Abstract
Cancer cells exist within a complex spatially structured ecosystem composed of resources and different cell types. As the selective pressures imposed by this environment determine the fate of cancer cells, an improved understanding of how this ecosystem evolves will better elucidate how tumors grow and respond to therapy. State of the art imaging methods can now provide highly resolved descriptions of the microenvironment, yielding the data required for a thorough study of its role in tumor growth and treatment resistance. The field of landscape ecology has been studying such species-environment relationship for decades, and offers many tools and perspectives that cancer researchers could greatly benefit from. Here, we discuss one such tool, species distribution modeling (SDM), that has the potential to, among other things, identify critical environmental factors that drive tumor evolution and predict response to therapy. SDMs only scratch the surface of how ecological theory and methods can be applied to cancer, and we believe further integration will take cancer research in exciting new and productive directions. Significance: Here we describe how species distribution modeling can be used to quantitatively describe the complex relationship between tumor cells and their microenvironment. Such a description facilitates a deeper understanding of cancers eco-evolutionary dynamics, which in turn sheds light on the factors that drive tumor growth and response to treatment.
Keywords: niche, habitat, species distribution modeling, ecology, microenvironment, immune system, immunotherapy, imaging
Introduction
The fate of a cancer cell depends on its ability to survive and replicate in its local microenvironment. If local conditions are inhospitable, there is Darwinian selection for cells that can either remodel the microenvironment, or adapt to hostile conditions. Remodeling may come in many forms, such as promoting angiogenesis or suppressing immune attack, while adaptation to hostile environments may include evolving the ability to tolerate high levels of acid, or “hiding” from predatory cytotoxic T-cells. In both cases, the dialog between cancer cells and the microenvironment can generate spatial heterogeneity, wherein cancer cells survive within distinct ecological niches.
The tumor microenvironment can be quantitatively described in great detail using a variety of modalities, including immunohistochemistry (IHC), immunofluorescence (IF), Magnetic resonance imaging (MRI), positron emission tomography (PET), and fluorescence in situ hybridization (FISH).1 However, despite the high quality and resolution of the data, tumors are typically described in terms of individual cell type abundances and, while the importance of space is often acknowledged, analysis of spatial heterogeneity is rare. Furthermore, little is understood about the dynamic aspects of these spatial relationships as current quantitative technologies are often destructive in nature. This is unfortunate, as a more dynamic and spatial view of cell-cell and cell-environment relationships has the potential to reveal how tumors adapt and survive, under diverse changing environments, which in turn may provide insights into how to control or eliminate tumors.
In ecological terms, the collection of various interacting cell types and environmental factors (e.g. oxygen, glucose, acid, growth factors, cytokines) that make up a functional tissue and dysfunctional tumor would be described as an ecosystem. The field of landscape ecology has been studying such multi-scale, high-dimensional, and spatially heterogeneous systems for decades.2,3 It is almost certain that cancer researchers will benefit from the approaches that have been, and continue to be, developed by landscape ecologists. In particular, ecologists have developed a wide variety of approaches to describe ecosystems. Adopting methods developed to describe scaling relationships, spatial landscape patterns, and species-environment relationships will allow cancer researchers to describe and compare tumors as entities, as opposed to examining cell types and environmental factors in isolation. Such an approach will provide a more complete and holistic view of the tumor ecosystem.
Of particular interest to the cancer community may be the approaches landscape ecology has developed to describe and predict how environmental factors determine the distribution of species. These approaches go under many names, including species distribution modeling (SDM), habitat suitability modeling, and niche modeling. A frequent application of these approaches is to determine which sort of environments support a species of interest, typically with an eye on management. As we will discuss in more detail below, if applied to cancer, SDMs have the potential to reveal new risk factors, and predict response to treatments.
Use of Habitat Models in Ecology
Species distribution modeling is a highly active field in ecology. In 2013, it was estimated that approximately 1000 papers with species distribution models were published each year.4 Some researchers use these models for the purpose of creating spatially-explicit maps of suitable habitat, which can be applied to managing threatened species, managing non-native invasive species, or implementing other kinds of spatial planning. SDMs are also used for identifying the environmental predictor variables that are most important for predicting the presence or abundance of target species. More recently, researchers have been applying SDMs to study changes in habitat suitability through time; these models are applied retrospectively5 and also to forecasting future changes.6
The ecological niche, a hypervolume in multivariate environmental space that depicts a species’ environmental limitations, is a central theme in SDMs.7 Models may be correlative or mechanistic in nature.8 In a correlative approach to species distribution modeling, field observations are used to statistically link species occurrence data with environmental data. These models begin with geo-referenced observations of a species and the environmental conditions (e.g., climate, soil) at those sites. The environmental conditions where the species is observed are inferred to be within that species’ tolerance range, and other sites with similar conditions are assumed to be similarly suitable. In contrast, mechanistic species distribution models differ from correlative models in that they consider how the environment constrains physiological performance.9 For example, the distribution of ectothermic (i.e., “cold-blooded”) species is limited by the range of temperatures under which they can survive.10 Suitable micro-climates for these species can be modeled in a spatially-explicit fashion using information such as solar radiation, topography, and vegetation cover.11
In addition to environmental variables, interactions with other species can also influence a species’ habitat selection. The presence of a mutualist might expand the environmental range of a species into otherwise unsuitable habitat, as in the case of some plants and their mutualistic mycorrhizal fungi.12 Conversely, the presence of competitors might restrict the distribution of a species beyond the limitations imposed by other environmental variables, as was seen for grazing mammals on African savannas.13 These interactions are known to be important but aren’t often included in habitat models; they might be particularly important for predicting future distributions under scenarios of climate change.14
Many different mathematical approaches have been applied to modeling species distributions. One early approach to SDMs was logistic regression, including stepwise approaches to identify the best predictor variables.15 Since then, methods have expanded to include classification and regression trees, genetic algorithms, maximum entropy methods, Bayesian approaches, and even agent-based models.16,17 A computer platform (BIOMOD) has been developed to compare and combine predictions from multiple modeling approaches, called “ensemble modeling.”18 A variety of SDM approaches can also be conducted using the popular R programming language.19
Species distribution models can use 3 kinds of species data: presence–absence, presence-background (if absences cannot be confirmed), and occupancy-detection (which incorporates the probability of detecting a species if it is present). The suitability of a particular modeling approach depends on the type of data that are available,20 Species distribution models also rely heavily on environmental data derived from geographic information science (GIS) and remote sensing.4 Species abundance models (SAMs) can be used for the same purposes, but use abundance, as discussed in21 SAMs may be a good option to study tumor habitats, given that cell segmentation and phenotyping is often performed using histology, therefore making this type of data somewhat easier to acquire.
Cancer Habitats
While much of cancer research has focused on quantifying the disease through the genomic lens, viewing the tumor as an ecosystem has been advocated for quite some time.22-31 This perspective argues that, while (epi-)genetic mutation is the source of variation, the environment is what imposes selection pressures, and therefore supervenes on the genotype. Furthermore, a single phenotype (e.g. drug resistance) can be encoded by multiple genotypes,32,33 therefore developing therapies which target the phenotype, not the genotype, may have a much greater impact. Before this can begin, however, a more detailed understanding of tumor-microenvironment relationships is required, especially if we hope to predict tumor eco-evolutionary dynamics. Application of SDMs to the tumor ecosystem presents an important opportunity to elucidate these relationships.
Since tumor biopsies and scans are routinely collected, the data required for SDMs is potentially both abundant and readily accessible.1 Multiplex immunohistochemistry, immunofluorescence imaging mass cytometry, and cyclic multiplex immunofluorescence (IF) can “stain” up to 37 and 50 cell markers on the same tissue, respectively,34-36 providing a highly detailed and spatially resolved description of the microenvironment. RNA in-situ imaging methods, reviewed in,37 can provide key information on cell behavior and phenotype, which can be an important component of SDMs, particularly ABMs.17 MRI and PET imaging have been used to define unique habitats within the tumor, such as necrosis or areas of high proliferation.38,39 Combined, these imaging modalities quantify numerous environmental variables, such as vasculature, hypoxia, acid, necrosis, growth factors, cytokines, etc…, that can predict the distribution of various cell types, such as tumor cells or immune cells.
Several studies have used the above imaging modalities to study and describe the tumor niche. In prostate, brain, and breast tumors, radiologically defined habitats have been used to detect hypoxic and/or necrotic niche, a prognostic marker for survival and response to treatment.40,41 Schürch et al. used the CO-Detection by indEXing (CODEX) platform to define 9 distinct cellular neighborhoods using 56 different cell markers, with the goal of developing a prognostic spatial signature of anti-tumor immune response.42 The immunoscore, used as a prognostic marker in colorectal cancer, is based on the numeration of 2 lymphocyte populations in tumor core versus the invasive margin43 In Gatenbee et al., several ecological methods were used to describe and compare the immune ecologies of colorectal adenomas and carcinomas to better understand what changes take place during the transition from benign to malignant colorectal cancer.44 Further, computational models have been used to simulate tumor-microenvironment eco-evolutionary dynamics, often with a focus on examining how the microenvironment shapes tumor evolution and response to therapy (recently reviewed in.24,45
Potential Applications of Habitat Modeling to Cancer
Species distribution models complement the current approaches cancer researchers are taking to study the tumor ecosystem. Because they are, at their core, statistical models, SDMs can be used to determine which environmental factors best predict the spatial distribution of cell types, such as tumor cells or cytotoxic T-cells. This can be accomplished by using information criteria to select the best SDM, revealing which factors play the largest role in creating suitable tumor or immune habitats.46 Such critical environmental factors may then serve as potential therapeutic targets, which when perturbed create inhospitable tumor habitats. Since habitats directly affect the cell phenotype, creation of such inhospitable habitats may have a larger impact on the tumor cells than targeted therapies that focus on specifically mutated populations.
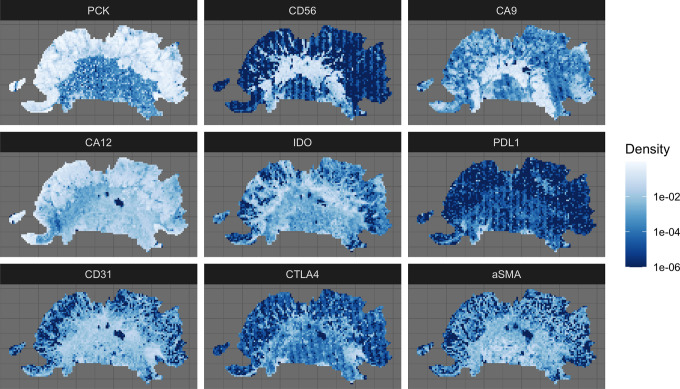
SDMs are not limited to static snapshots, but are also able to predict habitat suitability over time.47 Ecological forecasting uses SDMs to predict what would happen to a species distribution should the environment change. Figure 1 provides an illustrative example of how ecological forecasting using an SDM could be used to predict tumor response to an immunotherapy that facilitates invasion of cytotoxic T-cells into the tumor.
Figure 1.
Quadrat count data used to predict CD56 high regions of the tumor. PCK (Pan Cytokeratin) is a marker for tumor cells, and while it is not used in the SDM, it is shown to put the rest of the markers into context. CD56 is a marker for natural killer (NK), activated CD8 T cells, dendritic cells (DC); CA9 (carbonic anhydrase 9) and CA 12 are markers of hypoxia and low pH; IDO (Indoleamine 2,3-dioxygenase an immunosuppressive factor; PD-L1 (programmed death-protein 1 ligand) and CTLA-4 (cytotoxic T-lymphocyte-associated protein 4) are immune checkpoints; CD31 is a marker for endothelial cells; aSMA is a marker for cancer associated fibroblasts.
Since many primary tumors are resected during early treatment, tumor biopsies tend to be one off opportunities. Even in the metastatic setting, repeat biopsies are difficult to obtain, therefore temporal data of this kind can be hard to come by. However, tumor progression is often described in terms of sequential stages, progressing from homeostatic to benign to malignant to metastatic disease. Given tumors collected at different stages, temporal SDMs could potentially be used to study progression, potentially revealing which factors are driving tumorigenesis.
Example Application
In the following example, we use an SDM to get a better understanding of how the microenvironment shaped the immune response in a non-smoker with squamous cell carcinoma (SCC) of the head and neck. An increased understanding of the dialog between the microenvironment and immune response has potential significance for engineering effective immunotherapies.
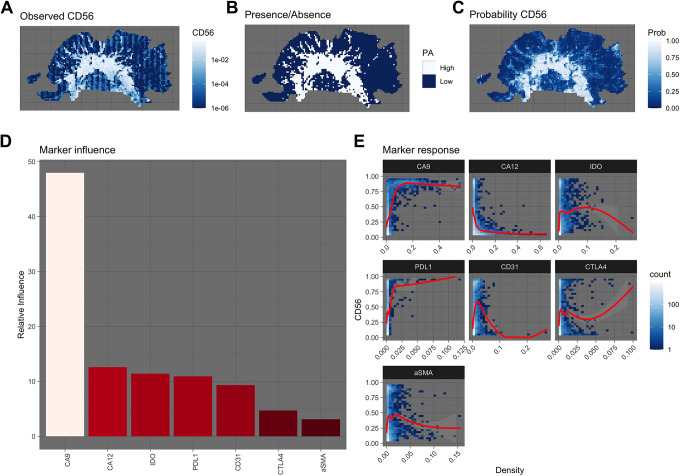
A slice from the patient’s biopsy was stained for 7 microenvironmental makers and an immune marker, CD56 (Figure 1). CD56 is the archetypal marker for natural killer (NK) cells, but is also found on other inflammatory cells, including gamma delta (γδ) T cells, activated CD8 T cells, and dendritic cells (DCs).48 After undergoing co-registration and stain segmentation, each image, scanned at 40x magnification, was divided into quadrats of 200 x 200 microns. Within each quadrat, the density of each marker was calculated by dividing the number of positive pixels by the quadrat area (in pixel units). The focus of the SDM was to estimate the degree to which each environmental factor affects the spatial distribution of CD56. As such, we labeled each quadrat as being either CD56 high or CD56 low (Figure 2). A quadrat was considered CD56 high if its CD56 density was greater than or equal to the 70th percentile of CD56 densities in all quadrats (Figure 2B).
Figure 2.
A Observed density of CD56. B Presence/absence map indicating high and low regions of CD56. C The trained SDM’s predicted probabilities of CD56 high regions. D The overall influence each marker has on the SDM’s ability to predict the location of CD56 high regions. E Response plot, showing how each marker influences the probability of there being a CD56 high region. Blues indicate the density of points, while the red line shows the trend line fit using a generalize additive model.
To estimate how the microenvironment determines the spatial distribution of CD56 high regions, we fit a boosted regression tree (BRT) to the data, using the R package, dismo.49,50 The optimal model was found using cross-validation, and the final model had an AUC (Area Under the Curve) ROC (Receiver Operating Characteristics) score of 0.88. This value indicates a fairly good fit to the data, as a value of 0.5 would mean the prediction is as good as a random guess, while a value of 1 would indicate a perfect fit.
Examining the relative importance of each microenvironmental variable in predicting the presence of CD56 reveals that CA9, a marker of hypoxia, is the dominant factor (Figure 2D). One can determine how each marker affects the distribution of CD56 high regions by examining the marker response functions, which show how the probability of a CD56 high region changes with marker density (Figure 2E). Here, the probability of a CD56 high region increases dramatically with CA9. Both of these observations are in line with experimental observations that hypoxia can favor the recruitment and survival of CD56 bright NK cells.51
This example illustrates how one might use an SDM to better understand how, and to what degree, the components of the microenvironment sculpt the anti-tumor immune response. However, application of SDMs to larger set of samples and markers would be needed to make more general predictions and gain further insights.
Conclusions
Oncology needs ecology. Imaging provides an overwhelming amount of multidimensional data, similar to what the field of landscape ecology has been working with for decades. The toolsets and approaches developed by ecologists can and should be adopted by cancer researchers to better describe and understand the tumor as a complex evolving system. The field of ecology may also benefit by investigating the tumor ecosystem. Due to the large amounts of available data, sophisticated experimental designs, and computational/mathematical models and theory, ecological studies of tumors may also be used to study fundamental ecological phenomenon, such as niche engineering, or succession.
While the evolutionary nature of cancer (initiation, progression and treatment) is becoming more readily accepted, the importance of viewing cancer through the lens of ecology still remains in its infancy. Here we have only scratched the surface of where landscape ecology can contribute to oncology, without a doubt there is so much more. As technology continues to enhance how we can measure cancer, not only at the genomic scale but across multiple spatial and temporal scales, the importance of cancer ecology will only continue to grow.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: CG and ARAA are supported by Physical Sciences Oncology Network (PSON) grant from the National Cancer Institute (Grant No. U54CA193489). ARAA also acknowledges support from the Cancer Systems Biology Consortium grant from the National Cancer Institute (Grant No. U01CA23238). ARAA also acknowledges support from the Moffitt Cancer Center of Excellence for Evolutionary Therapy.
ORCID iD: Chandler D. Gatenbee  https://orcid.org/0000-0002-9730-5964
https://orcid.org/0000-0002-9730-5964
Alexander R. A. Anderson  https://orcid.org/0000-0002-2536-4383
https://orcid.org/0000-0002-2536-4383
References
- 1. Lloyd M, Johnson J, Kasprzak A, Bui M. Image Analysis of the Tumor Microenvironment In R K. (Ed.), Systems Biology of Tumor Microenvironment. Advances in Experimental Medicine and Biology 2016;1–10:936:Springer [DOI] [PubMed] [Google Scholar]
- 2. Romme WH, Knight DH. Landscape diversity: the concept applied to Yellowstone Park. BioScience. 1982;32(8):664–670. doi:10.2307/1308816 [Google Scholar]
- 3. Wiens JA. Population responses to patchy environments. Ann Rev Ecol Syst. 1976;7(1):81–120. doi:10.1146/annurev.es.07.110176.000501 [Google Scholar]
- 4. Franklin J. Species distribution models in conservation biogeography: developments and challenges. Divers Distrib. 2013;19(10):1217–1223. doi:10.1111/ddi.12125 [Google Scholar]
- 5. Porzig EL, Seavy NE, Gardali T, Geupel GR, Holyoak M, Eadie JM. Habitat suitability through time: using time series and habitat models to understand changes in bird density. Ecosphere. 2014;5(2):art12 doi:10.1890/ES13-00166.1 [Google Scholar]
- 6. Kearney MR, Wintle BA, Porter WP. Correlative and mechanistic models of species distribution provide congruent forecasts under climate change. Conserv Lett. 2010;3(3):203–213. doi:10.1111/j.1755-263X.2010.00097.x [Google Scholar]
- 7. Sillero N. What does ecological modelling model? A proposed classification of ecological niche models based on their underlying methods. Ecol Model. 2011;222(8):1343–1346. 10.1016/j.ecolmodel.2011.01.018 [DOI] [Google Scholar]
- 8. Buckley LB, Urban MC, Angilletta MJ, Crozier LG, Rissler LJ, Sears MW. Can mechanism inform species’ distribution models? Ecol Lett. 2010;13(8):1041–1054. doi:10.1111/j.1461-0248.2010.01479.x [DOI] [PubMed] [Google Scholar]
- 9. Evans TG, Diamond SE, Kelly MW. Mechanistic species distribution modelling as a link between physiology and conservation. Conserv Physiol. 2015;3(1):cov056 doi:10.1093/conphys/cov056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deutsch CA, Tewksbury JJ, Huey RB, et al. Impacts of climate warming on terrestrial ectotherms across latitude. Proc Natl Acad Sci U S A. 2008;105(18):6668–6672. doi:10.1073/pnas.0709472105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bramer I, Anderson BJ, Bennie J, et al. Advances in monitoring and modelling climate at ecologically relevant scales In: Bohan DA, Dumbrell AJ, Woodward G, Jackson M, eds. Advances in Ecological Research. Academic Press; 2018: 101–161:58 Chap 3. [Google Scholar]
- 12. Peay KG. The mutualistic niche: mycorrhizal symbiosis and community dynamics. Ann Rev Ecol Evol Syst. 2016;47(1):143–164. doi:10.1146/annurev-ecolsys-121415-032100 [Google Scholar]
- 13. Mpakairi KS, Ndaimani H, Tagwireyi P, Gara TW, Zvidzai M, Madhlamoto D. Missing in action: species competition is a neglected predictor variable in species distribution modelling. PLoS One. 2017;12(7):e0181088 doi:10.1371/journal.pone.0181088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wisz MS, Pottier J, Kissling WD, et al. The role of biotic interactions in shaping distributions and realised assemblages of species: implications for species distribution modelling. Biol Rev. 2013;88(1):15–30. doi:10.1111/j.1469-185X.2012.00235.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Udevitz MS, Bloomfield P, Apperson CS. Prediction of the occurrence of four species of mosquito larvae with logistic regression on water-chemistry variables. Environ Entomol. 1987;16(1):281–285. doi:10.1093/ee/16.1.281 [Google Scholar]
- 16. Guisan A, Thuiller W. Predicting species distribution: offering more than simple habitat models. Ecol Lett. 2005;8(9):993–1009. doi:10.1111/j.1461-0248.2005.00792.x [DOI] [PubMed] [Google Scholar]
- 17. Semeniuk CAD, Musiani M, Marceau DJ. (October 10th 2011). In: Sofo (Ed.), Integrating Spatial Behavioral Ecology in Agent-Based Models for Species Conservation, Biodiversity. IntechOpen. [Google Scholar]
- 18. Thuiller W, Lafourcade B, Engler R, Araújo MB. BIOMOD—a platform for ensemble forecasting of species distributions. Ecography. 2009;32(3):369–373. doi:10.1111/j.1600-0587.2008.05742.x [Google Scholar]
- 19. Hijmans RJ, Phillips S, Leathwick J, Elit J. Dismo: species distribution modeling. 2017. https://cran.r-project.org/web/packages/dismo/index.html
- 20. Guillera-Arroita G, Lahoz-Monfort JJ, Elith J, et al. Is my species distribution model fit for purpose? Matching data and models to applications. Glob Ecol Biogeogr. 2015;24(3):276–292. doi:10.1111/geb.12268 [Google Scholar]
- 21. Johnston A, Fink D, Reynolds MD, et al. Abundance models improve spatial and temporal prioritization of conservation resources. Ecol Appl. 2015;25(7):1749–1756. doi:10.1890/14-1826.1 [DOI] [PubMed] [Google Scholar]
- 22. Alfarouk KO, Ibrahim ME, Gatenby RA, Brown JS. Riparian ecosystems in human cancers. Evol Appl. 2013;6(1):46–53. doi:10.1111/eva.12015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Basanta D, Anderson ARA. Exploiting ecological principles to better understand cancer progression and treatment. Interface Focus. 2013;3(4):20130020 doi:10.1098/rsfs.2013.0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Basanta D, Anderson ARA. Homeostasis back and forth: an ecoevolutionary perspective of cancer. Cold Spring Harb Perspect Med. 2017;7(9):a028332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Daoust SP, Fahrig L, Martin AE, Thomas F. From forest and agro-ecosystems to the microecosystems of the human body: what can landscape ecology tell us about tumor growth, metastasis, and treatment options? Evol Appl. 2013;6(1):82–91. doi:10.1111/eva.12031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gatenby RA. Population ecology issues in tumor growth. Cancer Res. 1991;51(10):2542–2547. [PubMed] [Google Scholar]
- 27. Lee HO, Silva AS, Concilio S, et al. Evolution of tumor invasiveness: the adaptive tumor microenvironment landscape model. Cancer Res. 2011;71(20):6327 doi:10.1158/0008-5472.CAN-11-0304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maley CC, Aktipis A, Graham TA, et al. Classifying the evolutionary and ecological features of neoplasms. Nat Rev Cancer(1474-1768 (Electronic)). [DOI] [PMC free article] [PubMed]
- 29. Pienta KJ, McGregor N, Axelrod R, Axelrod DE. Ecological therapy for cancer: defining tumors using an ecosystem paradigm suggests new opportunities for novel cancer treatments. Transl Oncol. 2008;1(4):158–164. doi:10.1593/tlo.08178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pienta KJ, McGregor N, Axelrod R, Axelrod DE. Ecological therapy for cancer: defining tumors using an ecosystem paradigm suggests new opportunities for novel cancer treatments. Transl Oncol. 2008;1(4):158–164. doi:10.1593/tlo.08178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang KR, Mooney SM, Zarif J, Coffey DS, Taichman RS, Pienta K. A cooperative pathway to enhance cancer cell fitness though ecosystem engineering. J Cell Biochem. 2014;115 doi:10.1002/jcb.24813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gerlee P, Kim E, Anderson ARA. Bridging scales in cancer progression: mapping genotype to phenotype using neural networks. Semin Cancer Biol. 2015;30:30–41. doi:10.1016/j.semcancer.2014.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi:10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chang Q, Ornatsky OI, Siddiqui I, Loboda A, Baranov VI, Hedley DW. Imaging mass cytometry. Cytometry A. 2017;91(2):160–169. doi:10.1002/cyto.a.23053 [DOI] [PubMed] [Google Scholar]
- 35. Goltsev Y, Samusik N, Kennedy-Darling J, et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174(4):968–981 e915 doi:10.1016/j.cell.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Remark R, Merghoub T, Grabe N, et al. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci Immunol. 2016;1(1):aaf6925 doi:10.1126/sciimmunol.aaf6925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pichon X, Lagha M, Mueller F, Bertrand E. A growing toolbox to image gene expression in single cells: sensitive approaches for demanding challenges. Mol Cell. 2018;71(3):468–480. doi:10.1016/j.molcel.2018.07.022 [DOI] [PubMed] [Google Scholar]
- 38. Chang YCC, Ackerstaff E, Tschudi Y, et al. Delineation of tumor habitats based on dynamic contrast enhanced MRI. Sci Rep. 2017;7(1):9746 doi:10.1038/s41598-017-09932-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gaw N, Hawkins-Daarud A, Hu LS, et al. Integration of machine learning and mechanistic models accurately predicts variation in cell density of glioblastoma using multiparametric MRI. Sci Rep. 2019;9(1):10063 doi:10.1038/s41598-019-46296-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chang YC, Ackerstaff E, Tschudi Y, et al. Delineation of tumor habitats based on dynamic contrast enhanced MRI. Sci Rep. 2017;7(1):9746 doi:10.1038/s41598-017-09932-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jardim-Perassi BV, Huang S, Dominguez-Viqueira W, et al. Multiparametric MRI and co-registered histology identify tumor habitats in breast cancer mouse models. Cancer Res. 2019;79(15):3952–3964. doi:10.1158/0008-5472.CAN-19-0213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schürch CM, Bhate SS, Barlow GL, et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. bioRxiv. 2019;743989 doi:10.1101/743989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Galon J, Mlecnik B, Bindea G, et al. Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J Pathol. 2014;232(2):199–209. doi:10.1002/path.4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gatenbee CD, Baker AM, Schenck RO, et al. Niche engineering drives early passage through an immune bottleneck in progression to colorectal cancer. bioRxiv. 2019;623959 doi:10.1101/623959 [Google Scholar]
- 45. Metzcar J, Wang Y, Heiland R, Macklin P. A review of cell-based computational modeling in cancer biology. JCO Clin Cancer Inform. 2019;3:1–13. doi:10.1200/CCI.18.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stevens BS, Conway CJ. Predicting species distributions: unifying model selection and scale optimization for multi-scale occupancy models. Ecosphere. 2019;10(5):e02748 doi:10.1002/ecs2.2748 [Google Scholar]
- 47. Rodríguez de Rivera Ó, López-Quílez A, Blangiardo M. Assessing the spatial and spatio-temporal distribution of forest species via bayesian hierarchical modeling. Forests. 2018;9(9):573. [Google Scholar]
- 48. Van Acker HH, Capsomidis A, Smits EL, Van Tendeloo VF. CD56 in the immune system: more than a marker for cytotoxicity? Front Immunol. 2017;8:892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Elith J, Leathwick JR. Boosted regression trees for ecological modeling. [DOI] [PubMed]
- 50. Elith J, Leathwick JR, Hastie T. A working guide to boosted regression trees. J Anim Ecol. 2008;77(4):802–813. [DOI] [PubMed] [Google Scholar]
- 51. Parodi M, Raggi F, Cangelosi D, et al. Hypoxia modifies the transcriptome of human NK cells, modulates their immunoregulatory profile, and influences NK cell subset migration. Front Immunol. 2018;9:2358. [DOI] [PMC free article] [PubMed] [Google Scholar]