Abstract
Long noncoding RNAs (lncRNAs) play important roles in regulating diverse cellular processes in the vessel wall, including atherosclerosis. RNA-Seq profiling of intimal lesions revealed a lncRNA, VINAS (Vascular INflammation and Atherosclerosis lncRNA Sequence), that is enriched in the aortic intima and regulates vascular inflammation. Aortic intimal expression of VINAS fell with atherosclerotic progression and rose with regression. VINAS knockdown reduced atherosclerotic lesion formation by 55% in LDL receptor–deficient (LDLR–/–) mice, independent of effects on circulating lipids, by decreasing inflammation in the vessel wall. Loss- and gain-of-function studies in vitro demonstrated that VINAS serves as a critical regulator of inflammation by modulating NF-κB and MAPK signaling pathways. VINAS knockdown decreased the expression of key inflammatory markers, such as MCP-1, TNF-α, IL-1β, and COX-2, in endothelial cells (ECs), vascular smooth muscle cells, and bone marrow–derived macrophages. Moreover, VINAS silencing decreased expression of leukocyte adhesion molecules VCAM-1, E-selectin, and ICAM-1 and reduced monocyte adhesion to ECs. DEP domain containing 4 (DEPDC4), an evolutionary conserved human ortholog of VINAS with approximately 74% homology, showed similar regulation in human and pig atherosclerotic specimens. DEPDC4 knockdown replicated antiinflammatory effects of VINAS in human ECs. These findings reveal a potentially novel lncRNA that regulates vascular inflammation, with broad implications for vascular diseases.
Keywords: Vascular Biology
Keywords: Atherosclerosis, Noncoding RNAs, endothelial cells
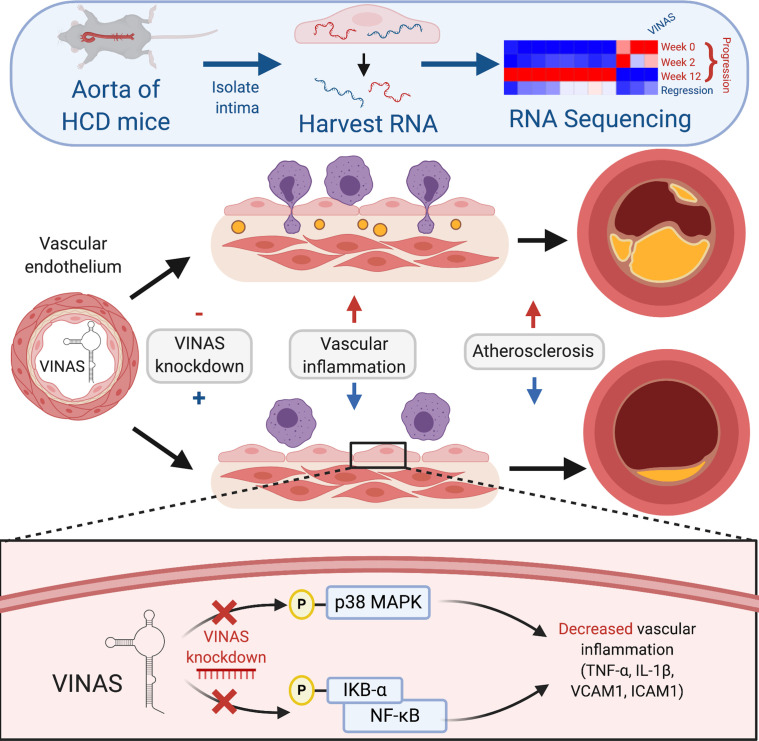
Neutralization of lncRNA VINAS or its human ortholog DEPDC4 limits the inflammatory response and atherosclerotic progression by regulating the NF-κB and MAPK signaling pathways.
Introduction
Accumulating studies highlight that inflammatory processes and traditional cardiac risk factors may cooperatively contribute to vascular disease leading to the development of cardiovascular events (1, 2). More than 150 years ago Virchow hypothesized involvement of inflammation in atherosclerosis (3). However, the CANTOS trial only recently confirmed in humans the inflammatory hypothesis of atherosclerosis, by showing that neutralization of the proinflammatory cytokine IL-1β reduced recurrent cardiovascular events independent of changes in serum lipid levels (4, 5).
Inflammation impairs endothelial functions. For example, in response to both biochemical (e.g., IL-1β, modified LDL) and biomechanical (e.g., disturbed blood flow) stimuli, endothelial activation occurs early in atherogenesis (6). Expression of adhesion molecules (e.g., VCAM-1, E-selectin, ICAM-1) and secretion of chemokines (e.g., MCP-1) facilitate the recruitment of leukocyte subsets into the vessel wall (7). Impaired endothelial barrier function accompanies vascular inflammation and atherosclerosis (1, 8). Similar to ECs, smooth muscle cells (SMCs) can also express a variety of adhesion molecules in response to cytokine stimulation to which monocytes and lymphocytes can adhere and migrate into the vessel wall (5, 9, 10). However, major mechanistic gaps remain in our understanding of regulatory pathways involved in homeostasis of the vessel wall in response to pathophysiological stimuli, contributing to the dearth of targeted therapeutics in a range of vascular disease states.
Recently, long noncoding RNAs (lncRNAs) have emerged as powerful regulators of nearly all biological processes by exerting epigenetic, transcriptional, or translational control of target genes owing to their polyvalent binding properties to RNA, DNA, and protein as well as acting as molecular sponges for other transcripts and miRNAs (11, 12). However, the role of lncRNAs in vascular inflammation and cardiovascular diseases is just now emerging (13). Identification of lncRNAs specifically expressed in the vascular intima of lesions during the progression of atherosclerosis may provide insight into their roles in atherogenesis and potentially uncover new insight into vascular inflammation in advanced lesions (14).
This study identifies lncRNA VINAS (Vascular INflammation and Atherosclerosis lncRNA Sequence) as a key regulator of vascular inflammation and atherosclerotic lesion formation. We further find that its human ortholog, DEP domain containing 4 (DEPDC4), was similarly expressed in atherosclerotic lesions and phenocopied effects on human endothelial cell (EC) inflammation. Collectively, these findings provide new insight into lncRNA-mediated control of inflammation in the vessel wall.
Results
Identification and characterization of VINAS lncRNA.
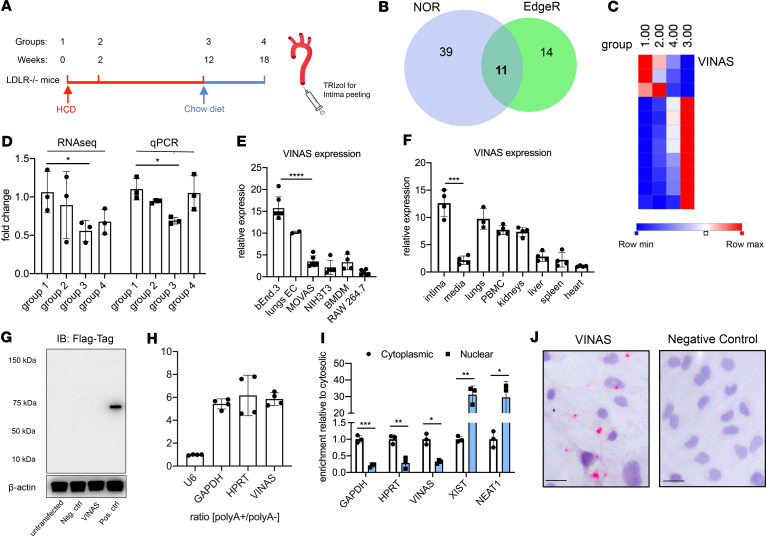
LDL receptor–deficient (B6.129S7-Ldlrtm1Her/J; LDLR–/–) male mice were placed on a high-cholesterol diet (HCD) for 0, 2, and 12 weeks (progression phases, groups 1–3) and subsequently placed on a chow diet for another 6 weeks in the fourth group (regression phase, Figure 1A). RNA was isolated from the aortic intima and RNA-Seq profiling revealed 11 differentially expressed lncRNAs (log2 fold change 1.5; FDR < 0.05) using EdgeR and no overlapping reads (NOR) algorithms (Figure 1B). Eight lncRNAs rose with atherosclerosis progression (group 3) and fell during regression (group 4), whereas only 3 lncRNAs decreased with atherosclerosis progression (Figure 1C). The lncRNA 1500026H17Rik showed the strongest decrease in group 3 (by 59%), while regaining initial levels with atherosclerosis regression as quantified by real-time quantitative PCR (RT-qPCR) (Figure 1, C and D). Because of its high regulability and participation in both vascular inflammation and atherosclerosis, as we show here, we have named this lncRNA VINAS (Vascular INflammation and Atherosclerosis lncRNA Sequence).
Figure 1. Identification of the lncRNA VINAS in lesional intima.
(A) RNA derived from aortic intima of LDLR–/– mice (n = 3; each sample represents RNA pooled from 2 mice) that were placed on a high-cholesterol diet (HCD) for 0 (group 1), 2 (group 2), 12 (group 3), and 18 weeks after 6 weeks of resumption of a normal chow diet (group 4). (B) Venn diagram displays significantly dysregulated lncRNAs in genome-wide RNA-Seq profiling using EdgeR and no-overlapping reads (NOR) showing intersecting hits (n = 11), uniquely identified in EdgeR (n = 14) or NOR (n = 39), (log2 fold change [1.5]; FDR < 0.05). (C) Heatmap for 11 lncRNAs that were dynamically regulated with progression and regression of atherosclerosis (n = 3). (D) RNA-Seq results for VINAS across groups 1–4 obtained by RNA-Seq analysis and verified by RT-qPCR (n = 3). (E) RT-qPCR expression analysis for VINAS in different cell types (n = 3). (F) VINAS expression in body organs and PBMCs of 24-week-old C57BL/6 mice (n = 4). (G) To test the coding potential, VINAS sequence was cloned upstream of 3xFlag-Tag cassette, transfected in HEK293T cells, and immunoblotted for Flag antibody. Positive control was provided with the kit (representative of 3 experiments). (H) RNA from mouse extracellular cells (ECs) was isolated for polyA+ and polyA– enriched RNA and analyzed by real-time quantitative PCR (RT-qPCR) (n = 3). (I) RT-qPCR analysis for RNA derived from mouse ECs separated into cytoplasmic and nuclear fractions and normalized to the cytoplasmic fraction (n = 3). (J) RNA in situ hybridization for negative control and VINAS probes on paraformaldehyde-fixed mouse ECs. Scale bar: 5 μm. Data represent the mean ± SD. Statistical differences were calculated using unpaired 2-tailed Student’s t test except for multiple comparisons (E and F) in which 1-way ANOVA with Bonferroni’s correction was used. *P < 0.05, **P < 0.01, ***P < 0.001.
Further experiments characterized arterial VINAS expression. VINAS expression was higher in ECs (isolated from lungs and b.End.3 cell line) compared with other cell types, such as vascular smooth muscle cells (VSMCs) (MOVAS cell line), NIH3T3 fibroblasts, bone marrow–derived macrophages (BMDMs), or the RAW 264.7 macrophage cell line (Figure 1E), and was broadly expressed in several other organs, with a strong enrichment in the aortic intima compared with the media in the vessel wall (Figure 1F). Our previous study verified the specificity of aortic intima isolation (15). Therefore, to test whether VINAS lncRNA encodes any protein or peptide, the VINAS sequence was cloned upstream of the p3xFLAG-CMV plasmid, transfected in HEK293 cells, and immunoblotted for FLAG Tag, yielding no detectable peptide or protein (Figure 1G). Additionally, VINAS was found to be polyadenylated (Figure 1H) and enriched in the cytosol, as observed by cellular fractionation and by RNA-ISH in mouse ECs (Figure 1, I and J).
VINAS regulates inflammation in vascular cells.
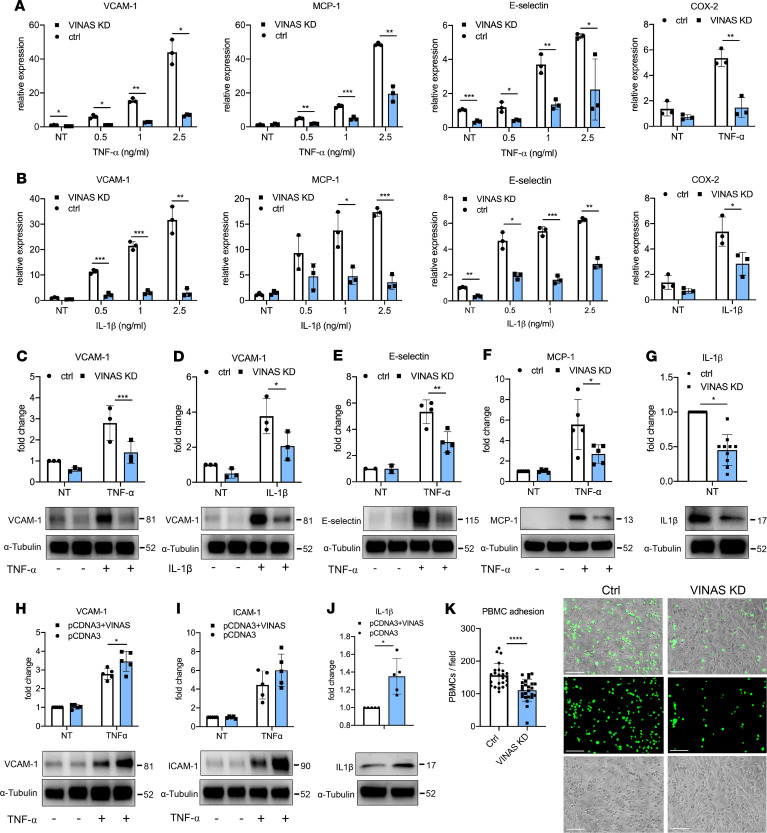
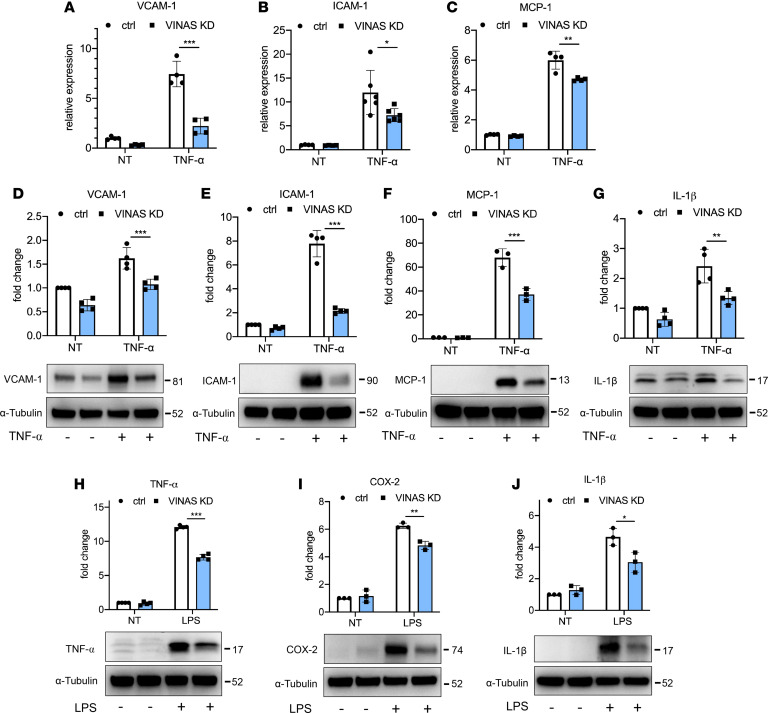
ECs participate pivotally in vascular inflammation and development of atherosclerosis. Because VINAS is enriched in ECs (Figure 1E), the potential phenotype of VINAS loss- and gain-of-function was assessed in mouse ECs. For the knockdown strategy, we designed 3 different locked nucleic acid (LNA) gapmeRs without any significant effect on apoptosis as observed by detection of Caspase 3 cleavage (Supplemental Figures 1B and 3; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.140627DS1) GapmeR 2 showed the highest silencing efficiency in a dose-dependent manner (Supplemental Figure 1C), and it was used throughout the study. The gapmeR-mediated knockdown of VINAS dramatically decreased the mRNA expression of the adhesion molecules VCAM-1 by 50%–95% and E-selectin by approximately 40%–65% in ECs activated with 0.5, 1, and 2.5 ng/mL TNF-α or IL-1β (Figure 2, A and B). In addition, VINAS knockdown in activated ECs reduced the mRNA expression of the chemokine MCP-1 by approximately 50%–80% and the inflammatory molecule COX-2 by approximately 40%–55% (Figure 2, A and B). Moreover, VINAS silencing produced similar effects at the protein level, decreasing VCAM-1 by 45%–55% after activation with 20 ng/mL TNF-α or IL-1β (Figure 2, C and D), and MCP-1, COX-2, and IL-1β by approximately 50% (Figure 2, E–G). Transfection with 2 different VINAS gapmeRs (gapmeR 1 and 3) produced comparable decreases in VCAM-1 and COX-2 in ECs activated with 20 ng/mL TNF-α (Supplemental Figure 1E). In contrast, VINAS overexpression using a pCDNA3 plasmid (Supplemental Figure 1D) had the opposite effect in mouse ECs, increasing the protein expression of VCAM-1 (20%), ICAM-1 (26%), and IL-1β (35%) (Figure 2, H–J). Because VINAS knockdown in ECs decreased the expression of VCAM-1 and E-selectin, 2 cell adhesion molecules known to mediate leukocyte adhesion to ECs, we assessed adhesion of PBMCs to EC monolayers in response to 10 ng/mL TNF-α stimuli. VINAS knockdown reduced PBMCs adherence to EC monolayers by 29% (P < 0.0001), verifying the functional importance of VINAS lncRNA in leukocyte-EC cellular interactions (Figure 2K). Further experiments assessed the antiinflammatory actions of VINAS in 2 other cell types that are enriched in atheroma: VSMCs and BMDMs. We observed similar effects of VINAS knockdown in the MOVAS SMC line, with reduced expression of VCAM-1 (70%), ICAM-1 (40%), and MCP-1 (22%) at the mRNA level and decreased protein expression of VCAM-1 (34%), ICAM-1 (72%), MCP-1 (22%), TNF-α (37%), and IL-1β (44%) after stimulation with 5 ng/mL TNF-α (Figure 3). Consistently, VINAS silencing also decreased COX-2 (19%), IL-1β (38%), and MCP-1 (37%) in primary BMDMs stimulated with 50 ng/mL LPS (Figure 3). Collectively, these findings indicate that VINAS broadly regulates inflammatory mediators in relevant cell types in the vessel wall. The stronger antiinflammatory phenotype observed in ECs compared with VSMCs and BMDMs correlated with the increased VINAS expression in ECs (Figure 1E).
Figure 2. VINAS regulates inflammatory markers in endothelial cells.
VINAS knockdown decreases the mRNA levels of VCAM-1, E-selectin, MCP-1, and COX2 in mouse ECs activated with TNF-α (A) and IL-1β (B); n = 3. VINAS silencing decreases the protein expression of VCAM-1 (C and D, n = 3), E-selectin (E, n = 4), MCP-1 (F, n = 5), and IL-1β (G, n = 10) in basal conditions or after activation with 20 ng/mL TNF- or IL-1β. VINAS overexpression increases the protein expression of VCAM-1 (H), ICAM-1 (I), and IL-1β (J) in mouse ECs not treated or activated with 20 ng/mL TNF-α (n = 5). (K) VINAS knockdown decreases the PBMCs’ adhesion to mouse ECs activated with TNF-α for 4 hours (5 ng/mL, representative of 3 experiments). Scale bar: 50 μm. Data represent the mean ± SD. Statistical differences were calculated using unpaired 2-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Figure 3. VINAS knockdown decreases inflammation in SMCs and BMDMs.
VINAS knockdown decreases mRNA levels of VCAM-1 (A, n = 4), ICAM-1 (B, n = 6), and MCP-1 (C, n = 4) in MOVAS smooth muscle cells (SMCs) stimulated with 5 ng/mL TNF-α. VINAS knockdown decreases protein expression of VCAM-1 (D, n = 4), ICAM-1 (E, n = 4), MCP-1 (F, n = 3), and IL-1β (G, n = 4) in MOVAS smooth muscle cells stimulated with 20 ng/mL TNF-α. VINAS knockdown decreases the protein expression of TNF-α (H), COX-2 (I), and IL-1β (J) in bone marrow–derived macrophages (BMDMs) stimulated with 50 ng/mL LPS (n = 3). Data represent the mean ± SD. Statistical differences were calculated using unpaired 2-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001.
VINAS regulates NF-κB and MAPK signaling pathways in ECs.
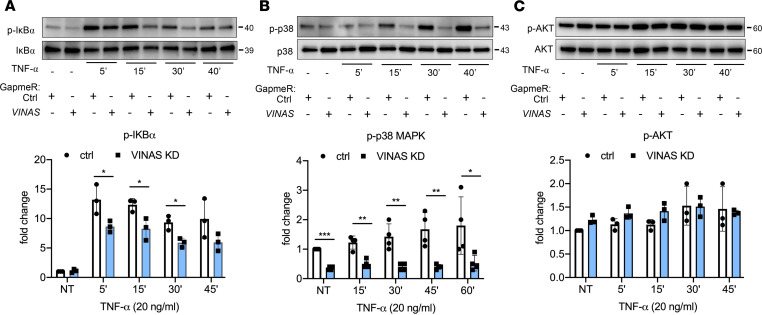
To identify potential signaling pathways subject to VINAS regulation, ECs transfected with VINAS gapmeRs were activated with 20 ng/mL TNF-α for 5–60 minutes and assessed for expression of key proinflammatory signaling pathways. Immunoblotting showed that VINAS knockdown significantly decreased the phosphorylation of IκBα in ECs activated with TNF-α (20 ng/mL) by 35%, 33%, and 37% after 5, 15, and 30 minutes, respectively (Figure 4A). In addition, silencing of VINAS in ECs reduced the phosphorylation of p38 MAPK by 55%–75% in the presence of TNF-α (20 ng/mL) for 15, 30, and 45 minutes (Figure 4B). Similar conditions were tested for AKT signaling pathway and showed no specific effect of VINAS silencing on AKT phosphorylation (Figure 4C). Taken together, these findings indicate that VINAS knockdown regulates predominantly the NF-κB and MAPK signaling pathways.
Figure 4. VINAS knockdown regulates NF-κB and p38 MAPK signaling pathways.
Mouse ECs were transfected with VINAS gapmeRs and activated with TNF-α (20 ng/mL) for 5, 15, 30, 45, and 60 minutes. Phosphorylation of IκBα (A, n = 3), p38 MAPK (B, n = 4), and AKT (C, n = 3) was assessed by Western blot. Data represent the mean ± SD. Statistical differences were calculated using 1-way ANOVA with Bonferroni’s correction. *P < 0.05, **P < 0.01, ***P < 0.001.
In vivo knockdown of VINAS markedly reduced atherosclerotic lesion formation by decreasing vascular inflammation.
To explore whether systemically delivered VINAS gapmeRs modulate atherosclerosis, LDLR–/– mice received i.v. injections of vehicle control or VINAS gapmeR (10 mg/kg/2 times weekly) over 12 weeks on an HCD (Figure 5A). After 12 weeks on an HCD, gapmeR-mediated silencing of VINAS reduced its expression in the aortic intima by 57% (Figure 5H) and in the media by 30% (Figure 5I).
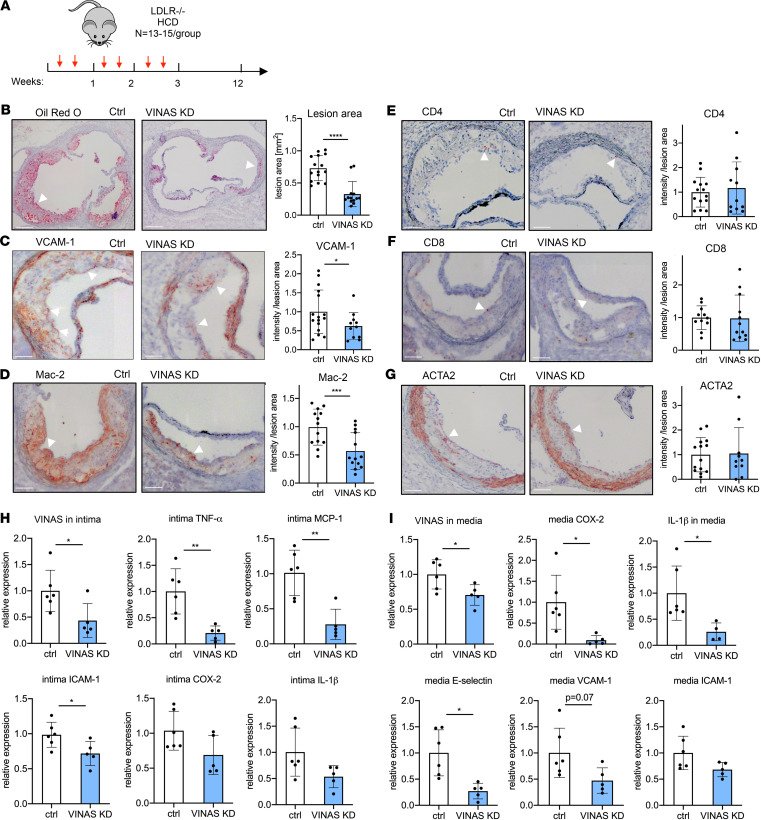
Figure 5. In vivo knockdown of VINAS inhibits atherosclerotic lesion formation by decreasing vascular inflammation.
(A) LDLR–/– mice were i.v. injected with vehicle control gapmeR (n = 15) or VINAS gapmeR (n = 13) twice per week (10 mg/kg/mouse/injection) and placed on an HCD for 12 weeks. Representative images and quantification for Oil Red O (scale bar: 400 μm) (B), VCAM-1 (C), Mac-2 (D), CD4+ (E), CD8+ (F), and ACTA2 (G) staining (arrowhead) of the aortic sinus of LDLR–/– HCD mice treated with control (n = 15) or MAARS (n = 13) gapmeRs for 12 weeks. Scale bar: 100 μm. VINAS silencing efficiency and expression of inflammatory markers was assessed by RT-qPCR in the intima (H) and media (I) fractions of the aortic arch from control gapmeR (n = 6) and VINAS gapmeR groups (n = 5). Data represent the mean ± SD. Statistical differences were calculated using unpaired 2-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Analysis of atherosclerotic lesion formation by Oil Red O (ORO) staining revealed a 55% decrease in lesion area in the aortic sinus after antagonism of VINAS (Figure 5B). Although VINAS knockdown was associated with a modest reduction in total cholesterol (22%), LDL (25%), HDL (6%), and triglycerides (7%) (Supplemental Figure 2A), the lesion areas as quantified by ORO staining remained 48% smaller in VINAS knockdown mice when examined in mice, with similar total cholesterol in both groups (Supplemental Figure 2B). Although approximately 8% of the atherosclerotic plaque reduction may be accounted for effects on cholesterol metabolism, it cannot account entirely for the marked reduction in atherosclerosis lesions following VINAS knockdown.
IHC staining revealed that VCAM-1 and the macrophage marker Mac-2 decreased by 38% and 43%, respectively, in the aortic sinus, indicating reduced vascular inflammation and macrophage accumulation in the vascular wall (Figure 5, C and D). No significant differences were observed for CD4+ or CD8+ T cells or VSMCs after normalization to lesion area (Figure 5, E–G). In vivo knockdown of VINAS in the aortic intima reduced the expression of inflammatory markers TNF-α, MCP-1, ICAM-1, COX-2, and IL-1β (Figure 5H). Moreover, VINAS knockdown in the aortic media decreased inflammatory effectors, such as COX-2, IL-1, E-selectin, VCAM-1, and ICAM-1 (Figure 5I). Although VINAS silencing also reduced circulating PBMCs (62%), it did not significantly alter mRNAs that encode the inflammatory mediators TNF-α, IL-1β, COX-2, and MCP-1 in these cells (Supplemental Figure 1F). Nor did VINAS knockdown alter the antiinflammatory Ly6Clo or the proinflammatory Ly6Cinterm or Ly6Chi fractions in the PBMCs as determined by flow cytometry (Supplemental Figure 1G). Overall, VINAS neutralization in LDLR–/– mice fed an HCD for 12 weeks muted atherosclerotic lesion formation in tandem with decreased inflammation.
DEPDC4 is a VINAS ortholog conserved in humans.
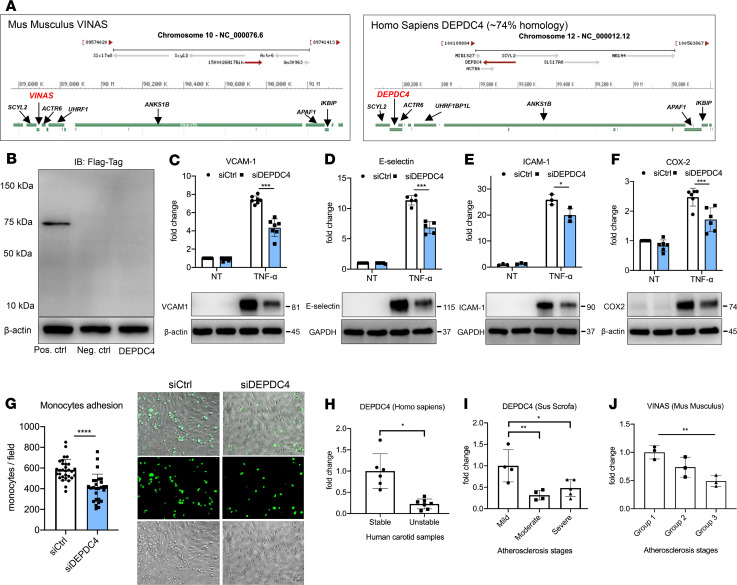
Although VINAS lncRNA is only present in the mouse genome, we observed that the genomic locus is largely conserved, with the genes SCYL2, ACTR6, and ANKS1B in the immediate proximity and the gene DEPDC4 in the same position as VINAS (Figure 6A). BLAST (http://www.ncbi.nlm.nih.gov/blast/) findings showed that DEPDC4 has an approximately 74% homology with VINAS in a region of 157–323 bp, depending on isoform variations (Supplemental Figure 4A). DEPDC4 is widely conserved across species, except for the mouse (Supplemental Figure 4A). To verify the coding probability, the DEPDC4 sequence was cloned upstream of the p3xFLAG-CMV plasmid, transfected in HEK293 cells, and immunoblotted for the FLAG Tag. The resulting immunoblot showed no detectable peptide or protein (Figure 6B). As with VINAS loss of function in mouse cells, DEPDC4 knockdown (Supplemental Figure 4B) induced an antiinflammatory program in HUVECs stimulated with TNF-α, decreasing the expression of VCAM-1 (42%), E-selectin (40%), and COX-2 (30%) (Figure 6, C–E). We then assessed adhesion of THP-1 monocytes to a monolayer of HUVECs in response to TNF-α stimulation. DEPDC4 silencing significantly decreased monocyte adherence to the EC monolayer by 30%, verifying the functional importance of DEPDC4 lncRNA in leukocyte-EC interactions (Figure 6F).
Figure 6. DEPDC4 is a human ortholog of VINAS.
(A) Illustration of the genomic locations of VINAS and DEPDC4 in the mouse and human chromosomes 10 and 12, respectively. (B) DEPDC4 does not encode for a protein or peptide. To test the coding potential, DEPDC4 sequence was cloned upstream of the 3xFlag-Tag cassette, transfected in HEK293T cells, and immunoblotted for Flag antibody; positive control was provided with the kit (n = 3 experiments). DEPDC4 silencing decreases the protein expression of VCAM-1 (C, n = 7), E-selectin (D, n = 5), and ICAM-1 (E, n = 3) COX-2 (F, n = 6) in HUVECs activated with 20 ng/mL TNF-α. (G) DEPDC4 knockdown decreases THP-1 monocyte adhesion to HUVEC monolayers activated with TNF-α for 4 hours (5 ng/mL, representative images and quantification of adhered monocytes). (H) RT-qPCR of DEPDC4 in human carotid arteries with stable (n = 6) or unstable (n = 7) atherosclerotic plaques. Scale bar: 50 μm. (I) Expression of DEPDC4 from RNA-Seq analyses of lesions with increasing severity of coronary atherosclerosis in Yorkshire pigs fed an HCD for 60 weeks (n = 4/group). (J) RT-qPCR of VINAS expression in aortic intima of LDLR–/– mice at 0, 2, and 12 weeks of an HCD (n = 3/group). Data represent the mean ± SD. Statistical differences were calculated using unpaired 2-tailed Student’s t test except for multiple comparisons (I and J) in which 1-way ANOVA with Bonferroni’s correction was used. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To assess the translational relevance of VINAS and DEPDC4 lncRNAs, RNA was isolated from human carotid atherosclerotic plaques with characteristics associated with stability or instability. DEPDC4 expression decreased by 77.4% in carotid arteries with plaques with unstable versus those with stable features (Figure 6G). To explore this expression pattern across species, we analyzed the RNA-Seq data from Yorkshire pigs that were placed for up to 60 weeks on an HCD and developed coronary atherosclerosis. Based on histopathological characterization, the coronary sections were separated into mild, intermediate, and severe groups for progression of atherosclerosis as previously described (15). Similar to VINAS regulation in LDLR–/– mice fed an HCD (Figure 1C), DEPDC4 decreased approximately 60% with disease progression in swine pigs fed an HCD (Figure 6H). Concordantly, in ECs stimulated with TNF-α VINAS and DEPDC4 expression also decrease after 4–8 hours and 16–24 hours, respectively (Supplemental Figure 1, H and I). In summary (Figure 7), these results demonstrate dynamic regulation of the lncRNA VINAS with atherosclerosis progression, that VINAS influences arterial inflammation, and that loss of function of VINAS’s evolutionary conserved lncRNA ortholog DEPDC4 exerts similar antiinflammatory effects.
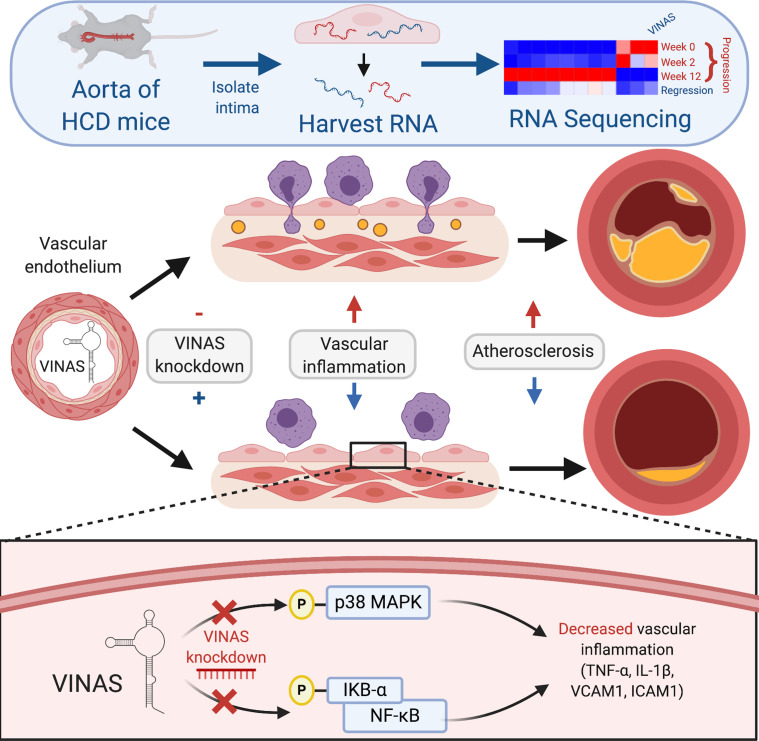
Figure 7. Summary of the role of lncRNA VINAS in vascular inflammation.
RNA-Seq profiling of intimal lesions revealed VINAS lncRNA that is enriched in the aortic intima, decreased with atherosclerotic progression, and increased with regression. VINAS knockdown decreased the expression of key inflammatory markers, NF-κB and MAPK signaling pathways, cell adhesion molecules, and the monocytes adhesion to ECs. In vivo VINAS knockdown reduced atherosclerotic lesion formation in LDLR–/– mice by decreasing vascular inflammation.
Discussion
Arterial inflammation occurs very early in atherogenesis and is associated with impairment of many salutary functions of the healthy endothelium. Accumulating studies point to lncRNAs as regulators of endothelial homeostasis, smooth muscle cell contractility, and macrophage-mediated inflammation in the vessel wall (11, 13, 15–18). This study provides evidence for the first time to our knowledge that the mouse-specific lncRNA VINAS and its human ortholog, DEPDC4, play important roles in vascular inflammation and atherogenesis.
Our study expands upon a growing body of literature implicating lncRNAs as pivotal regulators in the development and progression of atherosclerosis. Our group recently identified SNHG12 as an evolutionary conserved lncRNA that plays an important role in atherogenesis (15). SNHG12 mediated the interaction between DNA damage repair proteins DNA-PK and its binding partners Ku70 and Ku80. Following SNHG12 knockdown in LDLR–/– mice, atherosclerotic lesion area increased by 240%, with corresponding increases in markers of DNA damage and EC senescence (15). The lncRNAs LeXis and MeXis were identified as key regulators of cholesterol metabolism (19, 20). Both of these lncRNAs are transcriptionally regulated by the liver X receptor, a nuclear sterol receptor responsible for transcriptional control of genes involved with cholesterol metabolism. LeXis interacted with the ribonuclear protein RALY to aid in transcription of cholesterol metabolism genes in the liver, and in vivo delivery of LeXis using an adenoviral vector reduced aortic atherosclerosis in mice (21). MeXis altered ABCA1 expression via its binding partner DDX17, and genetic abrogation of MeXis increased serum cholesterol and atherosclerotic lesion area (20).
Similar to VINAS, several other lncRNAs regulate atherosclerosis by modulating inflammatory pathways. For example, the lncRNA NEXN-AS1 lies antisense to and increases the expression of NEXN, a protein that negatively regulates TLR4 and NF-κB signaling (22). Genetic depletion of NEXN-AS1 dramatically increased atherosclerosis in ApoE–/– mice, with concurrent increases in markers of vascular inflammation such as VCAM-1, ICAM-1, TNF-α, and MCP-1. Similarly, knockdown of lncRNA-FA2H-2 increased atherosclerotic plaque size and expression of inflammatory genes. Here, we show that VINAS plays an analogous role in inflammation and atherogenesis, albeit as a proinflammatory lncRNA in contrast to the antiinflammatory lncRNAs NEXN-AS1 or lncRNA-FA2H-2. In vivo delivery of VINAS-specific LNA gapmeRs markedly decreased the expression of important inflammatory mediators and cell adhesion molecules in the intima as well as the media of the aortic arch. VINAS silencing exerted strong antiinflammatory effects across different cellular constituents of the vessel wall, demonstrated by decreased key inflammatory effectors such as MCP-1, TNF-α, IL-1β, COX-2, and the leukocyte adhesion molecules VCAM-1, E-selectin, or ICAM-1, in both ECs and VSMCs (Figure 2 and Figure 3). The stronger antiinflammatory phenotype observed in ECs and the intima is likely attributed to increased VINAS silencing efficiency (Figure 5, H and I; and Supplemental Figure 1, B and C) coupled with the relatively higher expression of VINAS in ECs and intima (Figure 1, E and F) compared with the aortic media. Also, the aortic media is composed of more heterogeneity of cell types (e.g., fibroblasts, VSMCs, and immune cells), and VINAS expression is variable across these different cell types (Figure 1E).
Leukocyte adhesion to activated ECs overexpressing adhesion molecules such as VCAM-1 and E-selectin is among the earliest processes involved in atherosclerotic lesion initiation (23, 24). This study shows that VINAS knockdown in TNF-α–activated ECs significantly reduced monocyte adhesion to EC monolayers (Figure 2K). In line with this finding, in vivo VINAS knockdown decreased the staining of macrophage marker Mac-2 in the aortic root, suggesting a diminished macrophage accumulation in the vessel wall attributed to lower expression of cell adhesion molecules (Figure 5D). Macrophage polarization to a proinflammatory phenotype contributes to the progression and destabilization of atherosclerotic plaques. For example, symptomatic patients suffering from acute transient ischemic attacks with unstable plaques had a higher concentration of M1 proinflammatory macrophages in lesions compared with asymptomatic patients with stable plaques (25, 26). Although the M1/M2 macrophage dichotomy oversimplifies macrophage heterogeneity, an M1 proinflammatory macrophage predominance in atherosclerotic plaques associates with a higher incidence of ischemic stroke and increased lesional inflammation (27). Moreover, plaques from patients with recently symptomatic carotid disease have a predominance of M1 macrophages and higher lipid content than femoral plaques, consistent with a more unstable plaque (28). Although VINAS knockdown in BMDMs in vitro decreased the expression of MCP-1, IL-1β, and COX-2 (Figure 3K), there were no differences in these effectors or of Ly6C+ proinflammatory PBMCs in vivo, suggesting that the antiinflammatory effects of VINAS knockdown in vivo were likely driven more by affecting leukocyte adhesion molecules in intimal ECs (Figure 2 and Figure 3). Although the dominant impact of lncRNA VINAS knockdown is regulating inflammation in the vessel wall, with a 48% reduction in atherosclerotic plaque when cholesterol values are normalized between the groups (Supplemental Figure 2B), we cannot exclude a minor contribution to cholesterol metabolism.
Identification of the potential signaling pathways that lncRNAs regulate is critical from a therapeutic point of view. In some cases, deciphering the signaling pathway and its upstream or downstream regulators can indicate the mechanisms used by a specific lncRNA (29). In this study, VINAS and DEPDC4 knockdown in cytokine-activated ECs reduced the phosphorylation of IκBα and p38 MAPK while having no significant effects on phosphorylation of the AKT signaling pathway (Figure 4). Both the NF-κB and p38 MAPK inflammatory pathways serve as critical nodal points of regulation in atherosclerosis, particularly in the vascular endothelium (30–32). Previously, Gareus et al. demonstrated that endothelium-specific genetic depletion of IKKγ or IκBα, key signaling molecules in the NF-κB pathway, was sufficient to significantly reduce atherosclerosis in ApoE–/– mice (33). Systemic delivery of miRNAs that inhibit NF-κB activation in the vascular endothelium also reduced inflammation and atherosclerosis lesions in ApoE–/– mice (34). Similarly, p38 MAPK inhibitors decreased levels of systemic and vascular inflammation in both mouse models of atherosclerosis (35, 36) as well as humans with coronary artery disease (37, 38). Furthermore, Seeger et al. demonstrated that systemic p38 MAPK inhibition for 4 weeks reduced atherosclerotic lesion size by more than 50% (36). Our study extends these findings by showing that lncRNA VINAS is an important regulator of NF-κB and p38 MAPK signaling pathways and thus exerts considerable control over the development of vascular inflammation and atherosclerosis. The observed antiinflammatory phenotype induced by VINAS knockdown may inform the potential upstream mechanisms by which this lncRNA affects these inflammatory pathways. VINAS lncRNA is enriched in the cytosol, and its knockdown potently reduced the phosphorylation of p38 MAPK, a signaling pathway with its main effectors localized in the cytosol (39–43). Although cytosolic lncRNAs have been reported to interact with miRNAs by a bp-binding mechanism (44, 45), this competing endogenous RNA (ceRNA) hypothesis remains controversial in the field. An in vivo quantitative study showed that modulation of a miRNA target abundance is unlikely to cause significant effects on gene expression and metabolism through a ceRNA effect (46). Future studies exploring the candidate factor(s) mediating this inhibition of dual signaling pathways may further elucidate potential therapeutic targets for atherosclerosis and other chronic inflammatory disease states.
Finally, whereas lncRNAs are not typically as conserved across species compared with other noncoding RNAs, such as miRNAs, emerging studies demonstrate conservation via orthologous transcripts (20). Finding an evolutionary conserved transcript DEPDC4, a human ortholog of VINAS in humans with approximately 74% homology, exhibited regulation in human EC cells congruent to the effects of VINAS on mouse cells supports the human relevance of the present mouse findings. Consistent with VINAS regulation in atherosclerotic mice, DEPDC4 levels declined in coronary arteries of pigs with progression of atherosclerosis and in human carotid plaques with unstable characteristics (Figure 6, G–I). VINAS expression also decreases in ECs after 4 and 8 hours’ incubation with TNF-α, while returning to basal levels after 16–24 hours (Supplemental Figure 1H). In all our experiments, the cytokines were added to the cells at 48 hours after gapmeRs’ transfection, when the VINAS silencing efficiency was already achieved by approximately 90%. Hence, the potential VINAS downregulation after cytokines’ addition would be negligible. Several mechanisms can be responsible for the observed VINAS regulation, including compensatory or feedback mechanisms in response to stress induced by inflammatory stimuli. For instance, LPS induces inflammation via the NF-κB pathway. However, LPS also induces the synthesis of antiinflammatory cytokines such as IL-10 and IL-4, which in turn blocks NF-κB activation in a negative feedback mechanism (47, 48), suggesting that the upregulation of antiinflammatory genes is not always coincident with inflammatory state. Indeed, lncRNAs can be regulated as a negative feedback mechanism during inflammation. For example, LPS increases the expression of lncRNA Mirt2. However, lncRNA Mirt2 serves as a negative feedback regulator of excessive inflammation and reduces inflammation across different cell types (49). Interestingly, the IL-10 antiinflammatory phenotype is regulated by the ubiquitously expressed transcription factor Sp1 (50), which also has multiple binding sites in the VINAS and DEPDC4 promoters (Supplemental Table 2). Although we have not identified the exact mechanism for the upstream regulation of VINAS lncRNA, we cannot rule out the existence of a compensatory mechanism in response to proinflammatory stimuli. Future studies will need to assess the specific upstream mechanism of VINAS/DEPDC4 regulation at the promoter and the transcript levels and whether this is a regulatory effect on RNA stability or a compensatory mechanism in the cell.
In conclusion, the discovery of VINAS reported here extends the understanding of participation of lncRNAs in inflammatory signaling in general and in the pathogenesis of atherosclerosis and potentially other vascular diseases as well. Modulation of lncRNAs VINAS and DEPDC4 may facilitate fine-tuning of the inflammatory response in a range of chronic vascular diseases and perhaps of other organ systems as well.
Methods
RNA-Seq analysis.
RNA-Seq analysis was performed after ribodepletion and standard library construction using Illumina HiSeq2500 V4 2x100 PE (Genewiz). All samples were processed using an RNA-Seq pipeline implemented in the bcbio-nextgen project (https://bcbio-nextgen.readthedocs.org/en/latest/). Raw reads were examined for quality issues using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to ensure library generation and sequencing were suitable for further analysis. Trimmed reads were aligned to UCSC build mm10 of the mouse genome, augmented with transcript information from Ensembl release 79 using STAR (51). Alignments were checked for evenness of coverage, rRNA content, genomic context of alignments (for example, alignments in known transcripts and introns), complexity, and other quality checks using a combination of FastQC and Qualimap. Counts of reads aligning to known genes were generated by featureCounts (52). Differential expression at the gene level was called with EdgeR. The total gene hit counts and counts per million values were calculated for each gene, and downstream differential expression analysis between specified groups was performed using EdgeR and an adapted EdgeR algorithm, which excludes overlapping reads, called NOR. Genes with adjusted FDR of less than 0.05 and log2 fold change 1.5 were called as differentially expressed genes for each comparison. Mean quality score of all samples was 35.67 within a range of 40,000,000–50,000,000 reads per sample. All samples had at least 70% or more mapped fragments over the total. RNA-Seq data are available through the NCBI’s Gene Expression Omnibus database (GSE138219).
Polyadenylation.
RNA of 106 ECs was isolated using TRIzol reagent (Invitrogen) and resuspended in RNase-free water. Polyadenylated and nonpolyadenylated RNA were enriched with the polyA Spin mRNA isolation kit (New England Biolabs, S1560S) based on the manufacturer’s protocol. Real-time PCR (RT-qPCR) was performed with same input volume, independent of concentration, and normalized to nonpolyadenylated RNA fraction.
RNA-ISH.
Customized probe for VINAS was specifically developed to detect ENSMUST00000181598 (Advanced Cell Diagnostics). BMDMs were fixed in 4% paraformaldehyde, and the in situ hybridization protocol for cultured adherent cells was performed as described by the manufacturer (Basescope 2.5 HD Reagent Kit-Red; Advanced Cell Diagnostics).
Protein coding potential.
Transcripts for VINAS (1500026H17Rik, NCBI Ref. Seq. NR_130956.1, Ensemble ID ENSMUST00000181598) were synthesized by Genewiz. For in vitro validation of peptide coding potential, VINAS transcript was cloned upstream of p3xFLAG-CMV-14 expression vector (MilliporeSigma, E7908) using EcoRI restriction site. HEK293T cells (ATCC, CRL-11268) were transfected with 500 ng plasmid using Lipofectamine 2000 (Invitrogen), and protein lysate was isolated 72 hours after transfection, followed by immunoblotting for FLAG Tag (Cell Signaling Technology, 8146).
Molecular cloning for VINAS overexpression.
For overexpression studies, the VINAS transcript synthesized by Genewiz was cloned in a pCDNA.3 plasmid using the EcoRI restriction site. The integration was validated by DNA sequencing. For transfection studies in ECs, 0.25 μg plasmid/well (12-well plate) was used in combination with Lipofectamine 3000, according to the manufacturer’s instructions.
Cell culture and transfection.
Mouse ECs (b.End.3, ATCC, CRL-2299), MOVAS mouse aortic SMCs (ATCC, CRL-279), and RAW 264.7 cells (ATCC, TIB-71) were cultured in DMEM with 10% FBS and 1% penicillin/streptomycin. HUVECs (Lonza, cc-2159) were cultured in EC growth medium EGM-2 (Lonza, cc-3162). Cells passaged less than 7 times were used for all experiments. Bone marrow was isolated from the femur and tibia of C57BL/6 mice and cultured in IMDM supplemented with 10 ng/mL mouse macrophage colony stimulation factor (416ML, R&D Systems, Bio-Techne), 10% FBS, and 1% penicillin/streptomycin. Medium was changed every 2 days, and cells were used in experiments after 7–10 days in culture. Transfection was performed using Lipofectamine 3000 (Invitrogen, 11668-019) according to the manufacturer’s protocol, and customized gapmeRs for VINAS (QIAGEN, 25 nmol except when mentioned differently) or negative control 1 (QIAGEN). Cells were allowed to grow for 36 hours before treatment with recombinant human TNF-α (210-TA/CF, R&D Systems, Bio-Techne), IL-1β (401-ML, R&D Systems, Bio-Techne), or LPS (O26:B6 Escherichia coli; MilliporeSigma L2654) for various times, according to the experiment: Western blot, 16 hours; RT-qPCR, 6 hours.
Cell adhesion assay.
ECs grown in 24-well plates were transfected with gapmeRs. After 35 hours, 20 ng/mL TNF-α was added for 5 hours. PBMCs were isolated from C57BL/6 mice, washed, and suspended at 5 × 106 cells/mL in medium with 5 μM Calcein AM (C3100MP; Invitrogen). Cells were kept in an incubator containing 5% CO2 at 37°C for 30 minutes. The labeling reaction was stopped by the addition of the cell growth medium, and cells were washed with growth medium twice and resuspended in growth medium at 5 × 105 cells/mL. After 4 hours of TNF-α treatment, ECs were washed once with DMEM growth medium, and 500 μL Calcein AM–loaded PBMCs were added to each well. After 1 hour of incubation, nonadherent cells were removed carefully. Adherent cells were gently washed with prewarmed DMEM 4 times and were counted using a Nikon fluorescence microscope (Eclipse TE2000-U).
RNA isolation and RT-qPCR.
Tissues were homogenized using TissueLyser II (QIAGEN) according to the manufacturer’s instructions. For RNA isolation, TRIzol reagent (Invitrogen) or RNeasy kit (QIAGEN) was used based on the manufacturer’s protocol. Isolation of intimal RNA and subsequent RT-qPCR from aorta was performed as previously documented (34, 53).
Briefly, aortas were carefully flushed with PBS, followed by intima peeling using TRIzol reagent (Invitrogen, 15596018). TRIzol was flushed for 10 seconds, followed by a 10-second pause, then for another 10 seconds flushed, collected in an Eppendorf tube (~300–400 μL total), and snap frozen in liquid nitrogen. The intima-specific isolation was assessed by qPCR showing enrichment of endothelial marker CD31 and macrophage marker Mac2 in the intima fraction compared with the media/adventitia fraction as previously described (15). Subsequent RT-qPCR was performed using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, 4368813). GoTaq qPCR Master Mix (Promega, A6001) was used for RT-qPCR experiments. Expression of mRNAs and lncRNA expression levels were normalized to GAPDH, HPRT, or β-actin (Agilent, AriaMx Real-Time PCR System). Changes in expression were calculated using the ΔΔCt method. Primer sequences are described in Supplemental Table 1.
Cellular fractionation.
EC fractionation for cytoplasmic and nuclear fractions was performed using the Active Motif kit (catalog 40410) according to the manufacturer’s protocol. RNA was harvested as described previously and cleaned up using the RNeasy kit (QIAGEN). Equivalent RNA volumes of cytoplasmic and nuclear-associated RNA were converted to cDNA as described previously.
Western blot.
Proteins were isolated using RIPA buffer (Boston BioProducts, BP-115) with protease inhibitor (Roche, 4693132001) and phosphatase inhibitors (New England Biolabs, P0758L). Protein concentrations were determined using Pierce BCA assay (Thermo Fisher Scientific). A total of 20 μg protein were loaded per lane on a 4%–20% Mini-PROTEAN TGX Gel (Bio-Rad, 456-1096). Separated proteins were transferred to PVDF membranes using the Transfer Turbo Blot system (Bio-Rad) and Trans-Blot Turbo RTA Transfer Kit (Bio-Rad, 170-4272). The membrane was blocked with 5% nonfat milk in TBST for 1hour at room temperature. After blocking, the membrane was incubated overnight at 4°C with antibodies against Flag Tag (Cell Signaling Technology, 2368, 1:1000), GAPDH (Cell Signaling Technology, 2118, 1:4000), VCAM-1 (Cell Signaling Technology, sc-13160, 1:1000), ICAM-1 (R&D Systems, Bio-Techne, BBA3), IκBα (Cell Signaling Technology, 4812, 1:1000), β-actin (Cell Signaling Technology, 4970, 1:3000), and phospho-IκBα (Cell Signaling Technology, 2859, 1:1000), IL-1β (Abcam ab9722, 1:1000), MCP-1 (Abcam ab25124, 1:1000), COX-2 (Cell Signaling Technology 12282p), p-P38MAPK (Cell Signaling Technology 4511L, 1:1000), and P38 MAPK (Cell Signaling Technology 9212L, 1:1000). Quantification of protein bands was performed using a luminescent image analyzer (Bio-Rad, Chemidoc).
Immunohistology and characterization of atherosclerotic lesions.
To quantify atherosclerosis in LDLR–/– mice that were placed on a HCD (Research Diets Inc., D12108C), aortic roots and aortic arch were embedded in OCT and frozen at –80°C. Serial cryostat sections (6 μm) were prepared using tissue processor Leica CM3050. Lesion characterizations, including ORO staining of the thoracic-abdominal aorta and aortic root and staining for macrophages (anti-Mac2, BD Pharmingen, 553322, 1:900), T cells (anti-CD4, BD Pharmingen, 553043, 1:90; anti-CD8, Chemicon, CBL1318, 1:100), and VSMCs (SM-α-actin, MilliporeSigma, F-3777, 1:500), were performed as previously described (34, 54). The staining area was measured using Image-Pro Plus software, Media Cybernetics, and CD4+ and CD8+ cells were counted manually.
Intimal RNA isolation from aorta tissue.
Isolation of intimal RNA from aorta was performed as previously described (34, 53). Briefly, aortas were carefully flushed with PBS, followed by intima peeling using TRIzol reagent (Invitrogen, 15596018). TRIzol was flushed for 10 seconds followed by a 10-second pause, then for another 10 seconds flushed, collected in an Eppendorf tube (~300–400 μL total), and snap frozen in liquid nitrogen. The intima-specific isolation was assessed in a previous study (15) by qPCR showing enrichment of endothelial marker CD31 in the intima fraction compared with media/adventitia fraction.
Lipid profile analysis.
Lipid profile was measured as previously described (34). Briefly, triglyceride levels were determined using Infinity Triglycerides Liquid Stable Reagent (Thermo Fisher Scientific). Total cholesterol was measured using the Infinity Cholesterol Reagent (Thermo Fisher Scientific), and HDL-cholesterol was measured by colorimetric assay (BioAssay Systems, EnzyChrom HDL). LDL-cholesterol levels were calculated using the following formula: LDL = total cholesterol – HDL-cholesterol – (triglycerides/5). Standards were purchased from Pointe Scientific, Inc.
Pig atherosclerotic samples.
The study protocol included 15 male hypercholesterolemic Yorkshire swine (Pine Acres Farm) that were placed on an HCD for up to 60 weeks. Detailed sectioning of 3 mm coronary artery segments was performed so that the gene sequencing samples were derived from the exact same portions of the coronary artery plaques used for the histology and IHC analyses. Histology and IHC analyses included H&E, van Gieson elastin staining, α–smooth muscle actin, ORO staining, picrosirius red staining, and CD31 and CD45 cells as described previously (15, 55).
Human atherosclerotic specimens.
RNA was isolated from human carotid atherosclerotic lesions that were obtained from the Division of Cardiovascular Medicine, Brigham and Women’s Hospital in accordance with the IRB-approved protocol for use of discarded human tissues (protocol 2010-P-001930/2).
Statistics.
For illustration of differentially expressed genes, GraphPad Prism software (V.7.0a) was used.
Data are shown as the mean ± SD. Statistical differences were calculated using unpaired 2-tailed Student’s t test or 1-way ANOVA with Bonferroni’s correction for multiple comparisons. A P value of less than 0.05 was considered significant.
Study approval.
All protocols concerning animal use were approved by the IACUC at Brigham and Women’s Hospital and Harvard Medical School and conducted in accordance with the NIH’s Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). Studies were performed in LDLR–/– male mice (The Jackson Laboratory, stock 002207) or in C57BL/6 mice (Charles River Laboratories, strain code 027). The IRB approved the use of discarded human tissues (protocol 2010-P-001930/2).
Author contributions
MWF and VS conceived the hypothesis. VS, HZ, JBP, DY, and YT performed the experiments. VS, HZ, JBP, SH, GS, PS, PL, and MWF designed and interpreted the results. VS and MWF wrote the manuscript.
Supplementary Material
Acknowledgments
We would like to thank J. Hutchinson and L. Pantano (Harvard Chan Bioinformatics Core, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA) for assistance with RNA-Seq analysis. This work was supported by the NIH (HL115141, HL134849, HL148207, HL148355, HL153356 to MWF), the Arthur K. Watson Charitable Trust (to MWF), the Ralph and Marian Falk Medical Research Trust (to MWF), the American Heart Association (18SFRN33900144 and 20SFRN35200163 to MWF), and the Sarnoff Foundation Fellowship Award (JBP). PL received funding from the National Heart, Lung, and Blood Institute (R01HL080472), and American Heart Association (18CSA34080399).
Version 1. 10/06/2020
In-Press Preview
Version 2. 11/05/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, Simion et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2020;5(21):e140627.https://doi.org/10.1172/jci.insight.140627.
Contributor Information
Viorel Simion, Email: simiond.viorel@gmail.com.
Haoyang Zhou, Email: hzhou14@bwh.harvard.edu.
Jacob B. Pierce, Email: jacob.pierce@northwestern.edu.
Dafeng Yang, Email: yangdf0823@163.com.
Stefan Haemmig, Email: stefan.haemmig@bluewin.ch.
Yevgenia Tesmenitsky, Email: eugeniat_1@msn.com.
Galina Sukhova, Email: gsukhova@rics.bwh.harvard.edu.
Peter H. Stone, Email: pstone@bwh.harvard.edu.
Peter Libby, Email: plibby@bwh.harvard.edu.
Mark W. Feinberg, Email: mfeinberg@bwh.harvard.edu.
References
- 1.Libby P. Inflammation in atherosclerosis. Arterioscler Thromb Vasc Biol. 2012;32(9):2045–2051. doi: 10.1161/ATVBAHA.108.179705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolf D, Ley K. Immunity and inflammation in atherosclerosis. Circ Res. 2019;124(2):315–327. doi: 10.1161/CIRCRESAHA.118.313591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Virchow R. Cellular pathology. As based upon physiological and pathological histology. Lecture XVI--Atheromatous affection of arteries. 1858. Nutr Rev. 1989;47(1):23–25. doi: 10.1111/j.1753-4887.1989.tb02747.x. [DOI] [PubMed] [Google Scholar]
- 4.Ridker PM, et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377(12):1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 5.Nahrendorf M. Myeloid cell contributions to cardiovascular health and disease. Nat Med. 2018;24(6):711–720. doi: 10.1038/s41591-018-0064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Libby P. Mechanisms of acute coronary syndromes and their implications for therapy. N Engl J Med. 2013;368(21):2004–2013. doi: 10.1056/NEJMra1216063. [DOI] [PubMed] [Google Scholar]
- 7.Mullick AE, Soldau K, Kiosses WB, Bell TA, Tobias PS, Curtiss LK. Increased endothelial expression of Toll-like receptor 2 at sites of disturbed blood flow exacerbates early atherogenic events. J Exp Med. 2008;205(2):373–383. doi: 10.1084/jem.20071096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gimbrone MA, García-Cardeña G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res. 2016;118(4):620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doran AC, Meller N, McNamara CA. Role of smooth muscle cells in the initiation and early progression of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28(5):812–819. doi: 10.1161/ATVBAHA.107.159327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ley K, Miller YI, Hedrick CC. Monocyte and macrophage dynamics during atherogenesis. Arterioscler Thromb Vasc Biol. 2011;31(7):1506–1516. doi: 10.1161/ATVBAHA.110.221127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simion V, Haemmig S, Feinberg MW. LncRNAs in vascular biology and disease. Vascul Pharmacol. 2019;114:145–156. doi: 10.1016/j.vph.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aryal B, Suárez Y. Non-coding RNA regulation of endothelial and macrophage functions during atherosclerosis. Vascul Pharmacol. 2019;114:64–75. doi: 10.1016/j.vph.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haemmig S, Simion V, Feinberg MW. Long non-coding RNAs in vascular inflammation. Front Cardiovasc Med. 2018;5:22. doi: 10.3389/fcvm.2018.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haemmig S, Simion V, Yang D, Deng Y, Feinberg MW. Long noncoding RNAs in cardiovascular disease, diagnosis, and therapy. Curr Opin Cardiol. 2017;32(6):776–783. doi: 10.1097/HCO.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haemmig S, et al. Long noncoding RNA SNHG12 integrates a DNA-PK-mediated DNA damage response and vascular senescence. Sci Transl Med. 2020;12(531):eaaw1868. doi: 10.1126/scitranslmed.aaw1868. [DOI] [PubMed] [Google Scholar]
- 16.Leisegang MS, et al. Long noncoding RNA MANTIS facilitates endothelial angiogenic function. Circulation. 2017;136(1):65–79. doi: 10.1161/CIRCULATIONAHA.116.026991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahmoud AD, et al. The human-specific and smooth muscle cell-enriched LncRNA SMILR promotes proliferation by regulating mitotic CENPF mRNA and drives cell-cycle progression which can be targeted to limit vascular remodeling. Circ Res. 2019;125(5):535–551. doi: 10.1161/CIRCRESAHA.119.314876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stapleton K, et al. Novel long noncoding RNA, macrophage inflammation-suppressing transcript (MIST), regulates macrophage activation during obesity. Arterioscler Thromb Vasc Biol. 2020;40(4):914–928. doi: 10.1161/ATVBAHA.119.313359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sallam T, et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature. 2016;534(7605):124–128. doi: 10.1038/nature17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sallam T, et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat Med. 2018;24(3):304–312. doi: 10.1038/nm.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tontonoz P, Wu X, Jones M, Zhang Z, Salisbury D, Sallam T. Long noncoding RNA facilitated gene therapy reduces atherosclerosis in a murine model of familial hypercholesterolemia. Circulation. 2017;136(8):776–778. doi: 10.1161/CIRCULATIONAHA.117.029002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu YW, et al. Long noncoding RNA NEXN-AS1 mitigates atherosclerosis by regulating the actin-binding protein NEXN. J Clin Invest. 2019;129(3):1115–1128. doi: 10.1172/JCI98230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Cybulsky MI, Gimbrone MA, Libby P. An atherogenic diet rapidly induces VCAM-1, a cytokine-regulatable mononuclear leukocyte adhesion molecule, in rabbit aortic endothelium. Arterioscler Thromb. 1993;13(2):197–204. doi: 10.1161/01.ATV.13.2.197. [DOI] [PubMed] [Google Scholar]
- 24.Libby P. Mechanisms of acute coronary syndromes. N Engl J Med. 2013;369(9):883–884. doi: 10.1056/NEJMc1307806. [DOI] [PubMed] [Google Scholar]
- 25.de Gaetano M, Crean D, Barry M, Belton O. M1- and M2-type macrophage responses are predictive of adverse outcomes in human atherosclerosis. Front Immunol. 2016;7:275. doi: 10.3389/fimmu.2016.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams HJ, Fisher EA, Greaves DR. Macrophage differentiation and function in atherosclerosis: opportunities for therapeutic intervention? J Innate Immun. 2012;4(5–6):498–508. doi: 10.1159/000336618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho KY, et al. The phenotype of infiltrating macrophages influences arteriosclerotic plaque vulnerability in the carotid artery. J Stroke Cerebrovasc Dis. 2013;22(7):910–918. doi: 10.1016/j.jstrokecerebrovasdis.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 28.Shaikh S, Brittenden J, Lahiri R, Brown PA, Thies F, Wilson HM. Macrophage subtypes in symptomatic carotid artery and femoral artery plaques. Eur J Vasc Endovasc Surg. 2012;44(5):491–497. doi: 10.1016/j.ejvs.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Han J, et al. LncRNAs2Pathways: Identifying the pathways influenced by a set of lncRNAs of interest based on a global network propagation method. Sci Rep. 2017;7:46566. doi: 10.1038/srep46566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hajra L, Evans AI, Chen M, Hyduk SJ, Collins T, Cybulsky MI. The NF-kappa B signal transduction pathway in aortic endothelial cells is primed for activation in regions predisposed to atherosclerotic lesion formation. Proc Natl Acad Sci U S A. 2000;97(16):9052–9057. doi: 10.1073/pnas.97.16.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker RG, Hayden MS, Ghosh S. NF-κB, inflammation, and metabolic disease. Cell Metab. 2011;13(1):11–22. doi: 10.1016/j.cmet.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bäck M, Hansson GK. Anti-inflammatory therapies for atherosclerosis. Nat Rev Cardiol. 2015;12(4):199–211. doi: 10.1038/nrcardio.2015.5. [DOI] [PubMed] [Google Scholar]
- 33.Gareus R, et al. Endothelial cell-specific NF-kappaB inhibition protects mice from atherosclerosis. Cell Metab. 2008;8(5):372–383. doi: 10.1016/j.cmet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 34.Sun X, et al. Systemic delivery of microRNA-181b inhibits nuclear factor-κB activation, vascular inflammation, and atherosclerosis in apolipoprotein E-deficient mice. Circ Res. 2014;114(1):32–40. doi: 10.1161/CIRCRESAHA.113.302089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris JB, et al. p38 MAPK inhibition reduces aortic ultrasmall superparamagnetic iron oxide uptake in a mouse model of atherosclerosis: MRI assessment. Arterioscler Thromb Vasc Biol. 2008;28(2):265–271. doi: 10.1161/ATVBAHA.107.151175. [DOI] [PubMed] [Google Scholar]
- 36.Seeger FH, Sedding D, Langheinrich AC, Haendeler J, Zeiher AM, Dimmeler S. Inhibition of the p38 MAP kinase in vivo improves number and functional activity of vasculogenic cells and reduces atherosclerotic disease progression. Basic Res Cardiol. 2010;105(3):389–397. doi: 10.1007/s00395-009-0072-9. [DOI] [PubMed] [Google Scholar]
- 37.Sarov-Blat L, et al. Inhibition of p38 mitogen-activated protein kinase reduces inflammation after coronary vascular injury in humans. Arterioscler Thromb Vasc Biol. 2010;30(11):2256–2263. doi: 10.1161/ATVBAHA.110.209205. [DOI] [PubMed] [Google Scholar]
- 38.Elkhawad M, et al. Effects of p38 mitogen-activated protein kinase inhibition on vascular and systemic inflammation in patients with atherosclerosis. JACC Cardiovasc Imaging. 2012;5(9):911–922. doi: 10.1016/j.jcmg.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75(1):50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hommes DW, Peppelenbosch MP, van Deventer SJ. Mitogen activated protein (MAP) kinase signal transduction pathways and novel anti-inflammatory targets. Gut. 2003;52(1):144–151. doi: 10.1136/gut.52.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68(2):320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie Y, Xie K, Gou Q, Chen N. IκB kinase α functions as a tumor suppressor in epithelial-derived tumors through an NF-κB-independent pathway (Review) Oncol Rep. 2015;34(5):2225–2232. doi: 10.3892/or.2015.4229. [DOI] [PubMed] [Google Scholar]
- 44.Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018;9(7):722. doi: 10.1038/s41419-018-0759-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu DM, et al. LncRNA SNHG15 acts as a to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell Death Dis. 2018;9(10):947. doi: 10.1038/s41419-018-0975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54(5):766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iyer SS, Ghaffari AA, Cheng G. Lipopolysaccharide-mediated IL-10 transcriptional regulation requires sequential induction of type I IFNs and IL-27 in macrophages. J Immunol. 2010;185(11):6599–6607. doi: 10.4049/jimmunol.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukherjee S, Chen LY, Papadimos TJ, Huang S, Zuraw BL, Pan ZK. Lipopolysaccharide-driven Th2 cytokine production in macrophages is regulated by both MyD88 and TRAM. J Biol Chem. 2009;284(43):29391–29398. doi: 10.1074/jbc.M109.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du M, et al. The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat Commun. 2017;8(1):2049. doi: 10.1038/s41467-017-02229-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tone M, Powell MJ, Tone Y, Thompson SA, Waldmann H. IL-10 gene expression is controlled by the transcription factors Sp1 and Sp3. J Immunol. 2000;165(1):286–291. doi: 10.4049/jimmunol.165.1.286. [DOI] [PubMed] [Google Scholar]
- 51.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 53.Sun X, et al. MicroRNA-181b regulates NF-κB-mediated vascular inflammation. J Clin Invest. 2012;122(6):1973–1990. doi: 10.1172/JCI61495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao Z, et al. Kruppel-like factor KLF10 targets transforming growth factor-beta1 to regulate CD4(+)CD25(-) T cells and T regulatory cells. J Biol Chem. 2009;284(37):24914–24924. doi: 10.1074/jbc.M109.000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koskinas KC, et al. Synergistic effect of local endothelial shear stress and systemic hypercholesterolemia on coronary atherosclerotic plaque progression and composition in pigs. Int J Cardiol. 2013;169(6):394–401. doi: 10.1016/j.ijcard.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.