Abstract
Background
Gegen Qinlian (GGQL) decoction is a common Chinese herbal compound for the treatment of ulcerative colitis (UC). In this study, we aimed to identify its molecular target and the mechanism involved in UC treatment by network pharmacology and molecular docking. Material and Methods. The active ingredients of Puerariae, Scutellariae, Coptis, and Glycyrrhiza were screened using the TCMSP platform with drug‐like properties (DL) ≥ 0.18 and oral availability (OB) ≥ 30%. To find the intersection genes and construct the TCM compound-disease regulatory network, the molecular targets were determined in the UniProt database and then compared with the UC disease differential genes with P value < 0.005 and ∣log2 (fold change) | >1 obtained in the GEO database. The intersection genes were subjected to protein-protein interaction (PPI) construction and Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. After screening the key active ingredients and target genes, the AutoDock software was used for molecular docking, and the best binding target was selected for molecular docking to verify the binding activity.
Results
A total of 146 active compounds were screened, and quercetin, kaempferol, wogonin, and stigmasterol were identified as the active ingredients with the highest associated targets, and NOS2, PPARG, and MMP1 were the targets associated with the maximum number of active ingredients. Through topological analysis, 32 strongly associated proteins were found, of which EGFR, PPARG, ESR1, HSP90AA1, MYC, HSPA5, AR, AKT1, and RELA were predicted targets of the traditional Chinese medicine, and PPARG was also an intersection gene. It was speculated that these targets were the key to the use of GGQL in UC treatment. GO enrichment results showed significant enrichment of biological processes, such as oxygen levels, leukocyte migration, collagen metabolic processes, and nutritional coping. KEGG enrichment showed that genes were particularly enriched in the IL-17 signaling pathway, AGE-RAGE signaling pathway, toll-like receptor signaling pathway, tumor necrosis factor signaling pathway, transcriptional deregulation in cancer, and other pathways. Molecular docking results showed that key components in GGQL had good potential to bind to the target genes MMP3, IL1B, NOS2, HMOX1, PPARG, and PLAU.
Conclusion
GGQL may play a role in the treatment of ulcerative colitis by anti-inflammation, antioxidation, and inhibition of cancer gene transcription.
1. Background
Ulcerative colitis (UC) is chronic nonspecific enteritis with typical clinical manifestations of abdominal pain, diarrhea, and mucopurulent bloody stools; it mostly manifests as superficial ulcers occurring in the rectum and sigmoid colon but can also spread to the proximal colon and even the entire colon, causing permanent fibrosis and tissue damage. Despite improvements in treatment prospects, the incidence of long-term colectomy has not decreased over the last 10 years, and achieving mucosal healing early may be strongly associated with a reduced risk of future colectomy [1].
Gegen Qinlian (GGQL) decoction is from Treatise on Febrile and Miscellaneous Disease, and the monarch drug Pueraria has antipyretic and antidiarrheal properties; meanwhile, the subject drugs Scutellaria and Coptis have a function to eliminate dampness and heat and Glycyrrhiza can replenish qi.
A meta-analysis including 2028 patients defined that GGQL could improve clinical symptoms and reduce an endoscopic severity index (UCEIS) and recurrence rate; further, it also led to fewer adverse events whether it was used alone or in combination with Western medicine [2]. According to spectral efficiency studies, its main components—puerarin, daidzin, and coptisine—act through a synergistic relationship, but the specific mechanism of action is not clear.
Network pharmacology combines pharmacology, bioinformatics, and several other sciences with system network analysis and explains the multicomponent and multitarget drug treatment mechanism from the direction of gene distribution, molecular function, and signaling pathways by constructing a network related to “disease-phenotype-gene-drug,” which is suitable for the study of traditional Chinese medicine (TCM) compounds.
2. Materials and Methods
2.1. Screening of Active Ingredients and Target Genes
The active ingredients of the GGQL herbs, including Puerariae, Scutellariae, Coptis, and Glycyrrhiza, were screened using the TCMSP platform (http://lsp.nwu.edu.cn/tcmsp.php) with drug‐like properties (DL) ≥ 0.18 and oral bioavailability (OB) ≥ 30% as conditions. The predicted targets of the screened compounds were acquired from the DrugBank (https://www.drugbank.ca/) database and verified literature. Meanwhile, the UniProt database (https://www.uniprot.org/) was used for comparison of target information and gene name standardization.
2.2. Acquisition of Differential Genes
The genetic samples—Series: GSE38713—of UC patients and healthy people were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The script was run in Strawberry Perl-5.30.2.1 (Perl) software, and the gene probe names were annotated as gene symbols and grouped. The “limma” package was installed in the Perl software, and the sample values were corrected and subjected to log2 (logFC) transformation. Samples with P value < 0.005 and ∣log2 (fold change) | >1 were screened and considered to have statistically significant differential genes. The gene volcano map of the samples was generated, and the top 20 genes with the most significant up- and downregulation were selected to draw the heat map.
2.3. Traditional Chinese Medicine Compound-Disease Regulatory Network
The Perl software was used to acquire intersection genes of the disease differential genes and the target genes of TCM, as well as the active ingredients of TCM. Subsequently, the TCM compound-disease regulatory network was generated using the Cytoscape software.
2.4. Protein-Protein Interaction (PPI) Network and Topological Analysis
The “bisogenet, cytoNAC” package was installed in the Cytoscape 3.8.0 software, and the intersection genes were entered, and the parameter “homo sapiens” was selected. Data for constructing the PPI network were sourced from six main experimental research databases: Database of Interacting Proteins, Biological General Repository for Interaction Datasets, Human Protein Reference Database, IntAct molecular interaction database, Molecular INTeraction Database, and Biomolecular Interaction Network Database. The method “input nodes and its neighbors” was selected to obtain the PPI network and perform topological analysis based on its network centrality.
2.5. GO and KEGG Enrichment Analysis
The R package including “colorspace,” “stringi,” and “ggplot2” was installed in software R 4.0.0, and a bioconductor package that includes “DOSE,” “clusterProfiler,” and “enrichplot” was used for GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. The function “enrichGO” was used for GO enrichment analysis. The database was org. Hs .eg. db (DOI: 10.18129/http://b9.bioc.org.Hs.eg.db); the “enrichKEGG” function was applied for KEGG enrichment analysis, and the database was the KEGG database (https://www.kegg.jp/). As for the parameters of the two functions, species was set to “has,” and the filter values (i.e., P value and q-value) were set to 0.05. The first 20 enrichment results were visualized as a bar graph, and the KEGG regulatory network was generated by the Cytoscape 3.8.0 software.
2.6. Molecular Docking
The target genes involved in the first 10 pathways of the KEGG enrichment results were searched in the PDB database (https://www.rcsb.org), of which the 3D protein conformations with a crystal resolution of lower than 3 Å as determined by X crystal diffraction were acquired. The Mol2 format files of GGQL key active ingredients were downloaded from the TCMSP platform. The AutoDockTools 1.5.6 software was applied to process proteins as follows: separate proteins, add nonpolar hydrogen, calculate the Gasteiger charge, and assign the AD4 type, and set all the flexible bonds of small molecule ligands to be rotatable. According to the original ligand coordinates, the docking box was adjusted to include all protein structures. Meanwhile, the receptor protein was set to rigid docking, the genetic algorithm was selected, and the maximum number of evals was set as the medium. The docking results were obtained by running autogrid4 and autodock4, by which the binding energies were revealed. The partial diagram of molecular docking was then generated using the PyMol software.
3. Results
3.1. Screening of Active Ingredients and Target Genes
In the TCMSP database, GGQL contains 489 active ingredients in total, including 18 in Puerariae, 143 in Scutellariae, 48 in Coptis, and 280 in Glycyrrhiza. They were then screened with OB ≥ 30% and DL ≥ 0.18 as the conditions. Consequently, 146 active ingredients were obtained, including 4 in Puerariae, 36 in Scutellariae, 14 in Coptis, and 92 in Glycyrrhiza (Table 1). The targets were predicted by the DrugBank database and UniProt database. Eventually, 2660 targets—97 in Puerariae, 507 in Scutellariae, 287 in Coptis, and 1769 in Glycyrrhiza—were obtained.
Table 1.
The total available compounds of GGQL.
| Drug | ID | Compound | OB (%) | DL |
|---|---|---|---|---|
| Pueraria | MOL000392 | Formononetin | 69.67 | 0.21 |
| Pueraria | MOL000358 | Beta-sitosterol | 36.91 | 0.75 |
| Pueraria | MOL002959 | 3′-Methoxydaidzein | 48.57 | 0.24 |
| Pueraria | MOL003629 | Daidzein-4,7-diglucoside | 47.27 | 0.67 |
| Scutellaria | MOL001689 | Acacetin | 34.97 | 0.24 |
| Scutellaria | MOL000173 | Wogonin | 30.68 | 0.23 |
| Scutellaria | MOL000228 | (2R)-7-Hydroxy-5-methoxy-2-phenylchroman-4-one | 55.23 | 0.2 |
| Scutellaria | MOL002714 | Baicalein | 33.52 | 0.21 |
| Scutellaria | MOL002908 | 5,8,2′-Trihydroxy-7-methoxyflavone | 37.01 | 0.27 |
| Scutellaria | MOL002909 | 5,7,2,5-Tetrahydroxy-8,6-dimethoxyflavone | 33.82 | 0.45 |
| Scutellaria | MOL002910 | Carthamidin | 41.15 | 0.24 |
| Scutellaria | MOL002911 | 2,6,2′,4′-Tetrahydroxy-6′-methoxychaleone | 69.04 | 0.22 |
| Scutellaria | MOL002913 | Dihydrobaicalin_qt | 40.04 | 0.21 |
| Scutellaria | MOL002914 | Eriodyctiol (flavanone) | 41.35 | 0.24 |
| Scutellaria | MOL002915 | Salvigenin | 49.07 | 0.33 |
| Scutellaria | MOL002917 | 5,2′,6′-Trihydroxy-7,8-dimethoxyflavone | 45.05 | 0.33 |
| Scutellaria | MOL002925 | 5,7,2′,6′-Tetrahydroxyflavone | 37.01 | 0.24 |
| Scutellaria | MOL002926 | Dihydrooroxylin A | 38.72 | 0.23 |
| Scutellaria | MOL002927 | Skullcapflavone II | 69.51 | 0.44 |
| Scutellaria | MOL002928 | Oroxylin A | 41.37 | 0.23 |
| Scutellaria | MOL002932 | Panicolin | 76.26 | 0.29 |
| Scutellaria | MOL002933 | 5,7,4′-Trihydroxy-8-methoxyflavone | 36.56 | 0.27 |
| Scutellaria | MOL002934 | Neobaicalein | 104.34 | 0.44 |
| Scutellaria | MOL002937 | Dihydrooroxylin | 66.06 | 0.23 |
| Scutellaria | MOL000358 | Beta-sitosterol | 36.91 | 0.75 |
| Scutellaria | MOL000359 | Sitosterol | 36.91 | 0.75 |
| Scutellaria | MOL000525 | Norwogonin | 39.4 | 0.21 |
| Scutellaria | MOL000552 | 5,2′-Dihydroxy-6,7,8-trimethoxyflavone | 31.71 | 0.35 |
| Scutellaria | MOL000073 | Ent-epicatechin | 48.96 | 0.24 |
| Scutellaria | MOL000449 | Stigmasterol | 43.83 | 0.76 |
| Scutellaria | MOL001458 | Coptisine | 30.67 | 0.86 |
| Scutellaria | MOL001490 | Bis[(2S)-2-ethylhexyl] benzene-1,2-dicarboxylate | 43.59 | 0.35 |
| Scutellaria | MOL001506 | Supraene | 33.55 | 0.42 |
| Scutellaria | MOL002879 | Diop | 43.59 | 0.39 |
| Scutellaria | MOL002897 | Epiberberine | 43.09 | 0.78 |
| Scutellaria | MOL008206 | Moslosooflavone | 44.09 | 0.25 |
| Scutellaria | MOL010415 | 11,13-Eicosadienoic acid, methyl ester | 39.28 | 0.23 |
| Scutellaria | MOL012245 | 5,7,4′-Trihydroxy-6-methoxyflavanone | 36.63 | 0.27 |
| Scutellaria | MOL012246 | 5,7,4′-Trihydroxy-8-methoxyflavanone | 74.24 | 0.26 |
| Scutellaria | MOL012266 | Rivularin | 37.94 | 0.37 |
| Coptis | MOL001454 | Berberine | 36.86 | 0.78 |
| Coptis | MOL013352 | Obacunone | 43.29 | 0.77 |
| Coptis | MOL002894 | Berberrubine | 35.74 | 0.73 |
| Coptis | MOL002897 | Epiberberine | 43.09 | 0.78 |
| Coptis | MOL002903 | (R)-Canadine | 55.37 | 0.77 |
| Coptis | MOL002904 | Berlambine | 36.68 | 0.82 |
| Coptis | MOL002907 | Corchoroside A_qt | 104.95 | 0.78 |
| Coptis | MOL000622 | Magnograndiolide | 63.71 | 0.19 |
| Coptis | MOL000762 | Palmidin A | 35.36 | 0.65 |
| Coptis | MOL000785 | Palmatine | 64.6 | 0.65 |
| Coptis | MOL000098 | Quercetin | 46.43 | 0.28 |
| Coptis | MOL001458 | Coptisine | 30.67 | 0.86 |
| Coptis | MOL002668 | Worenine | 45.83 | 0.87 |
| Coptis | MOL008647 | Moupinamide | 86.71 | 0.26 |
| Glycyrrhiza | MOL001484 | Inermine | 75.18 | 0.54 |
| Glycyrrhiza | MOL001792 | DFV | 32.76 | 0.18 |
| Glycyrrhiza | MOL000211 | Mairin | 55.38 | 0.78 |
| Glycyrrhiza | MOL002311 | Glycyrol | 90.78 | 0.67 |
| Glycyrrhiza | MOL000239 | Jaranol | 50.83 | 0.29 |
| Glycyrrhiza | MOL002565 | Medicarpin | 49.22 | 0.34 |
| Glycyrrhiza | MOL000354 | Isorhamnetin | 49.6 | 0.31 |
| Glycyrrhiza | MOL000359 | Sitosterol | 36.91 | 0.75 |
| Glycyrrhiza | MOL003656 | Lupiwighteone | 51.64 | 0.37 |
| Glycyrrhiza | MOL003896 | 7-Methoxy-2-methyl isoflavone | 42.56 | 0.2 |
| Glycyrrhiza | MOL000392 | Formononetin | 69.67 | 0.21 |
| Glycyrrhiza | MOL000417 | Calycosin | 47.75 | 0.24 |
| Glycyrrhiza | MOL000422 | Kaempferol | 41.88 | 0.24 |
| Glycyrrhiza | MOL004328 | Naringenin | 59.29 | 0.21 |
| Glycyrrhiza | MOL004805 | (2S)-2-[4-Hydroxy-3-(3-methylbut-2-enyl)phenyl]-8,8-dimethyl-2,3-dihydropyrano[2,3-f]chromen-4-one | 31.79 | 0.72 |
| Glycyrrhiza | MOL004806 | Euchrenone | 30.29 | 0.57 |
| Glycyrrhiza | MOL004808 | Glyasperin B | 65.22 | 0.44 |
| Glycyrrhiza | MOL004810 | Glyasperin F | 75.84 | 0.54 |
| Glycyrrhiza | MOL004811 | Glyasperin C | 45.56 | 0.4 |
| Glycyrrhiza | MOL004814 | Isotrifoliol | 31.94 | 0.42 |
| Glycyrrhiza | MOL004815 | (E)-1-(2,4-Dihydroxyphenyl)-3-(2,2-dimethylchromen-6-yl)prop-2-en-1-one | 39.62 | 0.35 |
| Glycyrrhiza | MOL004820 | Kanzonol W | 50.48 | 0.52 |
| Glycyrrhiza | MOL004824 | (2S)-6-(2,4-Dihydroxyphenyl)-2-(2-hydroxypropan-2-yl)-4-methoxy-2,3-dihydrofuro[3,2-g]chromen-7-one | 60.25 | 0.63 |
| Glycyrrhiza | MOL004827 | Semilicoisoflavone B | 48.78 | 0.55 |
| Glycyrrhiza | MOL004828 | Glepidotin A | 44.72 | 0.35 |
| Glycyrrhiza | MOL004829 | Glepidotin B | 64.46 | 0.34 |
| Glycyrrhiza | MOL004833 | Phaseolinisoflavan | 32.01 | 0.45 |
| Glycyrrhiza | MOL004835 | Glypallichalcone | 61.6 | 0.19 |
| Glycyrrhiza | MOL004838 | 8-(6-Hydroxy-2-benzofuranyl)-2,2-dimethyl-5-chromenol | 58.44 | 0.38 |
| Glycyrrhiza | MOL004841 | Licochalcone B | 76.76 | 0.19 |
| Glycyrrhiza | MOL004848 | Licochalcone G | 49.25 | 0.32 |
| Glycyrrhiza | MOL004849 | 3-(2,4-Dihydroxyphenyl)-8-(1,1-dimethylprop-2-enyl)-7-hydroxy-5-methoxy-coumarin | 59.62 | 0.43 |
| Glycyrrhiza | MOL004855 | Licoricone | 63.58 | 0.47 |
| Glycyrrhiza | MOL004856 | Gancaonin A | 51.08 | 0.4 |
| Glycyrrhiza | MOL004857 | Gancaonin B | 48.79 | 0.45 |
| Glycyrrhiza | MOL004860 | Licorice glycoside E | 32.89 | 0.27 |
| Glycyrrhiza | MOL004863 | 3-(3,4-Dihydroxyphenyl)-5,7-dihydroxy-8-(3-methylbut-2-enyl)chromone | 66.37 | 0.41 |
| Glycyrrhiza | MOL004864 | 5,7-Dihydroxy-3-(4-methoxyphenyl)-8-(3-methylbut-2-enyl)chromone | 30.49 | 0.41 |
| Glycyrrhiza | MOL004866 | 2-(3,4-Dihydroxyphenyl)-5,7-dihydroxy-6-(3-methylbut-2-enyl)chromone | 44.15 | 0.41 |
| Glycyrrhiza | MOL004879 | Glycyrin | 52.61 | 0.47 |
| Glycyrrhiza | MOL004882 | Licocoumarone | 33.21 | 0.36 |
| Glycyrrhiza | MOL004883 | Licoisoflavone | 41.61 | 0.42 |
| Glycyrrhiza | MOL004884 | Licoisoflavone B | 38.93 | 0.55 |
| Glycyrrhiza | MOL004885 | Licoisoflavanone | 52.47 | 0.54 |
| Glycyrrhiza | MOL004891 | Shinpterocarpin | 80.3 | 0.73 |
| Glycyrrhiza | MOL004898 | (E)-3-[3,4-Dihydroxy-5-(3-methylbut-2-enyl)phenyl]-1-(2,4-dihydroxyphenyl)prop-2-en-1-one | 46.27 | 0.31 |
| Glycyrrhiza | MOL004903 | Liquiritin | 65.69 | 0.74 |
| Glycyrrhiza | MOL004904 | Licopyranocoumarin | 80.36 | 0.65 |
| Glycyrrhiza | MOL004905 | 3,22-Dihydroxy-11-oxo-delta(12)-oleanene-27-alpha-methoxycarbonyl-29-oic acid | 34.32 | 0.55 |
| Glycyrrhiza | MOL004907 | Glyzaglabrin | 61.07 | 0.35 |
| Glycyrrhiza | MOL004908 | Glabridin | 53.25 | 0.47 |
| Glycyrrhiza | MOL004910 | Glabranin | 52.9 | 0.31 |
| Glycyrrhiza | MOL004911 | Glabrene | 46.27 | 0.44 |
| Glycyrrhiza | MOL004912 | Glabrone | 52.51 | 0.5 |
| Glycyrrhiza | MOL004913 | 1,3-Dihydroxy-9-methoxy-6-benzofurano[3,2-c]chromenone | 48.14 | 0.43 |
| Glycyrrhiza | MOL004914 | 1,3-Dihydroxy-8,9-dimethoxy-6-benzofurano[3,2-c]chromenone | 62.9 | 0.53 |
| Glycyrrhiza | MOL004915 | Eurycarpin A | 43.28 | 0.37 |
| Glycyrrhiza | MOL004917 | Glycyroside | 37.25 | 0.79 |
| Glycyrrhiza | MOL004924 | (-)-Medicocarpin | 40.99 | 0.95 |
| Glycyrrhiza | MOL004935 | Sigmoidin B | 34.88 | 0.41 |
| Glycyrrhiza | MOL004941 | (2R)-7-Hydroxy-2-(4-hydroxyphenyl)chroman-4-one | 71.12 | 0.18 |
| Glycyrrhiza | MOL004945 | (2S)-7-Hydroxy-2-(4-hydroxyphenyl)-8-(3-methylbut-2-enyl)chroman-4-one | 36.57 | 0.32 |
| Glycyrrhiza | MOL004948 | Isoglycyrol | 44.7 | 0.84 |
| Glycyrrhiza | MOL004949 | Isolicoflavonol | 45.17 | 0.42 |
| Glycyrrhiza | MOL004957 | HMO | 38.37 | 0.21 |
| Glycyrrhiza | MOL004959 | 1-Methoxyphaseollidin | 69.98 | 0.64 |
| Glycyrrhiza | MOL004961 | Quercetin der. | 46.45 | 0.33 |
| Glycyrrhiza | MOL004966 | 3′-Hydroxy-4′-O-methylglabridin | 43.71 | 0.57 |
| Glycyrrhiza | MOL000497 | Licochalcone A | 40.79 | 0.29 |
| Glycyrrhiza | MOL004974 | 3′-Methoxyglabridin | 46.16 | 0.57 |
| Glycyrrhiza | MOL004978 | 2-[(3R)-8,8-Dimethyl-3,4-dihydro-2H-pyrano[6,5-f]chromen-3-yl]-5-methoxyphenol | 36.21 | 0.52 |
| Glycyrrhiza | MOL004980 | Inflacoumarin A | 39.71 | 0.33 |
| Glycyrrhiza | MOL004985 | Icos-5-enoic acid | 30.7 | 0.2 |
| Glycyrrhiza | MOL004988 | Kanzonol F | 32.47 | 0.89 |
| Glycyrrhiza | MOL004989 | 6-Prenylated eriodictyol | 39.22 | 0.41 |
| Glycyrrhiza | MOL004990 | 7,2′,4′-Trihydroxy-5-methoxy-3-arylcoumarin | 83.71 | 0.27 |
| Glycyrrhiza | MOL004991 | 7-Acetoxy-2-methylisoflavone | 38.92 | 0.26 |
| Glycyrrhiza | MOL004993 | 8-Prenylated eriodictyol | 53.79 | 0.4 |
| Glycyrrhiza | MOL004996 | Gadelaidic acid | 30.7 | 0.2 |
| Glycyrrhiza | MOL000500 | Vestitol | 74.66 | 0.21 |
| Glycyrrhiza | MOL005000 | Gancaonin G | 60.44 | 0.39 |
| Glycyrrhiza | MOL005001 | Gancaonin H | 50.1 | 0.78 |
| Glycyrrhiza | MOL005003 | Licoagrocarpin | 58.81 | 0.58 |
| Glycyrrhiza | MOL005007 | Glyasperin M | 72.67 | 0.59 |
| Glycyrrhiza | MOL005008 | Glycyrrhiza flavonol A | 41.28 | 0.6 |
| Glycyrrhiza | MOL005012 | Licoagroisoflavone | 57.28 | 0.49 |
| Glycyrrhiza | MOL005013 | 18α-Hydroxyglycyrrhetic acid | 41.16 | 0.71 |
| Glycyrrhiza | MOL005016 | Odoratin | 49.95 | 0.3 |
| Glycyrrhiza | MOL005017 | Phaseol | 78.77 | 0.58 |
| Glycyrrhiza | MOL005018 | Xambioona | 54.85 | 0.87 |
| Glycyrrhiza | MOL005020 | Dehydroglyasperin C | 53.82 | 0.37 |
| Glycyrrhiza | MOL000098 | Quercetin | 46.43 | 0.28 |
3.2. Differential Gene Screening
By comparing 15 normal samples with 30 disease samples in the GEO database, a total of 21,653 differential genes were acquired, including 9198 upregulated genes and 12,455 downregulated genes. After screening with a P value < 0.005 and ∣log2 (fold change) | >1, a total of 305 upregulated genes and 263 downregulated genes were obtained. As indicated by the gene volcano map (Figure 1), the differential genes in the disease samples display a normal distribution, with a larger number of significantly upregulated genes than significantly downregulated genes. The top 20 genes with the most significant upregulation and downregulation are presented in Figure 2 and Table 2.
Figure 1.
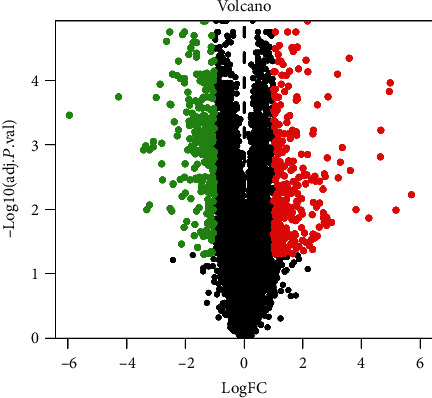
Gene volcano map shows the gene distribution in disease samples. Red and green represent upregulated genes (logFC > 0) and downregulated genes (logFC < 0), respectively, whereas black indicates no significant difference.
Figure 2.
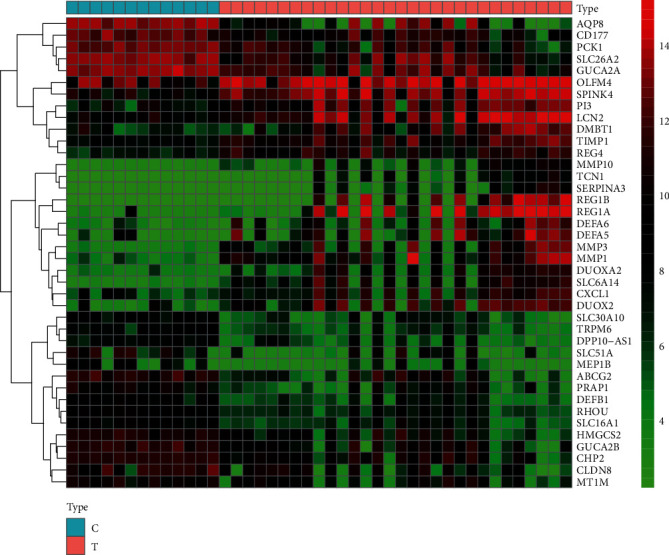
Gene heat map. In the gene heat map, red and green represent upregulated (logFC > 0) and downregulated (logFC < 0) genes in the sample, respectively, whereas black represents no significant difference. The first 13 samples were from healthy people, and the last 30 samples were from patients with ulcerative colitis.
Table 2.
The first 20 genes that are upregulated and downregulated.
| Gene names | LogFC | P value | Regulation direction |
|---|---|---|---|
| REG1A | 5.701613746 | 0.00052702 | Up |
| REG1B | 5.1898523 | 0.001131623 | Up |
| SLC6A14 | 4.991420548 | 8.72E − 07 | Up |
| MMP1 | 4.963627094 | 1.48E − 06 | Up |
| MMP3 | 4.668142374 | 1.70E − 05 | Up |
| DUOX2 | 4.65206741 | 7.48E − 05 | Up |
| DEFA5 | 4.261312381 | 0.001683584 | Up |
| DEFA6 | 3.819380723 | 0.001118108 | Up |
| DUOXA2 | 3.628039842 | 0.000154156 | Up |
| SPINK4 | 3.590142516 | 1.56E − 07 | Up |
| LCN2 | 3.354560231 | 4.42E − 05 | Up |
| PI3 | 3.28851345 | 0.000101703 | Up |
| MMP10 | 3.213180338 | 0.000222541 | Up |
| REG4 | 3.19089349 | 4.90E − 07 | Up |
| DMBT1 | 2.987003705 | 0.002100636 | Up |
| TIMP1 | 2.864973614 | 2.15E − 06 | Up |
| OLFM4 | 2.838270116 | 7.87E − 05 | Up |
| CXCL1 | 2.822789462 | 0.001668369 | Up |
| SERPINA3 | 2.811244589 | 0.002463382 | Up |
| TCN1 | 2.768089208 | 0.001564425 | Up |
| AQP8 | −5.946044173 | 6.72E − 06 | Down |
| ABCG2 | −4.278991642 | 2.21E − 06 | Down |
| PCK1 | −3.426343127 | 5.08E − 05 | Down |
| GUCA2B | −3.353230111 | 4.09E − 05 | Down |
| SLC51A | −3.319192058 | 0.001123545 | Down |
| PRAP1 | −3.226028948 | 5.10E − 05 | Down |
| CLDN8 | −3.223765137 | 0.000875494 | Down |
| SLC26A2 | −3.096598258 | 4.32E − 05 | Down |
| HMGCS2 | −3.089377316 | 3.28E − 05 | Down |
| SLC30A10 | −2.994061586 | 2.34E − 06 | Down |
| DEFB1 | −2.862625712 | 9.37E − 07 | Down |
| CHP2 | −2.806425883 | 0.000109215 | Down |
| CD177 | −2.802954854 | 3.61E − 05 | Down |
| GUCA2A | −2.776457399 | 0.000245692 | Down |
| TRPM6 | −2.636295344 | 3.93E − 08 | Down |
| RHOU | −2.531238896 | 1.72E − 08 | Down |
| DPP10-AS1 | −2.521195069 | 3.47E − 06 | Down |
| MT1M | −2.518431028 | 0.001039377 | Down |
| SLC16A1 | −2.492300121 | 3.72E − 06 | Down |
| MEP1B | −2.462876667 | 0.001188996 | Down |
3.3. Construction of the TCM Compound-Disease Regulatory Network
As listed in Table 3, there are 23 intersection genes (sorted by logFC). The targeting relationship between TCM active ingredients and intersection genes is presented by the TCM compound-disease regulatory network (Figure 3). Active ingredients of Glycyrrhiza and Scutellariae have the most amount of and related target genes, indicating that Glycyrrhiza and Scutellariae in GGQL are the most efficacious components. The active ingredients quercetin, kaempferol, wogonin, and stigmasterol are associated with 18, 5, 3, and 3 target genes, respectively. Therefore, they are classified as multitarget and multieffect compounds. The gene NOS2 is the gene associated with the highest number of active components, followed by PPARG and MMP1.
Table 3.
The 23 intersection genes sorted by logFC.
| Intersection gene | LogFC | P value |
|---|---|---|
| MMP1 | 4.963627094 | 1.48E − 06 |
| MMP3 | 4.668142374 | 1.69963E − 05 |
| DUOX2 | 4.65206741 | 7.47983E − 05 |
| PLAU | 2.323571552 | 0.0014514 |
| IL1B | 2.234056 | 0.006640247 |
| NOS2 | 2.061560817 | 0.003623843 |
| GJA1 | 2.011116812 | 0.000136087 |
| MMP9 | 1.867276075 | 0.006997334 |
| CXCL10 | 1.78792717 | 0.00738838 |
| CXCL11 | 1.728830977 | 0.005420693 |
| COL3A1 | 1.308597667 | 0.001234088 |
| PCOLCE | 1.293546472 | 0.006822444 |
| CAV1 | 1.2259564 | 0.00238233 |
| SLPI | 1.163710008 | 0.002878356 |
| F3 | 1.139466187 | 0.001216101 |
| SPP1 | 1.041938337 | 0.004669085 |
| COL1A1 | 1.022467924 | 0.002151629 |
| MAOA | −1.077796648 | 0.000138579 |
| ABAT | −1.190239356 | 9.18E − 06 |
| HMOX1 | −1.369875348 | 8.05E − 06 |
| PPARG | −1.377777361 | 6.87E − 06 |
| ADH1C | −2.038856081 | 0.002719353 |
| ABCG2 | −4.278991642 | 2.21E − 06 |
Figure 3.
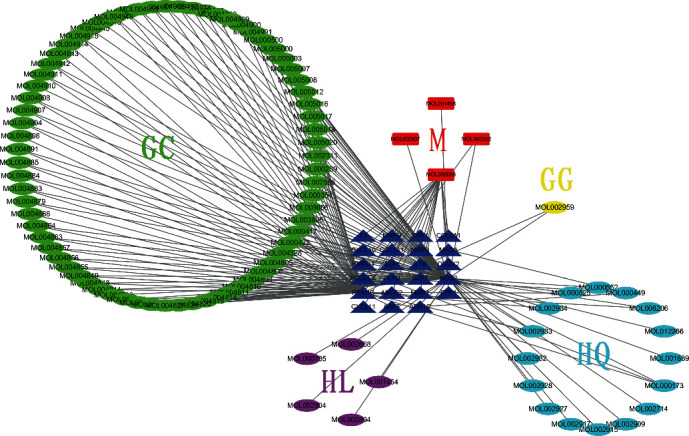
TCM compound-disease regulatory network. This network shows the targeted relationship between the active components of TCM and the intersection genes. Green GC represents Glycyrrhiza, yellow GG represents Pueraria, light blue HQ represents Scutellaria, purple HL represents Coptis, red M represents common components, and dark blue triangle represents intersection genes.
3.4. PPI Network and Topological Analysis
In the PPI network, the degree centrality (DC) of a node is simply the number of edges it has. The higher the degree, the more central the node is. The betweenness centrality (BC) captures how much a given node is in between others. Specifically, it is the ratio of the number of the shortest paths passing through the node to the total number of the shortest paths in the network. DC and BC reflect the influence of the corresponding node in the entire network. They describe the topological centrality based on the connectivity and controllability of the network. The combination of DC and BC values has been confirmed to be effective for screening reliable important proteins [3]. As shown in Figure 4, 830 protein nodes and 9689 edges were obtained for intersection genes. After screening with DC > 61 and a BC range of 0–113.2, the first 32 proteins are shown in Table 4 (in descending order of degree), with a total of 273 edges. Among the 32 proteins, nine proteins are predicted targets of the active ingredients, with their corresponding genes being EGFR, PPARG, ESR1, HSP90AA1, MYC, HSPA5, AR, AKT1, and RELA.
Figure 4.
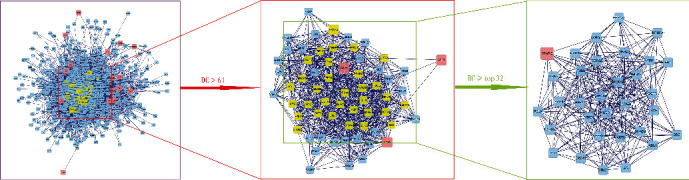
Topological analysis of the protein-protein interaction network. Herein, 830 protein nodes were obtained according to the intersection genes. After screening by DC > 61 for the first time, a total of 63 protein nodes were obtained, and the first 32 proteins were extracted according to BC for the second time.
Table 4.
Topological analysis results by degree—the first 32 proteins.
| Gene names | Annotation | Degree | Betweenness |
|---|---|---|---|
| NTRK1 | Neurotrophic receptor tyrosine kinase 1 | 239 | 79.2792394 |
| EGFR | Epidermal growth factor receptor | 170 | 57.27745249 |
| FN1 | Fibronectin | 169 | 36.34401789 |
| UBC | Ubiquitin C | 157 | 50.80248406 |
| PPARG | Peroxisome proliferator-activated receptor gamma | 153 | 37.99948007 |
| ESR1 | Estrogen receptor 1 | 150 | 113.2071674 |
| HSP90AA1 | Heat shock protein 90 alpha family class A member 1 | 132 | 103.9892463 |
| VCP | Valosin-containing protein | 121 | 53.86564087 |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 119 | 64.36717285 |
| MYC | v-myc avian myelocytomatosis viral oncogene homolog | 112 | 77.51063594 |
| HSPA5 | Heat shock protein family A (Hsp70) member 5 | 108 | 34.49237132 |
| NPM1 | Nucleophosmin | 106 | 63.99301971 |
| HSP90AB1 | Heat shock protein 90 alpha family class B member 1 | 104 | 52.53742327 |
| COPS5 | COP9 signalosome subunit 5 | 102 | 84.17298609 |
| EP300 | E1A-binding protein p300 | 98 | 59.70375531 |
| SRC | SRC protooncogene, nonreceptor tyrosine kinase | 94 | 52.01196867 |
| AR | Androgen receptor | 93 | 39.60526919 |
| BRCA1 | BRCA1, DNA repair associated | 92 | 37.41150073 |
| MDM2 | MDM2 protooncogene | 88 | 56.58152297 |
| AKT1 | AKT serine/threonine kinase 1 | 87 | 57.04410728 |
| CTNNB1 | Catenin beta 1 | 85 | 44.34296823 |
| HSPA4 | Heat shock protein family A (Hsp70) member 4 | 83 | 59.0919423 |
| EEF1A1 | Eukaryotic translation elongation factor 1 alpha 1 | 81 | 44.07432309 |
| SMAD2 | SMAD family member 2 | 78 | 38.64577814 |
| RELA | RELA protooncogene, NF-κB subunit | 77 | 40.34807667 |
| NFKB1 | Nuclear factor kappa B subunit 1 | 75 | 41.54441528 |
| TUBB | Tubulin beta class I | 72 | 85.09134103 |
| IKBKG | Inhibitor of nuclear factor kappa B kinase subunit gamma | 69 | 40.75351178 |
| HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | 67 | 45.38051383 |
| PRKDC | Protein kinase, DNA-activated, catalytic polypeptide | 66 | 45.37484081 |
| ABL1 | ABL protooncogene 1, nonreceptor tyrosine kinase | 65 | 34.38427075 |
| STUB1 | STIP1 homology and U-box-containing protein 1 | 64 | 41.4704054 |
3.5. Analysis of GO Function and KEGG Enrichment of Related Targets
GO enrichment analysis illustrates gene function on three levels: biological process (BP), cellular component (CC), and molecular function (MF). BP mainly involves aspects of response to oxygen levels, leukocyte migration, collagen metabolic process, and response to nutrients. CC is mainly related to the extracellular matrix, collagen-containing extracellular matrix, fibrillar collagen trimer, and banded collagen fibril. MF is mainly involved in serine-type endopeptidase activity, serine-type peptidase activity, serine hydrolase activity, CXCR chemokine receptor binding, platelet-derived growth factor binding, and cytokine activity (Figure 5). According to KEGG enrichment results, the mechanism of GGQL in treating UC is mainly concentrated in the IL-17 signaling pathway, relaxin signaling pathway, AGE-RAGE signaling pathway in diabetic complications, toll-like receptor signaling pathway, TNF signaling pathway, fluid shear stress and atherosclerosis, transcriptional misregulation in cancer, proteoglycans in cancer, rheumatoid arthritis, and prostate cancer (Figure 6). Genes associated with the greatest number of pathways were IL1B, MMP9, and MMP3 (Figure 7 and Table 5).
Figure 5.
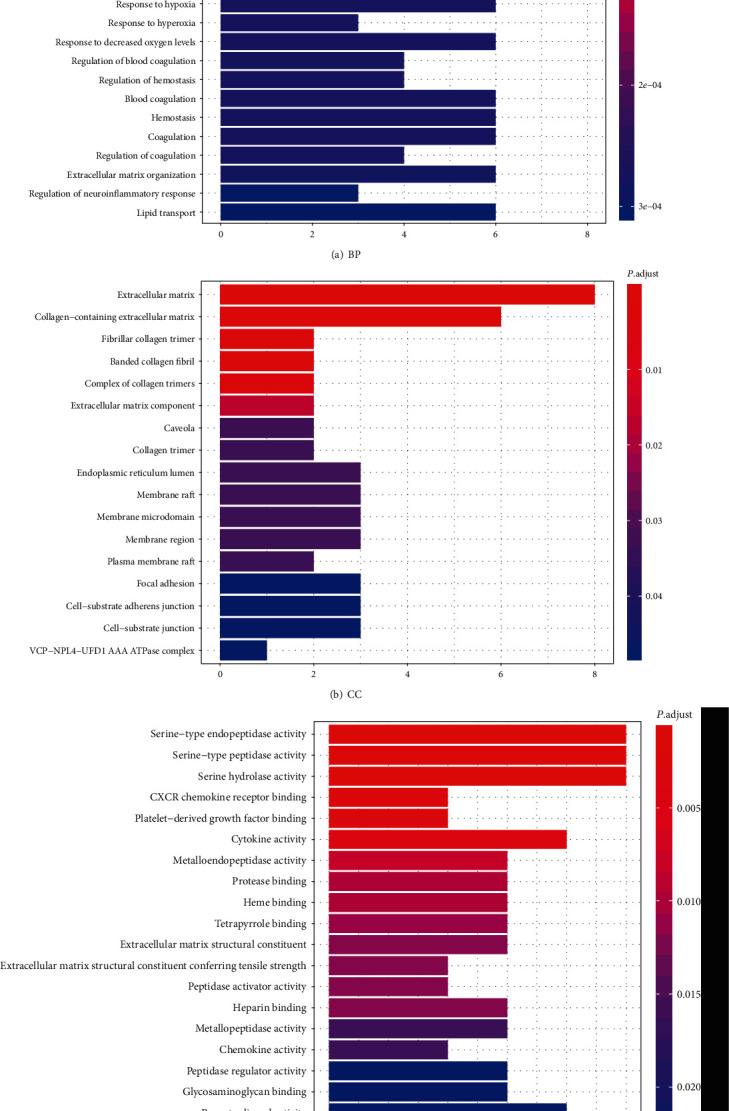
GO enrichment analysis of GGQL targets in treating UC. The horizontal axis of the BP, CC, and MF column represents the number of genes enriched in each item, and the color represents the enrichment significance based on the corrected P value.
Figure 6.
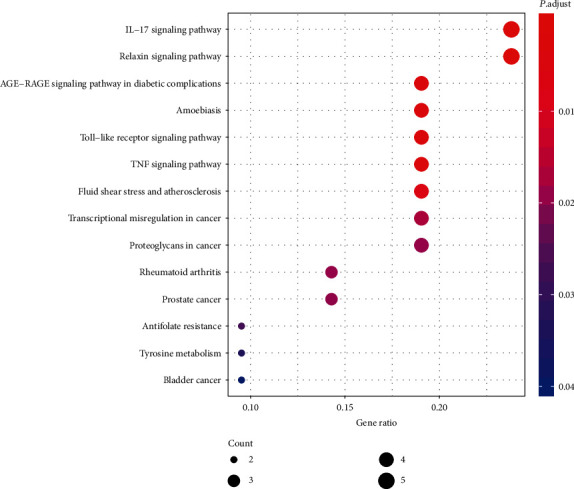
KEGG bubble. The horizontal axis of the KEGG bubble diagram represents the gene proportion enriched in each entry, and the vertical axis shows the enrichment degree according to the corrected P value.
Figure 7.
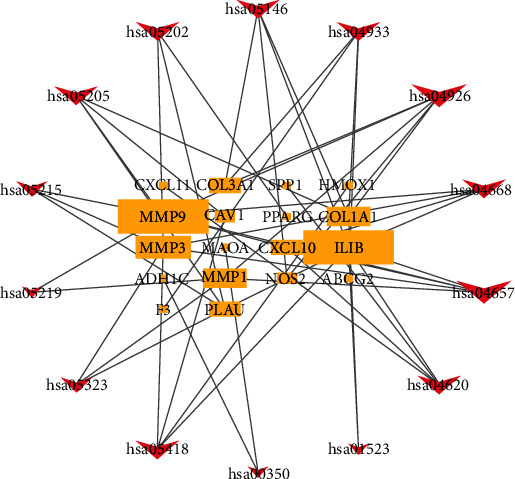
KEGG relational regulatory network. This network shows the relationship between the enriched 14 pathways and 18 genes, and the size of the graph shows the number of pathways or genes connected.
Table 5.
The enrichment pathways corresponding to intersection genes.
| Term | Description | Count | Gene ID |
|---|---|---|---|
| hsa04657 | IL-17 signaling pathway | 5 | MMP1/MMP3/MMP9/IL1B/CXCL10 |
| hsa04926 | Relaxin signaling pathway | 5 | NOS2/MMP1/MMP9/COL1A1/COL3A1 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 4 | F3/IL1B/COL1A1/COL3A1 |
| hsa05146 | Amoebiasis | 4 | NOS2/IL1B/COL1A1/COL3A1 |
| hsa04620 | Toll-like receptor signaling pathway | 4 | IL1B/CXCL11/CXCL10/SPP1 |
| hsa04668 | TNF signaling pathway | 4 | MMP3/MMP9/IL1B/CXCL10 |
| hsa05418 | Fluid shear stress and atherosclerosis | 4 | HMOX1/MMP9/CAV1/IL1B |
| hsa05202 | Transcriptional misregulation in cancer | 4 | PPARG/MMP3/PLAU/MMP9 |
| hsa05323 | Rheumatoid arthritis | 3 | MMP1/MMP3/IL1B |
| hsa05205 | Proteoglycans in cancer | 4 | PLAU/MMP9/CAV1/COL1A1 |
| hsa05215 | Prostate cancer | 3 | MMP3/PLAU/MMP9 |
| hsa01523 | Antifolate resistance | 2 | IL1B/ABCG2 |
| hsa00350 | Tyrosine metabolism | 2 | ADH1C/MAOA |
| hsa05219 | Bladder cancer | 2 | MMP1/MMP9 |
3.6. Molecular Docking
Molecular docking is a technique that mimics the interaction between small ligand molecules and receptor protein macromolecules, and the binding energy between the two counterparts can be calculated to predict their affinity. A binding energy lower than 0 indicates that the two molecules combine spontaneously and that smaller binding energies lead to more stable conformations. Most ingredients in GGQL can bind well with target genes, among which stigmasterol, coptisine, and berberine have the best binding properties (Table 6). The genes MMP3, IL1B, NOS2, HMOX1, PPARG, PLAU, and MMP1 can dock well with most active ingredients. Figure 8 illustrates some local structures of molecular docking in detail.
Table 6.
Binding energies of GGQL's key components to the target gene molecules.
| Key components | Binding energies (kcal/mol) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MMP3 | IL1B | NOS2 | HMOX1 | PPARG | PLAU | MMP1 | MMP9 | COCL10 | COL1A1 | COL3A1 | SPP1 | |
| Stigmasterol | −10.3 | −8.78 | −8.75 | −8.04 | −7.64 | −7.25 | −7.22 | −6.94 | −5.94 | −6.32 | −6.06 | −6.9 |
| Coptisine | −9.27 | −7.97 | −6.97 | −6.27 | −6.79 | −6.59 | −6.58 | −6.73 | −4.57 | −6.31 | −6.46 | −6.16 |
| Berberine | −9.81 | −7.71 | −6.92 | −6.36 | −6.57 | −7.29 | −6.81 | −6.43 | −4.84 | −5.63 | −5.89 | −6.01 |
| Liquiritigenin | −8.63 | −7.95 | −6.39 | −6.04 | −7.06 | −7.41 | −5.61 | −7.58 | −5.88 | −5.89 | −5.77 | −4.31 |
| Quercetin | −7.73 | −8.45 | −5.04 | −5.2 | −5.55 | −7.17 | −5.44 | −5.05 | −4.67 | −3.92 | −4.26 | −5.09 |
| Kaempferol | −8.37 | −7.89 | −5.82 | −6.59 | −6.2 | −6.7 | −4.79 | −5.09 | −5.38 | −4.03 | −5.37 | −5.16 |
| Wogonin | −8.96 | −7.42 | −5.98 | −6.23 | −5.91 | −6.89 | −4.74 | −5.72 | −4.48 | −4.9 | −5.22 | −5.34 |
| Baicalein | −8.41 | −7.23 | −6.59 | −5.27 | −6.67 | −7.28 | −5.65 | −7.12 | −5.19 | −4.62 | −6.16 | −5.57 |
| Puerarin | −6.46 | −7.19 | −4.95 | −4.77 | −4.93 | −4.43 | −4.07 | −3.37 | −2.73 | −3.28 | −4.39 | −3.92 |
| Daidzin | −8.7 | −6.76 | −6.33 | −4.15 | −4.52 | −6.01 | −4.27 | −6.65 | −4.07 | −3.4 | −4.56 | −4.67 |
| Epiberberine | −8.54 | −7.38 | −7.31 | −6.19 | −6.24 | −6.19 | −6.25 | −6.83 | −4.62 | −6.12 | −5.82 | −5.53 |
| Jatrorrhizine | −7.54 | −7.46 | −5.88 | −5.01 | −6.02 | −5.44 | −5.17 | −5.92 | −4.36 | −5.1 | −4.45 | −4.98 |
| Baicalin | −5.76 | −6.69 | −4.69 | −3.49 | −4.52 | −4.22 | −4.52 | −2.41 | −2.98 | −2.98 | −4.29 | −3.82 |
| Palmatine | −7.75 | −6.8 | −5.89 | −5.81 | −4.97 | −5.76 | −6.36 | −5.55 | −3.9 | −5.23 | −5.68 | −5.3 |
| Wogonoside | −7.68 | −8.5 | −5.99 | −4.83 | −5.22 | −6.5 | −4.73 | −3.13 | −5.36 | −3.38 | −4.49 | −5.53 |
Figure 8.
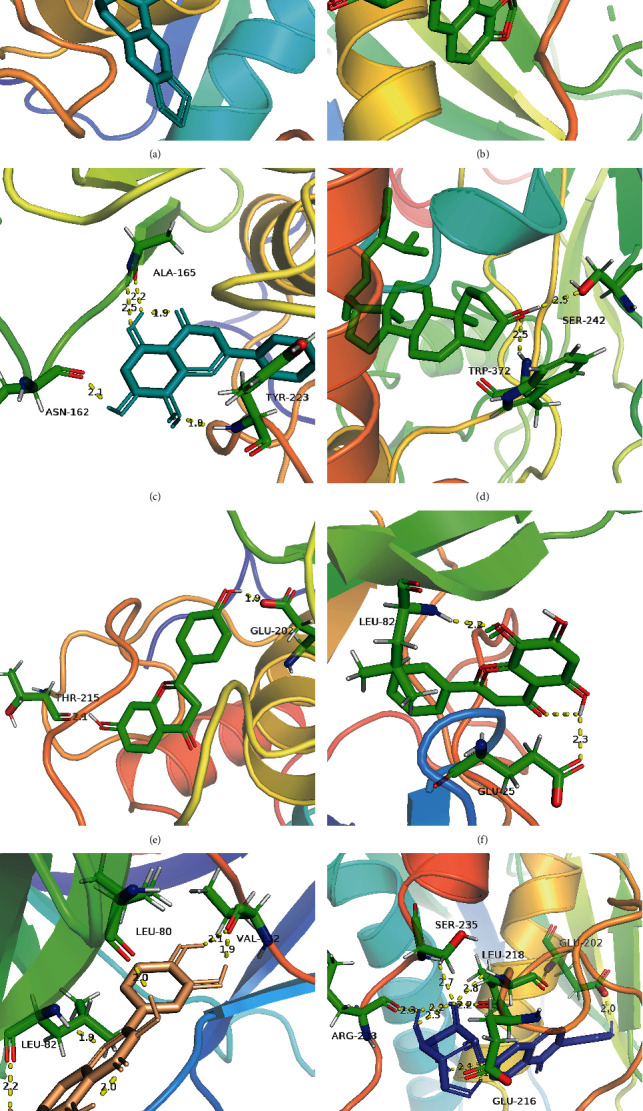
Partial diagram of molecular docking: (a) MMP3-berberine; (b) MMP3-coptisine; (c) MMP3-wogonin; (d) NOS2-stigmasterol; (e) MMP3-liquiritigenin; (f) IL1B-wogonin; (g) IL1B-quercetin; (h) MMP3-daidzin.
4. Discussion
In this study, a network pharmacological analysis was conducted on the medicinal ingredients of the four TCMs (Puerariae, Scutellariae, Coptis, and Glycyrrhiza) in GGQL and UC disease targets. Quercetin, kaempferol, and wogonin were identified as the active ingredients associated with the most targets. The results of molecular docking also verified that they have good binding properties with most target genes. Quercetin is a common flavonoid compound in nature. It is considered the most effective reactive oxygen species (ROS) scavenger and inhibits the production of several proinflammatory factors, such as TNF-α and NO. The antioxidant and anti-inflammatory advantages of quercetin in UC treatment have been confirmed by various in vivo and in vitro experiments [4]. Quercetin plays an anticancer role by reducing the activity of kinase MEK1, downregulating the cascade reaction of Raf and MAPK, and inhibiting telomerase [5]. Kaempferol is also a natural flavonoid, and its efficacy as an anti-inflammatory, antioxidant, and anticancer agent has been reported in the treatment of a variety of diseases, such as diabetes, obesity, and cancer (e.g., skin, liver, and colon cancers) [6]. Wogonin is the compound with the highest content in Scutellariae; it is quickly converted into metabolites, such as baicalin, after entering the bloodstream. Baicalin has been confirmed to significantly inhibit TLR4-induced increase in NF-κBp65 levels, reduce the activity of enzymes, such as MPO and COX-2, lower the production of factors, such as THF-α, IL-1β, IL-12, and IFN-γ, and regulate the Th17/Treg cell balance [7, 8]. The molecular docking results indicate that stigmasterol, coptisine, and berberine have superior affinities to the target genes MMP3, IL1B, NOS2, HMOX1, PPARG, and PLAU, and they are the effective ingredients with potent anti-inflammatory and antioxidant effects. Stigmasterol has been shown to inhibit the innate immune response induced by lipopolysaccharide in a mouse model [9]. Berberine exerts local anti-inflammatory effects by blocking the IL-6/STAT3/NF-κB signaling pathway. Meanwhile, it effectively enhances the expression of SIgA and lowers the expression of iNOS, MPO, and MDA [10].
A PPI topological analysis was performed for 23 intersection genes, revealing 32 strongly associated proteins, among which 9 proteins (EGFR, PPARG, ESR1, HSP90AA1, MYC, HSPA5, AR, AKT1, and RELA) are the predicted targets of TCMs. PPARG is also an intersection gene, and these 9 targets were speculated to be the key targets of GGQL in the treatment of UC.
UC is an inflammatory disease related to intestinal immune recognition disorders. The KEGG enrichment results indicate four inflammation-related pathways: IL-17 signaling pathway, AGE-RAGE signaling pathway in diabetic complications, toll-like receptor signaling pathway, and TNF signaling pathway. Meanwhile, further analysis revealed inflammation-related genes, such as IL1B, CXCL10, CXCL11, MMP9, MMP3, SPP1, NFKB1, IKBKG, and RELA. The first six genes are widely involved in IL-17, toll-like receptors, and TNF signaling pathways, while the latter three are participants in the NF-κB pathway, particularly the gene RELA, which encodes NF-κB p65. It is suggested by the results that both the treatment mechanism of GGQL and the pathogenesis of UC are related to inflammatory regulation. A meta-analysis showed that in allelic and dominant models, the genetic polymorphisms of IL-17A and IL-17F may increase the risk of UC occurrence. In addition, IL-17 levels in serum are significantly associated with the severity of UC [11]. Toll-like receptors (TLRs) are a group of transmembrane proteins widely distributed in immune cells, playing a key role in identifying invading pathogens and upregulating signals related to inflammatory cytokines and costimulatory molecules [12]. Tumor necrosis factor (TNF) not only is a potent proinflammatory mediator but also upregulates the production of ROS and RNS and exacerbates cell damage [13]. Anti-TNF therapy has been proven to quickly induce clinical and endoscopic remission in UC patients; however, its safety and related risks still require urgent attention [14]. NF-κB induces cytokine expression and neutrophil aggregation. It is often regarded as a sign and the central pathway of inflammatory response, while being involved in cancer development through various pathways [15]. In the classical NF-κB signaling pathway, TNF-α and IL-1 activate toll-like receptors (TLRs), followed by the activation of the IκB kinase complex, which can phosphorylate IκBα [15]. Advanced glycosylation end products (AGE) and IL-17 (highly expressed during UC activity) can also be used as mediators for NF-κB pathway activation [16, 17]. In this study, it is confirmed that the action mechanism of GGQL is related to the four pathways of inflammatory response, and the regulation of these pathways is linked to the transcription of NF-κB, indicating that GGQL plays its therapeutic role by inhibiting the inflammatory response mediated by the NF-κB signaling pathway. It has also been confirmed by other studies that baicalin and berberine exert inflammatory inhibition effects by downregulating the expression of proinflammatory cytokines (i.e., IL-1β, TNF-α, ICAM-1, TLR2, and TLR4) and inhibiting the NF-κBp65 signal transduction pathway [18, 19].
The excessive ROS in the intestinal mucosa can trigger inflammatory responses by inducing redox-sensitive signaling pathways and transcription factors (e.g., NF-κB, TNF-α, and AP-1). The inflammatory responses then lead to the generation of more ROS, forming a vicious circle between oxidative stress and inflammation [20]. The KEGG path map shows that there are three pathways related to oxidative stress: (1) relaxin signaling pathway, (2) fluid shear stress and atherosclerosis pathway that activates downstream second messengers of PI3K-AKT, thus inducing eNOS expression, and (3) AGE-RAGE signaling pathway in diabetic complications, in which the receptor RAGE activates NADPH oxidase 2, thus leading to excessive ROS production. PPARG, NOS2, and DUOX2 are the three key target genes closely related to oxidative stress. NOS2 and PPARG are the targets associated with most active ingredients. The results of molecular docking have also verified the effectiveness of the association. PPARG is an important gene for fatty acid metabolism and oxidation and insulin sensitization. It also inhibits the activation of NF-κB to exert anti-inflammatory effects. PPAR-γ has been proven to be a key receptor for 5-ASA to exert anti-inflammatory and antioxidant effects [21]. Relevant studies have found that the expression of PPAR-γ messenger RNA in the colonic mucosa of UC patients is impaired, accompanied by enhanced expression of the corresponding inflammatory factors (e.g., NF-κB). Partial PPAR-γ agonists may be a new target for UC treatment [22]. According to the results of differential genes, PPARG was downregulated. In GGQL, there are 67 compounds that have a regulatory effect on PPARG. NOS2 encodes the synthesis of inducible nitric oxide synthase (iNOS). The highly expressed iNOS in the inflamed mucosa plays a key role in oxidative stress-induced inflammation [23]. NOS activity in colon biopsies has been shown to be correlated with disease intensity [24]. DUOX2 participates in the regulation of hydrogen peroxide anabolism and mediates peroxidase activity on the mucosal surface. DUOXA2 is a partner of DUOX2. Significant upregulation was determined for DUOXA2, DUOX2, and NOS2, among the different genes. It implies that excessive production of ROS and RNS will bring oxidative damage to cellular components, including lipids, DNA, and proteins, thus causing damage to the mucosal barrier and leading to sustained inflammatory response [20]. In GGQL, 86 compounds, including quercetin and kaempferol, have been shown to have effects on NOS2 and DUOX2 by the TCM compound-disease regulatory network.
As suggested by the KEGG enrichment results, GGQL may prevent the occurrence of cancer in the following ways: transcriptional misregulation in cancer and proteoglycans in cancer. Persistent inflammatory response and oxidative stress state can promote the mutation of cancer genes. The abnormal expressions of NOS2, DUOX2/DUOXA2, ESR1, EGFR, MYC, and AKT1, which are being discussed in this study, have been confirmed to be related to cancer [25–28]. The expression of the protooncogene MYC is significantly upregulated in up to 70% of colorectal cancer (CRC) patients. The overexpression of its gene product, c-Myc, leads to the activation of downstream genes, DNA synthesis, cell proliferation, and chromosomal aberrations. These mechanisms ultimately lead to genomic instability and chemical resistance [29]. Encoded by the EGFR gene, EGFR is a transmembrane glycoprotein belonging to the ErbB family; it is reportedly excessively expressed in 85% of NSCLC cells and is associated with a poor prognosis. By regulating downstream signaling pathways, mainly PI3K/Akt and MAPK pathways, the activated EGFR leads to receptor dimerization and tyrosine autophosphorylation, which could result in aberrant proliferation in certain cells, such as NSCLC cells. MYC is a protooncogene that encodes transcription factors involved in basic cellular pathophysiological processes. Activation of MYC causes abnormal cell proliferation, regression, and redifferentiation of cancer cells and susceptibility to aurora kinase inhibition in SCLC cells [30]. Proteoglycans are the main components of the extracellular matrix. They participate in matrix remodeling in tumor cell growth and the formation of stromal vessels, thus affecting the response of tumor cells and tissues and regulating the cancer phenotype by influencing the signals within cancer cells [31].
Many other scholars' studies are consistent with our findings. Xu et al. found that GGQL decoction could increase the SOD activity and decrease the MDA and iNOS activities along with the reduction in TNF-α and IL-1β levels to take effect on the treatment of UC rats [18]. Li et al. revealed that GGQL could reduce the TLR4 expression and NF-κB activation along with several inflammatory cytokines such as TNF-α, IL-6, IL-1β, and IL-4 and NO [32].
This study shows that the mechanism of GGQL in UC treatment is related to its anti-inflammatory and antioxidative properties and inhibition of oncogene transcription. Meanwhile, the action targets are related to NOS2, PPARG, and MMP1. This study has not demonstrated the relationship between the mechanism of GGQL treatment and signaling pathways of relaxin, amoebiasis, and prostate cancer. Relevant studies on this are based on existing database information and lack experimental verification. Therefore, it is worthy of subsequent experimental analysis to confirm the reliability of the results.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Nos. 81603509 and 31600947) and Natural Science Foundation of Shandong Province (Nos. ZR2014HQ051 and ZR2016HM65).
Data Availability
The data that support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Authors' Contributions
Bin Yu was responsible for the conception and design. Meiqi Wei and He Li were responsible for the development of the methodology. Qifang Li, Qiao Yi, Qun Ma, and Ruining Xie were responsible for the acquisition of data. Rong Wang, Yuan Liu, Chao Wei, Canlei Zheng, Bing Sun, and Bingbing Li were responsible for the analysis and interpretation of data. Meiqi Wei, He Li, and Bin Yu were responsible for the writing, review, and/or revision of the manuscript. Meiqi Wei and He Li contributed equally to this work.
References
- 1.Fumery M., Singh S., Dulai P. S., Gower-Rousseau C., Peyrin-Biroulet L., Sandborn W. J. Natural history of adult ulcerative colitis in population-based cohorts: a systematic review. Clin Gastroenterol Hepatol. 2018;16(3):343–356.e3. doi: 10.1016/j.cgh.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan Y., Yi W., Huang H., Mei Z., Feng Z. Efficacy of herbal medicine (Gegen Qinlian decoction) on ulcerative colitis: a systematic review of randomized controlled trials. Medicine. 2019;98(52):p. e18512. doi: 10.1097/MD.0000000000018512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J., Peng W., Wu F.-X. Computational approaches to predicting essential proteins: A survey. PROTEOMICS - Clinical Applications. 2013;7(1-2):181–192. doi: 10.1002/prca.201200068. [DOI] [PubMed] [Google Scholar]
- 4.Sotnikova R., Nosalova V., Navarova J. Efficacy of quercetin derivatives in prevention of ulcerative colitis in rats. Interdisciplinary Toxicology. 2013;6(1):9–12. doi: 10.2478/intox-2013-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan F., Niaz K., Maqbool F., et al. Molecular targets underlying the anticancer effects of quercetin: an update. Nutrients. 2016;8(9):p. 529. doi: 10.3390/nu8090529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imran M., Rauf A., Shah Z. A., et al. Chemo-preventive and therapeutic effect of the dietary flavonoid kaempferol: a comprehensive review. PHYTOTHERAPY RESEARCH. 2019;33(2):263–275. doi: 10.1002/ptr.6227. [DOI] [PubMed] [Google Scholar]
- 7.Cui L., Feng L., Zhang Z. H., Jia X. B. The anti-inflammation effect of baicalin on experimental colitis through inhibiting TLR4/NF-κB pathway activation. INTERNATIONAL IMMUNOPHARMACOLOGY. 2014;23(1):294–303. doi: 10.1016/j.intimp.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Zou Y., Dai S.-X., Chi H.-G., et al. Baicalin attenuates TNBS-induced colitis in rats by modulating the Th17/Treg paradigm. ARCHIVES OF PHARMACAL RESEARCH. 2015;38(10):1873–1887. doi: 10.1007/s12272-014-0486-2. [DOI] [PubMed] [Google Scholar]
- 9.Antwi A. O., Obiri D. D., Osafo N., Forkuo A. D., Essel L. B. Stigmasterol inhibits lipopolysaccharide-induced innate immune responses in murine models. INTERNATIONAL IMMUNOPHARMACOLOGY. 2017;53:105–113. doi: 10.1016/j.intimp.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Zhu L., Gu P., Shen H. Protective effects of berberine hydrochloride on DSS-induced ulcerative colitis in rats. INTERNATIONAL IMMUNOPHARMACOLOGY. 2019;68:242–251. doi: 10.1016/j.intimp.2018.12.036. [DOI] [PubMed] [Google Scholar]
- 11.Li J., Tian H., Jiang H., Han B. Interleukin-17 SNPs and serum levels increase ulcerative colitis risk: a meta-analysis. WORLD JOURNAL OF GASTROENTEROLOGY. 2014;20(42):15899–15909. doi: 10.3748/wjg.v20.i42.15899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaisho T., Akira S. Toll-like receptors and their signaling mechanism in innate immunity. Acta Odontologica Scandinavica. 2009;59(3):124–130. doi: 10.1080/000163501750266701. [DOI] [PubMed] [Google Scholar]
- 13.Blaser H., Dostert C., Mak T. W., Brenner D. TNF and ROS crosstalk in inflammation. TRENDS IN CELL BIOLOGY. 2016;26(4):249–261. doi: 10.1016/j.tcb.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Pugliese D., Felice C., Papa A., et al. Anti TNF-α therapy for ulcerative colitis: current status and prospects for the future. Expert Review of Clinical Immunology. 2017;13(3):223–233. doi: 10.1080/1744666X.2017.1243468. [DOI] [PubMed] [Google Scholar]
- 15.Hoesel B., Schmid J. A. The complexity of NF-κB signaling in inflammation and cancer. Molecular Cancer. 2013;12(1):p. 86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tobon-Velasco J., Cuevas E., Torres-Ramos M. Receptor for AGEs (RAGE) as mediator of NF-kB pathway activation in neuroinflammation and oxidative stress. CNS & Neurological Disorders-Drug Targets. 2014;13(9):1615–1626. doi: 10.2174/1871527313666140806144831. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H., Xia B., Li J., et al. Expression and clinical significance of IL-17 and IL-17 receptor in ulcerative colitis. Journal of Huazhong University of Science and Technology-Medical Sciences. 2016;36(1):37–40. doi: 10.1007/s11596-016-1538-2. [DOI] [PubMed] [Google Scholar]
- 18.Xu B.-L., Zhang G.-J., Ji Y.-B. Active components alignment of Gegenqinlian decoction protects ulcerative colitis by attenuating inflammatory and oxidative stress. Journal of Ethnopharmacology. 2015;162:253–260. doi: 10.1016/j.jep.2014.12.042. [DOI] [PubMed] [Google Scholar]
- 19.Lu P. D., Zhao Y. H. Targeting NF-κB pathway for treating ulcerative colitis: comprehensive regulatory characteristics of Chinese medicines. Chinese Medicine. 2020;15(1) doi: 10.1186/s13020-020-0296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biasi F., Leonarduzzi G., Oteiza P. I., Poli G. Inflammatory bowel disease: mechanisms, redox considerations, and therapeutic targets. ANTIOXIDANTS & REDOX SIGNALING. 2013;19(14):1711–1747. doi: 10.1089/ars.2012.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rousseaux C., Lefebvre B., Dubuquoy L., et al. Intestinal antiinflammatory effect of 5-aminosalicylic acid is dependent on peroxisome proliferator-activated receptor-gamma. JOURNAL OF EXPERIMENTAL MEDICINE. 2005;201(8):1205–1215. doi: 10.1084/jem.20041948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubuquoy L., Jansson E. Å., Deeb S., et al. Impaired expression of peroxisome proliferator-activated receptor γ in ulcerative colitis. GASTROENTEROLOGY. 2003;124(5):1265–1276. doi: 10.1016/S0016-5085(03)00271-3. [DOI] [PubMed] [Google Scholar]
- 23.Senhaji N., Nadifi S., Zaid Y., et al. Polymorphisms in oxidative pathway related genes and susceptibility to inflammatory bowel disease. WORLD JOURNAL OF GASTROENTEROLOGY. 2017;23(47):8300–8307. doi: 10.3748/wjg.v23.i47.8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guihot G., Guimbaud R., Bertrand V., et al. Inducible nitric oxide synthase activity in colon biopsies from inflammatory areas: correlation with inflammation intensity in patients with ulcerative colitis but not with Crohn's disease. AMINO ACIDS. 2000;18(3):229–237. doi: 10.1007/s007260050020. [DOI] [PubMed] [Google Scholar]
- 25.Thomas D. D., Wink D. A. NOS2 as an emergent player in progression of cancer. ANTIOXIDANTS & REDOX SIGNALING. 2017;26(17):963–965. doi: 10.1089/ars.2016.6835. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Wang X., Xu L., Zhang Z., Wang F., Tang X. Investigation of Potential Genetic Biomarkers and Molecular Mechanism of Ulcerative Colitis Utilizing Bioinformatics Analysis. BioMed Research International. 2020;2020:9. doi: 10.1155/2020/4921387.4921387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B., Yan S., Qu L., Zhu J. Celecoxib enhances anticancer effect of cisplatin and induces anoikis in osteosarcoma via PI3K/Akt pathway. Cancer Cell International. 2017;17(1) doi: 10.1186/s12935-016-0378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akbari S., Hosseini M., Rezaei-Tavirani M., et al. Common and differential genetically pathways between ulcerative colitis and colon adenocarcinoma. Gastroenterology and hepatology from bed to bench. 2017;10:93–101. [PMC free article] [PubMed] [Google Scholar]
- 29.Elbadawy M., Usui T., Yamawaki H., Sasaki K. Emerging roles of C-Myc in cancer stem cell-related signaling and resistance to cancer chemotherapy: a potential therapeutic target against colorectal cancer. INTERNATIONAL JOURNAL OF MOLECULAR SCIENCES. 2019;20(9):p. 2340. doi: 10.3390/ijms20092340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhuang Z., Chen Q., Huang C., Wen J., Huang H., Liu Z. A Comprehensive Network Pharmacology-Based Strategy to Investigate Multiple Mechanisms of HeChan Tablet on Lung Cancer. Evidence-Based Complementary and Alternative Medicine. 2020;2020:17. doi: 10.1155/2020/7658342.7658342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theocharis A. D., Karamanos N. K. Proteoglycans remodeling in cancer: Underlying molecular mechanisms. Matrix Biology. 2019;75-76:220–259. doi: 10.1016/j.matbio.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 32.Li R., Chen Y., Shi M., et al. Gegen Qinlian decoction alleviates experimental colitis via suppressing TLR4/NF-κB signaling and enhancing antioxidant effect. PHYTOMEDICINE. 2016;23(10):1012–1020. doi: 10.1016/j.phymed.2016.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.


