Abstract
Background
Candida auris infections have recently emerged worldwide, and this species is highly capable of colonization and is associated with high levels of mortality. However, strain-dependent differences in colonization capabilities and virulence have not yet been reported.
Objectives
In the present study, we aimed to clarify the differences between clinically isolated invasive and non-invasive strains of C. auris.
Methods
We evaluated colonization, dissemination, and survival rates in wild C57BL/6J mice inoculated with invasive or non-invasive strains of C. auris under cortisone acetate immunosuppression, comparing with those of Candida albicans and Candida glabrata infections. We also evaluated the potency of biofilm formation.
Results
Stool fungal burdens were significantly higher in mice inoculated with the invasive strains than in those infected with the non-invasive strain. Along with intestinal colonization, liver and kidney fungal burdens were also significantly higher in mice inoculated with the invasive strains. In addition, histopathological findings revealed greater dissemination and colonization of the invasive strains. Regarding biofilm-forming capability, the invasive strain of C. auris exhibited a significantly higher capacity of producing biofilms. Moreover, inoculation with the invasive strains resulted in significantly greater loss of body weight than that noted following infection with the non-invasive strain.
Conclusions
Invasive strains showed higher colonization capability and rates of dissemination from gastrointestinal tracts under cortisone acetate immunosuppression than non-invasive strains, although the mortality rates caused by C. auris were lower than those caused by C. albicans.
Introduction
Candida species are common causative agents of bloodstream infections in hospitals, leading to high levels of mortality and morbidity [1,2]. There are two routes that Candida species follow for infecting the bloodstream as follows: exogenous or endogenous [3–6]. Endogenous bloodstream infections are considered to be caused by the invasion of Candida species colonizing in mucosa, on the other hands, exogenous infections are often caused as catheter-related bloodstream infection [6–8]. In particular, the previous report describes gastrointestinal tract is one of the primary areas of Candida species colonization and these species can invade and disseminate from this location, leading to bloodstream infection under specific conditions [6].
Candida auris was initially isolated and reported in a Japanese patient with chronic otitis media [9]. After the first isolation, C. auris infections were reported worldwide in places such as Korea, India, the United States, South America, and South Africa [10–16]. In several reports, C. auris bloodstream infections were associated with high mortality and morbidity, which indicated the high virulence of clinically isolated C. auris strains. In mouse experiments, C. auris strains clinically isolated from bloodstream infections were reportedly highly virulent, leading to severe infections and high mortality rates [17–19]. In these experiments, systemic candidiasis was caused by intravenous injection mimicking exogenous candidiasis; however, to the best of our knowledge, no model of endogenous candidiasis dissemination from the gastrointestinal tract caused by C. auris has yet been reported. C. auris was reported to colonize on mucocutaneous surfaces, especially skins, and healthcare environments, leading to bloodstream infections via catheters, although some articles have reported C. auris gastrointestinal colonization and the possibilities of dissemination [20–22]. Therefore, studies of capacity for colonization and dissemination from gastrointestinal tracts are necessary. In addition, 4 clades of C. auris; East Asian, South Asian, South America, and African clade; have been reported, and all C. auris isolates in Japan belonging to the East Asian clade were obtained from the patients with chronic otitis media [23]. However, C. auris strains found in other countries, belonging to the South Asian, South American, or African clade, were primarily isolated from bloodstream infections. Hence, differences in virulence and colonization capabilities between C. auris strains in Japan (non-invasive strains) and those from other countries (invasive strains) may exist, but research into these strain-specific differences is scarce.
In this report, we compared the capability for colonization and dissemination from gastrointestinal tracts between a non-invasive strain of C. auris isolated in Japan and invasive strains isolated in other countries using an endogenous candidiasis mouse model under cortisone acetate immunosuppression. We also evaluated the pathogenicity of C. auris strains compared with other pathogenic Candida species using survival analysis.
Material and methods
Mice
Male C57BL/6J mice, 6−7 weeks old, were purchased from Japan SLC, Inc. (Shizuoka, Japan) and maintained under specific pathogen-free conditions at the National Institute of Infectious Diseases in Japan. All experiments were reviewed and approved by the Animal Care and Use Committee of the National Institute of Infectious Diseases. All staffs including this investigation were sufficiently educated in animal care and handling before this procedure. Protocols were designed for minimizing animal suffering and limit the numbers used in experiments (Approval numbers:117139 and 119047).
Yeast strains and infection models
Two clinically isolated non-invasive strains of C. auris (JCM 15448: the first strain isolated in Japan [7], and TWCC 58362) and invasive strains obtained from the Center of Disease Control and Prevention AR Isolate Bank collection of C. auris isolates (AR0382, AR0383, AR0385, and AR0390) were used in this study (Table 1). Reference strains of Candida albicans (SC5314) and Candida glabrata (CBS 138) were used in the survival analysis.
Table 1. Candida auris strains used in the study.
| Strain | Clade | Origin |
|---|---|---|
| JCM 15448 | East Asian | Chronic otitis media |
| TWCC 58362 | East Asian | Chronic otitis media |
| AR0382 | South Asian | Bloodstream infection |
| AR0383 | African | Bloodstream infection |
| AR0385 | South American | Bloodstream infection |
| AR0390 | South Asian | Bloodstream infection |
For the Candida species colonization and dissemination mouse model, mice were administered with a cocktail of antibiotics in their drinking water for more than 1 week before inoculation with C. auris. The cocktail consisted of vancomycin (45 mg/L), gentamicin (35 mg/L), kanamycin (400 mg/L), metronidazole (215 mg/L), and colistin (850 U/L), as previously reported for the mouse model of Clostridioides difficile infection [24]. C. albicans and C. auris (AR0382, AR0383, AR 0385, and AR0390) were grown at 30°C and C. glabrata at 37°C for 2 days on yeast extract peptone dextrose (YPD) agar. C. auris (JCM 15448 and TWCC 58262) was grown at 30°C for 3 days on YPD agar. Next, C. albicans and C. auris were grown at 30°C and C. glabrata was grown at 37°C in YPD broth for 18−24 h. Following incubation, the yeasts were collected, washed, and resuspended in sterile normal saline at approximately 5.0 × 108−1.0 × 109 colony-forming units (CFU)/mL. Mice were inoculated with 100 μL (approximately 5.0 × 107–1.0 × 108 CFU/mouse) via a sterile feeding needle for instigating the colonization of the Candida species. One day previously, on the day, and 1 day after inoculation with the Candida species, cortisone acetate (225 mg/kg) was subcutaneously injected into each mouse, as previously reported for the mouse model of oropharyngeal candidiasis with modifications [25].
Evaluation of organ dissemination and gastrointestinal colonization
In these experiments, mice were divided into six groups as follows: C. auris JCM 15448, C. auris TWCC 58362, C. auris AR0382, C. auris AR0383, C. auris AR0385, or C. auris AR0390 inoculated groups. These mice were euthanatized 7 or 14 days after inoculation, followed by the aseptic collection of liver and kidneys. Prior to euthanasia, fresh stools from each mouse were collected for evaluating gastrointestinal colonization. The liver, kidneys, and stools were then homogenized in sterile PBS, and the homogenates were serially diluted. Liver and stool homogenates were plated on YPD agar with antibiotics (penicillin/streptomycin, to inhibit bacterial growth), and kidney homogenate was plated on YPD agar, both at 100 μL per plate. The fungal burden was determined by counting CFUs after 24−48 h of incubation at 30°C.
Histopathological analysis of organs from infected mice
For histopathological analysis, liver, kidneys, and colons were removed 14 days after C. auris inoculation and fixed in 10% neutral buffered formalin, dehydrated with ethanol, and embedded in paraffin following standard procedures. Tissue sections (4 μm) were mounted onto glass slides (Matsunami Glass, Osaka, Japan) and stained with hematoxylin and eosin or Grocott methenamine silver (GMS). After staining, histological examinations were performed using light microscopy.
Biofilm-forming assay
In this assay, invasive strains of C. auris (AR0382, AR0383, AR0385, or AR0390) were grown for 2 days and non-invasive strains of C. auris (JCM 15448 and TWCC 58362) for 3 days at 30°C on YPD agar. Each C. auris strain was then grown at 30°C in YPD broth for 18–24 h. After incubation, yeasts were collected, washed with sterile PBS, resuspended in YPD broth, and diluted to 1.0 × 108 cells/mL. First, 100 μL of this inoculum was added to each well of a 96-well plate and incubated for 4 h to allow the cells to adhere to the plastic surface, as previously described, with minor modifications [26,27]. In the negative control wells, 100 μL of YPD broth was added. After 4 hours of adhesion, the supernatant of each well was discarded to remove planktonic C. auris cells. After that, 100 μL of YPD broth were added to each well. The plate was then incubated for 24 h to allow C. auris to form a biofilm. After 24 h of incubation, the supernatant in each well was carefully removed and 150 μL of methanol were added to fix the biofilm. After methanol fixation, each well was washed three times with PBS and stained with 0.1% crystal violet for 60 min. After crystal violet staining, each well was washed several times with PBS. Lastly, the crystal violet bound to the C. auris biofilms was dissolved with 200 μL of methanol and the optimal density at 595 nm was measured using a microtiter plate reader.
Survival analysis
In this experiment, mice were divided into four groups as follows: C. auris JCM 15448, C. auris AR0382, C. albicans SC5314, or C. glabrata CBS 138 inoculated groups and survival rates were recorded for 4 weeks after inoculation. Mice were monitored at least once a day and the body weights of these inoculated mice were also monitored. Any mice that appeared to be unable to maintain standing position, loss of appetite and drinking, hunched position, or moribund (reduced activity, tachypnea, and poorly responsive to external stimuli) were humanely euthanized as soon as possible by the inhalation of carbon dioxide. All mice remaining at the end of the survival analysis were also euthanized by the inhalation of carbon dioxide. In total, 52 mice were used and divided into 4 groups in this analysis.
Statistical analysis
Continuous variables were compared among groups by the Kruskal-Wallis test and post hoc Dunn’s test for pair-wise comparisons. In the survival analysis, groups were compared using the log-rank (Mantel–Cox) test. Body weight changes during the survival analysis were compared using two-way ANOVA followed by post hoc Tukey’s multiple comparisons test. A P value of < 0.05 was considered significant for all tests. Statistical analyses were performed using GraphPad Prism, version 7 (GraphPad Software, La Jolla, CA, USA).
Results
Potency of gastrointestinal colonization of C. auris
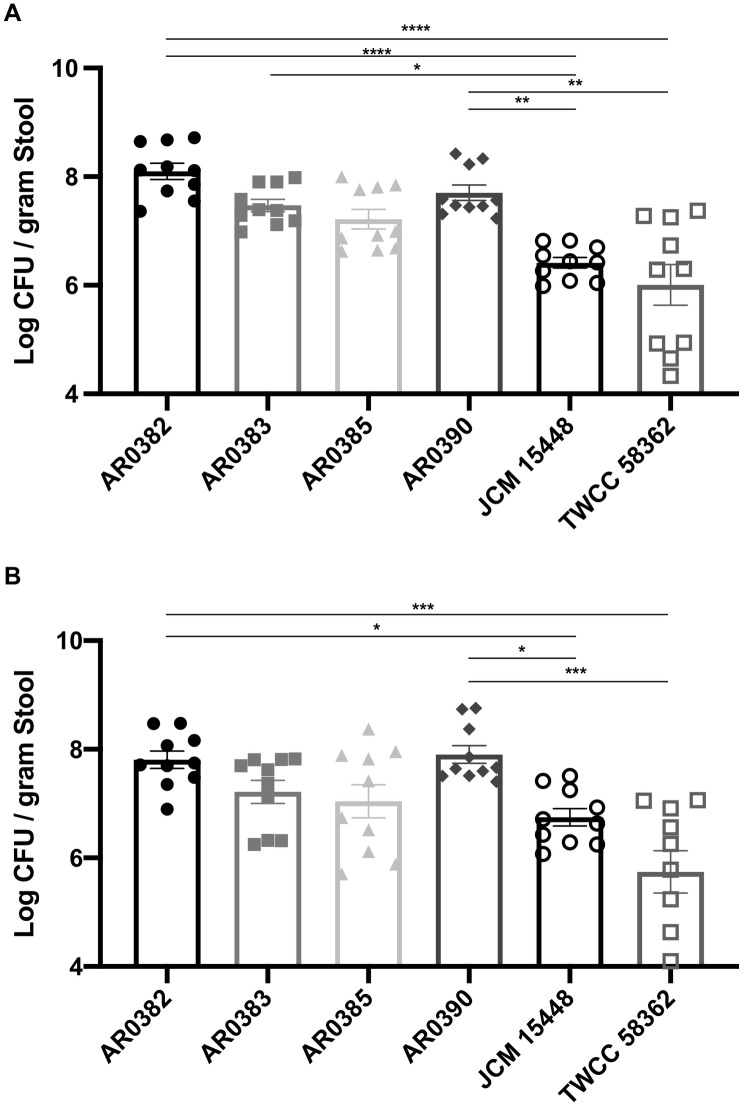
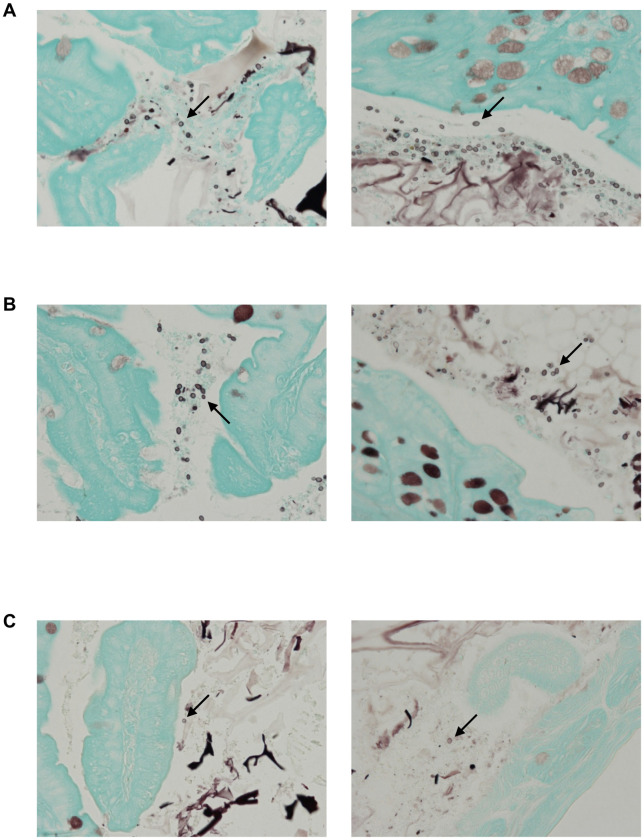
We intragastrically inoculated each mouse with non-invasive strains of C. auris JCM 15448 or C. auris TWCC 58362, or invasive strains AR0382, AR0383, AR0385, or AR0390. After inoculation, we evaluated the gastrointestinal colonization of each group by calculating stool CFUs. The fungal burden in the gastrointestinal tract 7 days after inoculation was significantly higher in mice infected with invasive strains, especially AR0382 or AR0390, compared with those infected with non-invasive strains (Fig 1A). Similarly, 14 days after inoculation, the stool fungal burden in each group inoculated with AR0382 or AR0390 was also significantly higher compared with that in the group infected with the non-invasive strain (Fig 1B). No significant differences were noted between mice inoculated with the invasive strains. Based on the above results, we histopathologically observed gastrointestinal colonization of murine colons 14 days after inoculation with non-invasive strains of C. auris JCM 15448, and invasive strains AR0382 or AR0390. In the colon, GMS staining demonstrated abundant levels of yeasts in the stools and on the surfaces of intestinal mucosa in mice inoculated with AR0382 or AR0390, but sparse levels in mice inoculated with JCM 15448 (Fig 2A–2C). Neither yeast invasion of the mucosa nor any filamentous forms were observed in any mice. These results demonstrated that each C. auris strain could colonize the gastrointestinal tracts following treatment of the mice with cortisone acetate and antibiotics. In addition, the invasive strains colonized the murine gastrointestinal tract more effectively than the non-invasive strain.
Fig 1. Stool fungal burden was significantly higher in mice infected by invasive strains of Candida auris strains than in those infected with the non-invasive strains.
(A, B) The fungal burdens in stools collected 7 days (A) and 14 days (B) after inoculation of invasive or non-invasive C. auris strains. Stool fungal burdens are shown as the Log CFU/g stool. All results are expressed as mean ± standard error of the mean from six independent experiments with a total of 10 stool samples per group. *** P < 0.001, **** P < 0.0001.
Fig 2. The invasive strains of Candida auris demonstrated greater colonization in murine intestines than the non-invasive strains.
(A–C) GMS staining of the colon 14 days after inoculation with the invasive strain AR0382 (A), the invasive strain AR0390 (B), and the non-invasive strain JCM 15448 (C). Arrows indicate yeast cells of C. auris.
Dissemination of C. auris from gastrointestinal tracts to organs
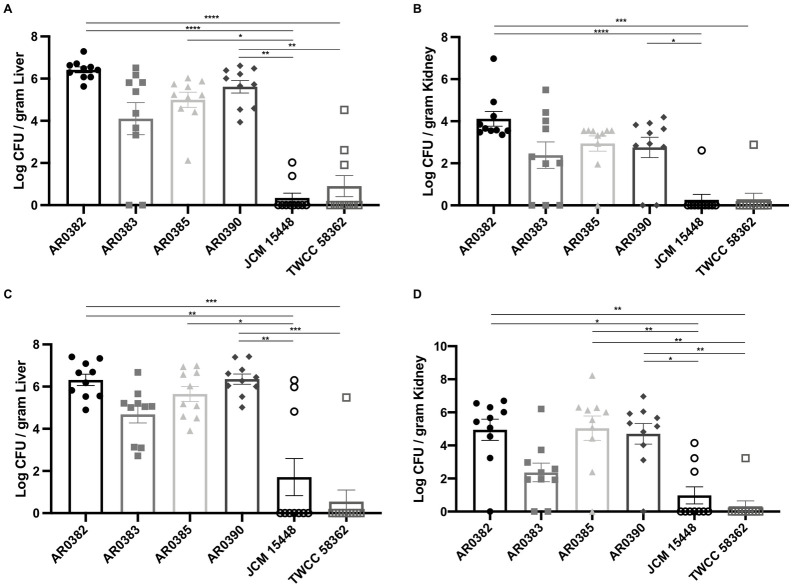
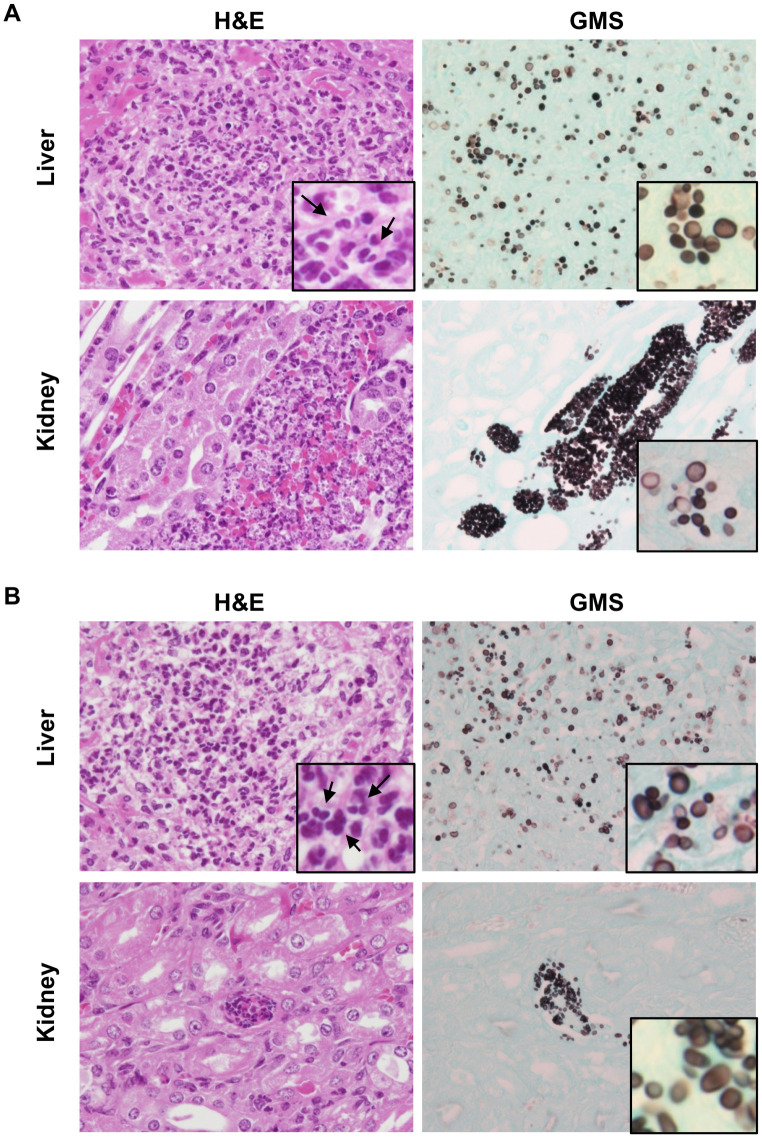
In addition to measuring gastrointestinal colonization, we also evaluated the dissemination of the yeasts from the gastrointestinal tracts. Seven days after inoculation, the fungal burdens of the liver and kidneys were significantly higher in mice infected with the invasive strains, especially AR0382 or AR0390, compared with those in mice infected with the non-invasive strains (Fig 3A and 3B). Moreover, there were no differences of organ disseminations among invasive strains. At 14 days after inoculation, the fungal burdens of the liver and kidneys of mice inoculated with the invasive strains were also significantly higher than those with the non-invasive strains (Fig 3C and 3D). No significant differences were noted between mice inoculated with the invasive strains. In some mice infected by the non-invasive strains, dissemination into liver or kidneys was found 7 or 14 days after inoculation; however, the dissemination rates in mice infected with this non-invasive strain were comparably small. In addition, on the basis of the results obtained from the dissemination experiments, we histopathologically evaluated the liver and kidneys of mice 14 days after inoculation with the invasive strains AR0382 or AR0390. The histological analyses revealed that round yeasts without hyphae formed a fungal mass in the organs of mice infected with AR0382 or AR0390 (Fig 4A and 4B). In the liver, focal necrosis with fungi and neutrophil infiltration was observed, although little inflammation was observed in the portal area. In the kidney, yeasts were detected in renal tubules with inflammatory cell infiltration. However, no yeast cells were detected in the liver and kidneys of mice infected with JCM 15448 (S1 Fig). Taken together, these results indicated that the invasive strains isolated from the bloodstream infections were more capable of colonizing and disseminating from the gastrointestinal tracts under cortisone acetate immunosuppression than the non-invasive strain isolated from the external ear canal.
Fig 3. The invasive strains of Candida auris demonstrated greater dissemination into organs than the non-invasive strains.
(A–D) The fungal burdens in the liver and kidneys 7 days (A, B) or 14 days (C, D) after inoculation with invasive or non-invasive C. auris strains. Liver and kidney fungal burdens are shown as the Log CFU/g liver and Log CFU/g kidney, respectively. All results are expressed as mean ± standard error of the mean from six independent experiments with a total of 10 tissue samples per group. * P < 0.05, *** P < 0.001, **** P < 0.0001.
Fig 4. The invasive strains of Candida auris showed massive fungal dissemination.
(A-B) Histopathological analysis of the liver and kidney of mice infected with invasive strains AR0382 (A) and AR0390 (B), 14 days after inoculation. Hematoxylin and eosin staining (left panels) and GMS staining (right panels) are shown. Original magnification: ×400 (inset: ×1000). Neutrophils are indicated by arrows (insets).
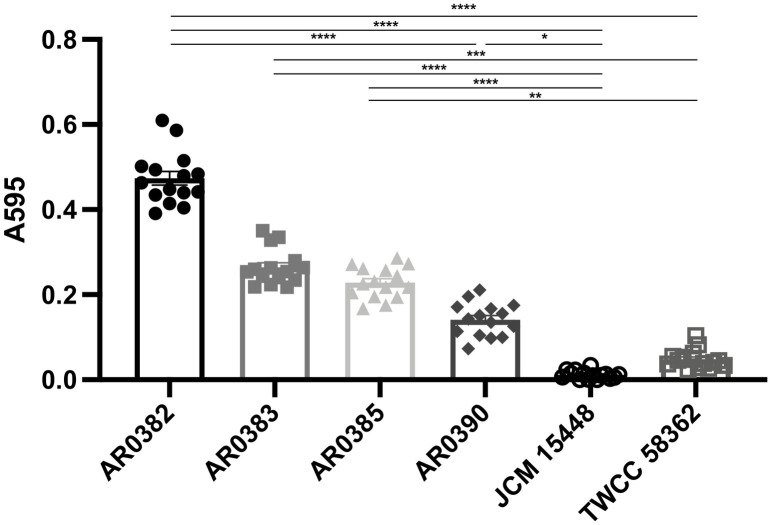
Biofilm-forming capability of C. auris
Given the results of colonization, dissemination, and histopathological analysis, we evaluated the biofilm-forming capability of each C. auris strain. In this analysis, the invasive strain of C. auris, AR0382, exhibited the highest levels of biofilm formation, and higher than the non-invasive C. auris strains (Fig 5). In addition, the other invasive strains were able to produce biofilms more effectively than the non-invasive strains. On the other hands, the non-invasive strain, especially JCM 15448, produced few biofilms. These results indicated that the invasive C. auris strains presented higher biofilm-forming capabilities, which may be associated with greater ability of colonizing the gastrointestinal tracts and disseminating to the organs.
Fig 5. The invasive strains of Candida auris showed higher biofilm-forming capacity than the non-invasive strains.
The biofilm values of each C. auris strain stained with crystal violet are shown. Biofilm values are given as optical density at 595nm. All results are expressed as mean ± standard error of the mean from three independent experiments with a total of 15 samples per group. ** P < 0.01, **** P < 0.0001.
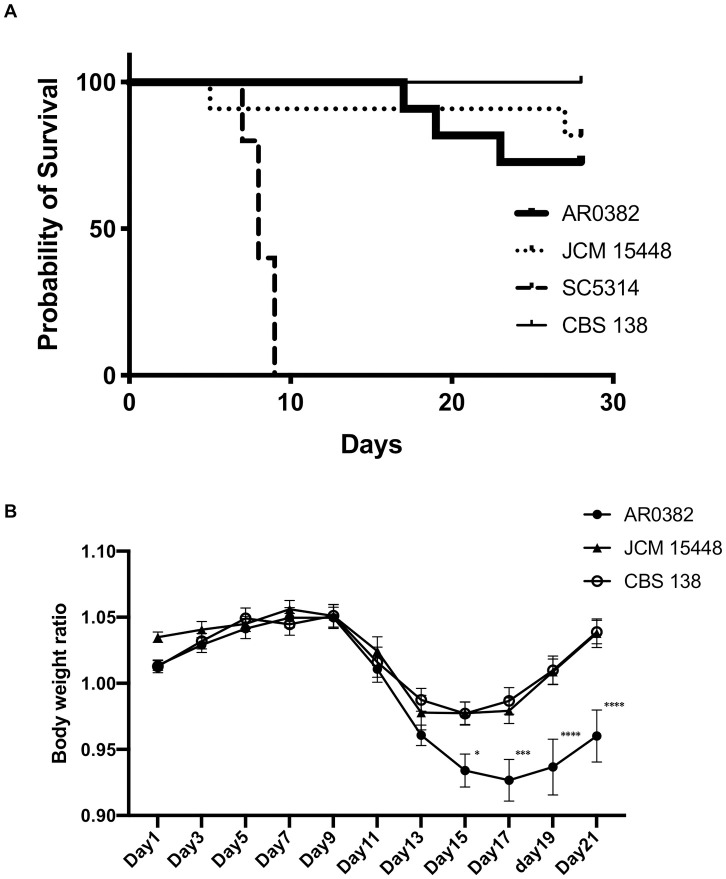
Survival analysis following dissemination of endogenous Candida species
Given the results of the organ CFU counts and histopathological analysis, we chose to use the invasive strain AR0382 and the non-invasive strain JCM 15448 in the survival analysis. In addition, we used C. albicans SC5314 and C. glabrata CBS 138 as comparisons. In this experiment, we observed and compared the survival rate and body weight changes after inoculation with each Candida species. All of the mice which reached the endpoint criteria were euthanized and no mice died before meeting criteria for euthanasia. The results demonstrated that the mortality rate was significantly higher in mice infected with C. albicans than those inoculated with other Candida species (P < 0.0001, Fig 6A). The weight loss of mice infected with C. auris AR0382 was significantly higher than that of those infected with C. auris JCM 15448 or C. glabrata (Day 15; P < 0.05, Day 17; P < 0.001, Days 19 and 21; P < 0.0001 Fig 6B). However, no significant differences were noted in the survival rates among groups other than those infected with C. albicans, despite the deaths of several mice following C. auris inoculation. Collectively, the invasive C. auris isolates were able to colonize and disseminate more effectively than the non-invasive C. auris isolates, leading to the higher virulence of invasive strains from the viewpoint of body weight changes. However, according to our endogenous Candida species dissemination mouse model, the mortality attributed to C. auris was lower than that for C. albicans, and no significant differences were noted among the strains of C. auris.
Fig 6. The invasive strains of Candida auris resulted in greater weight loss than the non-invasive strain and Candida glabrata despite of lower mortality rate compared with Candida albicans.
(A, B) The survival rates (A) and body weight changes (B) of mice inoculated with the invasive strain of C. auris AR0382, the non-invasive strain JCM 15448, the reference strain of C. albicans SC5314, and the reference strain of C. glabrata CBS 138 are shown. Body weight change results are expressed as mean ± standard error of the mean from three independent experiments including 16 C. auris–infected mice and 10 C. albicans of C. glabrata–infected mice. * P < 0.05, *** P < 0.001, **** P < 0.0001.
Discussion
Candida auris has recently been reported worldwide, and is believed to be a threat due to the potency of its multidrug-resistance and high virulence [11,15,16]. Globally, C. auris has been primarily isolated from patients with bloodstream infections, although all strains in Japan belonging to the East Asian clade were isolated from the external ear canals of infected patients. Therefore, it was hypothesized that there are differences in the virulence and colonization capabilities between the non-invasive isolates from Japan and the invasive strains isolated in other countries. In this study, we evaluated the capability of the colonization and dissemination of each C. auris strain using an endogenous dissemination mice model under treatment with cortisone acetate. The results demonstrated that the invasive strains, especially the South Asian clade, of C. auris were more effective at colonization and dissemination from the gastrointestinal tract than the non-invasive strains obtained from external ear canals.
During the analysis of fungal burden in organs and stools, both the rates of colonization in the gastrointestinal tract and dissemination to organs were significantly higher in mice infected with the invasive strains compared with those inoculated with the non-invasive strains. In addition to these results, the biofilm-forming capabilities of the invasive strains were shown to be higher than those of the non-invasive strain. Reports have indicated that the biofilms of C. albicans are associated with colonization on mucosal surfaces such as the oral cavity and vagina [28,29]. In addition, the biofilm is associated with resistance to the immune system and antifungal drugs [30–32]. Indeed, no reports have yet described the relationship between biofilm formation in C. auris and its potency in terms of colonization or virulence, although clinical data have suggested that biofilm formation is associated with C. albicans pathogenicity and patient deaths [33]. In our study, the highly effective formation of a biofilm by C. auris appeared to be associated with the capability to colonize the gastrointestinal tract, although further investigation of the direct relationship between biofilm formation and pathogenicity of C. auris is warranted. In addition, our investigation showed that C. auris isolates belonging to the same clade had different biofilm-forming capacities, however, there are no reports describing these differences. Further investigation is also necessary to clarify these differences.
Histopathological analyses revealed that the round cell form of C. auris was disseminated under cortisone acetate immunosuppression. In our experiments, the invasive strains of C. auris disseminated into organs including the liver and kidneys without establishing a filamentous form, which is accordant with previous reports [9,17,18]. A previous study demonstrated the ability of C. auris to generate a filamentous form under limited conditions, and this filamentous phenotype is associated with its invasive capacity [34]. However, our results did not demonstrate that C. auris had a filamentous phenotype in spite of confirmed colonization in colons and dissemination into organs, which implies that forming a filamentous phenotype is not the only factor associated with pathogenicity. In addition, the accumulation of neutrophils around the yeasts was histopathologically determined, which indicated that neutrophils play a role in protecting against C. auris invasion under cortisone acetate immunosuppression. However, a previous report revealed that C. auris can evade neutrophil attacks [35]. In addition, clinical investigations did not find that neutropenia was a common risk factor for infection of the bloodstream by C. auris [16,36]. Taken together, although neutrophils may be involved in defense against C. auris infection under certain conditions, it remains to be elucidated whether or not neutrophils are generally important factors in protecting patients against C. auris infection. Further investigation will be necessary for the clarification of the immunological response to systemic C. auris infection.
The survival analysis established that C. albicans is more pathogenic than C. auris. In our analysis, several mice infected by C. auris AR0382 died; however, the mortality rates of these mice were generally low. A number of previous reports have indicated a high rate of mortality following C. auris infection in experiments involving mice [17–19]. The reasons for the discrepancies between previous reports and our study may be due to differences in the mice strains used in the experiments, variances in the route of infection, and the type of immunosuppression used. As susceptibility to systemic C. auris infection differs between mice strains, we must note that strain C57BL/6J was used in the experiments reported in the present study, although ICR, BALB/c, or A/J strains were used in the studies reported elsewhere [17–19]. In addition, C57BL/6J mice have been reported to be more resistant to C. auris infection than A/J mice [19]. The differences in the infection route could be a more attributable point. Intravenous inoculation is considered to cause a more severe infection than other routes of exposure. Our experiments imitated endogenous dissemination from the gastrointestinal tract; therefore, the severity of the infection may have been lower than that observed in an intravenous infection model, leading to the lower rates of mortality noted in this study. The final point of difference was the type of immunosuppression implemented. Cortisone acetate was used in our experiments, although cyclophosphamide was used in most of the previous reports [18,19]. Cyclophosphamide can cause severe neutropenia, leading to high mortality rates. Conversely, cortisone acetate is considered to suppress cellular immunity but not cause neutropenia. The accumulation of neutrophils was noted in organs during our experiments, which implies that immune responses to C. auris remained, although their aptitude for phagocytosis or killing capability may have been suppressed.
There were some limitations encountered in our experiments. First, gastrointestinal colonization of C. auris was relatively rare compared with skin colonization because of poor growth under anaerobic conditions, therefore, the rate of endogenous candidiasis of C. auris was thought to be small [37]. However, some previous reports have described the gastrointestinal colonization of C. auris, and our investigations showed the possibilities of C. auris gastrointestinal colonization and dissemination under corticosteroid immunosuppression [21,22]. In addition, C. auris isolates could grow on agars under anaerobic conditions, although growth of C. auris were poor compared with C. albicans or C. glabrata (S2 Fig). These mouse models are not necessarily relevant to human events, however, the facts that C. auris could colonize and disseminate from gastrointestinal tracts under immunosuppressive conditions are thought to be significant. Based on our investigations, clinicians should pay attention to C. auris colonization and dissemination, especially in immunocompromised hosts. Second, we describe the differences in colonization ability and dissemination of non-invasive and invasive strains, although the detailed factors associated with these differences remain unknown. Previous reports have established that several genes and enzymes are associated with virulence in C. auris [37,38]; however, we did not investigate the differences of these virulence factors in this experiment and further investigations in this area are, therefore, warranted.
In summary, to the best of our knowledge, this is the first report to describe the gastrointestinal colonization and dissemination into organs of different strains of C. auris in a mouse model using cortisone acetate-induced immunosuppression. The invasive strains demonstrated greater capability for both colonization and dissemination from gastrointestinal tracts than the non-invasive strain in this model. The effect on mortality of invasive C. auris strains was lower than that of C. albicans; however, the capability for endogenous dissemination of C. auris from gastrointestinal tracts was thought to be a major factor in the invasive nature of this microorganism, especially in strains exhibiting multidrug resistance.
Supporting information
Histopathological analysis of the liver and kidney of mice infected with non-invasive strain JCM 15448, 14 days after inoculation. Hematoxylin and eosin staining (left panels) and GMS staining (right panels) are shown. Original magnification: ×200.
(TIF)
Anaerobic growth of Candida albicans (SC5314), Candida glabrata (CBS 138), and each strain of Candida auris on YPD agar, 7 days after inoculation is shown.
(TIF)
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
This work was supported in part by the Research Program on Emerging and Re-emerging Infectious Diseases of the Japan Agency for Medical Research and Development (AMED) under Grant Number (JP19fk0108045, JP19fk0108049, JP19fk0108094, and JP19fk0108104), and by JSPS KAKENHI Grant Number JP17K10040. JP19fk0108045, JP19fk0108049, JP19fk0108094, and JP17K10040 were given to YM. JP19fk0108104 was given to HK and HH. URL of AMED is https://www.amed.go.jp/en/index.html URL of JSPS KAKENHI is https://www.jsps.go.jp/english/index.html The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial Bloodstream Infections in US Hospitals: Analysis of 24,179 Cases from a Prospective Nationwide Surveillance Study. Clin Infect Dis. 2004; 39(3): 309–317. 10.1086/421946 [DOI] [PubMed] [Google Scholar]
- 2.Verfaillie C, Weisdorf D, Haake R, Hostetter M, Ramsay NKC, McGlave P. Candida infections in bone marrow transplant recipients. Bone Marrow Transplant. 1991; 8: 177–184. [PubMed] [Google Scholar]
- 3.Marr KA. Invasive Candida infections: the changing epidemiology. Oncology (Williston Park). 2004; 8: 9–14. [PubMed] [Google Scholar]
- 4.Bouza E, Muñoz P. Epidemiology of candidemia in intensive care units. Int J Antimicrob Agents. 2008; 32: S87–S91. 10.1016/S0924-8579(08)70006-2 [DOI] [PubMed] [Google Scholar]
- 5.Fridkin SK, Jarvis WR. Epidemiology of nosocomial fungal infections. Clin Microbiol Rev. 1996; 9(4): 499–511. 10.1128/CMR.9.4.499-511.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nucci M, Anaissie E. Revisiting the Source of Candidemia: Skin or Gut? Clin Infect Dis. 2001; 33(12): 1959–1967. 10.1086/323759 [DOI] [PubMed] [Google Scholar]
- 7.Poissy J, Damonti L, Bignon A, et al. Risk factors for candidemia: a prospective matched case-control study. Crit Care. 2020; 24: 109 10.1186/s13054-020-2766-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blumberg HM, Jarvis WR, Soucie JM, et al. Risk factors for candidal bloodstream infections in surgical intensive care unit patients: the NEMIS prospective multicenter study. The National Epidemiology of Mycosis Survey. Clin Infect Dis. 2001;33(2):177–186. 10.1086/321811 [DOI] [PubMed] [Google Scholar]
- 9.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53(1):41–44. 10.1111/j.1348-0421.2008.00083.x [DOI] [PubMed] [Google Scholar]
- 10.Kim M, Shin JH, Sung H, et al. Candida haemulonii and Closely Related Species at 5 University Hospitals in Korea: Identification, Antifungal Susceptibility, and Clinical Features. Clin Infect Dis. 2009;48(6):e57–e61. 10.1086/597108 [DOI] [PubMed] [Google Scholar]
- 11.Chowdhary A, Sharma C, Duggal S, et al. New Clonal Strain of Candida auris, Delhi, India. Emerg Infect Dis. 2013; 19(10): 1670–1673. 10.3201/eid1910.130393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarma S, Kumar N, Sharma S, et al. Candidemia caused by amphotericin B and Fluconazole resistant Candida auris. Indian J Med Microbiol. 2013; 31(1): 90 10.4103/0255-0857.108746 [DOI] [PubMed] [Google Scholar]
- 13.Kumar D, Banerjee T, Pratap CB, Tilak R. Itraconazole-resistant Candida auris with phospholipase, proteinase and hemolysin activity from a case of vulvovaginitis. J Infect Dev Ctries. 2015; 9(04): 435–437. 10.3855/jidc.4582 [DOI] [PubMed] [Google Scholar]
- 14.Lee WG, Shin JH, Uh Y, et al. First Three Reported Cases of Nosocomial Fungemia Caused by Candida auris. J Clin Microbiol. 2011; 49(9): 3139–3142. 10.1128/JCM.00319-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magobo RE, Corcoran C, Seetharam S, Govender NP. Candida auris–Associated Candidemia, South Africa. Emerg Infect Dis. 2014; 20(7): 1250–1251. 10.3201/eid2007.131765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin Infect Dis. 2017; 64(2): 134–140. 10.1093/cid/ciw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fakhim H, Vaezi A, Dannaoui E, et al. Comparative virulence of Candida auris with Candida haemulonii, Candida glabrata and Candida albicans in a murine model. Mycoses. 2018; 61(6): 377–382. 10.1111/myc.12754 [DOI] [PubMed] [Google Scholar]
- 18.Ben-Ami R, Berman J, Novikov A, et al. Multidrug-Resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis. 2017; 23(2): 195–203. 10.3201/eid2302.161486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xin H, Mohiuddin F, Tran J, Adams A, Eberle K. Experimental Mouse Models of Disseminated Candida auris Infection. mSphere. 2019; 4: e00339–19. 10.1128/mSphere.00339-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeffery-Smith A, Taori SK, Schelenz S, et al. Candida auris: a Review of the Literature. Clin Microbiol Rev. 2017; 31(1): e00029–17. 10.1128/CMR.00029-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian S, Rong C, Nian H, et al. First cases and risk factors of super yeast Candida auris infection or colonization from Shenyang, China. Emerg Microbes Infect. 2018; 7(1): 128 10.1038/s41426-018-0131-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welsh RM, Bentz ML, Shams A, et al. Survival, Persistence, and Isolation of the Emerging Multidrug-Resistant Pathogenic Yeast Candida auris on a Plastic Health Care Surface. J Clin Microbiol. 2017; 55(10): 2996–3005. 10.1128/JCM.00921-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekizuka T, Iguchi S, Umeyama T, et al. Clade II Candida auris possess genomic structural variations related to an ancestral strain. PLoS One. 2019; 14(10): e0223433 10.1371/journal.pone.0223433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Katchar K, Goldsmith JD, et al. A Mouse Model of Clostridium difficile–Associated Disease. Gastroenterology. 2008; 135(6): 1984–1992. 10.1053/j.gastro.2008.09.002 [DOI] [PubMed] [Google Scholar]
- 25.Solis NV, Filler SG. Mouse model of oropharyngeal candidiasis. Nat Protoc. 2012; 7(4): 637–642. 10.1038/nprot.2012.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peeters E, Nelis HJ, Coenye T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods. 2008; 72(2): 157–165. 10.1016/j.mimet.2007.11.010 [DOI] [PubMed] [Google Scholar]
- 27.Abe M, Nakamura S, Kinjo Y, et al. Efficacy of T-2307, a novel arylamidine, against ocular complications of disseminated candidiasis in mice. J Antimicrob Chemother. 2019; 74(5): 1327–1332. 10.1093/jac/dkz020 [DOI] [PubMed] [Google Scholar]
- 28.Williams DW, Jordan RPC, Wei X-Q, et al. Interactions of Candida albicans with host epithelial surfaces. J Oral Microbiol. 2013; 5(1): 22434 10.3402/jom.v5i0.22434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsui C, Kong EF, Jabra-Rizk MA. Pathogenesis of Candida albicans biofilm. Mobley H, ed. Pathog Dis. 2016; 74(4): ftw018 10.1093/femspd/ftw018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandra J, McCormick TS, Imamura Y, Mukherjee PK, Ghannoum MA. Interaction of Candida albicans with Adherent Human Peripheral Blood Mononuclear Cells Increases C. albicans Biofilm Formation and Results in Differential Expression of Pro- and Anti-Inflammatory Cytokines. Infect Immun. 2007; 75(5): 2612–2620. 10.1128/IAI.01841-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirschfeld J. Dynamic interactions of neutrophils and biofilms. J Oral Microbiol. 2014; 6(1): 26102 10.3402/jom.v6.26102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jabra-Rizk MA, Falkler WA, Meiller TF. Fungal Biofilms and Drug Resistance. Emerg Infect Dis. 2004; 10(1): 14–19. 10.3201/eid1001.030119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajendran R, Sherry L, Nile CJ, et al. Biofilm formation is a risk factor for mortality in patients with Candida albicans bloodstream infection—Scotland, 2012–2013. Clin Microbiol Infect. 2016; 22(1): 87–93. 10.1016/j.cmi.2015.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue H, Bing J, Zheng Q, et al. Filamentation in Candida auris, an emerging fungal pathogen of humans: passage through the mammalian body induces a heritable phenotypic switch. Emerg Microbes Infect. 2018; 7(1): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson CJ, Davis JM, Huttenlocher A, Kernien JF, Nett JE. Emerging Fungal Pathogen Candida auris Evades Neutrophil Attack. Alspaugh JA, ed. MBio. 2018; 9(4): e01403–18. 10.1128/mBio.01403-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chowdhary A, Anil Kumar V, Sharma C, et al. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis. 2014; 33(6): 919–926. 10.1007/s10096-013-2027-1 [DOI] [PubMed] [Google Scholar]
- 37.Day AM, McNiff MM, da Silva Dantas A, Gow NAR, Quinn J. Hog1 Regulates Stress Tolerance and Virulence in the Emerging Fungal Pathogen Candida auris. Mitchell AP, ed. mSphere. 2018; 3(5): e00506–18. 10.1128/mSphere.00506-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muñoz JF, Gade L, Chow NA, et al. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun. 2018;9(1):5346 10.1038/s41467-018-07779-6 [DOI] [PMC free article] [PubMed] [Google Scholar]