Abstract
Objective:
To study the relationship between long-chain non-coding RNA small nucleolar RNA host gene 16 (lncRNA SNHG16) polymorphisms and its interaction with environmental factors and susceptibility to colorectal cancer (CRC).
Methods:
Sanger sequencing was used to analyze genotypes of lncRNA SNHG16 gene rs7353, rs8038, and rs15278 sites. Multifactor dimensionality reduction was used to analyze interactions between lncRNA SNHG16 gene rs7353, rs8038, rs15278 sites, and environmental factors. Haploview 4.1 software was used to analyze linkage disequilibrium of lncRNA SNHG16 gene rs7353, rs8038, and rs15278 sites. Quantitative real-time polymerase chain reaction was used to analyze plasma lncRNA SNHG16 levels of CRC patients and control subjects.
Results:
Variation of the lncRNA SNHG16 gene rs7353 site A>G variation was associated with decreased CRC susceptibility (Odds ratio [OR] = 0.50, 95% confidence interval [CI]: 0.40–0.62, P < .01). The rs8038 site G>A and rs15278 site A>G variation were associated with increased CRC susceptibility (OR = 1.87, 95% CI: 1.47–2.36, P < .01). The rs15278 site G>A variation was associated with increased CRC susceptibility (OR = 2.24, 95% CI: 1.61–3.11, P < .01). Interaction combinations featuring age, rs7353, rs8038, and rs15278 single nucleotide polymorphism are 13.53 times more susceptible to CRC than other interactions (95% CI: 9.43–19.41, P < .01). The rs15278, rs8038, and rs7353 site AGA haplotypes were significantly associated with a decreased CRC risk (OR = 0.65, 95% CI: 0.48–0.88, P = .01), AAG haplotypes were significantly associated with an increased CRC risk (OR = 2.00, 95% CI: 1.27–3.17, P < .01). High lncRNA SNHG16 expression was associated with tumor progression in CRC patients (χ2 = 8.85, P = .03). The rs7353 site A>G variation caused a significant decrease in plasma lncRNA SNHG16 level (P < .01), while the rs8038 site G>A variation and rs15278 site A>G variation resulted in increased plasma lncRNA SNHG16 levels.
Conclusion:
Polymorphisms of lncRNA SNHG16 gene rs7353, rs8038, rs15278 loci and their interaction with age are significantly associated with CRC susceptibility.
Keywords: colorectal cancer, gene-environment interaction, long non-coding RNA, small nucleolar RNA host gene 16
1. Introduction
Colorectal cancer (CRC) is among the most common malignant tumors. It is the third most common tumor in both males and females; however, the incidence in males is significantly higher than that in females.[1] With economic and societal development and improvement of living standards, various environmental factors, including diet and lifestyle, are changing constantly, and the incidence of CRC has also shown an upward trend year on year.[2]
Studies have shown that CRC occurrence is affected by various factors including diet, lifestyle, genetic characteristics, and results from the combined action of genetic and environmental factors.[3–6] Studies have shown that many oncogenes, tumor-suppressor genes, microRNAs, and long-chain non-coding RNA (lncRNA) are closely related to CRC occurrence and development.[7–10] lncRNAs and miRNAs act as oncogenes or tumor-suppressor genes in the occurrence and development of tumors through the regulation of tumor cell proliferation, invasion, apoptosis, and drug resistance.[11–14]
The lncRNA small nucleolar RNA host gene 16 (SNHG16) is located on human chromosome 17q25.1 and was the first lncRNA to be detected in neuroblastoma.[15] Studies have shown that lncRNA SNHG16 is highly expressed in CRC, bladder cancer, and breast cancer tissues and participates in tumor invasion and metastasis.[11,16,17] In addition, in vitro studies have demonstrated that lncRNA SNHG16 may play a carcinogenic role in CRC, affecting the expression of genes related to lipid metabolism through ceRNA.[11]
However, at present, there is no research focusing on the correlation between lncRNA SNHG16 gene polymorphism and CRC susceptibility. In the present study, we selected the rs7353, rs8038, and rs15278 sites of the lncRNA SNHG16 gene for research. Selection of these sites was based on prediction by the lncRNASNP2 tool (http://bioinfo.life.hust.edu.cn/lncRNASNP#!/). The results showed that rs7353, rs8038, and rs15278 sites were all at sites which affected lncRNA structure and lncRNA binding to miRNA. Whether these key single nucleotide polymorphism (SNP) sites affect the role of lncRNA SNHG16 in CRC occurrence and development is worthy of study.
2. Materials and methods
2.1. Subjects
We enrolled 361 CRC patients treated in our hospital between January 2016 and October 2019, including 211 men and 150 women, aged 34 to 88 years, with an average age of 61.48 ± 10.70 years. CRC was confirmed in all patients by histopathology. In addition, 360 healthy subjects were randomly enrolled as the control group, including 215 males and 145 females, aged 35 to 87 years, with an average age of 61.96 ± 11.49 years. Patients with a history of other tumors were excluded. This study was approved by the Medical Ethics Committee of First People's Hospital of Yuhang District, Hangzhou, and all patients provided informed consent.
2.2. Genotype analysis of rs7353, rs8038, and rs15278 sites of lncRNA SNHG16 gene
We collected 5 ml of peripheral venous blood from subjects and used a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) to extract genomic DNA from peripheral blood mononuclear cells. DNA fragments containing SNP sites were obtained by PCR amplification. Primer information is as follows: rs7353:5′-ACT GTG CAA AGC CGT GTA GT-3′ (Forward primer), 5′-TCA GCT GAC GGT AGT TTC CC-3′ (Reverse primer); rs8038:5′-TGT CTG TTC TGT TGA TCA GAG GT-3′ (Forward primer), 5’-GCA CAC TTT ACA AGA GGG ATT GT-3’ (Reverse primer); rs15278:5′-TGA TTA TAC TCT GTT GGA AGA GCC T-3′ (Forward primer), 5’-ACT GGG GTT AAG CAT TAC TCT CTG-3′ (Reverse primer). PCR reaction system contained 2.0 μl 10 × buffer, 0.8 μl 25 mM Mg2+, 0.4 μl 2 mM dNTP, 0.4 μl forward primer/reverse primer, 30 ng genomic DNA, 0.15 μl 1U/μl Taq enzyme, and the remaining volume being ddH2O, adding up to a volume of 20 μl. The PCR amplification program was set to 95°C for 5 minutes; 95°C for 30 seconds; 60°C for 25 s; 72°C for 25 seconds, for a total of 30 cycles, then 72°C for 5 minutes. PCR products were then subjected to Sanger sequencing. The genotypes of the SNP loci were defined by comparison with the dbSNP database (https://www.ncbi.nlm.nih.gov/snp/) (Fig. 1).
Figure 1.
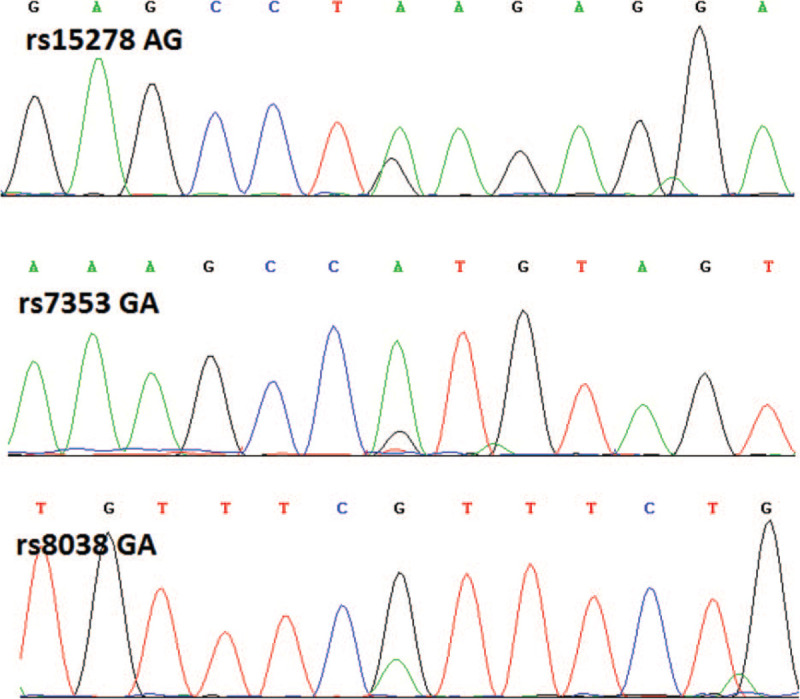
Sanger sequencing results.
2.3. Real-time fluorescence quantitative PCR (qRT-PCR) to detect lncRNA SNHG16 levels
Trizol (Invitrogen, Inc., Carlsbad, CA) was used to extract total RNA from plasma. Total RNA was used as a template to perform reverse transcription PCR and synthesize cDNA. A PrimeScript RT Reagent Kit (Takara, Bio Inc., Shiga, Japan) was used for this experiment. Using synthetic cDNA as a template and GAPDH as an internal control, qRT-PCR was performed on an ABI 7500 thermal cycler (Applied Biosystems) to detect lncRNA SNHG16 expression level in the plasma of subjects relative to GAPDH.
2.4. Statistical analysis
In this study, statistical analysis was performed using SPSS 22.0 software (SPSS Inc., Chicago). The χ2 test was used to assess whether the genotype of the SNP site of the lncRNA SNHG16 gene was in Hardy–Weinberg equilibrium. Continuous variables between groups were analyzed using t test and one-way analysis of variance and differences between categorical variables were analyzed using χ2 test. Logistic regression was used to analyze correlation between lncRNA SNHG16 gene SNP sites and CRC susceptibility risk. Odds ratio (OR) and 95% confidence interval (CI) were calculated, and adjusted by age, gender, body mass index (BMI), smoking history, drinking history and family tumor history. All statistical tests were 2-sided tests. In each case, P < .05 indicated a statistically significant difference.
3. Results
3.1. Clinical data of CRC patients and control group
General clinical data of the 361 CRC patients and 360 healthy subjects in this study are shown in Table 1. Statistical analysis demonstrated that there were no statistically significant differences in clinical data such as age, gender, BMI, smoking history, and drinking history between the two groups (P > .05). The proportion of patients with a family history of tumors was significantly higher in the CRC group than in the control group (P < .01). Among the 361 CRC patients, according to tumor stages grade, 33 cases were grade I, 164 were grade II, 129 were grade III and 35 were grade IV. Of the CRC cases, 208 were in colon and 153 were in rectum.
Table 1.
Comparison of clinical data between CRC patients and control group.
| Variables | CRC (n = 361) | Control (n = 360) | P |
| Age (years, mean ± SD) | 61.48 ± 10.70 | 61.96 ± 11.49 | .34 |
| Gender [n (%)] | .55 | ||
| Male | 211 (58.45%) | 215 (59.72%) | |
| Female | 150 (41.55%) | 145 (40.28%) | |
| BMI (kg/m2, mean ± SD) | 24.69 ± 2.61 | 24.70 ± 4.19 | .97 |
| Smoking [n (%)] | .85 | ||
| Yes | 109 (30.19%) | 111 (30.83%) | |
| No | 252 (69.81%) | 249 (69.17%) | |
| Drinking [n (%)] | .22 | ||
| Yes | 148 (41.00%) | 164 (45.56%) | |
| No | 213 (59.00%) | 196 (54.44%) | |
| Family history [n (%)] | <.01 | ||
| Yes | 56 (15.51%) | 29 (8.06%) | |
| No | 305 (84.49%) | 331 (91.94%) | |
| Tumor stages [n (%)] | |||
| I | 33 (9.14%) | ||
| II | 164 (45.43%) | ||
| III | 129 (35.73%) | ||
| IV | 35 (9.70%) | ||
| Tumor site [n (%)] | |||
| Colon | 208 (57.62%) | ||
| Rectum | 153 (42.38%) |
3.2. Correlation between lncRNA SNHG16 gene polymorphism and CRC
Genotypes of rs7353, rs8038, and rs15278 sites of the lncRNA SNHG16 gene in CRC patients and control group were analyzed in Hardy–Weinberg equilibrium (P > .05, Table 2). Analysis showed that AG genotype and GG genotype at the rs7353 site had a significantly lower risk of CRC compared with AA genotype (P < .01). Both the dominant (AG + GG vs AA) and recessive model (GG vs AA + AG) significantly reduced CRC risk (P < .01). The A>G allele variation at rs7353 caused a significant decrease in CRC susceptibility (OR = 0.50, 95% CI: 0.40–0.62, P < .01). The A>G allele variation at rs8038 significantly increased CRC susceptibility (OR = 1.87, 95% CI: 1.47–2.36, P < .01). Compared with the GG genotype, both GA and AA genotype carriers have a significantly increased risk of CRC, and both the dominant (GA + AA vs GG) and recessive model (AA vs GG + GA) have a significantly increased risk of CRC (P < .01).
Table 2.
Correlation between lncRNA SNHG16 genotype and allele frequency and CRC.
| CRC (n = 361) | Control (n = 360) | HWE p | OR (95% CI) | P | |
| rs7353 | 199 (55.12%) | 122 (33.80%) | 0.08/0.44 | 1.00 (reference) | |
| AA | 129 (35.73%) | 169 (46.81%) | 0.47 (0.34–0.65) | <.01 | |
| AG | 33 (9.14%) | 69 (19.11%) | 0.29 (0.18–0.47) | <.01 | |
| GG | 162 (44.88%) | 238 (65.93%) | 0.42 (0.31–0.56) | <.01 | |
| AG+GG | 328 (90.86%) | 291 (80.61%) | 1.00 (reference) | ||
| AA+AG | 33 (9.14%) | 69 (19.11%) | 0.42 (0.27–0.66) | <.01 | |
| GG | 527 (72.99%) | 413 (57.36%) | 1.00 (reference) | ||
| A | 195 (27.01%) | 307 (42.64%) | 0.50 (0.40–0.62) | <.01 | |
| G | |||||
| rs8038 | 156 (43.21%) | 226 (62.60%) | 0.19/0.71 | 1.00 (reference) | |
| GG | 171 (47.37%) | 117 (32.41%) | 2.12 (1.55–2.89) | <.01 | |
| GA | 34 (9.42%) | 17 (4.71%) | 2.90 (1.56–5.37) | <.01 | |
| AA | 205 (56.79%) | 134 (37.12%) | 2.22 (1.65–2.99) | <.01 | |
| AA+GA | 327 (90.58%) | 343 (95.01%) | 1.00 (reference) | ||
| GG+GA | 34 (9.42%) | 17 (4.71%) | 2.10 (1.15–3.83) | .02 | |
| AA | 483 (66.90%) | 569 (79.03%) | 1.00 (reference) | ||
| G | 239 (33.10%) | 151 (20.97%) | 1.87 (1.47–2.36) | <.01 | |
| A | |||||
| rs15278 | 254 (70.36%) | 305 (84.49%) | 0.08/0.08 | 1.00 (reference) | |
| AA | 92 (25.48%) | 50 (13.85%) | 2.21 (1.51–3.24) | <.01 | |
| AG | 15 (4.16%) | 5 (1.39%) | 3.60 (1.29–10.05) | .02 | |
| GG | 107 (29.64%) | 55 (15.24%) | 2.36 (1.62–3.37) | <.01 | |
| GG+AG | 346 (95.84%) | 355 (98.34%) | 1.00 (reference) | ||
| AA+AG | 15 (4.16%) | 5 (1.39%) | 3.08 (1.11–8.56) | .04 | |
| GG | 600 (83.10%) | 660 (91.67%) | 1.00 (reference) | ||
| A | 122 (16.90%) | 60 (8.33%) | 2.24 (1.61–3.11) | <.01 | |
| G |
3.3. Correlation between lncRNA SNHG16 gene polymorphism and the site and severity of CRC
Our analysis showed no significant correlation between tumor stage or site in patients with different genotypes of CRC in the rs7353, rs8038, and rs15278 sites of lncRNA SNHG16 gene (P > .05, Tables 3–5).
Table 3.
Correlation between tumor stages and tumor site of CRC patients with different genotypes of lncRNA SNHG16 gene rs7353 site.
| AA | AG | GG | χ2 | P | |
| Tumor stages | 4.21 | .65 | |||
| I | 21 (5.82%) | 11 (3.05%) | 1 (0.28%) | ||
| II | 89 (24.65%) | 57 (15.79%) | 18 (4.99%) | ||
| III | 67 (18.56%) | 51 (14.13%) | 11 (3.05%) | ||
| IV | 22 (6.09%) | 10 (2.77%) | 3 (0.83%) | ||
| Tumor site | 4.95 | .08 | |||
| Colon | 110 (30.47%) | 73 (20.22%) | 25 (6.93%) | ||
| Rectum | 89 (24.65%) | 56 (15.51%) | 8 (2.22%) |
Table 5.
Correlation between Tumor stages and Tumor site of CRC patients with different genotypes of lncRNA SNHG16 gene rs15278.
| AA | AG | GG | χ2 | p | |
| Tumor stages | 2.69 | 0.85 | |||
| I | 24 (6.65%) | 8 (2.22%) | 1 (0.28%) | ||
| II | 117 (32.41%) | 40 (11.08%) | 7 (1.94%) | ||
| III | 91 (25.21%) | 34 (9.42%) | 4 (11.11%) | ||
| IV | 22 (6.09%) | 10 (2.77%) | 3 (0.83%) | ||
| Tumor site | 2.93 | 0.23 | |||
| Colon | 153 (%) | 46 (12.74%) | 9 (2.49%) | ||
| Rectum | 101 (%) | 46 (12.74%) | 6 (1.66%) |
Table 4.
Correlation between tumor stages and tumor site of CRC patients with different genotypes of lncRNA SNHG16 gene rs8038 site.
| GG | GA | AA | χ2 | P | |
| Tumor stages | 9.54 | 0.15 | |||
| I | 12 (3.32%) | 20 (5.54%) | 1 (0.28%) | ||
| II | 70 (19.39%) | 75 (20.78%) | 19 (5.26%) | ||
| III | 62 (17.17%) | 54 (14.96%) | 13 (3.60%) | ||
| IV | 12 (3.32%) | 22 (6.09%) | 1 (0.28%) | ||
| Tumor site | 0.34 | 0.84 | |||
| Colon | 91 (25.21%) | 99 (27.42%) | 18 (4.99%) | ||
| Rectum | 65 (18.01%) | 72 (19.94%) | 16 (4.43%) |
3.4. Multifactor dimensionality reduction (MDR) analysis of the interaction between SNP of lncRNA SNHG16 gene and environmental factors
In this study, we used MDR to analyze the interaction between SNPs at rs7353, rs8038, and rs15278 sites of lncRNA SNHG16 gene and age, gender, BMI, smoking, drinking, and family history of subjects. Results from models under different interaction conditions are shown in Table 6. The cross-validation statistics of the interaction model including age, rs7353, rs8038, and rs152784 factors are: training balanced accuracy 0.7874, testing balanced accuracy 0.6172, cross-validation consistency 10/10, which is superior to other models. Therefore, it is regarded as the best model.
Table 6.
Information regarding the MDR fitted lncRNA SNHG16 gene SNP and environmental factor interaction model.
| Model | Training Balanced Accuracy | Testing Balanced Accuracy | Cross-validation Consistency |
| rs7353 | 0.6094 | 0.5354 | 5/10 |
| Age,rs7353 | 0.6801 | 0.5728 | 5/10 |
| Age,rs7353,rs8038 | 0.7402 | 0.595 | 9/10 |
| Age,rs7353,rs8038,rs15278 | 0.7874 | 0.6172 | 10/10 |
| Age,BMI,Smoking,rs7353,rs8038 | 0.8335 | 0.5547 | 6/10 |
| Age,Gender,Drinking,rs7353,rs8038,rs15278 | 0.8748 | 0.5282 | 3/10 |
| Age,Gender,BMI,Smoking,Drinking,rs7353,rs8038 | 0.9112 | 0.5156 | 8/10 |
| Age,Gender,BMI,Smoking,Drinking,rs7353,rs8038,rs15278 | 0.933 | 0.5183 | 9/10 |
| Age,Gender,BMI,Smoking,Drinking,Family history,rs7353,rs8038,rs15278 | 0.9435 | 0.5154 | 7/10 |
Subjects were classified according to the SNP-environment interaction combination of rs7353, rs8038, and rs15278 of the lncRNA SNHG16 gene. Estimated and tested results of the CRC risk interaction effects of training dataset statistics, testing dataset statistics, and whole dataset statistics are shown in Table 7. It is demonstrated that CRC susceptibility of the interactive combination of SNPs at age, rs7353, rs8038, and rs15278 is 13.53 times higher than the above combination (95% CI: 9.43–19.41, P < .01) (Table 7).
Table 7.
Estimation and test results of interaction between lncRNA SNHG16 gene SNP and environmental factors.
| Dataset statistics | χ2 | P | OR (95% CI) |
| Training dataset statistics | 217.86 | <.01 | 14.86 (10.07, 21.94) |
| Testing dataset statistics | 3.97 | .04 | 2.60 (1.01, 6.74) |
| Whole dataset statistics | 231.98 | <.01 | 13.53 (9.43, 19.41) |
3.5. Analysis of linkage disequilibrium
Using Haploview 4.1 software, linkage disequilibrium analysis of rs7353, rs8038, and rs15278 sites of lncRNA SNHG16 gene were performed (Fig. 2). Six haplotypes of each of rs15278, rs8038, and rs7353 were constructed, namely AGA, AGG, AAG, AAA, GGA, and GAA. Analysis revealed that AGA haplotype was significantly associated with decreased CRC risk (OR = 0.65, 95% CI: 0.48–0.88, P = .01), while AAG haplotype was significantly associated with increased CRC risk (OR = 2.00, 95% CI: 1.27–3.17, P < .01) (Table 8).
Figure 2.
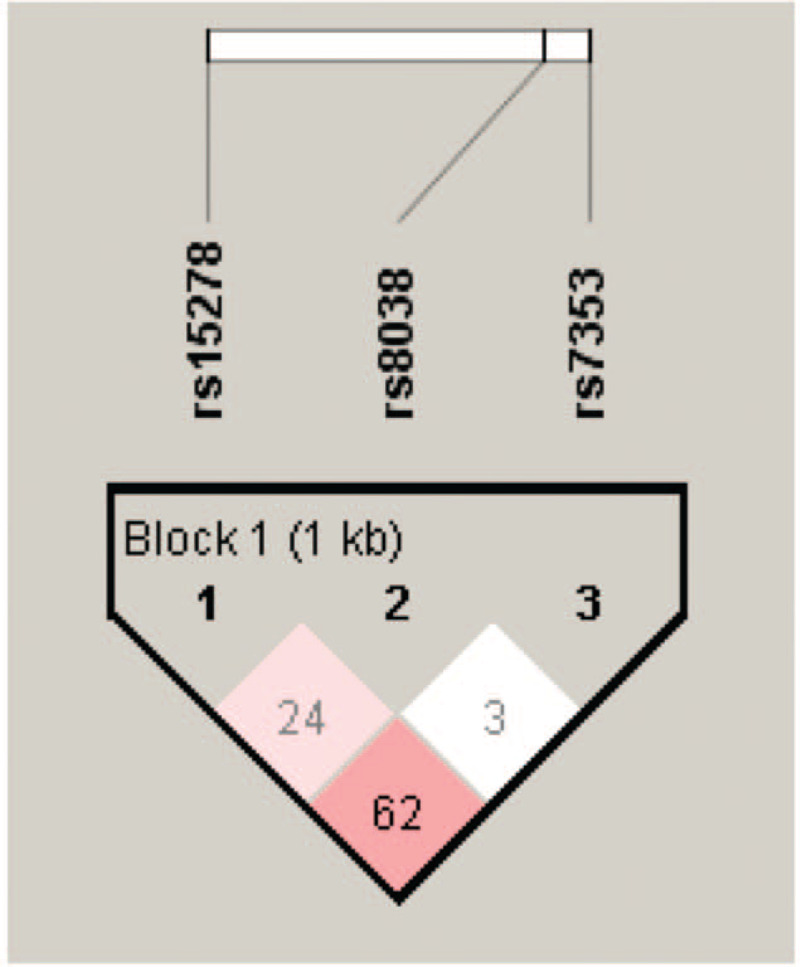
Linkage disequilibrium map of lncRNA SNHG16 gene rs7353, rs8038, rs15278 sites.
Table 8.
Analysis of the relationship between CRC risk and haplotype at rs7353, rs8038, rs15278 sites of lncRNA SNHG16 gene.
| Haplotype∗ | CRC (n = 361) | Control (n = 360) | OR (95% CI) | P-value |
| AGA | 152.0 | 148.7 | 0.65 (0.48–0.88) | 0.01 |
| AGG | 153.4 | 127.4 | 1.02 (0.75–1.38) | 0.96 |
| AAG | 18.8 | 31.8 | 2.00 (1.27–3.17) | <0.01 |
| AAA | 14.8 | 31.4 | 1.47 (0.91–2.39) | 0.15 |
| GGA | 11.6 | 11.2 | 1.09 (0.48–2.51) | 0.84 |
| GAA | 7.9 | 6.9 | 1.14 (0.41–3.19) | 0.80 |
3.6. Correlation between lncRNA SNHG16 gene polymorphism and plasma lncRNA SNHG16 level in CRC patients
We used qRT-PCR to analyze lncRNA SNHG16 expression in plasma of CRC patients. Taking the mean (3.5) as the dividing line, high and low lncRNA SNHG16 expression levels were defined as relative expression levels above and below 3.5, respectively. Analysis showed that high lncRNA SNHG16 expression was associated with tumor progression in CRC patients (c2 = 8.85, P = .03), but not with tumor site (c2 = 0.92, P = .34) (Table 9).
Table 9.
Correlation between lncRNA SNHG16 expression level and tumor stage and site in CRC patients.
| lncRNA SNHG16 High level (n = 129) | lncRNA SNHG16 Low level (n = 232) | χ2 | P | |
| Tumor stages | ||||
| I | 9 (6.98%) | 24 (10.34%) | 8.85 | 0.03 |
| II | 48 (37.21%) | 112 (48.28%) | ||
| III | 49 (37.98%) | 74 (31.90%) | ||
| IV | 23 (17.83%) | 22 (9.48%) | ||
| Tumor site | ||||
| Colon | 70 (54.26%) | 138 (59.48%) | 0.92 | 0.34 |
| Rectum | 59 (45.74%) | 94 (40.52%) |
3.7. Correlation between plasma lncRNA SNHG16 level and lncRNA SNHG16 gene polymorphism
Results of qRT-PCR showed that plasma lncRNA SNHG16 levels were higher in CRC patients than in the control group (P < .001, Fig. 3A). We used a receiver operating curve to evaluate the diagnostic value of lncRNA SNHG16 expression levels for CRC. Results showed that area under the curve was 0.7762 (95% CI: 0.7481–0.8092, P < .001), sensitivity was 67.5%, specificity was 72.58%, and the cut-off value was 2.45 (Fig. 3B). Further analysis showed that plasma level of lncRNA SNHG16 was highest in subjects with AA genotype at the rs7353 site, followed by AG genotype, and lowest in subjects with the GG genotype. analysis of variance results showed this was statistically significant (P < .01, Fig. 3C,F). There was also a statistically significant difference in plasma lncRNA SNHG16 levels among subjects with GG genotype, GA genotype, and AA genotype at the rs8038 site (P < .01, Fig. 3D,G) and among subjects with AA, AG, and GG genotypes at the rs15278 site (P < .01, Fig. 3E,H).
Figure 3.
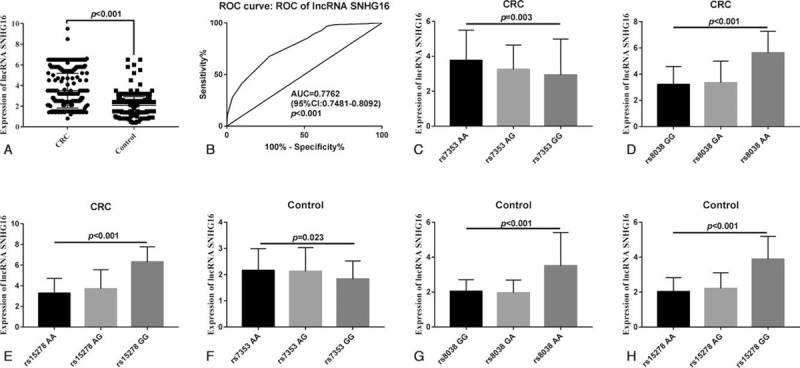
Correlation analysis of plasma lncRNA SNHG16 level and gene polymorphism. A. Comparison of plasma lncRNA SNHG16 levels between CRC patients and control group. B. Receiver operating curve evaluated the diagnostic value of different lncRNA SNHG16 expression levels for CRC. C. Comparison of plasma lncRNA SNHG16 levels in CRC patients with different genotypes at the rs7353 site. D. Comparison of plasma lncRNA SNHG16 levels in CRC patients with different genotypes at the rs8038 site. E. Comparison of plasma lncRNA SNHG16 levels in CRC patients with different genotypes at the rs15278 site. F. Comparison of plasma lncRNA SNHG16 levels in control group subjects with different genotypes at the rs7353 site. G. Comparison of plasma lncRNA SNHG16 levels in control group and CRC patients with different genotypes in the rs8038 site. analysis of variance was used to compare plasma lncRNA SNHG16 levels among subjects with different genotypes. CRC = colorectal cancer, lncRNA = long-chain non-coding RNA, SNHG16 = small nucleolar RNA host gene 16.
This shows that the A>G allele variation at rs7353 site resulted in a significant decrease in plasma lncRNA SNHG16 level in the subjects, and the rs8038 allele variation of G>A and the rs15278 allele variation of A>G allele both resulted in increased plasma lncRNA SNHG16 levels.
4. Discussion
In this study, we found that variation of the A>G allele at the rs7353 site in the lncRNA SNHG16 gene was associated with decreased CRC susceptibility. In addition, allele variation of G>A at the rs8038 site and A>G at the rs15278 site was associated with increased CRC susceptibility. MDR analysis also revealed that CRC susceptibility with a combination of age, rs7353, rs8038, and rs15278 SNPs was 13.53 times higher than that of the above combination. Furthermore, AGA haplotypes at rs15278, rs8038, and rs7353 sites were associated with decreased CRC risk, while AAG haplotype was associated with increased risk. It is therefore suggested that lncRNA SNHG16 gene polymorphism is significantly correlated with the occurrence of CRC. After analyzing plasma lncRNA SNHG16 levels, it was revealed that the A>G allele variation at the rs7353 site resulted in a significant decrease in subject plasma lncRNA SNHG16 level. The rs8038 site G>A variation and rs15278 site A>G variation both resulted in increased plasma lncRNA SNHG16 levels. Therefore, we speculate that polymorphisms of the rs7353, rs8038, and rs15278 sites of the lncRNA SNHG16 gene and interaction with age are significantly associated with CRC susceptibility. The lncRNA SNHG16 gene may be involved in CRC occurrence and development and is therefore a potential therapeutic target.
CRC is a common malignant tumor in humans, mainly caused by the combination of environmental and genetic factors.[18,19] In recent years, association studies, especially genome-wide association studies, have identified many SNPs associated with CRC. Considering most SNPs are within the intron region, their mechanism of action in Escherichia coli is currently unclear. With advancements in biological technology, it has been found that introns can participate in the regulation of gene expression through the transcription of noncoding RNA, of which lncRNA is the largest component. Thus, lncRNA has become a hot topic in research, with a focus on the relationship between lncRNA and malignant tumors. Using lncRNA as a new tumor marker and therapeutic target has shown good clinical application in the diagnosis and treatment of cancer.[20–25]
According to research, lncRNA SNHG16 is a carcinogenic lncRNA which participates in the development of a range of cancers through various mechanisms of action. For example, lncRNA SNHG16 was previously found to be upregulated in CRC and to affect genes related to lipid metabolism.[26] In vitro analysis suggests that regulation of lipid metabolism-related genes may be achieved through competitive endogenous RNA-related mechanisms.[11] However, there is no current research focusing on the correlation between lncRNA SNHG16 gene polymorphism and CRC occurrence and development.
In this study, we analyzed rs7353, rs8038, and rs15278 SNPs of the lncRNA SNHG16 gene, which is one of the innovations of this study, as no previous research has focused on these SNPs. The author speculates that these SNP sites may affect lncRNA SNHG16 structure and/or expression. Prediction results from the lncRNASNP2 tool show that the rs7353, rs8038, and rs15278 sites are either located on the miRNA binding site of lncRNA or change the structure of lncRNA. From the results of this study, SNPs at rs7353, rs8038, and rs15278 sites all affect the expression level of lncRNA SNHG16. The reason for this may be that SNPs changed the combination of lncRNA and miRNA, resulting in a change in lncRNA SNHG16 plasma level. The specific molecular mechanism underlying this is still unclear and is therefore a worthy direction for further study.
To evaluate the reliability of this study, we calculated the minimum required sample sizes based on rs7353, rs8038, and rs15278 data. The results showed that the minimum sample size required for CRC patients and control group were 142 cases, 142 cases; 207 cases, 207 cases; 232 cases, 232 cases. In this study, 361 CRC patients and 360 control subjects were included, which was significantly higher than the minimum sample size required for each, indicating that the results in this study are highly reliable.
MDR analysis of the correlation between susceptibility genes and the interaction between susceptibility genes and environmental factors and tumor susceptibility is a current direction of gene polymorphism research.[27,28] In this study, MDR analysis revealed that the combination of SNPs at age, rs7353, rs8038, and rs15278 sites was associated with increased risk of CRC susceptibility, further proving that CRC occurrence and development are determined by genetic and environmental factors.
There are still some limitations to this study. Firstly, it remains to be studied whether SNPs at rs7353, rs8038, and rs15278 affect targeted binding between lncRNA SNHG16 and miRNAs, and which types of miRNAs in particular. Furthermore, the mechanism of correlation between rs7353, rs8038, and rs15278 SNPs and lncRNA SNHG16 expression levels remains to be studied.
5. Conclusion
From this study, we found that rs7353, rs8038, and rs15278 SNPs of the lncRNA SNHG16 gene were significantly associated with CRC susceptibility.
Author contributions
Conceptualization: Li Zhou, Xuewei Gu.
Data curation: Li Zhou, Yuefeng Zhang, Xuewei Gu.
Formal analysis: Li Zhou, Yuefeng Zhang, Jianjiang Jin.
Funding acquisition: Xuewei Gu.
Investigation: Li Zhou, Jianjiang Jin.
Methodology: Li Zhou.
Project administration: Yuefeng Zhang.
Software: Li Zhou, Yuefeng Zhang, Jianjiang Jin.
Supervision: Xuewei Gu.
Validation: Jianjiang Jin.
Visualization: Yuefeng Zhang.
Writing – original draft: Li Zhou.
Writing – review & editing: Xuewei Gu.
Footnotes
Abbreviations: BMI = body mass index, CI = confidence interval, CRC = colorectal cancer, lncRNA = long-chain non-coding RNA, OR = Odds ratio, qRT-PCR = real-time fluorescence quantitative PCR, SNHG16 = small nucleolar RNA host gene 16.
How to cite this article: Zhou L, Zhang Y, Jin J, Gu X. Correlation between lncRNA SNHG16 gene polymorphism and its interaction with environmental factors and susceptibility to colorectal cancer. Medicine. 2020;99:48(e23372).
This work was supported by the grant Construction of Key Disciplines of Medical and Health Research in Yuhang District, Hangzhou (2016004).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
BMI = body mass index, CRC = colorectal cancer, SD = standard deviation.
CI = confidence interval, CRC = colorectal cancer, HWE = Hardy–Weinberg equilibrium, OR = odds ratio CRC = colorectal cancer, lncRNA = long-chain non-coding RNA, SNHG16 = small nucleolar RNA host gene 16..
CRC = colorectal cancer, lncRNA = long-chain non-coding RNA, SNHG16 = small nucleolar RNA host gene 16.
CRC = colorectal cancer, lncRNA = long-chain non-coding RNA, SNHG16 = small nucleolar RNA host gene 16.
CRC = colorectal cancer, lncRNA = long-chain non-coding RNA, SNHG16 = small nucleolar RNA host gene 16.
BMI = body mass index, lncRNA = long-chain non-coding RNA, MDR = multifactor dimensionality reduction, SNP = single nucleotide polymorphism.
CI = confidence interval, OR = odds ratio, lncRNA = long-chain non-coding RNA, SNHG16 = small nucleolar RNA host gene 16, SNP = single nucleotide polymorphism.
rs15278, rs8038, rs7353 site Haplotype.
CI = confidence interval, CRC = colorectal cancer, OR = odds ratio, SNHG16 = small nucleolar RNA host gene 16
CRC = colorectal cancer, SNHG16, Small nucleolar RNA host gene 16.
References
- [1].Aykut B, Ochs M, Radhakrishnan P, et al. EMX2 gene expression predicts liver metastasis and survival in colorectal cancer. BMC Cancer 2017;17:555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kuipers EJ, Grady WM, Lieberman D, et al. Colorectal cancer. Nat Rev Dis Primers 2015;1:15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Duraes RO, Berardinelli GN, da Costa AM, et al. Role of genetic ancestry in 1,002 Brazilian colorectal cancer patients from Barretos Cancer Hospital. Front Oncol 2020;10:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jedi M, Young GP, Pedersen SK, et al. Methylation and gene expression of BCAT1 and IKZF1 in colorectal cancer tissues. Clin Med Insights Oncol 2018;12:1179554918775064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang M, Miao F, Huang R, et al. RHBDD1 promotes colorectal cancer metastasis through the Wnt signaling pathway and its downstream target ZEB1. J Exp Clin Cancer Res 2018;37:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li B, Wang Z, Xie JM, et al. TIGAR knockdown enhanced the anticancer effect of aescin via regulating autophagy and apoptosis in colorectal cancer cells. Acta Pharmacol Sin 2019;40:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Okugawa Y, Toiyama Y, Toden S, et al. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut 2017;66:107–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Slattery ML, Herrick JS, Mullany LE, et al. The co-regulatory networks of tumor suppressor genes, oncogenes, and miRNAs in colorectal cancer. Genes Chromosomes Cancer 2017;56:769–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Amirkhah R, Farazmand A, Irfan-Maqsood M, et al. The role of microRNAs in the resistance to colorectal cancer treatments. Cell Mol Biol (Noisy-le-grand) 2015;61:17–23. [PubMed] [Google Scholar]
- [10].Chen DL, Chen LZ, Lu YX, et al. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis 2017;8:e3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Christensen LL, True K, Hamilton MP, et al. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol Oncol 2016;10:1266–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yan L, Yao J, Qiu J, et al. miRNA-495 suppresses proliferation and migration of colorectal cancer cells by targeting FAM83D. Biomed Pharmacother 2017;96:974–81. [DOI] [PubMed] [Google Scholar]
- [13].Deng B, Wang B, Fang J, et al. MiRNA-203 suppresses cell proliferation, migration and invasion in colorectal cancer via targeting of EIF5A2. Sci Rep 2016;6:28301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sam S, Sam MR, Esmaeillou M, et al. Effective targeting survivin, caspase-3 and MicroRNA-16-1 expression by Methyl-3-pentyl-6-methoxyprodigiosene triggers apoptosis in colorectal cancer stem-like cells. Pathol Oncol Res 2016;22:715–23. [DOI] [PubMed] [Google Scholar]
- [15].Yu M, Ohira M, Li Y, et al. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int J Oncol 2009;34:931–8. [DOI] [PubMed] [Google Scholar]
- [16].Duan W, Du L, Jiang X, et al. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget 2016;7:78850–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cai C, Huo Q, Wang X, et al. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun 2017;485:272–8. [DOI] [PubMed] [Google Scholar]
- [18].Jeon J, Du M, Schoen RE, et al. Determining risk of colorectal cancer and starting age of screening based on lifestyle, environmental, and genetic factors. Gastroenterology 2018;154:2152–64. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yang T, Li X, Farrington SM, et al. A systematic analysis of interactions between environmental risk factors and genetic variation in susceptibility to colorectal cancer. Cancer Epidemiol Biomarkers Prev 2020;29:1145–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47–62. [DOI] [PubMed] [Google Scholar]
- [21].Ling H, Vincent K, Pichler M, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 2015;34:5003–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Abdelmohsen K, Panda A, Kang MJ, et al. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell 2013;12:890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Anderson DM, Anderson KM, Chang CL, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015;160:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wei JT, Feng Z, Partin AW, et al. Can urinary PCA3 supplement PSA in the early detection of prostate cancer? J Clin Oncol 2014;32:4066–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yan X, Hu Z, Feng Y, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 2015;28:529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yang BY, Meng Q, Sun Y, et al. Long non-coding RNA SNHG16 contributes to glioma malignancy by competitively binding miR-20a-5p with E2F1. J Biol Regul Homeost Agents 2018;32:251–61. [PubMed] [Google Scholar]
- [27].Singh SA, Ghosh SK. Polymorphisms of XRCC1 and XRCC2 DNA repair genes and Interaction with environmental factors influence the risk of nasopharyngeal carcinoma in Northeast India. Asian Pac J Cancer Prev 2016;17:2811–9. [PubMed] [Google Scholar]
- [28].Fu D, Li P, Cheng W, et al. Impact of vascular endothelial growth factor gene-gene and gene-smoking interaction and haplotype combination on bladder cancer risk in Chinese population. Oncotarget 2017;8:22927–35. [DOI] [PMC free article] [PubMed] [Google Scholar]


