cloning method. Sequences of PCR primers are listed in Table S10. Both strands of inserts cloned into pSV2 vector are transcribed into dsRNA when introduced into the HT115 bacterial strain (Mainpal et al., 2011). RNAi was performed by the feeding method described by Timmons et al. (2001). For depleting PLP-1 by RNAi, L4 larvae were transferred to RNAi lawns and transferred to fresh RNAi lawns 24 h later. Embryos collected for 8-12 h on the fresh lawns were allowed to hatch and grow to adulthood. Phenotypes of these adults were scored. In the case of mex-3, pos-1 and spn-4, gravid hermaphrodites were placed on RNAi lawns for 8-12 h and transferred to fresh RNAi lawns. The worms were removed after they laid embryos for ∼8 h, and the embryos were counted. Embryos remaining after 24 h were counted again to determine the fraction of viable progeny.
Generation of anti-PLP-1 antibody
Polyclonal antibodies were raised in rabbits against bacterially-expressed fusion protein containing maltose-binding protein (MBP) and full-length PLP-1 protein (MBP::PLP-1). Coding sequences of PLP-1 were PCR-amplified from cDNA and inserted at the BamH1 site of pMAL-cE plasmid vector which expresses the inserted ORF as an MBP fusion (New England Biolabs). The resulting plasmid was introduced into the E. coli strain BL21(DE3). Expression and purification of MBP::PLP-1 were carried out as per standard protocols using IPTG to induce the expression of fusion protein, and deoxycholic acid for bacterial cell lysis (Sambrook et al., 1989). The MBP::PLP-1 fusion protein, which was mostly in the supernatant following centrifugation of the lysate, was purified by affinity chromatography using a HIS-select Cartridge following the manufacturer's protocols (Sigma-Aldrich, Cat. No. H8286).
Rabbit antiserum against MBP::PLP-1 was produced at a commercial facility (Bangalore Genei), and antibodies specific for PLP-1 were purified from the antiserum using bacterially-produced GST::PLP-1 as the affinity tag. For this, the bacterial cell lysate containing GST-PLP-1 was separated using SDS-PAGE and transferred to nitrocellulose membrane. Protein bands were visualized by staining with Amido Black, and the strip of membrane containing GST::PLP-1 was cut and incubated with the anti-MBP::PLP-1 antiserum, after washing off the dye. The bound anti-PLP-1 antibodies were eluted in 0.2 M glycine, 1 mM EGTA at pH 2.8. The pH of the eluate was adjusted to 7.0 by adding 0.1 volume of 1 M Tris base, and the eluate was stored at 4°C.
Generation of transgenic lines
Transgenic lines expressing PLP-1::GFP and mCherry::PLP-1 were generated by biolistic bombardment, as described previously (Jadhav et al., 2008; Praitis et al., 2001). The plasmid construct pLT2, used for introducing the plp-1:gfp transgene, carried the 2 kb genomic region immediately upstream of the plp-1 ORF, the plp-1 ORF (1 kb), the GFP coding sequences and 2 kb genomic region downstream of the plp-1 stop codon in the pBluescript backbone. In addition, this vector carried a 5.6 kb genomic fragment that rescues the unc-119(ed3) defect. The construct pLT5 used for introducing the mCherry:plp-1 transgene contained the pie-1 promoter and hip-1 3′ UTR sequences. In this construct, the coding sequences of mCherry were introduced at the N-terminus of PLP-1. At least three independent lines with similar expression patterns were obtained for both transgenes.
To produce PLP-1::GFP from the endogenous locus of plp-1, we inserted the coding sequences of GFP in-frame just before the plp-1 stop codon using the CRISPR/Cas9 method (Arribere et al., 2014). The sgRNA-expressing plasmids and the repair template carrying GFP coding sequences were prepared as described previously (Kumar and Subramaniam, 2018). Oligonucleotides used in both bombardment and CRISPR/Cas9 experiments are listed in Table S10.
Fluorescence microscopy
For visualization of chromatin in whole worms, worms were collected in PBST (PBS containing 0.1% Tween 20) and fixed in precooled methanol (−20°C) for at least 20 min. Fixed animals were incubated in methanol containing 25 ng/µl DAPI for 20 min in the dark at room temperature. Stained animals were then washed thrice in PBST and mounted in Vectashield antifading agent (Vector Laboratories). For the extrusion of adult germ lines, worms were anesthetized in 2 mM levamisole and cut just posterior to the pharynx using a 25 G injection needle on a microscope slide. Within 10 min of dissection, extruded germ lines were fixed in formaldehyde fixative (4% formaldehyde in PBS) for 1 h at room temperature. Germ lines were then washed thrice with PBST and incubated with PBS containing 25 ng/µl DAPI for 30 min.
For staining with affinity-purified anti-PLP-1 and anti-phosphohistone H3 antibodies (Sigma-Aldrich, H0412), germ lines dissected and fixed as above were washed twice with blocking buffer (PBS containing 0.1% bovine serum albumin and 0.1% Triton X-100) and incubated in the blocking buffer for 1 h at room temperature. Antibodies were diluted 1:1000 with blocking buffer and added to germ lines, and were incubated overnight at 4°C. Germ lines were then washed with blocking buffer and incubated with 1:50 diluted anti-rabbit secondary antibodies conjugated with FITC or rhodamine (Jackson ImmunoResearch, 711-095-152) for 7-8 h at 4°C. After washing thrice with blocking buffer, germ lines were mounted in Vectashield. Anti-PLP-1 antibody did not stain any noticeable structures in plp-1(ok2155) germ lines or embryos, confirming that the antibody is specific for PLP-1. For immunostaining of embryos, embryos were freeze-cracked using liquid nitrogen and fixed in pre-chilled (−20°C) methanol for 5 min at −20°C, followed by 4% formaldehyde for 20 min at room temperature. After blocking, the embryos were stained overnight with 1:10-diluted affinity-purified anti-PLP-1 antibodies in a humidity chamber. They were then washed thrice in blocking buffer, and stained with 1:500-diluted anti-rabbit secondary antibodies conjugated with Alexa Fluor 488 (Jackson ImmunoResearch, 711-545-152) for 6 h at room temperature.
Fluorescence signals from whole worms and germ lines prepared as above were observed using a Zeiss Axio Imager M2 fluorescence microscope and imaged using a Zeiss Axiocam 506 Mono CCD camera. Where necessary, images were deconvolved using the Deconvolution module of Zeiss Axiovision software.
Small RNA sequencing and data analysis
We picked 20-25 young adult stage worms from non-starved plates grown at 20°C into TRIzol reagent (Thermo Fisher Scientific). RNA was purified from TRIzol following standard guidelines from Johnstone (1999). Small RNAs were then purified using a mirVana kit through a modified protocol optimized for small sample input (Ambion, Thermo Fisher Scientific). Briefly, total RNA was mixed with mirVana lysis/binding buffer and homogenate buffer and incubated at room temperature for 5 min, and then small RNAs were purified through two rounds of alcohol precipitation. In the first, we added one third the volume of ethanol, centrifuged at 5000 rpm (2500 g) for 5 min, and then transferred the supernatant to a fresh tube. In the second round of extraction, we added one volume of isopropanol and centrifuged at 15,000 rpm (21,000 g) for 30 min. The purified small RNAs were treated with Cap-Clip enzyme (CellScript) to convert all 5′ ends to monophosphate, after which libraries were made using a TruSeq small RNA kit and sequenced on a miSeq instrument (Illumina).
For data analysis, reads were trimmed for adapter sequence at the 3′ end and aligned to the C. elegans protein coding (WS245) and ncRNA transcriptomes (ftp://ftp.wormbase.org/pub/wormbase/releases/WS245/). Counts of reads to each gene were generated using custom Python scripts.
Transcriptome sequencing
Total RNA was isolated from adult hermaphrodites using TRI reagent (Sigma-Aldrich) as per the manufacturer's protocols. Three biological replicates were used for transcriptome sequencing at a commercial facility (Macrogen). After removing potential DNA contamination by DNase treatment, polyA+ RNA was isolated using the TruSeq Stranded mRNA preparation kit (Illumina), which was then fragmented randomly and reverse-transcribed into cDNA. Following adaptor ligation to both ends, the cDNA was PCR-amplified and size-selected for fragments of 200-400 bp length. Both ends of these fragments were sequenced using a NovaSeq 6000 S4 Reagent kit in a NovaSeq 6000 NGS sequencing machine (Illumina) following the manufacturer's protocols. After removing low quality reads (Phred score <20), adapter sequences were trimmed using the Trimmomatic 0.38 program. Reads shorter than 36 bp were removed from the trimmed reads data. Trimmed reads were mapped to C. elegans reference genome data version WS264 (ftp://ftp.wormbase.org/pub/wormbase/releases/WS264/) using HISAT2 version 2.1.0 and Bowtie2 2.3.4.1. Reads were aligned into potential transcripts using StringTie version 1.3.4d. Transcripts corresponding to 15,316 genes were selected, after omitting the transcripts with read count 0 in more than two of the six samples [three biological replicates each of wild type and plp-1(ok2155)], for the differential gene expression analysis, which was performed using the DESeq2 package. Transcripts showing fold change ≥2 and nbinomWaldTest raw P-value <0.05 were selected as differentially expressed, and were annotated using the DAVID tool. Gene enrichment analyses were performed using the DAVID tool available at www.ncbi.nlm.nih.gov. Venn diagrams shown in Fig. 9 were generated using tools available at http://bioinformatics.psb.ugent.be/webtools/Venn/ and https://omics.pnl.gov/software/venn-diagram-plotter. Statistical significance of the overlaps was calculated using the tool available at http://nemates.org/MA/progs/overlap_stats.html.
Quantitation of transcripts using qRT-PCR
Total RNA was isolated from 100 1-day-old adult animals using TRI reagent (Sigma-Aldrich) and following manufacturer's protocols. The entire amount of RNA was used for cDNA synthesis using oligo dT primer and Revert Aid reverse transcriptase (Thermo Fisher Scientific) as per manufacturer's protocols. The final cDNA was diluted to 500 µl in TE buffer (10 mM Tris-HCl, pH 8.0 and 1 mM EDTA). The qPCR reactions were performed using a QuantStudio 7 flex real-time PCR system and the QuantStudio 6 and 7 flex real-time PCR system software V1.3 (Applied Biosystems). The qPCR reactions contained 1×SYBR Green master mix (Bio-Rad), 300 nM each of the two primers and 2 µl of the diluted cDNA; the PCR settings were: 95°C for 20 s; 40 cycles of 95°C for 2 s, 60°C for 25 s; followed by 60°C for 1 min and 95°C for 15 s. Relative quantities were calculated using ΔΔCt method and normalized using hip-1 and act-1 as internal controls. Values from three technical replicates each of three biological replicates were used for calculating the average fold change, and the statistical significance was calculated using Student's t-test (paired, two-tailed).
Supplementary Material
Acknowledgements
We thank Marc Ridyard and Eric Miska for the piRNA sensor strains SX1286, SX1287, SX1288 and SX2110, and Meetu Seth and Craig Mello for the strain WM250. Some of the C. elegans strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: R.V., L.T., K.S.; Methodology: R.V., L.T., L.W., A.F., K.S.; Validation: R.V., L.T., L.W., A.F., K.S.; Formal analysis: R.V., L.T., L.W., A.F., K.S.; Investigation: R.V., L.T., L.W., A.F., K.S.; Resources: A.F., K.S.; Writing - original draft: R.V., L.T., K.S.; Writing - review & editing: K.S.; Supervision: K.S.; Project administration: K.S.; Funding acquisition: K.S.
Funding
Research in the K.S. laboratory is supported by grants from the Department of Biotechnology, Ministry of Science and Technology, India (BT/PR29010/BRB/10/1694/2018) and the Science and Engineering Research Board, Department of Science and Technology, Ministry of Science and Technology, India (EMR/2016/002272). Work in the Fire laboratory is supported by National Institutes of Health grant R35GM130366. Deposited in PMC for release after 12 months.
Data availability
All raw small RNA and mRNA high-throughput sequencing data, and lists of identified C. elegans genes and read counts have been deposited in GEO under accession number GSE161011.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.195578.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.195578.reviewer-comments.pdf
References
- Ahringer J. and Kimble J. (1991). Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3′ untranslated region. Nature 349, 346-348. 10.1038/349346a0 [DOI] [PubMed] [Google Scholar]
- Ariz M., Mainpal R. and Subramaniam K. (2009). C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev. Biol. 326, 295-304. 10.1016/j.ydbio.2008.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X. H., Artiles K. L., Hartman P. S. and Fire A. Z. (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837-846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumiller V., Graebsch A., Kremmer E., Niessing D. and Förstemann K. (2012). Drosophila Pur-α binds to trinucleotide-repeat containing cellular RNAs and translocates to the early oocyte. RNA Biol. 9, 633-643. 10.4161/rna.19760 [DOI] [PubMed] [Google Scholar]
- Bagijn M. P., Goldstein L. D., Sapetschnig A., Weick E.-M., Bouasker S., Lehrbach N. J., Simard M. J. and Miska E. A. (2012). Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337, 574-578. 10.1126/science.1220952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M. K., Schedl T. B. and Kimble J. (1987). Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics 115, 107-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista P. J., Ruby J. G., Claycomb J. M., Chiang R., Fahlgren N., Kasschau K. D., Chaves D. A., Gu W., Vasale J. J., Duan S. et al. (2008). PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31, 67-78. 10.1016/j.molcel.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergemann A. D., Ma Z. W. and Johnson E. M. (1992). Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol. Cell. Biol. 12, 5673-5682. 10.1128/MCB.12.12.5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy A. A., Hammond S. M. and Hannon G. J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363-366. 10.1038/35053110 [DOI] [PubMed] [Google Scholar]
- Billi A. C., Fischer S. E. and Kim J. K. (2014). Endogenous RNAi pathways in C. elegans. In WormBook (ed. The C. elegans Research Community), pp. 1-49. Wormbook; 10.1895/wormbook.1.170.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Julicher F. and Hyman A. A. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729-1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeks R. J., Canman J. C., Gabriel W. N., Meyer N., Strome S. and Goldstein B. (2004). C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr. Biol. 14, 851-862. 10.1016/j.cub.2004.05.022 [DOI] [PubMed] [Google Scholar]
- Chepenik L. G., Tretiakova A. P., Krachmarov C. P., Johnson E. M. and Khalili K. (1998). The single-stranded DNA binding protein, Pur-α, binds HIV-1 TAR RNA and activates HIV-1 transcription. Gene 210, 37-44. 10.1016/S0378-1119(98)00033-X [DOI] [PubMed] [Google Scholar]
- Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J., van Wolfswinkel J. C., Chaves D. A., Shirayama M., Mitani S., Ketting R. F. et al. (2009). The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139, 123-134. 10.1016/j.cell.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine C. C., Batista P. J., Gu W., Claycomb J. M., Chaves D. A., Shirayama M. and Mello C. C. (2010). Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 107, 3588-3593. 10.1073/pnas.0911685107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel D. C. and Johnson E. M. (2018). PURA, the gene encoding Pur-alpha, member of an ancient nucleic acid-binding protein family with mammalian neurological functions. Gene 643, 133-143. 10.1016/j.gene.2017.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbinian N., Sawaya B. E., Khalili K., Jaffe N., Wortman B., Giordano A. and Amini S. (2001). Functional interaction between cyclin T1/cdk9 and Purα determines the level of TNFα promoter activation by Tat in glial cells. J. Neuroimmunol. 121, 3-11. 10.1016/S0165-5728(01)00372-1 [DOI] [PubMed] [Google Scholar]
- Dodson A. E. and Kennedy S. (2019). Germ Granules Coordinate RNA-Based Epigenetic Inheritance Pathways. Dev. Cell 50, 704-715.e4. 10.1016/j.devcel.2019.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy E. M. (1975). Germ plasm and the differentiation of the germ cell line. Int. Rev. Cytol. 43, 229-280. 10.1016/S0074-7696(08)60070-4 [DOI] [PubMed] [Google Scholar]
- Gallia G. L., Darbinian N., Jaffe N. and Khalili K. (2001). Single-stranded nucleic acid-binding protein, Purα, interacts with RNA homologous to 18S ribosomal RNA and inhibits translation in vitro. J. Cell. Biochem. 83, 355-363. 10.1002/jcb.1247 [DOI] [PubMed] [Google Scholar]
- Gent J. I., Lamm A. T., Pavelec D. M., Maniar J. M., Parameswaran P., Tao L., Kennedy S. and Fire A. Z. (2010). Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol. Cell 37, 679-689. 10.1016/j.molcel.2010.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson-Gurwitz A., Wang S., Sathe S., Green R., Yeo G. W., Oegema K. and Desai A. (2016). A Small RNA-Catalytic Argonaute Pathway Tunes Germline Transcript Levels to Ensure Embryonic Divisions. Cell 165, 396-409. 10.1016/j.cell.2016.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Shirayama M., Conte D. Jr., Vasale J., Batista P. J., Claycomb J. M., Moresco J. J., Youngman E. M., Keys J., Stoltz M. J. et al. (2009). Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36, 231-244. 10.1016/j.molcel.2009.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S., Bochner A. F., Pavelec D. M., Burkhart K. B., Harding S., Lachowiec J. and Kennedy S. (2008). An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321, 537-541. 10.1126/science.1157647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. (1986). Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics 114, 15-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N. N., Mootz D. E., Walhout A. J., Vidal M. and Hunter C. P. (2002). MEX-3 interacting proteins link cell polarity to asymmetric gene expression in Caenorhabditis elegans. Development 129, 747-759. [DOI] [PubMed] [Google Scholar]
- Ishidate T., Ozturk A. R., Durning D. J., Sharma R., Shen E.-Z., Chen H., Seth M., Shirayama M. and Mello C. C. (2018). ZNFX-1 functions within perinuclear nuage to balance epigenetic signals. Mol. Cell 70, 639-649.e6. 10.1016/j.molcel.2018.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav S., Rana M. and Subramaniam K. (2008). Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development 135, 1803-1812. 10.1242/dev.013656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. M., Kinoshita Y., Weinreb D. B., Wortman M. J., Simon R., Khalili K., Winckler B. and Gordon J. (2006). Role of Purα in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J. Neurosci. Res. 83, 929-943. 10.1002/jnr.20806 [DOI] [PubMed] [Google Scholar]
- Johnstone I. J. (1999). Molecular biology. In C. elegans A Practical Approach (ed. Hope I.), pp. 201-225. New York: Oxford University Press. [Google Scholar]
- Kadyk L. C. and Kimble J. (1998). Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125, 1803-1813. [DOI] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N. and Hirokawa N. (2004). Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513-525. 10.1016/j.neuron.2004.07.022 [DOI] [PubMed] [Google Scholar]
- Kawasaki I., Shim Y.-H., Kirchner J., Kaminker J., Wood W. B. and Strome S. (1998). PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635-645. 10.1016/S0092-8674(00)81605-0 [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Xu S., Montgomery M. K. and Fire A. (1997). Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146, 227-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R. F., Haverkamp T. H. A., van Luenen H. G. A. M. and Plasterk R. H. A. (1999). Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99, 133-141. 10.1016/S0092-8674(00)81645-1 [DOI] [PubMed] [Google Scholar]
- Ketting R. F., Fischer S. E., Bernstein E., Sijen T., Hannon G. J. and Plasterk R. H. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654-2659. 10.1101/gad.927801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili K., Del Valle L., Muralidharan V., Gault W. J., Darbinian N., Otte J., Meier E., Johnson E. M., Daniel D. C., Kinoshita Y. et al. (2003). Purα is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol. Cell. Biol. 23, 6857-6875. 10.1128/MCB.23.19.6857-6875.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson A. K., Egelhofer T., Rechtsteiner A. and Strome S. (2017). Germ granules prevent accumulation of somatic transcripts in the adult Caenorhabditis elegans germline. Genetics 206, 163-178. 10.1534/genetics.116.198549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G. A. and Subramaniam K. (2018). PUF-8 facilitates homologous chromosome pairing by promoting proteasome activity during meiotic entry in C. elegans. Development 145, dev163949 10.1242/dev.163949 [DOI] [PubMed] [Google Scholar]
- Lalani S. R., Zhang J., Schaaf C. P., Brown C. W., Magoulas P., Tsai A. C.-H., El-Gharbawy A., Wierenga K. J., Bartholomew D., Fong C.-T. et al. (2014). Mutations in PURA cause profound neonatal hypotonia, seizures, and encephalopathy in 5q31.3 microdeletion syndrome. Am. J. Hum. Genet. 95, 579-583. 10.1016/j.ajhg.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-C., Gu W., Shirayama M., Youngman E., Conte D. Jr. and Mello C. C. (2012). C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150, 78-87. 10.1016/j.cell.2012.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev I., Toker I. A., Mor Y., Nitzan A., Weintraub G., Antonova O., Bhonkar O., Ben Shushan I., Seroussi U., Claycomb J. M. et al. (2019). Germ granules govern small RNA inheritance. Curr. Biol. 29, 2880-2891.e4. 10.1016/j.cub.2019.07.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari R., Pushpa K. and Subramaniam K. (2016). A role for post-transcriptional control of endoplasmic reticulum dynamics and function in C. elegans germline stem cell maintenance. Development 143, 3097-3108. 10.1242/dev.134056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainpal R., Priti A. and Subramaniam K. (2011). PUF-8 suppresses the somatic transcription factor PAL-1 expression in C. elegans germline stem cells. Dev. Biol. 360, 195-207. 10.1016/j.ydbio.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D. and Seydoux G. (2008). 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18, 1476-1482. 10.1016/j.cub.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J. and Fire A. (2007). Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315, 241-244. 10.1126/science.1132839 [DOI] [PubMed] [Google Scholar]
- Parrish S. and Fire A. (2001). Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA 7, 1397-1402. [PMC free article] [PubMed] [Google Scholar]
- Pepper A. S., Killian D. J. and Hubbard E. J. (2003). Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163, 115-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M., Montgomery T. A., Breen P. C. and Ruvkun G. (2012). MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26, 1433-1444. 10.1101/gad.193904.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D. and Austin J. (2001). Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpa K., Kumar G. A. and Subramaniam K. (2013). PUF-8 and TCER-1 are essential for normal levels of multiple mRNAs in the C. elegans germline. Development 140, 1312-1320. 10.1242/dev.087833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. J., Svendsen J. M., Brown K. C., Montgomery B. E., Marks T. N., Vijayasarathy T., Parker D. M., Nishimura E. O., Updike D. L. and Montgomery T. A. (2020). Widespread roles for piRNAs and WAGO-class siRNAs in shaping the germline transcriptome of Caenorhabditis elegans. Nucleic Acids Res. 48, 1811-1827. 10.1093/nar/gkz1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V., Gil I. S., Ward S. and Kazmer K. (2004). Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131, 311-323. 10.1242/dev.00914 [DOI] [PubMed] [Google Scholar]
- Robert V. J., Mercier M. G., Bedet C., Janczarski S., Merlet J., Steve Garvis S., Ciosk R. and Francesca Palladino F. (2014). The SET-2/SET1 histone H3K4 methyltransferase maintains pluripotency in the Caenorhabditis elegans Germline. Cell Rep. 9, 443-450. 10.1016/j.celrep.2014.09.018 [DOI] [PubMed] [Google Scholar]
- Rogers A. K. and Phillips C. M. (2020). RNAi pathways repress reprogramming of C. elegans germ cells during heat stress. Nucleic Acids Res. 48, 4256-4273. 10.1093/nar/gkaa174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F. and Maniatis T. (1989). Molecular Cloning, A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schedl T. and Kimble J. (1988). fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119, 43-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Pitt J., Dennis S. and Priess J. R. (2010). Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development 137, 1305-1314. 10.1242/dev.044255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H.-C., Gu W., Ishidate T., Conte D. Jr. and Mello C. C. (2012). piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150, 65-77. 10.1016/j.cell.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Yan J., Pagano D. J., Dodson A. E., Fei Y., Gorham J., Seidman J. G., Wickens M. and Kennedy S. (2020). poly(UG)-tailed RNAs in genome protection and epigenetic inheritance. Nature 582, 283-288. 10.1038/s41586-020-2323-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K. L., Parrish S., Timmons L., Plasterk R. H. and Fire A. (2001). On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465-476. 10.1016/S0092-8674(01)00576-1 [DOI] [PubMed] [Google Scholar]
- Spike C. A., Bader J., Reinke V. and Strome S. (2008). DEPS-1 promotes P-granule assembly and RNA interference in C. elegans germ cells. Development 135, 983-993. 10.1242/dev.015552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S. and Wood W. B. (1982). Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 79, 1558-1562. 10.1073/pnas.79.5.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K. and Seydoux G. (2003). Dedifferentiation of primary spermatocytes into germ cell tumors in C. elegans lacking the pumilio-like protein PUF-8. Curr. Biol. 13, 134-139. 10.1016/S0960-9822(03)00005-8 [DOI] [PubMed] [Google Scholar]
- Tabara H., Hill R. J., Mello C. C., Priess J. R. and Kohara Y. (1999a). pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126, 1-11. [DOI] [PubMed] [Google Scholar]
- Tabara H., Sarkissian M., Kelly W. G., Fleenor J., Grishok A., Timmons L., Fire A. and Mello C. C. (1999b). The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123-132. 10.1016/S0092-8674(00)81644-X [DOI] [PubMed] [Google Scholar]
- Thivierge C., Makil N., Flamand M., Vasale J. J., Mello C. C., Wohlschlegel J., Conte D. Jr. and Duchaine T. F. (2011). Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nat. Struct. Mol. Biol. 19, 90-97. 10.1038/nsmb.2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D. L. and Fire A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103-112. 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Updike D. and Strome S. (2010). P granule assembly and function in Caenorhabditis elegans germ cells. J. Androl. 31, 53-60. 10.2164/jandrol.109.008292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaid S., Ariz M., Chaturbedi A., Kumar G. A. and Subramaniam K. (2013). PUF-8 negatively regulates RAS/MAPK signalling to promote differentiation of C. elegans germ cells. Development 140, 1645-1654. 10.1242/dev.088013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E. and Seydoux G. (2010). The C. elegans homolog of nucleoporin Nup98 is required for the integrity and function of germline P granules. Development 137, 1441-1450. 10.1242/dev.047654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G., Fields B. D., Spracklin G., Shukla A., Phillips C. M. and Kennedy S. (2018). Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 557, 679-683. 10.1038/s41586-018-0132-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Bernstein D. S., Kimble J. and Parker R. (2002). A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18, 150-157. 10.1016/S0168-9525(01)02616-6 [DOI] [PubMed] [Google Scholar]
- Witze E. S., Field E. D., Hunt D. F. and Rothman J. H. (2009). C. elegans pur alpha, an activator of end-1, synergizes with the Wnt pathway to specify endoderm. Dev. Biol. 327, 12-23. 10.1016/j.ydbio.2008.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolke U., Jezuit E. A. and Priess J. R. (2007). Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development 134, 2227-2236. 10.1242/dev.004952 [DOI] [PubMed] [Google Scholar]
- Yigit E., Batista P. J., Bei Y., Pang K. M., Chen C.-C. G., Tolia N. H., Joshua-Tor L., Mitani S., Simard M. J. and Mello C. C. (2006). Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127, 747-757. 10.1016/j.cell.2006.09.033 [DOI] [PubMed] [Google Scholar]
- Zetka M. C., Kawasaki I., Strome S. and Muller F. (1999). Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13, 2258-2270. 10.1101/gad.13.17.2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Pedigo N., Shenoy S., Khalili K. and Kaetzel D. M. (2005). Purα activates PDGF-A gene transcription via interactions with a G-rich, single-stranded region of the promoter. Gene 348, 25-32. 10.1016/j.gene.2004.12.050 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.