Abstract
Tregs require Foxp3 expression and induction of a specific DNA hypomethylation signature during development, after which Tregs persist as a self-renewing population that regulates immune system activation. Whether maintenance DNA methylation is required for Treg lineage development and stability and how methylation patterns are maintained during lineage self-renewal remain unclear. Here, we demonstrate that the epigenetic regulator ubiquitin-like with plant homeodomain and RING finger domains 1 (Uhrf1) is essential for maintenance of methyl-DNA marks that stabilize Treg cellular identity by repressing effector T cell transcriptional programs. Constitutive and induced deficiency of Uhrf1 within Foxp3+ cells resulted in global yet nonuniform loss of DNA methylation, derepression of inflammatory transcriptional programs, destabilization of the Treg lineage, and spontaneous inflammation. These findings support a paradigm in which maintenance DNA methylation is required in distinct regions of the Treg genome for both lineage establishment and stability of identity and suppressive function.
Keywords: Autoimmunity, Immunology
Keywords: Autoimmune diseases, Epigenetics, T cells
Maintenance DNA methylation at inflammatory gene loci is necessary to stabilize regulatory T cell identity and suppressive function during both development and lineage self-renewal.
Introduction
CD4+Foxp3+ Tregs prevent catastrophic inflammation by suppressing immune system activation and promoting self tolerance (1–3). The Foxp3 transcription factor serves as the Treg lineage–specifying marker, and Tregs require constitutive Foxp3 expression to maintain their identity and suppressive function. Mice lacking Tregs owing to a mutation in the Foxp3 gene exhibit the scurfy phenotype, succumbing to multiorgan lymphoproliferative inflammation approximately 4 weeks after birth (4, 5). Humans with FOXP3 mutations develop similar endocrine and enteral inflammation as part of immunodysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome (6). Treg dysfunction contributes to the pathogenesis of numerous autoimmune conditions, including systemic lupus erythematosus (7–11) and systemic sclerosis (12). In contrast, modern cancer immunotherapy blocks Treg-suppressive function to disinhibit effector T cell–mediated killing of malignant cells (13–15). Thus, mechanisms involved with both development and stability of the Treg lineage represent targets for therapies aimed at amelioration of autoimmune and malignant diseases (3, 16).
Tregs develop from CD4–single-positive autoreactive thymocytes that receive signals via CD28/Lck, the IL-2/CD25/Stat5 axis, and T cell receptor engagement by MHC self-peptide complexes (17). These events induce Foxp3 expression and independently establish a Treg-specific cytosine-phospho-guanine (CpG) hypomethylation pattern at certain genomic loci, including the Foxp3 locus and other loci whose gene products are important for Treg lineage identity and suppressive function (18–20). Consistent with the Treg requirement for CpG hypomethylation, pharmacologic inhibition of DNA methyltransferase activity is sufficient to induce Foxp3 expression in mature conventional CD4+ T cells and to potentiate Treg-suppressive function in multiple models of inflammation (21–24). In contrast, conditional constitutive genetic deletion of the DNA methyltransferase Dnmt1 but not Dnmt3a in developing Tregs diminishes their numbers and suppressive function (25). Treg-specific Dnmt1 deficiency decreases global methyl-CpG content while maintaining the Treg-specific CpG hypomethylation pattern at Foxp3. In the periphery and in vitro, TGF-β induces Foxp3 expression in naive CD4+ T cells to generate induced Treg (iTreg) cells, although these cells lack the stabilizing DNA hypomethylation landscape that defines thymus-derived (natural) Tregs (18, 26–28). A minor population of CD4+ T cells can promiscuously and transiently express Foxp3, a phenomenon that has been linked to a population of ex-Foxp3 cells that does not represent epigenetic reprogramming of mature Tregs (29, 30). Conversely, some thymic emigrants referred to as potential Tregs may harbor the Treg-specific CpG hypomethylation pattern, but fail to express Foxp3 (31). In adult mice, lineage-tracing studies determined that self-renewal of mature thymus-derived Tregs maintains the lineage even under inflammatory conditions (32). This self-renewal model presupposes that DNA methylation patterns are passed from parent to daughter cell during the lineage self-renewal process. Nevertheless, underlying mechanisms that regulate the stability of the mature Treg pool remain unclear, particularly the role of maintenance DNA methylation in lineage stability.
The epigenetic regulator ubiquitin-like with plant homeodomain and RING finger domains 1 (Uhrf1; also known as Np95 in mice and ICBP90 in humans) serves as a nonredundant adapter protein for Dnmt1 during S phase, recruiting Dnmt1 to hemimethylated DNA to ensure maintenance of CpG methylation patterns as part of multiprotein gene-repressive complexes (33–36). Uhrf1 also regulates de novo DNA methylation via recruitment of Dnmt3a and Dnmt3b to chromatin (37, 38). Global homozygous loss of Uhrf1 results in embryologic lethality (34), phenocopying of homozygous loss of Dnmt1 (39). Conditional loss of Uhrf1 in T cells using a Cd4-Cre driver leads to failure of colonic Treg proliferation and maturation in response to commensal bacterial colonization (40). Nevertheless, Uhrf1-deficient naive T cells generated using the Cd4-Cre system are able to suppress experimental colitis due to TGF-β–mediated conversion of these cells into an iTreg state (41). The specific role of Uhrf1-mediated maintenance of DNA methylation in Foxp3+ Treg development and stability of suppressive function remains unknown. Because of Uhrf1’s nonredundant role in recruiting Dnmt1 to hemimethylated DNA, we hypothesized that constitutive Treg-specific Uhrf1 deficiency would phenocopy Treg-specific Dnmt1 deficiency and result in failure of thymic Treg development. Additionally, based on pharmacologic studies showing augmentation of Foxp3 expression and Treg-suppressive function upon inhibition of DNA methyltransferase activity when administered to mature cell populations, we initially hypothesized that induction of Uhrf1 deficiency in mature Tregs would augment their suppressive function. Using conditional and chimeric knockout systems, we determined that Uhrf1-mediated DNA methylation is indeed required for thymic Treg development. In surprising contrast with our second hypothesis, we used an inducible conditional Uhrf1-knockout system to show that maintenance DNA methylation at inflammatory gene loci is essential for stabilizing the identity and suppressive function of mature Tregs.
Results
Treg-specific deletion of Uhrf1 results in a lethal inflammatory disorder.
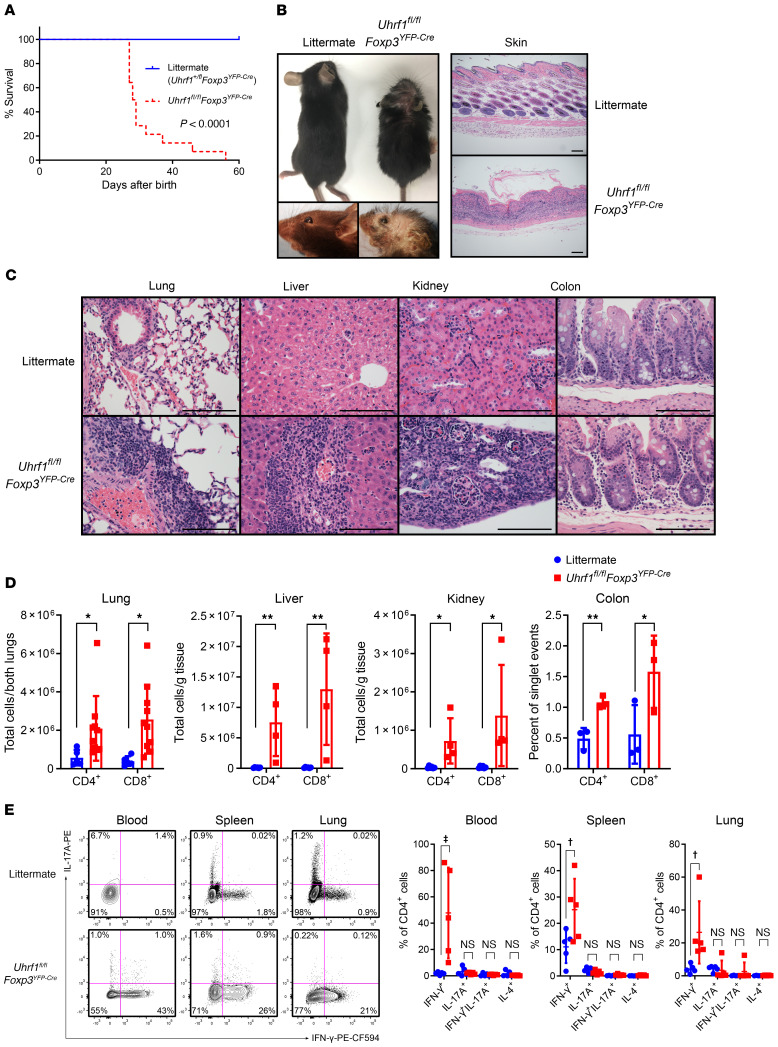
To test the necessity of maintenance DNA methylation in Treg development and function, we generated Treg-specific Uhrf1-deficient mice by crossing mice bearing loxP sequences at the Uhrf1 gene locus (Uhrf1fl/fl) (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI137712DS1) with mice expressing yellow fluorescent protein (YFP) and Cre recombinase driven by the Foxp3 promoter (Foxp3YFP-Cre) (42). Crosses of male Uhrf1+/flFoxp3YFP-Cre/Y mice with female Uhrf1fl/flFoxp3+/YFP-Cre mice generated F1 pups in statistically Mendelian ratios, although male Uhrf1fl/flFoxp3YFP-Cre/Y offspring were underrepresented at the time of genotyping (approximately 3 weeks of age) (Supplemental Figure 1B). Uhrf1fl/flFoxp3YFP-Cre mice appeared normal at birth, but then exhibited spontaneous mortality, with a median survival of 28.5 days (Figure 1A). Beginning at approximately 3 weeks of age, Uhrf1fl/flFoxp3YFP-Cre mice were smaller than littermate control mice (Uhrf1+/flFoxp3YFP-Cre) and displayed scaly skin with cratering and loss of fur (Supplemental Figure 1C and Figure 1B). Histological examination of the skin revealed infiltration of the dermis and subdermis by inflammatory cells, including lymphocytes and monocytes. Similarly to the skin, nearly every internal organ exhibited a mixed cellular infiltrate consisting predominantly of lymphocytes, but also monocytes and neutrophils (Figure 1C). This striking lymphocytic infiltrate consisted of both CD4+ and CD8+ T cells (Figure 1D), suggesting a lymphocytic inflammatory disorder reminiscent of the scurfy phenotype (4, 5).
Figure 1. Treg-specific Uhrf1-deficient mice spontaneously develop a fatal inflammatory disorder.
(A) Survival curves of littermate control (Uhrf1+/flFoxp3YFP-Cre, n = 9) and Uhrf1fl/flFoxp3YFP-Cre (n = 14) mice compared using the log-rank (Mantel-Cox) test. (B) Gross photographs of 3- to 4-week-old littermate and Uhrf1fl/flFoxp3YFP-Cre mice along with photomicrographs of skin. Scale bars: 100 μm. (C) Photomicrographic survey of organ pathology. Scale bars: 100 μm. (D) CD3ε+ T cell subsets in selected organs. For lung, n = 5 (littermate) and n = 10 (Uhrf1fl/flFoxp3YFP-Cre); for liver, n = 6 (littermate) and n = 4 (Uhrf1fl/flFoxp3YFP-Cre); for kidney, n = 6 (littermate) and n = 4 (Uhrf1fl/flFoxp3YFP-Cre); for colon, n = 3 (littermate and Uhrf1fl/flFoxp3YFP-Cre). (E) Cytokine profile of splenic CD3ε+CD4+ T cells following ex vivo stimulation with phorbol 12-myristate 13-acetate and ionomycin for 4 hours in the presence of brefeldin A. Representative contour plots and summary data are shown. n = 5 per group. Summary plots show all data points with mean and SD. *q < 0.05; **q < 0.01; †q < 0.001; ‡q < 0.0001; NS, not significant by the 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli with Q = 5%; exact q values are in Supplemental Data. See Supplemental Table 3 for fluorochrome abbreviations.
Treg-specific Uhrf1-deficient mice showed other signs of lymphocyte-driven immune system activation, including splenomegaly and splenic structural disarray characterized by architectural disruption and lymphoid hyperplasia (Supplemental Figure 1, D–H). CD3ε+CD4+ T cells in the spleen displayed an activated profile, exhibiting an increased frequency and total number of CD44hiCD62Llo effector T cells (Supplemental Figure 1I). Other secondary lymphoid organs also exhibited evidence of immune system activation, including replacement of lymph node germinal centers with a mixed cellular infiltrate and lymphocyte depletion of the thymus, which was also atrophic (Supplemental Figure 1, J–L). To better characterize this severe inflammation, we performed intracellular cytokine profiling of CD3ε+CD4+ T cells from the blood, spleen, and lung. These measurements revealed skewing toward a Th1 profile in Uhrf1fl/flFoxp3YFP-Cre mice, exemplified by a significant increase in the frequency of cells producing IFN-γ (Figure 1E). No bacteria were identified in the blood of 3- to 4-week-old mice by routine culture on Luria-Bertani (LB) agar. Collectively, Treg-specific deletion of Uhrf1 resulted in severe Th1-skewed inflammation, lymphocytic infiltration of essential organs, thymic atrophy, and mortality at approximately 3 to 4 weeks of age, suggesting insufficiency or dysfunction of Tregs as a result of Uhrf1 deficiency.
Uhrf1 deficiency results in failure of Tregs to persist after Foxp3 induction in the thymus.
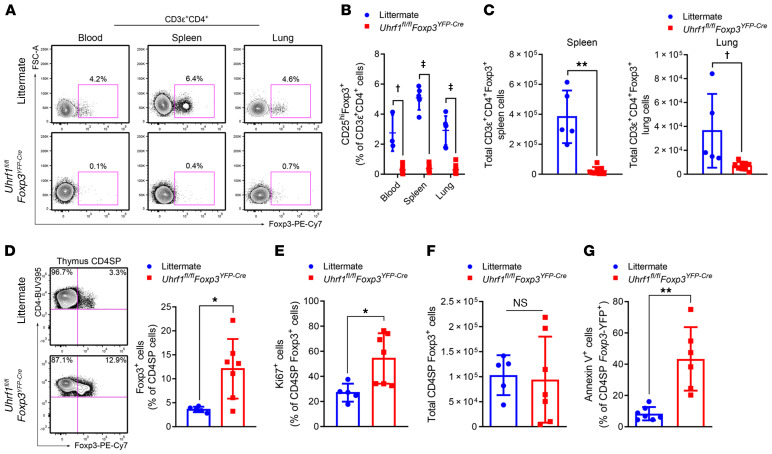
We next examined the CD3ε+CD4+Foxp3+ T cell compartment of Uhrf1fl/flFoxp3YFP-Cre mice to evaluate the effect of Uhrf1 deficiency on Tregs. Cre-mediated excision effectively deleted Uhrf1 and revealed the expected pattern of Uhrf1 genotypes within the sorted Foxp3-YFP– and Foxp3-YFP+ populations of CD3ε+CD4+ T cells from littermate control mice (Uhrf1+/flFoxp3YFP-Cre), confirming the Foxp3+ cell–exclusive nature of the Foxp3YFP-Cre construct when paired with a Uhrf1fl allele (Supplemental Figure 2A). At 3 to 4 weeks of age, Uhrf1fl/flFoxp3YFP-Cre mice exhibited a profound deficiency of Tregs, with Foxp3+ cells constituting less than 1% of CD3ε+CD4+ T cells in the blood, spleen, and lung (Figure 2A). Examination of Foxp3+ Tregs marked by high expression of CD25 (IL-2Rα) as a fraction of CD3ε+CD4+ T cells further highlighted the Treg deficiency in Uhrf1fl/flFoxp3YFP-Cre mice (Figure 2B). Total numbers of Foxp3+ Tregs were likewise diminished in the spleen and lung, representative of secondary lymphoid and parenchymal organs, respectively (Figure 2C).
Figure 2. Tregs are reduced in the periphery of 3- to 4-week-old Treg-specific Uhrf1-deficient mice.
(A) Foxp3+ cells as a frequency of CD3ε+CD4+ cells in blood, spleen, and lung shown as representative flow cytometry contour plots. (B) CD25hiFoxp3+ cells expressed as a percentage of CD3ε+CD4+ cells for blood, spleen, and lung. For blood, n = 3 (Uhrf1+/flFoxp3YFP-Cre littermate) and n = 7 (Uhrf1fl/flFoxp3YFP-Cre); for spleen, n = 5 (littermate) and n = 9 (Uhrf1fl/flFoxp3YFP-Cre); for lung, n = 6 (littermate) and n = 10 (Uhrf1fl/flFoxp3YFP-Cre). (C) Total spleen and lung CD3ε+CD4+Foxp3+ cells. For spleen, n = 5 (littermate) and 8 (Uhrf1fl/flFoxp3YFP-Cre); for lung, n = 5 (littermate) and 10 (Uhrf1fl/flFoxp3YFP-Cre). (D) Thymic Foxp3+ cell frequency. Representative flow cytometry contour plots of CD4–single-positive (SP) thymocytes from littermate and Uhrf1fl/flFoxp3YFP-Cre mice. Foxp3+ cells are shown as a percentage of the CD4SP population. n = 5 (littermate) and n = 7 (Uhrf1fl/flFoxp3YFP-Cre). (E) Proliferating (Ki67+) cells as a percentage of CD4SP Foxp3+ thymocytes. n = 5 (littermate) and 7 (Uhrf1fl/flFoxp3YFP-Cre). (F) Total thymic Tregs. n = 5 (littermate) and 7 (Uhrf1fl/flFoxp3YFP-Cre). (G) Apoptotic (annexin V+) cells as a percentage of CD4SP Foxp3+ thymocytes. n = 7 (littermate) and 6 (Uhrf1fl/flFoxp3YFP-Cre). Summary plots show all data points with mean and SD. *P < 0.05; **P < 0.01; †q or P < 0.001; ‡q < 0.0001, NS, not significant by the 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli with Q = 5% (B) or Mann-Whitney U test (C–G); exact q and P values are in Supplemental Data. FSC-A, forward scatter area. See Supplemental Table 3 for fluorochrome abbreviations.
The Foxp3YFP-Cre construct is first expressed at sustained levels at the thymic Treg stage of development (20). Accordingly, we examined the fraction of Foxp3+ cells among CD4–single-positive cells in the thymus and found that thymic Tregs were increased in frequency among Uhrf1fl/flFoxp3YFP-Cre mice (Figure 2D) and displayed increased proliferation (Figure 2E). Nevertheless, total thymic Treg numbers were similar between littermates and Uhrf1fl/flFoxp3YFP-Cre mice (Figure 2F), likely because of the severe thymic atrophy observed among Uhrf1fl/flFoxp3YFP-Cre animals (see Supplemental Figure 1, J–L). This observation excludes the possibility of thymic accumulation or trapping of Tregs as a cause of their peripheral deficiency. To examine the possibility that loss of Uhrf1 at the time of Foxp3 expression triggers apoptosis, we measured annexin V staining on thymic Tregs and found an increased frequency of annexin V+ thymic Tregs in Uhrf1fl/flFoxp3YFP-Cre mice compared with littermate control mice (Figure 2G). We then examined the genotype of the sorted Foxp3-YFP– and Foxp3-YFP+ populations from Uhrf1fl/flFoxp3YFP-Cre mice. The Foxp3-YFP+ population contained a Uhrf1-deleted sequence, confirming Cre-mediated deletion of Uhrf1 and excluding the possibility that the small population of Foxp3-YFP+ cells escaped Cre-mediated recombination (Supplemental Figure 2B). To our surprise, however, the Foxp3-YFP– population contained a minor Uhrf1-loxP signal and a dominant Uhrf1-deleted sequence, suggesting a population of ex-Foxp3 cells that once expressed the Foxp3YFP-Cre construct, but lost Foxp3 expression (and thus YFP+ status) shortly after the thymic Foxp3+ Treg stage of development. In summary, loss of Uhrf1 at the thymic Treg stage of development resulted in failure of Tregs to develop into a robust population, indicating the necessity of Uhrf1 in developmental stabilization of the Treg lineage.
Uhrf1 deficiency leads to a cell-autonomous and inflammation-independent decrease in Tregs without altering Foxp3 expression.
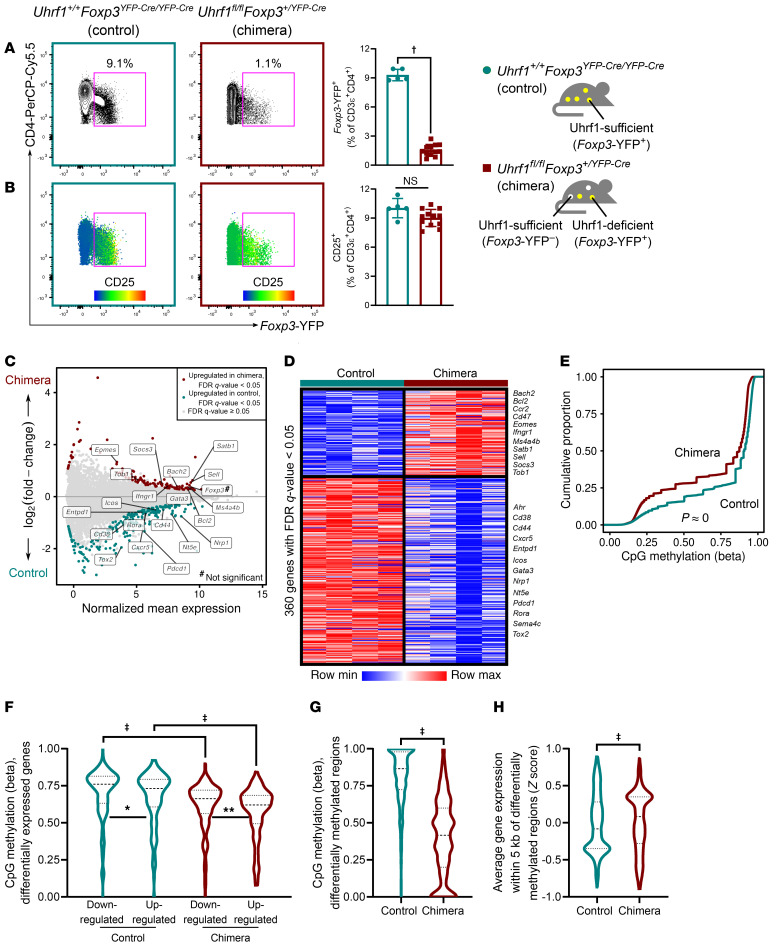
To establish that the lack of Tregs in Treg-specific Uhrf1-deficient animals is a cell-autonomous effect of Uhrf1 deficiency and not due to the profound inflammation observed in Uhrf1fl/flFoxp3YFP-Cre mice, we generated Uhrf1 chimeric knockout animals — female mice that are homozygous for the Uhrf1fl allele and heterozygous for the X-linked Foxp3YFP-Cre allele. Following random inactivation of the X chromosome in these mice, the Treg compartment contained a mix of Uhrf1-sufficient Foxp3-YFP– and Uhrf1-deficient Foxp3-YFP+ cells. Uhrf1 chimeric knockout mice (Uhrf1fl/flFoxp3+/YFP-Cre) displayed a significant decrease in Foxp3-YFP+ cells compared with control mice (Uhrf1+/+Foxp3YFP-Cre/YFP-Cre) (Figure 3A). Consistent with the chimeric construct, CD25 expression was distributed throughout the Foxp3-YFP– and small Foxp3-YFP+ compartments of Uhrf1 chimeric knockout animals but cosegregated with Foxp3-YFP expression in control mice (Figure 3B). Despite the dearth of Foxp3-YFP+ cells in Uhrf1 chimeric knockout animals, average per-cell Foxp3 protein expression was similar to that in control mice (Supplemental Figure 3A). Uhrf1 chimeric knockout mice appeared grossly normal, did not experience overt spontaneous inflammation, and did not display increased spleen mass, cellularity, or frequency of CD44hiCD62Llo effector T cells (Supplemental Figure 3, B–D). Taken together, these results demonstrate that Treg-specific Uhrf1 deficiency causes an inflammation-independent lack of Tregs in a cell-autonomous fashion. These data also suggest that Uhrf1-sufficient cells are able to fill the Treg niche in the presence of a population of Uhrf1-deficient cells.
Figure 3. Treg-specific Uhrf1 chimeric knockout mice reveal Treg-autonomous and inflammation-independent effects of Uhrf1 deficiency.
(A) Representative contour plots and quantification showing Foxp3-YFP+ cells as frequency of splenic CD3ε+CD4+ cells from 8-week-old female Uhrf1+/+ Foxp3YFP-Cre/YFP-Cre (control) and Uhrf1fl/flFoxp3+/YFP-Cre (chimeric) mice (schematic). (B) CD25 heatmap overlaid on populations shown in A; associated graph quantifies CD25+ cells as percentages of splenic CD3ε+CD4+ cells. (C) MA plot comparing gene expression of splenic CD3ε+CD4+CD25hiFoxp3-YFP+ cells from control mice (Uhrf1-sufficient cells) and Uhrf1 chimeric knockout mice (Uhrf1-deficient cells). (D) K-means clustering of differentially expressed genes scaled as Z score across rows. (E) Cumulative distribution function plot of 1.4 × 106 well-observed CpGs expressed as β scores, with 0 representing unmethylated and 1 representing fully methylated; a shift in the cumulative distribution function up and to the left represents relative hypomethylation. (F) CpG methylation at the loci of differentially expressed genes (defined as the gene body ± 2 kb). (G) CpG methylation at 1335 DMRs. (H) Average gene expression at 567 gene loci near DMRs. n = 5 (control) and 12 (chimera) for A and B and 4 mice per group for C–H. Summary plots show all data points with mean and SD; violin plots show median and quartiles. *q < 0.05; **q < 0.01; †P < 0.001; ‡q or P < 0.0001; NS, not significant by Mann-Whitney U test (A and B), a mixed-effects analysis with the 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli with Q = 5% (F), or Kolmogorov-Smirnov test for cumulative distributions (G and H); exact or asymptotic P and q values are in Supplemental Data. The asymptotic P value resulting from a Kolmogorov-Smirnov test for cumulative distributions is shown in E. See Supplemental Table 3 for fluorochrome abbreviations.
Foxp3+ cells from Uhrf1 chimeric knockout mice exhibit downregulation of genes associated with Treg-suppressive function and loss of DNA methylation while preserving Foxp3 locus expression and methylation patterning.
We took advantage of the Uhrf1 chimeric knockout system to investigate inflammation-independent mechanisms of impaired suppressive function in Uhrf1-deficient Tregs. Accordingly, we performed transcriptional profiling of CD3ε+CD4+CD25hiFoxp3-YFP+ cells isolated from control and Uhrf1 chimeric knockout mice. RNA-Seq followed by an unsupervised analysis revealed differential expression of 360 genes with a FDR q value of less than 0.05 (Figure 3, C and D). Genes that were downregulated in cells from chimeric knockout animals included those classically associated with Treg activation and suppressive function: Ahr, Cd38, Cd44, Entpd1, Icos, Gata3, Nrp1, Nt5e, Pdcd1, Sema4c, and Tox2. Genes upregulated in knockout cells included Bach2, Bcl2, Ccr2, Cd47, Eomes, Ifngr1, Ms4a4b, Satb1, Sell, Socs3, and Tob1 — many of which are associated with impaired Treg-suppressive function and gain of effector T cell function. Importantly, Foxp3 expression was not significantly affected by Uhrf1 deficiency (log2[fold-change] = 0.39, FDR q value = 0.06). Gene Set Enrichment Analysis using a list of genes canonically associated with Treg identity (43) revealed significant negative enrichment within cells from chimeric knockout mice (Supplemental Figure 4A). We noted that cells from chimeric knockout mice upregulated genes such as Eomes and Socs3 while downregulating Nrp1, a pattern associated with T cell exhaustion. Accordingly, we performed a simultaneous Gene Set Enrichment Analysis using a gene set characterizing the exhausted CD4+ T cell signature (44). To our surprise, there was significant negative enrichment for the exhausted signature (Supplemental Figure 4B). These findings in noninflamed Uhrf1 chimeric knockout mice confirm that Uhrf1 deficiency results in loss of the canonical Treg transcriptional program without significantly affecting levels of Foxp3 expression in Foxp3+ cells or inducing an exhaustion signature.
Genome-wide CpG methylation profiling with modified reduced representation bisulfite sequencing (mRRBS) revealed a striking global hypomethylation pattern within CD3ε+CD4+CD25hiFoxp3-YFP+ cells from Uhrf1 chimeric knockout mice compared with control mice (Figure 3E and Supplemental Figure 4C). CpG methylation was specifically reduced at the loci of differentially expressed genes in cells from chimeric knockout mice, particularly at upregulated loci (Figure 3F). We next used an unsupervised procedure to define 1,335 differentially methylated regions (DMRs); these DMRs were significantly hypomethylated in cells from chimeric knockout mice compared with control mice (Figure 3G). Gene expression was increased at loci near these DMRs (Figure 3H), consistent with loss of Uhrf1-mediated DNA methylation leading to derepression of these loci. Importantly, CpG methylation across Treg-specific super-enhancer elements and the super-enhancer at the Foxp3 locus, which contains the canonical Foxp3 conserved noncoding sequences (20), was unaffected by Uhrf1 deficiency (Supplemental Figure 4, D and E). Examination of gene loci that displayed hypomethylation and upregulation in knockout cells compared with control cells found genes associated with impaired Treg-suppressive function and gain of effector T cell function: Tob1 (45) and the promoter region of Ms4a4b (46) (Supplemental Figure 4, F and G). Altogether, deficiency of Uhrf1 in Tregs resulted in loss of the Treg lineage–defining transcriptional signature, disruption of DNA methylation patterning, and derepression of effector programs.
Uhrf1 is required for stability of Treg-suppressive function.
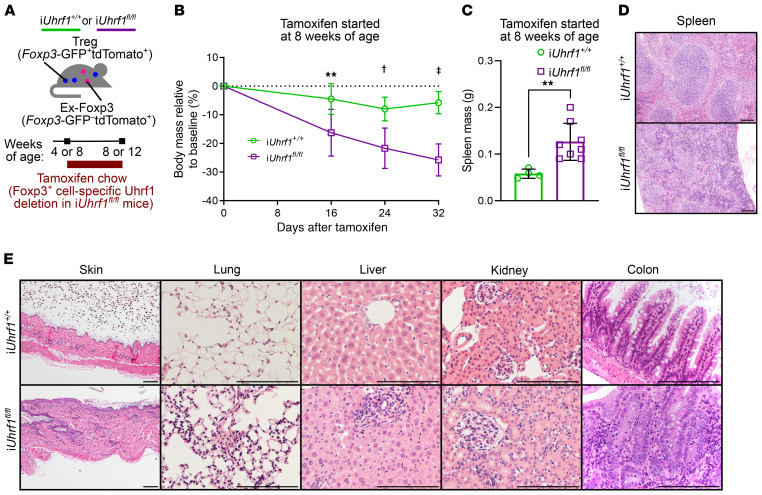
Thymus-derived Tregs self-renew to maintain the Treg lineage (32). Accordingly, we sought to understand how Treg-specific loss of Uhrf1 affects the stability of both the developing and mature Treg pools. To that end, we generated inducible, Treg-specific, Foxp3 lineage–traceable, Uhrf1-deficient mice (Figure 4A). These mice — which we refer to as iUhrf1fl/fl mice — are homozygous for the Uhrf1fl allele, an inducible Foxp3-Cre driver with a GFP label (Foxp3eGFP-CreERT2) and a loxP-flanked stop codon upstream of the red florescent protein tdTomato driven by a CAG promoter at the open ROSA26 locus (ROSA26SorCAG-tdTomato). We fed tamoxifen chow to juvenile 4-week-old iUhrf1fl/fl mice, which were still developing their Treg pool, as well as adult 8-week-old iUhrf1fl/fl mice that had established a mature Treg lineage with minimal input from thymic emigrants (32). Following tamoxifen administration, both 4- and 8-week-old iUhrf1fl/fl mice lost weight compared with iUhrf1+/+ control mice (Supplemental Figure 5A and Figure 4B), suggesting the onset of inflammation in both juvenile and adult iUhrf1fl/fl mice. As further evidence of inflammation, both 4- and 8-week-old iUhrf1fl/fl mice developed splenomegaly after 4 weeks of tamoxifen (Supplemental Figure 5B and Figure 4C). Similarly to Uhrf1fl/flFoxp3YFP-Cre mice, 8-week-old iUhrf1fl/fl mice fed tamoxifen for 4 weeks exhibited disrupted splenic architecture with distortion of germinal centers by lymphocyte expansion and infiltration with other inflammatory cells (Figure 4D). A histological survey of the skin and internal organs of these iUhrf1fl/fl mice revealed a phenotype reminiscent of the spontaneous lymphocytic inflammatory process observed in Uhrf1fl/flFoxp3YFP-Cre mice (Figure 4E), albeit less severe, as these iUhrf1fl/fl mice were not approaching the moribund status that Uhrf1fl/flFoxp3YFP-Cre mice displayed at 3 to 4 weeks of age.
Figure 4. Induced loss of Uhrf1 in Tregs results in spontaneous inflammation.
(A) Schematic of experimental design. (B) Body mass curves for iUhrf1+/+ and iUhrf1fl/fl mice initiated on tamoxifen chow at 8 weeks of age. n = 4 (iUhrf1+/+); n = 14 (iUhrf1fl/fl). (C) Spleen masses for iUhrf1+/+ and iUhrf1fl/fl mice initiated on tamoxifen at 8 weeks of age. n = 4 (iUhrf1+/+); n = 8 (iUhrf1fl/fl). (D) Photomicrographs of spleen pathology from 8-week-old mice after 4 weeks of tamoxifen. Scale bars: 100 μm. (E) Photomicrographic survey of organ pathology from 8-week-old mice after 4 weeks of tamoxifen. Scale bars: 100 μm. **P < 0.01; †P < 0.001; ‡P < 0.0001, 2-way ANOVA with Šidák’s post hoc testing for multiple comparisons (B) or Mann-Whitney U test (C); exact P values are in Supplemental Data. For the comparison between genotypes in B, F(DFn, DFd) = F(1, 42) = 51.4 with P < 0.0001.
Induction of Treg-specific Uhrf1 deficiency promotes generation of ex-Foxp3 cells in vivo.
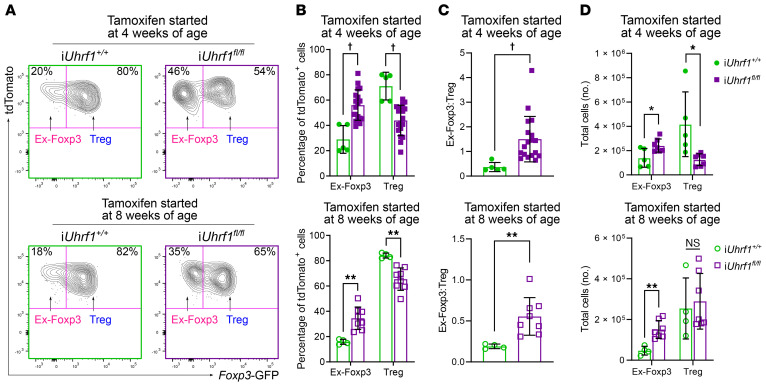
The iUhrf1fl/fl mice permitted tracking the Foxp3+ Treg lineage to determine whether loss of Uhrf1 promotes generation of ex-Foxp3 cells that would indicate loss of Foxp3+ Treg identity. Following tamoxifen administration to iUhrf1fl/fl mice, cells actively expressing Foxp3 were labeled with GFP, whereas tdTomato identified cells that had undergone Foxp3-Cre–mediated loss of Uhrf1 irrespective of active Foxp3 expression (see Figure 4A). TdTomato also serves as a Foxp3+ cell lineage tag in both iUhrf1+/+ and iUhrf1fl/fl mice, marking cells that have undergone Foxp3-Cre–mediated recombination of the ROSA26 locus. Approximately 0.5% of splenic CD3ε+CD4+ T cells expressed tdTomato in tamoxifen-naive mice; after 4 weeks of tamoxifen chow, labeling with tdTomato was nearly complete, with only 1% of splenic CD3ε+CD4+ T cells expressing Foxp3-GFP, but not the tdTomato label (Supplemental Figure 5C). The tdTomato label was specific to CD3ε+CD4+ T cells, with less than 0.25% of CD3ε–CD4– and CD3ε+CD4– cells expressing the tdTomato label in either iUhrf1+/+ or iUhrf1fl/fl mice (Supplemental Figure 5D). As reviewed in the Introduction, ex-Foxp3 cells detected in iUhrf1+/+ mice represent transient Foxp3 induction in a minor population of CD4+ T cells that does not represent epigenetic reprogramming of mature Tregs (29, 30). Thus, in iUhrf1fl/fl mice, any ex-Foxp3 cells detected in excess of those observed in control mice represent loss of Foxp3+ Treg lineage identity.
Using this system, we found that both 4- and 8-week-old iUhrf1fl/fl mice displayed a significant increase in the proportion of ex-Foxp3 (Foxp3-GFP–tdTomato+) cells as a fraction of labeled (tdTomato+) cells compared with iUhrf1+/+ mice (Figure 5, A and B). The ratio of ex-Foxp3 cells to Tregs was increased among both age groups, although 4-week-old iUhrf1fl/fl mice had a greater increase in the ratio than 8-week-old mice (Figure 5C), likely reflecting ongoing thymic Treg development at this age. Both 4- and 8-week-old mice exhibited an increase in total ex-Foxp3 cells after 4 weeks of tamoxifen; however, only 4-week-old mice exhibited a concomitant decrease in total Tregs, again indicating the deleterious effect of losing Uhrf1 on developing thymic Tregs before stabilization of a robust, self-renewing Treg population (Figure 5D).
Figure 5. Induced loss of Uhrf1 in Tregs results in generation of ex-Foxp3 cells.
(A) Representative flow cytometry contour plots gated on splenic CD3ε+CD4+tdTomato+ cells showing the percentage of ex-Foxp3 (Foxp3-GFP–) and Treg (Foxp3-GFP+) cells after 4 weeks of tamoxifen started at either 4 or 8 weeks of age. (B) Summary data of the percentage of ex-Foxp3 and Tregs within the tdTomato+ population for the experiments shown in A. (C) Ratio of ex-Foxp3 to Tregs after 4 weeks of tamoxifen started at either 4 or 8 weeks of age. For mice started on tamoxifen at 4 weeks of age, n = 5 (iUhrf1+/+) and n = 18 (iUhrf1fl/fl); for mice started on tamoxifen at 8 weeks of age, n = 4 (iUhrf1+/+) and n = 8 (iUhrf1fl/fl). (D) Total cell numbers of ex-Foxp3 and Tregs. For mice started on tamoxifen at 4 weeks of age, n = 5 (iUhrf1+/+) and n = 7 (iUhrf1fl/fl); for mice started on tamoxifen at 8 weeks of age, n = 4 (iUhrf1+/+) and n = 7 (iUhrf1fl/fl). *q < 0.05; **p or q < 0.01; †p or q < 0.001; NS, not significant by Mann-Whitney U test (C) or the 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli with Q = 5% (B and D); exact p and q values are in Supplemental Data.
We previously observed that loss of Treg-suppressive function results in slowed tumor growth (47). Accordingly, we evaluated the ability of iUhrf1fl/fl mice to grow B16 melanoma tumors. Compared with iUhrf1+/+ control mice, iUhrf1fl/fl mice exhibited slowed tumor growth and the beginning of tumor regression at 3 weeks after injection — the time when control mice required euthanasia due to tumor ulceration (Supplemental Figure 5, E–G). Flow cytometry analysis revealed a significant increase in the frequency of ex-Foxp3 cells within the tumors of iUhrf1fl/fl mice, which also exhibited a paucity of Tregs (Supplemental Figure 5H).
To determine whether Uhrf1 is necessary to induce Foxp3 expression during the generation of iTreg cells in vitro, we cultured naive CD4+ T cells from iUhrf1+/+ or iUhrf1fl/fl mice under either Th0 conditions (conventional) or Treg-skewing conditions (IL-2, TGF-β, and all-trans retinoic acid) in the presence of tamoxifen to delete Uhrf1 in iUhrf1fl/fl cells and induce the tdTomato Foxp3+ lineage tag in both genotypes. Induced Foxp3 expression occurred in a similar proportion of both iUhrf1+/+ and iUhrf1fl/fl cells (Supplemental Figure 5, I and J), confirming that Uhrf1 is dispensable for iTreg generation in vitro. No clear population of ex-Foxp3 cells emerged in culture; this observation suggests that Uhrf1 is likewise unnecessary for TGF-β–mediated persistence of iTregs.
Together, these data reveal that Uhrf1 is required for stabilization of Treg identity and suppressive function in vivo, as induced loss of Uhrf1 in Tregs led to spontaneous inflammation and generation of ex-Foxp3 cells in juvenile and adult mice. In contrast, Uhrf1 is not required for iTreg generation or persistence in vitro.
Ex-Foxp3 cells generated following loss of Uhrf1 exhibit a distinct inflammatory gene expression profile.
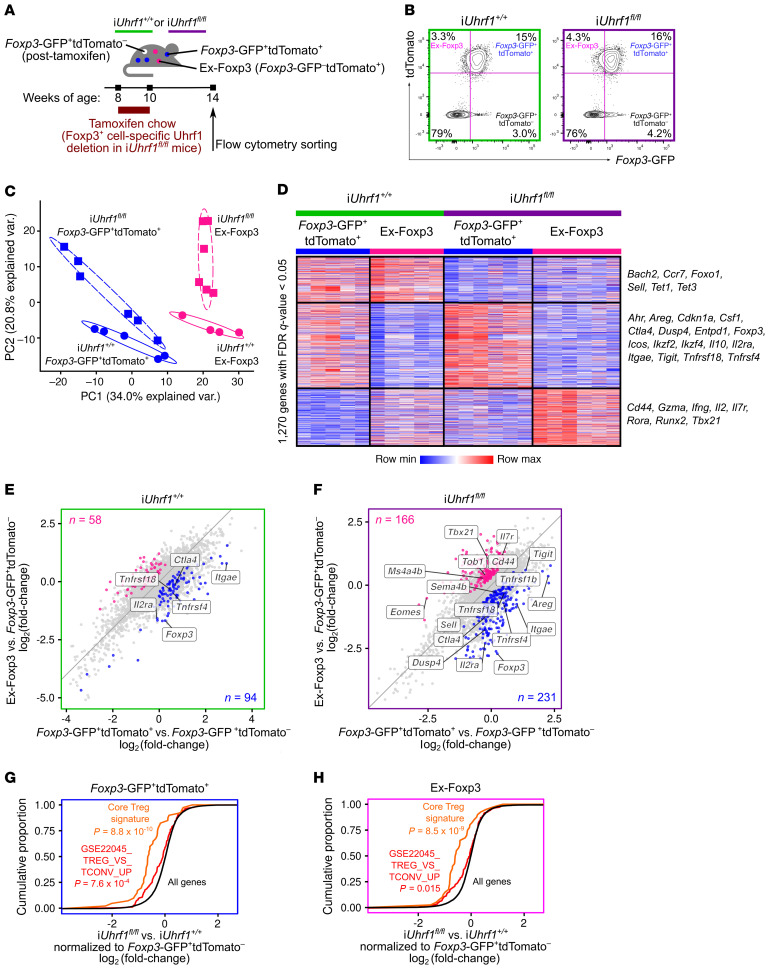
To define the molecular features of ex-Foxp3 cells resulting from loss of Uhrf1, we used a pulse-chase experimental design in which 8-week-old iUhrf1+/+ and iUhrf1fl/fl mice received tamoxifen for 2 weeks followed by standard chow for 4 weeks (Figure 6A). The pulse period allowed tracking of ex-Foxp3 cells; the chase period allowed accumulation of Uhrf1-sufficient (Foxp3-GFP+tdTomato–) cells that were able to suppress the spontaneous systemic inflammation observed in the extended pulse-only experiments (Figure 6B and Supplemental Figure 6, A–E). Of note, cell survival was similar between genotypes at the end of the 4-week chase period (see Figure 6B and Supplemental Figure 6D). Examination of the CD3ε+CD4+Foxp3– T cell compartment revealed an increased frequency of CD44hiCD62Llo cells in the spleens of iUhrf1fl/fl mice, consistent with the increased activation of ex-Foxp3 cells observed in iUhrf1fl/fl mice as revealed by the transcriptional profiling data below. Principal component analysis of 1270 differentially expressed genes with an FDR q value of less than 0.05 revealed distinct clusters based on cell type (Foxp3-GFP+tdTomato+ and ex-Foxp3) and genotype (iUhrf1+/+ and iUhrf1fl/fl) (Figure 6C). K-means clustering of these differentially expressed genes demonstrated a Uhrf1-dependent signature (top cluster), loss of the core Foxp3-dependent Treg gene signature among ex-Foxp3 cells of both genotypes (middle cluster), and a group of genes significantly upregulated in the ex-Foxp3 cells of iUhrf1fl/fl mice (bottom cluster) (Figure 6D and Supplemental Figure 6F). The top cluster contained genes encoding the DNA demethylases Tet1 and Tet3. The middle cluster largely reflected a Foxp3-dependent signature, although Il10 was upregulated in both the Foxp3-GFP+tdTomato+ and ex-Foxp3 populations of Uhrf1-deficient mice compared with control populations. Finally, the bottom cluster contained genes associated with a Th1-skewed effector T cell phenotype, including Gmza, Ifng, Il7r, and the master regulator of the Th1 lineage, Tbx21. These transcriptional profiles illuminate a gene expression signature characterized by increased activation and Th1 skewing of ex-Foxp3 cells generated following loss of Uhrf1.
Figure 6. Induction of Uhrf1 deficiency generates ex-Foxp3 cells with distinct inflammatory transcriptional programs.
(A) Schematic of the pulse-chase experimental design. (B) Representative contour plots of splenic live CD3ε+CD4+ cell subsets (see Supplemental Figure 6D for summary data). (C) Principal component analysis of 1270 differentially expressed genes identified from a generalized linear model and ANOVA-like testing with FDR q < 0.05. Ellipses represent normal contour lines with 1 SD probability. (D) K-means clustering of 1270 genes with an FDR q < 0.05 comparing the cell populations from C scaled as Z-score across rows. (E and F) Fold-change–fold-change plots for iUhrf1+/+ (E) and iUhrf1fl/fl (F) mice of Foxp3-GFP+tdTomato+ versus Foxp3-GFP+tdTomato– and ex-Foxp3 versus Foxp3-GFP+tdTomato– highlighting genes exhibiting an increase (q < 0.05) in Foxp3-GFP+tdTomato+ versus ex-Foxp3 (blue dots) and ex-Foxp3 versus Foxp3-GFP+tdTomato+ (pink dots). Numbers of differentially expressed genes are indicated. (G) Cumulative distribution function plots comparing Foxp3-GFP+tdTomato+ cells from iUhrf1+/+ versus iUhrf1fl/fl mice with each population normalized to the Foxp3-GFP+tdTomato– population sorted from their respective genotypes. The cumulative proportion of all genes (black), a Treg-defining gene set (43) (orange), and the GSE22045_TREG_VS_TCONV_UP gene set (48) (red) are shown. (H) Cumulative distribution function plots as in G comparing the ex-Foxp3 cells from iUhrf1+/+ versus iUhrf1fl/fl mice. n = 5 (iUhrf1+/+); n = 6 (iUhrf1fl/fl). P values resulting from Kolmogorov-Smirnov tests for cumulative distributions comparing all genes against either gene set are shown in G and H.
In contrast with the extended pulse experiment shown in Figure 4 and Figure 5, the frequency of each labeled population was similar between iUhrf1+/+ and iUhrf1fl/fl mice following the pulse-chase period (see Figure 6B and Supplemental Figure 6D). Therefore, we used the Uhrf1-sufficient Foxp3-GFP+tdTomato– cell population as an internal control for the transcriptional analysis of both iUhrf1+/+ and iUhrf1fl/fl mice. After normalizing to this Uhrf1-sufficient cell population within each genotype, the ex-Foxp3 cells of iUhrf1+/+ mice exhibited deficiency of the core Foxp3-dependent Treg signature (Figure 6E). The ex-Foxp3 cells of iUhrf1fl/fl mice also displayed loss of this core signature, but simultaneously upregulated the expression of genes classically associated with impaired Treg function and gain of conventional effector T cell function, including Tbx21, Eomes, Ms4a4b, Tob1, and Il7r in addition to evidence of activation (upregulation of Cd44 and downregulation of Sell) (Figure 6F). Consistent with the results of the Uhrf1 chimeric knockout experiments shown in Figure 3, examination of Foxp3-GFP+tdTomato+ cells from iUhrf1fl/fl versus iUhrf1+/+ mice with respect to previously annotated gene sets of core Treg identity (43) and Treg versus conventional T cell profiles (48) revealed loss of the core Treg signature and a shift toward a conventional effector T cell state (Figure 6G and Supplemental Figure 6G). Even when comparing ex-Foxp3 cells between genotypes and normalizing to the internal Uhrf1-sufficient Foxp3-GFP+tdTomato– cell population, iUhrf1fl/fl mice still exhibited a greater loss of Treg identity and skewing toward a conventional effector T cell state compared with iUhrf1+/+ mice (Figure 6H and Supplemental Figure 6H). To further explore these findings, we executed an unsupervised Gene Set Enrichment Analysis on the ranked gene list used to generate Figure 6, G and H (Foxp3-GFP+tdTomato+ and ex-Foxp3 cells of iUhrf1fl/fl versus iUhrf1+/+ normalized to their respective Foxp3-GFP+tdTomato– populations) against a comprehensive list of 4872 Immunologic Signature gene sets housed in the Molecular Signatures Database (49). These tests revealed 226 and 256 positively enriched gene sets for Foxp3-GFP+tdTomato+ and ex-Foxp3 cells, respectively, at an FDR q value of less than 0.25 (Supplemental Tables 1 and 2). Both analyses indicated a shift from Treg to conventional T cell signatures, and this signal was more robust in the ex-Foxp3 cell population. The ex-Foxp3 cell analysis revealed a gene set characterizing a shift from a naive to a Th1-skewed T cell signature in the top 10 gene sets when ranked by normalized enrichment score. Collectively, these unsupervised analyses demonstrate that loss of Uhrf1 results in generation of ex-Foxp3 cells displaying an excessively activated and Th1-skewed inflammatory profile.
Altered DNA methylation patterns underlie the gain of inflammatory signature and loss of Treg signature observed in Uhrf1-deficient ex-Foxp3 cells.
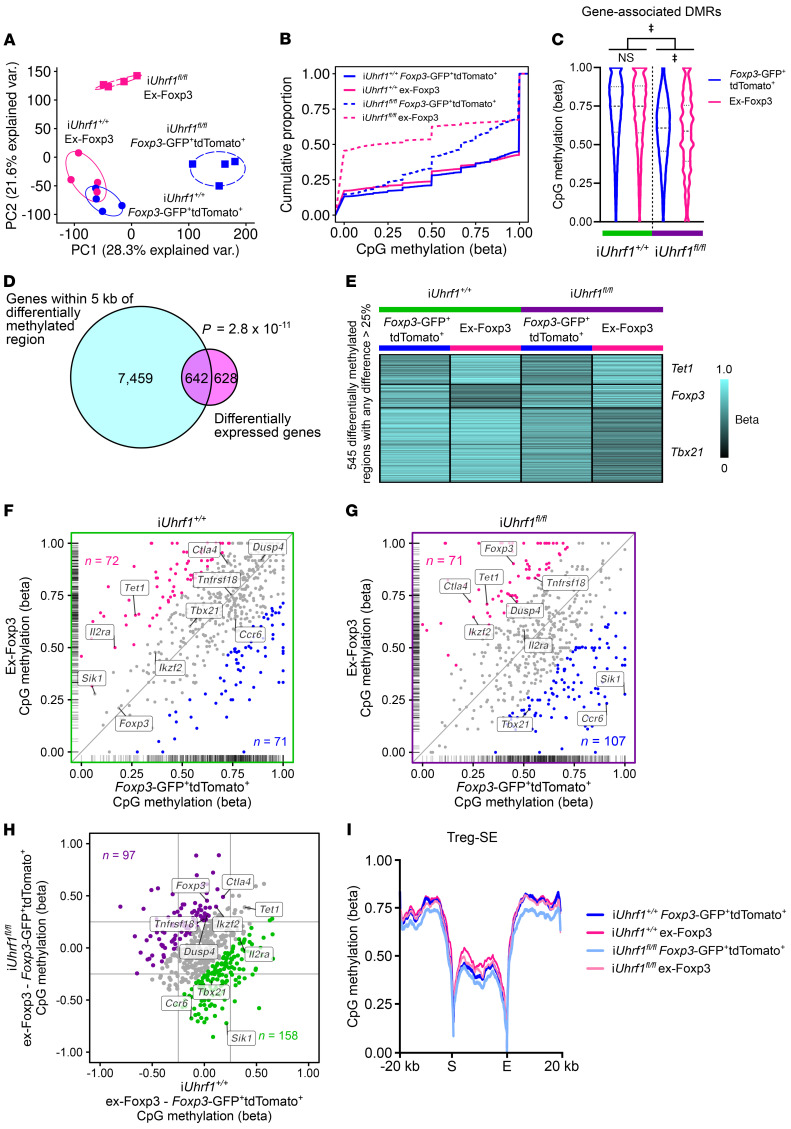
We performed genome-wide CpG methylation profiling on the sorted Foxp3-GFP+tdTomato+ and ex-Foxp3 cells from iUhrf1+/+ and iUhrf1fl/fl mice obtained following the pulse-chase experiment illustrated in Figure 6A. Principal component analysis of differentially methylated cytosines revealed nominal differences between the Foxp3-GFP+tdTomato+ and ex-Foxp3 cells from iUhrf1+/+ mice, but substantial spread between the same cell types obtained from iUhrf1fl/fl mice (Figure 7A). Similar to the findings from Uhrf1 chimeric knockout animals, Foxp3-GFP+tdTomato+ cells from iUhrf1fl/fl mice contained a global hypomethylation pattern that was further hypomethylated in ex-Foxp3 cells (Figure 7B). In contrast, Uhrf1-sufficient ex-Foxp3 cells exhibited only a minor degree of hypomethylation relative to Foxp3-GFP+tdTomato+ cells. To explore these differential methylation patterns in more detail, we performed an unsupervised procedure that revealed 17,249 DMRs that were within 5 kb of 8,101 gene bodies (inclusive). These putative regulatory elements did not exhibit differential methylation between Foxp3-GFP+tdTomato+ and ex-Foxp3 cells from iUhrf1+/+ mice; however, both cell types were hypomethylated in iUhrf1fl/fl mice, which displayed further reductions in methylation within ex-Foxp3 compared with Foxp3-GFP+tdTomato+ cells (Figure 7C). These DMRs were located near 642 differentially expressed genes (a majority of 1270), a finding that was unlikely due to chance alone (Figure 7D).
Figure 7. Altered DNA methylation patterns explain the transcriptional reprogramming of Uhrf1-deficient Treg and ex-Foxp3 cells.
(A) Principal component analysis of 202,525 differentially methylated CpGs with an FDR q < 0.05. Ellipses represent normal contour lines with 1 SD probability. (B) Cumulative distribution function plot of differentially methylated CpGs. (C) CpG methylation at 17,249 DMRs within 5 kb of gene bodies (inclusive). (D) Venn diagram of genes associated with DMR and differentially expressed genes. (E) K-means clustering of 545 DMRs with a difference of more than 25% between any groups and an FDR q < 0.05 that were grouped and then averaged by unique gene association. β Scores are scaled across rows. (F and G) Scatter plots comparing DMRs within Foxp3-GFP+tdTomato+ cells versus ex-Foxp3 cells from iUhrf1+/+ mice (F) and iUhrf1fl/fl mice (G). Interior axis ticks (rug plots) represent positions of DMRs on each axis. Colored points represent greater than 25% difference between ex-Foxp3 cells (pink) and Foxp3-GFP+tdTomato+ cells (blue) with the number of DMRs in each such subset shown. (H) Difference-difference plot comparing the difference in DMR methylation status between ex-Foxp3 and Foxp3-GFP+tdTomato+ cells in iUhrf1+/+ mice and iUhrf1fl/fl mice. Colored points represent greater than 25% difference between iUhrf1+/+ mice (green) and iUhrf1fl/fl mice (purple) with the number of DMRs in each such subset shown. (I) Metagene analysis of CpG methylation across the start (S) through the end (E) of Treg-specific super-enhancer elements (Treg-SE) as defined previously (20). Violin plots show median and quartiles. n = 4 mice per cell type for both genotypes. A hypergeometric P value is shown in D. ‡q < 0.0001; NS, not significant by a mixed-effects analysis with the 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli with Q = 5% (C); exact q values are in Supplemental Data.
We next examined DMRs found near differentially expressed genes and selected those regions with greater than 25% difference between groups. Functional enrichment analysis of these regions using the Genomic Regions Enrichment of Annotations Tool (GREAT) (50) and the Mouse Genome Informatics Phenotype ontology (51) revealed significant enrichment in phenotypes that are characterized by abnormal adaptive immunity and dysregulated T cell development and physiology (Supplemental Figure 7, A and B). We then averaged the methylation level of DMRs associated with unique genes and defined 3 k-means clusters (Figure 7E); the cluster structure mirrored the k-means transcriptional analysis shown in Figure 6D. Inspection of the 3 clusters revealed a large hypomethylated cluster peculiar to Uhrf1-deficient ex-Foxp3 cells that contained the master regulator of the Th1 lineage, Tbx21. Indeed, the DMR or DMRs associated with Tbx21 and other inflammatory genes were hypomethylated in Uhrf1-deficient but not Uhrf1-sufficient ex-Foxp3 cells (Figure 7, F–H, and Supplemental Figure 7C). Interestingly, DMRs near core Treg signature genes, including Foxp3, became methylated in Uhrf1-deficient but not Uhrf1-sufficient ex-Foxp3 cells (Supplemental Figure 7D). Because the primary mechanistic effect of Uhrf1 loss is hypomethylation, the observed hypermethylation of Treg signature genes appears to represent a secondary effect of derepressing the inflammatory program. Compared with the other populations, methylation across Treg-SE was greatest within the ex-Foxp3 cells of iUhrf1+/+ mice, likely reflecting their developmental origin as unstable Foxp3+ or potential Tregs that transiently expressed Foxp3 after thymic emigration (31) (Figure 7I). Altogether, genome-wide DNA methylation profiling and an unsupervised analytic approach revealed alterations in DNA methylation patterning that explained the transcriptional and phenotypic reprogramming that results from loss of Uhrf1.
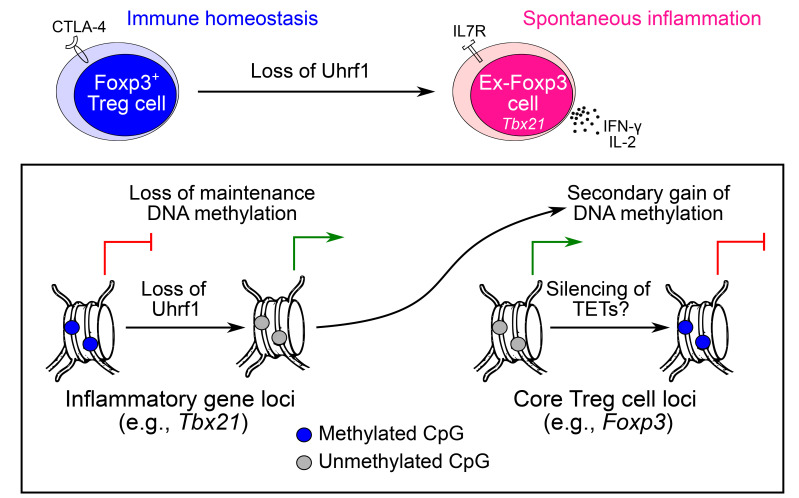
Collectively, our findings demonstrate that Foxp3+ Tregs require Uhrf1-mediated maintenance of DNA methylation at inflammatory gene loci for their development and functional stability (Supplemental Figure 7E).
Discussion
Tregs develop from the complex interplay of trans- and cis-regulatory mechanisms and are maintained as a stable, self-renewing population (18, 20, 28, 29, 32). Here, we determined that Tregs require maintenance DNA methylation mediated by the epigenetic regulator Uhrf1 for establishment of the Foxp3+ Treg lineage as well as stability of Treg identity and suppressive function. Loss of Uhrf1 at the Foxp3+ stage of thymic Treg development resulted in Treg deficiency, leading to a scurfy-like lethal inflammatory disorder. Studies using noninflamed Uhrf1 chimeric knockout animals revealed the cell-autonomous and Foxp3-independent requirement for Uhrf1-mediated DNA methylation in stabilizing the lineage by repressing effector T cell transcriptional programs. Induced deficiency of Uhrf1 in mature Tregs was sufficient to derepress loci-encoding inflammatory genes, resulting in loss of Treg identity and suppressive function, generation of inflammatory ex-Foxp3 cells, and spontaneous inflammation. Taken together, these results demonstrate the necessity of maintenance DNA methylation in stabilizing Treg identity and suppressive function during both development and self-renewal of the lineage in vivo. Our findings change the existing model of DNA hypomethylation as the dominant feature of the Treg epigenome by demonstrating an essential role for methylation-mediated silencing of effector programs in the development and stability of Treg lineage.
We found that Uhrf1 serves as an essential regulator that is required to maintain DNA methylation at defined genomic loci during both Treg development and self-renewal. Unlike Uhrf1fl/flCd4Cre mice with pan–T cell Uhrf1 deficiency that develop inflammation localized to the colon (40), Uhrf1fl/flFoxp3Cre mice in our study spontaneously developed widespread inflammation (the scurfy phenotype), indicating a fundamental role for Uhrf1-mediated maintenance of DNA methylation in stabilizing Treg identity after Foxp3 induction in the thymus. These findings suggest that Uhrf1 is required to maintain a specific DNA methylation landscape in pre-Treg (pre-Foxp3) thymocytes that differs from the landscape required after induction of thymic Foxp3 expression. In vitro, we found that Uhrf1 is dispensable for induction of Foxp3 expression and TGF-β–mediated persistence of iTregs, consistent with literature demonstrating that unlike thymic (natural) Tregs, alterations in DNA methylation are not necessary for iTreg generation (27, 28). Interestingly, the phenotype of Uhrf1-deficient Foxp3+ thymic cells resembled a described population of short-lived effector Tregs that lack CD25 and exhibit increased proliferative capacity and annexin V positivity (52, 53). Following thymic emigration, Treg lineage is remarkably stable with self-renewal across the life span (32). Indeed, the ex-Foxp3 cells that we observed in adult control animals did not display an inflammatory gene expression profile, but did exhibit increased super-enhancer methylation compared with Tregs, reflecting their developmental origin as either unstable Foxp3+ cells or potential Tregs as defined by Ohkura et al. (31). In contrast, the increased number of ex-Foxp3 cells generated following sustained loss of Uhrf1 expressed a Th1-like profile, confirming destabilization of the Treg lineage upon loss of maintenance DNA methylation. Data from our pulse-chase experiments revealed similar Treg frequencies in the spleen 4 weeks following induced loss of Uhrf1. Collectively, these results support that loss of maintenance DNA methylation causes Treg-intrinsic dysregulation of gene expression and subsequent loss of stability — rather than impaired cell survival — to result in immunopathology.
We propose a model in which Tregs require maintenance of DNA methylation at inflammatory gene loci for their development and stability (see Supplemental Figure 7E). In our model, loss of these marks results in derepression of inflammatory genes, including Tbx21. A secondary wave of methylation then represses core Treg loci, including Foxp3, to generate inflammatory ex-Foxp3 cells that contribute to spontaneous inflammation and tumor rejection. The genome-wide transcriptional and DNA methylation profiles of Uhrf1-deficient ex-Foxp3 cells support this mechanism, with hypomethylation of the Tbx21 locus and secondary methylation of the Foxp3 locus. How gain of the effector program results in silencing of Foxp3 and other Treg-defining loci remains an open question. Our data support the speculation that methylation-mediated silencing of TET enzyme expression in Uhrf1-deficient ex-Foxp3 cells contributes to gain of methylation at core Treg loci, ultimately downregulating their expression (54). Thus, the function of maintenance DNA methylation in Tregs is to prevent the adoption of an inflammatory program that results in loss of Foxp3 expression.
Our results inform the rational design of DNA methylating or demethylating agents for the purpose of immunomodulatory therapy (16). Numerous lines of evidence demonstrate that pharmacologic inhibition of DNA methyltransferase activity induces Foxp3 expression in CD4+ T cells and promotes Treg-suppressive function in adult mice that, by definition, harbor mature T cell populations (21–23, 55–57). Based on these studies, we initially hypothesized that loss of Uhrf1 in adult mice would promote mature Treg generation and augment suppressive function of Tregs. Nevertheless, our results show that adult mice with induced loss of Uhrf1 spontaneously develop widespread inflammation reminiscent of the developmental phenotype observed in the constitutive conditional knockout animals, scurfy mice, and humans with the autoimmune IPEX syndrome. Indeed, missense variants of UHRF1BP (Uhrf1-binding protein) are associated with the development of systemic lupus erythematosus (58), and epigenomic region variants within Tregs are linked to autoimmunity (59). Thus, our results suggest caution when applying pharmacologic approaches to epigenetic modification in immune-dysregulated conditions, such as autoimmune and malignant disorders.
Collectively, our findings reveal that Foxp3+ Tregs require maintenance DNA methylation mediated by the epigenetic regulator Uhrf1 for both development and stability of the lineage. Loss of Uhrf1-mediated DNA methylation during both development and self-renewal is sufficient to disrupt the lineage, generate inflammatory ex-Foxp3 cells, and cause inflammatory pathology. Because of the powerful role that DNA methylation programming plays in determining the stability of the Treg lineage, pharmacologic stabilization or disruption of the DNA methylation maintenance function of Uhrf1 could be explored to address inflammatory or malignant disorders, respectively.
Methods
Mice.
C57BL/6 Uhrf1fl/fl mice, which harbor loxP sequences flanking exon 4 (ENSMUSE00000139530) of the Uhrf1 gene (ENSMUSG00000001228) were a gift from the laboratory of Srinivasan Yegnasubramanian (Johns Hopkins University, Baltimore, Maryland, USA). Foxp3YFP-Cre (stock no. 016959), Foxp3eGFP-CreERT2 (stock no. 016961), and ROSA26SorCAG-tdTomato (Ai14, stock no. 007908) mice were obtained from The Jackson Laboratory. All animals were genotyped using services provided by Transnetyx Inc., with primers provided by The Jackson Laboratory or reported below. Experimental randomization and blinding were not performed due to obvious external phenotype; however, littermate controls were used whenever possible. Animals received water ad libitum, were housed at a temperature range of 20°C–23°C under 14-hour light/10-hour dark cycles, and received standard rodent chow except for animals that received chow containing tamoxifen (Envigo, 500 mg/kg of chow ≈ daily dose of 40–80 mg/kg) as noted.
Tissue and cell preparation.
Organs were prepared for histological assessment with sectioning and H&E staining as previously described (21, 47). For preparation of colonic tissue for flow cytometry, colons were collected from mice, flushed with 10 mL ice-cold PBS, and washed twice with room-temperature HBSS plus 2% FBS. Colons were then cut into 1 cm pieces, placed in a 50 mL conical tube containing 10 mL room-temperature HBSS, 2% FBS, and 2 mM EDTA, and incubated for 15 minutes at 37°C with rotation. Supernatant was discarded and the colon was washed twice with HBSS, cut into 2 mm pieces, placed in 10 mL of prewarmed RPMI plus 10% FBS, 1.5 mg/mL collagenase D (Roche, 11088866001), and 0.05 mg/mL collagenase V (MilliporeSigma, C9263) and incubated for 40 minutes with rotation. Digested colons were filtered through a 40 μm strainer into 10 mL of MACS buffer (Miltenyi Biotec, 130-091-221), centrifuged for 10 minutes at 445g, and resuspended in MACS buffer. Histological evaluations were performed in consultation with a veterinary pathologist through core services provided by the Northwestern University Mouse Histology and Phenotyping Laboratory.
Flow cytometry analysis and sorting.
Single-cell suspensions of visceral organs, blood, tumors, or cultured cells were prepared and stained for flow cytometry analysis and sorting as previously described (21, 47, 60–62) using the reagents and cytometer setup shown in Supplemental Table 3. Cell counts of single-cell suspensions were obtained using a hemocytometer with trypan blue exclusion or a Cellometer with AO/PI staining (Nexcelom Bioscience) before preparation for flow cytometry. Data acquisition for analysis was performed using a BD LSRFortessa or Symphony A5 instrument with FACSDiva software (BD). Cell sorting was performed using the 4-way purity setting on BD FACSAria SORP instruments with FACSDiva software. Fluorescence-minus-one controls were used. Analysis was performed with FlowJo, version 10.6.1, software. Dead cells were excluded using a viability dye for analysis and sorting.
Cytokine measurements.
Following lysis of erythrocytes, single-cell suspensions of spleen and lung tissue and blood at a concentration of 5 × 106 cells/mL were treated with RPMI plus 5 μL/mL Leukocyte Activation Cocktail with GolgiPlug (BD) or RPMI only and incubated for 4 hours at 37°C. After incubation, cells were spun down, washed, and resuspended in a viability dye (Fixable Viability Dye eFluor 506, eBioscience) for 30 minutes at 4°C and fixed and permeabilized with the Foxp3/Transcription Factor Staining Buffer Set (eBioscience). Cells were then stained with an antibody cocktail (Fc Block, Foxp3-PE-Cy7, CD4-PerCP-Cy5.5, IL-17A–PE, IFN-γ–PE-CF594, IL-4–APC; see Supplemental Table 3 for details) for 30 minutes at room temperature, washed, and resuspended in PBS with 0.5% BSA before flow cytometry analysis as above.
Annexin staining.
Single-cell suspensions were incubated with a UV-excitable viability dye (Molecular Probes) followed by surface staining as above and previously described (21, 47, 60, 61). Cells were then washed and resuspended in annexin-binding buffer (10 mM HEPES, 140 mM NaCl, and 2.4 mM CaCl2; pH 7.4) at a concentration of 1 × 106 cells/mL. Then 5 μL annexin V/Pacific blue conjugate (Invitrogen) per 100 μL of cell suspension was added and incubated for 15 minutes at room temperature and then washed with annexin-binding buffer before resuspension in PBS with 0.5% BSA for flow cytometry analysis as above.
B16 melanoma model.
100,000 B16-F10 cells (ATCC CRL-6475) were subcutaneously injected into the hair-trimmed flank of 8- to 12-week-old mice. All cells were determined to be free of Mycoplasma contamination before injection. Tumor progression was measured as previously described (47). A subset of tumors was resected postmortem for photographic and flow cytometry analysis.
In vitro Treg conversion assay.
Splenic naive CD4+ T cells from iUhrf1+/+ and iUhrf1fl/fl mice were isolated using the EasySep Mouse Naïve CD4+ T Cell Isolation Kit (STEMCELL Technologies) and resuspended at a concentration of 2 × 106 cells/mL in RPMI with 10% FBS, 2 mM l-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 100 μM MEM Non-Essential Amino Acid Solution (Gibco, Thermo Fisher Scientific), 50 μM β-mercaptoethanol, and 2000 U/mL IL-2. Cells were then plated in flat-bottom 96-well plates at a concentration of 1 × 105 cells per well with CD3- and CD28-coated beads (Treg Expansion Kit, Miltenyi Biotec) at a bead-to-cell ratio of 3:1 and 5 μM (Z)-4-hydroxytamoxifen (MilliporeSigma, H7904); some wells also contained ImmunoCult Mouse Treg Differentiation Supplement per the manufacturer’s recommendations (STEMCELL Technologies). Cells were incubated for 3 days to permit iTreg generation. After incubation, cells were spun down, washed, and resuspended in a viability dye (Fixable Viability Dye eFluor 506, eBioscience) for 30 minutes at 4°C. Cells were then stained with Fc Block and CD4-APC-eFluor780 for 20 minutes at 4°C, washed, and resuspended in PBS with 0.5% BSA before flow cytometry analysis as above.
Uhrf1 PCR.
DNA was extracted from sorted cells using the QIAGEN AllPrep DNA/RNA Micro Kit and quantified using a Qubit 3.0 fluorometer. PCR was performed using the following primers (IDT, standard desalting): forward (F) CTTGATCTGTGCCCTGCAT, reverse 1 (R1) ACCTCTGCTCTGATGGCTGT, and reverse 2 (R2) CCGAGGACACTCAAGAGAGC. As shown in Supplemental Figure 1A, the F and R1 primers flank the 5′ loxP site and produce a 198 bp band in WT mice and a 398 bp band in conditional (floxed) mice. The R2 primer sits downstream of the 3′ loxP site, resulting in a 247 bp band in cells that have undergone Cre-mediated excision of the floxed sequence. We combined 0.1 ng of template DNA with 10 μL iQ Supermix (Bio-Rad) and 2 μL each of 20 μM F primer, 10 μM R1 primer, and 10 μM R2 primer in a total reaction volume of 20 μL. A C1000 Touch Thermal Cycler (Bio-Rad) was programmed for 1 minute at 95°C; then 15 seconds at 95°C, 15 seconds at 65°C, and 30 seconds at 72°C for 35 cycles; and ending with 7 minutes at 72°C. We combined 20 μL of PCR product with 32 μL of Magbio High-Prep PCR beads (1.6× clean-up). After a 5-minute incubation, the tubes were placed on a magnet until the solution cleared. The supernatant was removed, the beads were washed twice with 80% ethanol, and the product was eluted in 20 μL water. The cleaned PCR product was run on an Agilent TapeStation 4200 using High Sensitivity D1000 ScreenTape and visualized using TapeStation Analysis Software A.02.01 SR1.
RNA-Seq.
Nucleic acid isolation, fragmentation, adapter ligation, and indexing were performed as previously described using the QIAGEN AllPrep DNA/RNA Micro Kit and the SMARTer Stranded Total RNA-Seq Kit, version 2 (Takara) (47, 63). Sequencing was performed on an Illumina NextSeq 500 instrument, employing single-end sequencing with the NextSeq 500/550 V2 High Output Reagent Kit (1 × 75 cycles) and targeting a read depth of at least 10 × 106 aligned reads per sample. Indexed samples were demultiplexed to fastq files with BCL2FASTQ, version 2.17.1.14, trimmed using Trimmomatic, version 0.38 (to remove end nucleotides with a phred score less than 30 while requiring a minimum length of 20 bp), and aligned to the mm10 (GRCm38) reference genome using TopHat, version 2.1.0 (47). Counts data for uniquely mapped reads over exons were obtained using SeqMonk, version 1.45.4, and filtered to protein-coding genes and genes with at least 1 count per million in at least 2 samples. Differential gene expression analysis was performed with the edgeR, version 3.24.3, R/Bioconductor package using R, version 3.5.1, and RStudio, version 1.1.447, as stated in the text and individual figure legends and as described previously (61, 64, 65). K-means clustering and heatmaps were generated using the Morpheus web interface (https://software.broadinstitute.org/morpheus/). Gene Set Enrichment Analysis was performed using the GSEA, version 4.0.3, GSEAPreranked tool (66) with genes ordered by log2(fold change) in average expression against cited gene sets or the Immunologic Signature gene sets housed in the Molecular Signatures Database of the Broad Institute (49).
mRRBS.
Genomic DNA was subjected to mRRBS as previously described (47, 61, 65, 67–70). Bisulfite conversion efficiency averaged 99.52% ± 0.052% (SD), as estimated by the measured frequency of unmethylated CpGs in λ-bacteriophage DNA (New England BioLabs, catalog N3013S) added at a 1:200 mass ratio to each sample. Processing and analysis were conducted as previously described using Trim Galore!, version 0.4.3, Bismark, version 0.16.3, and the DSS, version 2.30.1, R/Bioconductor package and quantified with the SeqMonk platform with the bisulphite feature methylation pipeline, all using the mm10 (GRCm38) reference genome. Well-observed CpGs were as previously defined (70). Multigroup comparisons at the single-CpG level were performed using a β-binomial regression model with an arcsine link function fitted using the generalized least square method and Wald-test FDR q value of less than 0.05. DMRs were defined using the callDMR function within the DSS R/Bioconductor package using a P value threshold of 0.05, a minimum length of 50 bp, a minimum significant CpG count of 2, merging DMR within 100 bp, and requiring 50% of the CpGs within a DMR to meet the significance threshold without a predefined methylation difference (delta). Functional enrichment analysis was performed using the GREAT tool (50) and the Mouse Genome Informatics Phenotype ontology (51) with the basal+extension association rule (constitutive 5 kb upstream and 1 kb downstream, up to 1000 kb max extension) against the whole genome as background. Metagene analysis was performed using SeqMonk’s quantitation trend plot function across Treg-specific super-enhancer elements (20) after liftover of coordinates from the mm9 to the mm10 reference genome (67).
Statistics.
P values and q values resulting from 2-tailed tests were calculated using statistical tests stated in the figure legends using GraphPad Prism, version 8.3.0, or R, version 3.5.1. Exact values are reported unless specified. Kolmogorov-Smirnov testing was performed using the base R ks.test function, which cannot return an exact P value in the presence of ties and instead returns an asymptotic P value. A P value or q value of less than 0.05 was considered significant. Central tendency and error are displayed as mean ± SD except as noted. Violin plots show median and quartiles. Numbers of biological and technical replicates are stated in the figures or accompanying legends. Histological and gel images are representative of at least 2 independent experiments. For next-generation sequencing experiments, indicated sample sizes were chosen to obtain a minimum of 106 unique CpGs per biological replicate, as modeled using the DSS statistical procedures (47, 61, 67–70). Computational analysis was performed using Genomics Nodes and Analytics Nodes on Quest, Northwestern University’s High-Performance Computing Cluster.
Data availability.
The raw and processed next-generation sequencing data sets were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE143974). Individual data points and detailed results of statistical tests are available in the Supplemental Data.
Code availability.
Code used for RNA-Seq processing is available from https://github.com/ebartom/NGSbartom Code used for mRRBS processing is available in the supplement to Singer et al. (70).
Study approval.
All animal experiments and procedures were conducted in accordance with the standards established by the United States Animal Welfare Act set forth in NIH guidelines and were approved by the IACUC at Northwestern University under protocol number IS00001624.
Author contributions
KAH, LMN, MATA, SEW, and BDS contributed to the conception, hypotheses delineation, and design of the study. KAH, LMN, MATA, KRA, SYC, HAV, YP, PC, MA, EMS, SEW, and BDS performed experiments, data acquisition, and analysis. KAH, LMN, SEW, and BDS wrote the manuscript or provided substantial involvement in its revision.
Supplementary Material
Acknowledgments
LMN was supported by NIH awards T32HL076139 and F32HL151127. MATA was supported by NIH award T32GM008152. KRA was supported by the David W. Cugell and Christina Enroth-Cugell Fellowship. BDS was supported by NIH awards K08HL128867, U19AI135964, and R01HL149883 and the Francis Family Foundation’s Parker B. Francis Research Opportunity Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding sources. We wish to acknowledge the Northwestern University Flow Cytometry Core Facility supported by CA060553. The BD FACSAria SORP system was purchased with the support of S10OD011996. We also wish to acknowledge the Northwestern University RNA-Seq Center/Genomics Lab of the Pulmonary and Critical Care Medicine and Rheumatology Divisions. Histology services were provided by the Northwestern University Mouse Histology and Phenotyping Laboratory, which is supported by P30CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. This research was supported in part through the computational resources and staff contributions provided by the Genomics Computer Cluster, which is jointly supported by the Feinberg School of Medicine, the Center for Genetic Medicine, and Feinberg’s Department of Biochemistry and Molecular Genetics, the Office of the Provost, the Office for Research, and Northwestern Information Technology. The Genomics Compute Cluster is part of Quest, Northwestern University’s high performance computing facility, with the purpose of advancing research in genomics.
Version 1. 09/08/2020
In-Press Preview
Version 2. 11/09/2020
Electronic publication
Version 3. 11/11/2020
Corrected X-axis label of Figure 6G.
Version 4. 12/01/2020
Print issue publication
Footnotes
Conflict of interest: BDS has a pending patent application: US patent application 15/542,380, “Compositions and methods to accelerate resolution of acute lung inflammation.”
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(12):6571–6587.https://doi.org/10.1172/JCI137712.
Contributor Information
Kathryn A. Helmin, Email: helmin@northwestern.edu.
Luisa Morales-Nebreda, Email: luisamorales@northwestern.edu.
Manuel A. Torres Acosta, Email: manuel.torres@northwestern.edu.
Kishore R. Anekalla, Email: kishore.anekalla@northwestern.edu.
Shang-Yang Chen, Email: shang-yangchen2020@u.northwestern.edu.
Hiam Abdala-Valencia, Email: h-abdala-valencia@northwestern.edu.
Yuliya Politanska, Email: yuliya.politanska@northwestern.edu.
Paul Cheresh, Email: p-cheresh@northwestern.edu.
Mahzad Akbarpour, Email: makbarpour@medicine.bsd.uchicago.edu.
Elizabeth M. Steinert, Email: elizabethsteinert@northwestern.edu.
Samuel E. Weinberg, Email: Samuel-Weinberg@northwestern.edu.
Benjamin D. Singer, Email: benjamin-singer@northwestern.edu.
References
- 1.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133(5):775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singer BD, King LS, D’Alessio FR. Regulatory T cells as immunotherapy. Front Immunol. 2014;5:46. doi: 10.3389/fimmu.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4(4):330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 5.Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27(1):68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 6.Wildin RS, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27(1):18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 7.Valencia X, Yarboro C, Illei G, Lipsky PE. Deficient CD4+CD25high T regulatory cell function in patients with active systemic lupus erythematosus. J Immunol. 2007;178(4):2579–2588. doi: 10.4049/jimmunol.178.4.2579. [DOI] [PubMed] [Google Scholar]
- 8.Valencia X, Lipsky PE. CD4+CD25+FoxP3+ regulatory T cells in autoimmune diseases. Nat Clin Pract Rheumatol. 2007;3(11):619–626. doi: 10.1038/ncprheum0624. [DOI] [PubMed] [Google Scholar]
- 9.Crispin JC, Martínez A, Alcocer-Varela J. Quantification of regulatory T cells in patients with systemic lupus erythematosus. J Autoimmun. 2003;21(3):273–276. doi: 10.1016/S0896-8411(03)00121-5. [DOI] [PubMed] [Google Scholar]
- 10.Miyara M, et al. Global natural regulatory T cell depletion in active systemic lupus erythematosus. J Immunol. 2005;175(12):8392–8400. doi: 10.4049/jimmunol.175.12.8392. [DOI] [PubMed] [Google Scholar]
- 11.Morales-Nebreda L, Alakija O, Ferguson KT, Singer BD. Systemic lupus erythematosus-associated diffuse alveolar hemorrhage: a case report and review of the literature. Clin Pulm Med. 2018;25(5):166–169. doi: 10.1097/CPM.0000000000000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antiga E, et al. Regulatory T cells in the skin lesions and blood of patients with systemic sclerosis and morphoea. Br J Dermatol. 2010;162(5):1056–1063. doi: 10.1111/j.1365-2133.2010.09633.x. [DOI] [PubMed] [Google Scholar]
- 13.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peggs KS, Quezada SA, Chambers CA, Korman AJ, Allison JP. Blockade of CTLA-4 on both effector and regulatory T cell compartments contributes to the antitumor activity of anti-CTLA-4 antibodies. J Exp Med. 2009;206(8):1717–1725. doi: 10.1084/jem.20082492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson TR, et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med. 2013;210(9):1695–1710. doi: 10.1084/jem.20130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morales-Nebreda L, Helmin KA, Singer BD. CoRESTed development of regulatory T cells. J Clin Invest. 2020;130(4):1618–1621. doi: 10.1172/JCI135713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordan MS, et al. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2(4):301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 18.Ohkura N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37(5):785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Morales-Nebreda L, McLafferty FS, Singer BD. DNA methylation as a transcriptional regulator of the immune system. Transl Res. 2019;204:1–18. doi: 10.1016/j.trsl.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitagawa Y, et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol. 2017;18(2):173–183. doi: 10.1038/ni.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singer BD, et al. Regulatory T cell DNA methyltransferase inhibition accelerates resolution of lung inflammation. Am J Respir Cell Mol Biol. 2015;52(5):641–652. doi: 10.1165/rcmb.2014-0327OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu CH, et al. DNA methyltransferase inhibitor promotes human CD4+CD25hFOXP3+ regulatory t lymphocyte induction under suboptimal TCR stimulation. Front Immunol. 2016;7:488. doi: 10.3389/fimmu.2016.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan MW, Chang CB, Tung CH, Sun J, Suen JL, Wu SF. Low-dose 5-aza-2’-deoxycytidine pretreatment inhibits experimental autoimmune encephalomyelitis by induction of regulatory T cells. Mol Med. 2014;20(1):248–256. doi: 10.2119/molmed.2013.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204(7):1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, et al. Foxp3+ T-regulatory cells require DNA methyltransferase 1 expression to prevent development of lethal autoimmunity. Blood. 2013;121(18):3631–3639. doi: 10.1182/blood-2012-08-451765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samstein RM, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151(1):153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lal G, Bromberg JS. Epigenetic mechanisms of regulation of Foxp3 expression. Blood. 2009;114(18):3727–3735. doi: 10.1182/blood-2009-05-219584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lal G, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182(1):259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyao T, et al. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 2012;36(2):262–275. doi: 10.1016/j.immuni.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 30.Zhou X, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10(9):1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38(3):414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Rubtsov YP, et al. Stability of the regulatory T cell lineage in vivo. Science. 2010;329(5999):1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317(5845):1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 34.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 35.Nishiyama A, et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013;502(7470):249–253. doi: 10.1038/nature12488. [DOI] [PubMed] [Google Scholar]
- 36.Kong X, et al. Defining UHRF1 domains that support maintenance of human colon cancer DNA methylation and oncogenic properties. Cancer Cell. 2019;35(4):633–648.e7. doi: 10.1016/j.ccell.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maenohara S, et al. Role of UHRF1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryos. PLoS Genet. 2017;13(10):e1007042. doi: 10.1371/journal.pgen.1007042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, et al. S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance. Cell Res. 2011;21(12):1723–1739. doi: 10.1038/cr.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 40.Obata Y, et al. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat Immunol. 2014;15(6):571–579. doi: 10.1038/ni.2886. [DOI] [PubMed] [Google Scholar]
- 41.Sun X, Cui Y, Feng H, Liu H, Liu X. TGF-β signaling controls Foxp3 methylation and T reg cell differentiation by modulating Uhrf1 activity. J Exp Med. 2019;216(12):2819–2837. doi: 10.1084/jem.20190550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28(4):546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 43.Hill JA, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27(5):786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 44.Crawford A, et al. Molecular and transcriptional basis of CD4(+) T cell dysfunction during chronic infection. Immunity. 2014;40(2):289–302. doi: 10.1016/j.immuni.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.May SL, et al. Nfatc2 and Tob1 have non-overlapping function in T cell negative regulation and tumorigenesis. PLoS ONE. 2014;9(6):e100629. doi: 10.1371/journal.pone.0100629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan Y, et al. TCR stimulation upregulates MS4a4B expression through induction of AP-1 transcription factor during T cell activation. Mol Immunol. 2012;52(2):71–78. doi: 10.1016/j.molimm.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weinberg SE, et al. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature. 2019;565(7740):495–499. doi: 10.1038/s41586-018-0846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonacci B, et al. Requirements for growth and IL-10 expression of highly purified human T regulatory cells. J Clin Immunol. 2012;32(5):1118–1128. doi: 10.1007/s10875-012-9701-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Godec J, et al. Compendium of immune signatures identifies conserved and species-specific biology in response to inflammation. Immunity. 2016;44(1):194–206. doi: 10.1016/j.immuni.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE, Mouse Genome Database Group The mouse genome database genotypes::phenotypes. Nucleic Acids Res. 2009;37(Database issue):D712–D719. doi: 10.1093/nar/gkn886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wing JB, et al. A distinct subpopulation of CD25- T-follicular regulatory cells localizes in the germinal centers. Proc Natl Acad Sci U S A. 2017;114(31):E6400–E6409. doi: 10.1073/pnas.1705551114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kornete M, Mason E, Istomine R, Piccirillo CA. KLRG1 expression identifies short-lived Foxp3+ Treg effector cells with functional plasticity in islets of NOD mice. Autoimmunity. 2017;50(6):354–362. doi: 10.1080/08916934.2017.1364368. [DOI] [PubMed] [Google Scholar]
- 54.Yang R, et al. Hydrogen sulfide promotes Tet1- and Tet2-Mediated Foxp3 demethylation to drive regulatory t cell differentiation and maintain immune homeostasis. Immunity. 2015;43(2):251–263. doi: 10.1016/j.immuni.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Polansky JK, et al. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38(6):1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 56.Varanasi SK, Reddy PBJ, Bhela S, Jaggi U, Gimenez F, Rouse BT. Azacytidine treatment inhibits the progression of herpes stromal keratitis by enhancing regulatory T cell function. J Virol. 2017;91(7):e02367-16. doi: 10.1128/JVI.02367-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu CJ, Yang CY, Chen YH, Chen CM, Chen LC, Kuo ML. The DNA methylation inhibitor 5-azacytidine increases regulatory T cells and alleviates airway inflammation in ovalbumin-sensitized mice. Int Arch Allergy Immunol. 2013;160(4):356–364. doi: 10.1159/000343030. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, et al. Two missense variants in UHRF1BP1 are independently associated with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 2011;12(3):231–234. doi: 10.1038/gene.2010.66. [DOI] [PubMed] [Google Scholar]
- 59.Ohkura N, et al. Regulatory T cell-specific epigenomic region variants are a key determinant of susceptibility to common autoimmune diseases. Immunity. 2020;52(6):1119–1132.e4. doi: 10.1016/j.immuni.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 60.Singer BD, et al. Flow-cytometric method for simultaneous analysis of mouse lung epithelial, endothelial, and hematopoietic lineage cells. Am J Physiol Lung Cell Mol Physiol. 2016;310(9):L796–L801. doi: 10.1152/ajplung.00334.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McGrath-Morrow SA, et al. DNA methylation regulates the neonatal CD4+ T-cell response to pneumonia in mice. J Biol Chem. 2018;293(30):11772–11783. doi: 10.1074/jbc.RA118.003589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tighe RM, et al. Improving the quality and reproducibility of flow cytometry in the lung. An official American Thoracic Society workshop report. Am J Respir Cell Mol Biol. 2019;61(2):150–161. doi: 10.1165/rcmb.2019-0191ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Misharin AV, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med. 2017;214(8):2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McGrath-Morrow SA, et al. Inflammation and transcriptional responses of peripheral blood mononuclear cells in classic ataxia telangiectasia. PLoS ONE. 2018;13(12):e0209496. doi: 10.1371/journal.pone.0209496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McGrath-Morrow SA, et al. DNA methylation and gene expression signatures are associated with ataxia-telangiectasia phenotype. Sci Rep. 2020;10(1):7479. doi: 10.1038/s41598-020-64514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walter JM, Helmin KA, Abdala-Valencia H, Wunderink RG, Singer BD. Multidimensional assessment of alveolar T cells in critically ill patients. JCI Insight. 2018;3(17):e123287. doi: 10.1172/jci.insight.123287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, et al. TET2 coactivates gene expression through demethylation of enhancers. Sci Adv. 2018;4(11):eaau6986. doi: 10.1126/sciadv.aau6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Piunti A, et al. CATACOMB: an endogenous inducible gene that antagonizes H3K27 methylation activity of Polycomb repressive complex 2 via an H3K27M-like mechanism. Sci Adv. 2019;5(7):eaax2887. doi: 10.1126/sciadv.aax2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singer BD. A practical guide to the measurement and analysis of DNA methylation. Am J Respir Cell Mol Biol. 2019;61(4):417–428. doi: 10.1165/rcmb.2019-0150TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed next-generation sequencing data sets were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE143974). Individual data points and detailed results of statistical tests are available in the Supplemental Data.