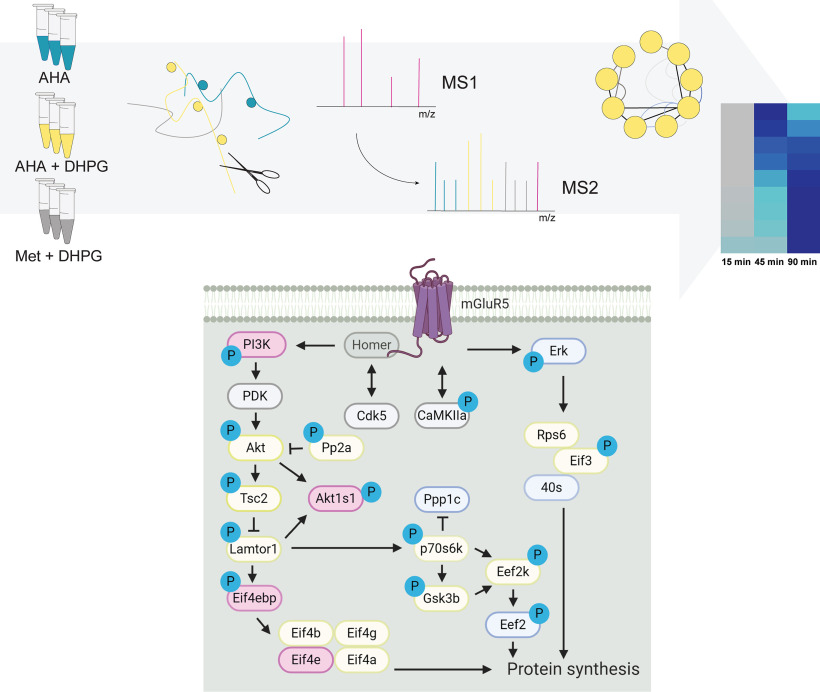
We integrated quantitative phosphoproteomics with the analyses of newly synthesized proteins in a pulsed labeling strategy in cultured hippocampal neurons stimulated with DHPG to study mGluR-induced signaling pathways. We identified and validated several kinases involved in mGluR activation, and constructed an overview of protein phosphorylation and synthesis, validating Intersectin-1 as a novel player in AMPA receptor endocytosis. This study revealed several new insights, and provides a rich resource into the molecular pathways downstream of group I mGluR activation.
Keywords: Protein synthesis, phosphoproteome, neurobiology, quantification, targeted mass spectrometry, translation, AMPA receptor, mGluR-LTD, synaptic plasticity, tandem mass tags
Graphical Abstract
Highlights
Integrated phosphoproteomics and analyses of newly synthesized proteins in neurons.
Resource of temporal mGluR-induced signaling pathways upon DHPG stimulation.
Validation of PKC, MAPK1, CAMKIIa, and CDK2 in mGluR-activation and signaling.
Validation of Intersectin-1 in DHPG-induced AMPAR internalization.
Abstract
At neuronal synapses, activation of group I metabotropic glutamate receptors (mGluR1/5) triggers a form of long-term depression (mGluR-LTD) that relies on new protein synthesis and the internalization of AMPA-type glutamate receptors. Dysregulation of these processes has been implicated in the development of mental disorders such as autism spectrum disorders and therefore merit a better understanding on a molecular level. Here, to study mGluR-induced signaling pathways, we integrated quantitative phosphoproteomics with the analyses of newly synthesized proteins via bio-orthogonal amino acids (azidohomoalanine) in a pulsed labeling strategy in cultured hippocampal neurons stimulated with DHPG, a specific agonist for group I mGluRs. We identified several kinases with important roles in DHPG-induced mGluR activation, which we confirmed using small molecule kinase inhibitors. Furthermore, changes in the AMPA receptor endocytosis pathway in both protein synthesis and protein phosphorylation were identified, whereby Intersectin-1 was validated as a novel player in this pathway. This study revealed several new insights into the molecular pathways downstream of group I mGluR activation in hippocampal neurons, and provides a rich resource for further analyses.
Activation of metabotropic glutamate receptors (mGluRs) initiates a broad array of signaling pathways that collectively modulate the efficiency of neuronal communication. mGluR-dependent signaling has been linked to cognitive functions such as attention, learning and memory, and disrupted mGluR signaling has been implicated in neurological disorders such as Fragile X Syndrome, mental retardation, schizophrenia, addiction and autism spectrum disorders (1–3). Group I mGluRs (mGluR1 and mGluR5), generally localized at the postsynaptic membrane, significantly contribute to synaptic function by modulating synaptic excitability, and inducing or facilitating different forms of synaptic plasticity (4–8). Probably the best characterized form of plasticity mediated by group I mGluRs is the long-term depression of synaptic strength referred to as mGluR-LTD (9). In contrast to NMDA receptor-dependent forms of LTD, the major mechanism of mGluR-LTD expression relies on the rapid and local synthesis of new proteins in dendrites (10), although not all forms of mGluR-LTD require protein synthesis (11, 12). Thus, defining the signaling pathways downstream of mGluR that control translational regulation, and identifying the proteins that are newly synthesized in response to mGluR activation, are important goals to better understand mGluR-dependent plasticity mechanisms.
Postsynaptic mGluRs canonically link to Gαq/11 G-proteins, which activate phospholipase C (PLC) to form diacylglycerol (DAG) and inositol tris-phosphate (IP3). IP3 in turn triggers the release of Ca2+ from internal stores, resulting in an increase in the Ca2+ concentration and activation of protein kinase C (PKC) (3). Apart from these pathways, mGluR stimulation has been found to activate a wide range of other downstream effectors, including c-Jun N-terminal kinase JNK1 (13), casein kinase 1, cyclin-dependent kinase 5 (CDK5) (14), and components of the ERK-MAPK (15–17), and PI3K-Akt-mTOR (18–20) signaling pathways. Induction of these latter two pathways are essential for the expression of mGluR-LTD, mainly because these converge on the regulation of translation initiation factors such as Mnk1, eIF4E and 4EBPs (18, 21). mGluR-LTD induces an acute wave of new protein synthesis that is required for the long-term reduction in surface α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPAR) underlying the depression of synaptic responses (10, 22). The generation of these “LTD proteins” is related to the rate of AMPAR endocytosis, and the rapid synthesis of proteins is required for the internalization of AMPARs and mGluR-LTD (23–30). However, it is unclear what other molecular processes are working in parallel to sustain mGluR-LTD. Thus, even though some of the key mechanisms underlying mGluR-LTD have been identified, characterizing the full repertoire of molecular events that are initiated by mGluR activation would greatly enhance our understanding of mGluR-LTD.
Here, to identify the phosphorylation dynamics initiated by mGluR activation in hippocampal neurons, we applied a phosphoproteomics approach using high-resolution LC–MS/MS. The sensitivity of our approach allowed us to profile multiple time points over the course of DHPG-induced mGluR activation. In addition, to identify newly synthesized proteins in response to mGluR activation, we used an azidohomoalanine (AHA) labeling strategy (31) in combination with tandem mass tag (TMT) labeling for accurate quantification of translated proteins. Based on the observed phosphorylation dynamics we identified several kinases important in the regulation of DHPG-mGluR signaling, which we confirmed using specific kinase inhibitors. Furthermore, we uncovered a broad spectrum of protein synthesis and phosphorylation dynamics in the AMPA receptor endocytosis pathway. We thereby highlight several novel insights into the signaling mechanisms upon mGluR activation and AMPAR internalization, and validated Intersectin-1 (Itsn1) to play an important role in mGluR-mediated AMPAR trafficking.
EXPERIMENTAL PROCEDURES
Ethics Statement
All animal experiments were performed in compliance with the guidelines for the welfare of experimental animals issued by the Government of The Netherlands. All animal experiments were approved by the Animal Ethical Review Committee (DEC) of Utrecht University.
Neuronal Cultures
Hippocampal cultures were prepared from embryonic day 18 (E18) rat brains as described in (99). Dissociated neurons were plated on poly-l-lysine (30 µg/ml) and laminin (2 µg/ml) at a density of 200,000 neurons per well. Cultures were grown in Neurobasal medium (NB) supplemented with B27, 0.5 mm glutamine, 12.5 μm glutamate, and penicillin/streptomycin at 37 °C/5% CO2. Neurons were transfected at DIV10-14 with indicated constructs using LipofectAMINE 2000 (Invitrogen, Carlsbad, CA) and experiments were performed 5–7 days later.
Phosphopeptide Analysis
Experimental Design and Statistical Rationale
Primary hippocampal neurons were stimulated with DHPG for 5, 10, or 20 min, or not stimulated (control, 0 min). This experiment was repeated three times, resulting in three biological replicate samples from separate cultures. No technical replicates were measured. The samples were grouped in triplicates (0 min, 5 min, 10 min, 20 min) and identifications were subsequently filtered for phosphosites having at least two valid values in at least one treatment group. Differences in intensity between different treatment groups was determined with an ANOVA using p < 0.05 as a significance threshold.
DHPG Stimulation and Protein Digestion
At DIV14-17 neurons were stimulated with 100 μm DHPG for 0, 5, 10, or 20 min. Neurons were washed three times with PBS and harvested directly in 8 M Urea lysis buffer supplemented with phosphatase inhibitor (PhosSTOP, Roche, Indianapolis, IN) and protease inhibitor (cOmplete mini EDTA-free, Roche, Liege, Belgium). Neurons were lysed at 4 °C with the Bioruptor Plus (Diagenode) by sonicating for 15 cycles of 30 s. Protein content was determined with a Pierce BCA protein quantification assay (Thermo Fisher, Bremen, Germany). Equal amounts of protein (45 µg of each sample) were heated at 95 °C for 5 min and then reduced (4 mm DTT) for 20 min at 56 °C and alkylated (8 mm IAA) for 25 min in the dark at room temperature. The proteins were the digested with Lys-C (1:75, Wako, Richmond, WA) for 4 h at 37 °C, after which the samples were diluted to a urea concentration of 2M and trypsin (1:50, Sigma Aldrich, Germany) was added overnight. The peptides were acidified to a total concentration of 1% Formic Acid (Merck, Etten-Leur, the Netherlands). Samples were cleaned up using OASIS sample cleanup cartridges (Waters) and dried in vacuo.
Phosphorylated Peptide Enrichment
Phosphorylated peptides were enriched using Fe(III)-NTA cartridges (Agilent technologies, Santa Clara, CA) in an automated fashion using the AssayMAP Bravo Platform (Agilent technologies). The cartridges were primed with 0.1% TFA in ACN and equilibrated with loading buffer (80% ACN/0.1% TFA). Samples were suspended in loading buffer and loaded onto the cartridge. The peptides bound to the cartridges were washed with loading buffer and the phosphorylated peptides were eluted with 1% ammonia directly into 10% formic acid, leading to an average phosphopeptide purity of 91%. Samples were dried in vacuo and stored at −80 °C until LC–MS/MS analysis.
Mass Spectrometry and Data-Acquisition
The phosphorylated peptide enriched samples were analyzed with an UHPLC 1290 system (Agilent technologies) coupled to an Orbitrap Q Exactive Plus mass spectrometer (Thermo Scientific). Before separation peptides were first trapped (Dr Maisch Reprosil C18, 3 μm, 2 cm × 100 μm) and then separated on an analytical column (Agilent Poroshell EC-C18, 2.7 μm, 50 cm × 75 μm). Trapping was performed for 10 min in solvent A (0.1% FA) and the gradient was as follows; 4–8% solvent B (0.1% FA in acetonitrile) in 2 min, 8–24% in 71 min, 24–35% in 16 min, 35–60% in 7 min, 60–100% in 2 min and finally 100% for 1 min. Flow was passively split to 300 nl/min. The mass spectrometer was operated in data-dependent mode. At a resolution of 35.000 m/z at 400 m/z, MS full scan spectra were acquired from m/z 375–1600 after accumulation to a target value of 3e6. Up to ten most intense precursor ions were selected for fragmentation. HCD fragmentation was performed at normalized collision energy of 25% after the accumulation to a target value of 5e4. MS/MS was acquired at a resolution of 17,500. Dynamic exclusion was enabled with an exclusion list of 500 and a duration of 18 s.
Data Analysis
RAW data files were processed with MaxQuant (v1.6.0.1 (32)) and MS2 spectra were searched with the Andromeda search engine against the TrEMBL protein database of Rattus Norvegicus (28,080 entries, downloaded 08/08/2017) spiked with common contaminants. Cystein carbamidomethylation was set as a fixed modification and methionine oxidation, protein N-term acetylation, and phosphorylation of serine, threonine, and tyrosine were set as variable modifications. Trypsin was specified as enzyme and up to two miss cleavages were allowed. Filtering was done at 1% false discovery rate (FDR) at the protein and peptide level. The mass tolerance was set to 4.5 ppm for the precursor ions and 20 ppm for the fragment ions. Label-free quantification (LFQ) was performed, and “match between runs” was enabled. The data were further processed using Perseus 1.6.0.7 (33), WebLogo (34, 35), MotifX (36, 37), and SynGO (38).
pAHA & TMT Labeling
Experimental Design and Statistical Rationale
Primary hippocampal neurons were stimulated with DHPG and subsequently incubated with l-AHA (n = 4, experimental condition). For each studied time point (15, 45, and 90 min), control experiments were included where either AHA was supplemented to unstimulated neurons, to define the set of proteins that are truly being translated in response to DHPG (n = 4), or neurons were stimulated with DHPG in methionine-supplemented media in the absence of AHA (n = 4), to control for nonspecific binding during the enrichment process. TMT reagents 126 – 13 °C were used to label the nine experimental conditions, and these were mixed in a 1:1 ratio. The TMT 131 label served as a reference pool between biological replicates and consisted of an equal mix of all nine experimental conditions of biological replicate 1, and was mixed in equal ratios with the other TMT labels. All biological replicates were injected twice (technical replicates) and searched as one result file. Only DHPG specific and AHA enriched proteins that had a higher relative expression than in the control conditions, in at least one of the time points, were taken for further analysis.
Stimulation and Lysis
DIV12 hippocampal neurons were incubated in NB media (Invitrogen life technologies) supplemented with B27, 0.5 μm glutamine and penicillin/streptomycin (supplemented NB) and either with 4 mm l-azidohomoalanine (l-AHA) (Bachem, St Helens, United Kingdom) or 4 mm l-Methionine (Sigma-Aldrich) and in parallel stimulated with 100 μm DHPG or vehicle for 5 min at 37 °C/5% CO2. Neurons were then moved into freshly supplemented NB media with either 4 mm l-AHA or 4 mm l-methionine and incubated at 37 °C/5% CO2 until the end of the experiment (15, 45, and 90 min after initial DHPG stimulation).
Harvest followed three washes with PBS, directly into urea lysis buffer (Click-it Protein enrichment kit, Invitrogen C10416) supplemented with protease inhibitor (cOmplete mini EDTA-free, Roche).
Enrichment and Digestion
Neurons were lysed at 4 °C with the Bioruptor Plus (Diagenode) by sonicating for 10 cycles of 30 s. Protein content was determined with a Pierce BCA protein quantification assay (Thermo Fisher). Newly synthesized proteins were then enriched from 100 µg of protein material per sample using the Click-it protein enrichment kit for chemistry capture of azide modified proteins (Invitrogen C10416) following the manufactures protocol with small modifications. In short, protein lysate volume was adjusted to equal protein input for each sample per biological replicate and final volume was adjusted by adding Milli-Q. Lysates were added to washed resin and was incubated end to end rotating overnight. Resins were washed with Milli-Q and SDS buffer was added. Proteins were reduced (1M DTT) for 15 min at 70 °C and alkylated (40 mm IAA) for 30 min in the dark at room temperature. The resins were then transferred to the supplied filter columns and extensively washed with subsequently SDS wash buffer, 8 M urea with 100 mm Tris pH 8, 20% ACN and 50 mm Ammonium Bicarbonate (AMBIC). Resins were transferred to a new tube and proteins were digested with 0.1 µg Lys-C for 2 h at 37 °C and 0.5 µg trypsin (Promega) overnight at 37 °C. Samples were centrifuged and the supernatant was taken. Sample cleanup was performed using the OASIS sample cleanup cartridges (Waters). Samples were dried in vacuo and stored at −80 °C.
TMT Labeling
TMT labeling was performed according to the manufacturer's instructions using the TMT10plex Isobaric Label Reagent Set (Thermo Scientific). In brief, samples were reconstituted in 100 µl 87.5% HEPES buffer pH 8.5/12.5% ACN, and TMT reagents in 41 µl anhydrous ACN. Full contents of the reagents were added to the samples and incubated at room temperature for one hour, after which the reaction was quenched with 5% hydroxylamine. Labeled mixtures were cleaned using the OASIS sample cleanup cartridges (Waters), dried in vacuo and stored at −80 °C until further processing.
Mass Spectrometry and Data-Acquisition
Fractions were reconstituted in 10% FA and analyzed in two technical replicates with a UHPLC 1290 system (Agilent technologies) coupled to an Orbitrap Q Exactive X mass spectrometer (Thermo Scientific). Peptides were trapped on an in house made trap column (Dr Maisch Reprosil C18 column, 3 μm, 2 cm × 100 μm) and separated on an analytical column (Agilent Poroshell EC-C18, 2.7 μm, 50 cm × 75 μm). Trapping was performed for 5 min in solvent A (0.1% FA) and separation was performed using a 85 min linear gradient from 15% to 45% solvent B. Flow was passively split to 300 nl/min. The mass spectrometer was operated in data-dependent mode. At a resolution of 60,000 at 200 m/z, MS full scan spectra were acquired from 375–1600 m/z after accumulation to a target value of 3e6. Up to 15 most intense precursor ions were selected for HCD fragmentation at a normalized collision energy of 32% after accumulation to a target value of 5e4. MS/MS was acquired at a resolution of 60,000, with a fixed first mass of 120 m/z. Dynamic exclusion was enabled with a duration of 12 s.
Data Analysis
RAW data files were processed using Thermo Proteome Discoverer (version 2.2.0.338) and Mascot search engine (v2.6.1), allowing for variable methionine oxidation, protein N-terminal acetylation, and methionine replacement by AHA. Carbamidomethylation of cysteines was set as a fixed modification. The protein database consisted of the TrEMBL protein database of Rattus Norvegicus (28,080 entries, downloaded 08/08/2017) spiked with common contaminants. Enzyme specificity was set for trypsin, with a maximum of two allowed missed cleavages. The precursor mass tolerance was 50 ppm, and fragment mass tolerance was set to 0.05 Da. TMT 10plex was set as quantification method, and only unique peptides were used for quantification. Normalization mode was disabled, and reporter abundances were based on signal to noise values in all cases. Results were filtered using a 1% FDR cutoff at the protein and peptide level.
Selective Reaction Monitoring
Spectral Library Generation
Spectral libraries were used to determine peptide fragmentation characteristics and their indexed retention time, which are key for the identification of peptides in the tier 2 SRM assay. The custom mix of heavy labeled peptides (JPT, Lelystad, the Netherlands) was mixed with iRT peptides (Biognosys), and analyzed using an Orbitrap Q-Exactive HF (Thermo Scientific). An unscheduled parallel reaction monitoring (PRM) method was use that scanned for the +2 and +3 charged peptides, including all possible methionine oxidations. Peptides were separated using a 2 h gradient and at least a 30 k resolution was used for the PRM assay, resulting in a minimum of 5 spectra per peptide. Raw files were analyzed using MaxQuant (version 1.6.10.43), carbamidomethyl cysteine as fixed modification, and the variable modifications serine/threonine/tyrosine phosphorylation, methionine oxidation, and isotope labels. The search results were filtered using a 1% FDR cut off, subsequently using Skyline (version 20.1.1.83) pseudo-MS2 spectra were generated which were used as the peptide library.
SRM Assay Development
The SRM assay was developed using previous described methods (39, 40). The assay was developed on a TSQ Altis (Thermo Scientific) and a TSQ Vantage (Thermo Scientific). In brief, the 6 most intense fragment ions from the library were used as initial transitions, to which essential transitions for identification were added, such as transitions that validate phosphorylation on specific residues. These transitions were used to optimize multiple parameters such as retention time and collision energy. Collision energy was optimized per transition using Skyline, with the TSQ Vantage CE formula as starting point (CE = 0.03 m/z + 2.905 for doubly charged precursors and CE = 0.038 m/z + 2.281 for precursor charges of three and higher), and optimized using steps of 1 voltage.
SRM LC–MS/MS Setup
Samples were analyzed on a TSQ Altis (Thermo Scientific) coupled to an UltiMate 3000 (Thermo Scientific), and an easy spray analytical column (ES802A, 25 cm, 75 mm ID PepMap RLSC, C18, 100 A˚, 2 mm particle size column (Thermo Scientific)). First, samples were reconstituted in 2% LC–MS grade formic acid, containing the heavy labeled peptides. Samples were loaded on a trap column (Acclaim™ PepMap™ 100 C18 HPLC Column 0.3x5mm with 5 μm particles (Thermo Scientific)) with Buffer A (0.1% FA), and subsequently separated using 0-35% buffer B (80%ACN, 0.1%FA) in 50 min at 300nL/min, followed by a 10 min column wash with 99% buffer B at 300nL/min. The TSQ Altis spray voltage was set at 1.9 kV and fragmented at 1.5 mTorr in the second quadrupole. The first quadrupole was set at 0.7 da FWHM, and the third quadrupole at 1.2 da FWHM. All transitions were measured with an optimized collision energy and a dwell time of 5 ms.
SRM Data Assessment
All experiments were analyzed using Skyline (version 20.1.1.83). Quality of the peptides was assessed mainly on the signal similarity between the heavy and the light peptides. Most important aspects were perfect co-elution, peak shape, and relative contributions of each transition between the heavy and the light peptide. A rdotp > 0.95 was maintained as an indicator of the similarity between the heavy and the light peptide. To normalize the endogenous peptides an in-house Python script compared the relative intensity of the heavy peptides between two time points. The relative difference was used as multiplier for the endogenous signal.
DNA Constructs
The Itsn1 miRNA knockdown construct was generated by annealing oligos containing the 21-nucleotide targeting sequences described in and ligating in the miRNA expression plasmids pSM155-GFP (provided by G. Du; University of Texas, Houston, TX) digested with BsmBI.
Kinase Inhibitor Assay
For the kinase inhibitor assays, neurons were pre-incubated with KN-93 (10 μm), staurosporine (1 μm), or roscovitine (20 μm) for 30 min before induction of mGluR-LTD. Neurons were stimulated with DHPG (100 μm) for 5 min, returned to original medium and fixed 30 min later. Blockers were present during the entire experiment.
Immunofluorescence and Confocal Microscopy
To induce mGluR-LTD, hippocampal neurons were stimulated with DHPG for 5 min and then returned to the original culture medium. After 30 min, neurons were fixed with 4% paraformaldehyde/4% sucrose in PBS (PBS) for 8–10 min. Fixed neurons were blocked with 10% normal goat serum in PBS for 30–60 min at room temperature, stained with rabbit anti-GluA1 (1:100; Calbiochem), or anti-ITSN1 and labeled with fluorescent goat anti-rabbit secondary antibodies. Confocal images were taken with a Zeiss LSM 710 with 63 × 1.40 oil objective. Images consist of a z-stack of 7–9 planes at 0.39 μm interval, and maximum intensity projections were generated for analysis and display. GluA1 cluster intensity was measured using the ParticleAnalyzer function in ImageJ and analyzed per region of interest.
RESULTS
Protein Synthesis upon Activation of mGluR via DHPG
To study the downstream effects of group I mGluR activation, primary hippocampal cultures were stimulated with (s)-3,5-dihydroxyphenylglycine (DHPG), a specific agonist of group I mGluRs. We confirmed that this induced a reduction in surface GluA1 expression (supplemental Fig. S1A) as has been established before (9, 41). To identify proteins that are synthesized de novo in response to mGluR activation by DHPG, we used a pulsed-AHA approach (31, 42). Cultured hippocampal neurons were stimulated with DHPG in the presence of the bio-orthogonal methionine analogue AHA to label newly synthesized proteins. Since these primary neuron cultures are limited in sample material, which poses a challenge in sensitivity to determine truly newly synthesized proteins, we decided to incorporate two control conditions. In one condition we performed DHPG stimulation in the presence of methionine to control for nonspecific binding during the enrichment process. Additionally, we included a control in which unstimulated neurons were supplemented with AHA, to define the set of proteins that are translated only in response to DHPG. To profile the temporal induction of protein synthesis, neurons were harvested and lysed at different time points up to 90 min after a 5-min DHPG stimulation (Fig. 1A). AHA-incorporated proteins were consecutively enriched via click-chemistry, digested by a combination of Lys-C and trypsin and analyzed using high-resolution nanoLC-MS/MS. Only DHPG specific and AHA enriched proteins that had a higher relative expression than in the control conditions, in at least one of the time points, were taken for further analysis. GO enrichment analysis against a full brain background using SynGO (38) confirmed enrichment of postsynaptic over presynaptic localization, as well as a clear enrichment in, among others, postsynaptic cytoskeleton and ribosome (Fig. 1B). Among the newly synthesized proteins are hallmark mGluR-LTD proteins involved in translation, such as translation initiation factor 4a (Eif4a2) and elongation factor 2 (Eef2) (22), which demonstrates the effectiveness of this experimental set up to identify protein translation upon DHPG-induced mGluR activation (Fig. 1C, supplemental Fig. S2B).
Fig. 1.
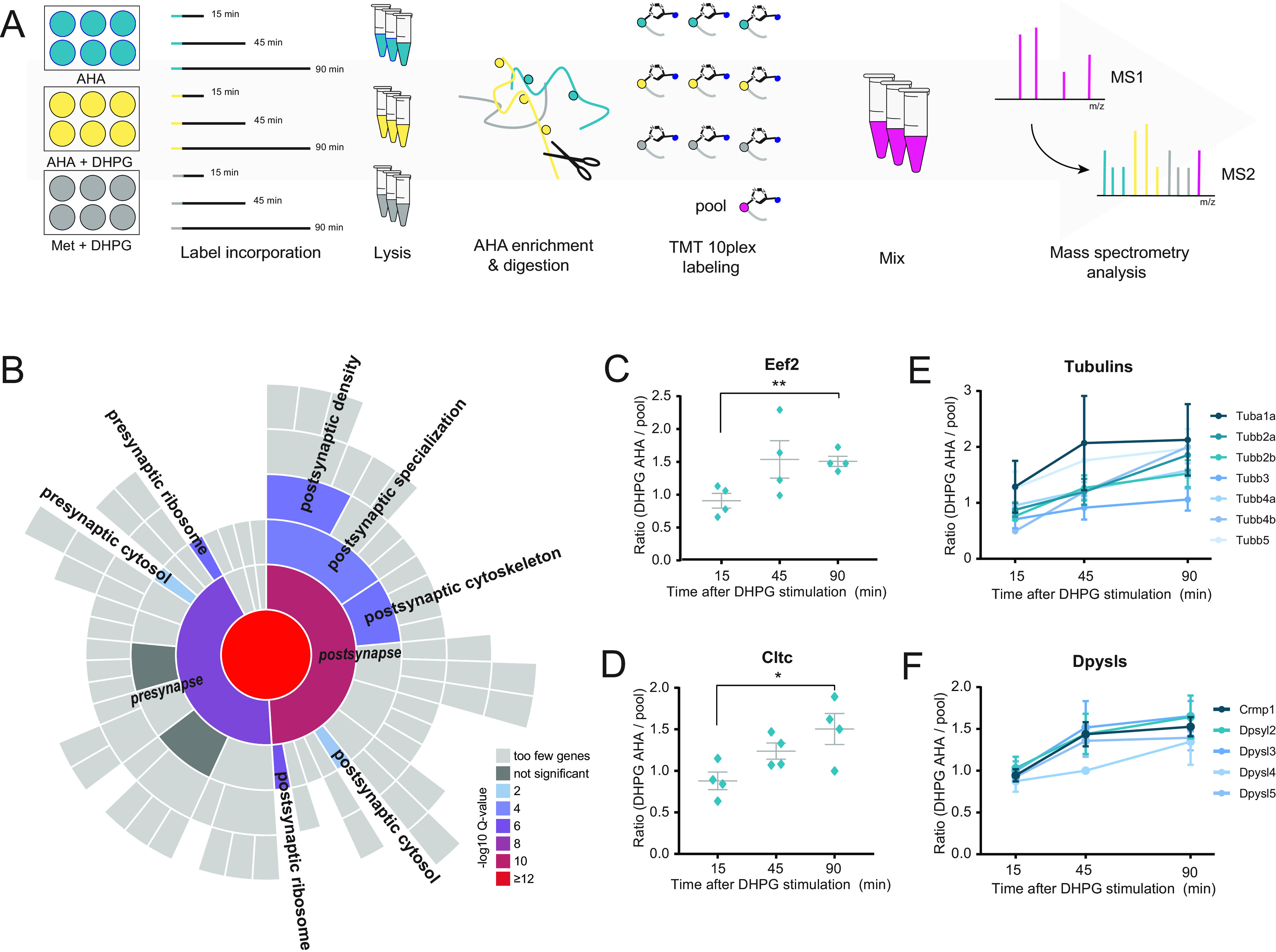
Identification of protein translation following DHPG-induced group I mGluR activation. A, Workflow: neurons were stimulated with DHPG for 5 min in combination with methionine (negative control, to control for nonspecific binding during the enrichment process, n = 4) or the methionine substitute AHA (n = 4). An extra control was included in which unstimulated neurons were supplemented with AHA, to define the set of proteins that are truly being translated in response to DHPG (n = 4). After DHPG removal, translation was followed for a total of 15, 45, and 90 min. Newly synthesized proteins were enriched, digested and labeled with TMT10plex. B, SynGO cellular location enrichment analysis of newly translated proteins (1% FDR) revealed enrichment of postsynaptic over presynaptic localization. C, Induction of mGluR-LTD using DHPG leads to identification of proteins involved in translation, such as Eef2, as well as (D) in receptor endocytosis, such as clathrin, showing increasing protein abundance over time. E, This enrichment and labeling approach reducibly identifies multiple members of the same protein families during mGluR-LTD, including subunits of both alpha and beta tubulins and (F) Dpysls. Data are represented as mean±S.E., * p < 0.05, p < 0.01, as determined with a one-way ANOVA.
DHPG-induced activation of group I mGluR stimulates augmented internalization of AMPARs (9, 10, 27, 29, 30). Accordingly, we found many newly synthesized proteins with functions related to receptor endocytosis and recycling. Examples include the adaptor protein complex AP-2 subunit alpha1 adaptin (Ap2a1), the GTPase Cdc42, and the neuronal migration protein doublecortin (Dcx), which is a known interactor of clathrin adaptor complexes (43) (supplemental Table S1). In line with this observation, we identified DHPG-dependent translation of the heavy chain of clathrin (Cltc), a key protein for the formation of coated vesicles, which was steadily being translated over all three time points (Fig. 1D, supplemental Fig. S2C).
In line with the findings that reorganization of the actin cytoskeleton underlies the expression of mGluR-LTD (44), and is for instance thought to cause the changes in spine morphology that are associated with mGluR-LTD (45), we observed several proteins with known roles in structural plasticity processes, including stathmins (Stmn1 and Stmn2), profilin (Pfn2), and neuromodulin (Gap43), as well as cofilin-2 (Cfl2) (supplemental Table S1). Interestingly, apart from these actin regulators we also observed profound translation of microtubule-associated proteins (Map2, Map6, Mapre1, Map1lc3b, and Mapt), as well as numerous subunits of both alpha and beta tubulins over all time points (Fig. 1E). Since the microtubule cytoskeleton is largely excluded from spines, these results suggest that postsynaptic mGluR activation has even wider effects, potentially reorganizing the microtubule cytoskeleton in the dendritic shaft to facilitate microtubule-based transport.
Interestingly, we also identified and measured the translation of several protein families and complexes over time, such as several proteasomal subunits, ribosomal proteins, 14-3-3 proteins, tubulins, and dihydropyrimidinase-related proteins, which are also known as the collapsing response mediator protein family (CRMPs) (Fig. 1F). These versatile proteins are involved in a variety of developmental and plasticity-related brain processes and have been shown to also localize in the PSD (46). Interestingly, the CRMP family of proteins were recently identified as some of the most stable, long-lived proteins in neurons (47).
To generate a more in-depth overview of the type of proteins being translated, and which processes they represent, we performed a k-means interaction-based clustering analysis using the protein-interaction database STRING (supplemental Table S1) (48). GO analysis of these resulted in the enrichment of six biological processes, visualized using the geneMANIA app in Cytoscape (49). This interaction-based clustering approach highlights interactions between proteins as identified by, among others, physical interactions, co-expression data and co-localization data, retrieved from a wide variety of assays, organisms, and cell types. With these combined data, we can identify groups of proteins with similar functions in relevant biological processes underlying mGluR signaling cascades. Fig. 2 displays the interaction network with representative proteins of each of these processes, as well as their z-score normalized expression levels over all pooled experimental conditions. As expected, we identify several protein clusters with an enrichment in biological processes related to protein translation. The first cluster contains proteins involved in the processing of mRNA, and includes several splicing factors (Srsf2 and Srsf3) and ribonucleoproteins (e.g. Snrnp70), as well as binding proteins (Nono and Pcbp2). The second cluster, with a clear enrichment in proteins regulating translation, includes translation initiation factor Eif4a2, and elongation factors (Eef1g, Eef2, and Tufm), as well as multiple ribosomal proteins, both from the 40S (e.g. Rps2 and Rps7) and 60S ribosome (e.g. Rpl3 and Rpl22). This suggests that mGluR group I activation initiates the formation of new ribosomes, presumably to meet the demands of an increased rate of protein synthesis. Cluster 3 contains proteins involved in protein folding, including several heat shock proteins (e.g. Hsp90aa1 and Hsp90ab1), as well as several subunits of the t-complex (e.g. Cct2 and Cct5), which are molecular chaperones in protein folding (50). These clusters, where protein translation and folding are the main biological processes, are part of what can be described as a 'first wave' of protein synthesis, which is initiated rapidly and already shows considerable new protein translation in the early time points (predominantly from the 45 min time point on). A 'second wave' of protein synthesis seems to be initiated at a later stage (45 to 90 min time point, as demonstrated in clusters 4 and 5), and contains proteins, among others, related to protein degradation. Recently, a dynamic interplay between protein translation and degradation has been described to be crucial for mGluR-LTD (51, 52). Inhibition of the proteasome rescued impairments in mGluR-LTD induction caused by blockage of protein translation (51), suggesting that a fine coordination between protein translation and degradation of proteins by the UPS is key for the efficient induction of mGluR-LTD. In line with this, we observed translation of many proteins involved in protein degradation in cluster 4, including several subunits of the proteasome (e.g. Psma2, Psma3, Psmb1, and Psmb7). Finally, we identified two protein clusters with a similar enrichment in GO terms, containing proteins involved in the organization of cellular components (cluster 5) and, more specifically, cytoskeleton organization (cluster 6). While both clusters contain multiple tubulins, cluster 5 also contains several 14-3-3 proteins, which have been studied extensively in relation to their function in the regulation of actin filaments (53). In addition to tubulins, cluster 6 also contains several actin regulating proteins (e.g. Arpc1a and Wdr1).
Fig. 2.
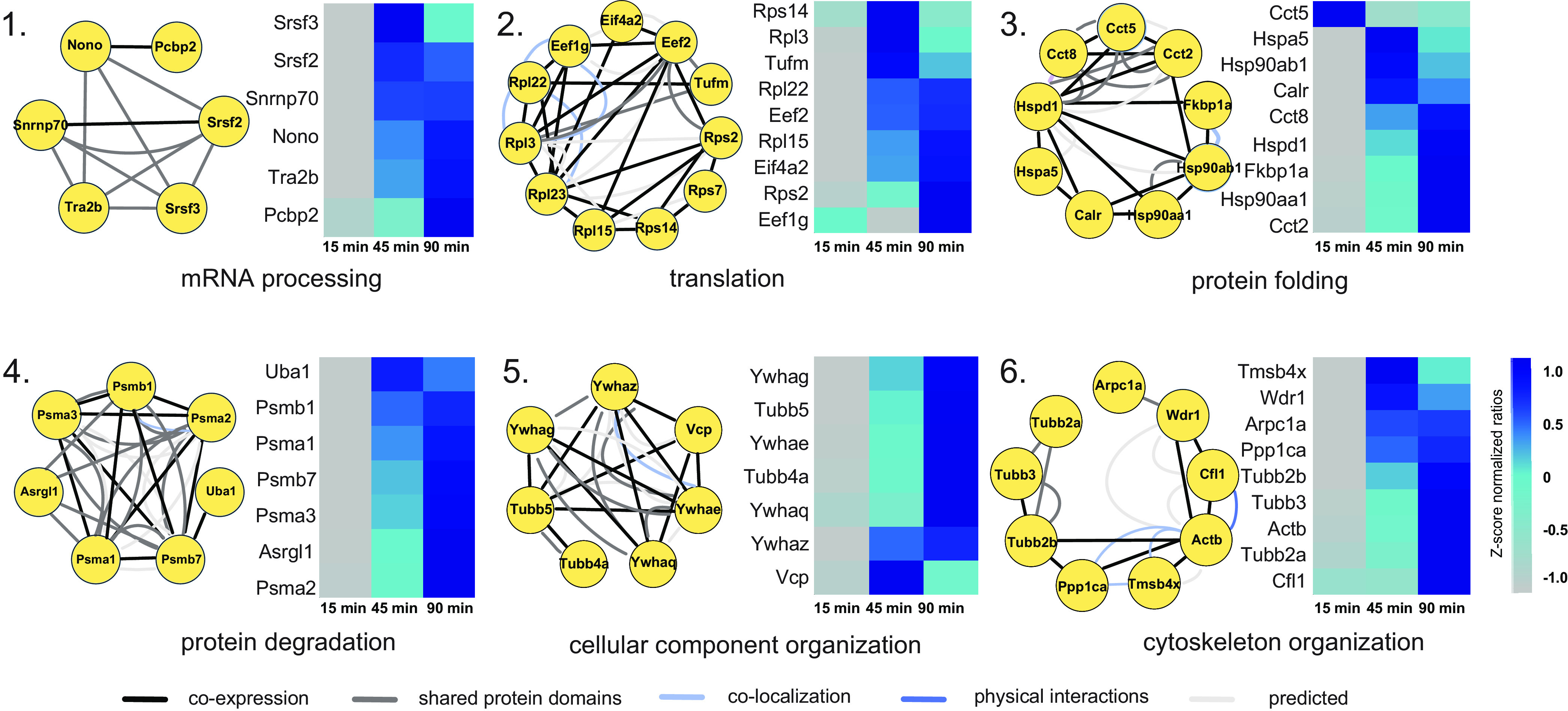
Interaction-based protein clusters with distinct biological processes involved in mGluR activation. GO enrichment analysis of k-means based interaction clusters resulted in six main clusters with a clear enrichment in a biological process. Proteins from each cluster are displayed with their known interaction profiles. Heatmaps represent the z-score normalized ratio DHPG AHA/pool over the three measured time points.
Phosphoproteomics of Group I mGluR Activation in Primary Neurons Reveals Defined Clusters of Phosphosite Regulation with a Strong Synaptic Signature
Due to the low stoichiometry of phosphorylation events in the proteome, phosphoproteomics analysis involves dedicated enrichment strategies. These strategies typically require milligrams of protein input material (54–56), hampering analysis of phosphorylation dynamics in primary neurons. Recently, we have shown that automated phosphopeptide enrichment using Fe(III)-IMAC cartridges on a Bravo AssayMap platform allows sensitive and reproducible enrichment of several thousands of unique phosphopeptides starting with only 1–10 µg of protein input material. Moreover, we showed that from a single neuronal culture plate, 200,000 cells delivering ∼50 µg of protein, we could identify biological relevant phosphorylation events among the ∼7000 observed phosphosites (57). This now allowed us to study phosphorylation events on multiple time points after DHPG stimulation in primary hippocampal neurons, without the need for combining extensive amounts of input material. The proteomics analysis of Fe(III)-IMAC enriched phosphorylated peptides was performed after stimulating rat hippocampal neurons for 0, 5, 10 or 20 min with DHPG. The applied workflow is outlined in Fig. 3A. This phosphoproteomics screen resulted in the identification of 17,556 phosphosites with a localization probability >0.75, of which 5423 could be quantified in at least two biological replicates in at least one experimental condition and were used for subsequent analysis (supplemental Table S2). Phosphopeptide abundance showed a normal distribution (supplemental Fig. S3A) and a high degree of overlap between the identified phosphoproteins from both the 5, 10 and 20 min DHPG-stimulated neurons could be observed compared with control (supplemental Fig. S3B). The distribution of phosphosites on serine, threonine and tyrosine residues is in line with previous publications (54, 57, 58) (supplemental Fig. S3C). The reproducibility of the experimental procedure was assessed between biological replicates, and between the different time points. A high degree of correlation between the biological replicates could be observed, suggesting that a small, but distinct subset of signaling pathways is activated by DHPG, consistent with the notion that DHPG activates local dendritic signaling pathways, rather than a robust, cell-wide response. Nevertheless, the DHPG-treated samples could still be clearly distinguished from the control samples (supplemental Fig. S3D). Furthermore, SynGO enrichment analysis of biological function compared with a full brain background resulted in a significant enrichment for PSD organization, regulation of postsynaptic neurotransmitter receptor activity and localization, and organization of the postsynaptic actin skeleton (supplemental Fig. S3E).
Fig. 3.
Quantitative phosphoproteomics of mGluR signaling in hippocampal neurons stimulated with DHPG. A, Quantitative phosphoproteomics workflow: samples were taken at 0, 5, 10 and 20 min after the addition of DHPG. B, Unsupervised clustering reveals five distinct clusters for the regulated phosphosites. C, GO-term enrichment analysis for molecular function (grey) and biological process (black). d–G, Normalized log2 intensities of regulated phosphosites upon DHPG stimulation. Data are represented as mean±S.E., * p < 0.05, ** p < 0.01, as determined with a one-way ANOVA.
To identify phosphosites regulated over time, a multiple-samples ANOVA was performed, resulting in a total of 504 phosphosites that were significantly regulated in response to DHPG (p < 0.05). Unsupervised fuzzy clustering of the proteins from which these regulated phosphosites originated revealed five distinct clusters (Fig. 3B) (59). The proteins grouped in cluster 1 are characterized by an increasing trend in phosphorylation and are enriched in multiple GO terms compared with the full phosphoproteome, such as actin binding and microtubule cytoskeleton organization (Fig. 3C). In cluster 2, showing a distinct pattern starting with immediate de-phosphorylation upon DHPG stimulation followed by re-phosphorylation at later time points, we observe enrichment in processes involved in signaling, such as protein kinase binding and serine/threonine kinase activity. Cluster 3, containing proteins involved in cytoskeleton organization and microtubule binding (e.g. Map1b and Mapt), shows initial phosphorylation followed by dephosphorylation at the later time points. Both clusters 4 and 5 show a steady trend in phosphorylation over all time points, where proteins are either steadily dephosphorylated over time (cluster 4), or phosphorylated over time (cluster 5). Both clusters are enriched for proteins associated with microtubule binding (e.g. Map2, Map4 and Macf1), small GTPase regulator activity, and more generally protein phosphorylation.
Taken together, we can use the phosphorylation patterns from these five clusters to generate a general timeline of biological processes, where some fast and short-acting processes such as protein kinase binding and kinase activity precede more general and longer-acting processes such as microtubule binding and cytoskeleton organization.
Phosphorylation Dynamics in Relation to DHPG-Activated Group I mGluRs
Regulation of major signaling pathways by kinases and phosphatases underlies various dynamic processes in cellular functioning, including mGluR-LTD (16, 18, 60–62). A selection of several kinases can influence important nodes of these signaling pathways. One kinase shown before to be important in mGluR-LTD is Ca2+/calmodulin-dependent protein kinase II (CaMKII). We found subunit CaMKIIβ to be dephosphorylated in our data set at S315 following DHPG stimulation. This is in contrast with observations in mature Purkinje cells, where DHPG stimulation drastically increased phosphorylation of S315, thereby decreasing its F-actin binding and bundling activity. In this system, CaMKIIβ was found to be directly phosphorylated by PKCɣ, via the mGluR IP3R1/Ca2+-dependent PKC pathway (63).
Next to CaMKII, we observed dephosphorylation, and thus activation, of Eef2 at T57, the major target of the Ca2+/calmodulin-dependent protein kinase eEF2K (64). Interestingly, one hour after DHPG stimulation, Eef2 phosphorylation has been shown to result in mGluR-LTD related protein translation of Arc/Arg3.1 and inhibition of global protein translation in mouse hippocampal slices (65).
Next to protein translation, several proteins involved in protein degradation showed regulation by phosphorylation upon DHPG stimulation. Among these are the T273 phosphosite of the Psmd1, and T9 of the Psmd2 regulatory subunits of the 26S proteasome, several other proteasome subunits (Psma5 and Psmc3), and several ubiquitin-related enzymes.
Multiple integral proteins of the PSD showed regulation at the phosphorylation level. For instance, the protein SynGAP1, an important negative regulator of AMPAR insertion at the membrane of the PSD (66), was significantly dephosphorylated over time at S773 (Fig. 3D). Phosphorylation of the S773 site alone has been shown to inhibit GAP activity, while concurrent phosphorylation of both S773 and S802 increased GAP activity (67). GAP activity is necessary for the inactivation of Ras and Rap, which are involved in AMPAR trafficking (67). Our data thus suggest that dephosphorylation of SynGAP1 at S773 in response to DHPG changes the Ras/Rap activation balance, perhaps promoting AMPAR endocytosis.
Significant changes in phosphorylation status were also found in several PDZ domain-containing proteins (supplemental Table S2). We observed alterations in phosphorylation in prominent synaptic scaffolding proteins such as Dlg2 (PSD-93), Dlg3 (SAP-102), Shank2 and Shank3. The latter was found to be phosphorylated already 5 mins after mGluR stimulation at S781 (Fig. 3E), a site that has not previously been identified in rat, but was recently shown to be induced by an LTP protocol in mouse PSD fractions (68). Other regulated PDZ proteins include microtubule-associated serine/threonine-protein kinase 2 (Mast2) at two distinct phosphorylation sites, as well as Rho GTPase-activating proteins Arhgap21 and Arhgap23. Also identified to be phosphorylated, but not found to be significantly regulated in these data set, is the PDZ scaffold protein GRIP, which is involved in AMPAR trafficking (69, 70).
Multiple proteins involved in endocytosis show significant regulation at the phosphorylation level, including Syndapin-1 (Pacsin1) and β-Pix (Arhgef7). Syndapin-1 becomes phosphorylated at the T181 site after DHPG stimulation (Fig. 3F). This phosphosite of Syndapin-1 is in the F-Bar domain of the protein and is important in neuronal membrane tubulation. The F-bar domain is involved in lipid binding and cytoskeleton reorganization (71). However, this specific phosphosite was not shown to be involved in the regulation of activity-dependent bulk endocytosis. More recently, it was shown that Syndapin-1 also interacts directly with Pick1 via its F-bar domain and that this interaction is important for AMPAR endocytosis in NMDAR-related cerebellar LTD (72). This might suggest a potential role for this T181 phosphosite in the Syndapin-1 and Pick1 binding following DHPG-induced group I mGluR activation, potentially influencing AMPAR endocytosis. β-Pix is a guanine nucleotide exchange factor and binds the p21–activated kinase Pak1. The site S71 of β-Pix is strongly conserved and has a possible role in its guanine exchange function (73). In our experiment, the phosphorylation pattern of S71 belongs to cluster 2, where after an initial rapid dephosphorylation, the phosphorylation state after 20 min of DHPG stimulation returns close to its initial level. Furthermore, two phosphosites (S623 and S1134) of Itsn1, a protein linked to receptor internalization, were significantly regulated upon DHPG stimulation (Fig. 3G). In neurons, Itsn1 was suggested to be involved in presynaptic vesicle recycling (74, 75), but was also shown to have a postsynaptic function in AMPAR internalization in invertebrates (76). The exact role of these phosphosites is still unknown, also in mGluR-LTD, which makes Itsn1 an interesting target for further study.
Activated Kinases upon Group I mGluR Stimulation with DHPG
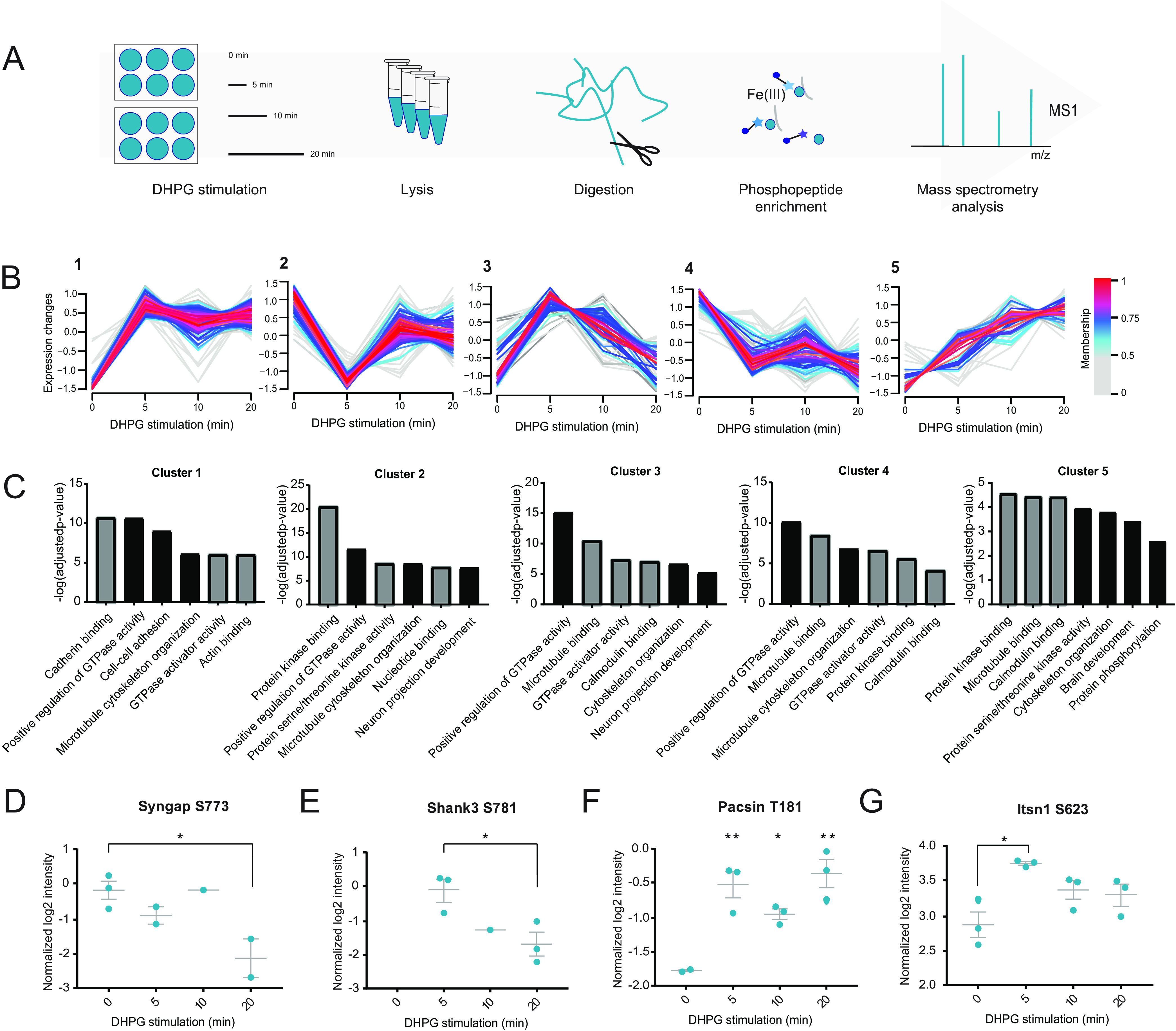
To assess if certain kinases are specifically involved in the phosphorylation events underlying DHPG-induced group I mGluR signaling, we next performed a phosphorylation site consensus motif analysis of the regulated phosphosites. This allowed us to extract multiple phosphorylation motifs (supplemental Fig. S5) resulting in four distinct typical kinase motif sequences linked to regulated proteins in our data set (Fig. 4A). The first and most pronounced motif is the proline directed motif at the +1 position, which is a known recognition motif of the cyclin-dependent kinases (CDKs) (77). Moreover, we identified the threonine directed TPxK motif, which is a known substrate binding site of Cdk5. The third motif is the RxxS motif, which is part of the known targeting sequence for the MAPK-activated protein Kinases (MKs) (78, 79). A double MK2/3 knockout mouse model showed impaired mGluR-LTD and GluA1 endocytosis, indicating a regulatory role for these kinases in the process. This RxxS motif is also described as a consensus motif for CaMKII kinases, as is the KxxS motif, which is also visible as the fourth most dominant motif. To experimentally validate the activation of these kinases, as well as kinases associated to mGluR5 activation and AMPAR internalization, we performed quantitative selected reaction monitoring (SRM) analysis of activation sites of several of these kinases (39). With this analysis we were unable to detect the activation site of CDK, however, could clearly show increased phosphorylation of the respective activation sites of CAMKIIa (T285), ERK2/MAPK1 (T182 and Y184) and PKCα (S225 and T227) upon DHPG stimulation (Fig. 4B). Next, we used well-characterized pharmacological inhibitors to specifically block the activity of CDKs (roscovitine), or CaMKII (KN-93) before stimulation with DHPG in hippocampal cultures. Confirming the predictions, we found that pre-incubation with roscovitine or KN-93 blocked the DHPG-induced reduction in surface GluA1 levels (Fig. 4C, 4D). On the other hand, pre-incubation with staurosporine, blocking PKC activity did not prevent mGluR-induced GluA1 internalization. These experiments thus confirmed our prediction that the activity of CDKs and CaMKII underlie the DHPG-induced AMPAR internalization.
Fig. 4.
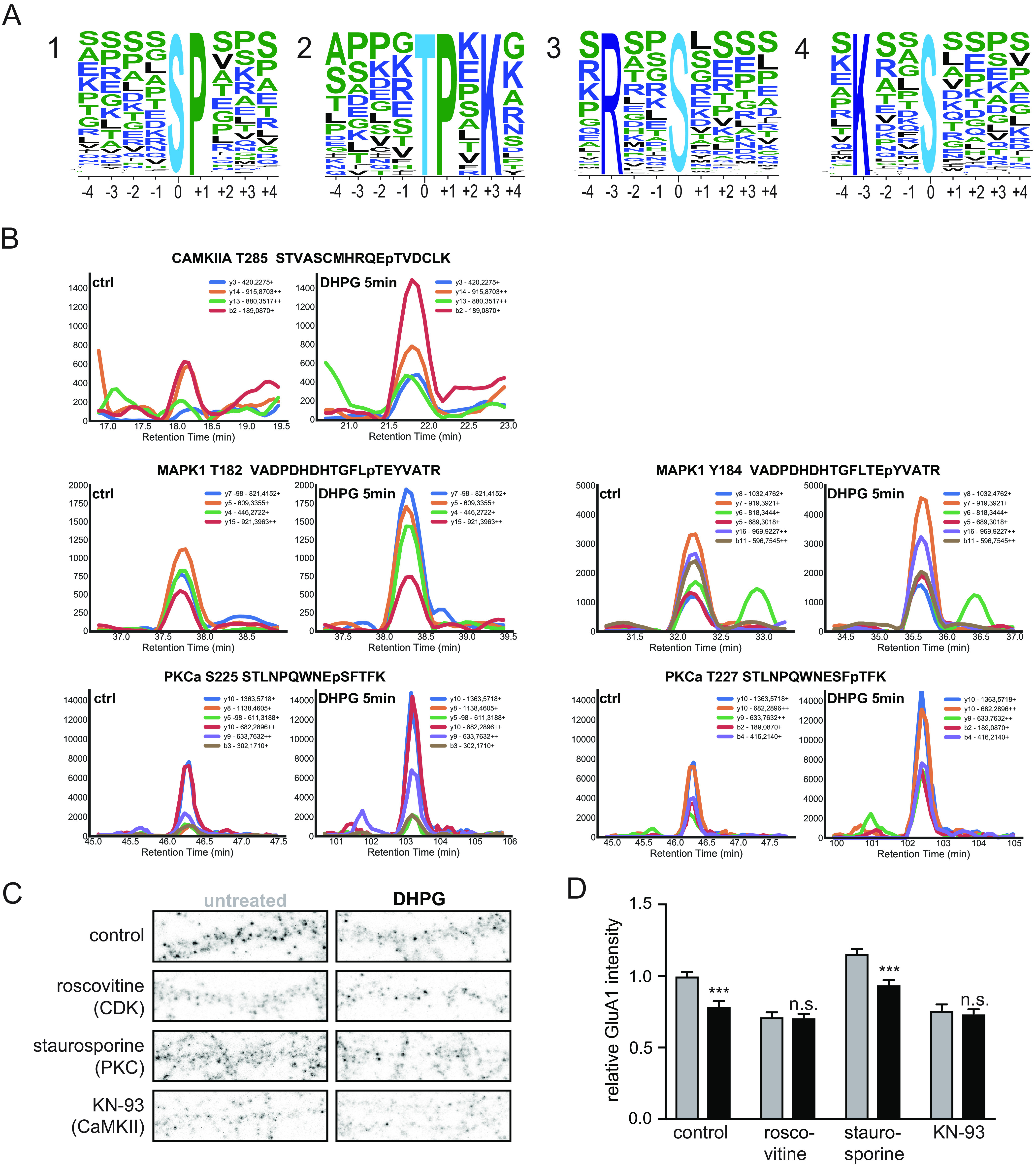
Kinases involved in DHPG-activated mGluR-LTD. A, MotifX sequence motifs of regulated phosphosites (p < 0.01) implicated in mGluR-LTD. B, Representative SRM traces of known kinases downstream of mGluR5 activation by DHPG. A clear increase in phosphorylation was observed after 5 min of DHPG stimulation compared with control for several activation sites of PKCa, CaMKIIa and MAPK1. C, Immunostaining of GluA1 subunits at the cell surface after control or DHPG treatment, pre-incubateed with control (untreated, n = 19)) or 3 different kinase inhibitors (Roscovitine (CDKs, n = 15), staurosporine (PKC, n = 12) and KN-93 (CaMKII, n = 12)). D, Relative quantification of cell surface GluA1 intensity; pre-incubation with roscovitine (n = 13) or KN-93 (n = 12) blocked DHPG-induced reduction in surface GluA1 levels in contrary to staurosporine (n = 12) pre-incubation which did not prevent mGluR-induced GluA1 internalization. Data are represented as mean±S.E. *** p < 0.001, n.s. not significant, as determined with a one-way ANOVA.
Further analysis of the most distinguished and characteristic kinase motifs, such as the TPxK motif known for Cdk1, Cdk2, and Cdk5, matched 99 of our significantly regulated phosphosites (supplemental Table S2). Here, we could find back known Cdk5 signaling proteins, such as the cyclin-dependent kinase 5 activator 1 (or p35), a neuron specific activator of Cdk5, the membrane associated cytoskeleton protein Amphiphysin, and the microtubule-associated protein Map2. The same group also contains other Maps, including Map1b and Mapt, as well as other cytoskeleton related proteins (i.e. members of the Dpysl family). Other interesting phosphorylation sites from our generated list of potential Cdk substrates include Aak1 T338, which is reported to be phosphorylated by Cdk1 (phosphosite plus). Aak1 in turn phosphorylates AP2 subunits 2m1 and 2m2 (also both found regulated by phosphorylation in our data set). Based on downstream substrate candidates presented here, Cdk's seem to be involved in initiation of AMPAR endocytosis, next to their known role in cytoskeleton reorganization (80, 81).
Newly Translated Proteins Regulated by Phosphorylation
Fifteen proteins were found to be both newly synthesized and regulated by phosphorylation upon DHPG stimulation (supplemental Table S2). These proteins include several proteins involved in translation and protein synthesis and degradation, as well as microtubule-associated protein 6 (Map6), neuronal migration protein doublecortin (Dcx) and previously discussed elongation factor (Eef2). It also contains neuromodulin (Gap43), a PKC substrate that is known for its presynaptic role in NMDAR-mediated LTD (82). More recent studies however, have shown that neuromodulin is also abundantly expressed postsynaptically, where it has to be cleaved by Caspase-3 to regulate LTD (83). Interestingly, two of its regulated phosphosites (T95 and S96) are near a Caspase-3 cleavage site. Dephosphorylation of two amino acids in this cleavage domain could very well result in a conformational change, making it more accessible to caspase cleavage.
We validated changes in phosphorylation of two of these proteins, and their three significantly regulated phosphosites. We confirmed dephosphorylation of Stmn1 S62 after 5 min of DHPG stimulation using SRM (supplemental Fig. S4A), which matches the unsupervised clustering and GO enrichment profile of this microtubule destabilizing protein (cluster 2, Fig. 3B, 3C). The same holds true for Dcx S331 and S337, (supplemental Fig. S4B, S4C), a microtubule-associated protein found in cluster 3 (Fig. 3B, 3C). Both Dcx and Stmn1 are proteins in cytoskeleton and cellular component organization, which are predominantly synthesized in the second wave of protein translation (clusters 5 and 6, Fig. 2). These data again highlight the distinct timing of events with a fast response through phosphorylation at 5 min followed by translation of the same proteins after ∼45 min. Integration of the phosphoproteomics and protein synthesis datasets, in combination with the data generated from the kinase motif analysis, resulted in a global view of molecular events triggered by group I mGluR activation in neurons (Fig. 5).
Fig. 5.
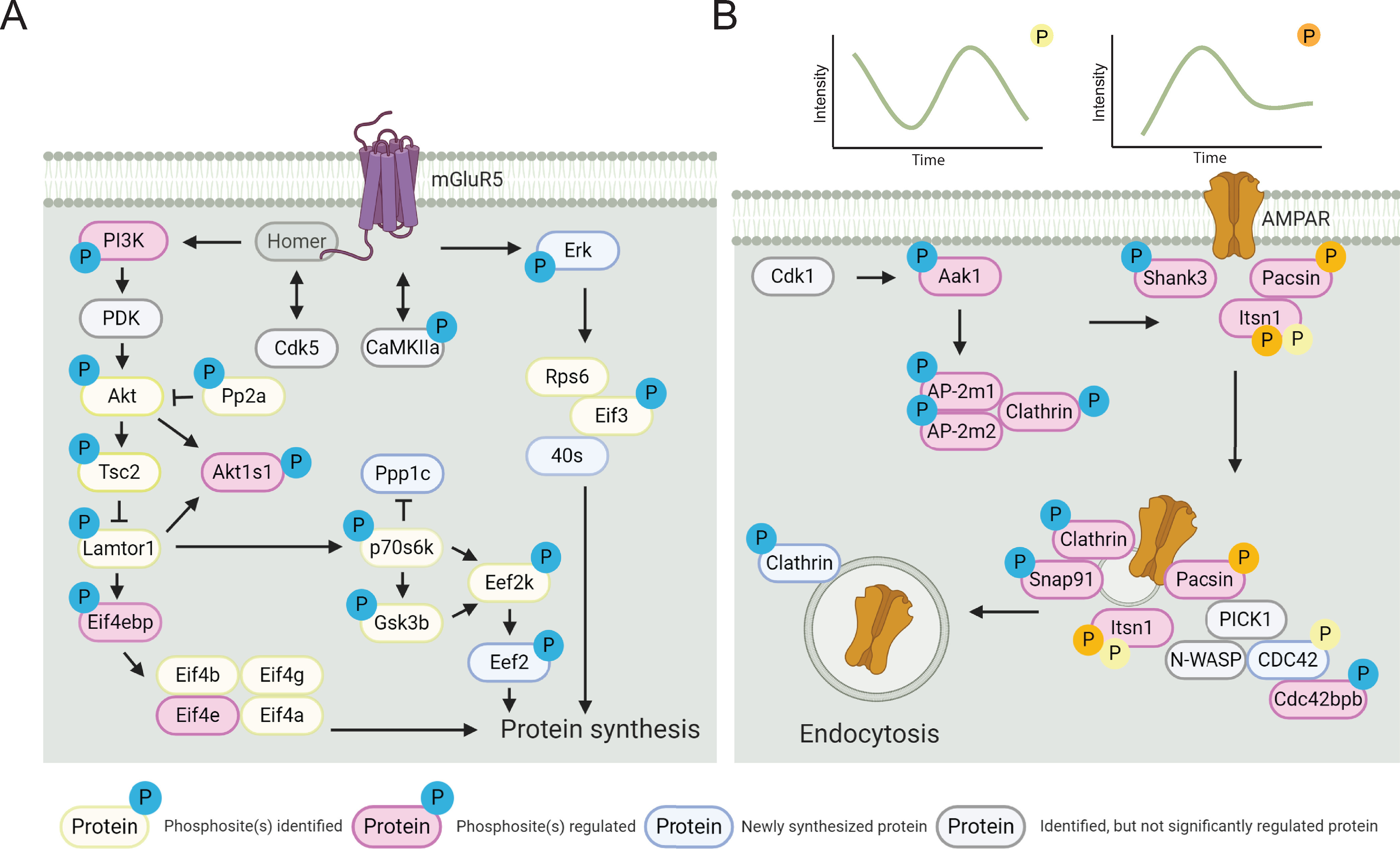
Pathways involved in DHPG-induced mGluR activation and AMPAR internalization. A, Detailed overview of the molecular pathways involved in DHPG-induced mGluR signaling identified in the (phospho)proteomics experiments. B, Visualization of phosphorylated proteins shows the activity of several signaling cascades facilitating and activating AMPAR recycling and internalization. The relative quantification of these phosphorylation events (upper panel) shows distinct phosphorylation patterns over time. Created with BioRender.
Itsn1 is Essential for DHPG-Induced AMPAR Internalization
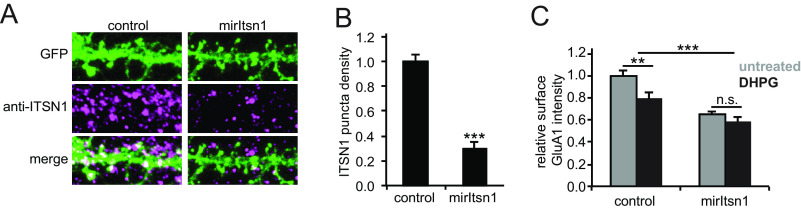
In the phosphorylation data we identified that Itsn1 phosphorylation, a guanine exchange factor (GEF) for the GTPase Cdc42 (84), was regulated at two phosphosites (S623 and S1134) upon DHPG stimulation (Fig. 3D). To test whether Itsn1 also has a functional role in mGluR-mediated AMPAR internalization, we transfected neurons with a miRNA-based knockdown construct targeting both the long and short forms of Itsn1 (mirItsn1) (85). Immunostaining of endogenous Itsn1 in control neurons showed a punctate pattern, as described before (85), and confirmed significant depletion of Itsn1 in mirItsn1-transfected neurons (Fig. 6A, 6B). Interestingly, surface GluA1 expression was significantly reduced in Itsn1 knockdown neurons under basal conditions, indicating that Itsn1 is involved in the regulation of AMPAR surface expression. Furthermore, we found that the reduction in GluA1 surface levels in response to DHPG was severely affected in Itsn1 knockdown neurons (Fig. 6C), further indicating that Itsn1 contributes to stimulated AMPAR trafficking. The presented experimental confirmation of candidate regulators in AMPAR internalization underlines the strength of this quantitative and high-resolution proteomics approach.
Fig. 6.
Itsn1 is essential for DHPG-induced AMPAR internalization. A, Immunostaining of GluA1 of transfected neurons with a miRNA-based knockdown construct targeting both the long and short forms of Itsn1 (mirItsn1) or control transfection. B, Immunostaining of endogenous Itsn1 confirms significant depletion of Itsn1 in mirItsn1-transfected neurons. C, Surface expression of GluA1 was significantly reduced in Itsn1 knockdown neurons under basal conditions and GluA1 surface levels in response to DHPG are severely affected in Itsn1 knockdown neurons. Data are represented as mean±S.E. ** p < 0.01, *** p < 0.001, n.s. not significant, as determined with a one-way ANOVA.
DISCUSSION
Here we used a combination of pulsed AHA and TMT labeling approaches to study protein synthesis in response to group I mGluR activation. This combination ensured for enrichment and MS-based relative quantification of labeled and thus newly synthesized proteins. This enrichment method has been shown to be applicable to different types of cultured cells, including primary cultures like neurons (31, 42, 86, 87). A major advantage of the use of these pulsed labeling approaches in combination with MS analysis is the possibility to identify and quantify subtle alterations in expression of proteins amidst the background of a steady state proteome, even of low abundant proteins. This approach led to the identification of 273 newly synthesized proteins upon mGluR activation, containing both known and novel proteins linked to mGluR-LTD. Here, we identified several proteins previously shown to be involved in mGluR-LTD, such as ERK1 (88) and CaMKII, and proteins implicated in endocytosis such as Cdc42 (8) and clathrin. Unfortunately, we did not identify some other known mGluR-LTD markers including Arc/Arg3.1, Step or Ophn1 (9, 22), which might be caused by their expression abundance, the chosen time points of analysis, or the nature of the enrichment method. This could potentially be improved by higher affinity click chemistry enrichment techniques or fractionation before nanoLC-MS/MS analysis. For instance, CaMKIIa was identified as a newly synthesized protein after DHPG stimulation in the proteomics data set but was not included for further analysis due to missing quantitative values. Further, we observed translation of several proteins that are themselves involved in protein synthesis, such as translation initiation and elongation factors, and multiple ribosomal proteins, but also proteins involved in proteasomal degradation, e.g. multiple proteasome subunits and enzymes involved in ubiquitination. These findings hint to the importance of regulating the fine balance between translation and degradation of proteins involved in the induction of mGluR-LTD (51).
Functional clustering of translated proteins DHPG stimulation led to the identification of known mGluR-LTD proteins, and highlighted several functionally relevant proteins with potential roles in mGluR-LTD related processes. Importantly, these functional families of proteins are not limited to the provided clusters, since these are solely based on annotated proteins with known functions or interactions. For instance, the splicing factors in cluster 1 (Fig. 2), could potentially be supplemented with less-studied family members, including Srsf1, Srsf4, and Srsf7. This also holds true for, among others, proteasomal subunits and ribosomal proteins. These extended clusters and their corresponding heatmaps can be found in supplemental Fig. S6.
We complemented the protein translation data set information on the phosphorylation events that potentially drive the signaling pathways downstream of group I mGluR. These events are usually very fast, to allow the cell to rapidly respond to external signals and precede protein translation. This makes phosphorylation likely the first step in the induction of mGluR stimulated signaling pathways, and it could therefore yield valuable information on the underlying molecular events, including mGluR-LTD-associated receptor dynamics. Here, we confirmed the role of CaMKII in mGluR-induced AMPAR internalization in both our sequence motif analysis and kinase inhibitor assay on GluA1 internalization. Multiple studies have shown the importance of kinase and phosphatase activity in the induction and maintenance of mGluR-LTD in general (16, 18, 61), emphasizing the importance of phosphorylation in this dynamic process. Involvement of serine/threonine kinase activity has been studied intensively, and have been shown to be prominently involved in the activation of protein translation via the PI3K/Akt and subsequently Tsc and mTOR pathways, while tyrosine phosphatases and kinases are believed to be responsible for AMPAR tagging for internalization, and subsequent degradation (90, 91). Contrary to the intracellular signaling pathways that generally follow Gaq/11 protein stimulation, DHPG-LTD has repeatedly been shown to activate G-protein-independent signaling pathways (92, 93). In line with these findings, we find little phosphorylation, or translation, of proteins of the PLC, DAG, and PKC pathways leading to intracellular calcium release. Furthermore, the kinase inhibitor assay confirmed that inhibition of PKC activity does not influence DHPG-induced GluA1 internalization. This is in line with previous research, which also showed that DHPG induced mGluR-LTD is not dependent on PKC activity (93). Alternatively, we found significant regulation of CDK-type kinases. Cdk5 is a known regulator of mGluR5 activation, as it controls phosphorylation of the binding site of the adaptor protein Homer to the proline-rich C terminus of group I mGluRs. Via this mechanism, Cdk5 activation is negatively correlated with mGluR5 activation (94). Moreover, hippocampal slices treated with a Cdk4 inhibitor showed impaired DHPG-induced LTD (95), suggesting that at least two prominent members of the CDK family have functions in synaptic plasticity processes in hippocampal neurons. Our motif analysis indeed confirmed a role for CDK-type kinases in mGluR-LTD, as further demonstrated using the Cdk1, 2 and 5 specific inhibitor Roscovitine, which inhibited GluA1 internalization after DHPG stimulation, however, does not fully distinguish the individual roles of Cdk1, 2 and 5. This also holds true for KN-93, which is does not only block CamKII, but also open voltage-gated potassium channels.
Next to kinase-specific signaling functions, clear regulation of cytoskeleton elements by phosphorylation was observed (Fig. 3), suggesting that group I mGluR activation can induce cytoskeleton reorganization. Most prominently, alterations in phosphorylation status of cytoskeletal regulators were observed, some of which were described before (45, 96), suggesting acute reorganization suggesting acute microtubule reorganization of both the actin and microtubule cytoskeleton. Indeed, mGluR-induced actin dynamics have been shown to be involved in regulating spine morphology (45). The effect of mGluR activation on microtubule-associated proteins suggest that mGluR activation can have even wider effects on the microtubule cytoskeleton in the dendritic shaft. Potentially, this could facilitate transport of components of the protein synthesis and degradation machinery. Next to cytoskeleton related processes, GO term analysis on molecular functions of the identified phosphorylated proteins yielded enrichment of regulatory activity of small GTPases. Several members of the Ras GTPase superfamily were found to be newly synthesized upon group I mGluR activation, including members of the Rab and Ran subfamilies, Cdc42, and several of their interacting proteins. Recently, studies have been performed on some small GTPases in relation to several types of synaptic plasticity, shedding light on the possible importance of these types of molecules in mGluR-LTD as well. Overall, these and other data provide evidence for the possible importance of small GTPases in mGluR-LTD, and should be followed up further (97, 98).
Itsn1 has not been subjected to extensive analysis in the context of synaptic plasticity before, although it was shown to influence trafficking of the AMPAR subunit GluA1 in C. Elegans (76). Here, showed a role for Itsn1 in GluA1 trafficking at dendritic spines in mammals as well. Interestingly, knockdown of Itsn1 reduced the expression of GluA1 in dendritic spines, even in the absence of DHPG. Importantly, the induction of AMPAR internalization upon DHPG stimulation was not observed in the knockdown neurons, emphasizing a central role of Itsn1 in basal and activity-induced GluA1 internalization. The exact mechanism by which Itsn1 influences receptor trafficking remains to be studied further. An interesting question is whether Itsn1 exerts its functionality mostly via posttranslational modifications such as the here identified phosphorylation sites, or via one of its interacting domains with other proteins. Previous research showed that its DH domain was critical for Cdc42 activation, and its SH3 domain for N-WASP interaction (84). Although one of our identified phosphorylation sites falls outside of these regions (S623 is in the coiled coil part of the protein), the second one, S1134, falls within the N-WASP interacting SH3 domain.
In conclusion, we were able to construct a comprehensive map of signaling and translational events upon group I mGluR activation by integrating significant changes in regulation of protein phosphorylation and translation. Over time, we could monitor activation of several signaling pathways, as well as up-regulation of protein signaling complexes involved in clathrin-mediated endocytosis. This multipronged analysis revealed several novel players in mGluR-stimulated AMPAR internalization, of which we could validate the involvement of Itsn1. We anticipate that our quantitative data set on protein phosphorylation and translation in response to group I mGluR activation can be used as a rich resource for further analyses.
DATA AVAILABILITY
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD014043. The MS SRM data have been deposited to the ProteomeXchange Consortium via Panorama Public (https://panoramaweb.org/23Gtf2.url) the with the data set identifier PXD020777.
Supplementary Material
Footnotes
This article contains supplemental data.
Funding and additional information—A.F.M.A. is supported by the Netherlands Organization for Scientific Research (NWO) through a VIDI grant (723.012.102). This research was part of the Netherlands X-omics Initiative and partially funded by NWO, project 184.034.019. H.D.M. is supported by the NWO through a ALW-Open grant (ALWOP.191), by the European Research Council (ERC starting Grant 71601) and received support from a FEBS Return-to-Europe fellowship and a NARSAD Young Investigator Award. C.C.H. is supported by the NWO (NWO-ALW-VICI 865.10.010), the Netherlands Organization for Health Research and Development (ZonMW-TOP 91213017 and 91215084) and the European Research Council (ERC) (ERC Consolidator Grant 617050).
Conflict of interest—The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Abbreviations—The abbreviations used are:
- mGluRs
- metabotropic glutamate receptors
- PLC
- phospholipase C
- DAG
- diacylglycerol
- AMPAR
- α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors
- TMT
- tandem mass tag.
REFERENCES
- 1. Bhakar A. L., Dolen G., and Bear M. F. (2012) The pathophysiology of fragile X (and what it teaches us about synapses). Annu. Rev. Neurosci. 35, 417–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nicoletti F., Bockaert J., Collingridge G. L., Conn P. J., Ferraguti F., Schoepp D. D., Wroblewski J. T., and Pin J. P. (2011) Metabotropic glutamate receptors: from the workbench to the bedside. Neuropharmacology 60, 1017–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Niswender C. M., and Conn P. J. (2010) Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol. 50, 295–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bashir Z. I., Bortolotto Z. A., Davies C. H., Berretta N., Irving A. J., Seal A. J., Henley J. M., Jane D. E., Watkins J. C., and Collingridge G. L. (1993) Induction of LTP in the hippocampus needs synaptic activation of glutamate metabotropic receptors. Nature 363, 347–350 [DOI] [PubMed] [Google Scholar]
- 5. Bellone C., Luscher C., and Mameli M. (2008) Mechanisms of synaptic depression triggered by metabotropic glutamate receptors. Cell. Mol. Life Sci. 65, 2913–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bortolotto Z. A., Bashir Z. I., Davies C. H., and Collingridge G. L. (1994) A molecular switch activated by metabotropic glutamate receptors regulates induction of long-term potentiation. Nature 368, 740–743 [DOI] [PubMed] [Google Scholar]
- 7. Hu J.-H., Park J. M., Park S., Xiao B., Dehoff M. H., Kim S., Hayashi T., Schwarz M. K., Huganir R. L., Seeburg P. H., Linden D. J., and Worley P. F. (2010) Homeostatic scaling requires group I mGluR activation mediated by Homer1a. Neuron 68, 1128–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Popkirov S. G., and Manahan-Vaughan D. (2011) Involvement of the metabotropic glutamate receptor mGluR5 in NMDA receptor-dependent, learning-facilitated long-term depression in CA1 synapses. Cereb. Cortex 21, 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luscher C., and Huber K. M. (2010) Group 1 mGluR-dependent synaptic long-term depression: mechanisms and implications for circuitry and disease. Neuron 65, 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huber K. M., Kayser M. S., and Bear M. F. (2000) Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science 288, 1254–1257 [DOI] [PubMed] [Google Scholar]
- 11. Moult P. R., Correa S. A., Collingridge G. L., Fitzjohn S. M., and Bashir Z. I. (2008) Co-activation of p38 mitogen-activated protein kinase and protein tyrosine phosphatase underlies metabotropic glutamate receptor-dependent long-term depression. J. Physiol. 586, 2499–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nosyreva E. D., and Huber K. M. (2005) Developmental switch in synaptic mechanisms of hippocampal metabotropic glutamate receptor-dependent long-term depression. J. Neurosci. 25, 2992–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li X.-M., Li C.-C., Yu S.-S., Chen J.-T., Sabapathy K., and Ruan D.-Y. (2007) JNK1 contributes to metabotropic glutamate receptor-dependent long-term depression and short-term synaptic plasticity in the mice area hippocampal CA1. Eur. J. Neurosci. 25, 391–396 [DOI] [PubMed] [Google Scholar]
- 14. Liu F., Ma X. H., Ule J., Bibb J. A., Nishi A., DeMaggio A. J., Yan Z., Nairn A. C., and Greengard P. (2001) Regulation of cyclin-dependent kinase 5 and casein kinase 1 by metabotropic glutamate receptors. Proc. Natl. Acad. Sci. U S A 98, 11062–11068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bolshakov V. Y., Carboni L., Cobb M. H., Siegelbaum S. A., and Belardetti F. (2000) Dual MAP kinase pathways mediate opposing forms of long-term plasticity at CA3-CA1 synapses. Nat. Neurosci. 3, 1107–1112 [DOI] [PubMed] [Google Scholar]
- 16. Gallagher S. M., Daly C. A., Bear M. F., and Huber K. M. (2004) Extracellular signal-regulated protein kinase activation is required for metabotropic glutamate receptor-dependent long-term depression in hippocampal area CA1. J. Neurosci. 24, 4859–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rush A. M., Wu J., Rowan M. J., and Anwyl R. (2002) Group I metabotropic glutamate receptor (mGluR)-dependent long-term depression mediated via p38 mitogen-activated protein kinase is inhibited by previous high-frequency stimulation and activation of mGluRs and protein kinase C in the rat dentate gyrus in. J. Neurosci. 22, 6121–6128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hou L., and Klann E. (2004) Activation of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J. Neurosci. 24, 6352–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ronesi J. A., and Huber K. M. (2008) Homer interactions are necessary for metabotropic glutamate receptor-induced long-term depression and translational activation. J. Neurosci. 28, 543–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rong R., Ahn J.-Y., Huang H., Nagata E., Kalman D., Kapp J. A., Tu J., Worley P. F., Snyder S. H., and Ye K. (2003) PI3 kinase enhancer-Homer complex couples mGluRI to PI3 kinase, preventing neuronal apoptosis. Nat. Neurosci. 6, 1153–1161 [DOI] [PubMed] [Google Scholar]
- 21. Banko J. L., Hou L., Poulin F., Sonenberg N., and Klann E. (2006) Regulation of eukaryotic initiation factor 4E by converging signaling pathways during metabotropic glutamate receptor-dependent long-term depression. J. Neurosci. 26, 2167–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Waung M. W., and Huber K. M. (2009) Protein translation in synaptic plasticity: mGluR-LTD, Fragile X. Curr. Opin. Neurobiol. 19, 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Purgert C. A., Izumi Y., Jong Y.-J. I., Kumar V., Zorumski C. F., and O'Malley K. L. (2014) Intracellular mGluR5 can mediate synaptic plasticity in the hippocampus. J. Neurosci. 34, 4589–4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakase I., Kobayashi N. B., Takatani-Nakase T., and Yoshida T. (2015) Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic Ras expression potentiates cellular uptake efficacy of exosomes. Sci. Rep 5, 10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. MacGillavry H. D., Song Y., Raghavachari S., and Blanpied T. A. (2013) Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron 78, 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shepherd J. D., and Huganir R. L. (2007) The cell biol. of synaptic plasticity: AMPA receptor trafficking. Annu. Rev. Cell Dev. Biol. 23, 613–643 [DOI] [PubMed] [Google Scholar]
- 27. Davidkova G., and Carroll R. C. (2007) Characterization of the role of microtubule-associated protein 1B in metabotropic glutamate receptor-mediated endocytosis of AMPA receptors in hippocampus. J. Neurosci. 27, 13273–13278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nadif Kasri N., Nakano-Kobayashi A., and Van Aelst L. (2011) Rapid synthesis of the X-linked mental retardation protein OPHN1 mediates mGluR-dependent LTD through interaction with the endocytic machinery. Neuron 72, 300–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Waung M. W., Pfeiffer B. E., Nosyreva E. D., Ronesi J. A., and Huber K. M. (2008) Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron 59, 84–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y., Venkitaramani D. V., Gladding C. M., Zhang Y., Kurup P., Molnar E., Collingridge G. L., and Lombroso P. J. (2008) The tyrosine phosphatase STEP mediates AMPA receptor endocytosis after metabotropic glutamate receptor stimulation. J. Neurosci. 28, 10561–10566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eichelbaum K., Winter M., Berriel Diaz M., Herzig S., and Krijgsveld J. (2012) Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat. Biotechnol. 30, 984–990 [DOI] [PubMed] [Google Scholar]
- 32. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 33. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 34. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schneider T. D., and Stephens R. M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chou M. F., and Schwartz D. (2011) Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinforma. 13, 15–24 [DOI] [PubMed] [Google Scholar]
- 37. Schwartz D., and Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 38. Koopmans F., van Nierop P., Andres-Alonso M., Byrnes A., Cijsouw T., Coba M. P., Cornelisse L. N., Farrell R. J., Goldschmidt H. L., Howrigan D. P., Hussain N. K., Imig C., de Jong A. P. H., Jung H., Kohansalnodehi M., Kramarz B., Lipstein N., Lovering R. C., MacGillavry H., Mariano V., Mi H., Ninov M., Osumi-Sutherland D., Pielot R., Smalla K.-H., Tang H., Tashman K., Toonen R. F. G., Verpelli C., Reig-Viader R., Watanabe K., van Weering J., Achsel T., Ashrafi G., Asi N., Brown T. C., De Camilli P., Feuermann M., Foulger R. E., Gaudet P., Joglekar A., Kanellopoulos A., Malenka R., Nicoll R. A., Pulido C., de Juan-Sanz J., Sheng M., Südhof T. C., Tilgner H. U., Bagni C., Bayés À., Biederer T., Brose N., Chua J. J. E., Dieterich D. C., Gundelfinger E. D., Hoogenraad C., Huganir R. L., Jahn R., Kaeser P. S., Kim E., Kreutz M. R., McPherson P. S., Neale B. M., O'Connor V., Posthuma D., Ryan T. A., Sala C., Feng G., Hyman S. E., Thomas P. D., Smit A. B., and Verhage M. (2019) SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron 103, 217–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmidlin T., Debets D. O., van Gelder C. A. G. H., Stecker K. E., Rontogianni S., van den Eshof B. L., Kemper K., Lips E. H., van den Biggelaar M., Peeper D. S., Heck A. J. R., and Altelaar M. (2019) High-throughput assessment of kinome-wide activation states. Cell Syst. 9, 366–374.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. de Graaf E. L., Kaplon J., Mohammed S., Vereijken L. A. M., Duarte D. P., Redondo Gallego L., Heck A. J. R., Peeper D. S., and Altelaar A. F. M. (2015) Signal transduction reaction monitoring deciphers site-specific PI3K-mTOR/MAPK pathway dynamics in oncogene-induced senescence. J. Proteome Res. 14, 2906–2914 [DOI] [PubMed] [Google Scholar]
- 41. Anggono V., and Huganir R. L. (2012) Regulation of AMPA receptor trafficking and synaptic plasticity. Curr. Opin. Neurobiol. 22, 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dieterich D. C., Link A. J., Graumann J., Tirrell D. A., and Schuman E. M. (2006) Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. U S A 103, 9482–9487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Friocourt G., Chafey P., Billuart P., Koulakoff A., Vinet M. C., Schaar B. T., McConnell S. K., Francis F., and Chelly J. (2001) Doublecortin interacts with μ subunits of clathrin adaptor complexes in the developing nervous system. Mol. Cell. Neurosci. 18, 307–319 [DOI] [PubMed] [Google Scholar]
- 44. Sanderson T. M., Hogg E. L., Collingridge G. L., and Corrêa S. A. L. (2016) Hippocampal metabotropic glutamate receptor long-term depression in health and disease: focus on mitogen-activated protein kinase pathways. J. Neurochem. 139, 200–214 [DOI] [PubMed] [Google Scholar]
- 45. Zhou Z., Hu J., Passafaro M., Xie W., and Jia Z. (2011) GluA2 (GluR2) regulates metabotropic glutamate receptor-dependent long-term depression through N-cadherin-dependent and cofilin-mediated actin reorganization. J. Neurosci. 31, 819–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bayés A., Collins M. O., Croning M. D. R., van de Lagemaat L. N., Choudhary J. S., and Grant S. G. N. (2012) Comparative study of human and mouse postsynaptic proteomes finds high compositional conservation and abundance differences for key synaptic proteins. PLoS ONE. 7, e46683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heo S., Diering G. H., Na C. H., Nirujogi R. S., Bachman J. L., Pandey A., and Huganir R. L. (2018) Identification of long-lived synaptic proteins by proteomic analysis of synaptosome protein turnover. Proc. Natl. Acad. Sci. U S A 115, E3827–E3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Montojo J., Zuberi K., Rodriguez H., Kazi F., Wright G., Donaldson S. L., Morris Q., and Bader G. D. (2010) GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics 26, 2927–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Frydman J. (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70, 603–647 [DOI] [PubMed] [Google Scholar]
- 51. Klein M. E., Castillo P. E., and Jordan B. A. (2015) Coordination between Translation and Degradation Regulates Inducibility of mGluR-LTD. Cell Rep. 10, 1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ramachandran K. V., and Margolis S. S. (2017) A mammalian nervous-system-specific plasma membrane proteasome complex that modulates neuronal function. Nat. Struct. Mol. Biol. 24, 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sluchanko N. N., and Gusev N. B. (2010) 14-3-3 proteins and regulation of cytoskeleton. Biochemistry. Mosc. 75, 1528–1546 [DOI] [PubMed] [Google Scholar]
- 54. Humphrey S. J., Azimifar S. B., and Mann M. (2015) High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 33, 990–995 [DOI] [PubMed] [Google Scholar]
- 55. Villén J., and Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zarei M., Sprenger A., Metzger F., Gretzmeier C., and Dengjel J. (2011) Comparison of ERLIC-TiO2, HILIC-TiO2, and SCX-TiO2 for global phosphoproteomics approaches. J. Proteome Res. 10, 3474–3483 [DOI] [PubMed] [Google Scholar]
- 57. Post H., Penning R., Fitzpatrick M. A., Garrigues L. B., Wu W., MacGillavry H. D., Hoogenraad C. C., Heck A. J. R., and Altelaar A. F. M. (2017) Robust, sensitive, and automated phosphopeptide enrichment optimized for low sample amounts applied to primary hippocampal neurons. J. Proteome Res. 16, 728–737 [DOI] [PubMed] [Google Scholar]
- 58. Ye J., Zhang X., Young C., Zhao X., Hao Q., Cheng L., and Jensen O. N. (2010) Optimized IMAC-IMAC protocol for phosphopeptide recovery from complex biological samples. J. Proteome Res. 9, 3561–3573 [DOI] [PubMed] [Google Scholar]
- 59. Rigbolt K. T., Vanselow J. T., and Blagoev B. (2011) GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol. Cell. Proteomics 10, O110.007450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Collingridge G. L., Peineau S., Howland J. G., and Wang Y. T. (2010) Long-term depression in the CNS. Nat. Rev. Neurosci. 11, 459–473 [DOI] [PubMed] [Google Scholar]
- 61. Gladding C. M., Fitzjohn S. M., and Molnár E. (2009) Metabotropic glutamate receptor-mediated long-term depression: molecular mechanisms. Pharmacol. Rev. 61, 395–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Moult P. R., Gladding C. M., Sanderson T. M., Fitzjohn S. M., Bashir Z. I., Molnar E., and Collingridge G. L. (2006) Tyrosine phosphatases regulate AMPA receptor trafficking during metabotropic glutamate receptor-mediated long-term depression. J. Neurosci. 26, 2544–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sugawara T., Hisatsune C., Miyamoto H., Ogawa N., and Mikoshiba K. (2017) Regulation of spinogenesis in mature Purkinje cells via mGluR/PKC-mediated phosphorylation of CaMKIIβ. Proc. Natl. Acad. Sci 114, E5256–E5265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hizli A. A., Chi Y., Swanger J., Carter J. H., Liao Y., Welcker M., Ryazanov A. G., and Clurman B. E. (2013) Phosphorylation of eukaryotic elongation factor 2 (eEF2) by cyclin A–cyclin-dependent kinase 2 regulates its inhibition by eEF2 kinase. Mol. Cell Biol. 33, 596–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Park S., Park J. M., Kim S., Kim J.-A., Shepherd J. D., Smith-Hicks C. L., Chowdhury S., Kaufmann W., Kuhl D., Ryazanov A. G., Huganir R. L., Linden D. J., and Worley P. F. (2008) Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron 59, 70–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rumbaugh G., Adams J. P., Kim J. H., and Huganir R. L. (2006) SynGAP regulates synaptic strength and mitogen-activated protein kinases in cultured neurons. Proc. Natl. Acad. Sci. U S A 103, 4344–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Walkup W. G., Washburn L., Sweredoski M. J., Carlisle H. J., Graham R. L., Hess S., and Kennedy M. B. (2015) Phosphorylation of synaptic GTPase-activating protein (synGAP) by Ca2+/calmodulin-dependent protein kinase II (CaMKII) and cyclin-dependent kinase 5 (CDK5) alters the ratio of its GAP activity toward Ras and Rap GTPases. J. Biol. Chem. 290, 4908–4927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li J., Wilkinson B., Clementel V. A., Hou J., O'Dell T. J., and Coba M. P. (2016) Long-term potentiation modulates synaptic phosphorylation networks and reshapes the structure of the postsynaptic interactome. Sci. Signal. 9, rs8. [DOI] [PubMed] [Google Scholar]
- 69. Derkach V. A., Oh M. C., Guire E. S., and Soderling T. R. (2007) Regulatory mechanisms of AMPA receptors in synaptic plasticity. Nat. Rev. Neurosci. 8, 101–113 [DOI] [PubMed] [Google Scholar]
- 70. Henley J. M., and Wilkinson K. A. (2016) Synaptic AMPA receptor composition in development, plasticity and disease. Nat. Rev. Neurosci. 17, 337–350 [DOI] [PubMed] [Google Scholar]
- 71. Quan A., Xue J., Wielens J., Smillie K. J., Anggono V., Parker M. W., Cousin M. A., Graham M. E., and Robinson P. J. (2012) Phosphorylation of syndapin I F-BAR domain at two helix-capping motifs regulates membrane tubulation. Proc. Natl. Acad. Sci. U S A 109, 3760–3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Anggono V., Koç-Schmitz Y., Widagdo J., Kormann J., Quan A., Chen C.-M., Robinson P. J., Choi S.-Y., Linden D. J., Plomann M., and Huganir R. L. (2013) PICK1 interacts with PACSIN to regulate AMPA receptor internalization and cerebellar long-term depression. Proc. Natl. Acad. Sci. U S A 110, 13976–13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mayhew M. W., Jeffery E. D., Sherman N. E., Nelson K., Polefrone J. M., Pratt S. J., Shabanowitz J., Parsons J. T., Fox J. W., Hunt D. F., and Horwitz A. F. (2007) Identification of phosphorylation sites in betaPIX and PAK1. J. Cell Sci. 120, 3911–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pechstein A., Bacetic J., Vahedi-Faridi A., Gromova K., Sundborger A., Tomlin N., Krainer G., Vorontsova O., Schäfer J. G., Owe S. G., Cousin M. A., Saenger W., Shupliakov O., and Haucke V. (2010) Regulation of synaptic vesicle recycling by complex formation between intersectin 1 and the clathrin adaptor complex AP2. Proc. Natl. Acad. Sci. U S A 107, 4206–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yu Y., Chu P.-Y., Bowser D. N., Keating D. J., Dubach D., Harper I., Tkalcevic J., Finkelstein D. I., and Pritchard M. A. (2008) Mice deficient for the chromosome 21 ortholog Itsn1 exhibit vesicle-trafficking abnormalities. Hum. Mol. Genet. 17, 3281–3290 [DOI] [PubMed] [Google Scholar]
- 76. Glodowski D. R., Chen C. C., Schaefer H., Grant B. D., and Rongo C. (2007) RAB-10 regulates glutamate receptor recycling in a cholesterol-dependent endocytosis pathway. Mol. Biol. Cell 18, 4387–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Errico A., Deshmukh K., Tanaka Y., Pozniakovsky A., and Hunt T. (2010) Identification of substrates for cyclin dependent kinases. Adv. Enzyme Regul. 50, 375–399 [DOI] [PubMed] [Google Scholar]
- 78. Gaestel M. (2006) MAPKAP kinases - MKs - two's company, three's a crowd. Nat. Rev. Mol. Cell Biol. 7, 120–130 [DOI] [PubMed] [Google Scholar]
- 79. Stokoe D., Caudwell B., Cohen P. T., and Cohen P. (1993) The substrate specificity and structure of mitogen-activated protein (MAP) kinase-activated protein kinase-2. Biochem. J 296, 843–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Plattner F., Giese K. P., and Angelo M. (2008) Involvement of Cdk5 in Synaptic Plasticity, and Learning and Memory. in Cyclin Dependent Kinase 5 (Cdk5) 227–260 Springer US; 10.1007/978-0-387-78887-6_16 [DOI] [Google Scholar]
- 81. Contreras-Vallejos E., Utreras E., Bórquez D. A., Prochazkova M., Terse A., Jaffe H., Toledo A., Arruti C., Pant H. C., Kulkarni A. B., and González-Billault C. (2014) Searching for novel Cdk5 substrates in brain by comparative phosphoproteomics of wild type and Cdk5−/− Mice. PLoS ONE. 9, e90363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ramakers G. M., Heinen K., Gispen W. H., and de Graan P. N. (2000) Long term depression in the CA1 field is associated with a transient decrease in pre- and postsynaptic PKC substrate phosphorylation. J. Biol. Chem. 275, 28682–28687 [DOI] [PubMed] [Google Scholar]
- 83. Han M.-H., Jiao S., Jia J.-M., Chen Y., Chen C. Y., Gucek M., Markey S. P., and Li Z. (2013) The novel caspase-3 substrate Gap43 is involved in AMPA receptor endocytosis and long-term depression. Mol. Cell. Proteomics 12, 3719–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hussain N. K., Jenna S., Glogauer M., Quinn C. C., Wasiak S., Guipponi M., Antonarakis S. E., Kay B. K., Stossel T. P., Lamarche-Vane N., and McPherson P. S. (2001) Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat. Cell Biol. 3, 927–932 [DOI] [PubMed] [Google Scholar]
- 85. Thomas S., Ritter B., Verbich D., Sanson C., Bourbonnière L., McKinney R. A., and McPherson P. S. (2009) Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J. Biol. Chem. 284, 12410–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kenney J. W., Genheden M., Moon K.-M., Wang X., Foster L. J., and Proud C. G. (2016) Eukaryotic elongation factor 2 kinase regulates the synthesis of microtubule-related proteins in neurons. J. Neurochem. 136, 276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schanzenbächer C. T., Sambandan S., Langer J. D., and Schuman E. M. (2016) Nascent proteome remodeling following homeostatic scaling at hippocampal synapses. Neuron 92, 358–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Thomas G. M., and Huganir R. L. (2004) MAPK cascade signalling and synaptic plasticity. Nat. Rev. Neurosci. 5, 173–183 [DOI] [PubMed] [Google Scholar]
- 89. Ramakers G. J. A., Wolfer D., Rosenberger G., Kuchenbecker K., Kreienkamp H.-J., Prange-Kiel J., Rune G., Richter K., Langnaese K., Masneuf S., Bösl M. R., Fischer K.-D., Krugers H. J., Lipp H.-P., van Galen E., and Kutsche K. (2012) Dysregulation of Rho GTPases in the alphaPix/Arhgef6 mouse model of X-linked intellectual disability is paralleled by impaired structural and synaptic plasticity and cognitive deficits. Hum. Mol. Genet. 21, 268–286 [DOI] [PubMed] [Google Scholar]
- 90. Bidinosti M., Botta P., Krüttner S., Proenca C. C., Stoehr N., Bernhard M., Fruh I., Mueller M., Bonenfant D., Voshol H., Carbone W., Neal S. J., McTighe S. M., Roma G., Dolmetsch R. E., Porter J. A., Caroni P., Bouwmeester T., Lüthi A., and Galimberti I. (2016) CLK2 inhibition ameliorates autistic features associated with SHANK3 deficiency. Science 351, 1199–1203 [DOI] [PubMed] [Google Scholar]
- 91. O'Connor E. C., Bariselli S., and Bellone C. (2014) Synaptic basis of social dysfunction: a focus on postsynaptic proteins linking group-I mGluRs with AMPARs and NMDARs. Eur. J. Neurosci. 39, 1114–1129 [DOI] [PubMed] [Google Scholar]
- 92. Fitzjohn S. M., Palmer M. J., May J. E., Neeson A., Morris S. A., and Collingridge G. L. (2001) A characterisation of long-term depression induced by metabotropic glutamate receptor activation in the rat hippocampus in vitro. J. Physiol. (Lond) 537, 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Schnabel R., Kilpatrick I. C., and Collingridge G. L. (1999) An investigation into signal transduction mechanisms involved in DHPG-induced LTD in the CA1 region of the hippocampus. Neuropharmacology 38, 1585–1596 [DOI] [PubMed] [Google Scholar]
- 94. Hu J.-H., Yang L., Kammermeier P. J., Moore C. G., Brakeman P. R., Tu J., Yu S., Petralia R. S., Li Z., Zhang P.-W., Park J. M., Dong X., Xiao B., and Worley P. F. (2012) Preso1 dynamically regulates group I metabotropic glutamate receptors. Nat. Neurosci. 15, 836–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Li C., Li X., Chen W., Yu S., Chen J., Wang H., and Ruan D. (2007) The different roles of cyclinD1-CDK4 in STP and mGluR-LTD during the postnatal development in mice hippocampus area CA1. BMC Dev. Biol. 7, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nakamura Y., Wood C. L., Patton A. P., Jaafari N., Henley J. M., Mellor J. R., and Hanley J. G. (2011) PICK1 inhibition of the Arp2/3 complex controls dendritic spine size and synaptic plasticity. EMBO J. 30, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Barnes S. A., Wijetunge L. S., Jackson A. D., Katsanevaki D., Osterweil E. K., Komiyama N. H., Grant S. G. N., Bear M. F., Nägerl U. V., Kind P. C., and Wyllie D. J. A. (2015) Convergence of hippocampal pathophysiology in Syngap+/- and Fmr1-/y Mice. J. Neurosci. 35, 15073–15081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zheng N., Jeyifous O., Munro C., Montgomery J. M., and Green W. N. (2015) Synaptic activity regulates AMPA receptor trafficking through different recycling pathways. Elife 4, e06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Esteves da Silva M., Adrian M., Schätzle P., Lipka J., Watanabe T., Cho S., Futai K., Wierenga C. J., Kapitein L. C., and Hoogenraad C. C. (2015) Positioning of AMPA receptor-containing endosomes regulates synapse architecture. Cell Reports 13, 933–943 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD014043. The MS SRM data have been deposited to the ProteomeXchange Consortium via Panorama Public (https://panoramaweb.org/23Gtf2.url) the with the data set identifier PXD020777.